Abstract
Ensuring data quality and proper instrument functionality is a prerequisite for scientific investigation. Manual quality assurance is time-consuming and subjective. Metrics for describing liquid chromatography mass spectrometry (LC–MS) data have been developed; however, the wide variety of LC–MS instruments and configurations precludes applying a simple cutoff. Using 1150 manually classified quality control (QC) data sets, we trained logistic regression classification models to predict whether a data set is in or out of control. Model parameters were optimized by minimizing a loss function that accounts for the trade-off between false positive and false negative errors. The classifier models detected bad data sets with high sensitivity while maintaining high specificity. Moreover, the composite classifier was dramatically more specific than single metrics. Finally, we evaluated the performance of the classifier on a separate validation set where it performed comparably to the results for the testing/training data sets. By presenting the methods and software used to create the classifier, other groups can create a classifier for their specific QC regimen, which is highly variable lab-to-lab. In total, this manuscript presents 3400 LC–MS data sets for the same QC sample (whole cell lysate of Shewanella oneidensis), deposited to the ProteomeXchange with identifiers PXD000320–PXD000324.
Keywords: mass spectrometry, liquid chromatography, quality control
Introduction
Determining whether an instrument is operating within acceptable performance metrics is an essential step during scientific investigation.1,2 A variety of methods are commonly used in liquid chromatography mass spectrometry (LC–MS),3 with many groups routinely utilizing a common sample which is regularly analyzed, for example, weekly. Samples range from simple proteins and mixtures to whole cell-lysates.4 In addition to regularly running a quality control (QC) sample, recent research has investigated analysis methods and metrics for assessing data quality. Rudnick et al. defined 46 “NIST metrics” to quantify the performance of various LC–MS aspects, such as chromatography, ion source, dynamic sampling, and peptide identifications.2,5 These metrics are largely dependent on MS/MS data and their subsequent peptide identifications. The Quameter software package proposed an additional set of metrics, which are not dependent on MS/MS identifications.6
The NIST and Quameter efforts focused on generating a comprehensive set of metrics for monitoring the various aspects of system performance. When employing the metrics to monitor the day-to-day operation of an LC–MS system, it is important to note that some metrics are heavily dependent on the specific instrument and configuration. What is an acceptable value for one setup may be wholly unacceptable for another. Thus, a critical shortcoming of current metric sets is that they produce an array of values and leave interpretation to the user. Specifically, there is no guidance as to when the performance of the LC–MS system drops below acceptable limits. Given the large number of metrics available, routine interpretation of the metrics and quality assurance is typically determined via a subjective weighting of a subset of the metrics. The specific subset of metrics, and associated acceptable values, varies from lab to lab and among individual operators.
As pointed out by Tabb, there remains a need for “decision support tools for the interpretation of metrics derived from data”.1 A variety of statistical methods may be used to explore and model how the multivariate metrics predict data quality, including principal component analysis,7 classification8 and regression and model selection techniques.9,10 Leveraging a large corpus of manually curated QC data, we developed a Lasso logistic regression classifier (LLRC) that predicts, with high sensitivity and specificity, whether a QC data set is in or out of control. This signature, which is a composite of LC–MS performance metrics, is more robust than any single metric itself. As performance can be viewed as a continuum, the LLRC model natively computes a quality score within the range 0–1. The trained model then identifies a cutoff for the dichotomous classification that achieves the highest sensitivity and specificity. An important feature of the LLRC is that it differentiates between false positive and false negative errors. These errors have distinct real-life implications on data generation and use. Therefore, the penalties associated with these errors are separately defined, which allows the classifier to be more responsive in the balance between sensitivity and specificity.11 Finally, we present the software used to create the classifier, so that other laboratories can create a classifier specific to their QC regimen, instrumentation, and needs.
Experimental Procedures
Sample Preparation
A Shewanella oneidensis MR-1 lysate digest is used as a quality control sample in our laboratory to provide sufficient proteomic complexity to assess both LC and MS performance. The cultures (7 L) were grown in fed-batch mode using a Bioflow 3000 model fermentor (New Brunswick, Inc.) and allowed to achieve steady state before sampling and harvesting. The medium was HBa MR-1 with 0.5 mL/L of 100 mM ferric NTA, 1 mL/L of 1 mM Na2SeO4, and 1 mL/L of 3 M MgCl2·6H2O as well as vitamins, minerals, and amino acids. The cultures used O2 as the terminal electron acceptor from a house air source (5 L/min flow rate) and were maintained at 20% dissolved oxygen (DO). Other monitored parameters included maintaining a pH of 7.00 (±0.03), a constant agitation of the culture at 5000 rpm, and temperature of 30 °C. These samples were pelleted (11900g for 8 min at 4 °C) and frozen at −80 °C until processing for proteomic analysis. Enough material was prepared in this manner to provide QC samples for the last four years. The majority of the culture is still waiting to be used.
Cells were then lysed by homogenizing the cells with 0.1 mm zirconia/silica beads in the Bullet Blender (Next Advance, Averill Park NY) speed 8 for 3 min. Samples were then immediately placed on ice to inhibit proteolysis and then transferred, and the beads were rinsed with 100 μL of 100 mM Na4HCO3 and 1 mL of 100 mM ammonium bicarbonate. A BCA protein assay (Thermo Scientific, San Jose CA) was performed to determine concentration. Samples were denatured by adding urea to 7 M and reduced by dithiothreitol (DTT) to a concentration of 5 mM. The samples were then incubated for 30 min at 60 °C, and then diluted 10-fold with 100 mM Na4HCO3. CaCl2 was added to a concentration of 1 mM. Next, trypsin (Affymetrix, Santa Clara CA) was added in a ratio 1:50 trypsin/protein and incubated at 37 °C for 3 h. Samples were desalted by 100 mg DSC-18 columns (Sigma Aldrich). Each column was conditioned with 3 mL of MeOH and rinsed with 2 mL of 0.1% TFA in H2O. Peptides were then loaded on the resin and washed with 4 mL of 95:5 H2O/ACN with 0.1% TFA. Peptides were eluted with 1 mL 80:20 ACN/H2O with 0.1% TFA. Collected sample was concentrated via SpeedVac (Thermo Scientific, San Jose CA), and then the samples were transferred to ultracentrifuge tubes and ultracentrifuged at 100 000 rpm for 10 min at 4 °C, and the supernatant was drawn off and pooled. Final concentration was determined by peptide BCA (Thermo Scientific, San Jose CA). Samples were brought to a concentration of 0.5 μg/μL with H2O (MilliPore, Billerica MA) and divided into aliquots for injections on the HPLC.
The HPLCs used to run the samples were built in-house utilizing various commercial pumps, valves, and auto samplers, all of which were coordinated by a custom software packaged called LCMSnet. The data sets analyzed for this paper were run using LC columns that were 75 μm inner diameter, and either 30 or 65 cm in length. These LC columns were packed in house with Phenomenex Jupiter C18 3 μm porous beads. The flow rate was 300 nL/min. Both 60 and 100 min acquisitions were used. Mobile phase A is 0.1% formic acid in H2O and mobile phase B is 0.1% formic acid in acetonitrile. The 100 min gradient was delivered by starting at 5% mobile phase B and advancing to 8%, 12%, 35%, 60%, and 75% at times (in minutes) 2, 20, 75, 97, 100 respectively. The times were scaled proportionally to deliver the same gradient in 60 min. Typically 2.5 μg of Shewanella digest was loaded to the head of the column or to a trapping column. Although operating conditions varied by capabilities of each instrument, typical conditions for each are as follows. The LTQ was run in data-dependent MS-MS mode, selecting the top 10 parent ions from each survey scan. The Exactive runs were high resolution MS only with the target resolution set to 100 000. The LTQ-Orbitrap and the Velos-Orbitrap instruments were typically set to have a high resolution survey scan of 60 000 resolution followed by the top 6 or 10 data-dependent MS–MS scans, respectively. Because of the diversity of data sets used in this study, this is not a comprehensive list of conditions. Data are presented from four classes of instruments from Thermo Scientific: the LTQ linear ion trap, Exactive, LTQ-Orbitrap, and Velos Orbitrap platforms. There are 1150 testing and training data sets: 224 on LTQ instruments, 85 on Exactive instruments, 380 on LTQ-Orbitrap instruments, and 461 on Velos-Orbitrap instruments (see Table 1).
Table 1. Number of Manually Curated and Noncurated Data Sets for Each Instrument Platform.
| manually curated data sets |
||||
|---|---|---|---|---|
| instrument | number of good/OK datasets | number of poor data sets | total | number of noncurated data sets |
| Exactive | 66 | 19 | 85 | 225 |
| LTQ IonTrap | 123 | 101 | 224 | 243 |
| LTQ Orbitrap | 257 | 123 | 380 | 461 |
| Velos Orbitrap | 339 | 122 | 461 | 1321 |
| total | 785 | 365 | 1150 | 2250 |
For each QC data set, the NIST and Quameter metrics were calculated exactly as described in the original publications.5,6 The Quameter metrics were calculated using the software from the Tabb group (http://fenchurch.mc.vanderbilt.edu/software.php); the NIST metrics were calculated using in-house software, SMAQC, which is posted on our github repository (https://github.com/PNNL-Comp-Mass-Spec). For data from the Exactive instrument class, no MS-MS data were collected, and therefore any metrics relating to MS/MS data were omitted. Peptide identifications were performed with MSGF+ (version v9593, 05/06/2013). The protein sequence database was the Shewanella oneidensis MR1 proteome supplemented with trypsin and keratin sequences. Relevant search parameters were tryptic specificity (semi tryptic allowed), no static modifications, dynamic methionine oxidation. Precursor and fragment mass tolerance were dependent upon instrument type. High resolution instruments used a 20 ppm tolerance, low resolution data used 2.5 Da. All of the data has been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with the data set identifiers PXD000320, PXD000321, PXD000322, PXD000323, and PXD000324. This includes the instrument .RAW files, the MS-GF+ peptide identifications in .mzIdentML format,12 .mgf converted spectra files, and the spreadsheet containing the metrics for every data set.
Expert Annotation
The data sets were manually reviewed by three expert instrument operators (30+ years of combined LC–MS experience) using an in-house graphical user interface viewer. This viewer contained the base peak chromatogram, total ion current chromatogram, plots of both the top 50 000 and top 500 000 LC–MS detected features, and the number of peptides identified. In the first round, 1150 data sets were manually curated as “good”, “okay”, or “poor” and used to develop the classifier. In cases where the assessors disagreed (∼5–10%), the majority opinion was taken for the curated value. Moreover, the “okay” value was used to denote the wide range of performance, which, although not optimal, was still acceptable. For the validation, an additional 1321 data sets classified with the statistical model from which a subset of 100 data sets were chosen for manual curation (Supplemental Files 1 and 2, Supporting Information).
Data quality assessment requires knowledge of the conditions under which the QC was run to properly account for the variety of run parameters, for example, high resolution MS only, high resolution MS low resolution MS/MS, high resolution MS high resolution MS/MS, and low resolution MS low resolution MS/MS. Data-dependent acquisition regimes varied from MS/MS of top 3, top 6, and top 10 most intense peaks from the survey MS scan. Fragmentation methods included high energy collisional dissociation (HCD) and collision induced dissociation (CID). Resolution ranged from 1000 to 100 000 based on instrument capabilities and settings to meet the needs of ongoing experiments. Run times were either 60 or 100 min, and LC column lengths varied accordingly. Each of these criteria is considered to determine what constitutes an acceptable (in control) data set. For example, under identical mass spectrometer conditions, an acceptable 60 min HPLC run would provide results that would be unacceptable (out of control) for a 100 min run.
Training Statistical Models
Multivariate statistical techniques7 were applied to the data to identify which metrics might be useful to understand the quality of a given data set and to develop a model to predict the quality of future data sets. Principal component analysis (PCA)7 was applied on the NIST and Quameter metrics, 87 in all, using the PCA package in the R statistical programming language. PCA was used to motivate the design of statistical classification models. There are a wide variety of classification algorithms.7,8 We applied a few of them, including classification and regression tree algorithms (CART), linear discriminant analysis (LDA), and logistic regression augmented by a classification threshold. The most effective was logistic regression,9 an approach for predicting a binary response using a linear combination of continuous and/or categorical predictor variables. The goal for the classifier is to identify data sets that are out of control. To that end, the binary response was coded as a “0” for a data set that was in control (annotated as “good” or “OK”) and “1” for a data set that was out of control (annotated as “poor”). The linear component of the logistic regression model can be expressed as
where i, (i = 1, ..., N) indexes the data sets, β0 is the intercept, β1 is a vector of coefficients (one for each of the NIST and Quameter quality metrics included in the model), and xi is a vector representing the quality metrics included in the model. The probability of the binomial response is modeled by the logistic function:
where π (xi) estimates the probability of data set i being out of control.
An important step in any regression model is determining the set of variables that best predict the response. We used the Lasso approach,9,10,13−15 a model selection technique that accounts for collinearity among the predictors while selecting a subset of the predictor variables that results in the “best” regression model. In this case, “best” is defined as the subset of variables whose coefficients maximize a penalized log-likelihood function (see Appendix).
The inputs to the Lasso logistic regression model include the complete set of possible quality metrics, the binary expert annotations (0 = “good” or “OK”, 1 = “poor”), and λ, a regularization parameter that simultaneously restricts the size of model coefficients and the number of quality metrics included in the model. We developed a separate Lasso logistic regression model for each of the four instruments types. We used a threshold parameter, τ, to make a definitive prediction as to whether a data set was good or poor. Specifically, we classify data set i as out of control if π(xi) > τ and in control otherwise. We will refer to the combination of the logistic regression and the threshold, τ, as the Lasso logistic regression classifier (LLRC).
When predicting the quality of a data set, the LLRC will either predict a data set correctly, or will make either a false positive or false negative error. A false positive error occurs when the LLRC predicts the data set to be out of control, when it was annotated as in control. The false negative error occurs when the LLRC predicts the data set as being in control when it was annotated as out of control. The rates of these errors depend on the threshold criteria and are inversely related (as one goes up, the other generally goes down). To this end, a loss function is constructed that reflects the consequences we attribute to the two types of errors. When the LLRC predicts a data set correctly, zero loss occurs. When the LLRC produces a false positive, a loss of 1 occurs, and a false negative receives a loss of κ ≥ 1. Hence, the consequences (or cost) of a false negative are κ times greater than those of a false positive.
Cross validation was employed to determine the optimal λ and τ that minimize the expected loss. Cross validation was performed on the 1150 testing/training data sets by randomly dividing the data set into five equal parts. Four parts were then combined to train the LLRC which was subsequently used to predict the “held-out” part. This continues such that each part was predicted from a model trained by the other four parts. The entire process is discussed in detail in the Appendix. An LLRC was fit separately for each instrument platform.
The R package used in the creation of the LLRC, and a tutorial for its use is available for download at http://omics.pnl.gov/software/MSDataQualitySignatures.php.
Results
One critical setting where subjective or time-consuming manual data analysis can lead to systemic problems is in the evaluation of quality control (QC) data sets. In an effort to automate the assessment of QC data sets, we selected a testing/training sample of 1150 data sets (see Table 1) and manually annotated them as being “good,” “OK”, or “poor.” These data sets originated from four types of mass spectrometers and are all replicate analyses of the same whole-cell lysate of S. oneidensis (see Experimental Procedures). In addition to the manual rating, all computational metrics from NIST and Quameter were calculated for each data set.5,6 Initial analyses of the 1150 data sets using PCA showed a separation by quality rating and also by instrument type (Figure 1). This finding confirmed the need to build classifiers for each instrument separately. The separation by quality rating indicates that statistical classification approaches that rely on multiple variables are likely to be successful, to some degree, in predicting data quality (Supplemental Figure 1, Supporting Information).
Figure 1.
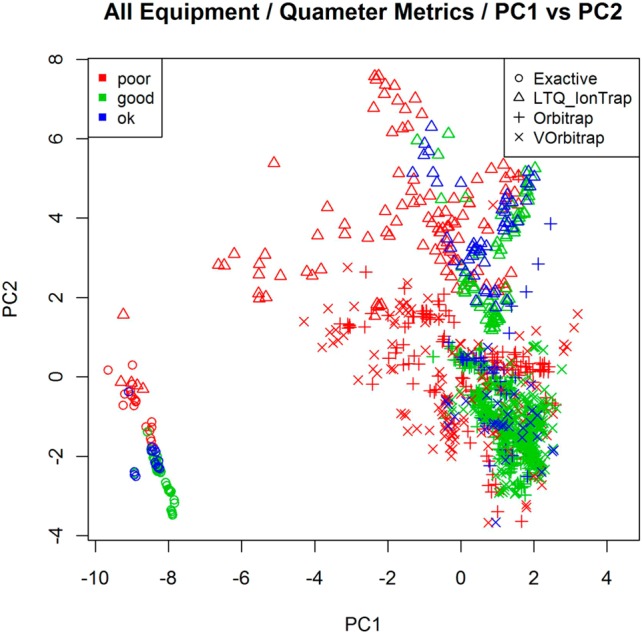
Principal component analysis of curated data. Each data point on the plot represents a single QC data set and is identified by its instrument class (the symbol) and its manually curated quality (the color). The first two principal components explain about 40% of the variability in the original data. Only the Quameter data quality metrics could be calculated for Exactive data, as it lacks MS2 fragmentation scans.
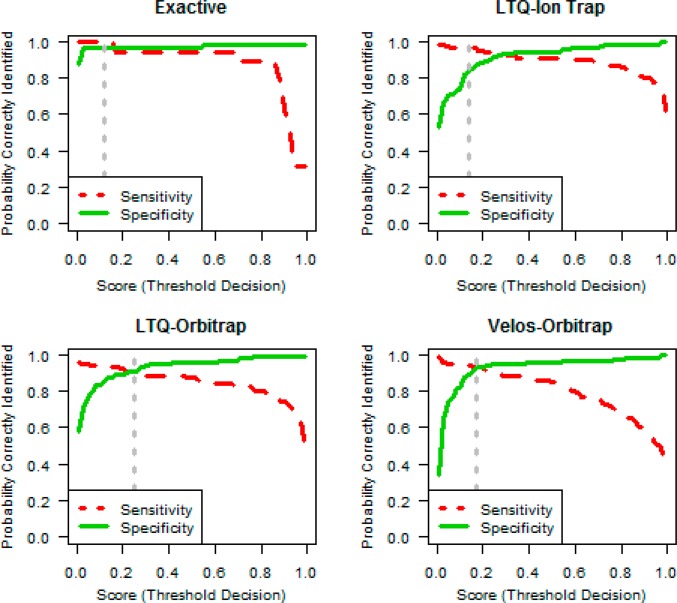
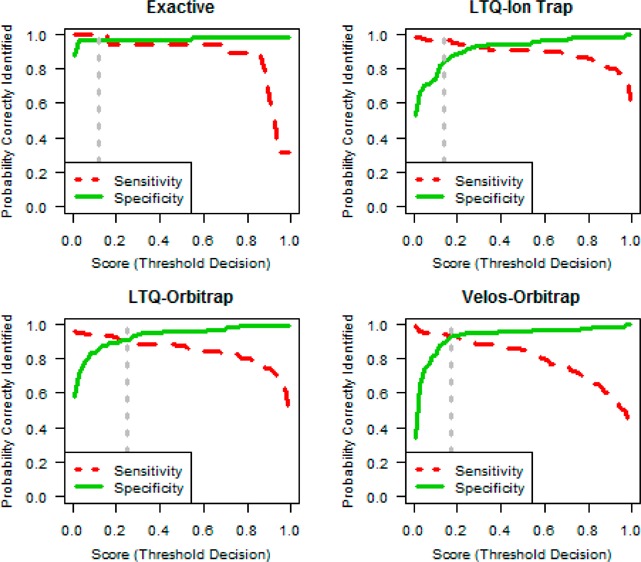
The primary goal was to build a classifier that accurately predicts the data set quality, specifically when a data set is out of control (poor). We utilized a Lasso logistic regression classifier (LLRC) which simultaneously restricts the size of model coefficients and the number of quality metrics included the model (see Experimental Procedures). Importantly, the model does not treat false negatives and false positives as equally deleterious events. The different impact of each error type is reflected in the practical implication of day-to-day instrument operation. While false positives incur extra work and inconvenience for operators to review and overturn the decision, the false-negative error is of much greater consequence; if the classifier fails to predict an out of control data set, a problem in the LC–MS system may not be discovered and poor quality data will continue to be produced. Optimal parameter values were obtained by using cross validation to minimize a loss function that treated false negatives as five times worse than false positives. The resulting LLRCs for each of the four instrument types are shown in Table 2. The classifier output is natively a value within the range 0–1. This reflects the continuum of performance of real LC–MS systems in day-to-day operation. Converting this value into the binary classification is accomplished by applying a threshold cutoff. Figure 2 shows the sensitivity and specificity as the threshold value varies for each instrument platform. The dashed gray line on each plot in Figure 2 indicates the optimal threshold that minimizes the loss function. Mathematical and algorithmic details for obtaining the LLRC are provided in the Appendix.
Table 2. Classification Measures from the LLRC Models.
| instrument | loss function parameter (κ) | optimal lambda (λ) | optimal threshold (τ) | sensitivity (%) | specificity (%) |
|---|---|---|---|---|---|
| Exactive | 5 | 0.05 | 0.12 | 100 | 97.0 |
| LTQ IonTrap | 5 | 0.13 | 0.14 | 97.0 | 84.6 |
| LTQ-Orbitrap | 5 | 0.05 | 0.25 | 89.4 | 91.4 |
| Velos-Orbitrap | 5 | 0.09 | 0.17 | 93.4 | 92.9 |
Figure 2.
Sensitivity and specificity trade-off. The 1150 curated data sets are shown according to their classification from the cross-validation results. Data are separated by instrument class and run through their separate classifier models. We define sensitivity as the probability of correctly classifying an out of control (or poor) data set, equal to 1 minus the false negative rate. Specificity is the proportion of good data sets that are correctly classified and is equal to 1 minus the false positive rate (see Experimental Procedures). The dotted vertical line indicates the optimal threshold, τ, which balances the cost of a false positive or false negative classification error.
The final step was to develop an LLRC for predicting the quality of future data sets on each platform. These classifiers were created by training on all the available data using the optimal parameter values previously obtained. In developing the LLRC, the Lasso method selected between 2 and 12 quality metrics to be included in the LLRC for each of the four instrument types. These metrics are listed in Table 3. The LLRC utilized a different number of metrics to classify data from each of the different instrument types. The number of metrics used seems to correlate with how well separated the data sets appeared in the PCA analysis (Supplemental Figure 1, Supporting Information). Exactive data, which required only two metrics, was the most distinctly separated by the first two principle components. In contrast, Orbitrap data required 12 metrics and was visibly less clearly separated in the PCA analysis. It is important to note that many of the NIST and Quameter quality metrics are highly correlated with each other. Although Lasso selected a certain subset of these metrics, it is likely there are other subsets that would have performed similarly had they been selected. Consequently, one cannot directly compare the subset of metrics chosen for one instrument class to another, and we do not claim that the metrics in Table 3 constitute the best set.
Table 3. Quality Metrics Selected by Lasso for Inclusion in the Logistic Regression Models.
| Exactive | LTQ IonTrap | LTQ-Orbitrap | Velos-Orbitrap |
|---|---|---|---|
| MS1_TIC_Q2 | XIC_WideFrac | XIC_WideFrac | XIC_WideFrac |
| MS1_Density_Q1 | MS2_4B | XIC_Height_Q4 | MS2_4A |
| P_2C | MS1_TIC_Change_Q2 | MS2_4B | |
| MS1_Density_Q2 | P_2B | ||
| DS_1A | |||
| DS_2A | |||
| IS_1A | |||
| IS_3A | |||
| MS2_1 | |||
| MS2_4A | |||
| MS2_4B | |||
| P_2B |
Table 2 shows the sensitivity and specificity at the optimal threshold for each instrument platform. The sensitivity for each instrument category ranged from 100% for Exactive to 89.4% for LTQ-Orbitrap. The specificity ranged from 97% for Exactive to 84.6% for LTQ Ion trap (which had a very high sensitivity of 97%). A value of τ that performs well with both sensitivity and specificity is preferable. However, if one measure is more important in a specific cost/benefit scenario, the threshold will adjust to reflect the trade-offs captured in the loss function. This is reflected in the LTQ Ion trap sensitivity and specificity calculations. Sensitivity was more desirable, and, therefore, resulted in a threshold with a larger discrepancy between sensitivity and specificity. Moreover, the expected rates of false positives and false negative are implicitly considered as part of the optimization.
Comparison to Single Metric Classifiers
The LLRC is a composite classifier, in that it relies on multiple metrics to determine the quality of each data set. Some instrument operators may rely only on a single metric to assess data quality. Commonly used single metrics include: total tryptic peptide count (P_2A), distinct tryptic peptide count (P_2C), and the absolute intensities of peaks in the TIC (MS1_2B). In Table 4, we present a comparison of the LLRC to these single measures of quality for Velos-Orbitrap data sets. The table illustrates the proportion of false positives and false negatives that would occur when using the LLRC, as well as the four aforementioned metrics, each applied individually with their own threshold. As before, the loss function used to optimize the LLRC parameters assumed that false negatives are 5 times worse than false positives, resulting in only 7.1% (24) false positives and 6.6% (8) false negatives (see Table 4). To create an equitable comparison, the threshold for each individual metric was selected to achieve a false negative rate of 0.066 (equal to that of the LLRC). The LLRC had a false positive rate of 7.1%, while the single metrics resulted in considerably worse false positive rates ranging between 32.3% and 73.5% (see Table 4). The LLRC kept the false negative rate low (i.e., high sensitivity to detecting out-of-control data sets) while also controlling the false positive rate. To frame this comparison in real-world terms, let us compare LLRC to the P_2C single metric classifier in terms of the total number of data sets that would require validation. Our comparison depends on two assumptions: both classifiers have the same sensitivity (0.934), and if a classifier predicts a data set is poor it will be curated. Consequently, if we use the P_2C classifier instead of the LLRC, we would expect to have to validate an additional (0.323–0.071) × π × 100% of the data sets. Here, π is the unknown fraction of data sets that truly are out of control. So, in a group of 1000 data sets, if π were 0.15, we would expect to have to curate 38 additional data sets. Although the time spent in curating a single data set is typically small (1–2 min), there is a distinct advantage for automating curation and avoiding manual revalidation. Individually, the 38 additional manual analyses may not take much time, but the repetitive monotony increases the propensity for operator error and drifting subjectivity.
Table 4. False Positive (Specificity) and False Negative (Sensitivity) of LLRC versus Single Metrics for Velos-Orbitrapa.
| metric | LLRC | P_2C | P_2A | MS1_2B |
|---|---|---|---|---|
| false positive (specificity) | 0.071 (0.929) | 0.323 (0.677) | 0.386 (0.614) | 0.735 (0.265) |
| false negative (sensitivity) | 0.066 (0.934) | 0.066 (0.934) | 0.066 (0.934) | 0.066 (0.934) |
The sensitivity is held constant at 0.934 to show the sensitivity for any single metric compared to the LLRC. From Rudnick et al.5 P_2C is the total unique tryptic peptide identifications; P_2A is the total spectrum identifications; MS1_2B is the median TIC.
Validation of Classifier
To further validate the LLRC, we predicted the data quality of an additional 1321 noncurated Velos-Orbitrap data sets using the optimal LLRC trained on all the data (denoted f* in the Appendix). We randomly (but systematically) selected 100 of these classified data sets such that the probabilities estimated by the logistic regression were evenly spaced across the range of predicted probabilities. These data sets were manually curated and compared to the LLRC predictions. The LLRC achieved a sensitivity of 91.9% and a specificity of 81.6%. The sensitivity was quite similar to the cross validation estimates (shown in Table 2), but the specificity was 11% lower than what was observed in cross validation. Notwithstanding this reduction in specificity, it is noteworthy that sensitivity was preserved, especially because sensitivity is more important than specificity; that is, a false negative error is considerably worse than the false positive error.
Discussion
In this manuscript, we present a very large corpus of LC–MS/MS replicates using four different instrument classes. The primary purpose for which we use the data is to build a classifier for automatically detecting out-of-control data sets and therefore poor instrument performance. We envision, however, that this corpus could be used for a myriad of other bioinformatics and biostatistics applications. With data presented on four different classes of instruments, an obvious application would be the development of MS/MS scoring functions, using this data to understand how fragmentation differs between instrument types. A second type of analysis could be to understand the qualitative presence/absence of various peptides observed across a large number of replicates. The missing data problem is an important issue for quantitative proteomics, and this corpus would allow for rigorous understanding of the scope of the problem and potentially causes for sporadic peptide observation. Another application that we envision is the investigation of unidentified or unattributed spectra. With thousands of LC–MS/MS data sets, there are a very large number of fragmentation spectra for which there is not a confident identification. Of those unidentified species, many are fragmented in multiple data sets; spectrum averaging or other methods could be utilized to obtain a confident identification. For these and doubtless many other bioinformatics explorations, we have posted the data to public repositories and encourage researchers to use them as a resource. We note that the original intent of our research (classifying bad data sets) necessitated the publication of LC–MS/MS data that would normally not be published. We openly acknowledge that some of the data are of low quality, as was required for our goals. Therefore, in secondary analyses of these data, we encourage users to carefully consider whether such data are appropriate for their application, and consult Supplementary Files 1 and 2, Supporting Information for a list of data that is considered out-of-control.
The presented approach demonstrates how the Lasso logistic regression classifier (LLRC) approach can leverage a collection LC–MS performance metrics to accurately predict the quality of LC–MS data sets. We note that our classifier models may not be directly usable by those whose LC–MS instrumentation or QC regime is different from ours. However, the described methodology and accompanying software can be applied by others who want to develop an automated QC analysis. Given the diverse QC samples, instruments and even goals of various research laboratories and core facilities, we strongly recommend that each group train their own classifier. Our research shows that a statistically trained composite metric is dramatically more effective than any single metric. Thus, this work describes an automated process that will greatly improve the use of quality metrics.
While the approach produces a dichotomous prediction (i.e., in or out of control) for each data set, operators may find it more useful to omit the final classification step and base their assessments on the probability scores alone. These scores, which range in the unit interval, may be useful in making decisions about borderline cases. Similarly, adding an explicit borderline class would be a valuable future direction. The LLRC may also provide the underpinnings for a more comprehensive statistical model designed to identify the onset of operational drift. Such a methodology would alert the instrument operator to more closely monitor the system. A final extension would be to cluster and classify different types of malfunction in the LC–MS system. Understanding the particular subsets of the quality metrics that correspond to out-of-control conditions in LC or MS would assist operators in diagnosing malfunctions. Moreover, new and more specific metrics could be created to specifically target different types of malfunction. Such classifications would provide immense utility for day-to-day instrument operation.
Availability and Supporting Data
All data used for this project are available at http://omics.pnl.gov and have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository16 with the data set identifiers PXD000320, PXD000321, PXD000322, PXD000323, and PXD000324. This includes representative instrument raw files and the complete list of QC metrics for all data sets (Supplemental Files 1 and 2, Supporting Information).
Acknowledgments
We wish to thank Professor Jack Hagemeister and his team: Jenny Williams, Michael Piggott, Cameron Ackerman, Winston McCracken, Aaron Cain, Matt Klug, Shane Crowley, Tanner Jump, and Trevor Owen from the Electrical Engineering and Computer Science Department at Washington State University for the implementation of the NIST metrics. They would also like to thank David Tabb for helpful discussion of the Quameter metrics.
Portions of the research described in this paper were funded by the Signature Discovery Initiative (http://signatures.pnnl.gov) at Pacific Northwest National Laboratory, conducted under the Laboratory Directed Research and Development Program at PNNL. Portions of this work were supported by a grant (U24-CA-160019) from the National Cancer Institute Clinical Proteomic Tumor Analysis Consortium (CPTAC). Portions of this research were supported by the National Institutes of Health: the National Institute of General Medical Sciences (P41 GM103493), National Institute of Allergy and Infectious Diseases (Y1-AI-8401), and the Department of Energy Office of Biological and Environmental Research Genome Sciences Program under the Pan-Omics project. S.H.P. acknowledges funding from a U.S. Department of Energy Early Career award. Mass spectrometry datasets were collected in the Environmental Molecular Science Laboratory, a U.S. Department of Energy (DOE) national scientific user facility at Pacific Northwest National Laboratory (PNNL) in Richland, WA. Battelle operates PNNL for the DOE under contract DE-AC05-76RLO01830.
Glossary
Abbreviations
- PCA
principal components analysis
- QC
quality control
- LLRC
Lasso logistic regression classifier
Appendix
Identifying the best LLRC model requires choosing the values of a variety of parameters that satisfy defined optimization criteria. Let yi = 1 if the true status of data set i is out of control and let yi = 0 if it is in control. Let πi = π(xi, β0, β1, λ) represent the probability that data set i is out of control. This probability is a function of the quality metrics, xi, the intercept β0, the regression coefficients, β1 = (β1, ..., βp)T, and the regularization parameter, λ. Now let ŷl ≡ f(xi, β̂0, β̂1, λ, τ) = I{π̂i>τ} denote the status of the data set predicted by the LLRC, where, once again, ŷi = 1 if data set i is predicted to be out of control, and 0 if in control. The “hat” notation above a parameter (or symbol) designates a numeric value of the parameter that has been estimated from data. In Lasso, the estimates of the regression parameters, β̂0 and β̂1, are chosen to maximize the penalized log likelihood function
| 1 |
for a given value of λ, a regularization parameter. The log likelihood function is given by
| 2 |
We
obtained the estimates of λ and τ using 5-fold cross
validation.10 Let δ: {1, ..., N} → {1, 2, 3, 4, 5} map the data set i into one of five randomly chosen, nonoverlapping, and exhaustive
partitions of the N data sets. Let f–k(xi, β̂0, β̂1, λ, τ) represent
the classifier whose regression parameter estimates are obtained by
maximizing (1) using all the data except the kth
partition. Now define the loss function, L(y, ŷ, κ) according to the
following matrix: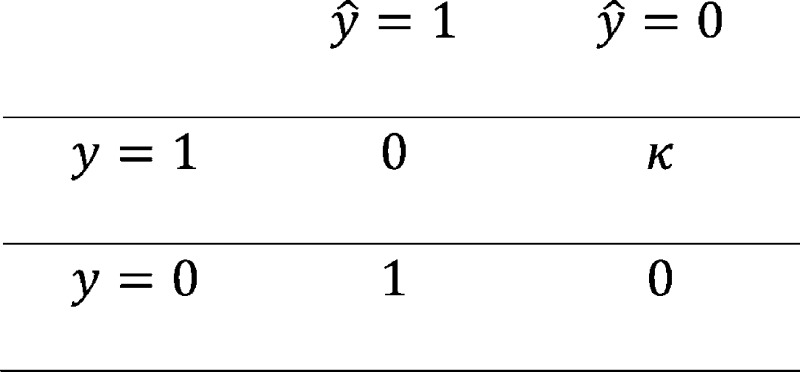
The parameter κ defines the trade-off between false positives and false negatives. For example, κ = 5 indicates it is five times more costly (or 5 times worse, in some sense) to call a poor data set “good” than to call a good data set “poor.”
The optimal values of λ and τ can be obtained by minimizing the expected loss over the data:
| 3 |
and thus
| 4 |
The optimal classifier (according to the objective functions defined by (1) and (3)) that would be used for future data sets is given by f* = f (x, β̂0, β̂1, λ̂, τ̂), where the final estimates β̂0 and β̂1 are obtained using λ̂ and maximizing (1) for all the data. We arrived at estimates for λ and τ by calculating (3) over a grid λ × τ where λ = {0.05 to 0.15 by 0.02} and τ = {0.01 to 0.99 by 0.01}.
We now describe the entire process algorithmically as follows:
(1) Randomly divide the data into five partitions, {1, 2, 3, 4, 5}.
(2) Define a sequence of λ values and a sequence of τ values and construct a grid of the two sequences, λ × τ.
(3) For each (λ × τ) in the grid λ × τ:
(a) For each training partition ({1, 2, 3, 4}, {1, 2, 3, 5}, {1, 2, 4, 5}, {1, 3, 4, 5}, and {2, 3, 4, 5}), fit the logistic regression model; that is, obtain the estimates β̂0 and β̂1 by maximizing the penalized log likelihood function, (1), using the current value of λ.
(b) For each data set i, calculate the predicted quality, ŷi = f–δ(i) (xi, β̂0, β̂1, λ, τ), using the current values of λ and τ and the parameter estimates β̂0 and β̂1 that were obtained from the four partitions that did not include data set i.
(c) For each data set, calculate the loss as defined by the loss matrix above.
(d) Calculate the expected loss, V(λ, τ) by averaging the loss values over all the data sets.
(4) Identify the (λ, τ) pair in the grid λ × τ that has the lowest V(λ, τ). This pair, (λ̂, τ̂), is the “optimal” estimate of the regularization parameter λ and the threshold τ.
(5) Having identified (λ̂, τ̂), obtain new estimates β̂0 and β̂1 using λ̂ and all the data. This final LLRC model, f* = f (x, β̂0, β̂1, λ̂, τ̂), can be used to classify future data sets.
The calculation of sensitivity and specificity in Tables 2 and 4 and Figure 2 is similar to (3), except an indicator loss function is used, and it is averaged over a subset of the data. For sensitivity, or the true positive rate (TPR), we have
| 5 |
and for specificity, the true negative rate (TNR):
| 6 |
Supporting Information Available
Representative instrument raw files and the complete list of QC metrics for all data sets. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Tabb D. L. Quality assessment for clinical proteomics. Clin. Biochem. 2013, 46, 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulovich A. G.; Billheimer D.; Ham A. J.; Vega-Montoto L.; Rudnick P. A.; et al. Interlaboratory study characterizing a yeast performance standard for benchmarking LC-MS platform performance. Mol. Cell. Proteomics 2010, 9, 242–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke M. M.; Waters K. M.; Metz T. O.; Jacobs J. M.; Sims A. C.; et al. Improved quality control processing of peptide-centric LC-MS proteomics data. Bioinformatics 2011, 27, 2866–2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong C. C.; Cociorva D.; Miller C. A.; Schmidt A.; Monell C.; et al. Proteomics of Pyrococcus furiosus (Pfu): Identification of Extracted Proteins by Three Independent Methods. J. Proteome Res. 2013, 12, 763–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudnick P. A.; Clauser K. R.; Kilpatrick L. E.; Tchekhovskoi D. V.; Neta P.; et al. Performance metrics for liquid chromatography-tandem mass spectrometry systems in proteomics analyses. Mol. Cell Proteomics 2010, 9, 225–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z. Q.; Polzin K. O.; Dasari S.; Chambers M. C.; Schilling B.; et al. QuaMeter: multivendor performance metrics for LC-MS/MS proteomics instrumentation. Anal. Chem. 2012, 84, 5845–5850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rencher A. C.Methods of Multivariate Analysis; John Wiley & Sons, Inc.: Hoboken, NJ, 2003. [Google Scholar]
- Hand D. J.Construction and Assessment of Classification Rules; Wiley: New York, 1997. [Google Scholar]
- David W. Hosmer J, Lemeshow S, Sturdivant R. X.. Applied Logistic Regression; Wiley: New York, 2013. [Google Scholar]
- Hastie T. J., Tibshirani R. J., Friedman J. J. H.. The Elements of Statistical Learning; Springer-Verlag: New York, 2009. [Google Scholar]
- Bramwell D. An introduction to statistical process control in research proteomics. J. Proteomics 2013, 95, 3–21. [DOI] [PubMed] [Google Scholar]
- Kim S.; Mischerikow N.; Bandeira N.; Navarro J. D.; Wich L.; Mohammed S.; Heck A. J.; Pevzner P. A. The generating function of CID, ETD, and CID/ETD pairs of tandem mass spectra: applications to database search. Mol. Cell. Proteomics 2010, 12, 2840–2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T. T.; Chen Y. F.; Hastie T.; Sobel E.; Lange K. Genome-wide association analysis by lasso penalized logistic regression. Bioinformatics 2009, 25, 714–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakitsch B.; Lippert C.; Stegle O.; Borgwardt K. A Lasso multi-marker mixed model for association mapping with population structure correction. Bioinformatics 2013, 29, 206–214. [DOI] [PubMed] [Google Scholar]
- Liu J.; Huang J.; Ma S.; Wang K. Incorporating group correlations in genome-wide association studies using smoothed group Lasso. Biostatistics 2013, 14, 205–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcaino J. A.; Cote R. G.; Csordas A.; Dianes J. A.; Fabregat A.; et al. The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 2013, 41, D1063–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.