Summary
In Caulobacter crescentus, the actin homologue MreB is critical for cell shape maintenance. Despite the central importance of MreB for cell morphology and viability, very little is known about MreB-interacting factors. Here, we use an overexpression approach to identify a novel MreB interactor, MbiA. MbiA interacts with MreB in both biochemical and genetic assays, colocalizes with MreB throughout the cell cycle, and relies on MreB for its localization. MbiA over-expression mimics the loss of MreB function, severely perturbing cell morphology, inhibiting growth and inducing cell lysis. Additionally, mbiA deletion shows a synthetic growth phenotype with a hypomorphic allele of the MreB interactor RodZ, suggesting that these two MreB-interacting proteins either have partially redundant functions or participate in the same functional complex. Our work thus establishes MbiA as a novel cell shape regulator that appears to function through regulating MreB, and opens avenues for discovery of more MreB-regulating factors by showing that overexpression screens are a valuable tool for uncovering potentially redundant cell shape effectors.
Introduction
The bacterial cytoskeleleton plays a vital role in many cellular functions, particularly those that determine the spatial architecture of the cell (reviewed in Graumann, 2007). MreB is a polymer-forming ATPase homologous to eukaryotic actin (van den Ent et al., 2001; Salje et al., 2011). It is found in the genomes of most rod-shaped bacteria, and its loss is either lethal or conditionally lethal in a variety of phylogenetically diverse organisms (van den Ent et al., 2001; Jones et al., 2001; Figge et al., 2004; Gitai et al., 2004; Bendezú and de Boer, 2008). In Caulobacter crescentus, MreB is required for viability in all conditions examined. When MreB is perturbed by genetic depletion or treatment with MreB-targeting drugs, cells become wider, lose viability and undergo lysis (Figge et al., 2004; Gitai et al., 2004; 2005; Takacs et al., 2010). Based on the severe morphological phenotypes of MreB disruption in Caulobacter and other bacteria, MreB is thought to be critical for coordinating the remodelling of the peptidoglycan cell wall. Consistently, Caulobacter MreB localizes to the sites of new peptidoglycan synthesis, appearing as a patchy structure early in the cell cycle and condensing into a mid-cell ring during the assembly of the division apparatus (de Pedro et al., 1997; Figge et al., 2004; Gitai et al., 2004; Aaron et al., 2007; Divakaruni et al., 2007).
MreB and actin are homologous proteins that both fulfil structural roles in the cell. However, while actin is known to have a vast multitude of different regulators and effectors spanning at least 70 distinct families (reviewed in Pollard et al., 2000 and Staiger and Blanchoin, 2006), there is remarkably little information about MreB-interacting factors that either function upstream of MreB to regulate its activities or downs tream of MreB to modulate its activities. Of the potential downstream effectors, MreB has been shown to interact with several proteins involved in peptidoglycan precursor biosynthesis and cell wall remodelling (Carballido-López et al., 2006; Kawai et al., 2009; White et al., 2010; Gaballah et al., 2011). The only definitive upstream MreB regulators known are the Escherichia coli-specific toxins YeeV and CptA, which inhibit MreB polymerization in the absence of their cognate antitoxins (Tan et al., 2011; Masuda et al., 2012). Additionally, MreB directly interacts with the essential transmembrane protein RodZ and the translation elongation factor EF-Tu, but the significance of these interactions remains unclear (Bendezú et al., 2009; Defeu Soufo et al., 2010; van den Ent et al., 2010). Many behaviours of MreB filaments likely require modes of regulation not covered by the known interacting factors, including nucleation of de novo polymer formation, maintenance of a pool of unpolymerized monomers, dynamic localization, and the response to environmental factors such as starvation, especially considering the energy cost of MreB’s ATPase activity (Esue et al., 2005). Thus, identification of additional MreB interactors is crucial to advancing the understanding of MreB function.
In this paper, we report the identification of a novel MreB-interacting factor, MbiA, which colocalizes with MreB in an MreB-dependent fashion. We show that MbiA exhibits both genetic and biochemical interactions with MreB. Finally, the overexpression of MbiA severely perturbs MreB localization, cell shape and cell viability.
Results
MbiA colocalizes with MreB and requires MreB for localization
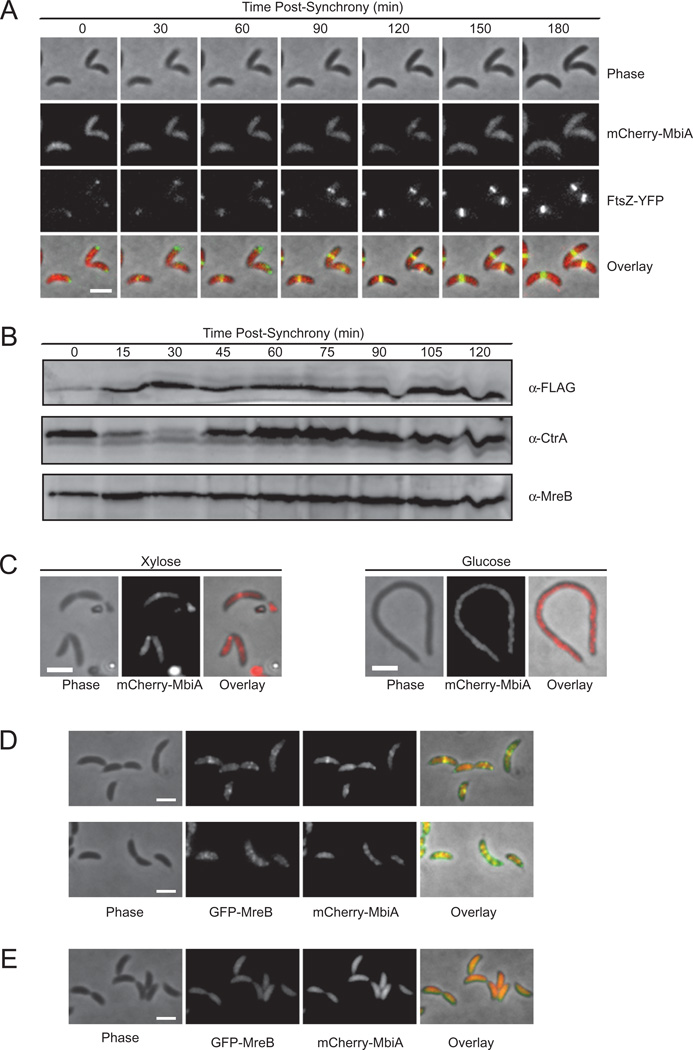
In a visual screen of the C. crescentus localizome library (Werner et al., 2009), we identified a previously uncharacterized protein, CC1438 (from here on referred to as MbiA for MreB Interactor A), whose subcellular distribution resembled that of MreB. MbiA is a small, 100-amino acid, predicted cytoplasmic protein conserved in a subset of α-Proteobacteria. None of its homologues have a known function, although some show distant similarity to GIY. YIG family of endonucleases. Like MreB, MbiA formed an extended structure of stripes or patches early in the cell cycle and condensed into a mid-cell band shortly after the appearance of the FtsZ protein at the nascent division site (Fig. 1A). Subsequently, the MbiA and FtsZ rings colocalized until MbiA dispersed from mid-cell slightly before the completion of cytokinesis.
Fig. 1. Cell cycle dependence and localization of MbiA.
A. Time-lapse microscopy of ZG691 expressing mCherry-MbiA and FtsZ-YFP. FtsZ-YFP was induced with xylose for 1 h, and mCherry-MbiA was induced with vanillate for 2 h. Cells are shown at the indicated times after synchronization. PYE agarose pads supplemented with vanillate were used for microscopy. The images, from top to bottom, are phase contrast, mCherry-MbiA fluorescence, FtsZ-YFP fluorescence, and an overlay of the three images.
B. Immunoblot with anti-FLAG, anti-CtrA and anti-MreB antibodies of the synchronized ZG694 cells expressing MbiA-FLAG as a native-site fusion at the indicated time points after synchronization.
C. Phase-contrast (left), mCherry-MbiA fluorescence (middle), and phase/fluorescence overlay (right) of ZG693 after 4 h of FtsZ depletion (Glucose) or with continued FtsZ expression (Xylose) in M2G or M2X respectively.
D. Colocalization of GFP-MreB and mCherry-MbiA in a double-labelled strain ZG692 grown in PYE supplemented with xylose and vanillate. The images, from left to right, are phase contrast, GFP-MreB fluorescence, mCherry-MbiA fluorescence, and an overlay of the three channels.
E. Colocalization of GFP-MreB and mCherry-MbiA in ZG692 grown in PYE supplemented with xylose and vanillate before treatment with 50 µg ml−1 of A22 on the agarose pad only. The images, from left to right, are phase contrast, GFP-MreB fluorescence, mCherry-MbiA fluorescence, and an overlay of the three channels. All scale bars are 2 µm.
Given the similarity of the localization patterns exhibited by MbiA and MreB (Figge et al., 2004; Gitai et al., 2004; Goley et al., 2011), we tested whether the mid-cell ring structure of MbiA, like that of MreB, formed in an FtsZ-dependent manner. Upon depletion of FtsZ, MbiA failed to organize into a discrete band, but retained some structure, as evident from its uneven, patchy distribution within the cell (Fig. 1C). Therefore, MbiA and MreB share a common mid-cell localization determinant.
In order to determine if MreB and MbiA colocalize with each other, as implied by their individual localization patterns, we constructed a double-labelled strain expressing GFP-MreB from the xylose promoter and mCherry-MbiA from the vanillate promoter. Indeed, MbiA colocalized with MreB both as a mid-cell band in early pre-divisional and stalked cells and as an extended patchy structure in the late pre-divisional and swarmer cells (Fig. 1D). Moreover, MbiA localization was MreB-dependent, as the disruption of the MreB filaments by treatment with A22, a small-molecule inhibitor of MreB polymerization (Gitai et al., 2005; Bean et al., 2009), led to a concomitant dispersal of MbiA throughout the cytoplasm (Fig. 1E). The MbiA-mCherry fusion integrated at the native mbiA locus recapitulated both the colocalization with MreB and the response to A22 treatment (Fig. S1A and B). Since MbiA-mCherry behaves like wild-type MbiA in overexpression assays (see below), this evidence suggests that MbiA directly or indirectly interacts with MreB and relies on it to determine its proper localization. To our knowledge, this is the only other Caulobacter protein, besides RodZ (Alyahya et al., 2009), known to both colocalize with MreB and depend on it for localization.
MbiA overexpression perturbs cell shape, cell viability and MreB localization
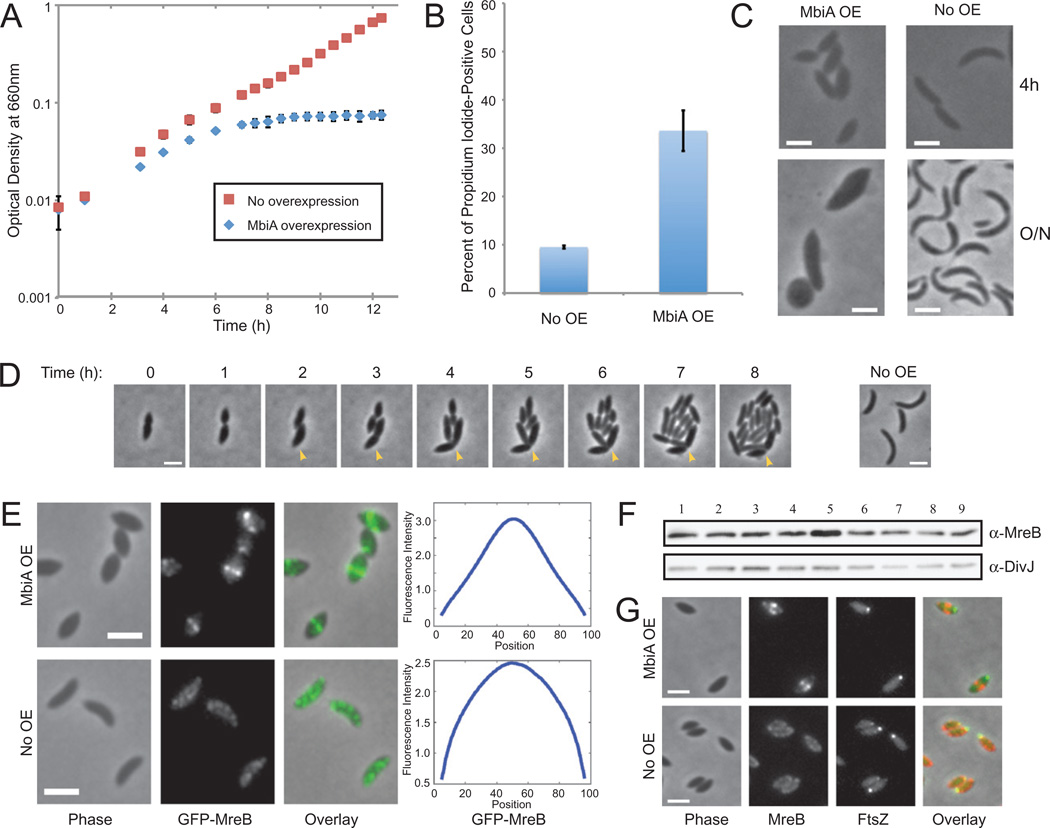
In order to characterize the cellular function of MbiA, we examined the effects of its overexpression in Caulobacter. Cells overexpressing either the Gateway-cloned MbiA from the xylose promoter or the untagged MbiA from the vanillate promoter on a high copy-number plasmid displayed reduced growth and stopped growing altogether by 7 h after the onset of induction (Figs 2A and S2A). Interestingly, by 5 h of MbiA overexpression, cells lost proper cell shape, becoming wider (Figs 2C and S2C). The increase in cell width was even more pronounced after overnight overexpression. This phenotype is very similar to the loss of function of MreB, which also leads to widening of the cells and inhibits cell growth (Figge et al., 2004; Gitai et al., 2004; 2005).
Fig. 2. Phenotypes of MbiA overexpression.
A. Growth curves of Caulobacter cells carrying pXyl::mbiA grown in PYE supplemented with either xylose (MbiA OE) or glucose (No OE). Normalized results of two experiments are presented.
B. Proportion of propidium iodide-positive cells carrying pXyl::mbiA after 7 h of growth in PYE with glucose (No OE) or xylose (MbiA OE).
C. Phase contrast images of cells carrying pXyl::mbiA and grown in PYE with xylose (MbiA OE) or glucose (No OE) for 4 h or overnight (around 24 h).
D. Cells carrying pXyl::mbiA grown in PYE with xylose for 8 h were placed on PYE pads with glucose to repress MbiA overexpression and imaged at indicated times after the transfer. Arrowhead indicates the progeny of the recovering cell that fails to divide. The panel at the right shows an isogenic strain grown in PYE with glucose and imaged at the 0 h time point.
E. Phase contrast, GFP-MreB fluorescence, and overlay images of swarmer cells immediately after synchronization and chemical fixation. LS3814 cells carrying (MbiA OE) or not carrying (No OE) pXyl::MbiA were grown in PYE with xylose. The plots at the right represent the average fluorescence intensity (in arbitrary units) at each position along the longitudinal axis (in % cell length) for the entire population of swarmers. n = 1126 for MbiA OE and n = 859 for No OE.
F. Immunoblot with anti-MreB and anti-DivJ antibodies of cells expressing either wild-type levels, increased levels, or no MbiA. Lane 1: CB15N; Lane 2: ΔmbiA (ZG708); Lane 3: MT196 with xylose; Lane 4: MT196 with glucose; Lane 5: MT196 pXyl::mbiA with xylose; Lane 6: MT196 pXyl::mbiA with glucose; Lane 7: CB15N pBV-mbiA (vanillate-inducible overexpression vector) with vanillate; Lane 8: CB15N pBV-mbiA without vanillate; Lane 9: CB15N pBVMCS-4 (empty vector) with vanillate. All cells were grown in PYE for 5 h.
G. Colocalization of mCherry-MreB and FtsZ-CFP in ZG695 swarmers carrying (MbiA OE) or not carrying (No OE) pXyl::MbiA. Cells were grown in PYE with xylose, inducing FtsZ-CFP for 1 h with 50 µM vanillate, and the images were taken immediately after synchronization and chemical fixation. Phase contrast, mCherry-MreB fluorescence, FtsZ-CFP fluorescence, and overlay images are provided. All scale bars are 2 µm.
Prolonged depletion of MreB eventually causes cell lysis (Figge et al., 2004), so the appearance of phase-bright lysed cells upon MbiA overexpression led us to hypothesize that MbiA overexpression might also be bacteriocidal. After 8 h of MbiA overexpression, the fraction of cells that stained positively with the membrane-impermeable DNA-binding dye propidium iodide greatly increased (Fig. 2B). This result shows that MbiA overexpression compromises membrane integrity, which is consistent with its culmination in cell death. Moreover, the number of colony-forming units per unit of optical density significantly decreased upon induction of MbiA overexpression, confirming that MbiA overexpression is bacteriocidal (Fig. S2B). Nevertheless, when MbiA overexpression was halted prior to cell death, a fraction of cells was capable of recovering proper morphology after a few divisions, even if their shape was already abnormally wide at the on set of the recovery process (Figs 2D and S3).
Since MbiA overexpression leads to morphology and viability phenotypes similar to those of the loss of MreB, we examined the effect of MbiA overexpression on MreB localization. Interestingly, MbiA overexpression did not abolish MreB localization, but instead led to a novel phenotype in which MreB partially or completely condensed near the centre of the cell (Fig. S2D). In order to remove the confounding variable of cell cycle-dependent changes in localization, we chose to specifically analyse swarmer cells. At this stage of the cell cycle, uninduced control cells exhibited extended, patchy MreB localization. In contrast, upon MbiA overexpression, MreB condensed towards the middle of the cell, sometimes forming bands reminiscent of those seen in stalked cells (Fig. 2E). Quantitative analysis of GFP-MreB fluorescence over the length of the cell confirmed that MbiA overexpression leads to the condensation of the MreB structure.
MreB condensation did not result from altered levels of the MreB protein, as one of the two MbiA overexpression vectors had no effect on MreB levels (Fig. 2F). Although the other MbiA overexpression strain showed a slight increase in MreB levels, we confirmed that MreB overexpression alone failed to recapitulate the MbiA overexpression phenotypes (Fig. S4).
Since MreB condensation into a tight band normally requires FtsZ ring formation (Figge et al., 2004), we tested if MbiA-induced MreB condensation could be explained by mislocalization of FtsZ. In wild-type swarmer cells, fluorescent FtsZ fusions localize as a single focus at the non-flagellated pole (Thanbichler and Shapiro, 2006). In swarmer cells overexpressing MbiA, FtsZ similarly exhibited unipolar localization, even when MreB formed a mid-cell band (Fig. 2G). Consequently, the MbiA-induced MreB condensation appears to be an FtsZ-independent process.
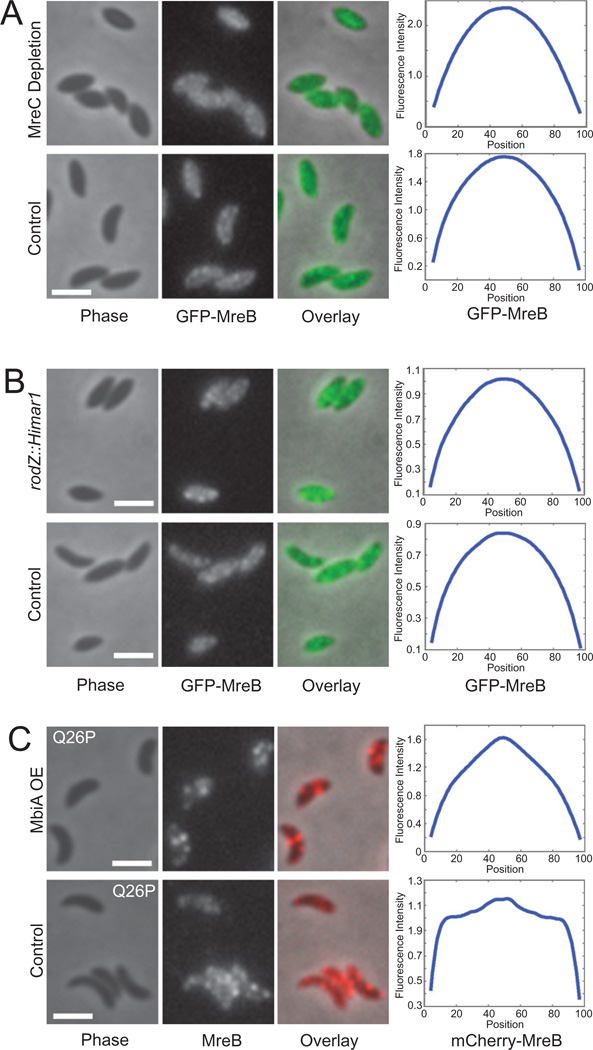
Finally, in order to test if the MbiA-induced change in MreB localization is a secondary effect of the altered cell shape, we examined MreB localization in swarmer cells depleted for MreC (Dye et al., 2005; Divakaruni et al., 2007) or carrying a hypomorphic rodZ mutation, rodZ:: Himar1 (Alyahya et al., 2009). Under both of these conditions, MreB maintained the extended, patchy localization seen in wild-type cells despite the aberrant cell morphology, confirming that MreB condensation is not a necessary consequence of the increase in cell width (Fig. 3A and B).
Fig. 3. Genetic examination of MreB localization in swarmer cells.
A. Localization of GFP-MreB in swarmers of ZG498 grown in PYE with either glucose (MreC Depletion) or xylose (Control; induction of a complementing copy of MreC-RFP1) overnight and subcultured into respective media supplemented with vanillate 5 h prior to synchronization and chemical fixation. Phase contrast, GFP-MreB fluorescence, and overlay images are provided. The plots at the right represent the average fluorescence intensity (arbitrary units) with respect to position (in % cell length) along the long axis of the cell. n = 713 for Depletion and n = 912 for Control.
B. Localization of GFP-MreB in rodZ::Himar1 hypomorph (ZG706) and wild-type (ZG472) swarmer cells grown in PYE with vanillate and imaged immediately after synchronization and chemical fixation. The phase contrast, GFP-MreB fluorescence, and overlay images as well as the averaged fluorescence profiles for the populations are presented. n = 495 for rodZ::Himar1 and n = 452 for wild-type.
C. Localization of mCherry-MreBQ26P in ZG707 swarmer cells carrying either pBV-mbiA (MbiA OE) or pBVMCS-4 (No OE) and grown in PYE with xylose and vanillate. Cells are shown immediately after synchronization and chemical fixation. The images are phase contrast, mCherry-MreB fluorescence, and an overlay of the two channels. All scale bars are 2 µm. The averaged fluorescence profiles are provided on the right. n = 979 for MbiA OE and n = 653 for No OE.
MreB condensation is a unique phenotype of MbiA overexpression
Because MbiA overexpression affects both cell shape and the localization of MreB, we examined the consequences of MbiA overexpression for the localization of other proteins involved in peptidoglycan synthesis and cell shape maintenance. In particular, we looked at the localization of the cell shape determinant MreC and the transpeptidase PBP2, both of which are required for rod shape (Dye et al., 2005; Wagner et al., 2005, Divakaruni et al., 2007; Bendezú and de Boer, 2008), as well as the peptidoglycan precursor biosynthesis enzyme MurG and the putative peptidoglycan peptidase DipM, both of which arrive at the incipient division plane concurrently with MreB (Aaron et al., 2007; Goley et al., 2010; 2011; Poggio et al., 2010). None of these proteins was mislocalized by MbiA overexpression (Fig. S2E–H). Thus, MbiA overexpression appears to affect the localization of MreB, but not that of other prominent members of the peptidoglycan synthesis machinery.
We also examined whether the partial condensation exhibited by MreB in the presence of MbiA overexpression is related to the MreB condensation that occurs in the course of the normal cell cycle shortly after the swarmer-to-stalked cell transition (Figge et al., 2004; Gitai et al., 2004; Goley et al., 2011). This was a particularly tempting hypothesis due to the finding that MbiA protein levels appear to be slightly lower in swarmer cells and increase during the differentiation into stalked cells (Fig. 1B). In order to test this hypothesis, we took advantage of Q26P, a previously characterized MreB point mutant that fails to condense into a ring (Aaron et al., 2007; Dye et al., 2011). As previously reported, a Q26P merodiploid strain carrying both mCherry-fused and unlabelled copies of the mutant mreB exhibited an extended patchy pattern of localization with polar foci at all stages of the cell cycle. However, in the presence of MbiA overexpression, the mutant MreB still condensed into partial or complete rings, largely excluding the protein from the cell poles (Fig. 3C). Consequently, MbiA-mediated partial condensation of MreB is either independent of the usual pathway of MreB condensation or MbiA overexpression is epistatic to the effects of the Q26P mutation.
MbiA overexpression phenotypes can be suppressed by mutations in mreB
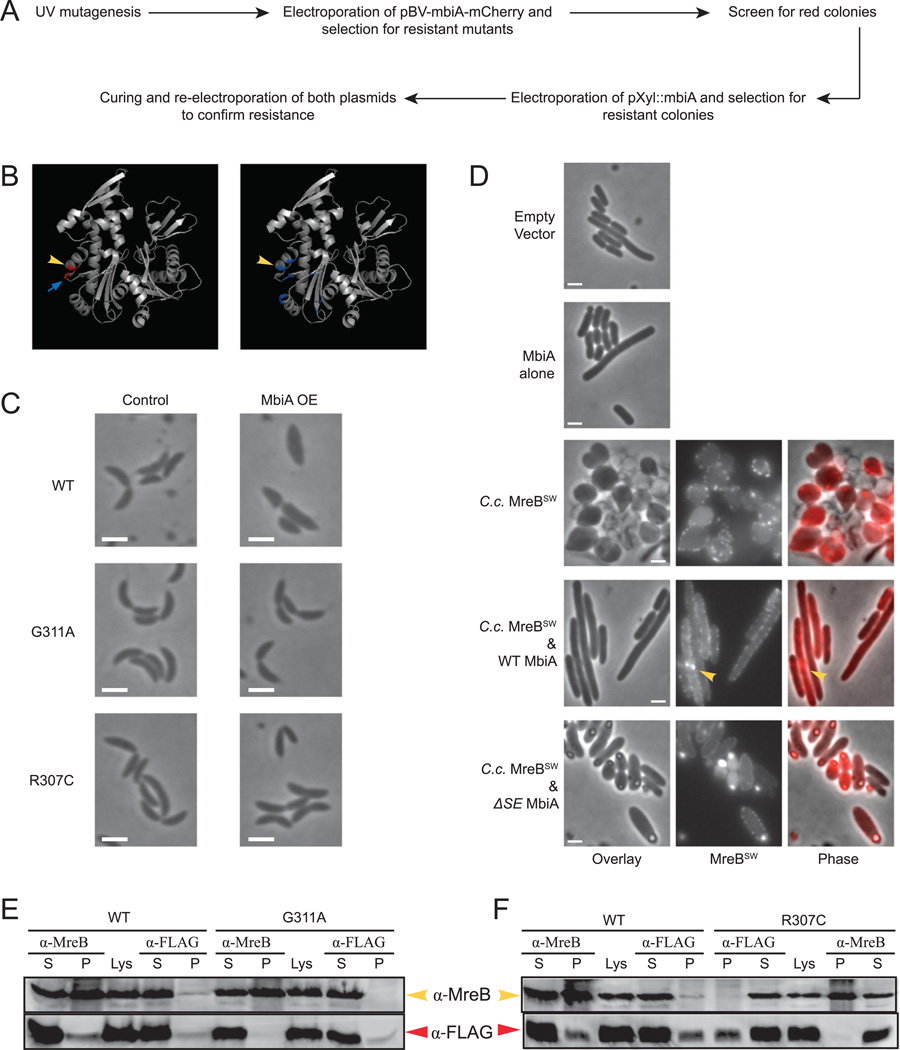
In order to uncover the mechanism of MbiA action, we conducted a suppressor screen for mutants resistant to MbiA overexpression. The design of the screen is presented in Fig. 4A. First, CB15N wild-type cells were mutagenized with UV radiation and transformed with a high copy-number plasmid encoding an MbiA-mCherry fusion under the control of a vanillate-inducible promoter. Cells were selected for growth in the presence of vanillate, and the resulting colonies were screened for red fluorescence to ensure continued fusion expression. We then transformed the cells grown from the red colonies with a non-compatible high copy-number vector encoding MbiA under the control of a xylose-inducible promoter and selected for mutants that could grow in the presence of xylose. This strategy yielded many plasmid-borne non-functional MbiA alleles (Fig. S5) as well as two chromosomal mutants capable of maintaining either of the two MbiA overexpression plasmids.
Fig. 4. Genetic and biochemical analyses of the interaction between MbiA and MreB.
A. Schematic diagram of the genetic screen for chromosomal mutants resistant to MbiA overexpression.
B. Crystal structures of Thermotoga maritima MreB based on (van den Ent et al., 2001). In the left panel, residues corresponding to the mutations from MbiA-resistant Caulobacter mutants are shown in red. In the right panel, residues predicted to be important for MreB–RodZ interaction (van den Ent et al., 2010) are shown in blue. Yellow arrowheads indicate the R307 residue, and the blue arrow points at G311.
C. Phase contrast images of CB15N and the two MbiA-resistant mutants (ZG696 and ZG697) carrying pBV-mbiA-mCherry grown for 5 h in PYE either with vanillate (MbiA OE) or without vanillate (No OE).
D. Phase contrast images of DH5α E. coli cells carrying pTrc99a empty vector or pTrc-mbiA expression plasmid. Additionally, phase contrast, MreBSW fluorescence, and overlay images of DH5α expressing Caulobacter MreB sandwich fusion alone (pTrc-MRM), with wild-type MbiA (pTrc-mbiA-MRM) or with ΔSE MbiA (pTrc-mbiAΔSE-MRM). Cells were grown in LB and induced with 1 mM IPTG for 2 h. The arrowhead indicates the bright bundle of MreBSW. All scale bars are 2 µm.
E. Immunoblot of co-immunoprecipitation experiment between MreB and MbiA-FLAG in CB15N or G311A cells carrying pBV-mbiA-FLAG plasmid. The immunoprecipitation was performed with the antibodies indicated above the corresponding lanes. The supernatant (S) and the pellet (P) from the co-immunoprecipitation as well as the original lysate (Lys) were loaded on the gel. The immunoblot was performed with the antibodies indicated at the right.
F. Immunoblot of co-immunoprecipitation experiment between MreB and MbiA-FLAG in CB15N or R307C cells carrying pBV-mbiA-FLAG plasmid. The immunoprecipitation was performed with the antibodies indicated above the corresponding lanes. The supernatant (S) and the pellet (P) from the co-immunporecipitation as well as the original cell lysate (Lys) are shown. The immunoblot was performed with the antibodies indicated at the left.
We hypothesized that MreB might be the direct target of MbiA. Therefore, we sequenced the mreB gene in the two chromosomal suppressors. Indeed, both mutants had mutations in helix 11 of MreB corresponding to residues G311A and R307C respectively (Fig. 4B). In order to confirm that these mutations were truly responsible for the resistance to MbiA overexpression, the mreB gene from the mutants was transduced into a clean CB15N background. All transductants tested were resistant to both of the overexpression vectors. Additionally, when the mutations were introduced into a wild-type genetic background by double recombination, the resulting strains were immune to MbiA overexpression (data not shown). The experiments described below were performed with the transductants unless stated otherwise.
The suppressor mutations not only suppressed the lethality of MbiA overexpression, but also the cell width phenotype. While MbiA overexpression caused a substantial increase in cell width of wild-type cells, both G311A and R307C cells remained visibly unchanged (Figs 4C and S6A). Additionally, overexpression of MbiA in the mutants did not slow down cell growth compared to the uninduced controls (data not shown). Therefore, MbiA phenotypes appear to be completely reversed by mutations in MreB, suggesting that MbiA normally mediates its effects through a direct interaction with MreB. Furthermore, although MbiA normally colocalizes with MreB in an MreB-dependent manner, its localization in the two suppressor mutants became completely diffuse (Fig. 5B), supporting the conclusion that these alleles of MreB can no longer interact with MbiA.
Fig. 5. Phenotypic analysis of the MbiA-resistant mutants.
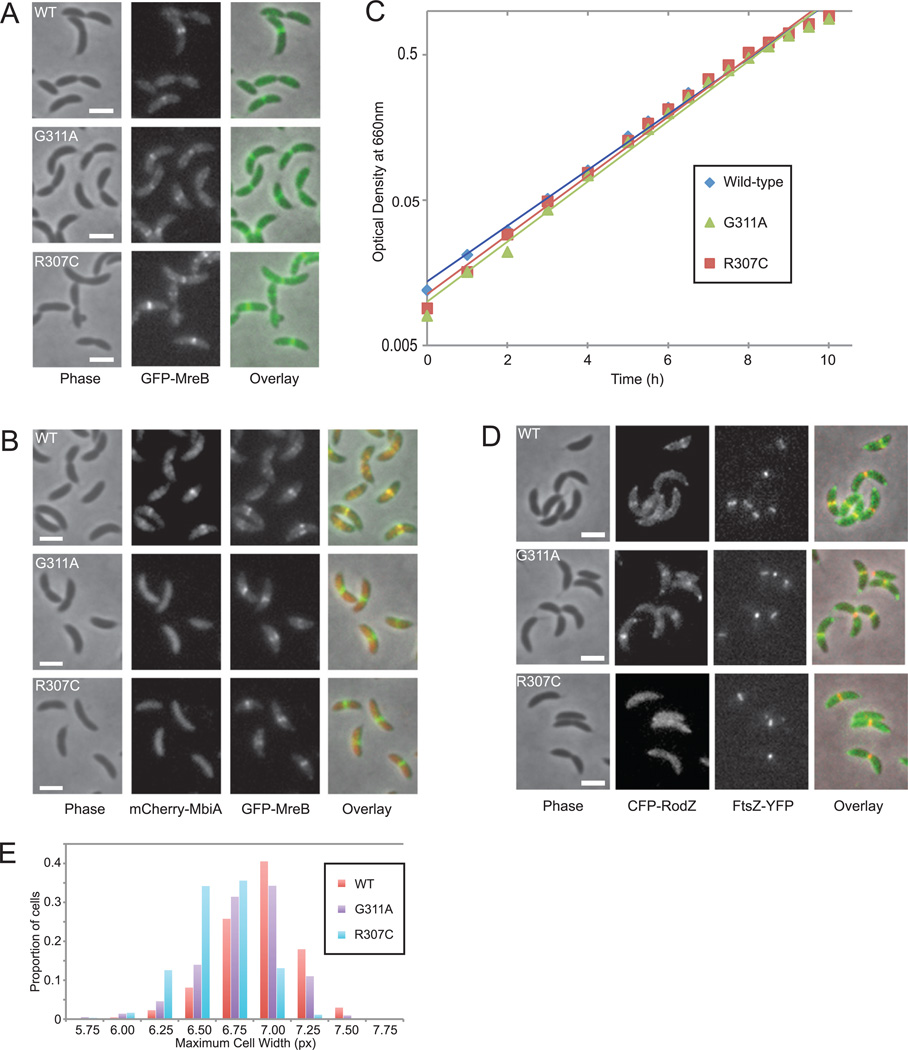
A. Localization of GFP-MreB in wild-type (WT) (LS3814), G311A (ZG698) and R307C (ZG699) cells grown in PYE with xylose. The native site and the fluorescent copies of mreB in the mutants both contain the indicated mutation. Phase contrast, GFP-MreB fluorescence, and overlay images are shown.
B. Colocalization of mCherry-MbiA and GFP-MreB in double-labelled WT (ZG692), G311A (ZG700), and R307C (ZG701) strains grown in PYE with xylose and vanillate. Phase contrast, mCherry-MbiA fluorescence, GFP-MreB fluorescence, and overlay images are provided.
C. Growth curves of CB15N, G311A (ZG696), and R307C (ZG697) grown in PYE. A representative result of several experiments is presented. The lines represent exponential best-fit regressions.
D. Colocalization of CFP-RodZ and FtsZ-YFP in double-labelled WT (ZG703), G311A (ZG704) and R307C (ZG705) strains grown in PYE with xylose. FtsZ-YFP was induced with 1 mM vanillate for 1 h. Phase contrast, CFP-RodZ fluorescence, FtsZ-YFP fluorescence, and overlay images are shown. All scale bars are 2 µm.
E. Histogram of maximum cell widths in CB15N, G311A (ZG696), and R307C (ZG697) cells measured with the Microbe Tracker program (Sliusarenko et al., 2011). n = 1188 for CB15N; n = 1353 for G311A; and n = 1005 for R307C.
Both R307 and G311 are surface-exposed residues, which could allow them to participate in protein–protein interaction by constituting a binding site, especially given their spatial proximity to each other. Interestingly, Q298 of Thermotoga maritima MreB, the residue equivalent to R307 of Caulobacter, has previously been implicated in the MreB–RodZ interaction (van den Ent et al., 2010; Fig. 4B). This finding raises the possibility that MbiA might either competitively inhibit RodZ binding or regulate the function of the MreB–RodZ complex in some other way. Due to the high background of the available fusions, we were unable to determine whether MbiA overexpression compromised RodZ localization or merely altered it in a manner similar to MbiA overexpression’s effect on MreB. However, examination of the R307C mutant showed extremely poor efficiency of RodZ localization, which barely rose above the background levels of fluorescence (Figs 5D and S6B). In contrast, both the G311A mutant and wild-type cells exhibited strong RodZ localization at the old pole and at the division plane, in agreement with previously published results (Alyahya et al., 2009).
Surprisingly, cells carrying the R307C allele were also visibly thinner than either wild-type or G311A mutant cells (Figs 4C and 5A). This observation was confirmed by quantitative analysis of cell width, which indicated a statistically significant difference between the wild-type width of 6.81 ± 0.30 px and the width of R307C mutant cells, 6.50 ± 0.25 px (P-value < 0.001, n = 1188 for CB15N, n = 1005 for R307C, Fig. 5E). Since the cell width of the G311A mutant was more similar to that of wild-type CB15N, the morphological phenotype of R307C is likely a secondary effect of the mreB mutation, unrelated to the function of MbiA. However, given that the loss of either MreB or RodZ leads to increased cell width, it is interesting that a mutation that reduces the efficiency of RodZ colocalization with MreB causes cell width to decrease (Figge et al., 2004; Gitai et al., 2004; Alyahya et al., 2009).
Neither the morphological changes nor the efficiency of MreB binding to MbiA and RodZ appear to play a critical role in cell growth, as the division time of the two suppressor mutants was not significantly different from that of wild-type cells (Fig. 5C). Additionally, neither mutation altered the localization of MreB (Fig. 5A), suggesting that a functional interaction with MbiA is unnecessary for proper MreB localization.
MbiA interacts with MreB biochemically
Since MreB and MbiA interact genetically, we sought to test if this interaction is direct. In a phylogenetically distinct organism, E. coli, which does not encode an mbiA homologue, overexpression of MbiA from an IPTG-inducible promoter failed to significantly affect cell morphology (Figs 4D and S7B). Expression of the Caulobacter MreB internal mCherry sandwich fusion (MreB’-mCherry-’MreB or MreBSW), on the other hand, caused E. coli cells to become round and lyse, while MreBSW primarily localized to the periphery in distinct foci. The phenotypes of MreBSW heterologous expression were robustly suppressed by the co-expression of MbiA. E. coli cells co-expressing MreBSW and MbiA exhibited cell width closer to that of wild type E. coli and displayed slight filamentation. In these cells, MreBSW localized either as an extended, helix-like structure or as bright strands that might correspond to MreB filament bundles (Fig. 4D arrowhead).As a caveat, the presence of the mbiA gene upstream of mreB on the overexpression vector lowered the MreBSW expression levels in these cells (data not shown). To make sure that this change in the expression level was not responsible for the observed suppression of the MreB overexpression phenotype, MreBSW was co-expressed with MbiAΔSE and MbiAY71N, mutant versions of MbiA that were isolated in the suppressor screen (Fig. S5) and do not affect Caulobacter viability upon overexpression. In the presence of these MbiA mutants, the cell width phenotype of the MreBSW overexpression was only mildly ameliorated (Fig. 4D and data not shown). Similarly, the G311A and R307C variants of MreBSW produced abnormally wide E. coli cells even when co-expressed with wild-type MbiA (Fig. S7B and data not shown). These results imply that MbiA is capable of interacting with MreB in a heterologous system, in the absence of any other Caulobacter proteins.
Finally, we sought to confirm the interaction between MbiA and MreB with a biochemical assay. Lysates from wild-type, MreBG311A, and MreBR307C cells overexpressing an MbiA-FLAG fusion from a high copy-number vector were subjected to immunoprecipitation with either α-MreB or α-FLAG antibodies. In the wild-type lysate, α-MreB antibody was capable of co-immunoprecipitating MbiA-FLAG with MreB and, conversely, α-FLAG antibody was capable of co-immunoprecipitating MreB with MbiA-FLAG (Fig. 4E and F). However, in the control lysates derived from the MreB mutants that suppressed MbiA overexpression, α-MreB antibody only precipitated MreB and failed to co-immunoprecipitate MbiA-FLAG, while α-FLAG antibody only precipitated MbiA-FLAG and failed to co-immunoprecipitate MreB. To confirm that the observed MbiA–MreB interaction was not forced by MbiA-FLAG overexpression, the immunoprecipitation was repeated with lysates from the strains encoding MbiA-FLAGx2 fusion at the native site locus. The α-FLAG antibody was found to precipitate MreB with MbiA-FLAGx2 and the α-MreB antibody was found to precipitate MbiA-FLAGx2 from wild-type, but not from the control G311A, lysate (Fig. S7A). Together with the evidence presented above, these results suggest that MbiA and MreB interact in wild-type cells, and that this interaction requires the G311 and R307 residues of MreB.
mbiA deletion has a synthetic phenotype with rodZ
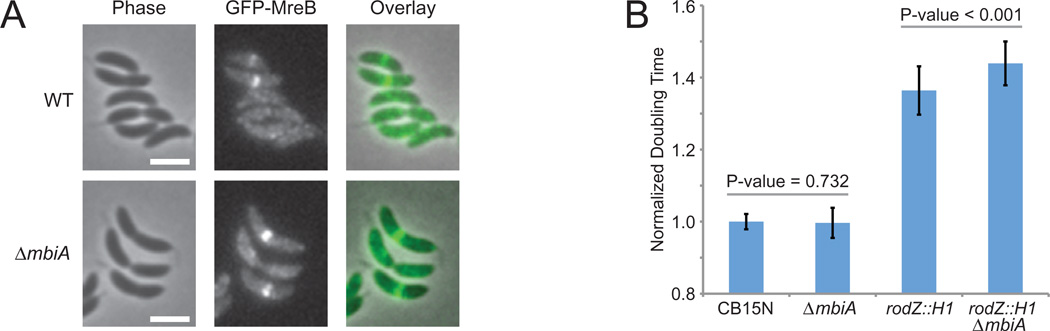
Seeking to further elucidate the role of MbiA in the MreB-directed peptidoglycan synthesis pathway, we examined the phenotypic effects of deleting the mbiA gene. ΔmbiA cells grew at the same rate as wild-type Caulobacter (Figs 6B and S8A; n = 22 cultures for each strain). This is consistent with the observation that the mreB mutations that abolished MbiA binding did not significantly perturb cell growth. If MreB is the sole target of MbiA, then removal of MbiA should not have a more severe phenotype than that observed with the MreB mutants that fail to bind MbiA. Consistently, ΔmbiA did not significantly affect either cellular morphology or MreB localization (Figs 6A and S8C).
Fig. 6. Phenotypic analysis of mbiA deletion.
A. Localization of GFP-MreB in wild-type (WT) (LS3814) and ΔmbiA (ZG709) strains grown in PYE with xylose. Phase contrast, GFP-MreB, and overlay images are provided. The scale bars are 2 µm.
B. Average growth rates of CB15N, ΔmbiA (ZG708), rodZ::Himar1 (CJW2537) and rodZ::Himar1 ΔmbiA (ZG710). The growth measurements were performed using a Synergy HT microplate reader, and the cells were grown in PYE (CB15N and ZG708) or PYE with kanamycin (CJW2537 and ZG710). The data shown are normalized to wild-type and represent the results of one experiment with 22 replicates of each strain for CB15N and ZG708 and the results of two experiments with 88 total replicates of each strain for CJW2537 and ZG710.
Since the mbiA deletion failed to produce an overt phenotype by itself, we examined its phenotype in a sensitized genetic background. Specifically, the partial overlap between the MreB binding sites for MbiA and RodZ suggested that MbiA and RodZ might function in either a cooperative or competitive manner. We found that mbiA deletion reproducibly caused a small, but statistically significant synthetic reduction in the growth rate of the rodZ::Himar1 hypomorphic strain (Figs 6B and S8B; n = 88 cultures for each strain). Therefore, it appears that rather than competitively inhibit the MreB–RodZ interaction, MbiA might either positively contribute to the functionality of the MreB–RodZ complex or act in a pathway parallel to RodZ.
Discussion
Eukaryotic actin is regulated at multiple levels, including monomer pool maintenance, nucleation, bundling, nucleotide exchange, filament elongation, stabilization, capping and severing (Pollard et al., 2000). These activities are mediated by a diverse set of interacting factors spanning at least 70 different families (Staiger and Blanchoin, 2006). Therefore, the paucity of known interactors of its prokaryotic homologue MreB is particularly surprising. Here, we identified a novel MreB interacting factor, MbiA. MbiA overexpression causes mislocalization of the MreB structure to a fully or partially condensed mid-cell ring and mimics the loss of MreB by increasing cell width, lowering growth rate and eventually leading to lysis. MbiA appears to act directly on MreB, since both proteins reciprocally co-immunoprecipitate with each other, can interact in vivo in a heterologous system, and mutations in MreB that abolish MbiA binding suppress the effects of MbiA overexpression. In contrast to the known E. coli MreB regulators YeeV and CptA (Tan et al., 2011; Masuda et al., 2012), mbiA does not appear to be in an operon with an antitoxin, so MbiA is unlikely to represent a toxin of a toxin-antitoxin system. Moreover, there are no essential sequence elements in the vicinity of the mbiA locus, pointing to the absence of a potential RNA antitoxin (Christen et al., 2011).
How does MbiA modulate MreB to affect cell shape?
MbiA seems to control cell morphogenesis in Caulobacter by acting on MreB, but how? One possibility is that the binding of MbiA to MreB competitively or allosterically inhibits the binding of MreB to downstream components of the peptidoglycan remodelling machinery. Overexpression of MbiA failed to affect the localization of either peptidoglycan biosynthesis pathway enzyme MurG [which is known to interact with MreB (White et al., 2010; Gaballah et al., 2011)], the morphogenesis factor MreC (Lee and Stewart, 2003; Dye et al., 2005; Bendezú and de Boer, 2008), or the primary transpeptidase associated with the elongation mode of cell growth, PBP2 (de Pedro et al., 2001; Divakaruni et al., 2007; Uehara and Park, 2008). However, the maintenance of proper localization does not necessarily imply the maintenance of proper function. We were also unable to conclusively determine whether MbiA overexpression affects the localization of the MreB-binding protein RodZ (Shiomi et al., 2008; Alyahya et al., 2009; Bendezú et al., 2009), which was a particularly tempting candidate since the RodZ binding site of MreB is adjacent to the MbiA interaction site (van den Ent et al., 2010). However, RodZ overexpression fails to suppress MbiA overexpression (Fig. S9), so the genetic evidence argues against the idea that MbiA competitively inhibits MreB–RodZ binding. mbiA deletion also exhibits a synthetic growth phenotype with a rodZ hypomorphic mutation. Thus, the data point to a role of MbiA in the MreB-mediated peptidoglycan synthesis that is distinct from inhibition of RodZ.
Alternatively, MbiA could directly modulate MreB architecture, which would explain the effects of MbiA overexpression on MreB localization. The mislocalization of MreB could lead to aberrant cell growth by either sequestering MreB in a non-functional state or recruiting the MreB-directed growth machinery to an ectopic site. For example, MbiA could act as a nucleation or elongation factor and selectively promote MreB nucleation or elongation only at mid-cell. Conversely, it could act as a depolymerization factor akin to eukaryotic ADF/cofilin (Pollard et al., 2000) and selectively destabilize MreB filaments at the poles. We view these possibilities as unlikely because they would require MbiA to directly impart spatial information to the system, either acting exclusively at the mid-cell in the case of a nucleator or an elongation factor or at the poles in the case of a depolymerization factor. However, MbiA localization was shown to be completely MreB-dependent, as MbiA fails to localize after A22 treatment. Alternatively, MbiA could act as a bundling factor akin to eukaryotic α-actinin (Sjöblom et al., 2008) and mediate the collapse of an extended helix-like structure into a more condensed one by increasing the lateral interaction between MreB filaments. This MbiA-dependent condensation of MreB could potentially be related to the condensation of MreB into a mid-cell ring that occurs in the course of a normal cell cycle. However, the ability of MreB to form a mid-cell band in ΔmbiA mutant suggests that other factors must also contribute to the normal cell cycle-dependent MreB condensation.
The in-depth analysis of the potential involvement of MbiA in MreB filament bundling, nucleation, elongation or destabilization awaits the development of an in vitro system for examining Caulobacter MreB filaments. Previous attempts to purify Caulobacter MreB in our lab failed due to poor solubility of the protein. However, the recent discovery of an amphipathic helix and a membrane-interacting loop in MreB homologues of Gram-negative bacteria (Salje et al., 2011) opens new avenues for the development of in vitro MreB polymerization assays with solubilized protein lacking the membrane-interacting sites.
Overexpression as a valuable tool for cell shape studies
Although MbiA overexpression leads to a dramatic phenotype, the mbiA deletion in the wild-type background fails to produce an overt effect either on MreB localization, cell growth rate or cell morphology. ΔmbiA does show a synthetic growth phenotype with a RodZ hypomorph, highlighting its involvement in the essential MreB-mediated peptidoglycan synthesis pathway, but the mildness of this phenotype suggests either the existence of redundant factors or the necessity of MbiA only under specific conditions. A preliminary screen of ≈ 12 000 transposon mutants for synthetic lethality with ΔmbiA failed to yield a synthetic lethal partner. However, considering the small size of mbiA itself, the hypothetical gene encoding an alternative factor would likely also be small, thus lowering its chances of being disrupted by a transposon. Therefore, we cannot discard the possibility of an alternative pathway existing. Additionally, ΔmbiA does not exhibit altered growth rate in minimal medium compared to wild-type cells, and MbiA levels are not significantly different in stationary phase compared to the exponential phase, suggesting that nutrient limitation and stationary phase are not conditions at which MbiA becomes critical for growth.
MbiA is not the only protein to have a dramatic overexpression phenotype, but a mild deletion phenotype. In fact, the analysis of the ASKA overexpression library in E. coli showed that 51% of the genes caused a severe growth phenotype upon overexpression (Kitagawa et al., 2005). Conversely, the systematic deletion study of E. coli Open Reading Frames conducted during the construction of the Keio collection led to the identification of only 303 essential genes (Baba et al., 2006), which is merely 7.6% of all genes encoded by this organism. Specifically in the field of cell shape, both of the previously known MreB regulators, the toxins YeeV and CptA, dramatically hinder cell growth upon overexpression without being essential for cell viability (Baba et al., 2006; Tan et al., 2011; Masuda et al., 2012). As these results and our characterization of MbiA indicate, genes that are sufficient, but not necessary, for regulating cell shape are missed in traditional deletion studies and transposon screens due to either redundancy or testing under conditions where those genes or the encoded proteins are not induced or activated. Although overexpression screens have been used to identify factors involved in a variety of different processes, they have not been systematically employed to study cell shape. MbiA is not a part of a toxin-antitoxin system, so the targeted approach used to discover YeeV and CptA would not have identified it either. Moreover, given the low degree of conservation of MbiA, YeeV and CptA, homology searches might not be an effective method of discovering MreB regulators. Thus our results indicate that overexpression studies provide a valuable tool for uncovering novel effectors of cell shape.
Experimental procedures
Strains, media and DNA manipulations
Caulobacter crescentus cells were grown in peptone yeast medium (PYE) or M2-glucose (M2G) medium at 30°C. The antibiotics were used at the following concentrations: 5 µg ml−1 kanamycin (25 µg ml−1 on plates), 1 µg ml−1 oxytetracycline (2 µg ml−1 on plates), 5 µg ml−1 streptomycin, 2.5 µg ml−1 gentamycin (5 µg ml−1 on plates), 1.5 µg ml−1 chloramphenicol, 50 µg ml−1 A22. Induction was typically achieved with 0.3% xylose or 1 mM vanillate for 5 h unless mentioned otherwise in the text. Glucose and sucrose were also used at the concentration of 0.3%.
The bacterial strains and plasmids used in this work are listed in Tables S1 and S2 respectively. Additionally, the primers used for construction of plasmids that were created specifically for this work are listed in Table S3. Plasmids were introduced into Caulobacter cells by electroporation. Transductions were achieved with φCr30 bacteriophage in accordance with the standard procedure (Ely and Johnson, 1977). The cloning of the plasmids and the construction of the strains used in this work are described in Supporting Information. Gateway cloning employed the LR and BP Clonase II enzyme kits (Invitrogen) as was outlined in the respective protocols provided by the company. Cloning into the pCR-BLUNT II-TOPO vector was accomplished using Zero Blunt Topo Cloning Kit (Invitrogen) in accordance with the manufacturer’s protocol.
Microscopy
Cells were immobilized on 1% agarose pads and imaged with a Nikon 90i microscope equipped with a Nikon Plan Apo 100x/1.4 phase contrast objective, a Rolera XR cooled CCD camera, and NIS Elements software. When necessary, cells were synchronized prior to visualization according to the standard protocol for Caulobacter synchronization (Ely, 1991) and/or chemically fixed by 10 min incubation with 1% paraformaldehyde. The images were processed with NIS Elements, Fiji and Adobe Photoshop. Cell shape analysis on the images was conducted with the Microbe Tracker Matlab-based software package (Sliusarenko et al., 2011). When propidium iodide was used to determine cell viability, it was added to an exponentially growing culture 1.5 h in advance of imaging at the concentration of 5 µM, and the cells were washed once with an equal volume of PYE prior to visualization.
Cell growth experiments
Bacterial cell growth for the provided growth curves was monitored by taking optical density readings at 660 nm wavelength on aliquots of Caulobacter cultures grown at 30°C in PYE. The number of colony forming units was determined by serial dilution of growing cultures and plating on PYE plates without antibiotics. In order to ensure reproducibility of results, the growth curve experiments comparing CB15N with ΔmbiA strains and rodZ::Himar1 with rodZ::Himar1 ΔmbiA strains were conducted in a 96-well format using a Synergy HT Microplate Reader equipped with Gen5 software (BioTek). These cells were grown in PYE (or PYE with kanamycin, in the case of the rodZ::Himar1 derivatives) at 30°C with continuous shaking at the medium setting, and the optical density measurements at 660 nm were taken every 15 min in the course of 24 h. Each culture was layered with mineral oil to prevent evaporation of the medium during the experiment.
Suppressor screen
The suppressor screen was conducted as described in the text. In short, 5 ml of saturated CB15N Caulobacter wild-type cell culture were transferred into an empty Petri dish and submitted to ultraviolet radiation in the Stratalinker UV Crosslinker (Stratagene) at the energy setting of 15 mJ cm−2. Cells were then subcultured into separate tubes at a 1:25 dilution and allowed to grow overnight. The MbiA overexpression plasmid pBV-mbiA-mCherry was then electroporated into each of the resulting cultures, and the cells were plated on PYE plates supplemented with gentamycin and vanillate. The colonies obtained from the initial selection were screened for mCherry fluorescence via a Petri dish imager that was designed and assembled by our lab. The imager uses a 100-watt mercury lamp as an illumination source, a 562/4 nm (centre wavelength/bandpass) filter (Semrock) for excitation, and a 624/40 nm filter (Semrock) for emission. The images of the plates were acquired with a Canon EOS T1i digital camera with an 18–55 mm f/ 3.5–5.6 lens (Canon), and the fluorescent colonies were grown in liquid PYE culture with gentamycin and vanillate to confirm MbiA resistance. The resulting cultures were then electroporated with pXyl::mbiA plasmid, plating on PYE plates supplemented with oxytetracycline and xylose. The MbiA resistance of the resulting colonies was confirmed by curing the pXyl::mbiA plasmid and re-electroporating both the pXyl::mbiA and pBV-mbiA-mCherry overexpression vectors. The two isolated chromosomal mutations were transduced into un-mutagenized CB15N after integration of the pNPTS-mreB vector into their respective mreB loci for the use as a selective marker. The vector was subsequently cured out of the transductants through counterselection with sucrose, and the presence of the expected mutations was confirmed by PCR and sequencing. The images showing the mutation sites and the RodZ-interacting residues (van den Ent et al., 2010) on the MreB crystal structure were produced with the PyMOL software (DeLano Scientific LLC) based on the structure published in van den Ent et al. (2001).
Immunoblotting and immunoprecipitation
To assess MreB levels in Caulobacter strains not expressing, expressing, or overexpressing MbiA, the cultures were normalized by optical density before running on SDS-PAGE. Samples were probed with primary antibodies against MreB (1:5000) and DivJ (1:5000) and the secondary horseradish peroxidase-conjugated donkey anti-rabbit antibody (GE Healthcare; 1:5000 dilution). For the analysis of MbiA expression as a function of the cell cycle, cells expressing an MbiA-FLAG native site fusion were synchronized as for microscopy, and the swarmers diluted into fresh PYE medium and allowed to grow at 30°C. Equal-volume aliquots of the culture were collected at indicated time points, immediately centrifuged, and resuspended in the SDS-PAGE running buffer. Samples were run on SDS-PAGE and blotted with primary antibodies against FLAG (Sigma; 1:5000 dilution), MreB (1:10 000), and CtrA (1:10 000) and the secondary alkaline phosphatase-conjugated goat anti-rabbit antibody (GE Healthcare; 1:10 000 dilution).
The co-immunoprecipitation protocol was modified from (Lin and Grossman, 1998). Five hundred millilitres of Caulobacter cells overexpressing MbiA-FLAG fusion were grown to OD660 of 0.4 and induced with 1 mM vanillate for 2 h. Cells were then washed with 50 ml of PBS and treated with 1 mM Dithiobis (succinimidyl propinionate) in PBS at room temperature for 30 min. The cross-linking reaction was quenched by incubation with 167 mM Tris, pH 7, for 10 min. The rest of the procedure was performed at 4°C. The cross-linked cells were centrifuged, resuspended in 3 ml of Lysis Buffer (50 mM Tris, pH 7.0; 150 mM NaCl; 1 mM EDTA; 1% Triton X100; 2 mM PMSF; 1 tablet per 10 ml Roche cOmplete Mini EDTA-free Protease Inhibitor Cocktail), and lysed with French Press at 12 000 psi. Cell debris was removed by centrifugation in a bench-top centrifuge at 13 200 r.p.m. for 30 min. The lysate was then incubated for 1 h with 35 µl of Protein A agarose slurry (Invitrogen) per millilitre of lysate for 1 h on a shaker. After removal of the agarose beads by centrifugation for 1 min at 10 000 r.p.m., the cleared lysate was incubated with primary antibodies against FLAG (1:100) or MreB (1:333) for 2 h, then 35 µl ml−1 of Protein A agarose was added and incubated for 1 h. The precipitant was washed three times with equal volumes of IP Buffer (100 mM Tris, pH 7.0; 300 mM NaCl; 1 mMEDTA;2%Triton X100;1 mMPMSF) and two times with equal volumes of TE Buffer, centrifuging each time for 1 min at 10 000 r.p.m. The pellet was finally resuspended in 50 µl of SDS-PAGE running buffer, and cross-linking was reversed by incubation with 1.14 M β-mercaptoethanol for 5 min. The immunoblots were probed with primary antibodies against FLAG (Sigma; 1:5000 dilution) and MreB (1:10 000) and the secondary alkaline phosphatase-conjugated anti-rabbit antibody (GE Healthcare, 1:10 000 dilution). The protocol was slightly modified for co-immunoprecipitation from a strain expressing the native site MbiA-FLAGx2 fusion by omitting the induction step, using 50 ml ml−1 of Protein A agarose, 1:166 dilution of α-MreB, and 1:83 dilution of α-FLAG for immunoprecipitation, and blotting the membrane with the α-FLAG antibody at the 1:3750 dilution.
Supplementary Material
Acknowledgements
We thank the members of the Gitai lab for helpful discussions. We also thank John Werner for reagents and the identification of MbiA as a localized protein, Natalie Dye for generating and sharing the fluorescent fusions to PBP2 and MreC, and Sven van Teeffelen for help with the quantitative analysis of MreB localization. This work was supported in part by a New Innovator Award (DP2OD004389) to Z. G.
Footnotes
Supporting information
Additional supporting information may be found in the online version of this article.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Aaron M, Charbon G, Lam H, Schwarz H, Vollmer W, Jacobs-Wagner C. The tubulin homologue FtsZ contributes to cell elongation by guiding cell wall precursor synthesis in Caulobacter crescentus . Mol Microbiol. 2007;64:938–952. doi: 10.1111/j.1365-2958.2007.05720.x. [DOI] [PubMed] [Google Scholar]
- Alyahya SA, Alexander R, Costa T, Henriques AO, Emonet T, Jacobs-Wagner C. RodZ, a component of the bacterial core morphogenic apparatus. Proc Natl Acad Sci USA. 2009;106:1239–1244. doi: 10.1073/pnas.0810794106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2:2006. doi: 10.1038/msb4100050. 0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bean GJ, Flickinger ST, Westler WM, McCully ME, Sept D, Weibel DB, Amann KJ. A22 disrupts the bacterial actin cytoskeleton by directly binding and inducing a low-affinity state in MreB. Biochemistry. 2009;48:4852–4857. doi: 10.1021/bi900014d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendezú FO, de Boer PA. Conditional lethality, division defects, membrane involution, and endocytosis in mre and mrd shape mutants of Escherichia coli . J Bacteriol. 2008;190:1792–1811. doi: 10.1128/JB.01322-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendezú FO, Hale CA, Bernhardt TG, de Boer PA. RodZ (YfgA) is required for proper assembly of the MreB actin cytoskeleton and cell shape in E. coli . EMBO J. 2009;28:193–204. doi: 10.1038/emboj.2008.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carballido-López R, Formstone A, Li Y, Ehrlich SD, Noirot P, Errington J. Actin homolog MreBH governs cell morphogenesis by localization of the cell wall hydrolase LytE. Dev Cell. 2006;11:399–409. doi: 10.1016/j.devcel.2006.07.017. [DOI] [PubMed] [Google Scholar]
- Christen B, Abeliuk E, Collier JM, Kalogeraki VS, Passarelli B, Coller JA, et al. The essential genome of a bacterium. Mol Syst Biol. 2011;7:528. doi: 10.1038/msb.2011.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Defeu Soufo HJ, Reimold C, Linne U, Knust T, Gescher J, Graumann PL. Bacterial translation elongation factor EF-Tu interacts and colocalizes with actin-like MreB protein. Proc Natl Acad Sci USA. 2010;107:3163–3168. doi: 10.1073/pnas.0911979107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divakaruni AV, Baida C, White CL, Gober JW. The cell shape proteins MreB and MreC control cell morphogenesis by positioning cell wall synthetic complexes. Mol Microbiol. 2007;66:174–188. doi: 10.1111/j.1365-2958.2007.05910.x. [DOI] [PubMed] [Google Scholar]
- Dye NA, Pincus Z, Theriot JA, Shapiro L, Gitai Z. Two independent spiral structures control cell shape in Caulobacter . Proc Natl Acad Sci USA. 2005;102:18608–18613. doi: 10.1073/pnas.0507708102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dye NA, Pincus Z, Fisher IC, Shapiro L, Theriot JA. Mutations in the nucleotide binding pocket of MreB can alter cell curvature and polar morphology in Caulobacter . Mol Microbiol. 2011;81:368–394. doi: 10.1111/j.1365-2958.2011.07698.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ely B. Genetics of Caulobacter crescentus . Methods Enzymol. 1991;204:372–384. doi: 10.1016/0076-6879(91)04019-k. [DOI] [PubMed] [Google Scholar]
- Ely B, Johnson RC. Generalized transduction in Caulobacter crescentus . Genetics. 1977;87:391–399. doi: 10.1093/genetics/87.3.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Ent F, Amos LA, Löwe J. Prokaryotic origin of the actin cytoskeleton. Nature. 2001;413:39–44. doi: 10.1038/35092500. [DOI] [PubMed] [Google Scholar]
- van den Ent F, Johnson CM, Persons L, de Boer P, Löwe J. Bacterial actin MreB assembles in complex with cell shape protein RodZ. EMBO J. 2010;29:1081–1090. doi: 10.1038/emboj.2010.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esue O, Cordero M, Wirtz D, Tseng Y. The assembly of MreB, a prokaryotic homolog of actin. J Biol Chem. 2005;280:2628–2635. doi: 10.1074/jbc.M410298200. [DOI] [PubMed] [Google Scholar]
- Figge RM, Divakaruni AV, Gober JW. MreB, the cell shape-determining bacterial actin homologue, co-ordinates cell wall morphogenesis in Caulobacter crescentus . Mol Microbiol. 2004;51:1321–1332. doi: 10.1111/j.1365-2958.2003.03936.x. [DOI] [PubMed] [Google Scholar]
- Gaballah A, Kloeckner A, Otten C, Sahl HG, Henrichfreise B. Functional analysis of the cytoskeleton protein MreB from Chlamydophila pneumoniae . PLoS ONE. 2011;6:e25129. doi: 10.1371/journal.pone.0025129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitai Z, Dye N, Shapiro L. An actin-like gene can determine cell polarity in bacteria. Proc Natl Acad Sci USA. 2004;101:8643–8648. doi: 10.1073/pnas.0402638101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitai Z, Dye NA, Reisenauer A, Wachi M, Shapiro L. MreB actin-mediated segregation of a specific region of a bacterial chromosome. Cell. 2005;120:329–341. doi: 10.1016/j.cell.2005.01.007. [DOI] [PubMed] [Google Scholar]
- Goley ED, Comolli LR, Fero KE, Downing KH, Shapiro L. DipM links peptidoglycan remodeling to outer membrane organization in Caulobacter . Mol Microbiol. 2010;77:56–73. doi: 10.1111/j.1365-2958.2010.07222.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goley ED, Yeh YC, Hong SH, Fero MJ, Abeliuk E, McAdams HH, Shapiro L. Assembly of the Caulobacter cell division machine. Mol Microbiol. 2011;80:1680–1698. doi: 10.1111/j.1365-2958.2011.07677.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graumann PL. Cytoskeletal elements in bacteria. Annu Rev Microbiol. 2007;61:589–618. doi: 10.1146/annurev.micro.61.080706.093236. [DOI] [PubMed] [Google Scholar]
- Jones LJ, Carballido-López R, Errington J. Control of cell shape in bacteria: helical, actin-like filaments in Bacillus subtilis . Cell. 2001;104:913–922. doi: 10.1016/s0092-8674(01)00287-2. [DOI] [PubMed] [Google Scholar]
- Kawai Y, Daniel RA, Errington J. Regulation of cell wall morphogenesis in Bacillus subtilis by recruitment of PBP1 to the MreB helix. Mol Microbiol. 2009;71:1131–1144. doi: 10.1111/j.1365-2958.2009.06601.x. [DOI] [PubMed] [Google Scholar]
- Kitagawa M, Ara T, Arifuzzaman M, Ioka-Nakamichi T, Inamoto E, Toyonaga H, Mori H. Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): unique resources for biological research. DNA Res. 2005;12:291–299. doi: 10.1093/dnares/dsi012. [DOI] [PubMed] [Google Scholar]
- Lee JC, Stewart GC. Essential nature of the mreC determinant of Bacillus subtilis . J Bacteriol. 2003;185:4490–4498. doi: 10.1128/JB.185.15.4490-4498.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin DC, Grossman AD. Identification and characterization of a bacterial chromosome partitioning site. Cell. 1998;92:675–685. doi: 10.1016/s0092-8674(00)81135-6. [DOI] [PubMed] [Google Scholar]
- Masuda H, Tan Q, Awano N, Yamaguchi Y, Inouye M. A novel membrane-bound toxin for cell division, CptA (YgfX), inhibits polymerization of cytoskeletal proteins, FtsZ and MreB, in Escherichia coli . FEMS Microbiol Lett. 2012;328:174–181. doi: 10.1111/j.1574-6968.2012.02496.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Pedro MA, Quintela JC, Höltje JV, Schwarz H. Murein segregation in Escherichia coli . J Bacteriol. 1997;179:2823–2834. doi: 10.1128/jb.179.9.2823-2834.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Pedro MA, Donachie WD, Höltje JV, Schwarz H. Constitutive septal murein synthesis in Escherichia coli with impaired activity of the morphogenetic proteins RodA and penicillin-binding protein 2. J Bacteriol. 2001;183:4115–4126. doi: 10.1128/JB.183.14.4115-4126.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poggio S, Takacs CN, Vollmer W, Jacobs-Wagner C. A protein critical for cell constriction in the Gram-negative bacterium Caulobacter crescentus localizes at the division site through its peptidoglycan-binding LysM domains. Mol Microbiol. 2010;77:74–89. doi: 10.1111/j.1365-2958.2010.07223.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD, Blanchoin L, Mullins RD. Molecular mechanisms controlling actin filament dynamics in non-muscle cells. Annu Rev Biophys Biomol Struct. 2000;29:545–576. doi: 10.1146/annurev.biophys.29.1.545. [DOI] [PubMed] [Google Scholar]
- Salje J, van den Ent F, de Boer P, Löwe J. Direct membrane binding by bacterial actin MreB. Mol Cell. 2011;43:478–487. doi: 10.1016/j.molcel.2011.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiomi D, Sakai M, Niki H. Determination of bacterial rod shape by a novel cytoskeletal membrane protein. EMBO J. 2008;27:3081–3091. doi: 10.1038/emboj.2008.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjöblom B, Salmazo A, Djinović-Carugo K. Alpha-actinin structure and regulation. Cell Mol Life Sci. 2008;65:2688–2701. doi: 10.1007/s00018-008-8080-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sliusarenko O, Heinritz J, Emonet T, Jacobs-Wagner C. High-throughput, subpixel precision analysis of bacterial morphogenesis and spatio-temporal dynamics. Mol Microbiol. 2011;80:612–627. doi: 10.1111/j.1365-2958.2011.07579.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staiger CJ, Blanchoin L. Actin dynamics: old friends with new stories. Curr Opin Plant Biol. 2006;9:554–562. doi: 10.1016/j.pbi.2006.09.013. [DOI] [PubMed] [Google Scholar]
- Takacs CN, Poggio S, Charbon G, Pucheault M, Vollmer W, Jacobs-Wagner C. MreB drives de novo rod morphogenesis in Caulobacter crescentus via remodeling the cell wall. J Bacteriol. 2010;192:1671–1684. doi: 10.1128/JB.01311-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Q, Awano N, Inouye M. YeeV is an Escherichia coli toxin that inhibits cell division by targeting the cytoskeletal proteins, FtsZ and MreB. Mol Microbiol. 2011;79:109–118. doi: 10.1111/j.1365-2958.2010.07433.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanbichler M, Shapiro L. MipZ, a spatial regulator coordinating chromosome segregation with cell division in Caulobacter . Cell. 2006;126:147–162. doi: 10.1016/j.cell.2006.05.038. [DOI] [PubMed] [Google Scholar]
- Uehara T, Park JT. Growth of Escherichia coli: significance of peptidoglycan degradation during elongation and septation. J Bacteriol. 2008;190:3914–3922. doi: 10.1128/JB.00207-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner JK, Galvani CD, Brun YV. Caulobacter crescentus requires RodA and MreB for stalk synthesis and prevention of ectopic pole formation. J Bacteriol. 2005;187:544–553. doi: 10.1128/JB.187.2.544-553.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner JN, Chen EY, Guberman JM, Zippilli AR, Irgon JJ, Gitai Z. Quantitative genome-scale analysis of protein localization in an asymmetric bacterium. Proc Natl Acad Sci USA. 2009;106:7858–7863. doi: 10.1073/pnas.0901781106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White CL, Kitich A, Gober JW. Positioning cell wall synthetic complexes by the bacterial morphogenetic proteins MreB and MreD. Mol Microbiol. 2010;76:616–633. doi: 10.1111/j.1365-2958.2010.07108.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.