Abstract
The origins of the First Americans remain contentious. Although Native Americans seem to be genetically most closely related to east Asians1–3, there is no consensus with regard to which specific Old World populations they are closest to4–8. Here we sequence the draft genome of an approximately 24,000-year-old individual (MA-1), from Mal’ta in south-central Siberia9, to an average depth of 13. To our knowledge this is the oldest anatomically modern human genome reported to date. The MA-1 mitochondrial genome belongs to haplogroup U, which has also been found at high frequency among Upper Palaeolithic and Mesolithic European hunter-gatherers10–12, and the Y chromosome of MA-1 is basal to modern-day western Eurasians and near the root of most Native American lineages5. Similarly, we find autosomal evidence that MA-1 is basal to modern-day western Eurasians and genetically closely related to modern-day Native Americans, with no close affinity to east Asians. This suggests that populations related to contemporary western Eurasians had a more north-easterly distribution 24,000 years ago than commonly thought. Furthermore, we estimate that 14 to 38% of Native American ancestry may originate through gene flow from this ancient population. This is likely to have occurred after the divergence of Native American ancestors from east Asian ancestors, but before the diversification of Native American populations in the New World. Gene flow from the MA-1 lineage into Native American ancestors could explain why several crania from the First Americans have been reported as bearing morphological characteristics that do not resemble those of east Asians2,13. Sequencing of another south-central Siberian, Afontova Gora-2 dating to approximately 17,000 years ago14, revealed similar autosomal genetic signatures as MA-1, suggesting that the region was continuously occupied by humans throughout the Last Glacial Maximum. Our findings reveal that western Eurasian genetic signatures in modern-day Native Americans derive not only from post-Columbian admixture, as commonly thought, but also from a mixed ancestry of the First Americans.
In 2009 we visited Hermitage State Museum in St. Petersburg, Russia, and sampled skeletal remains of a juvenile individual (MA-1) from the Mal’ta Upper Palaeolithic site in south-central Siberia. Mal’ta, located along the Belaya River near Lake Baikal, was excavated between 1928 and 1958 (ref. 9) and yielded a plethora of archaeological finds including 30anthropomorphic Venus figurines, which are rare for Siberia but found at a number of Upper Palaeolithic sites across western Eurasia15–17 (Fig. 1a and Supplementary Information, section 1). Accelerator mass spectrometry (AMS) 14C dating of MA-1 produced an age of 20,240 ± 60 14C years before present or24,423–23,891calendar years before present (cal. bp) (Supplementary Information, section 2).
Figure 1.
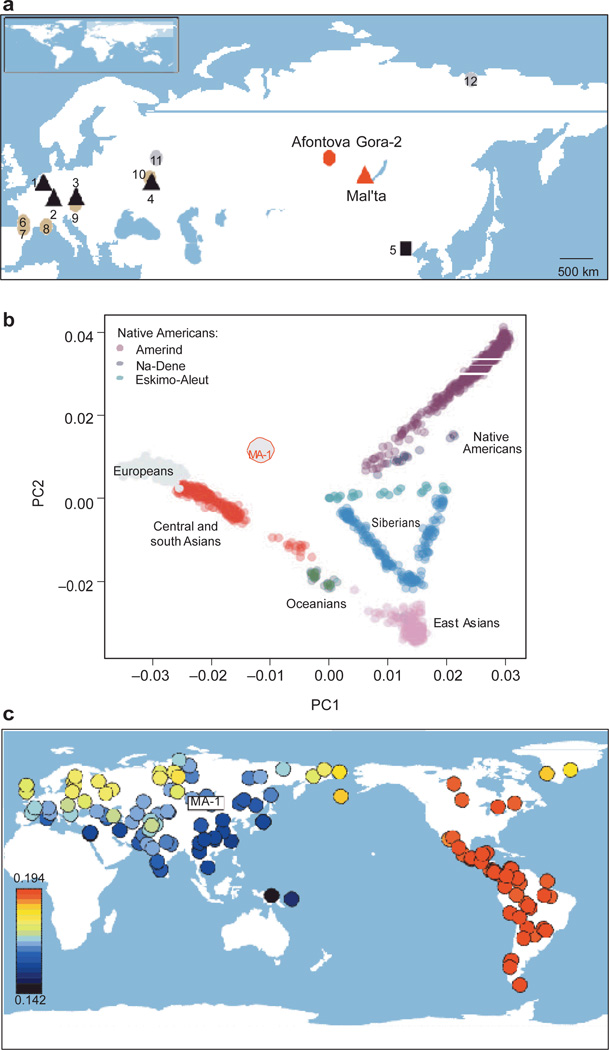
Sample locations and MA-1 genetic affinities. a, Geographical locations of Mal’ta and Afontova Gora-2 in south-central Siberia. For reference, Palaeolithic sites with individuals belonging to mtDNA haplogroup U are shown (red and black triangles): 1, Oberkassel; 2, Hohle Fels; 3, Dolni Vestonice; 4, Kostenki-14. A Palaeolithic site with an individual belonging to mtDNA haplogroup B is represented by the square: 5, Tianyuan Cave. Notable Palaeolithic sites with Venus figurines are marked by brown circles: 6, Laussel; 7, Lespugue; 8, Grimaldi; 9, Willendorf; 10, Gargarino. Other notable Palaeolithic sites are shown by grey circles: 11, Sungir; 12, Yana RHS. b, PCA (PC1 versus PC2) of MA-1 and worldwide human populations for which genomic tracts from recent European admixture in American and Siberian populations have been excluded19. c, Heat map of the statistic f3(Yoruba; MA-1, X) where X is one of 147 worldwide non-African populations (standard errors shown in Supplementary Fig. 21). The graded heat key represents the magnitude of the computed f3 statistics.
DNA from 0.15 g of bone from MA-1 was sequenced to an average depth of 13 (Supplementary Information, section 3). From one library (referred to as MA-1_1stextraction in Supplementary Information, section 3.1), approximately 17% of the total reads generated mapped uniquely to the human genome, in agreement with good DNA preservation (see Supplementary Information Table 2). Low contamination rates were inferred for both mitochondrial DNA (mtDNA) (1.1%) and the X chromosome (1.6 to 2%; MA-1 is male) (Supplementary Information, section 5). The overall error rate for the data set was estimated to be 0.27%, with the most dominant errors being transitions typical of ancient DNA damage deriving from post-mortem deamination of cytosine18 (Supplementary Information, section 6.1).
Phylogenetic analysis of the MA-1 mtDNA genome (76.6X) places it within mtDNA haplogroup U without affiliation to any known subclades, implying a lineage that is rare or extinct in sampled modern populations (Supplementary Information, section 7 and Supplementary Fig. 4a). Present-day distribution of haplogroup U encompasses a large area including North Africa, the Middle East, south and central Asia, western Siberia and Europe (Supplementary Fig. 4b), although it is rare or absent east of the Altai Mountains; that is, in populations living in the region surrounding Mal’ta. Haplogroup U has also been found at high frequency (>80%) in ancient hunter-gatherers from Upper Palaeolithic and Mesolithic Europe10–12. Our result therefore suggests a connection between pre-agricultural Europe and Upper Palaeolithic Siberia. The Y chromosome of MA-1 was sequenced to an average depth of 1.5X, with coverage across 5.8 million bases. Acknowledging the low depth of coverage, we determined the most likely phylogenetic affiliation of the MA-1 Y chromosome to a basal lineage of haplogroup R (Supplementary Information, section 8 and Supplementary Fig. 5a). The extant sub-lineages of haplogroup R show regional spread patterns within western Eurasia, south Asia and also extend to the Altai region in southern Siberia (Supplementary Fig. 5b). The sister lineage to these extant sub-lineages of haplogroup R, haplogroup Q, is the most common haplogroup in Native Americans5 and it was recently shown that, in Eurasia, haplogroup Q lineages closest to Native Americans are found in southern Altai7.
To get an overview of the genomic signature of MA-1, we conducted principal component analysis (PCA) using a large data set from worldwide human populations for which genomic tracts of recent European admixture in American and Siberian populations have been excluded19 (Supplementary Information, section 10). In the first two principal components, MA-1 is intermediate between modern western Eurasians and Native Americans, but distant from east Asians (Fig. 1b). To investigate the relationship of MA-1 to global human populations in further detail, we used the f-statistics framework20 to compute an ‘outgroup’ f3-statistic, which is expected to be proportional to the amount of shared genetic history between MA-1 and each of 147 non-African populations from a large worldwide human single-nucleotide polymorphism (SNP) array data set (see Supplementary Information, section 14.2 for details on the f3-statistics). We find that genetic affinity to MA-1 is greatest in two regions: first, the Americas; and second, northeast Europe and northwest Siberia, with north-to-south latitudinal clines in shared drift with MA-1 in both Europe and Asia (Fig. 1c and Supplementary Figs 21 and 22). Notably, the lack of genetic affinity between MA-1 and most populations in south-central Siberia today suggests that there was substantial gene flow into the region after the Last Glacial Maximum (LGM), mostly probably from east Asian sources (Supplementary Information, section 9.1.3).
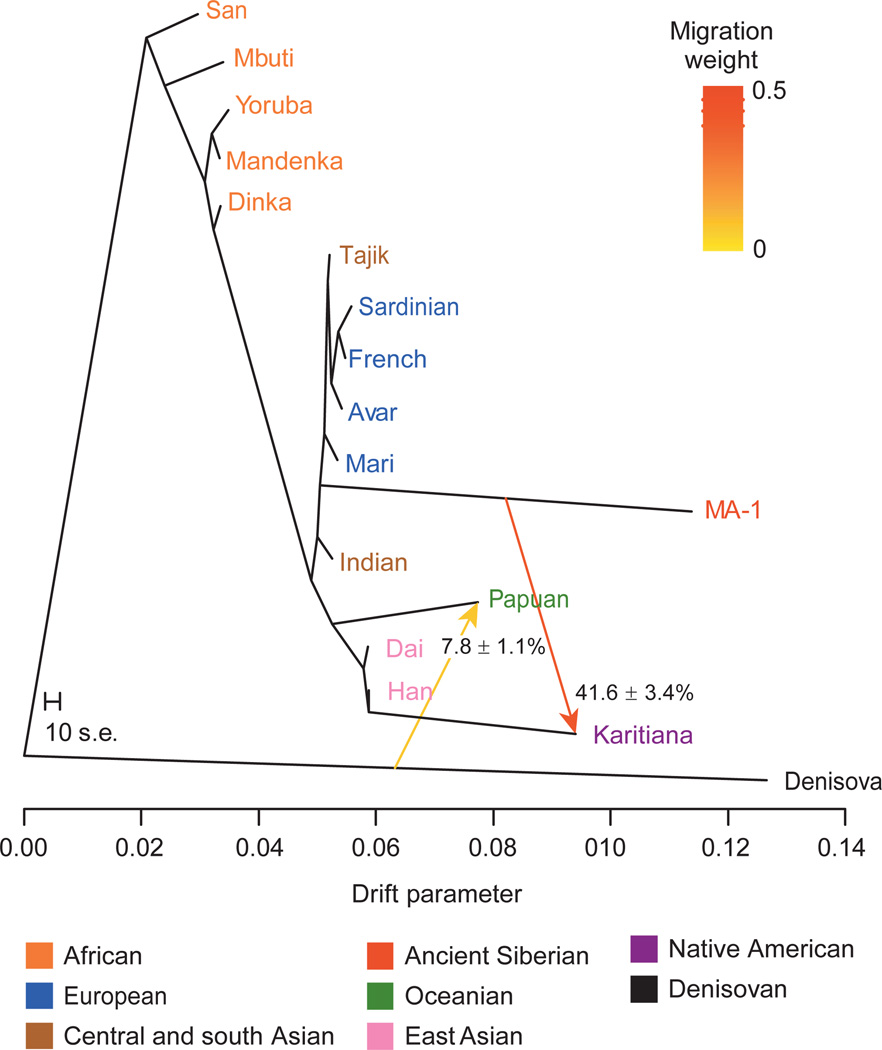
We reconstructed admixture graphs using TreeMix21 to relate the population history of MA-1 to 11 modern genomes from worldwide populations22, 4 new genomes from Eurasia (Mari, Avar, Indian and Tajik ancestry) and the Denisova genome22 (Supplementary Information, section 11). The maximum-likelihood population tree inferred without admixture events places MA-1 on a branch that is basal to western Eurasians (Supplementary Fig. 12). However, a significant residual was observed between the empirical covariance for MA-1 and Karitiana, a Native American population, and the covariance predicted by the tree model (Supplementary Fig. 12). Consequently, gene flow between these lineages was inferred in all graphs incorporating two or more migration events (Fig. 2 and Supplementary Fig. 13). Bootstrap support for the migration edge from MA-1 to Karitiana, rather than from Karitiana to MA-1, was 99% in this analysis.
Figure 2.
Admixture graph for MA-1 and 16 complete genomes. An admixture graph with two migration edges (depicted by arrows) was fitted using TreeMix21 to relate MA-1 to 11 modern genomes from worldwide populations22, 4 modern genomes produced in this study (Avar, Mari, Indian and Tajik), and the Denisova genome22. Trees without migration, graphs with different number of migration edges, and residual matrices are shown in Supplementary Information, section 11. The drift parameter is proportional to 2Ne generations, where Ne is the effective population size. The migration weight represents the fraction of ancestry derived from the migration edge. The scale bar shows ten times the average standard error (s.e.) of the entries in the sample covariance matrix. Note that the length of the branch leading to MA-1 is affected by this ancient genome being represented by haploid genotypes.
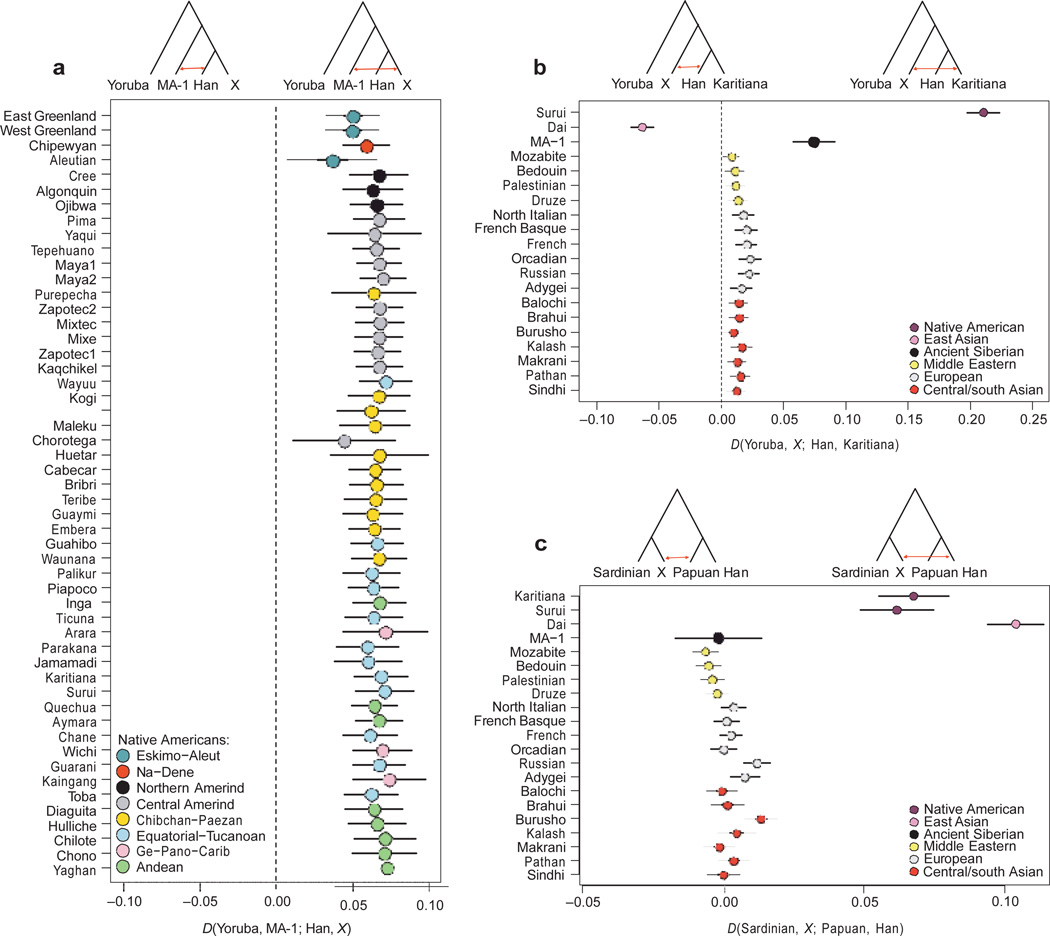
We investigated further the population history of MA-1 by conducting sequence read-based D-statistic tests23 on proposed tree-like histories comprising MA-1 and combinations of 11 modern genomes (Supplementary Information, section 13). In agreement with the TreeMix results, these tests reject the tree ((X, Han), MA-1) where X represents Avar, French, Indian, Mari, Sardinian and Tajik, consistent with the MA-1 lineage sharing more recent ancestry with the western Eurasian branch after the split of Europeans and east Asians (Supplementary Table 13). This result also holds true when the Han Chinese is replaced with Dai, another east Asian population (Supplementary Table 13). Notably, we can also reject the tree ((Han, Karitiana), MA-1) (Z 5 10.8), suggesting gene flow between MA-1 and ancestral Native Americans, in accordance with the admixture graphs (Supplementary Table 13). This result is consistent with allele frequency-based D-statistic tests20 on SNP arrays for 48 Native American populations of entirely First American ancestry19, indicating that all tested populations are equally related to MA-1 and that the admixture event occurred before the population diversification of the First American gene pool (Fig. 3a, Supplementary Information, section 14.4 and Supplementary Fig. 24).
Figure 3.
Evidence of gene flow from a population related to MA-1 and western Eurasians into Native American ancestors. Allele frequency-based D-statistic tests20 of the forms. a, D (Yoruba, MA-1; Han, X), where X represents modern-day populations from North and South America. The D-statistic is significantly positive for all the tests, providing evidence for gene flow between Native American ancestors and the MA-1 population lineage; however, it is not informative with respect to the direction of gene flow. b, D (Yoruba, X; Han, Karitiana), where X represents non-African populations. Since all of the 17 tested western Eurasian populations are closer to Karitiana than to Han Chinese, the most parsimonious explanation is that Native Americans have western Eurasian-related ancestry. c, D (Sardinian, X; Papuan, Han), where X represents non-African populations. MA-1 is not significantly closer to Han Chinese than to Papuans, which is compatible with MA-1 having no Native American-related admixture in its ancestry. Thick and thin error bars correspond to 1 and 3 standard errors of the D-statistic, respectively.
The genetic affinity between Native Americans and MA-1 could be explained by gene flow after the split between east Asians and Native Americans, either from the MA-1 lineage into Native American ancestors or from Native American ancestors to the ancestors of MA-1. However, MA-1, at approximately 24,000 cal. bp, pre-dates time estimates of the Native American–east Asian population divergence event24,25. This presents little time for the formation of a diverged Native American gene pool that could have contributed ancestry to MA-1, suggesting gene flow from the MA-1 lineage into Native American ancestors. Such gene flow should also be detectable using modern-day western Eurasian populations in place of MA-1. Consistent with this, D-statistic tests estimated from outgroup-ascertained SNP data20 reveal significant evidence (Z == 3) for Middle Eastern, European, central Asian and south Asian populations being closer to Karitiana than to Han Chinese20 (Fig. 3b and Supplementary Information, section 14.5). Similar signals were also observed when we replaced modern-day Han Chinese with data from chromosome 21 from a 40,000-yearold east Asian individual (Tianyuan Cave, China), which has been found to be ancestral to modern-day Asians and Native Americans26 (Supplementary Information, section 14.5). Thus, if the gene flow direction was from Native Americans into western Eurasians it would have had to spread subsequently to European, Middle Eastern, south Asian and central Asian populations, including MA-1 before 24,000 years ago. Moreover, as Native Americans are closer to Han Chinese than to Papuans (Fig. 3c), Native American-related gene flow into the ancestors of MA-1is expected to result in MA-1 also being closer to Han Chinese than to Papuans. However, our results suggest that this is not the case (D (Papuan, Han; Sardinian, MA-1) = 20.002 ± 0.005 (Z = 20.36)), which is compatible with all or almost all of the gene flow being into Native Americans (Supplementary Information, section 14.6). Similar results are obtained when MA-1 is replaced with most modern-day western Eurasian populations, except populations with recent admixture from east Asia (Russian, Adygei and Burusho) and Africa (Middle Eastern populations) (Fig. 3c). The most parsimonious explanation for these results is that Native Americans have mixed origins, resulting from admixture between peoples related to modern-day east Asians and western Eurasians. Admixture graphs fitted with MixMapper27 model Karitiana as having 14–38% western Eurasian ancestry and 62–86% east Asian ancestry, but we caution that these estimates assume unadmixed ancestral populations (Supplementary Information, section 12).
Importantly, in addition to the low contamination rates and rare or extinct uniparental lineages, we exclude modern DNA contamination as being the source of the observed population affinities of MA-1 for three reasons. First, we corrected the sequence read-based D-statistics tests for differing amounts of contamination, using a European individual as the contamination source (Supplementary Information, section 13.5). We find similar outcomes for corrected and uncorrected tests (Supplementary Fig. 20), even when contamination levels larger than that estimated for MA-1 are considered, confirming that our results are not affected by contamination from a European source. Second, restricting the PCA to sequences with evidence of post-mortem degradation gives results that are comparable with those using the complete data set (Supplementary Information, section 15). Finally, the genome sequence of the researcher (Indian ancestry) who carried out DNA extraction and library preparation of MA-1 enables us to exclude the researcher as a source of contamination (Supplementary Information, sections 11 and 13). In addition, we exclude post-Columbian European admixture (after 1492 ad) as an explanation for the genetic affinity between MA-1 and Native Americans for three reasons. First, for SNP array-based analyses, we take recent European admixture into account by using a data set masked for inferred admixed genomic regions19. Second, allele frequency-based D-statistic tests20 show that all 48 tested modern-day populations with First American ancestry19 are equally related to MA-1 within the resolution of our data (Supplementary Information, section 14.4), which would not be expected if the signal was driven by recent European admixture. Third, MA-1 is closer to Native Americans than any of the 15 tested European populations (Supplementary Information, section 14.8).
Human dispersals in northeast Asia immediately before and after the LGM are most likely to have led to the settlement of Beringia, and ultimately the Americas28. As MA-1 pre-dates the LGM, we investigated whether the genetic composition of southern Siberia changed during the LGM by generating a low-coverage data set (~0.1X) of a post-LGM individual from Afontova Gora-2 (AG-2) (ref. 14), located on the western bank of the Enisei River in south-central Siberia (Fig. 1a). We obtained a direct AMS 14C date of 13,810 ± 35 14C years before present or 17,075–16,750 cal. bp for AG-2 (Supplementary Information, section 2). Despite substantial present-day DNA contamination in this sample (Supplementary Information, section 5), we find that AG-2 shows close similarity to the genetic profile of MA-1 on a PCA (Supplementary Information, section 15 and Supplementary Fig. 29) and is significantly closer to Karitiana than to Han (D (Yoruba, AG-2; Han, Karitiana) = 0.078 ± 0.004, Z = 19.9) (Supplementary Information, section 15). We observe consistent results when restricting analyses to sequences with evidence of post-mortem degradation (Supplementary Information, section 15 and Supplementary Fig. 29), implying that southern Siberia may have experienced genetic continuity through the environmentally harsh LGM.
Our study has four important implications. First, we find evidence that contemporary Native Americans and western Eurasians share ancestry through gene flow from a Siberian Upper Palaeolithic population into First Americans. Second, our findings may provide an explanation for the presence of mtDNA haplogroup X in Native Americans, which is related to western Eurasians but not found in east Asian populations29. Third, such an easterly presence in Asia of a population related to contemporary western Eurasians provides a possibility that non-east Asian cranial characteristics of the First Americans13 derived from the Old World via migration through Beringia, rather than by a trans-Atlantic voyage from Iberia as proposed by the Solutrean hypothesis30. Fourth, the presence of an ancient western Eurasian genomic signature in the Baikal area before and after the LGM suggests that parts of south-central Siberia were occupied by humans throughout the coldest stages of the last ice age.
METHODS
Samples
A humerus (MA-1) from Mal’ta and a humerus (AG-2) from Afontova Gora-2 were sampled at the Hermitage Museum, St. Petersburg, Russia in 2009 for ancient DNA analysis and accelerator mass spectrometry (AMS) 14C dating. In addition, four modern human samples (Avar, Mari, Tajik and Indian) were obtained for genome sequencing in accordance with informed consent requirements for human demographic studies. Ethical approval for genome sequencing of the above four modern samples was acquired from The National Committee on Health Research Ethics, Denmark (H-3-2012-FSP21).
Radiocarbon dating
AMS 14C dating was carried out on the two ancient bone samples following standard protocols31,32 (Supplementary Information, section 2). Contemporary 14C standards included National Bureau of Standards Oxalic Acid-I and ANU sucrose. Respective chemistry and combustion backgrounds were determined by using .70,000-year-old collagen isolated from the fossil Eschrichtius robustus (grey whale)32,33 and Sigma Aldrich L-Alanine (catalogue number A7627). The graphitized samples and standards were analysed at the University of California-Irvine WM Keck Carbon Cycle Accelerator Mass Spectrometry Laboratory (UCIAMS). The 14C dates were calibrated using OxCal 4.2 (ref. 34) and the INTCAL09 data set35.
Genome sequencing and read processing
DNA extractions and library constructions for the ancient samples were performed in a laboratory facility dedicated to the analysis of ancient DNA (Centre for GeoGenetics, Copenhagen). Bone powder from MA-1 and AG-2 (149 mg and 119 mg, respectively) was extracted using a silica spin-column protocol11,36,37 (Supplementary Information, section3.1.1). Undigested pellets were subject to another round of digestion. Blood samples from one individual each of Avar, Mari and Tajik ancestry were extracted using standard protocol38 (Supplementary Information, section 3.2.2). A saliva sample from an individual of Indian ancestry was extracted using a prepITNL2P extraction kit (DNA Genotek) (Supplementary Information, section 3.2.2). Illumina libraries were constructed on the ancient and modern extracts (Supplementary Information, sections 3.1.2 and 3.2.3). The protocols outlined in the kit manuals (GS FLX Titanium Rapid Library Preparation Kit, 454 Life Sciences, Roche, Branford, CO and NEBNext DNA Sample Prep Master Mix Set 2, New England Biolabs, E6070) as well as in a previous paper39 were followed. Equimolar pools of the ancient (100 cycles, single-read mode) and modern libraries (100 cycles, paired-end mode) were sequenced on the llumina HiSeq 2000 at the Danish National High-Throughput DNA Sequencing Centre. The ancient libraries were sequenced to near-saturation.
Read processing was performed on the ancient and modern genomes produced in this study as well as previously published genomes (Supplementary Information, sections 4.1 and 4.2). The latter genomes included 11 high-coverage modern genomes22, one low-coverage Cambodian genome40, and the Denisovan22 and Tianyuan26 ancient data sets. All sequences were trimmed using Adapter Removal41 and mapped to the human reference genome builds hg18 and 37.1 using the Burrows-Wheeler Aligner (BWA)42. The seed length option was disabled for ancient reads to optimize the mapping efficiency43. Polymerase chain reaction (PCR) duplicates were removed using Picard Mark Duplicates (http://picard.sourceforge.net). All modern samples (except the Cambodian genome) and the Denisova individual were genotyped using samtools mpileup and bctools44, and filtered to achieve a high-confidence SNP set (Supplementary Information, section4.2). Only bi-allelic sites were included when producing the final call set and the individual calls were merged to a final set using Genome Analysis Toolkit (GATK) CombineVariants-2.5-2 (ref. 45).
Contamination and error rate estimation
Mitochondrial DNA (mtDNA) contamination rates for MA-1 and AG-2 were estimated by identifying consensus calls in the ancient mtDNA data set that are private or near-private to the ancient individual (at an allele frequency of less than 1% in a set of 311 modern human mtDNA genomes)46 (Supplementary Information, section 5.1). The near-private consensus alleles and potential contaminating reads at these positions were counted, and a 95% confidence interval was obtained assuming that the allele observed in each read is a random outcome of drawing one of two alleles (endogenous and contaminant). Positions with a depth of less than 103 were excluded, as were positions where the consensus allele was either C or G in a transition polymorphism, as these are sensitive to post-mortem nucleotide misincorporations. A phred-scaled base quality of 30 was required.
As we found both individuals (MA-1 and AG-2) to be males by comparing the number of alignments to the X and Y chromosomes47 (Supplementary Information, section 4.3), it was possible to obtain X chromosome-based contamination estimates using previously published methods48 (Supplementary Information, section 5.2). These estimates were based on a fixed set of SNPs known to be polymorphic in European HapMap phase II release 27 data49. This SNP data set was pruned such that polymorphic sites were more than 10 bases apart. The same HapMap data was used for estimating allele frequencies in Europeans. The MA-1 and AG-2 data sets were filtered to remove: regions homologous between the X and Y chromosomes; reads mapping non-uniquely to multiple regions of the genome with more than 98% identity; reads with mapping quality score less than 30 and base quality score less than 20; and sites with a read depth of less than 3 (or 2 depending on library depth) or above 40.
The error rates for the sequenced ancient and modern libraries were estimated using a method similar to a previously published method40 that makes use of a high quality genome (Supplementary Information, section 6.1). The estimates were based on the rationale that any given human sample should have the same expected number of derived alleles compared to some outgroup, in this case the chimpanzee, panTro2, from the multiway alignment hg19 multiz46. The numbers of derived alleles were counted from the high-quality genome (individual NA06985 from the 1000 Genomes Project Consortium50) and the error rate estimates were based on the assumption that any excess of derived alleles (compared to the high quality genome) observed in our sample is due to errors. The overall error rates were estimated using a method of moment estimator, while the type specific error rates were estimated using a maximum likelihood approach. The model and the estimation methods are described in detail elsewhere39. All reads with a mapping quality score less than 30 and all bases with a base quality scoreless than20 were excluded. mtDNA and Y-chromosome haplogroup determination. Sequence reads from MA-1 were mapped to the revised Cambridge Reference Sequence (rCRS, NC_012920.1) and filtered for PCR duplicates and paralogues, requiring a minimum mapping quality of 25 (Supplementary Information, section 4.1). A file of variants filtered for a minimum depth of 10, was generated (Supplementary Information, section 7). Indels were excluded from the analysis. mtDNA sequences from the individual Dolni Vestonice 14 (DV-14; GenBank accession number KC521458), basal to the extant mtDNA haplogroup U5 (ref. 12), was included in the analysis for comparison. Both the MA-1 and DV-14 mtDNA sequences were analysed for the presence of diagnostic mutations of the major sub-haplogroups of extant mtDNA haplogroup U lineages, using information from mtDNA tree Build 15 (Sept 30, 2012)51. A phylogenetic tree including all major extant branches of mtDNA haplogroup U was built, with the age estimates (kilo years ± s.d.) of the different sub-haplogroups52 (Supplementary Fig. 4a). To show the present spread of haplogroup U and its different sub-haplogroups, the average frequencies, divided into four frequency classes, were calculated in regional groups, using a data set consisting of approximately 30,000 partial mtDNA genomes (references in Supplementary Information, section 7).
Owing to low depth of coverage of the MA-1 individual, genotyping at each site on the Y chromosome was performed by selecting the allele with the highest frequency of bases with a base quality of 13 or higher (Supplementary Information, section 8). A multi-fast a file was generated from the variable positions on the Y chromosomes available from 24 Complete Genomics public genomes53. SNPs were filtered for quality (using the threshold VQHIGH as defined by Complete Genomics), with tri-allelic positions excluded and only Y-chromosome regions determined as phylogenetically informative being used54. This yielded a final data set of 22,492 positions that was merged with MA-1 Y chromosome data. A neighbour joining tree with default parameters in MEGA phylogenetic software55 was constructed (Supplementary Fig. 5a). Phylogenetically informative positions and their state in MA-1 were then determined to confirm the placement of MA-1 on the tree. Non-informative positions, including those with more than four Ns in the public data set, were excluded (633 positions). Moreover, the following positions were also excluded which were: in reference state in all individuals, including MA-1 (7,172 positions); N in MA-1 and either N or reference state among the rest of the individuals (9,682 positions); ‘N-ref’, those with only N or reference state among all individuals (586 positions), and ‘N-alt’, positions with alternative alleles, but difficult to classify (11 positions); ‘reference-specific’ (79 positions); and ‘recurrent’ (28 positions). This resulted in 4,301 positions being retained that were classified according to their haplogroup affiliations. Among those phylogenetically informative positions, 1,889 non-N positions were retrieved from MA-1.
Principal component analysis
A single read was sampled from each position in the MA-1 data set, which overlapped with SNPs in a data set compiled from a previous paper19 in which the authors had used local ancestry inference to mask segments of European and African ancestry in Siberian and Native American populations56–59 (Supplementary Information, section 10). A phred-scaled mapping quality of 30 and base quality score of30 was required in the sequence data for a haploid genotype to be called, and reads with indels were excluded. SNPs with minor allele frequency of, 1% in the total data set were removed. To reduce the effect of nucleotide misincorporations, the first and last three bases of each sequence read in the MA-1 data were excluded. SNPs where there was no information from MA-1 were excluded, and a single haploid genotype was randomly sampled from each modern individual to match the single-pass nature of the shotgun data60. PCA was performed on various population subsets separately using EIGENSOFT 4.0 (ref. 61), removing one SNP from each pair for which linkage disequilibrium exceeded a low arbitrary threshold (r2 − 0.2). Transition SNPs, where the ancient individual displayed a T or an A62, as well as triallelic SNPs, were excluded.
To look more closely at the genetic affinities of AG-2 to modern-day populations, data from non-African populations59,63,64 were used as a reference panel and PCA was performed as detailed above (Supplementary Information, section 15). To compare the PCA results from MA-1 and AG-1, Procrustes transformation was performed as described in a previous paper62, rotating the PC1–PC2 configurations obtained for the two individuals to the configuration obtained using only the reference panel (Supplementary Information, section 15). The analysis was repeated using only those sequences which displayed a C R T mismatch consistent with post-mortem ancient DNA nucleotide misincorporations (PMD) in the first five bases of the sequence read (requiring a base quality of at least 30) (Supplementary Information, section 15).
Admixture graph inference
To infer admixture graphs, a total of 17 individuals were used: the archaic Denisova genome22; 11 present-day individuals22; the 4 novel genomes from this study (Supplementary Information, section 4.2); and the MA-1 genome (Supplementary Information, section 11). Haploid genotypes from MA-1 were added to variants identified in the other individuals, as in the PCA analysis to alleviate the increased rate of errors in low-coverage ancient DNA sequence data. If multiple sequence reads overlapped a position, one read was randomly sampled23. This avoids biasing for, or against, heterozygotes and renders the MA-1 data haploid. All transition SNPs were excluded and MA-1 sequence reads with a mapping quality less than 30 and bases with base quality less than 30 were discarded. Positions at which there was no data from one of the individuals in the analysis were also excluded. This resulted in a final count of 156,250 SNPs for the main analysis. TreeMix21 (version 1.12) was used to build ancestry graphs assuming 0 to 10 migration edges, the placement and weight of each being optimized by the algorithm. TreeMix was run using the ‘-global’ option, which corresponds to performing a round of global rearrangements of the graph after initial fitting. Sample size correction was also disabled, as all the populations consisted of single individuals (‘-noss’). Standard errors were estimated in blocks with 500 SNPs in each. For those analyses that included one or more a priori specified events, a round of optimization was performed on the original migration edge (option ‘-climb’).
Admixture graphs relating MA-1 to modern groups were also inferred using MixMapper v1.0 (ref. 27) (Supplementary Information, section12). A scaffold tree was constructed using four African genomes (San, Yoruba, Mandenka, Dinka), and Sardinian and Han22 genomes, to which MA-1 and other genomes were fitted. All transitions were excluded, and standard errors of the f-statistics were estimated using 500 bootstrap replicates over 50 blocks of the autosomal genome.
D-statistics
To investigate the relationship between MA-1 and a number of modern populations, a sequence read-based D-statistic test (‘ABBA-BABA test’), equivalent to previously published tests23,40, was applied to sequencing data from a single genome from each of the populations of interest (Supplementary Information, section 13). MA-1 and 11 high-coverage present-day genomes were included in this test. For the chimpanzee outgroup, the multiway alignment, which includes both chimpanzee and human (pantro2 from the hg19 multiz46), was used. The data were filteredasfollowsbeforecalculatingthesequenceread-basedD-statistic39. First, all reads with mapping quality below 30 were removed. Subsequently, bases of low quality were removed by dividing all bases into eight base categories: A, C, G, T on the plus strand and A, C, G, T on the minus strand. The lowest-scoring 50% of bases from each of the eight categories were then discarded. More specifically, within each base category, we found the highest base quality score, Q, for which less than half of the bases in the base category had a quality score smaller than Q. We then removed all bases with quality score smaller than Q, and randomly sampled and removed bases with quality score equal to Q until 50% of the bases from the base category had been removed in total. The data were filtered separately for each of the eight base categories to avoid bias in the test in case of significant difference in the base quality between the categories. After filtering, a single base was sampled at each site for each individual in order to avoid introducing bias due to differences in sequencing depth. Finally, all sites containing transitions were removed. Based on the filtered data, D-statistics were calculated and to assess if these were significantly different from 0, standard errors and Z scores were obtained using a method knownas‘delete-mJackknifefor unequal m’, with a block size of 5 mega bases65.
For genotype data from SNP arrays we computed an allele frequency-based D-statistic test, which is a generalization of the sequence read-based test (Supplementary Information, section 14.3). We used previously presented estimators20,66, obtaining standard errors using a block jackknife procedure over 5-megabase blocks in the genome, except for the tests with the Tianyuan data (chromosome 21), in which case we used 100-kb blocks to increase power. Two main data sets were used: first, a published SNP data set (364,470SNPs) masked for European and African ancestries in Siberian and Native American populations19, which was merged with additional data from Finnish populations63; and second, SNPs ascertained in San and Yoruban individuals and typed in worldwide populations20. As the San and Yoruba populations are approximate outgroups to non-African populations, this data are unbiased for all comparisons between non-Africans. Transition SNPs were included but the first and last three bases of each sequence read were excluded since the majority of nucleotide misincorporations occur at the ends of ancient DNA templates (Supplementary Information, section 6.2). For other tests, (in Supplementary Information, section 14), SNP data described in Supplementary Table 11 were used. We sampled a single read at each position from the MA-1 data as in the principal component analysis.
Outgroup f3-statistics
Classical measures of pair wise genetic distance, such as Wright’s fixation index FST, are sensitive to genetic drift that has occurred since the divergence of the two test populations. If such lineage-specific genetic drift differs between populations that share an equal amount of genetic history with an ancient individual, the ancient individual would be observed as being closer to the modern populations with the least degree of historical genetic drift using distance-based methods such at FST. To circumvent these issues and obtain a statistic that is informative of the genetic relatedness between a particular sample and each candidate population in a reference set, an ‘outgroup f3-statistic’ was computed (Supplementary Information, section 14.2). The expected value of the f3-statistic20, f3 (Outgroup; A, B), equals the sum of expected squared change in allele frequency (normalized for heterozygosity in the outgroup) due to genetic drift on the path in the population tree from the outgroup to the root and from the root to the ancestor of populations A and B. As genetic drift in the lineage specific to the outgroup is expected to be constant regardless of which populations A and B are used (in the absence of gene flow), the remaining variation between statistics will depend on how much genetic history is shared between populations A and B. We used Yoruba as an outgroup to non-African populations and computed the statistic f3(Yoruba; MA-1, X) to investigate the shared history of MA-1 and a set of 147 worldwide candidate populations (as X) obtained by merging several data sets (Supplementary Figs 21 and 22), and we corroborated major patterns using SNPs from a San individual from southern Africa (Supplementary Information, section 14).
Supplementary Material
Acknowledgements
We thank the Hermitage State Museum for providing access to the Mal’ta and Afontova Gora-2 human remains. We also thank the Danish National High-Throughput DNA Sequencing Centre and T. Reisberg for technical assistance. This work was supported by the Danish National Research Foundation and the Lundbeck Foundation (E.W. and M.R.) and the Arctic Social Sciences Program, National Science Foundation (grant PLR-1003725 to K.E.G.). R.V.,M.M., M.K., E.M., K.T., S.Ro. and R.M. were supported by the European Regional Development Fund (European Union) through the Centre of Excellence in Genomics to Estonian Biocentre and University of Tartu and Estonian Basic Research grant SF0270177As08. M.M. thanks the Estonian Science Foundation grant no. 8973 and Baltic-American Freedom Foundation Research Scholarship program and M.I.V. thanks the Government of Russian Federation grant no. 14.B25.31.0033 (to E. I. Rogaev). M.D. was supported by the US National Science Foundation (grant DBI-1103639). Computational analyses were carried out at the High Performance Computing Center, University of Tartu, and the Swedish National Infrastructure for Computing (SNIC-UPPMAX, project b2012063).
Footnotes
Supplementary Information is available in the online version of the paper.
Author Contributions E.W. and K.E.G. conceived the project. E.W. headed the project. E.W. and M.R. designed the experimental research project setup. S.D. and K.E.G. provided access to the Mal’ta and Afontova Gora-2 samples, and K.E.G. provided archaeological context for the samples. T.W.S. Jr performed AMS dating. E.B. and O.B. (Tajik individual), E.K. and S.L. (Mari and Avar individuals) provided modern DNA extracts for complete genome sequencing. E.K. and S.L. (Kazakh, Kirghiz, Uzbek and Mari individuals), L.P.O. (Selkup individuals), S.A.F. (Even, Dolgan and Yakut individuals) and M.I.V. (Altai individuals) provided access to modern DNA extracts for genotyping. R.V. carried out Illumina chip analysis on modern samples. P.F.C. performed DNA extraction from the Indian individual. M.R. performed the ancient extractions and library constructions on the modern and ancient samples —the latter with input from L.O. M.R. coordinated the sequencing. M.R. and S.Ra. performed mapping of MA-1 and AG-2 data sets with input from L.O. S.Ra., T.S.-P. and S.B. provided super-computing resources, developed the next-generation sequencing pipeline and performed mapping and genotyping for all the modern genomes. M.R. performed DNA damage analysis with input from L.O. M.M. performed the admixture analysis.M.M.,E.M., K.T. and R.V. performed the mtDNA analysis.M.M.,M.K., S.Ro.,T.K., E.X. and R.M. performed the Y-chromosome analysis. A.A. and I.M. performed the autosomal contamination estimates, error rate estimates, D-statistics tests based on sequence reads and ngsAdmix analyses. P.S. performed biological sexing, mtDNA contamination estimates, PCA, TreeMix, MixMapper, D-statistic tests based on allele frequencies, f3-statistics and phenotypic analyses, and analysis of AG-2 using nucleotide misincorporation patterns under the supervision of R.N. and M.J. M.R., P.S. and E.W. wrote the majority of the manuscript with critical input from R.N., M.J., M.M., K.E.G., A.A., I.M. and M.D. M.M., A.A. and I.M.
Author Information Sequence data for MA-1 and AG-2, produced in this study, are available for download through NCBI SRA accession number SRP029640. Data from the Illumina genotyping analysis generated in this study are available through GEO Series accession number GSE50727; PLINK files can be accessed from http://www.ebc.ee/free_data. In addition, the above data and alignments for the published modern genomes, Denisova genome, Tianyuan individual and the two ancient genomes are available at http://www.cbs.dtu.dk/suppl/malta. Raw reads and alignments for the four modern genomes sequenced in this study are available for demographic research under data access agreement with E.W.
The authors declare no competing financial interests.
References
- 1.Turner CG. Advances in the dental search for native american origins. Acta Anthropogenet. 1984;8:23–78. [PubMed] [Google Scholar]
- 2.Hubbe M, Harvati K, Neves W. Paleoamerican morphology in the context of European and East Asian Pleistocene variation: implications for human dispersion into the New World. Am. J. Phys. Anthropol. 2011;144:442–453. doi: 10.1002/ajpa.21425. [DOI] [PubMed] [Google Scholar]
- 3.Schurr T. The peopling of the New World: perspectives from molecular anthropology. Annu. Rev. Anthropol. 2004;33:551–583. [Google Scholar]
- 4.O’Rourke DH, Raff JA. The human genetic history of the Americas: the final frontier. Curr. Bio. 2010;20:R202–R207. doi: 10.1016/j.cub.2009.11.051. [DOI] [PubMed] [Google Scholar]
- 5.Lell JT, et al. The dual origin and siberian affinities of native american Y chromosomes. Am. J. Hum. Genet. 2002;70:192–206. doi: 10.1086/338457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Starikovskaya EB, et al. Mitochondrial DNA diversity in indigenous populations of the southern extent of Siberia, and the origins of Native American haplogroups. Ann. Hum. Genet. 2005;69:67–89. doi: 10.1046/j.1529-8817.2003.00127.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dulik MC, et al. Mitochondrial DNA and Y chromosome variation provides evidence for a recent common ancestry between Native American and Indigenous Altaians. Am. J. Hum. Genet. 2012;90:229–246. doi: 10.1016/j.ajhg.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Regueiro M, Alvarez J, Rowold D, Herrera RJ. On the origins, rapid expansion and genetic diversity of Native Americans from hunting-gatherers to agriculturalists. Am. J. Phys. Anthropol. 2013;150:333–348. doi: 10.1002/ajpa.22207. [DOI] [PubMed] [Google Scholar]
- 9.Gerasimov MM. The Archaeology and Geomorphology of Northern Asia: Selected Works 5–32. University of Toronto Press; 1964. [Google Scholar]
- 10.Bramanti B, et al. Genetic discontinuity between local hunter-gatherers and central Europe’s first farmers. Science. 2009;326:137–140. doi: 10.1126/science.1176869. [DOI] [PubMed] [Google Scholar]
- 11.Malmström H, et al. Ancient DNA reveals lack of continuity between Neolithic hunter-gatherers and contemporary Scandinavians. Curr. Biol. 2009;19:1758–1762. doi: 10.1016/j.cub.2009.09.017. [DOI] [PubMed] [Google Scholar]
- 12.Fu Q, et al. A revised timescale for human evolution based on ancient mitochondrial genomes. Curr. Biol. 2013;23:553–559. doi: 10.1016/j.cub.2013.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Owsley DW, Jantz RL. Claiming the Stones-Naming the Bones: Cultural Property and the Negotiation of National and Ethnic Identity. Getty Research Institute; 2002. [Google Scholar]
- 14.Astakhov SN. Paleolit Eniseia: Paleoliticheskie Stoianki Afontovoi Gore v G. Krasnoiarske. Evropaiskii Dom; 1999. [Google Scholar]
- 15.Gamble C. Interaction and alliance in Palaeolithic society. Man (Lond) 1982;17:92–107. [Google Scholar]
- 16.Abramova Z. L’art Paléolithique d’Europe Orientale et de Sibérie. Jérôme Millon; 1995. [Google Scholar]
- 17.White R. The women of Brassempouy: a century of research and interpretation. J. Archaeol. Method and Theory. 2006;13:250–303. [Google Scholar]
- 18.Hansen AJ, Willerslev E, Wiuf C, Mourier T, Arctander P. Statistical evidence for miscoding lesions in ancient DNA templates. Mol. Biol. Evol. 2001;18:262–265. doi: 10.1093/oxfordjournals.molbev.a003800. [DOI] [PubMed] [Google Scholar]
- 19.Reich D, et al. Reconstructing Native American population history. Nature. 2012;488:370–374. doi: 10.1038/nature11258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Patterson N, et al. Ancient admixture inhuman history. Genetics. 2012;192:1065–1093. doi: 10.1534/genetics.112.145037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pickrell JK, Pritchard JK. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet. 2012;8:e1002967. doi: 10.1371/journal.pgen.1002967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meyer M, et al. A high-coverage genome sequence from an archaic Denisovan individual. Science. 2012;338:222–226. doi: 10.1126/science.1224344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Green RE, et al. A draft sequence of the Neandertal genome. Science. 2010;328:710–722. doi: 10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gutenkunst RN, Hernandez RD, Williamson SH, Bustamante CD. Inferring the joint demographic history of multiple populations from multidimensional SNP frequency data. PLoS Genet. 2009;5:e1000695. doi: 10.1371/journal.pgen.1000695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wall JD, et al. Genetic variation in Native Americans, inferred from latino SNP and resequencing data. Mol. Biol. Evol. 2011;28:2231–2237. doi: 10.1093/molbev/msr049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fu Q, et al. DNA analysis of an early modern human from Tianyuan Cave, China. Proc. Natl Acad. Sci. USA. 2013;110:2223–2227. doi: 10.1073/pnas.1221359110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lipson M, et al. Efficient moment-based inference of admixture parameters and sources of gene flow. Mol. Biol. Evol. 2013 doi: 10.1093/molbev/mst099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goebel T. Pleistocene human colonization of siberia and peopling of the Americas: an ecological approach. Evol. Anthropol. 1999;8:208–227. [Google Scholar]
- 29.Brown MD, et al. mtDNA haplogroup X: an ancient link between Europe/Western Asia and North America? Am. J. Hum. Genet. 1998;63:1852–1861. doi: 10.1086/302155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bradley B, Stanford D. The North Atlantic ice-edge corridor: a possible Palaeolithic route to the New World. World Archaeol. 2004;36:459–478. [Google Scholar]
- 31.Stafford TW, Jr, Jull AJT, Brendel K, Duhamel R, Donahue D. Study of bone radiocarbon dating accuracy at the University of Arizona NSF accelerator facility for radioisotope analysis. Radiocarbon. 1987;29:24–44. [Google Scholar]
- 32.Stafford TW, Jr, Brendel K, Duhamel R. Radiocarbon, 13C and 15N analysis of fossil bone: removal of humates with XAD-2 resin. Geochim. Cosmochim. Acta. 1988;52:2257–2267. [Google Scholar]
- 33.Stafford TW, Jr, Hare PE, Currie L, Jull AJT, Donahue D. Accelerator radiocarbon dating at the molecular level. J. Archaeol. Sci. 1991;18:35–72. [Google Scholar]
- 34.Ramsey CB. Bayesian analysis of radiocarbon dates. Radiocarbon. 2009;51:337–360. [Google Scholar]
- 35.Reimer PJ, et al. IntCal09 and Marine09 radiocarbon age calibration curves, 0–50,000 years cal bp. Radiocarbon. 2009;51:1111–1150. [Google Scholar]
- 36.Yang DY, Eng B, Waye JS, Dudar JC, Sanders SR. Technical note: improved DNA extraction from ancient bones using silica-based spin columns. Am. J. Phys. Anthropol. 1998;105:539–543. doi: 10.1002/(SICI)1096-8644(199804)105:4<539::AID-AJPA10>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 37.Svensson EM, et al. Tracing genetic change over time using nuclear SNPs in ancient and modern cattle. Anim. Genet. 2007;38:378–383. doi: 10.1111/j.1365-2052.2007.01620.x. [DOI] [PubMed] [Google Scholar]
- 38.Powell R, Gannon F. Purification of DNA by phenol extraction and ethanol precipitation. Oxford Practical Approach Series. 2002 http://fds.oup.com/www.oup.co.uk/pdf/pas/9v1-7-3.pdf. [Google Scholar]
- 39.Orlando L, et al. Recalibrating Equus evolution using the genome sequence of an early Middle Pleistocene horse. Nature. 2013;499:74–78. doi: 10.1038/nature12323. [DOI] [PubMed] [Google Scholar]
- 40.Reich D, et al. Genetic history of an archaic hominin group from Denisova Cave in Siberia. Nature. 2010;468:1053–1060. doi: 10.1038/nature09710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lindgreen S. AdapterRemoval: easy cleaning of next-generation sequencing reads. BMC Res. Notes. 2012;5:337. doi: 10.1186/1756-0500-5-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schubert M, et al. Improving ancient DNA read mapping against modern reference genomes. BMC Genomics. 2012;13:178. doi: 10.1186/1471-2164-13-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nature Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Krause J, et al. A complete mtDNA genome of an early modern human from Kostenki, Russia. Curr. Biol. 2010;20:231–236. doi: 10.1016/j.cub.2009.11.068. [DOI] [PubMed] [Google Scholar]
- 47.Skoglund P, Storå J, Götherström A, Jakobsson M. Accurate sex identification in ancient human remains using DNA shotgun sequencing. J. Archaeol. Sci. 2013;40:4477–4482. [Google Scholar]
- 48.Rasmussen M, et al. An Aboriginal Australian genome reveals separate human dispersals in Asia. Science. 2011;334:94–98. doi: 10.1126/science.1211177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Frazer KA, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.The 1000 Genomes Project Consortium. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Van Oven M, Kayser M. Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum. Mutat. 2009;30:E386–E394. doi: 10.1002/humu.20921. [DOI] [PubMed] [Google Scholar]
- 52.Behar DM, et al. A “Copernican” reassessment of the human mitochondrial DNA tree from its root. Am. J. Hum. Genet. 2012;90:675–684. doi: 10.1016/j.ajhg.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Drmanac R, et al. Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science. 2010;327:78–81. doi: 10.1126/science.1181498. [DOI] [PubMed] [Google Scholar]
- 54.Wei W, et al. A calibrated human Y-chromosomal phylogeny based on resequencing. Genome Res. 2013;23:388–395. doi: 10.1101/gr.143198.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tamura K, et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hancock AM, et al. Adaptations to climate-mediated selective pressures in humans. PLoS Genet. 2011;7:e1001375. doi: 10.1371/journal.pgen.1001375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rasmussen M, et al. Ancient human genome sequence of an extinct Palaeo-Eskimo. Nature. 2010;463:757–762. doi: 10.1038/nature08835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.International HapMap 3 Consortium. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li JZ, et al. World wide human relationships inferred from genome-wide patterns of variation. Science. 2008;319:1100–1104. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- 60.Skoglund P, Jakobsson M. Archaic human ancestry in East Asia. Proc. Natl Acad. Sci. USA. 2011;108:18301–18306. doi: 10.1073/pnas.1108181108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Patterson N, Price AL, Reich D. Population structure and Eigen analysis. PLoS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Skoglund P, et al. Origins and Genetic legacy of Neolithic farmers and hunter-gatherers in Europe. Science. 2012;336:466–469. doi: 10.1126/science.1216304. [DOI] [PubMed] [Google Scholar]
- 63.Surakka I, et al. Founder population-specific HapMap panel increases power in GWA studies through improved imputation accuracy and CNV tagging. Genome Res. 2010;20:1344–1351. doi: 10.1101/gr.106534.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.International HapMap3 Consortium. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Busing FMTA, Meijer E, Van der Leeden R. Delete-m Jackknife for Unequal m. Stat. Comput. 1999;9:3–8. [Google Scholar]
- 66.Durand EY, Patterson N, Reich D, Slatkin M. Testing for ancient admixture between closely related populations. Mol. Biol. Evol. 2011;28:2239–2252. doi: 10.1093/molbev/msr048. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.