Abstract
The constitutive androstane receptor (CAR; NR1I3) regulates the expression of genes involved in xenobiotic metabolism. Alternative splicing of the human CAR gene yields an array of mRNAs that encode structurally diverse proteins. One form of CAR, termed CAR2, contains an additional four amino acids (SPTV) that are predicted to reshape the ligand-binding pocket. The current studies show a marked, ligand-independent, CAR2-mediated transactivation of reporters containing optimal DR-3, DR-4, and DR-5 response elements, and reporters derived from the natural CYP2B6 and CYP3A4 gene promoters. Overexpression of the RxRα ligand binding domain was critical for achieving these effects. CAR2 interaction with SRC-1 was similarly dependent on the coexpression of RxRα. Mutagenesis of Ser233 (SPTV) to an alanine residue yielded a receptor possessing higher constitutive activity. Alternatively, mutating Ser233 to an aspartate residue drastically reduced the transactivation capacity of CAR2. The respective abilities of these mutagenized forms of CAR2 to transactivate a DR-4 × 3 reporter element correlated with their ability to interact with RxRα and to recruit SRC-1 in a ligand-regulated manner. Together, these results demonstrate a robust RxRα-dependent recruitment of coactivators and transactivation by CAR2. In addition, CAR2 displays novel dose responses to clotrimazole and androstanol compared with the reference form of the receptor while at the same time retaining the ability to bind CITCO. This result supports a hypothesis whereby the four-amino-acid insertion in CAR2 structurally modifies its ligand binding pocket, suggesting that CAR2 is regulated by a set of ligands distinct from those governing the activity of reference CAR.
The constitutive androstane receptor (CAR; NR1I3) is a nuclear hormone receptor that is predominantly expressed in the liver (Baes et al., 1994; Wei et al., 2002). It has been implicated in the metabolism of xenobiotics and drugs (Wei et al., 2000; Zhang et al., 2002; Wang et al., 2004b), carcinogens (Xie et al., 2003), steroids (Xie et al., 2003), heme (Huang et al., 2003; Xie et al., 2003; Huang et al., 2004a), bile acids (Guo et al., 2003; Saini et al., 2004; Zhang et al., 2004), and thyroid hormone (Maglich et al., 2004). Furthermore, there is evidence that CAR activity impinges on cholesterol homeostasis (Kocarek and Mercer-Haines, 2002; Wang et al., 2003b) and signaling pathways that control food consumption (Qatanani et al., 2004). In large part, the effects that CAR exerts on these processes are dependent on the receptor’s ability to modulate hepatic gene expression (Maglich et al., 2002; Ueda et al., 2002). The battery of CAR target genes include members of all three phases of xeno/endobiotic metabolism and clearance, such as certain cytochrome P450, UDP-glucuronosyltransferase, sulfotransferase, glutathione transferase, aldehyde dehydrogenase, and avidin-biotinylated enzyme complex transporter families (Maglich et al., 2002; Ueda et al., 2002). Thus far, CAR response elements have been mapped in a number of the corresponding human genes, including CYP2B6 (Sueyoshi et al., 1999; Wang et al., 2003a), CYP3A4 (Goodwin et al., 2002), CYP3A5 (Burk et al., 2004), CYP2C8 (Ferguson et al., 2005) CYP2C9 (Ferguson et al., 2002; Gerbal-Chaloin et al., 2002), CYP2C19 (Chen et al., 2003), UGT1A1 (Sugatani et al., 2001), MDR1 (Burk et al., 2005), and ALAS1 (Podvinec et al., 2004).
The regulation of CAR activity is complex and still poorly understood. Most studies of CAR regulation have focused on mouse CAR. In mouse, CAR is localized cytosolically in the absence of inducer, such as the prototypical inducer phenobarbital (Kawamoto et al., 1999; Zelko et al., 2001). Recent studies have identified a number of CAR interacting proteins that complex with the cytosolic receptor, including two heat shock proteins that may function to anchor CAR to the cytoskeleton (Kobayashi et al., 2003; Yoshinari et al., 2003). Upon exposure to an inducing agent, CAR is released from this complex by a mechanism that probably involves protein phosphatase 2A, in turn accumulating in the nucleus, where the receptor heterodimerizes with RxRα and subsequently interacts with coregulators such as SRC-1 (Makinen et al., 2002) to regulate target genes. It is currently unknown whether CAR activity is governed similarly in human hepatocytes although the available evidence supports such a hypothesis (Pascussi et al., 2000; Maglich et al., 2003; Wang et al., 2004a). It is odd that most of the inducing agents that act through the CAR signaling pathway do not interact directly with the receptor (Moore et al., 2000; Zhang et al., 2002; Huang et al., 2004a). Only a limited number of CAR ligands have been identified that regulate the receptor through interaction with its ligand binding pocket. These include clotrimazole (Moore et al., 2000; Makinen et al., 2002; Moore et al., 2002), 5β-pregnane-3,20-dione (Moore et al., 2000), CITCO (Maglich et al., 2003), androstanol (Forman et al., 1998), androstenol (Forman et al., 1998), 17α-ethynyl-3,17β-estradiol (Makinen et al., 2002), 1,4-bis[2-(3,5-dichloropyridyloxy)]benzene (Tzameli et al., 2000), and meclizine (Huang et al., 2004b).
We and others have recently described a number of mRNA splice variants of the human CAR gene that potentially represent a large expansion of the CAR proteome (Auerbach et al., 2003; Savkur et al., 2003; Arnold et al., 2004; Jinno et al., 2004; Lamba et al., 2004). One of the variant forms results from the use of an alternative splice acceptor site in intron 6, leading to the insertion of 12 additional nucleotides. The resultant mRNA has been reported to make up 6 to 10% of the total CAR transcript in human liver (Jinno et al., 2004). This transcript encodes a protein containing an additional four amino acids (SPTV) that are predicted to extend helix 6 of the ligand binding domain and potentially affect the structure of the ligand binding pocket (Auerbach et al., 2003; Savkur et al., 2003). We term this form of the receptor CAR2 [CAR1 being the reference form of the receptor (Baes et al., 1994)]. CAR2 retains a limited ability to transactivate CAR-responsive reporters (Auerbach et al., 2003; Arnold et al., 2004; Jinno et al., 2004), a result that correlates with a reduced affinity for RxRα and in turn a compromised ability to interact with DNA (Auerbach et al., 2003; Arnold et al., 2004). Ligand studies of CAR2 demonstrated that clotrimazole deactivated the receptor, whereas CITCO produced a weak, albeit significant activation of CAR2 (Jinno et al., 2004)—a result that is contrary to mammalian two-hybrid studies published separately (Arnold et al., 2004). It is noteworthy that in transfected mouse hepatocytes, nuclear translocation of CAR2 is not observed after CITCO treatment (Jinno et al., 2004).
Studies presented here now demonstrate that CAR2 constitutively transactivates DR-3, DR-4, and DR-5 nuclear receptor response elements in addition to the endogenously encountered PBREM, CYP2B6-XREM, and CYP3A4-XREM luciferase reporters. The CAR inverse agonists androstanol and clotrimazole differentially repress CAR2 compared with CAR1 at varying doses. CAR2 also seems to retain the ability to bind CITCO. Furthermore, we report that RxRα cotransfection greatly enhances the ability of CAR2 to transactivate reporter constructs through a mechanism that is highly dependent on the ability of the two receptors to heterodimerize and requires the DNA binding domains of both receptors together with the AF2 domain of CAR2. Mammalian 2-hybrid studies revealed that in contrast to CAR1, the ability of CAR2 to interact with coactivators is largely dependent on RxRα. Finally, mutagenesis studies conducted on Ser233 of CAR2 reveal that this site plays an important role in the ability of the receptor to transactivate reporters, heterodimerize with RxRα, and recruit SRC-1.
Materials and Methods
Chemicals
Clotrimazole and 5α-androstan-3α-ol were obtained from Sigma (St Louis, MO). CITCO was purchased from BIOMOL Research Laboratories (Plymouth Meeting, PA). Dimethyl sulfoxide (DMSO) was purchased from EM Scientific (Gibbstown, NJ). Primers for polymerase chain reaction and EMSA were purchased from Integrated DNA Technologies (Coralville, IA).
Plasmids
Polymerase chain reaction based cloning was done with Accu-POL DNA polymerase (GeneChoice, Frederick, MD). The primary structures of all of the resulting plasmid constructs were verified by DNA sequencing. The sequences cloned into the respective expression vectors represented only the protein coding regions and were preceded by a Kozak sequence (Kozak, 1987). Mutagenesis was performed using the indicated primers (Tables 1–4) and the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) according to the manufacturer’s protocol. Before transfection, plasmids were prepped using the Quantum Prep Plasmid Maxiprep Kit (Bio-Rad Laboratories, Hercules, CA). All RXR clones referred to in this manuscript were derived from human RxRα.
TABLE 1. pTracer-CMV2 (Invitrogen).
This plasmid contains a CMV promoter that yields robust expression of cloned sequences when transfected into mammalian cells. This plasmid is referred to in this text as “CMV2.”
| Name | Primers | Accession Number |
Source | Amino Acids |
Restriction Sites |
|---|---|---|---|---|---|
| CAR1 | NM_005122 | Human liver | 1–348 | EcoR1/EcoRV | |
| cDNA | |||||
| Forward | GATCGAATTCGTCATGGCCAGTAGGGAAGATGAG | ||||
| Reverse | GATCGATATCTCAGCTGCAGATCTCCTGGAGCCAG | ||||
| CAR2 | NA | Human liver cDNA | 1–352 | EcoR1/EcoRV | |
| Forward | GATCGAATTCGTCATGGCCAGTAGGGAAGATGAG | ||||
| Reverse | GATCGATATCTCAGCTGCAGATCTCCTGGAGCCAG | ||||
| CAR2-A | NA | CMV2 clone | 1–352 S233A | EcoR1/EcoRV | |
| Forward | GATCGAATTCGTCATGGCCAGTAGGGAAGATGAG | ||||
| Reverse | GATCGATATCTCAGCTGCAGATCTCCTGGAGCCAG | ||||
| Mutagenesis Forward | GAAGATGGAGCCCGTGTAGCTCCCACAGTGGGGTTCCA | ||||
| Mutagenesis Reverse | TGGAACCCCACTGTGGGAGCTACACGGGCTCCATCTTC | ||||
| CAR2-D | NA | CMV2 clone | 1–352 S233D | EcoR1/EcoRV | |
| Forward | GATCGAATTCGTCATGGCCAGTAGGGAAGATGAG | ||||
| Reverse | GATCGATATCTCAGCTGCAGATCTCCTGGAGCCAG | ||||
| Mutagenesis Forward | GAAGATGGAGCCCGTGTAGATCCCACAGTGGGGTTCCA | ||||
| Mutagenesis Reverse | TGGAACCCCACTGTGGGATCTACACGGGCTCCATCTTC |
NA, not available.
TABLE 4. pGL3-Basic (Promega).
The pGL3 basic vector was engineered with the thymidine kinase core promoter as described previously to generate a TK-luc reporter (Auerbach et al., 2003). The DR-1×3 through DR-5×3 reporters were made with complimentary primers that were annealed and blunt-end–ligated into the Sma1 site upstream of the TK promoter.
| DR-1×3 | |
| Forward | TCAGTTCACAGTTCACAGTTCACAGTTCACAGTTCACAGTTCAGA |
| Reverse | TCTGAACTGTGAACTGTGAACTGTGAACTGTGAACTGTGAACTGA |
| DR-2×3 | |
| Forward | TCAGTTCAGCAGTTCAGCAGTTCAGCAGTTCAGCAGTTCAGCAGTTCAGA |
| Reverse | TCTGAACTGCTGAACTGCTGAACTGCTGAACTGCTGAACTGCTGAACTGA |
| DR-3×3 | |
| Forward | TCAGTTCAGGCAGTTCAGGCAGTTCAGGCAGTTCAGGCAGTTCAGGCAGTTCAGA |
| Reverse | TCTGAACTGCCTGAACTGCCTGAACTGCCTGAACTGCCTGAACTGCCTGAACTGA |
| DR-4×3 | |
| Forward | GATCAGTTCATGGCAGTTCATGGCAGTTCATGGCAGTTCATGGCAGTTCATGGCAGTTCAGATC |
| Reverse | GATCTGAACTGCCATGAACTGCCATGAACTGCCATGAACTGCCATGAACTGCCATGAACTGATC |
| DR-5×3 | |
| Forward | TCAGTTCACTGGCAGTTCACTGGCAGTTCACTGGCAGTTCACTGGCAGTTCACTGGCAGTTCAGA |
| Reverse | TCTGAACTGCCAGTGAACTGCCAGTGAACTGCCAGTGAACTGCCAGTGAACTGCCAGTGAACTGA |
| 2B6-XREM-PBREMa | |
| Forward | GATCGGTACCAGACTGTGCCAGATTGCACAACAC |
| Reverse | GATCGCTAGCCCACGAGGAGAGGACCAACAAAG |
| 3A4-XREM-pER6b | |
| pER6 Forward | GATCGAATTCTAAGAACCCAGAACCCTTGGAC |
| pER6 Reverse | GATCCTCGAGTGTGCTCTGCCTGCAGTTGGAA |
| XREM Forward | GATCGGTACCGTCCCAATTAAAGGTCATAAAG |
| XREM Reverse | GATCGAATTCCTCGTCAACAGGTTAAAGGAG |
| PBREM | c |
A polymerase chain reaction amplicon was generated from human genomic DNA that contained the 2B6 XREM sequences recently described (Wang et al., 2003a). The amplicon was ligated upstream of the TK promoter using the KpnI and NheI restriction sites.
Amplicons encompassing the proximal (p) ER-6 (Barwick et al., 1996) and distal XREM (Goodwin et al., 1999) sequences in the CYP3A4 promoter were amplified separately. Individual amplicons were digested with EcoRI, purified, and ligated. The ligation was then amplified with the XREMFP and pER6RP. The product from this second amplification was then blunt-end–ligated into the SmaI site upstream of the thymidine kinase promoter.
The PBREM-TK-Luc reporter was described previously (Auerbach et al., 2003).
Cell Culture
COS-1 cells (simian virus-40 –transformed green monkey kidney cells) were maintained and transfected in Dulbecco’s modified Eagle’s medium with 10% fetal bovine serum, 2 mM l-glutamine, 10 mM HEPES, 0.15% sodium bicarbonate, 50 units/ml penicillin G, and 50 µg/ml streptomycin. Human hepatoma HepG2 cells were maintained and transfected in minimal essential medium; all other components remained the same except for the addition of 1.0 mM sodium pyruvate. All cell culture reagents were purchased from Invitrogen (Carlsbad, CA).
Reporter and Mammalian Two-Hybrid Assays
All transfections using COS-1 cells for luciferase reporter assays were performed in a 48-well format. On the morning of day 1, cells were plated to approximately 50,000 cells per well. While the cells were attaching, DNA transfection mixtures were assembled using Fugene6 transfection reagent (Roche Applied Science, Indianapolis, IN). All transfections using HepG2 cells for reporter assays were performed in a 48-well format using Lipofectamine LTX reagent (Invitrogen). In this case, DNA transfection mixtures were assembled and placed in the individual wells, and the cells were then plated directly onto the DNA mixture at approximately 100,000 cells per well. In general, for assays involving standard reporters (not two-hybrid), 25 ng of CMV2 or CMV2-CAR expression plasmid, 25 ng of pcDNA3.1 or 3.1-RXR expression plasmid, 100 ng of luciferase reporter, and 10 ng of pRL-CMV (for transfection normalization; Promega, Madison, WI). All mammalian two-hybrid assays were performed with 40 ng of pVP16 expression plasmid, 10 ng of pM (GAL4) expression plasmid, 100 ng of pFR-luc reporter, and 10 ng of pRL-CMV. When the pcDNA3.1 expression plasmid, containing RXR-LBD, was incorporated in to the two-hybrid assay, 10 ng of was used. In all transfections, the transfection reagent was used at a ratio of 1:3 (micrograms of DNA to microliters of transfection reagent) as recommended in the manufacturer’s protocol. Within a given experiment, all transfections contained the same total amount of DNA. At the time of transfection (within 1–6 h after plating), cells were approximately 80% confluent and had initiated cell division (in the case of COS-1 cells). The following day (16–18 h after transfection), cells were treated with chemical agents as indicated in the figures. If chemical treatment was not performed, cells were lysed and assayed 24 h after transfection. In all treatments, DMSO levels never exceeded 0.1% (v/v). On day 3 (24 h after chemical treatment), cells were washed with PBS and luciferase assays were performed using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI) and a Veritas Microplate Luminometer (Turner Biosystems, Sunnyvale, CA). Luciferase assay and stop and glow reagents were diluted with 1× Tris-buffered saline, pH 8.0, to a 0.5× final concentration. All other aspects of the assay were performed in accordance to the manufacturer’s protocol. Dilution of luciferase reagent had no effect on normalized luciferase values.
Statistical Analysis
Quantitative data were examined by analysis of variance. Unless stated otherwise significance was declared if p < 0.01. Data are expressed as means ± S.D. (n = 4).
Results
CAR2 Activated Various Response Elements in Reporter Assays, an Effect That Was Greatly Enhanced by Coexpression of RxRα
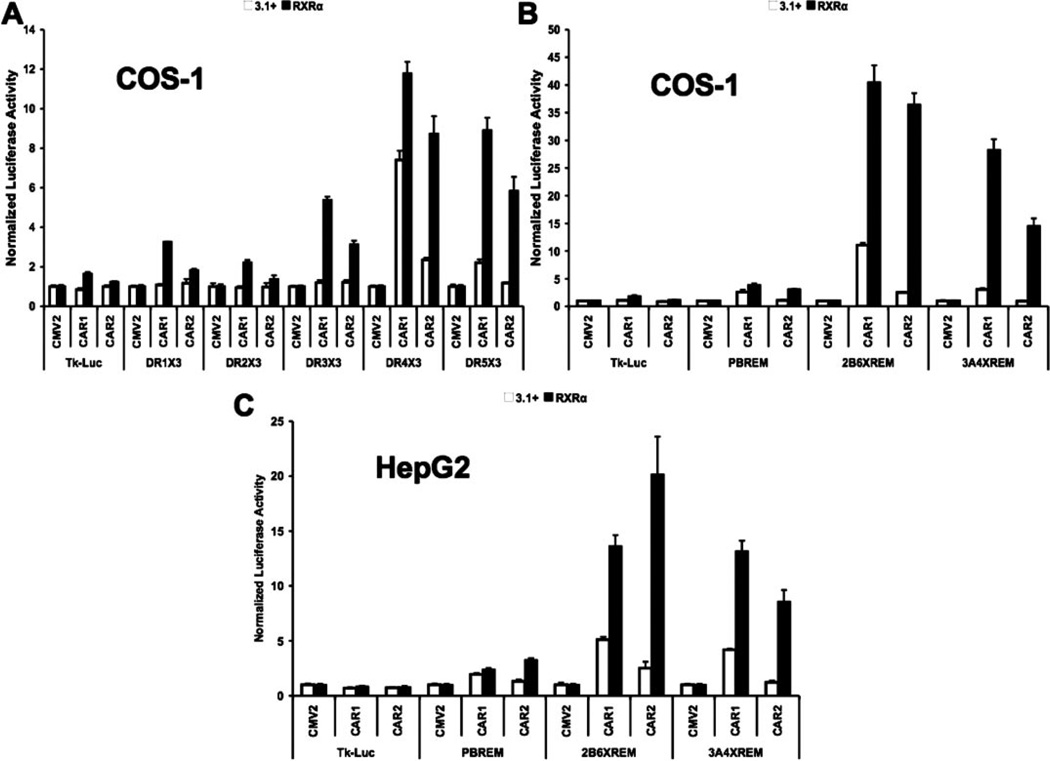
Using luciferase reporters designed to preferentially interact with CAR/RXR heterodimers (Frank et al., 2003), CAR2 produced a statistically significant transactivation of a DR-4 × 3 reporter in the absence of cotransfected RxRα (Fig. 1A). This effect was greatly enhanced by overexpression of RxRα. Furthermore, the inclusion of RxRα allowed CAR2 to also transactivate DR-3 and DR-5 reporter constructs. In parallel assays, CAR1 significantly transactivated the DR-4 and DR-5 reporters even in the absence of cotransfected RxRα. The overexpression of RxRα led to a more permissive activation by CAR1, enabling the receptor to induce all of the direct repeat reporters and enhancing its overall transactivation potential (Fig. 1A).
Fig. 1.
CAR2 transactivation potential is greatly enhanced by RxRα. Transfection assays were performed in COS-1 or HepG2 cells with plasmids indicated/illustrated in the figure and as described under Materials and Methods. Experiments were performed in the presence or absence of cotransfected RxRα. Data are presented as normalized and adjusted luciferase values in which the activity of the 3.1 (or RxRα)/CMV2 group is adjusted to 1 for each respective response element. Each data point represents the mean (± S.D.) of four separate transfections. A, transactivation of multiple DR response elements by CAR1 and CAR2 in the presence and absence of RxRα in COS-1 cells. B, transactivation of multiple endogenous response elements by CAR1 and CAR2 in the presence and absence of RxRα in COS-1 cells. C, transactivation of multiple endogenous response elements by CAR1 and CAR2 in the presence and absence of RxRα in HepG2 cells.
The DR reporters used in Fig. 1A contain three copies of an optimal DR element, a situation that does not exist in the human genome. To test the question of whether or not CAR2 is capable of transactivation via the more degenerate elements found in the promoters of CAR target genes, we created reporters containing previously identified CAR responsive sequences, the promoters of the CYP2B6 and CYP3A4 genes, both of these included an upstream enhancer sequence termed the XREM (Goodwin et al., 1999; Sueyoshi et al., 1999; Goodwin et al., 2002; Wang et al., 2003a). We also included the CYP2B6 promoter without the XREM sequence (PBREM) (Fig. 1B). The PBREM reporter is weakly yet significantly activated by CAR1 (2.6-fold) in the absence of cotransfected RxRα; CAR2 did not activate the reporter under these conditions. Overexpression of RxRα allowed for stronger activation by CAR1 and induction by CAR2 on a level comparable with CAR1 (3.0- and 3.8-fold for CAR2 and CAR1, respectively). Both CAR1 and CAR2 were able to activate the 2B6-XREM reporter in the absence of RxRα (11.1- and 2.5-fold, respectively), but only CAR1 was able to activate the 3A4-XREM reporter without cotransfected RxRα (3.1-fold). The addition of RxRα greatly enhanced the ability of both CAR variants to transactivate these two reporter constructs. In the case of CAR1, the activity on the 2B6-XREM reporter increased to 40.4-fold over basal an amount 3.6 times greater than seen without RxRα. Activity on the 3A4-XREM reporter increased to 28.2-fold over basal or 9.1 times greater than what was seen without the addition of RxRα. CAR2 activities exhibited even greater returns from the addition of RxRα, activity on the 2B6-XREM reporter increased to 36.4-fold over basal, 14.6 times greater than without RxRα, and activity on the 3A4-XREM reporter went from undetected to 14.4-fold over basal (Fig. 1B).
Figure 1, A and B, was generated in transformed green monkey kidney COS-1 cells. These cells are desirable cell models because of their lack of CAR expression and their ease of transfection. However, because CAR is mainly expressed in the liver, we decided to confirm our results for the endogenous promoters in the human hepatoma cell line HepG2 (Fig. 1C). The results shown in Fig. 1C are in good agreement with those of Fig. 1B. The most notable difference was that the overall activity was decreased particularly in the groups with cotransfected RxRα. The activity of CAR2 on the PBREM was almost identical to that seen in COS-1 cells, whereas CAR1 activities were slightly decreased. Activity on the 2B6-XREM reporter by CAR1, in the absence and presence of RxRα, was 5.1- and 13.6-fold over basal, respectively; the comparable values for CAR2 were 2.5-and 20.1-fold over basal. In this case, RxRα increased CAR1 activity by 2.7-fold and CAR2 activity by 8-fold. For the 3A4-XREM reporter, CAR1 activity in the absence of RxRα was 4.2-fold over basal and 13.2-fold in the presence of RxRα 3.2 times greater than without. CAR2 activity was undetectable in the absence of RxRα; addition of RxRα to the system resulted in activities that increased up to 8.5-fold over basal.
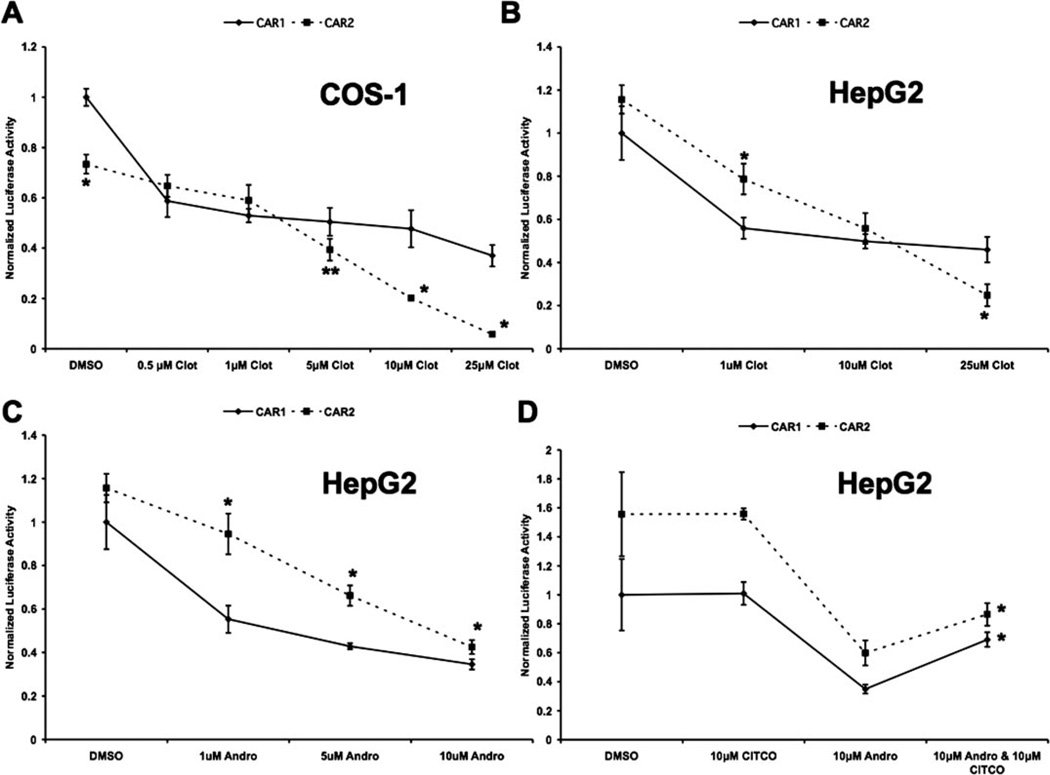
CAR2 Displayed Altered Activity in Response to Different Concentrations of the Inverse Agonists Clotrimazole and Androstanol Compared with CAR1 and Retained the Capacity to Bind CITCO
The four amino acid insertion of CAR2 is within the vicinity of the ligand-binding pocket. To test whether or not this insertion would alter ligand binding, we investigated the effects of the inverse agonists clotrimazole and androstanol and the CAR ligand CITCO on CAR1 and CAR2. All the transfections performed for Fig. 2 were carried out using the 2B6-XREM reporter, RxRα, and CAR1 or CAR2. Treatments were applied 18 h after transfection and cells were harvested 24 h after treatment. All data were normalized to the expression of Renilla reniformis luciferase and adjusted such that activity of the CAR1/DMSO group in each panel was equal to 1.
Fig. 2.
CAR2 displays altered activity in response to different concentrations of the inverse agonists clotrimazole and androstanol compared with CAR1 and retains the capacity to bind CITCO. Transfection assays were performed in COS-1 or HepG2 cells. All transfections were conducted on the 2B6-XREM reporter and included RxRα and either CAR1 or CAR2. Treatments were administered 18 h after transfection and the cells were harvested 24 h after treatment. Data are presented as normalized luciferase values adjusted so that the CAR1/DMSO group in each panel is equal to 1. Each data point represents the mean (± S.D.) of four separate transfections. A, inverse agonism of CAR1 and CAR2 in response to clotrimazole in COS-1 cells. B, inverse agonism of CAR1 and CAR2 in response to clotrimazole in HepG2 cells. C, inverse agonism of CAR1 and CAR2 in response to 5α-androstan-3α-ol in HepG2 cells. Asterisks denote data points at which CAR2 is significantly different from CAR1 at a specific concentration of chemical. (mean ± S.D., n = 4, *, p < 0.01;**, p < 0.05) D, reversal of androstanol’s inverse agonism by CITCO. The asterisks indicate that the activity of CAR1 and CAR2 is significantly greater in the groups treated with both androstanol and CITCO compared with the respective androstanol-only groups (mean ± S.D., n = 4;*, p < 0.01), clot (clotrimazole), Andro (5α-androstan-3α-ol), and CITCO.
In Fig. 2A, CAR1 and CAR2 activity in COS-1 cells was tested with increasing concentrations of clotrimazole. In the DMSO control group, the activity of CAR2 was 73.4% of CAR1 activity. At 0.5 µM clotrimazole, CAR1 activity was markedly reduced, and then only modestly decreased in inhibition across the rest of the concentrations tested. In contrast, CAR2 was only weakly inhibited at lower concentrations; its activity equaled CAR1 at 0.5 and 1.0 µM. At higher doses, CAR2 activity decreased much more rapidly than CAR1. At 5 µM CAR2, activity again drops below CAR1 and continues to decrease up to 25 µM, at which point it is strongly repressed, exhibiting only 15.8% of the activity of CAR1 at that concentration. To further investigate the effect of clotrimazole as well as another CAR1 inverse agonist, androstanol, similar experiments were conducted in HepG2 cells (Fig. 2, B and C). A comparable trend was seen under these conditions, such that the response of CAR1 was almost maximal by 1 µM clotrimazole, whereas CAR2 inhibition at 1 µM was significantly less than that observed for CAR1. However, as the concentration of this agent was increased, CAR2 activities exhibited greater levels of repression. It is noteworthy that the inverse agonism of CAR2 caused by high concentrations of clotrimazole was considerably less in HepG2 cells than in COS-1.
No known ligands of hCAR are able to increase its activity above constitutive levels. However, there are compounds that activate hCAR by interacting with its ligand binding domain and inducing nuclear translocation. CITCO is a well characterized ligand that activates hCAR in this manner (Maglich et al., 2003). It is difficult to study ligands that work through this mechanism because the nuclear translocation of CAR does not recapitulate in transfected cell systems where CAR spontaneously accumulates in the nucleus. To see whether CAR2 could interact with CITCO, we decided to test whether CITCO could reverse the inverse agonism of androstanol through competition. The data presented in Fig. 2D demonstrate that 10 µM CITCO alone exerts no effect on CAR1 or CAR2 and that 10 µM androstanol repressed the activity of both receptors. When administered together, the activity of both receptors was significantly greater than that seen with androstanol alone.
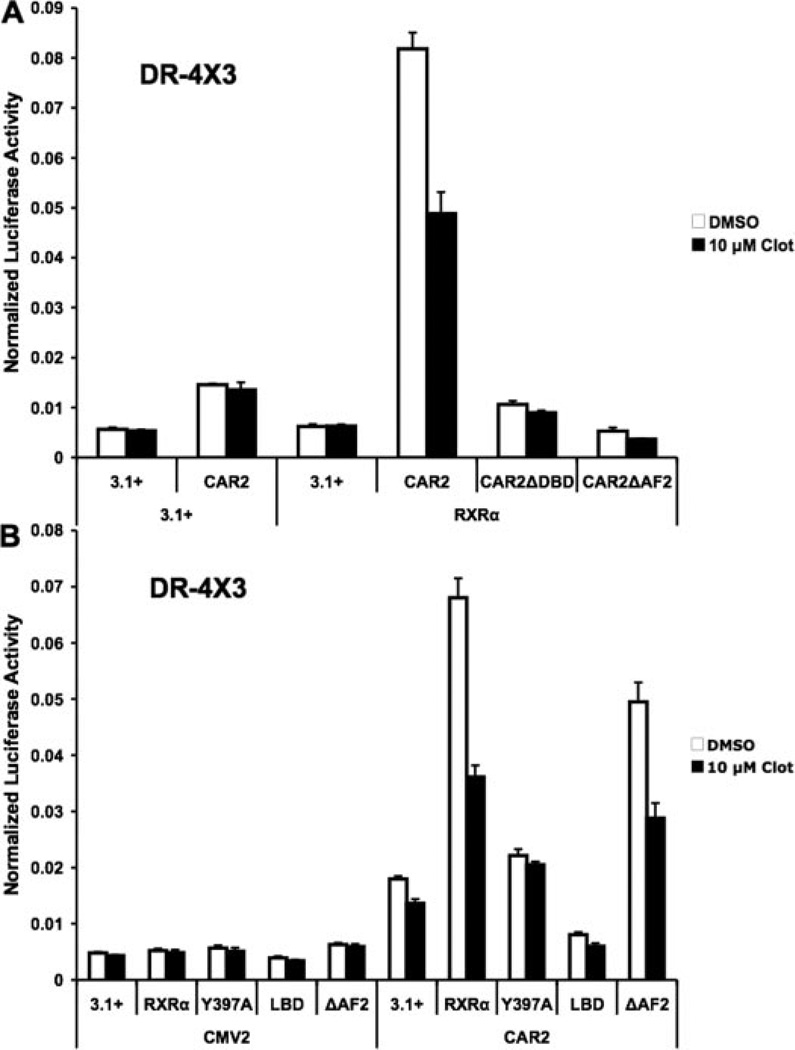
Transactivation of the DR-4 × 3 Reporter Required the CAR2 AF-2 and DBD, and Enhancement of CAR2 Activity by RxRα Was Dependent on Heterodimerization and the DBD of RxRα
Using an EMSA protocol, we previously demonstrated a weak interaction of a GST-CAR2/GST-RxRα complex with a CYP2B6 NR1 probe (Auerbach et al., 2003). However, additional EMSA studies using nuclear extracts from transfected COS-1 cells failed to demonstrate CAR2-DNA binding under conditions that produced robust DNA interaction of CAR1 (data not shown). This finding is in agreement with other published results (Arnold et al., 2004). Therefore, the question remains as to the mechanism of CAR2 transactivation. We generated CAR2 DNA constructs that contained deletions of sequences representing the DBD and AF-2 receptor regions. Cotransfection of these constructs in combination with RxRα led to a complete loss of CAR2-mediated transactivation of the DR-4 × 3 reporter (Fig. 3A). These findings suggest that CAR2 is directly interacting with DNA in a cellular context and mediating transactivation by AF-2-dependent coactivator recruitment.
Fig. 3.
Enhancement of CAR2 activity by RxRα is dependent on heterodimerization with RxRα and the AF-2 domain of CAR2. Transfection assays were performed in COS-1 cells with plasmids indicated/illustrated in the figure and as described under Materials and Methods. Chemical treatments indicated in the figure were done 18 h after transfection, and the cells were harvested 24 h after treatment. All experiments were conducted using the DR-4 × 3 reporter construct. Data are presented as normalized luciferase values. Each data point represents the mean (± S.D.) of four separate transfections. A, transactivation of the DR-4 × 3 response element by various forms of CAR2 in the presence and absence of RxRα and 10 µM clotrimazole. B, transactivation of the DR-4 × 3 response element by CAR2 in the presence of different forms of RxRα and 10 µM clotrimazole. In A, the full-length CAR2 expression plasmid encodes a N-terminal hemagglutinin-tagged form of the receptor. The epitope tag does not affect the activity of the receptor (data not shown).
A similar set of experiments were designed to test the influence of different forms of RxRα on CAR2 activity and are presented in Fig. 3B. Cotransfection of a heterodimerization-deficient form of RxRα (Y397A) (Vivat-Hannah et al., 2003) yielded a level of activation that was statistically significant (p < 0.01) compared with the CAR2/3.1+ control group, however; the magnitude of this activation was considerably less than that seen with wild-type RxRα. Deletion of the DBD (LBD) of RxRα inhibited CAR2 transactivation relative to transfection of CAR2 alone. Deletion of the RxRα AF-2 sequence did not greatly affect the ability of RxRα to enhance CAR2 activity. Together, these data suggest that RxRα must heterodimerize with CAR2 to facilitate its interaction with DNA and, furthermore, that the interaction of RxRα with coactivators through its AF-2 domain is not essential to attain CAR2-dependent transactivation.
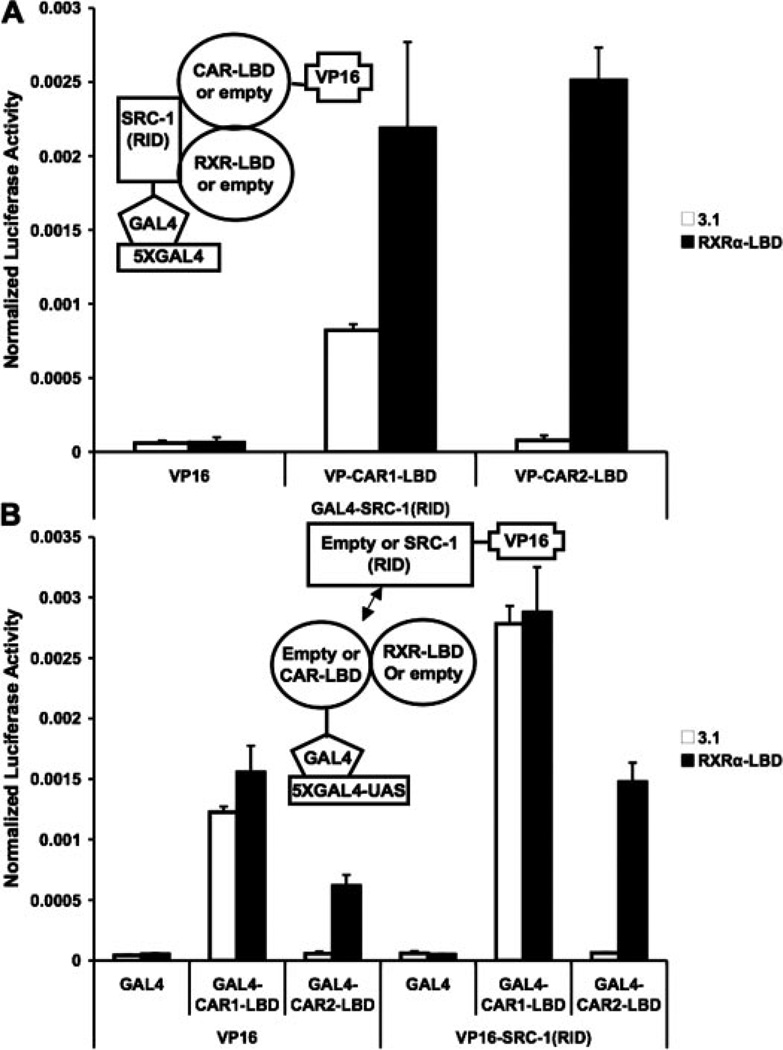
The LBD of RxRα Facilitated the Interaction of CAR2 with the RID (Receptor Interaction Domain) of SRC-1
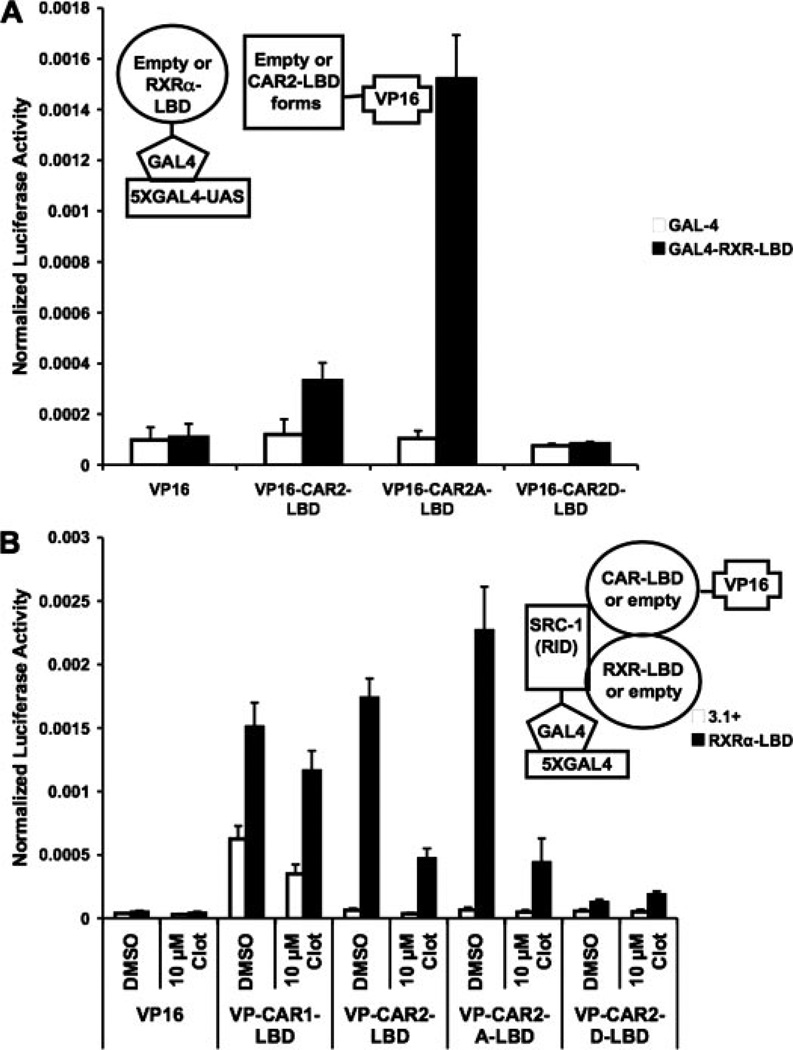
Considering the noted influence of RxRα on CAR2 activity and previously documented effects of RxRα on CAR1 interaction with coactivators (Dussault et al., 2002), two hybrid experiments were performed to assess the effect of RxRα on CAR2/coactivator interaction. The results presented in Fig. 4A demonstrate a strong, RxRα-dependent interaction of CAR2 with GAL4-SRC-1 (RID). In the case of CAR1, inclusion of RxRα yielded only a modest increase in the interaction between it and SRC-1. In other experiments, it was determined that VP16-CAR1 or -2 did not interact with unfused (empty) GAL4 (data not shown).
Fig. 4.
CAR2 recruitment of SRC-1 is RxRα-dependent. Mammalian two-hybrid experiments were performed in COS-1 cells with plasmids indicated in the figures/illustrations and as described under Materials and Methods. Data are presented as normalized and adjusted luciferase values in which the activity of the VP16 (empty)/3.1 (empty)/DMSO (A) or GAL4/VP16/DMSO (B) data points are adjusted to 1. Each data point represents the mean (± S.D.) of four separate transfections. A, SRC-1 recruitment of CAR1 or CAR2 in the presence and absence of RxRα. B, CAR1 and CAR2 recruitment of SRC-1 in the presence and absence of RxRα.
To further verify the results in Fig. 4A, an experiment was performed in which CAR1 or 2-LBD and SRC-1 (RID) were inserted into the GAL4 and VP16 vectors, respectively (Fig. 4B). Transactivation by the LBD of CAR2 was observed only in the presence of overexpressed RxRα-LBD. Cotransfection of VP16-SRC-1 (RID) further enhanced the RxRα-dependent transactivation by CAR2. It is noteworthy that the inclusion of RxRα had no effect on the ability of the GAL4-CAR1 to transactivate the reporter or its ability to recruit VP16 via the SRC-1 (RID) construct in these experiments. This result is a stark contrast to those observed with CAR2.
Mutation of the Ser233 Site in CAR2 Modified Receptor Activity
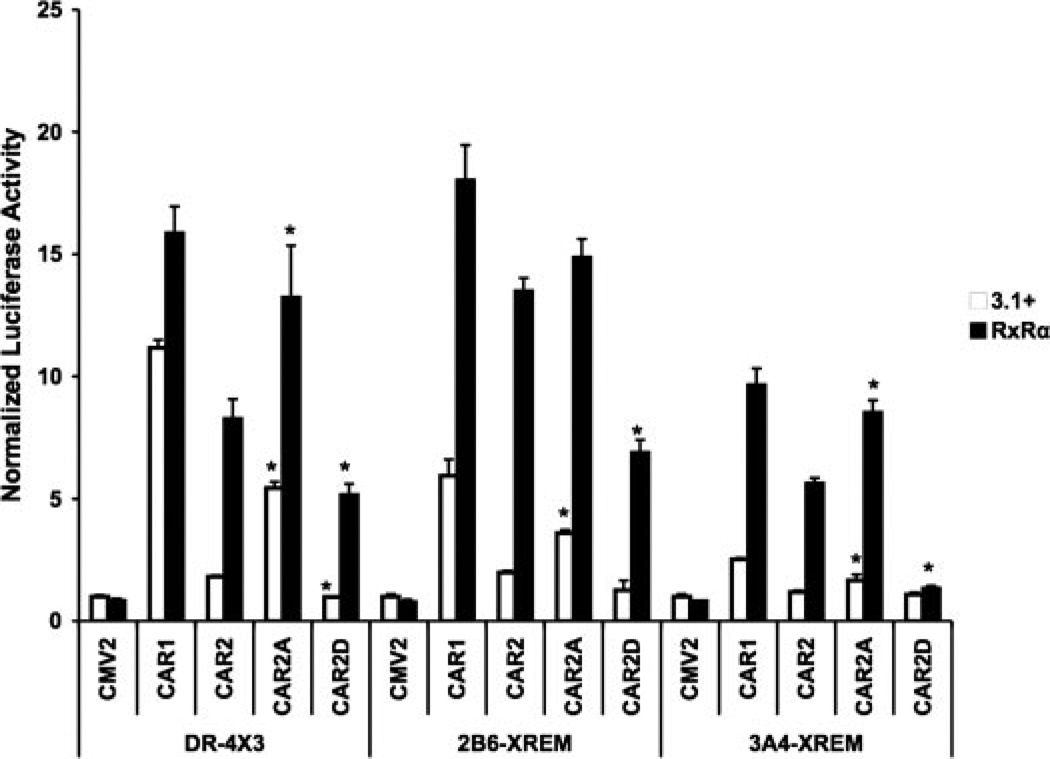
In our previous study (Auerbach et al., 2003), we identified a serine residue, Ser233, in the inserted SPTV sequence of CAR2 as a putative target of phosphorylation. To determine the potential impact of Ser233 phosphorylation, we mutated the residue either to an alanine (CAR2-A) to prevent phosphorylation or to an aspartate (CAR2-D) to mimic constitutive phosphorylation. CAR2-A produced a significantly greater transactivation response than CAR2, on all reporters and in all experimental contexts tested, with the exception of the 2B6-XREM reporter in the presence of RxRα cotransfection (Fig. 5). CAR2-D exhibited a compromised ability to transactivate relative to CAR2. The latter effect was most robust when using the CYP3A4 reporter in the presence of RxRα over expression. The data in Fig. 5 also show that CAR2-A acts more like the reference form of the receptor (CAR1) in its ability to transactivate the three reporter constructs.
Fig. 5.
Mutation of the Ser233 site in CAR2 modifies its transactivation potential. Transfection assays were performed in COS-1 cells with plasmids indicated/illustrated in the figure and as described under Materials and Methods. Transfections were performed in the presence or absence of cotransfected RxRα. Data are presented as normalized and adjusted luciferase values in which the activity of the 3.1/CMV2 group is adjusted to 1 for each respective response element. Each data point represents the mean (± S.D.) of four separate transfections. Figure 5 shows transactivation of the DR-4 × 3, 2B6-XREM and the 3A4-XREM reporters by CAR1, CAR2, and the CAR2 mutants S233A and S233D in the presence and absence of RxRα. Within group comparisons, data points are denoted with an asterisk to indicate that they deviate significantly from the relevant control. For example, on the DR4 × 3 reporter, CAR2A/3.1+ is compared with CAR2/3.1+ and found to have a level of activation significantly greater, the same is found comparing the CAR2A/RxRα group with the CAR2/RxRα group. This process of comparing the CAR2 mutants back with the wild-type CAR2 construct, with or without RxRα, is repeated for each reporter (mean ± S.D., n = 4; p < 0.01).
To further characterize the effects of these mutations, we employed them in our mammalian two-hybrid system to assess their effects on RxRα heterodimerization and SRC-1 recruitment (Fig. 6). In Fig. 6A, the ligand-binding domain of RxRα and the CAR2, CAR2A, and CAR2D ligand binding domains were fused into the GAL4 and VP16 vectors, respectively. CAR2 had a limited ability to interact with RxRα, whereas CAR2A displayed a greatly enhanced interaction and CAR2D had no ability to interact with RxRα. In Fig. 6B, we tested the ability of these mutants to recruit the RID of SRC-1 in the presence and absence of RxRα and 10 µM clotrimazole. As expected, none of the CAR2 constructs were able to transactivate the reporter very well in the absence of RxRα. When RxRα was included in the assay, both CAR2 and CAR2A were able to recruit SRC-1 in an equal capacity; however, CAR2D displayed little ability to interact with SRC-1 under these conditions. Furthermore, 10 µM clotrimazole was much more disruptive of this process on CAR2 than it was for CAR1. This result is in strong agreement with the result presented in Fig. 2A.
Fig. 6.
Effects of Ser233 mutagenesis on heterodimerization and SRC-1 recruitment. Mammalian two-hybrid experiments were performed in COS-1 cells with plasmids indicated in the figures/illustrations and as described under Materials and Methods. Chemical treatments indicated in the figure were done 18 h after transfection and the cells were harvested 24 h after treatment. Data are presented as normalized luciferase values. Each data point represents the mean (± S.D.) of four separate transfections. A, interaction between CAR2 and its S233A and S233D mutants with RxRα. B, interaction between CAR2 and its S233A and S233D mutants with SRC-1 in the presence and absence of RxRα and clot (clotrimazole).
Discussion
CAR2 contains a four-amino-acid insertion (SPTV) in its LBD. Initial studies of the variant receptor demonstrated a severely compromised capacity for transactivation (Auerbach et al., 2003; Arnold et al., 2004; Jinno et al., 2004). Here, we show that cotransfection of RxRα markedly enhances the constitutive activity of CAR2 in reporter assays. The enhancement of CAR2 transactivation seems independent of an RxRα-mediated interaction of the receptor with coactivators. We also assessed the ability of the known CAR ligands clotrimazole and androstanol to function as inverse agonists of CAR2 at various concentrations and found marked differences in the response of CAR2 to these compounds compared with CAR1. Further studies also demonstrated that CITCO reverses the inverse agonism of androstanol on both CAR1 and CAR2, providing strong evidence that CITCO is a CAR2 ligand. Mammalian two-hybrid studies showed that the ability of CAR2 to recruit coactivators has a stronger dependence on RxRα than CAR1 and that CAR2 displays a compromised ability to heterodimerize with RxRα. This later effect can be circumvented in part by mutating the Ser233 residue of CAR2 to an alanine and is completely ablated by changing the same residue to an aspartate. The S233D mutation also lost all ability to recruit SRC-1. Finally, and in agreement with the COS-1 dose-response data, the recruitment of SRC-1 by CAR2 was abrogated more effectively by 10 µM clotrimazole than for CAR1.
It was reported previously that mouse CAR interaction with coactivators was enhanced by the overexpression of RxRα (Dussault et al., 2002). This phenomenon has also been observed with FXR (Pineda, 2004). In the case of CAR, it was suggested that RxRα produces an allosteric effect on the receptor and was not directly involved in coactivator recruitment (Dussault et al., 2002). Data presented in this study for human CAR are in agreement with the mouse report (Dussault et al., 2002) and suggest that RxRα, independent of its AF-2 domain, allosterically modifies the activity of CAR2, without directly interacting with coactivator when in complex with CAR2. Therefore, these data imply that the AF-2 function of CAR2, and not that of RxRα, mediates coactivator recruitment, a conclusion that is further supported by the results demonstrating the complete ablation of CAR2 activity after a deletion of its AF-2 motif.
During the course of these studies, we noticed that CAR2 was being repressed more strongly by higher concentrations of clotrimazole than those observed for CAR1. To further investigate this, CAR1 and CAR2 were both assayed for activity on the 2B6-XREM promoter with multiple concentrations of clotrimazole and androstanol. The results of Fig. 2 demonstrate that CAR2 exhibits an altered response to inverse agonists compared with CAR1. Further studies also revealed that CAR2 retains the ability to interact with CITCO, a well characterized CAR1 ligand. Therefore, it is conceivable that CAR2 may be regulated by a distinct yet overlapping set of ligands in comparison with CAR1, a concept that is supported by molecular modeling approaches indicating that the differential splicing of CAR2 results in an insertion of four amino acids (SPTV) in close proximity to the receptor ligand binding pocket (Auerbach et al., 2003). Large chemical screens should reveal whether unique pharmacophores exist that specifically modulate the CAR2 receptor and its respective biology. This process is complicated by CAR’s unique regulation, allowing certain ligands to act as agonists, such as has been shown with 1,4-bis[2-(3,5-dichloropyridyloxy)]benzene on mCAR (this effect has not yet been seen on hCAR) (Forman et al., 1998), inverse agonists, as well as general activators of receptor translocation, such as phenobarbital.
Why is transfection of RxRα necessary in COS-1 cells that already express levels of the receptor detectable by Western immunoblotting? The RXR antibody we have used (ΔN197; Santa Cruz Biotechnologies, Santa Cruz, CA) is not RXR isotype-selective. Isotype-selective heterodimerization has already been documented for nurr1 (Sacchetti et al., 2002) and vitamin D receptor (Kephart et al., 1996). In our studies, certain receptors, such as PXR and FXR, are unaffected by overexpression of RxRα in COS-1 cells (S. S. Auerbach, J. G. De Keyser, and C. J. Omiecinski, unpublished data). Perhaps PXR and FXR preferentially interact with an abundant isotype of RXR expressed in COS-1 cells, whereas CAR more selectively heterodimerizes with RxRα that is not expressed in high abundance in these cells. Furthermore, data from HepG2 cells (Fig. 1C) showed that CAR1 and CAR2 had similar transactivation potentials in the absence of cotransfected RxRα compared with COS-1 cells (Fig. 1B). The addition of exogenous RxRα yielded a greater increase in activity in COS-1 cells than in HepG2 cells. In our experience, COS-1 cells have transfection efficiencies 5- to 10-fold greater (based on the activity of R. reniformis luciferase) than that seen in HepG2 cells. The effect of cotransfected RxRα is compared within the same cell line in contrast to transfection efficiency that is compared across lines. Therefore, it would be expected that if everything else were equal, the maximal activity would be lower in HepG2 cells because of the lower transfection efficiency but that the percentage of increase imparted by cotransfected RxRα should be equal between the two cell lines. It is possible that HepG2 cells express more RxRα compared with the other RXR isotypes, creating an environment more favorable for CAR activity compared with COS-1 cells. This idea is further supported by the data from Fig. 2A. The strong repression of CAR2 by clotrimazole in COS-1 cells was surprising and may indicate that a combination of an unfavorable RXR isotype background acts synergistically with CAR2’s strong dependence on RxRα for coactivator recruitment, essentially silencing the receptor. Finally, it seems reasonable to speculate that heterodimeric complexes composed of different RXR isotypes may preferentially interact with distinct subsets of DNA response elements. If this hypothesis is correct, expression ratios of RXR isotypes in hepatocytes may significantly influence expression of drug metabolizing enzymes. Consistent with this idea, ablation of RxRα in mouse hepatocytes substantially reduces CAR-mediated gene expression, although other RXR isotypes are expressed in the liver (Mangelsdorf et al., 1992; Cai et al., 2003).
CAR1 and CAR2 probably coexist within the human liver and may have negative or synergistic effects on each other. Over the course of these studies, we also performed cotransfection experiments to investigate the activity of the 2B6-XREM reporter in response to varying ratios of CAR1 to CAR2, with concomitant treatment by DMSO, androstanol, clotrimazole, and CITCO (data not shown). The results of these studies showed no differences between groups that contained CAR1 and CAR2 compared with each receptor alone. These results do not rule out the possibility that CAR1 and CAR2 interact in vivo, in that our studies were conducted on an idealized reporter, containing endogenous promoter sequences known to be highly responsive to CAR and removed from their native chromatin environment. In the context of a full-length endogenous promoter, it is conceivable that there are many situations in which gene regulation could be modified through the combined effect of the two isoforms. Furthermore, endogenous expression levels of CAR are likely much less than those achieved in the transfection experiments conducted here. Therefore, it is possible that the high concentrations achieved in the in vitro assays mask certain potential effects. Further experiments will be required to more thoroughly determine potential interactions between the CAR isoforms.
At this point, it is unclear whether the activities of CAR, or the respective CAR variants, may be subject to regulation through differential phosphorylation. In our studies, we used site-specific mutagenesis to evaluate the potential contributions of differential phosphorylation of the Ser233 residue in CAR2. We observed significant differences in transactivation potential among the modified CAR2 proteins. One potential explanation for these differences among CAR-2, CAR2-A, and CAR2-D is that Ser233 serves as a site of phosphorylation functioning to negatively regulate the activity of CAR2 by modifying its ability to heterodimerize with RxRα and engage in subsequent coactivator recruitment. This hypothesis is consistent with the transactivation capacities of the different CAR2 forms: with CAR2-A > CAR2 > CAR2-D. This pattern of activity matches the ability of the receptors to interact with RxRα and to bind coactivator in an RxRα-dependent fashion. Despite the intriguing nature of these results, it is still premature to suggest that CAR2 is regulated by phosphorylation in vivo. However, it is clear that the amino acid constituency of the inserted SPTV sequence in CAR2 imparts unique functional attributes to the receptor in both its ability to heterodimerize with RxRα and its response to inverse agonists. The biological implications of these findings, in vivo, will require additional investigations.
Current estimates place the number of human genes at 20,000 to 25,000, approximately equal to the number of genes in Arabidopsis thaliana and only approximately 4 times more than the number of genes found in Saccharomyces cerevisiae (Blencowe, 2006). A possible explanation for increased complexity in higher organisms could be their more extensive use of alternative splicing. An estimated 40 to 60% of all human genes undergo alternative splicing (Modrek and Lee, 2002). On the other hand, S. cerevisiae has been shown to use very few alternative-splicing events (Sapra et al., 2004). The CAR gene has been shown to undergo extensive alternative splicing. Along with the pregnane X receptor, CAR mediates a defense against potentially toxic exogenous and endogenous chemicals, along with an ever-growing list of other roles. Regulation of CAR activity is complex and poorly understood. The studies presented here indicate a number of biological differences between the reference form of CAR and an alternatively spliced isoform, CAR2. Alternative splicing of the CAR gene may be an evolutionarily derived mechanism that allows it to produce an array of proteins that are able to carry out separate tasks by using subtle differences in their regulation and interactions with chemical activators.
TABLE 2. pcDNA3.1(+) (Invitrogen).
This plasmid contains a CMV promoter that yields robust expression of cloned sequences when transfected into mammalian cells. This plasmid is referred to in the text as “3.1.”
| Name | Primers | Accession Number |
Source | Amino Acids | Restriction Sites |
|---|---|---|---|---|---|
| HA-CAR2 | NA | CMV2 clone | HA-Tag+ 1–352 |
EcoR1/Xba1 | |
| Forward | GGAATTCGCCACCATGGCATACCCATACGATGTTCCAGATTACGCTATGGCCAGTAGGGAAGATGAGC | ||||
| Reverse | BGH | ||||
| CAR2-LBD | NA | CMV2 clone | 80–352 | EcoR1/Xba1 | |
| Forward | GATCGAATTCGCCACCATGGTACTGTCGGCAGAAGCCC | ||||
| Reverse | BGH | ||||
| CAR2-ΔAF2 | NA | CMV2 clone | 1–344 | EcoR1/Xba1 | |
| Forward | GATCGAATTCGTCATGGCCAGTAGGGAAGATGAG | ||||
| Reverse | GTCTAGACTACATCATGGCAGACAGGCCCTG | ||||
| RXRα | NM_002957 | CMV2 clone described previouslya | 1–462 | EcoR1/Xba1 | |
| Forward | GATCGAATTCGCCGCCATGGACACCAAACATTTCCTG | ||||
| Reverse | GATCTCTAGACTAAGTCATTTGGTGCGGCGC | ||||
| RXRα-LBD | NA | CMV2 clone described previouslya | 198–462 | EcoR1/Xba1 | |
| Forward | GGAATTCGCCGCCATGGGCATGAAGCGGGAAG | ||||
| Reverse | GATCTCTAGACTAAGTCATTTGGTGCGGCGC | ||||
| RXRα-ΔAF2 | NA | CMV2 clone described previouslya | 1–449 | EcoR1/Xba1 | |
| Forward | GATCGAATTCGCCGCCATGGACACCAAACATTTCCTG | ||||
| Reverse | GATCTCTAGATTAGGTGTCAATGGGTGTGTCCC | ||||
| RXRα-Y397A | NA | CMV2 clone described previouslya; based on mutagenized mouse cloneb |
1–462 | EcoR1/Xba1 | |
| Forward | GATCGAATTCGCCGCCATGGACACCAAACATTTCCTG | ||||
| Reverse | GATCTCTAGACTAAGTCATTTGGTGCGGCGC | ||||
| Mutagenesis Forward | GGAGAAGGTCGCTGCGTCCTTGGAGGCCTACTGCAAG | ||||
| Mutagenesis Reverse | CTTGCAGTAGGCCTCCAAGGACGCAGCGACCTTCTCC |
TABLE 3. pM and pVP16 (BD Biosciences).
In-frame cloning of sequences into pM and VP16 generate fusion proteins of the GAL4-DNA binding domain and the VP16 activation domain, respectively. These plasmids are used in combination to access interaction of proteins in mammalian cells. The multiple cloning sites of these two plasmids are identical; therefore, cloned sequences are interchangeable. The luciferase reporter employed in the mammalian two-hybrid assays is pFR-Luc (Stratagene)
| Name | Primers | Accession Number | Source | Amino Acids | Restriction sites |
|---|---|---|---|---|---|
| CAR1-LBD | NA | CMV2 clone | 80–348 | EcoR1/Xba1 | |
| Forward | GATCGAATTCGCCACCATGGTACTGTCGGCAGAAGCCC | ||||
| Reverse | BGH | ||||
| CAR2-LBD | NA | CMV2 clone | 80–352 | EcoR1/Xba1 | |
| Forward | GATCGAATTCGCCACCATGGTACTGTCGGCAGAAGCCC | ||||
| Reverse | BGH | ||||
| CAR2 | NA | CMV2 clone | 1–352 | EcoR1/EcoRV | |
| Forward | GATCGAATTCGTCATGGCCAGTAGGGAAGATGAG | ||||
| Reverse | GATCGATATCTCAGCTGCAGATCTCCTGGAGCCAG | ||||
| hCAR2-A | NA | CMV2 clone | 1–352 | EcoR1/EcoRV | |
| Forward | GATCGAATTCGTCATGGCCAGTAGGGAAGATGAG | ||||
| Reverse | GATCGATATCTCAGCTGCAGATCTCCTGGAGCCAG | ||||
| Mutagenesis Forward | GAAGATGGAGCCCGTGTAGCTCCCACAGTGGGGTTCCA | ||||
| Mutagenesis Reverse | TGGAACCCCACTGTGGGAGCTACACGGGCTCCATCTTC | ||||
| hCAR2-D | NA | CMV2 clone | 1–352S S223A | EcoR1/EcoRV | |
| Forward | GATCGAATTCGTCATGGCCAGTAGGGAAGATGAG | ||||
| Reverse | GATCGATATCTCAGCTGCAGATCTCCTGGAGCCAG | ||||
| Mutagenesis Forward | GAAGATGGAGCCCGTGTAGAT CCCACAGTGGGGTTCCA | ||||
| Mutagenesis Reverse | TGGAACCCCACTGTGGGATCTACACGGGCTCCATCTTC | ||||
| RXRα -LBD | NA | CMV2 clone described previouslya | 198–462 S223D | EcoR1/Xba1 | |
| Forward | GGAATTCGCCGCCATGGGCATGAAGCGGGAAG | ||||
| Reverse | GATCTCTAGACTAAGTCATTTGGTGCGGCGC | ||||
| RXRα | NM_002957 | CMV2 clone described previouslya | 1–462 | EcoR1/Xba1 | |
| Forward | GATCGAATTCGCCGCCATGGACACCAAACATTTCCTG | ||||
| Reverse | GATCTCTAGACTAAGTCATTTGGTGCGGCGC | ||||
| SRC-1 RID | NM_003743 | Human liver cDNA | 570–780 | EcoR1/Xba1 | |
| Forward | GATCGAATTCCCTAGCAGATTAAATATACAACCAG | ||||
| Reverse | GATCTCTAGATCACATCTGTTCTTTCTTTTCCACTT |
NA, not available.
Acknowledgments
We are grateful to Denise Weyant and Shengzhong Su for their skilled technical assistance and are thankful to Jack Vanden Heuvel and Eric Tien for gifts of plasmids and advice with respect to the mammalian two-hybrid assay.
This work was supported by Grant GM66411 from the National Institute of General Medical Sciences (to C.J.O.) and National Institute of Environmental Health Sciences Training Grant ES07032 (to S.S.A.).
ABBREVIATIONS
- NR
nuclear receptor
- CAR
constitutive androstane receptor
- RXR
retinoid X receptor
- FXR
farnesoid X receptor
- DBD
DNA-binding domain
- LBD
ligand-binding domain
- DR
direct repeat
- PBREM
phenobarbital response enhancer module
- XREM
xenobiotic response enhancer module
- CMV
cytomegalovirus
- VP16
virus protein 16
- EMSA
electrophoretic mobility shift assay
- CITCO
6-(4-chlorophenyl: imidazo[2, 1-b]thiazole-5-carbaldehyde O-(3, 4-dichlorobenzyl)oxime
- h
human
- m
mouse
- SRC-1
steroid receptor coactivator 1
- bp
base pair(s)
- DMSO
dimethyl sulfoxide
- AF-2
activation function 2
- RID
receptor interaction domain.
References
- Arnold KA, Eichelbaum M, Burk O. Alternative splicing affects the function and tissue-specific expression of the human constitutive androstane receptor. Nucl Recept. 2004;2:1. doi: 10.1186/1478-1336-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auerbach SS, Ramsden R, Stoner MA, Verlinde C, Hassett C, Omiecinski CJ. Alternatively spliced isoforms of the human constitutive androstane receptor. Nucleic Acids Res. 2003;31:3194–3207. doi: 10.1093/nar/gkg419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baes M, Gulick T, Choi HS, Martinoli MG, Simha D, Moore DD. A new orphan member of the nuclear hormone receptor superfamily that interacts with a subset of retinoic acid response elements. Mol Cell Biol. 1994;14:1544–1551. doi: 10.1128/mcb.14.3.1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barwick JL, Quattrochi LC, Mills AS, Potenza C, Tukey RH, Guzelian PS. Transspecies gene transfer for analysis of glucocorticoid-inducible transcriptional activation of transiently expressed human CYP3A4 and rabbit CYP3A6 in primary cultures of adult rat and rabbit hepatocytes. Mol Pharmacol. 1996;50:10–16. [PubMed] [Google Scholar]
- Blencowe BJ. Alternative splicing: new insights from global analyses. Cell. 2006;126:37–47. doi: 10.1016/j.cell.2006.06.023. [DOI] [PubMed] [Google Scholar]
- Burk O, Arnold KA, Geick A, Tegude H, Eichelbaum M. A role for constitutive androstane receptor in the regulation of human intestinal MDR1 expression. Biol Chem. 2005;386:503–513. doi: 10.1515/BC.2005.060. [DOI] [PubMed] [Google Scholar]
- Burk O, Koch I, Raucy J, Hustert E, Eichelbaum M, Brockmoller J, Zanger UM, Wojnowski L. The induction of cytochrome P450 3A5 (CYP3A5) in the human liver and intestine is mediated by the xenobiotic sensors pregnane X receptor (PXR) and constitutively activated receptor (CAR) J Biol Chem. 2004;279:38379–38385. doi: 10.1074/jbc.M404949200. [DOI] [PubMed] [Google Scholar]
- Cai Y, Dai T, Ao Y, Konishi T, Chuang KH, Lue Y, Chang C, Wan YJ. Cytochrome P450 Genes are differentially expressed in female and male hepatocyte retinoid X receptor alpha-deficient mice. Endocrinology. 2003;144:2311–2318. doi: 10.1210/en.2002-0129. [DOI] [PubMed] [Google Scholar]
- Chen Y, Ferguson SS, Negishi M, Goldstein JA. Identification of constitutive androstane receptor and glucocorticoid receptor binding sites in the CYP2C19 promoter. Mol Pharmacol. 2003;64:316–324. doi: 10.1124/mol.64.2.316. [DOI] [PubMed] [Google Scholar]
- Dussault I, Lin M, Hollister K, Fan M, Termini J, Sherman MA, Forman BM. A structural model of the constitutive androstane receptor defines novel interactions that mediate ligand-independent activity. Mol Cell Biol. 2002;22:5270–5280. doi: 10.1128/MCB.22.15.5270-5280.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson SS, Chen Y, LeCluyse EL, Negishi M, Goldstein JA. Human CYP2C8 is transcriptionally regulated by the nuclear receptors constitutive androstane receptor, pregnane x receptor, glucocorticoid receptor, and hepatic nuclear factor 4α. Mol Pharmacol. 2005;68:747–757. doi: 10.1124/mol.105.013169. [DOI] [PubMed] [Google Scholar]
- Ferguson SS, LeCluyse EL, Negishi M, Goldstein JA. Regulation of human CYP2C9 by the constitutive androstane receptor: discovery of a new distal binding site. Mol Pharmacol. 2002;62:737–746. doi: 10.1124/mol.62.3.737. [DOI] [PubMed] [Google Scholar]
- Forman BM, Tzameli I, Choi HS, Chen J, Simha D, Seol W, Evans RM, Moore DD. Androstane metabolites bind to and deactivate the nuclear receptor CAR-beta. Nature (Lond) 1998;395:612–615. doi: 10.1038/26996. [DOI] [PubMed] [Google Scholar]
- Frank C, Gonzalez MM, Oinonen C, Dunlop TW, Carlberg C. Characterization of DNA complexes formed by the nuclear receptor constitutive androstane receptor. J Biol Chem. 2003;278:43299–43310. doi: 10.1074/jbc.M305186200. [DOI] [PubMed] [Google Scholar]
- Gerbal-Chaloin S, Daujat M, Pascussi JM, Pichard-Garcia L, Vilarem MJ, Maurel P. Transcriptional regulation of CYP2C9 gene. Role of glucocorticoid receptor and constitutive androstane receptor. J Biol Chem. 2002;277:209–217. doi: 10.1074/jbc.M107228200. [DOI] [PubMed] [Google Scholar]
- Goodwin B, Hodgson E, D’Costa DJ, Robertson GR, Liddle C. Transcriptional regulation of the human CYP3A4 gene by the constitutive androstane receptor. Mol Pharmacol. 2002;62:359–365. doi: 10.1124/mol.62.2.359. [DOI] [PubMed] [Google Scholar]
- Goodwin B, Hodgson E, Liddle C. The orphan human pregnane X receptor mediates the transcriptional activation of CYP3A4 by rifampicin through a distal enhancer module. Mol Pharmacol. 1999;56:1329–1339. doi: 10.1124/mol.56.6.1329. [DOI] [PubMed] [Google Scholar]
- Guo GL, Lambert G, Negishi M, Ward JM, Brewer HB, Jr, Kliewer SA, Gonzalez FJ, Sinal CJ. Complementary roles of farnesoid X receptor, pregnane X receptor, and constitutive androstane receptor in protection against bile acid toxicity. J Biol Chem. 2003;278:45062–45071. doi: 10.1074/jbc.M307145200. [DOI] [PubMed] [Google Scholar]
- Huang W, Zhang J, Chua SS, Qatanani M, Han Y, Granata R, Moore DD. Induction of bilirubin clearance by the constitutive androstane receptor (CAR) Proc Natl Acad Sci USA. 2003;100:4156–4161. doi: 10.1073/pnas.0630614100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Zhang J, Moore DD. A traditional herbal medicine enhances bilirubin clearance by activating the nuclear receptor CAR. J Clin Investig. 2004a;113:137–143. doi: 10.1172/JCI200418385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Zhang J, Wei P, Schrader WT, Moore DD. Meclizine is an agonist ligand for mouse constitutive androstane receptor (CAR) and an inverse agonist for human CAR. Mol Endocrinol. 2004b;18:2402–2408. doi: 10.1210/me.2004-0046. [DOI] [PubMed] [Google Scholar]
- Jinno H, Tanaka-Kagawa T, Hanioka N, Ishida S, Saeki M, Soyama A, Itoda M, Nishimura T, Saito Y, Ozawa S, Ando M, Sawada J. Identification of novel alternative splice variants of human constitutive androstane receptor and characterization of their expression in the liver. Mol Pharmacol. 2004;65:496–502. doi: 10.1124/mol.65.3.496. [DOI] [PubMed] [Google Scholar]
- Kawamoto T, Sueyoshi T, Zelko I, Moore R, Washburn K, Negishi M. Phenobarbital-responsive nuclear translocation of the receptor CAR in induction of the CYP2B gene. Mol Cell Biol. 1999;19:6318–6322. doi: 10.1128/mcb.19.9.6318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kephart DD, Walfish PG, DeLuca H, Butt TR. Retinoid X receptor isotype identity directs human vitamin D receptor heterodimer transactivation from the 24-hydroxylase vitamin D response elements in yeast. Mol Endocrinol. 1996;10:408–419. doi: 10.1210/mend.10.4.8721985. [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Sueyoshi T, Inoue K, Moore R, Negishi M. Cytoplasmic accumulation of the nuclear receptor CAR by a tetratricopeptide repeat protein in HepG2 cells. Mol Pharmacol. 2003;64:1069–1075. doi: 10.1124/mol.64.5.1069. [DOI] [PubMed] [Google Scholar]
- Kocarek TA, Mercer-Haines NA. Squalestatin 1-inducible expression of rat CYP2B: evidence that an endogenous isoprenoid is an activator of the constitutive androstane receptor. Mol Pharmacol. 2002;62:1177–1186. doi: 10.1124/mol.62.5.1177. [DOI] [PubMed] [Google Scholar]
- Kozak M. An analysis of 5-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamba JK, Lamba V, Yasuda K, Lin YS, Assem M, Thompson E, Strom S, Schuetz EG. Expression of CAR splice variants in human tissues and their functional consequences. J Pharmacol Exp Ther. 2004;311:811–821. doi: 10.1124/jpet.104.069310. [DOI] [PubMed] [Google Scholar]
- Maglich JM, Parks DJ, Moore LB, Collins JL, Goodwin B, Billin AN, Stoltz CA, Kliewer SA, Lambert MH, Willson TM, Moore JT. Identification of a novel human constitutive androstane receptor (CAR) agonist and its use in the identification of CAR target genes. J Biol Chem. 2003;278:17277–17283. doi: 10.1074/jbc.M300138200. [DOI] [PubMed] [Google Scholar]
- Maglich JM, Stoltz CM, Goodwin B, Hawkins-Brown D, Moore JT, Kliewer SA. Nuclear pregnane X receptor and constitutive androstane receptor regulate overlapping but distinct sets of genes involved in xenobiotic detoxification. Mol Pharmacol. 2002;62:638–646. doi: 10.1124/mol.62.3.638. [DOI] [PubMed] [Google Scholar]
- Maglich JM, Watson J, McMillen PJ, Goodwin B, Willson TM, Moore JT. The nuclear receptor CAR is a regulator of thyroid hormone metabolism during caloric restriction. J Biol Chem. 2004;279:19832–19838. doi: 10.1074/jbc.M313601200. [DOI] [PubMed] [Google Scholar]
- Makinen J, Frank C, Jyrkkarinne J, Gynther J, Carlberg C, Honkakoski P. Modulation of mouse and human phenobarbital-responsive enhancer module by nuclear receptors. Mol Pharmacol. 2002;62:366–378. doi: 10.1124/mol.62.2.366. [DOI] [PubMed] [Google Scholar]
- Mangelsdorf DJ, Borgmeyer U, Heyman RA, Zhou JY, Ong ES, Oro AE, Kakizuka A, Evans RM. Characterization of three RXR genes that mediate the action of 9-cis retinoic acid. Genes Dev. 1992;6:329–344. doi: 10.1101/gad.6.3.329. [DOI] [PubMed] [Google Scholar]
- Modrek B, Lee C. A genomic view of alternative splicing. Nat Genet. 2002;30:13–19. doi: 10.1038/ng0102-13. [DOI] [PubMed] [Google Scholar]
- Moore LB, Maglich JM, McKee DD, Wisely B, Willson TM, Kliewer SA, Lambert MH, Moore JT. Pregnane X receptor (PXR), constitutive androstane receptor (CAR), and benzoate X receptor (BXR) define three pharmacologically distinct classes of nuclear receptors. Mol Endocrinol. 2002;16:977–986. doi: 10.1210/mend.16.5.0828. [DOI] [PubMed] [Google Scholar]
- Moore LB, Parks DJ, Jones SA, Bledsoe RK, Consler TG, Stimmel JB, Goodwin B, Liddle C, Blanchard SG, Willson TM, Collins JL, Kliewer SA. Orphan nuclear receptors constitutive androstane receptor and pregnane X receptor share xenobiotic and steroid ligands. J Biol Chem. 2000;275:15122–15127. doi: 10.1074/jbc.M001215200. [DOI] [PubMed] [Google Scholar]
- Pascussi JM, Gerbal-Chaloin S, Fabre JM, Maurel P, Vilarem MJ. Dexamethasone enhances constitutive androstane receptor expression in human hepatocytes: consequences on cytochrome P450 gene regulation. Mol Pharmacol. 2000;58:1441–1450. doi: 10.1124/mol.58.6.1441. [DOI] [PubMed] [Google Scholar]
- Pineda TI, Freedman LP, Garabedian MJ. Identification of DRIP205 as a coactivator for the farnesoid X receptor. J Biol Chem. 2004;279:36184–36191. doi: 10.1074/jbc.M405126200. [DOI] [PubMed] [Google Scholar]
- Podvinec M, Handschin C, Looser R, Meyer UA. Identification of the xenosensors regulating human 5-aminolevulinate synthase. Proc Natl Acad Sci USA. 2004;101:9127–9132. doi: 10.1073/pnas.0401845101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qatanani M, Wei P, Moore DD. Alterations in the distribution and orexigenic effects of dexamethasone in CAR-null mice. Pharmacol Biochem Behav. 2004;78:285–291. doi: 10.1016/j.pbb.2004.04.001. [DOI] [PubMed] [Google Scholar]
- Sacchetti P, Dwornik H, Formstecher P, Rachez C, Lefebvre P. Requirements for heterodimerization between the orphan nuclear receptor Nurr1 and retinoid X receptors. J Biol Chem. 2002;277:35088–35096. doi: 10.1074/jbc.M205816200. [DOI] [PubMed] [Google Scholar]
- Saini SP, Sonoda J, Xu L, Toma D, Uppal H, Mu Y, Ren S, Moore DD, Evans RM, Xie W. A novel constitutive androstane receptor-mediated and CYP3A-independent pathway of bile acid detoxification. Mol Pharmacol. 2004;65:292–300. doi: 10.1124/mol.65.2.292. [DOI] [PubMed] [Google Scholar]
- Sapra AK, Arava Y, Khandelia P, Vijayraghavan U. Genome-wide analysis of pre-MRNA splicing: intron features govern the requirement for the second-step factor, Prp17 in Saccharomyces cerevisiae and Schizosaccharomyces pombe. J Biol Chem. 2004;279:52437–52446. doi: 10.1074/jbc.M408815200. [DOI] [PubMed] [Google Scholar]
- Savkur RS, Wu Y, Bramlett KS, Wang M, Yao S, Perkins D, Totten M, Searfoss G, III, Ryan TP, Su EW, Burris TP. Alternative splicing within the ligand binding domain of the human constitutive androstane receptor. Mol Genet Metab. 2003;80:216–226. doi: 10.1016/j.ymgme.2003.08.013. [DOI] [PubMed] [Google Scholar]
- Sueyoshi T, Kawamoto T, Zelko I, Honkakoski P, Negishi M. The repressed nuclear receptor CAR responds to phenobarbital in activating the human CYP2B6 gene. J Biol Chem. 1999;274:6043–6046. doi: 10.1074/jbc.274.10.6043. [DOI] [PubMed] [Google Scholar]
- Sugatani J, Kojima H, Ueda A, Kakizaki S, Yoshinari K, Gong QH, Owens IS, Negishi M, Sueyoshi T. The phenobarbital response enhancer module in the human bilirubin UDP-glucuronosyltransferase UGT1A1 gene and regulation by the nuclear receptor CAR. Hepatology. 2001;33:1232–1238. doi: 10.1053/jhep.2001.24172. [DOI] [PubMed] [Google Scholar]
- Tzameli I, Pissios P, Schuetz EG, Moore DD. The xenobiotic compound 1,4-bis[2-(3,5-dichloropyridyloxy)]benzene is an agonist ligand for the nuclear receptor CAR. Mol Cell Biol. 2000;20:2951–2958. doi: 10.1128/mcb.20.9.2951-2958.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda A, Hamadeh HK, Webb HK, Yamamoto Y, Sueyoshi T, Afshari CA, Lehmann JM, Negishi M. Diverse roles of the nuclear orphan receptor CAR in regulating hepatic genes in response to phenobarbital. Mol Pharmacol. 2002;61:1–6. doi: 10.1124/mol.61.1.1. [DOI] [PubMed] [Google Scholar]
- Vivat-Hannah V, Bourguet W, Gottardis M, Gronemeyer H. Separation of retinoid X receptor homo- and heterodimerization functions. Mol Cell Biol. 2003;23:7678–7688. doi: 10.1128/MCB.23.21.7678-7688.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Faucette S, Moore R, Sueyoshi T, Negishi M, LeCluyse E. Human constitutive androstane receptor mediates induction of CYP2B6 gene expression by phenytoin. J Biol Chem. 2004a;279:29295–29301. doi: 10.1074/jbc.M400580200. [DOI] [PubMed] [Google Scholar]
- Wang H, Faucette S, Sueyoshi T, Moore R, Ferguson S, Negishi M, LeCluyse EL. A novel distal enhancer module regulated by pregnane X receptor/constitutive androstane receptor is essential for the maximal induction of CYP2B6 gene expression. J Biol Chem. 2003a;278:14146–14152. doi: 10.1074/jbc.M212482200. [DOI] [PubMed] [Google Scholar]
- Wang H, Faucette SR, Moore R, Sueyoshi T, Negishi M, LeCluyse EL. Human CAR mediates induction of CYP2B6 gene expression by phenytoin. J Biol Chem. 2004b;279:29295–29301. doi: 10.1074/jbc.M400580200. [DOI] [PubMed] [Google Scholar]
- Wang X, Le RI, Nicodeme E, Li R, Wagner R, Petros C, Churchill GA, Harris S, Darvasi A, Kirilovsky J, Roubertoux PL, Paigen B. Using advanced intercross lines for high-resolution mapping of HDL cholesterol quantitative trait loci. Genome Res. 2003b;13:1654–1664. doi: 10.1101/gr.1185803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei P, Zhang J, Dowhan DH, Han Y, Moore DD. Specific and overlapping functions of the nuclear hormone receptors CAR and PXR in xenobiotic response. Pharmacogenomics J. 2002;2:117–126. doi: 10.1038/sj.tpj.6500087. [DOI] [PubMed] [Google Scholar]
- Wei P, Zhang J, Egan-Hafley M, Liang S, Moore DD. The nuclear receptor CAR mediates specific xenobiotic induction of drug metabolism. Nature (Lond) 2000;407:920–923. doi: 10.1038/35038112. [DOI] [PubMed] [Google Scholar]
- Xie W, Yeuh MF, Radominska-Pandya A, Saini SP, Negishi Y, Bottroff BS, Cabrera GY, Tukey RH, Evans RM. Control of steroid, heme, and carcinogen metabolism by nuclear pregnane X receptor and constitutive androstane receptor. Proc Natl Acad Sci USA. 2003;100:4150–4155. doi: 10.1073/pnas.0438010100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshinari K, Kobayashi K, Moore R, Kawamoto T, Negishi M. Identification of the nuclear receptor CAR:HSP90 complex in mouse liver and recruitment of protein phosphatase 2A in response to phenobarbital. FEBS Lett. 2003;548:17–20. doi: 10.1016/s0014-5793(03)00720-8. [DOI] [PubMed] [Google Scholar]
- Zelko I, Sueyoshi T, Kawamoto T, Moore R, Negishi M. The peptide near the C terminus regulates receptor CAR nuclear translocation induced by xenochemicals in mouse liver. Mol Cell Biol. 2001;21:2838–2846. doi: 10.1128/MCB.21.8.2838-2846.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Huang W, Chua SS, Wei P, Moore DD. Modulation of acetaminopheninduced hepatotoxicity by the xenobiotic receptor CAR. Science (Wash DC) 2002;298:422–424. doi: 10.1126/science.1073502. [DOI] [PubMed] [Google Scholar]
- Zhang J, Huang W, Qatanani M, Evans RM, Moore DD. The constitutive androstane receptor and pregnane X receptor function coordinately to prevent bile acid-induced hepatotoxicity. J Biol Chem. 2004;279:49517–49522. doi: 10.1074/jbc.M409041200. [DOI] [PubMed] [Google Scholar]