Abstract
• Premise of the study: For cultivar classification, identification, and genetic improvement, microsatellite markers were developed to analyze the genetic diversity of androdioecious Osmanthus fragrans cultivars.
• Methods and Results: Fifteen microsatellite markers were developed from sequences downloaded from the National Center for Biotechnology Information, which included two with null alleles. These primers were screened on 62 typical androdioecious O. fragrans cultivars belonging to four groups (Asiaticus, Albus, Luteus, and Aurantiacus). The number of alleles ranged from two to six, with a mean of 3.7 per locus. The observed and expected heterozygosities ranged from 0.1000 to 0.9091 and from 0.1287 to 0.9167, respectively. Results from structure analyses indicated that Asiaticus and Albus were genetically mixed, and Luteus and Aurantiacus were partially genetically differentiated.
• Conclusions: These markers will be useful for genetic study of androdioecious O. fragrans cultivars and facilitate cultivar classification, particularly for the cultivar groups Luteus and Aurantiacus.
Keywords: androdioecy, genetic diversity, microsatellite markers, Oleaceae, Osmanthus fragrans
Osmanthus fragrans (Thunb.) Lour. (Oleaceae), a valuable fragrant plant, is found to be functionally androdioecious (the presence of males and hermaphrodites in a population) (Hao et al., 2011). As one of the top 10 traditional flowers in China, O. fragrans has been cultivated for about 2500 yr and more than 120 cultivars have been identified. These cultivars are categorized into four groups (Asiaticus, Albus, Luteus, and Aurantiacus) based on morphological and phenological traits (e.g., flower color, peduncle, and flowering period), and there are male and hermaphroditic cultivars in each group (Xiang and Liu, 2008). Moreover, it is thought that the cultivar groups Asiaticus and Albus are less differentiated from wild O. fragrans than the other two groups, based on morphological features and research data (Xiang and Liu, 2008).
Several dominant molecular markers have been used for cultivar identification and classification of O. fragrans (Xiang and Liu, 2008; Yuan et al., 2011). However, codominant microsatellite markers, which have become preferred markers as they are polymorphic, highly abundant, analytically simple, and transferable, have not been reported in O. fragrans cultivars. In this study, microsatellite markers were developed to analyze the genetic diversity of androdioecious O. fragrans cultivars, which will provide new molecular tools for cultivar classification, identification, and genetic improvement.
METHODS AND RESULTS
Through careful field investigation, O. fragrans cultivars and their genders were identified during the 2009 to 2011 flowering seasons. A total of 62 typical O. fragrans cultivars (nine hermaphrodites and 53 males) and six closely related species that were used as outgroup taxa were collected (Appendix 1). Genomic DNA was extracted from young and fully expanded leaves of study materials using the cetyltrimethylammonium bromide (CTAB) method (Doyle and Doyle, 1987). Microsatellite sequences of O. fragrans were downloaded from the National Center for Biotechnology Information (NCBI, http://www.ncbi.nlm.nih.gov/nuccore), then analyzed with the Simple Sequence Repeat Identification Tool (SSRIT; http://www.gramene.org/gramene/searches/ssrtool) to identify simple sequence repeat (SSR) loci with a minimum length of 10 bp for all repeats. Twenty-nine SSR sequences were selected, and flanking primer sets were designed using the software Primer 5.0 (Clarke and Gorley, 2001). Primers had an optimum length of 22 nucleotides (18 bp minimum, 27 bp maximum) and CG contents ranged from 20% to 80%. The designed primers were synthesized at Generay Biotech Co. Ltd. (Shanghai, China), and 15 microsatellite markers were selected based on amplification and reproducibility in all accessions (Table 1). The final 12.5-μL reaction volume for PCR contained 6.25 μL 2× Taq Master mix (100 U/mL Taq polymerase, 400 μM dNTPs, and 4 mM MgCl2 [Generay Biotech Co. Ltd.]), 0.3 μM of each forward and reverse primer (Generay Biotech Co. Ltd.), and 20 ng of DNA template. Amplification was performed with a 5-min initial denaturation at 94°C, followed by 35 cycles of 94°C for 30 s, annealing at 46–52°C for 30 s, and an extension at 72°C for 30 s. A final extension was performed at 72°C for 8 min. A pBR322 DNA-MspI digest marker (Tiangen, Beijing, China) yielding 26 fragments from nine to 622 bp was used as the molecular size standard. PCR products were separated on 8% denaturing polyacrylamide gels and stained with a silver-staining method.
Table 1.
Characteristics of 15 Osmanthus fragrans microsatellite loci.
| Locus | Ta (°C) | Primer sequences (5′–3′) | Repeat motif | Size range (bp) | GenBank accession no. |
| OFP001 | 50 | F: GGAAGCACCACCATAAGC | (TG)13(AG)9 | 106–148 | GU980659 |
| R: AGCAACAGTACCCAGGAG | |||||
| OFP002 | 46 | F: TTGCATCTTCATTTTACA | (CT)17 | 110–190 | GU980660 |
| R: ATGGAAGATAATGAACAA | |||||
| OFP003 | 50 | F: AGTCAGGGGTATTCCAGG | (CA)11(TA)8 | 123–155 | GU980661 |
| R: AAGCCCAAAGTATGTTCC | |||||
| OFP004 | 50 | F: CTGCCTCTTCTTCTGCTC | (CT)20 | 94–138 | GU980662 |
| R: CACGAACTATCACAATATGTG | |||||
| OFP005 | 50 | F: AACATGATATTCTTGGAG | (AC)23 | 176–194 | GU980663 |
| R: GTTTTGCCTTAGGGTTAG | |||||
| OFP006 | 52 | F: CCAAAGCCATCACATACC | (AG)8(AG)18 | 125–170 | GU980664 |
| R: CAAGGAGACCTACCCACT | |||||
| OFP008 | 50 | F: GAGACAGGCATAAACTCT | (CT)17(GT)10GCGT(GC)6 | 76–160 | GU980666 |
| R: TAGCACTCAATCACTTCG | |||||
| OFP016 | 46 | F: TATTCACCAGCAGAGGAG | (GC)6(AC)10(AT)5 | 170–188 | GU980674 |
| R: AGTTGCTTGTAGAAATGG | |||||
| OFP019 | 52 | F: TCAGTGAATGCCTGTGCT | (AG)20 | 93–117 | GU980677 |
| R: ACCCTTTCTTCTGTGCTT | |||||
| OFP020 | 48 | F: TTGTTTTCTCCCTCTTCC | (TC)11 | 111–123 | GU980678 |
| R: TTCGGTTGTAATGGTTGT | |||||
| OFP022* | 46 | F: CCTTTCTTTCCCTTCTGT | (CCT)5TCTC(CTT)16 | 136–170 | GU980680 |
| R: GAGCCATCGTTGTACTTG | |||||
| OFP023 | 50 | F: TTGGTGGTGCTGGGAAGA | (TC)11 | 82–106 | GU980681 |
| R: GTGCCAAACTACCTAACCA | |||||
| OFP024 | 50 | F: CGCACAGAACAGCTCATA | (AC)8 | 180–216 | GU980682 |
| R: GGAGAATAATTTGGTGGC | |||||
| OFP028 | 50 | F: TAGCTTATGCATTGAGTG | (AC)13 | 181–227 | GU980686 |
| R: AAAACCACAGGTAGATGA | |||||
| OFP029* | 50 | F: CGTCCCTGTTTATGTTGT | (AG)14 | 200–242 | GU980687 |
| R: AGGTTAGTGATGCTGCTA |
Note: Ta = annealing temperature.
Loci OPF022 and OPF029 showed evidence of null alleles.
The presence of null alleles was tested with the program MICRO-CHECKER 2.2.3 (van Oosterhout et al., 2004), which suggested loci OPF022 and OPF029 showed evidence of null alleles. These loci were excluded from subsequent data analyses. POPGENE version 1.32 (Yeh et al., 1999) was used to calculate the number of alleles per locus, observed heterozygosity, and expected heterozygosity. The number of alleles ranged from two to six, with a mean of 3.7 per locus. The observed and expected heterozygosities ranged from 0.1000 to 0.9091 and from 0.1287 to 0.9167, respectively (Table 2).
Table 2.
Results of screening of 13 microsatellite loci in Osmanthus fragrans cultivars.
| Asiaticus group (N = 13) | Albus group (N = 20) | Luteus group (N = 15) | Aurantiacus group (N = 14) | ||||||
| Locus | A | Ho | He | Ho | He | Ho | He | Ho | He |
| OFP001 | 5 | 0.8571 | 0.7582 | 0.7117 | 0.8125 | 0.3571 | 0.6640 | 0.9091 | 0.7316 |
| OFP002 | 6 | 0.4545 | 0.3680 | 0.4444 | 0.6016 | 0.5000 | 0.6429 | 0.6429 | 0.5582 |
| OFP003 | 6 | 0.3333 | 0.3007 | 0.2632 | 0.2447 | 0.5333 | 0.4667 | 0.3571 | 0.4735 |
| OFP004 | 4 | 0.8133 | 0.7645 | 0.7895 | 0.6586 | 0.7857 | 0.6058 | 0.8000 | 0.6737 |
| OFP005 | 2 | 0.2500 | 0.2333 | 0.2222 | 0.2032 | 0.1333 | 0.1287 | 0.3571 | 0.3042 |
| OFP006 | 3 | 0.1429 | 0.3626 | 0.1508 | 0.1538 | 0.2308 | 0.2185 | 0.2500 | 0.2417 |
| OFP008 | 5 | 0.8182 | 0.5411 | 0.5882 | 0.4902 | 0.6000 | 0.5421 | 0.3000 | 0.7053 |
| OFP016 | 2 | 0.4000 | 0.3556 | 0.2105 | 0.1935 | 0.1538 | 0.1477 | 0.1667 | 0.1594 |
| OFP019 | 3 | 0.5833 | 0.5627 | 0.2778 | 0.4841 | 0.5333 | 0.4805 | 0.3571 | 0.3201 |
| OFP020 | 2 | 0.1250 | 0.1854 | 0.1333 | 0.1287 | 0.1000 | 0.2684 | 0.2857 | 0.2637 |
| OFP023 | 4 | 0.3333 | 0.5217 | 0.6316 | 0.5249 | 0.2667 | 0.2391 | 0.3571 | 0.3254 |
| OFP024 | 3 | 0.5642 | 0.8333 | 0.5917 | 0.6316 | 0.4000 | 0.3425 | 0.6154 | 0.4800 |
| OFP028 | 3 | 0.5368 | 0.7273 | 0.6306 | 0.4595 | 0.6667 | 0.4891 | 0.5181 | 0.9167 |
Note: A = number of alleles; He = expected heterozygosity; Ho = observed heterozygosity; N = number of cultivars.
The genetic structure of study materials was inferred using the program STRUCTURE 2.3.1 (Pritchard et al., 2000), with a burn-in length of 30,000 followed by 500,000 cycles, and each run was iterated five times. The number of subgroups (K) was determined to be K = 4 using Evanno’s method (Evanno et al., 2005). The results of the structure analyses are presented in Fig. 1. The outgroup (represented in Fig. 1 in yellow) was genetically distinct from all O. fragrans cultivar groups. Cultivar groups Luteus and Aurantiacus were somewhat genetically differentiated (mainly represented in Fig. 1 in red and blue, respectively), but gene exchange was evident among many cultivars (indicated in Fig. 1 by the presence of the same color in different groups) and was extensive for Asiaticus and Albus cultivars. The results indicate that Asiaticus and Albus were genetically mixed and incompletely differentiated. Thus, the cultivar groups Asiaticus and Albus possibly have diverged more recently, as they were less genetically differentiated, while the cultivar groups Luteus and Aurantiacus, which displayed greater genetic differentiation, might have diverged earlier. In sum, the molecular results provide some support for the morphological classification of O. fragrans cultivars groups Luteus and Aurantiacus (Xiang and Liu, 2008).
Fig. 1.
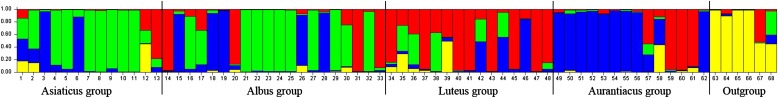
Estimated genetic structure of study materials for K = 4 obtained with the STRUCTURE program.
CONCLUSIONS
The 13 microsatellite markers developed for O. fragrans are highly polymorphic and informative. These loci will be useful for genetic study of androdioecious O. fragrans cultivars and for cultivar classification, particularly for cultivar groups Luteus and Aurantiacus. They also hold potential for further genetic study of O. fragrans cultivars.
Appendix 1.
List of Osmanthus fragrans cultivars and outgroup species analyzed in this study.a
| Code | Cultivar | Accession no. | Gender | Collection site | Geographical coordinates |
| Asiaticus group | |||||
| 1 | ‘Danzhuang’ | JH004 | Hermaphrodite | Jinhua, Zhejiang | 29°07′N, 119°39′E |
| 2 | ‘Yuegui’ | XN002 | Hermaphrodite | Xianning, Hubei | 29°50′N, 114°20′E |
| 3 | ‘Daye Sijigui’ | LY006 | Male | Liyang, Jiangsu | 31°26′N, 119°29′E |
| 4 | ‘Tianxiang Taige’ | JH001 | Male | Jinhua, Zhejiang | 29°07′N, 119°39′E |
| 5 | ‘Xiaoye Fodingzhu’ | CD002 | Male | Chengdu, Sichuan | 30°40′N, 104°01′E |
| 6 | ‘Chenghuang Sijigui’ | CD004 | Male | Chengdu, Sichuan | 30°40′N, 104°01′E |
| 7 | ‘Rixianggui’ | CD001 | Male | Chengdu, Sichuan | 30°40′N, 104°01′E |
| 8 | ‘Juye Sijigui’ | CQ001 | Male | Chongqing | 29°35′N, 106°28′E |
| 9 | ‘Daye Fodingzhu’ | CD003 | Male | Chengdu, Sichuan | 30°40′N, 104°01′E |
| 10 | ‘Yuntian Caigui’ | CQ002 | Male | Chongqing | 29°35′N, 106°28′E |
| 11 | ‘Pixian Caigui’ | CQ003 | Male | Chongqing | 29°35′N, 106°28′E |
| 12 | ‘Sijigui’ | NJ001 | Male | Nanjing, Jiangsu | 32°00′N, 118°48′E |
| 13 | ‘Tiannv Sanhua’ | JH002 | Male | Jinhua, Zhejiang | 29°07′N, 119°39′E |
| Albus group | |||||
| 14 | ‘Changgengbai’ | XN026 | Male | Xianning, Hubei | 29°50′N, 114°20′E |
| 15 | ‘Baijie’ | CZ021 | Male | Xianning, Hubei | 29°50′N, 114°20′E |
| 16 | ‘Kuoye Zaoyingui’ | NJ006 | Male | Nanjing, Jiangsu | 32°00′N, 118°48′E |
| 17 | ‘Yinsu’ | XN005 | Male | Xianning, Hubei | 29°50′N, 114°20′E |
| 18 | ‘Boye Yingui’ | XN034 | Male | Xianning, Hubei | 29°50′N, 114°20′E |
| 19 | ‘Chiye Yingui’ | XN008 | Male | Xianning, Hubei | 29°50′N, 114°20′E |
| 20 | ‘Juban’ | XN037 | Male | Xianning, Hubei | 29°50′N, 114°20′E |
| 21 | ‘Zie’ | XN018 | Male | Xianning, Hubei | 29°50′N, 114°20′E |
| 22 | ‘Kuoye Ziyingui’ | WH003 | Hermaphrodite | Wuhan, Hubei | 30°37′N, 114°08′E |
| 23 | ‘Yulinglong’ | JH006 | Male | Jinhua, Zhejiang | 29°07′N, 119°39′E |
| 24 | ‘Chuiban’ | CZ022 | Male | Jinhua, Zhejiang | 29°07′N, 119°39′E |
| 25 | ‘Zaoyingui’ | NJ004 | Male | Nanjing, Jiangsu | 32°00′N, 118°48′E |
| 26 | ‘Jiulonggui’ | CD005 | Male | Chengdu, Sichuan | 30°40′N, 104°01′E |
| 27 | ‘Wanyingui’ | CZ019 | Male | Changzhou, Jiangsu | 31°46′N, 119°56′E |
| 28 | ‘Qiuyun’ | CZ025 | Male | Xianning, Hubei | 29°50′N, 114°20′E |
| 29 | ‘Jiangnan Liren’ | XN030 | Male | Xianning, Hubei | 29°50′N, 114°20′E |
| 30 | ‘Yinxing’ | XN035 | Male | Xianning, Hubei | 29°50′N, 114°20′E |
| 31 | ‘Ziyingui’ | NJ007 | Hermaphrodite | Nanjing, Jiangsu | 32°00′N, 118°48′E |
| 32 | ‘Liuyegui’ | XN005 | Male | Xianning, Hubei | 29°50′N, 114°20′E |
| 33 | ‘Changye Bizhu’ | CZ009 | Hermaphrodite | Jinhua, Zhejiang | 29°07′N, 119°39′E |
| Luteus group | |||||
| 34 | ‘Zijingui’ | NJ007 | Hermaphrodite | Chengdu, Sichuan | 30°40′N, 104°01′E |
| 35 | ‘Susheng Jingui’ | CD008 | Male | Chengdu, Sichuan | 30°40′N, 104°01′E |
| 36 | ‘Changbing Jingui’ | CD008 | Male | Chengdu, Sichuan | 30°40′N, 104°01′E |
| 37 | ‘Wandianjin’ | JH008 | Male | Jinhua, Zhejiang | 29°07′N, 119°39′E |
| 38 | ‘Chuizhihuang’ | JH009 | Male | Jinhua, Zhejiang | 29°07′N, 119°39′E |
| 39 | ‘Congzhongxiao’ | JH011 | Male | Jinhua, Zhejiang | 29°07′N, 119°39′E |
| 40 | ‘Xiaoye Zijingui’ | JH012 | Hermaphrodite | Jinhua, Zhejiang | 29°07′N, 119°39′E |
| 41 | ‘Lihuang’ | JH010 | Male | Jinhua, Zhejiang | 29°07′N, 119°39′E |
| 42 | ‘Yuanban Jingui’ | CZ013 | Male | Changzhou, Jiangsu | 31°46′N, 119°56′E |
| 43 | ‘Zuiyun’ | XNQS005 | Hermaphrodite | Xianning, Hubei | 29°50′N, 114°20′E |
| 44 | ‘Xiaojinling’ | JH013 | Male | Jinhua, Zhejiang | 29°07′N, 119°39′E |
| 45 | ‘Boye Jingui’ | NJ009 | Male | Nanjing, Jiangsu | 32°00′N, 118°48′E |
| 46 | ‘Jinqiugui’ | NJ008 | Male | Nanjing, Jiangsu | 32°00′N, 118°48′E |
| 47 | ‘Qiugui’ | XN016 | Male | Xianning, Hubei | 29°50′N, 114°20′E |
| 48 | ‘Huangchuan Jingui’ | XNXY007 | Hermaphrodite | Xianning, Hubei | 29°50′N, 114°20′E |
| Aurantiacus group | |||||
| 49 | ‘Zuijihong’ | CZ032 | Male | Changzhou, Jiangsu | 31°46′N, 119°56′E |
| 50 | ‘Chiye Dangui’ | CZ007 | Male | Changzhou, Jiangsu | 31°46′N, 119°56′E |
| 51 | ‘Suzhou Qiancheng’ | CZ009 | Male | Changzhou, Jiangsu | 31°46′N, 119°56′E |
| 52 | ‘Pingmaihong’ | CZ010 | Male | Changzhou, Jiangsu | 31°46′N, 119°56′E |
| 53 | ‘Yingye Dangui’ | CZ014 | Male | Changzhou, Jiangsu | 31°46′N, 119°56′E |
| 54 | ‘Xionghuanggui’ | CD009 | Male | Chengdu, Sichuan | 30°40′N, 104°01′E |
| 55 | ‘Zhusha Dangui’ | CZ017 | Male | Changzhou, Jiangsu | 31°46′N, 119°56′E |
| 56 | ‘Pucheng Dangui’ | PC001 | Male | Pucheng, Fujian | 27°55′N, 118°32′E |
| 57 | ‘Moye Dangui’ | XNQS003 | Male | Xianning, Hubei | 29°50′N, 114°20′E |
| 58 | ‘Boye Dangui’ | CQ004 | Male | Chongqing | 29°35′N, 106°28′E |
| 59 | ‘Dahua Dangui’ | CZ039 | Male | Changzhou, Jiangsu | 31°46′N, 119°56′E |
| 60 | ‘Hangzhou Dangui’ | JH014 | Male | Jinhua, Zhejiang | 29°07′N, 119°39′E |
| 61 | ‘Xiaoye Dangui’ | CZ018 | Male | Changzhou, Jiangsu | 31°46′N, 119°56′E |
| 62 | ‘Zhuangyuanhong’ | CZ003 | Male | Changzhou, Jiangsu | 31°46′N, 119°56′E |
| Outgroup | |||||
| 63 | O. cooperi | NJ010 | Nanjing, Jiangsu | 32°00′N, 118°48′E | |
| 64 | O. heterophyllus ‘Goshiki’ | NJ011 | Nanjing, Jiangsu | 32°00′N, 118°48′E | |
| 65 | O. heterophyllus | NJ012 | Nanjing, Jiangsu | 32°00′N, 118°48′E | |
| 66 | O. fordii | JH016 | Jinhua, Zhejiang | 29°07′N, 119°39′E | |
| 67 | O. serrulatus | JH015 | Jinhua, Zhejiang | 29°07′N, 119°39′E | |
| 68 | O. armatus | JH017 | Jinhua, Zhejiang | 29°07′N, 119°39′E |
All the cultivars, their genders, and the outgroup species were identified with the help of Prof. Xiang Qi Bai, the international cultivar registration authority for Osmanthus. Voucher specimens of all the cultivars and species with their accession numbers were deposited in the herbarium of Nanjing Forestry University (NF).
LITERATURE CITED
- Clarke K. R., Gorley R. N. 2001. PRIMER v5: User manual/tutorial. PRIMER-E Ltd. , Plymouth, United Kingdom [Google Scholar]
- Doyle J. J., Doyle J. L. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin 19: 11–15 [Google Scholar]
- Evanno G., Regnaut S., Goudet J. 2005. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Molecular Ecology 14: 2611–2620 [DOI] [PubMed] [Google Scholar]
- Hao R. M., Zhao H. B., Wang J. H., Zhou L. H. 2011. Observation and study on breeding system of wild Osmanthus fragrans. Journal of Plant Resources and Environment 20: 17–24 (in Chinese with English abstract) [Google Scholar]
- Pritchard J. K., Stephens M., Donnelly P. 2000. Inference of population structure using multilocus genotype data. Genetics 155: 945–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Oosterhout C., Hutchinson W. F., Wills D. P. M., Shipley P. 2004. MICRO-CHECKER: A software for identifying and correcting genotyping errors in microsatellite data. Molecular Ecology Notes 4: 535–538 [Google Scholar]
- Xiang Q. B., Liu Y. L. 2008. An illustrated monograph of the sweet Osmanthus cultivars in China. Zhejiang Science and Technology Publishing House, Hangzhou, China [Google Scholar]
- Yeh F. C., Yang R. C., Boyle T. 1999. POPGENE: Microsoft Windows–based freeware for population genetic analysis. Release 1.31. University of Alberta, Edmonton, Canada [Google Scholar]
- Yuan W. J., Han Y. J., Dong M. F., Shang F. D. 2011. Assessment of genetic diversity and relationships among Osmanthus fragrans cultivars using AFLP markers. Electronic Journal of Biotechnology 14(1). Website http://dx.doi.org/10.2225/vol14-issue1-fulltext-9 [accessed 17 April 2013].


