Abstract
SAMHD1 is a GTP-activated nonspecific dNTP triphosphohydrolase that depletes dNTP pools in resting CD4+ T cells and macrophages and effectively restricts infection by HIV-1. We have designed a nonsubstrate dUTP analogue with a methylene bridge connecting the α phosphate and 5′ carbon that potently inhibits SAMHD1. Although pppCH2dU shows apparent competitive inhibition, it acts by a surprising allosteric mechanism that destabilizes active enzyme tetramer.
SAMHD1 is a homotetrameric human enzyme with nonspecific dNTP triphosphohydrolase activity that serves to deplete the dNTP substrates of reverse transcriptase in myeloid lineage target cells of HIV-1 such as resting T cells, macrophages, and dendritic cells, resulting in viral restriction.1,2 Paradoxically, SAMHD1-related suppression of HIV infection in dendritic cells may prevent their activation of CD4+ T cells through the IFN1-toll receptor pathway, thereby preventing the stimulation of a strong adaptive immune response by the host.3,4 Thus, inhibitors of SAMHD1 could serve as useful tool compounds to explore host adaptive immune responses in these immune cells.5
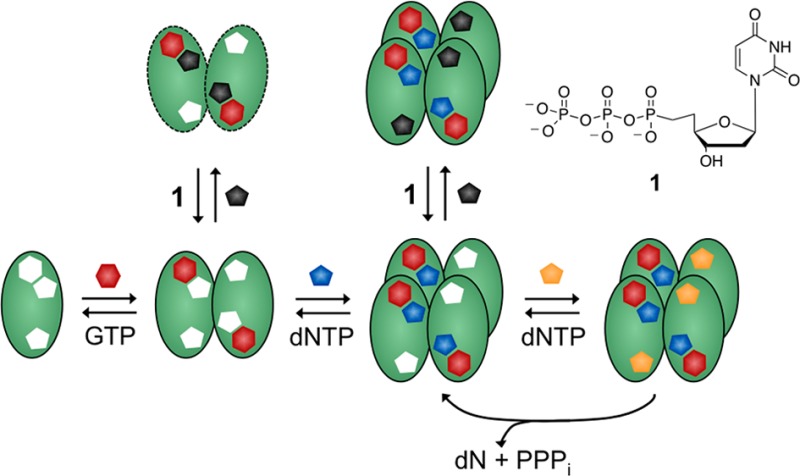
SAMHD1 shows a complex activation mechanism. The enzyme displays ordered essential activation by binding of GTP to a guanine nucleotide-specific activator site (A1) on each monomer which induces dimerization,6 followed by binding of any dNTP to a second nonspecific activator site on each subunit (A2) as well as the substrate sites.7 Thus, the active tetramer contains four occupied pairs of activator sites (A1A2)4 and four catalytic sites.7,8 This mechanism of activation, which involves coupling of nucleotide binding energy to drive formation of the active tetramer, presents multiple sites for the rational design of small molecule modulators of the enzyme activity, including subunit interfaces. In this report we describe the synthesis and characterization of a dUTP-derived inhibitor of SAMHD1 that acts by the surprising mechanism of preventing tetramer association.
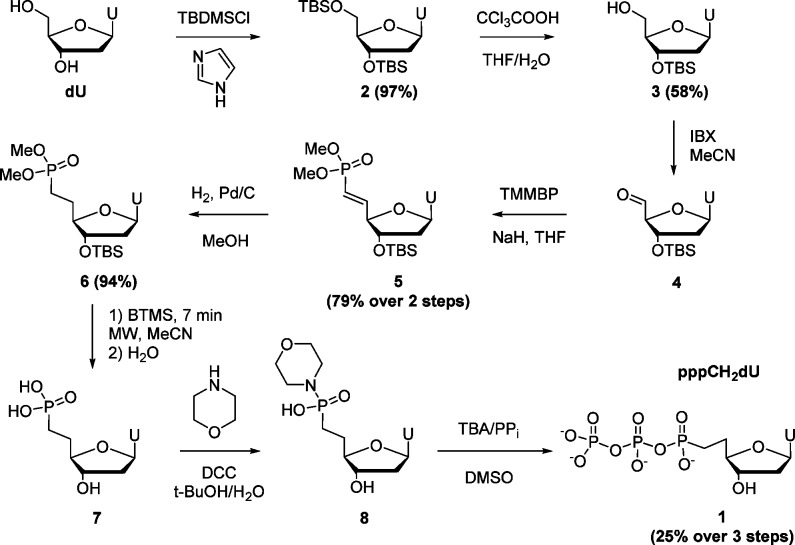
As a mechanistic prelude to our studies, we first ascertained whether SAMHD1 catalyzes attack of water at the α phosphate or the 5′ carbon of the nucleotide sugar. Reactions of the self-activating substrate dGTP were performed in 16O- and 18O-substituted water, and the masses of the triphosphate and nucleoside products were determined using mass spectrometry (Figure S1). Exclusive incorporation of 18O into the triphosphate product confirms that nucleophilic attack occurs at the α phosphate and that the leaving group is the sugar 5′ alcohol. It follows that if the 5′ oxygen is replaced by a nonhydrolyzable moiety9,10 (such as a methylene group), enzymatic cleavage of dNTP would be prevented. We therefore synthesized a 5′ methylene dUTP (pppCH 2dU, 1) by the route outlined in Scheme 1.
Scheme 1. Synthesis of pppCH2dU (1).
Briefly, intermediate 3 was prepared by protection of the 3′- and 5′-hydroxyl groups of deoxyuridine (dU) with tert-butyldimethylsilyl chloride (TBDMSCl)11,12 followed by selective removal of the 5′-silyl group from the product 2 with trichloroacetic acid.12,13 Adapting the procedure of Schinazi,12 alcohol 3 was then oxidized with 2-iodoxybenzoic acid, and the resulting aldehyde 4 was reacted with the sodium salt of tetramethyl methylenebis(phosphonate) (TMMBP) carbanion, leading to formation of the alkene 5, predominantly as the trans-isomer. Compound 5 was hydrogenated in the presence of Pd/C, affording the 3′-TBS protected dimethyl ester of pCH2dU, 6.
The methyl- and t-butyldimethylsilyl-protecting groups were removed by bromotrimethylsilane (BTMS),14,15 making possible facile hydrolysis of the resulting trimethylsilyl ester14,15 of 6 to pCH2dU, 7. It was previously reported16 that conventional BTMS deprotection of the diethyl ester of 7 resulted in significant anomerization of the nucleotide (β:α anomer ratio 2:3). In contrast, we found that rapid microwave-assisted BTMS silyldealkylation17 (MeCN, 60 °C, 7 min, MW), followed by hydrolysis with H2O provided phosphonic acid 7 with minimal anomerization (β:α anomer ratio 8:1) (Figure S14; the ratios are based on values obtained after conversion to 1). Although it was also possible to prepare 7 free of α-anomer by HPLC purification, the 7 as obtained was more conveniently first converted to the corresponding dUTP analogue 1 (via coupling of morpholidate 8 with tributylammonium pyrophosphate (TBA/PPi) in DMSO),18,19 followed by isolation of the pure β-anomer 1 by dual HPLC purification.17 The structures of 1 and 5–7 were confirmed by 1H, 31P, and 13C NMR and by HRMS.
We evaluated whether 1 was a substrate of SAMHD1 by incubating 1 mM 1 with SAMHD1 in the presence and absence of GTP activator (Figure S2). Compound 1 was found to be completely unreactive even after 24 h incubation with SAMHD1 regardless of whether GTP activator was present. For comparison, we also evaluated the reactivity of dGTPαS (Figure S2), which is known to occupy all activator and substrate sites of SAMHD1 based on structural studies.8 Unlike 1, dGTPαS (racemic) was slowly hydrolyzed both in the absence and presence of GTP activator (t1/2 ∼ 24 h), indicating that this analogue is not only a slow substrate but also an activator for its own hydrolysis. Compound 1 is the first completely nonreactive substrate analogue of SAMHD1. The inhibition mechanism of 1 was investigated using dUTP as the substrate (Figure 1a). The entire data set was globally fitted to a competitive inhibition model to obtain the inhibition constant Ki = 80 ± 6 μM. Thus, 1 appeared to follow simple competitive inhibition with respect to dUTP with a Ki value that was a surprising 20-fold lower than the Km for the substrate dUTP.6
Figure 1.
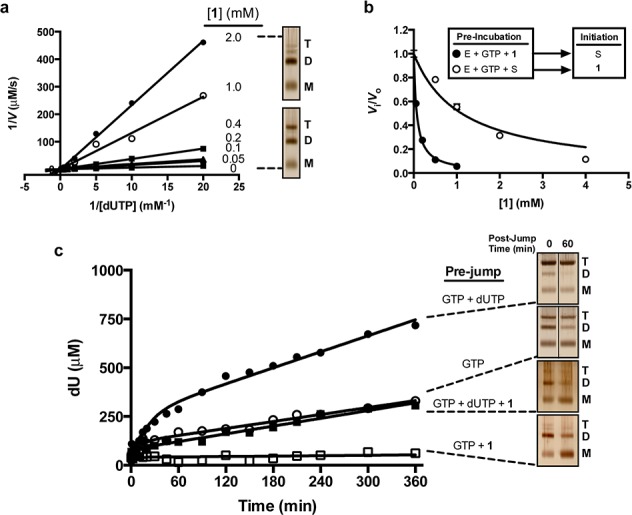
Inhibition by 1. (a) Double-reciprocal plot of dUTP hydrolysis in the presence of various fixed concentrations of 1 as indicated. The reactions contain a saturating concentration of GTP activator (5 mM). The entire data were fitted globally to a competitive inhibition model to obtain the inhibition constant (Ki = 80 ± 6 μM). The two right panels show silver stained polyacrylamide gel images of reactions (with and without 2 mM 1) that were quenched with the cross-linking agent glutaradehyde. The monomer (M), dimer (D), and tetramer (T) cross-linked forms of SAMHD1 are indicated. (b) Order of addition experiments in which 1 is added before or after substrate (see text). (c) DJK and corresponding cross-linking (DJXL) experiment to evaluate the effects of 1 on the activity and oligomeric state of SAMHD1 (see text). The various pre-jump conditions are listed above the dotted lines and the post-jump reactions contained 1 mM dUTP in all cases. The post-jump delay times at which 50 mM glutaraldehyde was added are indicated above the gel images and the monomer (M), dimer (D), and tetramer (T) cross-linked forms are indicated.
Since the Ki of 1 was much less than the Km for dUTP and dUTP acts as a coactivator by binding to the A2 sites,6 we tested whether 1 might target the A2 site but fail to activate. (The possibility that 1 binds antagonistically with GTP to the A1 site is excluded because this site is highly specific for guanine nucleotides and this type of inhibition would not appear as competitive.)6 Our approach took advantage of the previous finding that the A1 and A2 sites become inaccessible to free ligands after formation of the tetramer, but that the catalytic site remained accessible.6 Thus, we reasoned that order-of-addition experiments where 1 was added before or after dUTP and GTP might differentiate between inhibition caused by binding of 1 to the A2 or catalytic site or both (Figure 1b). To test for binding of 1 to the catalytic site, a reaction of SAMHD1 with GTP and dUTP was initiated and allowed to proceed for 2.5 min to ensure occupation of the A1 and A2 sites with GTP and dUTP and the sequestration of these sites from free ligand. Then the reactions were spiked with various concentrations of 1 and the subsequent rate of dUTP consumption was followed over time (Figures 1b and S3a). The Ki = 670 ± 110 μM that was obtained under these conditions was similar to the 1.5 mM Km for dUTP, suggesting that when the A2 site was sequestered, 1 targeted the catalytic site.6 In contrast, when GTP and 1 were preincubated with SAMHD1 before addition of dUTP, a Ki = 52 ± 5 μM was obtained, which is 30-fold lower than the Km for dUTP6 (Figures 1b and S3b). We conclude that 1 targets both the A2 and catalytic sites, but its potency largely arises from tight binding to the A2 site.
Since 1 seemed to behave as a dUTP analogue, we wondered why low concentrations of 1 that only populated the A2 site gave rise to inhibition rather than activation. Our initial hypothesis was that 1 might be antagonistic to formation of active tetramer. This was confirmed by glutaraldehyde cross-linking in the presence and absence of 1 under the conditions of the inhibition experiment in Figure 1a.6 Separation of the cross-linked forms by denaturing gel electrophoresis with visualization by silver staining showed that 2 mM 1 completely prevented tetramer formation in the presence of 1 mM dUTP and 5 mM GTP (Figure 1a).
To further explore this surprising mechanism, we employed our recently developed dilution-jump method that allows investigation of the activity and stability of various oligomeric forms of SAMHD1.6 This approach involved the formation of complexes of SAMHD1 with activators and/or dNTPs at high concentration (pre-jump condition) followed by rapid 100-fold dilution into a solution with low activator and/or substrate dNTP composition (post-jump condition). The post-jump assay took two general forms: kinetic and structural. In the structural mode the oligomeric state(s) that were present at the time of dilution as well as their rate of decay in a given post-jump condition, were probed using glutaraldehyde cross-linking (DJXL). The dilution-jump kinetic (DJK) mode queried the time-dependent changes in catalytic activity after dilution into a post-jump solution containing a radiolabeled substrate ([5-3H] dUTP).
The DJXL experiment assessed the effect of 1 on the oligomeric states of SAMHD1. Without 1 in the pre-jump solution, the presence of [dUTP + GTP] resulted in the efficient formation of tetramer (Figure 1c), while in the absence of these nucleotides only the monomer and dimer forms of SAMHD1 were present.6 The tetramer generated in the pre-jump persisted for hours even in the absence of GTP activator in the post-jump, confirming our previous findings that the tetramer is long-lived.6 When 1 was included in the pre-jump solution, tetramer formation was completely abrogated as judged by glutaraldehyde cross-linking, and the resulting monomer and dimer forms persisted in the post-jump reaction for at least 1 h (Figure 1c).
The DJK method probed the effect of 1 on the post-jump enzymatic activity. The pre-jump solution consisted of SAMHD1, GTP activator, and dUTP substrate in the absence or presence of 1. The post-jump solution contained [5-3H] dUTP substrate but no GTP activator. In the absence of 1 in the pre-jump, SAMHD1 showed a burst-phase for dUTP hydrolysis in the post-jump that decayed exponentially and eventually yielded a linear steady-state rate that persisted for hours (Figure 1c).6 We have attributed the burst-decay period (t1/2 ∼ 10 min) to the decay of SAMHD1 from a highly active form generated in the pre-jump solution to a less active but long-lived form that persists throughout the steady-state period even in the absence of GTP activator.6 When 1 (5 mM) was added to the pre-jump solution, the post-jump burst-decay amplitude was almost abolished, and the subsequent linear steady-state rate was reduced indefinitely even though the diluted concentration of pppCH2dU in the post-jump was 1/20 that of the dUTP substrate (Figure 1c).
In conclusion, although we initially viewed 1 as an excellent substrate mimic, the above data require that 1 binds to SAMHD1 in a manner that is disruptive to both tetramer formation and catalytic activity (Figure 2). The data suggest an unusual inhibition mechanism that takes advantage of the ordered essential activation mechanism and the ligand specificities of the A1, A2, and catalytic sites.6,7 Antagonistic binding of 1 to the A2 sites provides a reasonable explanation for the unexpected long-term inhibition of tetramer formation caused by 1 in the DJXL experiment (Figure 1c). That is, when 1 binds to the A2 sites in the pre-jump solution, it disrupts the ordered-essential binding of GTP and dUTP, and since GTP is absent in the post-jump, tetramer reassembly is not possible.
Figure 2.
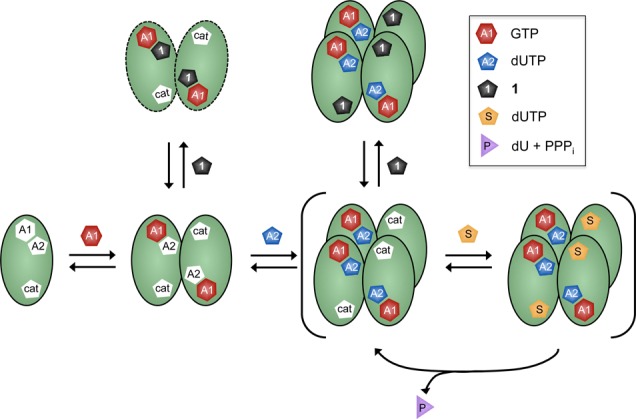
Double hit inhibition mechanism of 1. Compound 1 intercepts the ordered-essential activation mechanism of SAMHD1 by binding to the A2 activator site in a nonproductive manner that cannot lead to tetramer formation (Ki = 50 μM). In addition, 1 binds to the catalytic site and competitively inhibits substrate binding. Binding of the GTP activator to the A1 site is required to assemble dimers and to form the A2 site. The scheme also depicts the previously determined mechanistic feature where SAMHD1 turns over without release of bound activator nucleotides (brackets).
These findings reveal an unexpected diversity of inhibition mechanisms for dNTP analogues of SAMHD1. While the slow substrate and inhibitor dGTPαS occupies all activator and substrate sites of SAMHD1 and induces tetramerization (Figure S4),6,81 interferes with the ordered activation mechanism and prevents tetramerization. The antithetical inhibitory mechanisms of these two analogues and the multiple ligand binding pockets on SAMHD1 indicate that this enzyme is highly amenable to rational design of inhibitors and activators that could modulate its activity in cells. Such ligands could have research or therapeutic utility given the emerging role of SAMHD1 in autoimmune and chronic inflammatory diseases20 and its established role in restricting viral infection of antigen-presenting cells.21
Acknowledgments
This work was supported by US National Institute of Health grants T32GM008763, T32GM080189, GM056834 (J.T.S.), GM103853 (N.N.B.) and U19CA177547 (C.E.McK.). C.E.McK. thanks Dr. Samuel Wilson for suggesting this collaboration, UCR High Resolution Mass Spectrometry Facility and Ron New for performing HRMS.
Supporting Information Available
Mechanistic mass spectometry, synthetic details and spectroscopic data for 1, inertness of 1 in the presence of SAMHD1, and kinetics for inhibition of SAMHD1 by 1. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Goldstone D. C.; Ennis-Adeniran V.; Hedden J. J.; Groom H. C. T.; Rice G. I.; Christodoulou E.; Walker P. A.; Kelly G.; Haire L. F.; Yap M. W.; de Carvalho L. P. S.; Stoye J. P.; Crow Y. J.; Taylor I. A.; Webb M. Nature 2011, 480, 379. [DOI] [PubMed] [Google Scholar]
- Lahouassa H.; Daddacha W.; Hofmann H.; Ayinde D.; Logue E. C.; Dragin L.; Bloch N.; Maudet C.; Bertrand M.; Gramberg T.; Pancino G.; Priet S.; Canard B.; Laguette N.; Benkirane M.; Transy C.; Landau N. R.; Kim B.; Margottin-Goguet F. Nat. Immunol. 2012, 13, 223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manel N.; Littman D. R. Cell 2011, 147, 271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luban J. Cell Host Microbe 2012, 12, 408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manel N.; Hogstad B.; Wang Y.; Levy D. E.; Unutmaz D.; Littman D. R. Nature 2010, 467, 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen E. C.; Seamon K. J.; Cravens S. L.; Stivers J. T. Proc. Natl. Acad. Sci. U.S.A. 2014, 111, E1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C.; Gao W.; Zhao K.; Qin X.; Zhang Y.; Peng X.; Zhang L.; Dong Y.; Zhang W.; Li P.; Wei W.; Gong Y.; Yu X.-F. Nat. Commun. 2013, 42722E1843. [DOI] [PubMed] [Google Scholar]
- Ji X.; Wu Y.; Yan J.; Mehrens J.; Yang H.; Delucia M.; Hao C.; Gronenborn A. M.; Skowronski J.; Ahn J.; Xiong Y. Nat. Struct. Mol. Biol. 2013, 20, 1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upton T. G.; Kashemirov B. A.; McKenna C. E.; Goodman M. F.; Prakash G. K. S.; Kultyshev R.; Batra V. K.; Shock D. D.; Pedersen L. C.; Beard W. A.; Wilson S. H. Org. Lett. 2009, 11, 1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlain B. T.; Batra V. K.; Beard W. A.; Kadina A. P.; Shock D. D.; Kashemirov B. A.; McKenna C. E.; Goodman M. F.; Wilson S. H. ChemBioChem 2012, 13, 528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitzer R.; Kloiber K.; Tollinger M.; Kreutz C. ChemBioChem 2011, 12, 2007. [DOI] [PubMed] [Google Scholar]
- Pradere U.; Amblard F.; Coats S. J.; Schinazi R. F. Org. Lett. 2012, 14, 4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moharram S.; Zhou A.; Wiebe L. I.; Knaus E. E. J. Med. Chem. 2004, 47, 1840. [DOI] [PubMed] [Google Scholar]
- McKenna C. E.; Higa M. T.; Cheung N. H.; McKenna M. C. Tetrahedron Lett. 1977, 155. [Google Scholar]
- McKenna C. E.; Schmidhauser J. J. Chem. Soc., Chem. Commun. 1979, 739. [Google Scholar]
- Van Poecke S.; Sinnaeve D.; Martins J. C.; Balzarini J.; Van Calenbergh S. Nucleosides Nucleotides Nucleic Acids 2012, 31, 256. [DOI] [PubMed] [Google Scholar]
- Mustafa D. A.; Kashemirov B.; McKenna C. E. Tetrahedron Lett. 2011, 52, 2285. [Google Scholar]
- Moffat J. G.; Khorana H. G. J. Am. Chem. Soc. 1961, 83, 649. [Google Scholar]
- McKenna C. E.; Kashemirov B. A.; Upton T. G.; Batra V. K.; Goodman M. F.; Pedersen L. C.; Beard W. A.; Wilson S. H. J. Am. Chem. Soc. 2007, 129, 15412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrendt R.; Schumann T.; Gerbaulet A.; Nguyen L. A.; Schubert N.; Alexopoulou D.; Berka U.; Lienenklaus S.; Peschke K.; Gibbert K.; Wittmann S.; Lindemann D.; Weiss S.; Dahl A.; Naumann R.; Dittmer U.; Kim B.; Mueller W.; Gramberg T.; Roers A. Cell Rep. 2013, 4, 689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puigdomènech I.; Casartelli N.; Porrot F.; Schwartz O. J. Virol. 2013, 87, 2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.