Abstract

Although fucose-α(1-2)-galactose (Fucα(1-2)Gal)-containing glycans have been implicated in cognitive processes such as learning and memory, their precise molecular mechanisms are poorly understood. Here we employed the use of multivalent glycopolymers to enable the first proteome-wide identification of weak affinity, low abundance Fucα(1-2)Gal glycan-binding proteins (GBPs). Biotin-terminated glycopolymers containing photoactivatable cross-linking groups were designed to capture and enrich GBPs from rat brain lysates. Candidate proteins were tested for their ability to bind Fucα(1-2)Gal, and the functional significance of the interaction was investigated for the synaptic vesicle protein SV2a using a knockout mouse system. The results suggest a role for SV2a-Fucα(1-2)Gal interactions in SV2a trafficking and synaptic vesicle recycling. More broadly, our studies outline a general chemical approach for the systems-level discovery of novel GBPs.
Glycan-binding interactions play important roles in many complex physiological processes, including the immune response, tumor metastasis, and viral infection.1 The distinct arrays of glycans presented on cell surfaces are recognized by various glycan-binding proteins (GBPs) or lectins. The ability of GBPs to bind and often cluster glycan-presenting receptors can have important consequences for processes such as cell adhesion, migration, and gene regulation.1,2
Despite their importance, mammalian GBPs that interact with specific glycans of interest have been challenging to identify.3 The weak affinities of many glycan–protein interactions (Kassoc = 103–106 M–1) have complicated efforts to capture and study endogenous GBPs. Most of the well-characterized lectins are derived from plants (e.g. concanavalin A) or were discovered through classical biochemical purification or structural homology to known lectins. Notably, general systems-level approaches for the proteome-wide identification of GBPs have been lacking. The development of such methods is critical for elucidating the structure–function relationships of carbohydrates and understanding the diverse roles of GBPs. Here we address these challenges through the synthesis and characterization of chemical probes for the discovery of novel mammalian GBPs.
We focused on targeting neuronal GBPs that interact with glycans containing the fucose-α(1-2)-galactose (Fucα(1-2)Gal) motif. Fucα(1-2)Gal is found on the nonreducing termini of many glycans and has been implicated in neuronal development, learning, and memory.4 For example, treatment of animals with 2-deoxy-d-galactose (2-dGal), a compound that disrupts the formation of Fucα(1-2)Gal linkages, caused reversible amnesia4a and interfered with the maintenance of long-term potentiation,4b an electrophysiological model of learning and memory. Moreover, previous work from our laboratory has suggested the existence of both Fucα(1-2)Gal GBPs and glycoproteins as well as their involvement in neurite outgrowth and synaptogenesis.4c,4d Understanding the molecular mechanisms underlying the activity of Fucα(1-2)Gal sugars will require identification of the key molecular components involved. However, no Fucα(1-2)Gal GBPs have been identified from the mammalian brain.
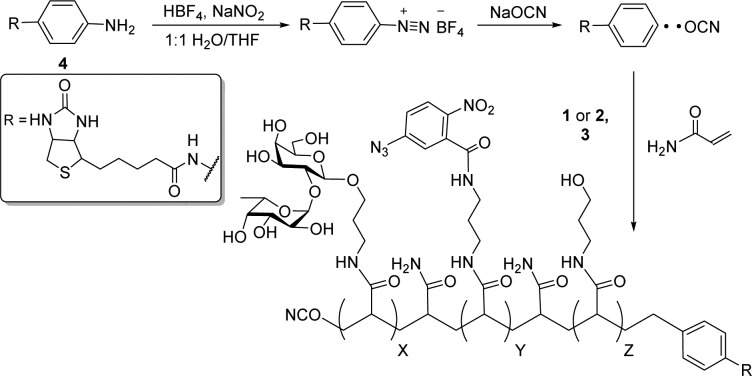
Our early attempts to isolate Fucα(1-2)Gal GBPs employed monovalent ligands conjugated to agarose beads with or without photoactivatable cross-linking groups. Although a few candidate GBPs were successfully detected, this approach failed to capture sufficient quantities of protein for mass spectrometry (MS) analysis. To overcome these challenges, we designed synthetic glycopolymers that contain several key elements (Figure 1). First, we exploited multivalent sugar epitopes to augment weak glycan–GBP interactions.5 Second, we incorporated a photoreactive nitrophenylazide moiety into the glycopolymer to enable covalent cross-linking of the associated proteins. Lastly, we end-functionalized the glycopolymers with a biotin handle to facilitate affinity enrichment and identification of the GBPs.
Figure 1.
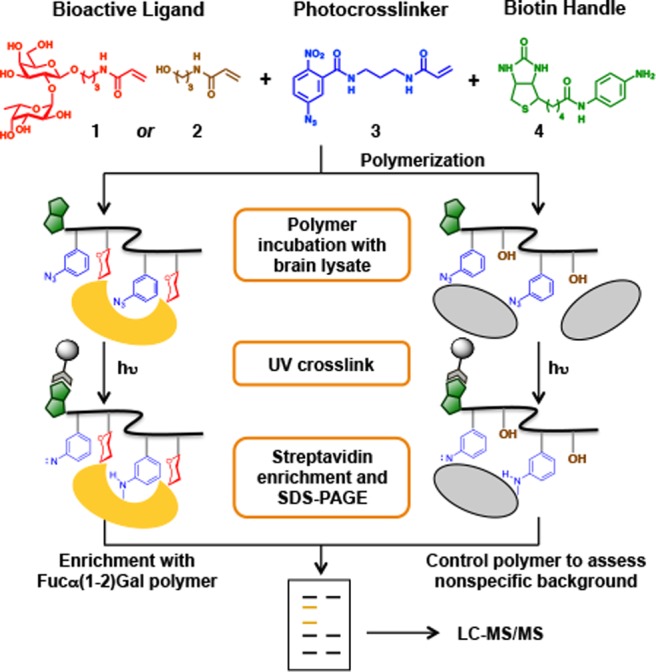
Strategy for the systems-level, proteome-wide identification of GBPs.
To synthesize the glycopolymers, we explored cyanoxyl (OCN)-mediated free radical polymerization chemistry because it affords water-soluble polymers of controlled length and narrow polydispersity.6 The Fucα(1-2)Gal epitope or a control ligand lacking the disaccharide was incorporated into acryloyl-functionalized monomers 1 and 2, respectively (Figure 1). We chose a nitrophenylazide cross-linking agent for monomer 3 because this group has been successfully applied to proteins at membrane interfaces.7 For end-labeling the polymers with a biotin moiety, we employed the arylamine initiator 4 developed by Chaikof et al. (6b)
Fucα(1-2)Gal monomer 1 was prepared using several one-pot, multistep reactions (Scheme S1). Briefly, disaccharide 5(8) was treated with (phenylthio)trimethylsilane in the presence of zinc iodide, deprotected with TBAF, and the resulting C-6 hydroxyl group acetylated to afford thioglycoside 6. Although coupling of 3-azido-1-propanol to this thioglycoside using NIS/AgOTf in CH2Cl2 yielded an anomeric α/β mixture, acetonitrile as the solvent promoted formation of the β-linked glycoside 7 as the major product in 64% yield, presumably due to nitrilium ion coordination in the axial position.9 Catalytic hydrogenation of 7 over Pd/C gave the amine intermediate, which was directly transformed without further purification into the corresponding acrylamide. Finally, facile deacetylation under Zemplén conditions afforded the desired disaccharide 1 in 90% yield. Nitrophenylazide monomer 3 (Scheme S2) and arylamine initiator 4(6b) were readily synthesized using standard procedures.
With the monomers in hand, we generated glycopolymers 9–11 using acrylamide as the comonomer for the polymer backbone (Scheme 1 and Table 1). Treatment of 4 with HBF4 and NaNO2 in degassed 1:1 H2O/THF gave the arenediazonium cation, which upon reaction with NaOCN at 60 °C generated the biotinyl aryl radical in situ to serve as the initiator. Addition of disaccharide monomer 1 and acrylamide (1:4 molar ratio) generated polymer 9 with desirable sugar density and water solubility. Using the same sugar to acrylamide ratio, we also copolymerized monomer 1 with nitrophenylazide monomer 3 (1:3 molar ratio) to give polymer 10. Furthermore, we synthesized the control polymer 11 from 2 and 3, wherein each disaccharide unit was replaced by 2 equiv of 3-hydroxypropyl units (1:6 molar ratio). Characterization of the polymers by size-exclusion chromatography-multiangle light scattering (SEC-MALS) revealed narrow polydispersity index (PDI) values (∼1.2) and number-average molecular weights (Mn) of 23–26 kDa. We also estimated an average polymer composition of up to 30 disaccharides per chain by comparing the integrated signal from the phenyl protons to that of the fucose methyl protons by 1H NMR (see SI for details). Importantly, the relative ratios of ligand (1 or 2) to cross-linking agent (3) could be tuned to reproducibly control the glycan/cross-linker content of the polymer.
Scheme 1. Synthesis of Biotin-Functionalized, Photoactivatable Glycopolymers.
Table 1. Polymers Generated via OCN-Mediated Radical Polymerization.
| pol | monomer | [3]0/[1 or 2]0a | xb | yb | zb | Mnc | PDIc |
|---|---|---|---|---|---|---|---|
| 9 | 1 | – | 19 | – | – | 26.3 | 1.17 |
| 10 | 1 and 3 | 1/3 | 30 | 9 | – | 23.9 | 1.22 |
| 11 | 2 and 3 | 1/6 | – | 7 | 42 | 23.0 | 1.17 |
Initial ratio of cross-linker [3]0 to ligand [1 or 2]0.
Cross-linker and ligand content in the resulting polymer were estimated by 1H NMR (see SI for details).
Mn in kDa and PDI values were determined by SEC-MALS.
We first validated our strategy using the well-established plant lectin Ulex europaeus agglutinin I (UEAI), which binds with weak affinity to the Fucα(1-2)Gal epitope (IC50 = 0.73 mM).10 UEAI conjugated to fluorescein was incubated with polymer 10 or 11 for 3 h at 37 °C and subsequently exposed to 365 nm light for 15 min at 4 °C. The cross-linked proteins were isolated by streptavidin affinity chromatography, resolved by SDS-PAGE, and detected by in-gel fluorescence imaging. We found that polymer 10, but not polymer 11, captured UEAI, indicating selective recognition of the glycan moiety (Figure 2A). Moreover, photo-cross-linking of the polymer led to a 2-fold increase in UEAI capture. These results validate the polymer design and demonstrate that synthetic glycopolymers can be used to capture lectins with weak glycan binding affinities.
Figure 2.
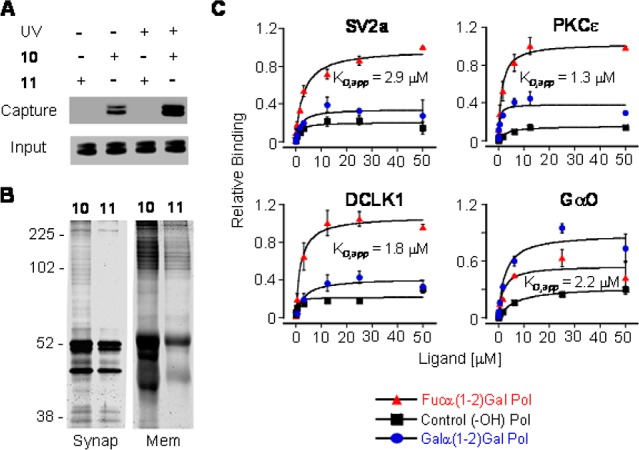
(A) Selective capture of UEAI by 10. (B) Representative silver-stained gel of GBPs isolated from synaptosomal and membrane fractions. (C) Relative binding profiles of candidate GBPs (n = 3).
Next, we investigated whether polymer 10 could be utilized to identify novel, endogenous Fucα(1-2)Gal GBPs from mammalian tissues. Various subcellular fractions from rat brain were incubated with polymer 10 or 11, cross-linked by UV irradiation, and isolated as described above. The captured proteins were resolved by SDS-PAGE, and the entire lanes of proteins captured either by 10 or 11 were cut into 20 similarly sized gel pieces. All gel pieces were individually subjected to in-gel proteolytic digestion and liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis. We observed enhanced, specific protein capture by 10 compared to 11 in both synaptosomal and membrane fractions (Figure 2B). Individual proteins were identified using highly stringent criteria, and after filtering out any proteins present in both sample sets, 44 candidate GBPs were identified from six total experiments. Interestingly, the GBPs represented a broad range of functions, including proteins involved in signal transduction, protein trafficking, protein scaffolding, and metabolism (Table S1).
As Fucα(1-2)Gal carbohydrates have been implicated in synaptic regulation and plasticity, we focused our attention on GBPs associated with those functions. Four proteins were selected for further investigation: double cortin-like kinase 1 (DCLK1), protein kinase C epsilon subunit (PKCε), guanine nucleotide-binding protein G(o) alpha subunit (GαO), and synaptic vesicle glycoprotein 2a (SV2a).11 Although DCLK1, PKCε, and GαO are found in the cytoplasm, they also undergo translocation to the plasma membrane.12 Activity-dependent secretion of these proteins could provide a mechanism for interaction with extracellular Fucα(1-2)Gal sugars. Indeed, secretion of cytosolic galectin 1, a lectin that binds β-galactosides, into the extracellular space has been reported to mediate T cell clearance during tumor progression.13 SV2a could engage Fucα(1-2)Gal sugars in synaptic vesicles or at the extracellular surface after synaptic vesicle exocytosis. We assessed the binding affinities of the candidate GBPs using enzyme-linked lectin assays. Each protein was immobilized on a microtiter plate and incubated with varying concentrations of a biotinylated Fucα(1-2)Gal glycopolymer. Bound polymer was detected using streptavidin conjugated to horseradish peroxidase. For comparison, we also examined the binding of a corresponding biotinylated glycopolymer bearing Galα(1-2)Gal epitopes. All four proteins interacted with the Fucα(1-2)Gal glycopolymer in a dose-dependent manner, albeit with different affinities (Figure 2C). Furthermore, SV2a, PKCε, and DCLK1 showed greater (2.5–2.8-fold) binding to Fucα(1-2)Gal compared to Galα(1-2)Gal glycopolymers, suggesting that the terminal fucose residue was important for recognition. In contrast, GαO exhibited similar binding affinity toward both Fucα(1-2)Gal and Galα(1-2)Gal glycopolymers (KD,app = 2.2 and 2.1 μM, respectively). Together, these results demonstrate that all four candidate GBPs recognize Fucα(1-2)Gal sugars and that our glycopolymer-based approach can successfully identify novel GBPs.
As the mere presence of a glycan-protein interaction is not sufficient to establish biological function, elucidating the physiological roles of GBPs remains a major challenge. To assess whether the interaction of Fucα(1-2)Gal sugars with GBPs might have important functional consequences, we studied SV2a. SV2a is a 12-transmembrane glycoprotein that has no close homology to any known animal lectins. The trafficking of SV2a between secretory vesicles and synapses is critical for activity-dependent neurotransmission.11c,11d To investigate the role of Fucα(1-2)Gal sugars in SV2a trafficking, we utilized genetically altered mice lacking FUT1 and FUT2, the fucosyltransferase genes responsible for Fucα(1-2)Gal biosynthesis.14 We examined whether expression of Fucα(1-2)Gal sugars was regulated by FUT1 or FUT2 in the mouse cortex using our recently reported chemoenzymatic strategy.15 This approach exploits an exogenous bacterial glycosyltransferase BgtA to transfer a non-natural N-azidoacetylgalactosamine (GalNAz) moiety onto the C-3 position of galactose in Fucα(1-2)Gal glycans (Figure S1). Subsequent bioorthogonal reaction with alkyne-functionalized fluorophores or biotin enables rapid, sensitive detection of the labeled Fucα(1-2)Gal glycans. We observed significantly reduced expression of Fucα(1-2)Gal glycans on cortical glycoproteins from FUT2 knockout mice (FUT2–/–) compared to WT littermate controls (Figures 3A and S2). In contrast, FUT1 knockout mice (FUT1–/–) showed a lower reduction in the overall levels of Fucα(1-2)Gal protein fucosylation. These results suggest that FUT2 is primarily responsible for Fucα(1-2)Gal biosynthesis in the mouse cortex.
Figure 3.

(A) Fucα(1-2)Gal expression on glycoproteins in FUT1–/– and FUT2–/– mice. (B) Enrichment of SV2a in the membrane fraction of FUT2–/– mice (n = 3, *P < 0.02). (C) Increased colocalization (yellow) of SV2a and syt1 puncta along neuronal processes in FUT2–/– cortical neurons (n = 70–80, **P < 0.005). (D) Increased cell surface biotinylation of SV2a in FUT2–/– cortical neurons (n = 4, *** P < 0.05). See SI for experimental details.
Trafficking of SV2a to the membrane influences synaptic vesicle exocytosis, also known as neurotransmitter release. Following neurotransmitter release, the endocytic reuptake of SV2a from the membrane into synaptic vesicles is critical for the proper regeneration of synaptic vesicles. To investigate whether Fucα(1-2)Gal glycans influence SV2a trafficking, we examined the distribution of SV2a at the membrane in cultured cortical neurons from WT, FUT1–/–, or FUT2–/– mice. Notably, we found that FUT2–/– cortical neurons exhibited a 2-fold increase in membrane-associated SV2a compared to WT or FUT1–/– cortical neurons, as determined by subcellular fractionation and quantitative Western blot analysis (Figure 3B). In contrast, the levels of synaptophysin (syp), another synaptic vesicle protein, remained unchanged.
SV2a also regulates expression and endocytosis of synaptotagmin (syt1), a calcium sensor protein required for vesicle fusion during neurotransmitter release.11d We next assessed the effect of Fucα(1-2)Gal glycan loss on SV2a-mediated endocytosis of syt1 by measuring the colocalization of SV2a and syt1 puncta at synapses in neurons from WT and FUT2–/– mice. The density of colocalized SV2a and syt1 puncta increased by 1.6-fold in FUT2–/– neurons compared to WT neurons, consistent with reduced endocytic reuptake (Figures 3C and S3).
Finally, analysis of SV2a trafficking requires measurement of the SV2a subpopulation exposed to the cell surface during synaptic vesicle cycling. We therefore conducted a cell-surface biotinylation assay, in which living neurons were incubated with a non-cell permeable, reactive biotin molecule to label surface proteins. Following immunoprecipitation of SV2a, we analyzed the immunoprecipitated protein by both streptavidin and Western blotting to measure the amount of SV2a on the neuronal surface compared to the total amount of SV2a precipitated. We found that the level of SV2a on the cell surface was 1.9-fold greater in FUT2–/– neurons compared to WT neurons (Figure 3D). Thus, we show using three independent methods that SV2a localization at the membrane is disrupted in FUT2 knockout mice, strongly suggesting that Fucα(1-2)Gal sugars influence SV2a function. Based on these results, we propose that the interaction of SV2a with a Fucα(1-2)Gal glycoprotein on the cell surface influences the endocytic reuptake of SV2a into synaptic vesicles. As FUT2–/– mice have reduced Fucα(1-2)Gal sugars on their glycoproteins, this interaction is disrupted in FUT2–/– neurons, attenuating the reuptake of SV2a into vesicles and increasing the localization of SV2a at the cell surface. As such, Fucα(1-2)Gal glycans may serve as a sorting signal for SV2a endocytosis during synaptic vesicle recycling. Interestingly, while SV2a itself is a glycoprotein, we found that SV2a is unlikely to be modified by Fucα(1-2)Gal glycans, as assessed by blotting of rat brain SV2a with UEAI (Figure S4). Together with the in vitro binding data, our results provide support for an important functional interaction between SV2a and Fucα(1-2)Gal sugars.
In summary, we have designed and synthesized glycopolymers as chemical tools for the identification of novel GBPs. These studies represent, to our knowledge, the first application of synthetic glycopolymers for the systems-level, proteomic profiling of GBPs. Notably, our polymer design enables the enrichment of endogenous GBPs that would be otherwise difficult to identify. Indeed, the present studies represent the culmination of a challenging, 12 year effort to identify Fucα(1-2)Gal GBPs. Using this approach, we discovered the first mammalian GBPs that recognize Fucα(1-2)Gal-containing glycans, and we demonstrate that the interaction of SV2a with Fucα(1-2)Gal sugars may have important functional consequences for SV2a trafficking and synaptic vesicle recycling. Future studies with the GBPs identified herein will continue to provide important insights into the molecular mechanisms underlying these important sugars. Moreover, we anticipate that this approach can be readily extended to discover novel GBPs for many different glycan classes.
Acknowledgments
We thank Drs. J. Lowe and S. Domino for the FUT KO mice, Dr. S. Bajjalieh for the SV2a plasmid, Dr. J. Gleeson for the DCLK1 plasmid, Dr. T. Kosaza for GαO protein, Dr. M. Shahgoli in the CCE Division Mass Spectrometry Facility, and Dr. C. Krishnamurthy for BgtA enzyme. This work was supported by the NIH (R01 GM084724).
Supporting Information Available
Experimental details and data. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- a Rabinovich G. A.; Croci D. O. Immunity 2012, 36, 322. [DOI] [PubMed] [Google Scholar]; b Hudak J. E.; Canham S. M.; Bertozzi C. R. Nat. Chem. Biol. 2014, 10, 69. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Miwa H. E.; Song Y.; Alvarez R.; Cummings R. D.; Stanley P. Glycoconj. J. 2012, 29, 608. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Borsig L.; Wong R.; Hynes R. O.; Varki N. M.; Varki A. Proc. Natl. Acad. Sci. U.S.A. 2002, 99, 2193. [DOI] [PMC free article] [PubMed] [Google Scholar]; e Wang C. C.; Chen J. R.; Tseng Y. C.; Hsu C. H.; Hung Y. F.; Chen S. W.; Chen C. M.; Khoo K. H.; Cheng T. J.; Cheng Y. S. E.; Jan J. T.; Wu C. Y.; Ma C.; Wong C. H. Proc. Natl. Acad. Sci. U.S.A. 2009, 106, 18137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dam T. K.; Brewer C. F. Glycobiology 2010, 20, 270. [DOI] [PubMed] [Google Scholar]
- Paulson J. C.; Blixt O.; Collins B. E. Nat. Chem. Biol. 2006, 2, 238. [DOI] [PubMed] [Google Scholar]
- a Bullock S.; Potter J.; Rose S. P. R. J. Neurochem. 1990, 54, 135. [DOI] [PubMed] [Google Scholar]; b Krug M.; Jork R.; Reymann K.; Wagner M.; Matthies H. Brain. Res. 1991, 540, 237. [DOI] [PubMed] [Google Scholar]; c Kalovidouris S. A.; Gama C. I.; Lee L. W.; Hsieh-Wilson L. C. J. Am. Chem. Soc. 2005, 127, 1340. [DOI] [PubMed] [Google Scholar]; d Murrey H. E.; Gama C. I.; Kalovidouris S.; Luo W. I.; Hsieh-Wilson L. C. Proc. Natl. Acad. Sci. U.S.A. 2006, 103, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Gestwicki J. E.; Strong L. E.; Kiessling L. L. Chem. Biol. 2000, 7, 583. [DOI] [PubMed] [Google Scholar]; b Lee R. T.; Lee Y. C. Glycoconj. J. 2000, 17, 543. [DOI] [PubMed] [Google Scholar]
- a Grande D.; Guerrero R.; Gnanou Y. J. Polym. Sci., Part A: Polym. Chem. 2005, 43, 519. [Google Scholar]; b Sun X. L.; Faucher K. M.; Grande D.; Chaikof E. L. J. Am. Chem. Soc. 2002, 124, 7258. [DOI] [PubMed] [Google Scholar]
- Gubbens J.; Ruijter E.; de Fays L. E. V.; Damen J. M. A.; de Kruijff B.; Slijper M.; Rijkers D. T. S.; Liskamp R. M. J.; de Kroon A. I. P. M. Chem. Biol. 2009, 16, 3. [DOI] [PubMed] [Google Scholar]
- Wegmann B.; Schmidt R. R. Carbohydr. Res. 1988, 184, 254. [Google Scholar]
- Braccini I.; Derouet C.; Esnault J.; de Penhoat C. H.; Mallet J. M.; Michon V.; Sinay P. Carbohydr. Res. 1993, 246, 23. [Google Scholar]
- Hindsgaul O.; Khare D. P.; Bach M.; Lemieux R. U. Can. J. Chem. 1985, 63, 2653. [Google Scholar]
- a Deuel T. A.; Liu J. S.; Corbo J. C.; Yoo S. Y.; Rorke-Adams L. B.; Walsh C. A. Neuron 2006, 49, 41. [DOI] [PubMed] [Google Scholar]; b Chou W. H.; Wang D.; McMahon T.; Qi Z. H.; Song M.; Zhang C.; Shokat K. M.; Messing R. O. J. Neurosci. 2010, 30, 13955. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Janz R.; Goda Y.; Geppert M.; Missler M.; Südhof T. C. Neuron. 1999, 24, 1003. [DOI] [PubMed] [Google Scholar]; d Yao J.; Nowack A.; Gardner R. G.; Kensel-Hammes P.; Bajjalieh S. M. J. Neurosci. 2010, 30, 5569. [DOI] [PMC free article] [PubMed] [Google Scholar]; e Nakata H.; Kozasa T. Mol. Pharmacol. 2005, 67, 695. [DOI] [PubMed] [Google Scholar]
- a Shin E.; Kashiwagi Y.; Kuriu T.; Iwasaki H.; Tanaka T.; et al. Nat. Commun. 2013, 4, 1440. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Chen Y.; Cantrell A. R.; Messing R. O.; Scheuer T.; Catterall W. A. J. Neurosci. 2005, 25, 507. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Yuyama K.; Sekino-Suzuki N.; Sanai Y.; Kasahara K. J. Biol. Chem. 2007, 282, 26392. [DOI] [PubMed] [Google Scholar]
- a He J.; Baum L. G. J. Biol. Chem. 2004, 279, 4705. [DOI] [PubMed] [Google Scholar]; b Perillo N. L.; Pace K. E.; Seilhamer J. J.; Baum L. G. Nature 1995, 378, 736. [DOI] [PubMed] [Google Scholar]
- Domino S. E.; Zhang L.; Gillespie P. J.; Saunders T. L.; Lowe J. B. Mol. Cell. Biol. 2001, 21, 8336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaubard J.-L.; Krishnamurthy C.; Yi W.; Smith D. F.; Hsieh-Wilson L. C. J. Am. Chem. Soc. 2012, 134, 4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.