Abstract
SUMMARY
Introduction: The majority of nosocomial fungal infections are caused by Candida spp. where C. albicans is the species most commonly identified. Molecular methods are important tools for assessing the origin of the yeasts isolated in hospitals.
Methods: This is a study on the genetic profifiles of 39 nosocomial clinical isolates of C. albicans using two typing methods: random amplifified polymorphic DNA (RAPD) and microsatellite, two different primers for each technique were used.
Results: RAPD provided 10 and 11 different profiles with values for SAB of 0.84 ± 0.126 and 0.88 ± 0.08 for primers M2 and P4, respectively. Microsatellite using two markers, CDC3 and HIS3, allowed the observation of six and seven different alleles, respectively, with combined discriminatory power of 0.91.
Conclusions: Although genetic variability is clear, it was possible to identify high similarity, suggesting a common origin for at least a part of isolates. It is important to emphasize that common origin was proven from yeasts isolated from colonization (urine, catheter or endotracheal secretions) and blood culture from the same patient, indicating that the candidemia must have started from a site of colonization. The combination of RAPD and microsatellite provides a quick and efficient analysis for investigation of similarity among nosocomial isolates of C. albicans.
Keywords: Candida albicans, Microsatellite, RAPD, Nosocomial infection
Abstract
RESUMO
Introdução: A maioria das infecções fúngicas hospitalares são causadas por Candida spp. e C. albicans é a espécie mais comumente identificada. Métodos moleculares são ferramentas importantes para a avaliação da origem das leveduras isoladas em hospitais.
Métodos: Este é um estudo sobre o perfil genético de 39 isolados clínicos nosocomiais de C. albicans através das técnicas de RAPD e microssatélite, foram usados dois diferentes iniciadores para cada técnica.
Resultados: RAPD forneceu 10 e 11 diferentes perfis com valores de SAB 0,84 ± 0,126 e 0,88 ± 0,08 para os primers M2 e P4, respectivamente. A análise de microssatélites, usando os marcadores CDC3 e HIS3 permitiu a observação de seis e sete diferentes alelos respectivamente, com poder discriminatório combinado de 0,91.
Conclusões: Embora seja clara a variabilidade genética, foi possível identificar alta similaridade, sugerindo origem comum para pelo menos parte deles. É importante enfatizar que foi comprovada origem comum de leveduras isoladas de colonização (urina, cateter ou secreção orotraqueal) e hemocultura do mesmo paciente, indicando que a candidemia deve ter iniciado a partir de um sítio de colonização. A combinação das técnicas RAPD e microssatélites fornece uma análise rápida e eficiente para investigação de similaridade entre isolados nosocomiais de C. albicans.
INTRODUCTION
The incidence of fungal infections in hospitals is increasing substantially in different parts of the world. This is due, among other factors, to the increase of immunocompromised patients undergoing invasive treatments and prolonged hospital stay 27 . Candida spp. is the most frequent genus of fungal infections, in the U.S. it is the fourth agent of bloodstream infections, which are considered severe and with attributed mortality rates of 30-60% 4 8 18 . In Brazil, the incidence of candidemia in teaching hospitals is of 2.49 cases per 1,000 admissions 14 , which represents the values 6.6-12.5 times higher than those reported in some European countries 14 . C. albicans is still the most frequently isolated species of both superficial and invasive infections in Brazil as well as in the world 3 9 .
Despite the evidence for transmission of Candida spp. by direct or indirect contact and evidence of cross-infection by health care workers, little is known about the origin of clinical isolates of C. albicans. Information on molecular epidemiology has great relevance for the clinical management, treatment and epidemiology of recurrent infections, especially among critically ill patients 10 11 30 . This context led to the proposal of a variety of molecular typing techniques aiming at distinguishing isolates of Candida spp. from different sources. Among others, the best known are: multilocus sequence typing (MLST), pulsed-field gel electrophoresis (PFGE), duplex PCR, restriction fragment length polymorphisms (RFLP), randomly amplified polymorphic DNA (RAPD) and microsatellites 5 13 21 26 31 34 38 .
The microsatellite analysis is a technique which has been recently used for genotyping C. albicans 1 6 7 . It is formed by short tandem repeats of two to six nucleotides known to be highly polymorphic, generating a characteristic profile of different alleles for a given locus. Due to its high discriminatory potential, this approach allows studies of nosocomial transmission routes 6 32 33 . Microsatellite has been used to investigate the molecular profile of C. albicans from healthy individuals 15 , HIV sero-positive 26 and with recurrent vulvovaginal candidiasis 32 .
Among the currently available to C. albicans genotyping techniques, RAPD is relatively cost-effective and it offers similar resolving power to electrophoretic karyotyping. These characteristics together with dendrograms of genetic relatedness among C. albicans isolates has significantly advanced lineage studies over progressive infective episodes or during asymptomatic carriage 16 36 . In addition, RAPD has been used to investigate infections caused by identical or similar strains 35 , emergence of resistance strains during antifungal therapy 20 26 , colonization patterns of yeast strains in different clinical situations, and microevolution of strains within a particular species 20 .
Thus, the aim of this study was to determine the genetic relatedness of C. albicans from hospitalized patients by using the RAPD and microsatellites assays.
MATERIAL AND METHODS
Microorganisms: A total of 39 strains of C. albicans isolated from different sources of patients hospitalized at University Hospital of Maringa in 2009, were used in this study. The yeasts were screened in chromogenic media CHROMagar® Candida and identified by conventional phenotypic methods (germ tube, microculture in cornmeal agar supplemented with 1% Tween 80, auxanogram and zymogram test) 39 .
Yeasts were taken from the following sources: urine 51.5% (N = 20), blood 20.5% (N = 8), catheter tip 15.5% (N = 6), orotracheal discharge 10% (N = 4) and peritoneal fluid 2.5% (N = 1). Regarding hospital sectors, 49% (N = 19) were isolated from adult Intensive Care Unit (ICU), 18% (N = 7) medical clinic, 13% (N = 5) pediatrics, 7.5% (N = 3) Neonatal Intensive Care Unit (NICU), 5% (N = 2) surgical clinic and 7.5% (N = 3) pediatric ICU.
DNA Extraction: The yeast strains were grown overnight at 25 °C using Sabouraud Dextrose Broth (SDB, Difco, USA) and genomic DNA extracted as described by CHONG et al. 12 . The concentration (260 nm) and purity (260/280) of the genomic DNA obtained were determined by optical density in a spectrophotometer, the visualization was made out in agarose gel at 0.7% with 1X TBE buffer (Tris-base 90 mmol l–1, Boric acid 90 mmol l–1, EDTA 2 mmol l–1 pH 8.0).
Nested-PCR: Identification of yeasts was confirmed by Nested-PCR which comprised two amplification stages, according to LUO & MITCHELL 23 . Briefly, primers that amplified Internal Transcribed Spacer (ITS) fragments of DNAr and identified the genus Candida were used in the first reaction. Amplification of species specific primers was used in the second reaction.
RAPD (Random Amplified Polymorphic DNA): RAPD was performed using the kit Ready-To-Go RAPD Analysis Beads ® (Amersham Biosciences Corporation, Piscataway, NJ, USA) as described by the manufacturer. The RAPD reactions were performed by adding 30 ng of genomic DNA, one µmol l–1 oligonucleotide and water for a final volume of 25 µL to each tube containing Ready-To-Go beads. The oligonucleotides used were M2 (5′-CTTGATTGCC-3′) 25 and P4 (5′-AAGAGCCCGT-3′ - Analysis Kit Ready-To-Go/RAPD Beads). The reaction was conducted in a in a Eppendorf Mastercycler Gradient Thermocycler® as follows: 95 °C for five min, followed by 45 cycles consisting of 95 °C for one min, 36 °C for one min and 72 °C for five min. Control tubes without template DNA were included in each run and reproducibility of the method was checked by repeating the amplification using different DNA extractions from two isolates and at least three different days.
The PCR products were electrophoresed in 2% agarose gel (w/v) in 1X TBE buffer at 150 volts for three hours. Amplicons in the gel were stained with ethidium bromide (0.5 mg mL–1) and visualized under UV transillumination (UVP Bioimaging Systems, Upland, CA®). The RAPD profiles were analyzed using Bionumerics® software version 4.6 (Applied Maths®).
The similarity was verified by the coefficient (SAB) between each pair of standards for A and B isolates and calculated with the formula SAB = 2E / (2E + a + b), where E is the number of common bands in the patterns A and B, a is the number of bands with an a pattern and no B correlated patterns, and b is the number of bands with B pattern and no correlation in pattern A. From the similarity matrix, the units were grouped by UPGMA (Unweighted Pair-Group Method with Arithmetical Average). An SAB value of 1.00 indicates that the pattern of bands for line A is identical to B; values between 0.80 to 0.99 represent very similar clinical isolates but not identical, and may suggest microevolution of a single strain; SAB values less than 0.80 represent independent lines 12 .
Microsatellites: Samples were genotyped using two microsatellite markers, CDC3 and HIS3, whose primer sequences were shown in Table 1, and all technical procedure was performed as described by BOTTEREL et al. 7 . The amplification products were analyzed by electrophoresis in polyacrylamide gel at 8% (w/v) in 1X TBE buffer for five hours at 140 volts. For the determination of the sizes of the fragments we used the molecular size marker 25 bp (Invitrogen®). After the run, the gel was stained with ethidium bromide (0.5 mg mL–1) and photodocumented under UV transillumination (UVP Bioimaging Systems, Upland, CA®). The size of the amplified fragments was determined by image analysis software LabImage 1D (Loccus Biotech®).
Table 1. Primers used for genotyping of C. albicans isolates by Microsatellite and RAPD.
| Locus (GenBank access number), chromosome | Primer | Nucleotide sequence (forward and reverse) | |
|---|---|---|---|
| Microsatellite | CDC3 (Z25869), chromosome 1 | CDC3 | 5′-CAGATGATTTTTTGTATGAGAAGAA-3′ |
| 5′-CAGTCACAAGATTAAAATGTTCAAG-3′ | |||
| HIS3 (AF006605), chromosome 2 | HIS3 | 5′-TGGCAAAAATGATATTCCAA-3′ | |
| 5′-TACACTATGCCCCAAACACA-3′ | |||
| RAPD | - | P4 | 5′-AAGAGCCCGT-3′ |
| - | M2 | 5′-CTTGATTGCC-3′ |
The results were expressed according to the tested locus name and size of the two alleles observed in base pairs. The reproducibility of this step was ensured by the inclusion of analysis of a strain of C. albicans ATCC 38696 which provided repeatable and consistent results with those obtained by BOTTEREL et al. 7 .
RESULTS
Analysis by Nested-PCR: Amplifications with primers ITS1/ITS4 resulted in patterns of bands with 500 bp identifying Candida spp. The species-specific primers provided amplification of fragments with approximately 272 bp, thus confirming the classic identification that the isolated are indeed C. albicans species.
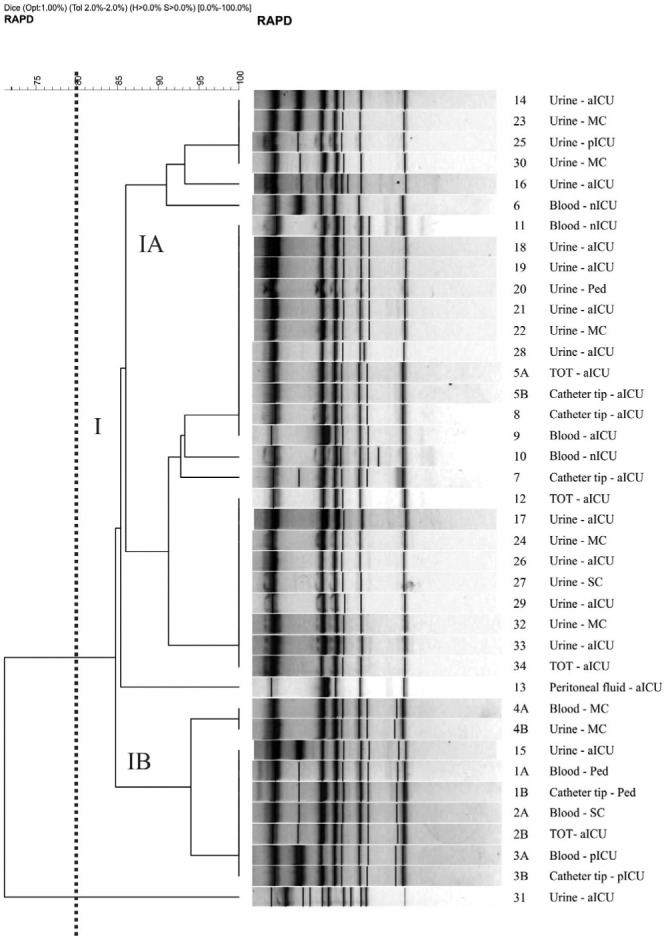
RAPD profiles: Analysis using primer M2 demonstrated the formation of 10 profiles with values of 0.84 ± 0.126 for SAB (Fig. 1). Three groups (I, II and III) were formed with 67% similarity between them. Group I consisted of four subgroups (IA, IB, IC and ID), which clustered approximately 70% of the isolates with similarity of 80%. Primer P4 generated 11 different profiles, with a SAB value of 0.88 ± 0.08. There was the formation of only one cluster, containing 95% of the isolates with approximately 85% similarity (Fig. 2).
Fig. 1. Dendrogram generated by amplification of primer M2 and by UPGMA grouping, in which SAB was calculated by the coefficient of Dice for 39 C. albicans isolates. Vertical line divides dendrogram as from the 80% similarity level; the four sub-groups (IA, IB, IC and ID) gather almost 70% of samples. In the samples identification the equal number and different letter mean same patient. SC: Surgery clinic; aICU: adult ICU; pICU: pediatry ICU; nICU: neonatal ICU; MC: Medical clinic; Ped: Pediatrics.
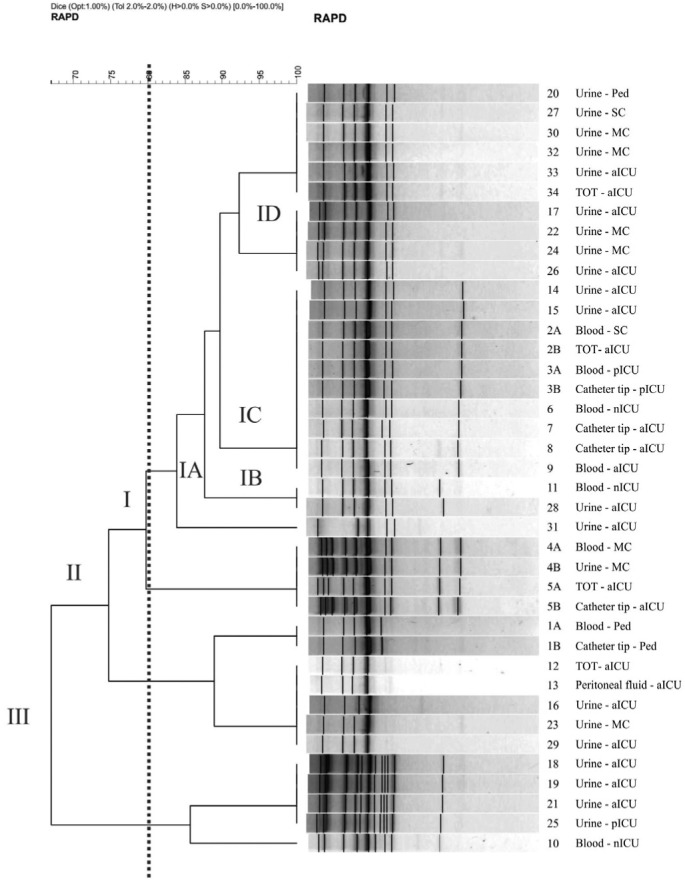
Fig. 2. Dendrogram generated from the amplification by primer P4 and by UPGMA grouping, in which SAB was calculated by the coefficient of Dice for 39 C. albicans isolates. Vertical line divides dendrogram as from 80% similarity level, in which Group I gathers 95% of samples. In the samples identification the equal number and different letter mean same patient. SC: Surgery clinic; aICU: adult ICU; pICU: pediatry ICU; nICU: neonatal ICU; MC: Medical clinic; Ped: Pediatrics.
Microsatellites: For all isolates tested we obtained products characteristic of microsatellite amplification. One or two PCR fragments by locus were produced for each isolate, since C. albicans is diploid, and each fragment was defined as an allele. The observed differences in size of alleles are attributed to the different numbers of repeats of microsatellites. The strains with two PCR products were typed as heterozygous, while those who had a single amplification product were considered homozygous.
The analysis of independent 39 isolates showed that all microsatellite loci were polymorphic, evidencing between six and seven alleles, and eight and nine different genotypes for the CDC3 and HIS3 primers respectively (Table 2). The discriminatory power (DP) was calculated for each marker according to the Simpson index:
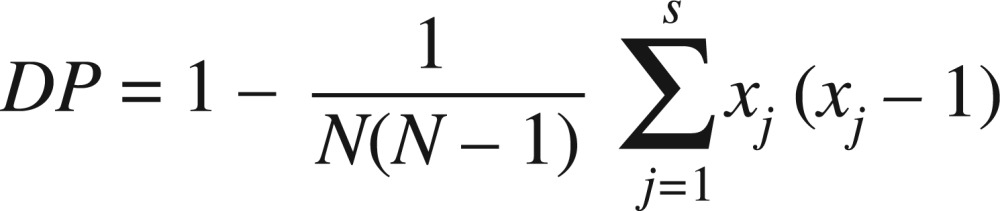 |
where N is the number of strains, s is the total number of different genotypes, and nj is the number of strains of genotype j 22 . The results showed that CDC3 was the microsatellite with the highest DP value (0.85), while HIS3 presented the lowest DP value (0.90). When the two markers are combined, the DP was 0.91. An index over or greater than 0.90 is desirable if the typing results are to be interpreted with confidence 22 .
Table 2. Origin of 39 Candida albicans isolates and their respective genotypes determined by microsatellite analysis.
| Samples | Origin of isolates | Primer CDC3 | Primer HIS3 | N | Genotypes | |||
|---|---|---|---|---|---|---|---|---|
| H. U. | Source | Allele 1 (bp) | Allele 2 (bp) | Allele 1 (bp) | Allele 2 (bp) | |||
| 2A | SC | Blood | 129 | 125 | 150 | 194 | 2 | A |
| 2B | aICU | TOT | 129 | 125 | 150 | 194 | ||
| 07 | aICU | Catheter tip | 121 | 117 | 154 | 154 | 1 | B |
| 3B | pICU | Catheter tip | 129 | 125 | 150 | 162 | 2 | C |
| 3A | pICU | Blood | 129 | 125 | 150 | 162 | ||
| 08 | aICU | Catheter tip | 125 | 117 | 162 | 162 | 9 | D |
| 11 | nICU | Blood | 125 | 117 | 162 | 162 | ||
| 32 | MC | Urine | 125 | 117 | 162 | 162 | ||
| 33 | aICU | Urine | 125 | 117 | 162 | 162 | ||
| 20 | Pediatrics | Catheter tip | 125 | 117 | 162 | 162 | ||
| 34 | aICU | TOT | 125 | 117 | 162 | 162 | ||
| 21 | aICU | Urine | 125 | 117 | 162 | 162 | ||
| 19 | aICU | Urine | 125 | 117 | 162 | 162 | ||
| 10 | nICU | Blood | 125 | 117 | 162 | 162 | ||
| 1B | Pediatrics | Catheter tip | 137 | 121 | 158 | 162 | 6 | E |
| 23 | MC | Urine | 137 | 121 | 158 | 162 | ||
| 30 | MC | Urine | 137 | 121 | 158 | 162 | ||
| 14 | aICU | Urine | 137 | 121 | 158 | 162 | ||
| 15 | aICU | Urine | 137 | 121 | 158 | 162 | ||
| 1A | Pediatrics | Blood | 137 | 121 | 158 | 162 | ||
| 4A | MC | Blood | 129 | 121 | 158 | 158 | 4 | F |
| 4B | MC | Urine | 129 | 121 | 158 | 158 | ||
| 5B | aICU | Catheter tip | 129 | 121 | 158 | 158 | ||
| 5A | aICU | TOT | 129 | 121 | 158 | 158 | ||
| 09 | aICU | Blood | 117 | 113 | 150 | 162 | 2 | G |
| 13 | aICU | Peritoneal fluid | 117 | 113 | 150 | 162 | ||
| 31 | aICU | Urine | 121 | 121 | 154 | 174 | 2 | H |
| 06 | nICU | Blood | 121 | 121 | 154 | 174 | ||
| 22 | MC | Urine | 121 | 117 | 166 | 166 | 2 | I |
| 12 | aICU | TOT | 121 | 117 | 166 | 166 | ||
| 17 | aICU | Urine | 125 | 125 | 166 | 166 | 1 | J |
| 16 | aICU | Urine | 125 | 125 | 162 | 162 | 2 | K |
| 18 | aICU | Urine | 125 | 125 | 162 | 162 | ||
| 25 | pICU | Urine | 129 | 121 | 150 | 150 | 1 | L |
| 24 | MC | Urine | 129 | 121 | 162 | 162 | 5 | M |
| 28 | aICU | Urine | 129 | 121 | 162 | 162 | ||
| 29 | aICU | Urine | 129 | 121 | 162 | 162 | ||
| 27 | SC | Urine | 129 | 121 | 162 | 162 | ||
| 26 | aICU | Urine | 129 | 121 | 162 | 162 | ||
SC: Surgery clinic; aICU (Intensive Care Unit): adult ICU; pICU: pediatric ICU; nICU: neonatal ICU; MC: medical clinic. N: number of genotype. H.U.: Hospital Unit; TOT: endotracheal aspirate.
The 39 samples were recovered from 34 patients because in five of them the same species was isolated from two different sites. A comparison of the genetic profile by microsatellite and RAPD (Table 3) showed total identity between these pairs.
Table 3. Similarity by Random Amplified Polymorphic DNA and genotype by Microsatellite of Candida albicans isolated in two sources from a same patient.
| Patient | Source | Genotype | RAPD | ||
|---|---|---|---|---|---|
| CDC3 (bp) | HIS3 (bp) | SAB Primer M2 | SAB Primer P4 | ||
| 1 | 1B– Catheter tip | 137:121 | 158:162 | 1.00 | 1.00 |
| 1A – Blood | 137:121 | 158:162 | 1.00 | 1.00 | |
| 2 | 2B – TOT | 129:125 | 150:194 | 1.00 | 1.00 |
| 2A – Blood | 129:125 | 150:194 | 1.00 | 1.00 | |
| 3 | 3B – Catheter tip | 129:125 | 150:162 | 1.00 | 1.00 |
| 3A – Blood | 129:125 | 150:162 | 1.00 | 1.00 | |
| 4 | 4B – Urine | 129:121 | 158:158 | 1.00 | 1.00 |
| 4A – Blood | 129:121 | 158:158 | 1.00 | 1.00 | |
| 5 | 5A – TOT | 129:121 | 158:158 | 1.00 | 1.00 |
| 5B – Catheter tip | 129:121 | 158:158 | 1.00 | 1.00 | |
TOT: endotracheal aspirate.
DISCUSSION
RAPD and microsatellite analysis were able to show similarity among C. albicans isolates recovered from a hospital. Microsatellite analysis supplied a good DP with markers used, it allowed formation of various genotypes grouping samples, confirming the similarity between them, which reinforces the interpretation of the data found in RAPD. Additionally, it was possible to prove the high similarity (100%) of the same yeast which was from colonization (urinary catheter, tracheal secretions) and later detected in blood cultures from the same patient.
The RAPD results showed SAB values of 0.84 ± 0.126 for the primer M2 and SAB 0.88 ± 0.08 for P4 (Fig. 1 and 2). It is important to highlight that the strains that are considered identical by a primer are not always necessarily also considered identical or belong to the same cluster when analyzed by another primer. This should be referred as a limitation of the technique, nevertheless according to CHONG et al. 12 the values found, in RAPD, indicate high similarity between the isolates.
In microsatellite analysis we were able to verify the presence of six different alleles with the primer CDC3 and seven alleles with primer HIS3, of which 113bp, 117bp, 125bp, 150bp, 154bp, 158bp and 162bp have already been recognized by other authors 1 6 7 . These primers amplify microsatellite regions highly polymorphic for C. albicans. Moreover, these regions are stable over generations and were chosen because they are located on different chromosomes, which increase the chances of finding polymorphisms 7 . The discriminatory power (DP) found using markers CDC3 and HIS3, was 0.85 and 0.90 respectively. These results and especially the combined value of DP (0.91) are considered by several authors as reliable studies of molecular typing 7 22 . The data presented in Table 2 show the prevalence of genotypes (D, E, F, M), however, there is no relation with sites of isolation of yeasts. This type of observation has already been described in another study using the same genotyping technique 2 . Finally, by putting together epidemiological data (Fig. 1 and 2, Table 2), it is possible to observe the formation of groups with high similarity (90-100%). These are mostly from patients hospitalized in ICU where the evidence of common origin is of great importance. According to AL-KARAAWI et al. 2 , the clinical isolates of C. albicans tend to be genetically similar to each other if they were isolates from patients with a similar profile, as those interned in ICU. CHAVES et al. 10 recently showed that candidemic patients had highly related microsatellites genotype in colonizing and bloodstream isolates. However, it should be noted that the detection of yeasts highly similar in our study was not associated with hospital unit. The same profile was found in various hospital areas such as pediatric and internal medicine. These data reinforce that most C. albicans infections are from endogenous sources. They should also suggest that these strains may be circulating in the various units, but not characterizing the occurrence of outbreaks.
Although the infection of different patients from different sectors with yeasts of the same genetic profile insinuates cross-transmission 17 19 , high similarity among samples suggests an adaptation to the environmental conditions, thus characterizing microevolutions 28 . Five (14.70%) of all patients enrolled in this study are particularly interesting since C. albicans were isolated from different sites. In all cases the analysis confirmed that the clinical isolates were identical to each other (Table 3) indicating the migration of yeasts from colonization (urine catheters, tracheal secretion) into the blood, suggesting the source of systemic infection. This result indicates that each isolated pair has genotypic identity, suggesting clonal origin. This fact has been demonstrated by molecular typing, in several studies 10 24 29 37 and helps confirm that previous colonization is an important predisposing factor for systemic infection.
Despite the small number of samples analyzed, this study contributes with the understanding on epidemiology of fungal infections in hospitals. The analyzed data allow us to conclude that both techniques generated reproducible profiles showing similarity among the isolates. These techniques are suitable for epidemiological molecular studies of C. albicans and can be applied in larger populations. The good performance of these techniques allows its use for genotyping of outbreaks of hospital origin or not, and characterization of isolates from different sites, including recurrent infections such as vulvovaginal candidiasis and investigations before and after treatments. Knowledge of the relationship of clinical isolates involved in infections is extremely important for the development and application of the correct therapeutic strategy and to better understand the epidemiology of these infections.
REFERENCES
- 1.Adachi H, Shimizu K, Hattori H, Tanaka R, Chibana H, Takagi Y, et al. Genotyping of Candida albicans by fragment analysis of microsatellites combined with 25S rDNA and RPS-based strategies. Nihon Ishinkin Gakkai Zasshi. 2009;50:167–74. doi: 10.3314/jjmm.50.167. [DOI] [PubMed] [Google Scholar]
- 2.Al-Karaawi ZM, Manfredi M, Waugh ACW, McCullough MJ, Jorge J, Scully C, et al. Molecular characterization of Candida spp. isolated from the oral cavities of patients from diverse clinical settings. Oral Microbiol Immunol. 2002;17:44–9. doi: 10.1046/j.0902-0055.2001.00081.x. [DOI] [PubMed] [Google Scholar]
- 3.Araújo SM, Fontes CJ, Leite Júnior DP, Hahn RC. Fungal agents in different anatomical sites in public health services in Cuiabá, state of Mato Grosso, Brazil. Rev Inst Med Trop Sao Paulo. 2012;54:5–10. doi: 10.1590/s0036-46652012000100002. [DOI] [PubMed] [Google Scholar]
- 4.Asmundsdottir LR, Erlendsdottir H, Haraldsson G, Guo H, Xu J, Gottfredsson M. Molecular epidemiology of candidemia: evidence of clusters of smoldering nosocomial infections. Clin Infect Dis. 2008;47:17–24. doi: 10.1086/589298. [DOI] [PubMed] [Google Scholar]
- 5.Ben Abdeljelil J, Saghrouni F, Emira N, Valentin-Gomez E, Chatti N, Boukadida J, et al. Molecular typing of Candida albicans isolates from patients and health care workers in a neonatal intensive care unit. J Appl Microbiol. 2011;111:1235–49. doi: 10.1111/j.1365-2672.2011.05121.x. [DOI] [PubMed] [Google Scholar]
- 6.Beretta S, Fulgencio JP, Enache-Angoulvant A, Bernard C, El Metaoua S, Ancelle T, et al. Application of microsatellite typing for the investigation of a cluster of cases of Candida albicans candidaemia. Clin Microbiol Infect. 2006;12:674–6. doi: 10.1111/j.1469-0691.2006.01438.x. [DOI] [PubMed] [Google Scholar]
- 7.Botterel F, Desterke C, Costa C, Bretagne S. Analysis of microsatellite markers of Candida albicans used for rapid typing. J Clin Microbiol. 2001;39:4076–81. doi: 10.1128/JCM.39.11.4076-4081.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bougnoux ME, Kac G, Aegerter P, d'Enfert C, Fagon JY, CandiRea Study Group. Candidemia and candiduria in critically ill patients admitted to intensive care units in France: incidence, molecular diversity, management and outcome. Intensive Care Med. 2008;34:292–9. doi: 10.1007/s00134-007-0865-y. [DOI] [PubMed] [Google Scholar]
- 9.Chang MR, Correia FP, Costa LC, Xavier PCN, Palhares DB, Taira DL, et al. Candida bloodstream infection: data from a teaching hospital in Mato Grosso do Sul, Brazil. Rev Inst Med Trop Sao Paulo. 2008;50:265–8. doi: 10.1590/s0036-46652008000500003. [DOI] [PubMed] [Google Scholar]
- 10.Chaves GM, Santos FP, Colombo AL. The persistence of multifocal colonisation by a single ABC genotype of Candida albicans may predict the transition from commensalism to infection. Mem Inst Oswaldo Cruz. 2012;107:198–204. doi: 10.1590/s0074-02762012000200008. [DOI] [PubMed] [Google Scholar]
- 11.Chong PP, Abdul Hadi SR, Lee YL, Phan CL, Tan BC, Ng KP, et al. Genotyping and drug resistance profile of Candida spp. in recurrent and one-off vaginitis, and high association of non-albicans species with non-pregnant status. Infect Genet Evol. 2007;7:449–56. doi: 10.1016/j.meegid.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 12.Chong PP, Lee YL, Tan BC, Ng KP. Genetic relatedness of Candida strains isolated from women with vaginal candidiasis in Malaysia. J Med Microbiol. 2003;52:657–66. doi: 10.1099/jmm.0.04973-0. [DOI] [PubMed] [Google Scholar]
- 13.Chávez-Galarza J, Pais C, Sampaio P. Microsatellite typing identifies the major clades of the human pathogen Candida albicans . Infect Genet Evol. 2010;10:697–702. doi: 10.1016/j.meegid.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 14.Colombo AL, Nucci M, Park BJ, Nouér SA, Arthington-Skaggs B, Da Matta DA, et al. Epidemiology of candidemia in Brazil: a nationwide sentinel surveillance of candidemia in eleven medical centers. J Clin Microbiol. 2006;44:2816–23. doi: 10.1128/JCM.00773-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dalle F, Dumont L, Franco N, Mesmacque D, Caillot D, Bonnin P, et al. Genotyping of Candida albicans oral strains from healthy individuals by polymorphic microsatellite locus analysis. J Clin Microbiol. 2003;41:2203–5. doi: 10.1128/JCM.41.5.2203-2205.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dassanayake RS, Samaranayake LP. Amplification-based nucleic acid scanning techniques to assess genetic polymorphism in Candida . Crit Rev Microbiol. 2003;29:1–24. doi: 10.1080/713610402. [DOI] [PubMed] [Google Scholar]
- 17.De Pinho Resende JC, Franco GR, Rosa CA, Hahn RC, Hamdan JS. Phenotypic and genotypic identification of Candida spp. isolated from hospitalized patients. Rev Iberoam Micol. 2004;21:24–8. [PubMed] [Google Scholar]
- 18.DiNubile MJ, Lupinacci RJ, Strohmaier KM, Sable CA, Kartsonis NA. Invasive candidiasis treated in the intensive care unit: observations from a randomized clinical trial. J Crit Care. 2007;22:237–44. doi: 10.1016/j.jcrc.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 19.Eggimann P, Garbino J, Pittet D. Epidemiology of Candida species infections in critically ill non-immunosuppressed patients. Lancet Infect Dis. 2003;3:685–702. doi: 10.1016/s1473-3099(03)00801-6. [DOI] [PubMed] [Google Scholar]
- 20.Enger L, Joly S, Pujol C, Simonson P, Pfaller M, Soll DR. Cloning and characterization of a complex DNA fingerprinting probe for Candida parapsilosis . J Clin Microbiol. 2001;39:658–69. doi: 10.1128/JCM.39.2.658-669.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ge SH, Xie J, Xu J, Li J, Li DM, Zong LL, et al. Prevalence of specific and phylogenetically closely related genotypes in the population of Candida albicans associated with genital candidiasis in China. Fungal Genet Biol. 2011;49:86–93. doi: 10.1016/j.fgb.2011.10.006. [DOI] [PubMed] [Google Scholar]
- 22.Hunter PR, Gaston MA. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J Clin Microbiol. 1988;26:2465–6. doi: 10.1128/jcm.26.11.2465-2466.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luo G, Mitchell TG. Rapid identification of pathogenic fungi directly from cultures by using multiplex PCR. J Clin Microbiol. 2002;40:2860–5. doi: 10.1128/JCM.40.8.2860-2865.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marco F, Lockhart SR, Pfaller MA, Pujol C, Rangel-Frausto MS, Wiblin T, et al. Elucidating the origins of nosocomial infections with Candida albicans by DNA fingerprinting with the complex probe Ca3. J Clin Microbiol. 1999;37:2817–28. doi: 10.1128/jcm.37.9.2817-2828.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Melo AS, de Almeida LP, Colombo AL, Briones MR. Evolutionary distances and identification of Candida species in clinical isolates by randomly amplified polymorphic DNA (RAPD) Mycopathologia. 1998;142:57–66. doi: 10.1023/a:1006998325716. [DOI] [PubMed] [Google Scholar]
- 26.Metzgar D, Van Belkum A, Field D, Haubrich R, Wills C. Random amplification of polymorphic DNA and microsatellite genotyping of pre-and posttreatment isolates of Candida spp. from human immunodeficiency virus-infected patients on different fluconazole regimens. J Clin Microbiol. 1998;36:2308–13. doi: 10.1128/jcm.36.8.2308-2313.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patterson TF. Advances and challenges in management of invasive mycoses. Lancet. 2005;366:1013–25. doi: 10.1016/S0140-6736(05)67381-3. [DOI] [PubMed] [Google Scholar]
- 28.Pires-Gonçalves RH, Miranda ET, Baeza LC, Matsumoto MT, Zaia JE, Mendes-Giannini MJS. Genetic relatedness of commensal strains of Candida albicans carried in the oral cavity of patients' dental prosthesis users in Brazil. Mycopathologia. 2007;164:255–63. doi: 10.1007/s11046-007-9052-5. [DOI] [PubMed] [Google Scholar]
- 29.Pittet D, Monod M, Suter PM, Frenk E, Auckenthaler R. Candida colonization and subsequent infections in critically ill surgical patients. Ann Surg. 1994;220:751–8. doi: 10.1097/00000658-199412000-00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Richards MJ, Edwards JR, Culver DH, Gaynes RP. Nosocomial infections in medical intensive care units in the United States. Crit Care Med. 1999;27:887–92. doi: 10.1097/00003246-199905000-00020. [DOI] [PubMed] [Google Scholar]
- 31.Robles JC, Koreen L, Park S, Perlin DS. Multilocus sequence typing is a reliable alternative method to DNA fingerprinting for discriminating among strains of Candida albicans . J Clin Microbiol. 2004;42:2480–8. doi: 10.1128/JCM.42.6.2480-2488.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sampaio P, Gusmão L, Alves C, Pina-Vaz C, Amorim A, Pais C. Highly polymorphic microsatellite for identification of Candida albicans strains. J Clin Microbiol. 2003;41:552–7. doi: 10.1128/JCM.41.2.552-557.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sampaio P, Gusmão L, Correia A, Alves C, Rodrigues AG, Pina-Vaz C, et al. New microsatellite multiplex PCR for Candida albicans strain typing reveals microevolutionary changes. J Clin Microbiol. 2005;43:3869–76. doi: 10.1128/JCM.43.8.3869-3876.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmid J, Tortorano AM, Jones G, Lazzarini C, Zhang N, Bendall M, et al. Increased mortality in young candidemia patients associated with presence of a Candida albicans general-purpose genotype. J Clin Microbiol. 2011;49:3250–6. doi: 10.1128/JCM.00941-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shin JH, Kook H, Shin DH, Hwang TJ, Kim M, Suh SP, et al. Nosocomial cluster of Candida lipolytica fungemia in pediatric patients. Eur J Clin Microbiol. 2000;19:344–9. doi: 10.1007/s100960050491. [DOI] [PubMed] [Google Scholar]
- 36.Soll DR. The ins and outs of DNA fingerprinting the infectious fungi. Clin Microbiol Rev. 2000;13:332–70. doi: 10.1128/cmr.13.2.332-370.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tay ST, Na SL, Chong J. Molecular differentiation and antifungal susceptibilities of Candida parapsilosis isolated from patients with bloodstream infections. J Med Microbiol. 2009;58:185–91. doi: 10.1099/jmm.0.004242-0. [DOI] [PubMed] [Google Scholar]
- 38.Vaz C, Sampaio P, Clemons KV, Huang YC, Stevens DA, Pais C. Microsatellite multilocus genotyping clarifies the relationship of Candida parapsilosis strains involved in a neonatal intensive care unit outbreak. Diagn Microbiol Infect Dis. 2011;71:159–62. doi: 10.1016/j.diagmicrobio.2011.05.014. [DOI] [PubMed] [Google Scholar]
- 39.Yarrow D. Methods for the isolation, maintenance and identification of yeasts. In: Kurtzman CP, Fell JW, editors. The yeast, a taxonomic study. New York: Elsevier; 1998. pp. 77–100. [Google Scholar]


