Abstract
SUMMARY
This study evaluated the applicability of kDNA-PCR as a prospective routine diagnosis method for American tegumentary leishmaniasis (ATL) in patients from the Instituto de Infectologia Emílio Ribas (IIER), a reference center for infectious diseases in São Paulo - SP, Brazil. The kDNA-PCR method detected Leishmania DNA in 87.5% (112/128) of the clinically suspected ATL patients, while the traditional methods demonstrated the following percentages of positivity: 62.8% (49/78) for the Montenegro skin test, 61.8% (47/76) for direct investigation, and 19.3% (22/114) for in vitro culture. The molecular method was able to confirm the disease in samples considered negative or inconclusive by traditional laboratory methods, contributing to the final clinical diagnosis and therapy of ATL in this hospital. Thus, we strongly recommend the inclusion of kDNA-PCR amplification as an alternative diagnostic method for ATL, suggesting a new algorithm routine to be followed to help the diagnosis and treatment of ATL in IIER.
Keywords: American tegumentary leishmaniasis, Diagnostic, kDNA-PCR, Leishmania (Viannia) braziliensis
Abstract
RESUMO
Este estudo avaliou a aplicabilidade do kDNA-PCR como método de rotina para diagnóstico de leishmaniose tegumentar americana (ATL) no Instituto de Infectologia Emílio Ribas (IIER), São Paulo, SP, Brasil. O método kDNA-PCR detectou DNA de Leishmania em 87,5% (112/128) dos pacientes com suspeita de ter leishmaniose e, os métodos tradicionais apresentaram as seguintes porcentagens de positividade: 62,8% (49/78) para o teste de Montenegro, 61,8% (47/76) para a pesquisa direta e 19,3% (22/114) para cultura in vitro. O método molecular confirmou a doença em amostras negativas ou inconclusivas pelos métodos laboratoriais tradicionais e, mostrou-se capaz de auxiliar na identificação de infecções causadas pela espécie Leishmania (V.) braziliensis. Além disso, a revisão dos prontuários médicos confirmou a importância do método PCR-RFLP no diagnóstico final de ATL, prognóstico e escolha do tratamento. Assim, recomendamos a inclusão do PCR como método diagnóstico de ATL na rotina hospitalar, e sugerimos um fluxograma para solicitação de exames laboratoriais.
INTRODUCTION
American tegumentary leishmaniasis (ATL) presents different clinical manifestations in Latin America, including mucosal leishmaniasis (ML), cutaneous localized leishmaniasis (CL), disseminated leishmaniasis (LCD), and diffuse leishmaniasis (DCL). These manifestations are caused by seven different species of Leishmania: Leishmania (Leishmania) amazonensis, L. (Viannia) braziliensis, L. (V.) guyanensis, L. (V.) lainsoni, L. (V.) naiffi, L. (V.) lindenberg, and L. (V.) shawi 7 24 . Currently, the diagnosis of ATL is based mainly on a clinical examination, epidemiological data, and complementary laboratory methods, including direct investigation (DI), Montenegro skin test (MST), and in vitro culture 7 . Nevertheless, these classical diagnostic methods are often time consuming; require an expert in microscopy; have low sensitivity and/or specificity and can be influenced by the age of infection and quality of sampling 7 . Furthermore, these methods are unable to differentiate between the seven species of parasite involved, which can be considered an important failure for the disease prognosis and choice of an appropriate treatment 9 32 . VOLPINI et al., 2004 41 described the potential of a PCR-RFLP method performed with primers specific for kinetoplast DNA (kDNA) of Leishmania genus, followed by HaeIII restriction enzyme digestion, for detection of L. (V.) braziliensis in infected DNA samples. This species is reported to be the major causative agent of ML in Latin America; and it is estimated that 3-5% of CL patients progress to ML form when infected with this species 7 . The specificity of these specific primers for the Leishmania genus has been reported in previous studies and no cross-reaction was observed for paracoccidioidomycosis, histoplasmosis, cutaneous tuberculosis, and candidiasis 3 , squamous cell carcinoma, sporotrichosis, leprosy, lentigo, pyodermitis and vascular ulcer 17 .
The identification of L. (V.) braziliensis in ATL suspected patients is important for elaboration of accurate prognoses and adequate therapy to prevent the resurgence of lesions, and the progression of the disease 7 24 . Thus, the aim of this study was to evaluate the applicability of the kDNA-PCR followed by restriction enzyme analyze method for routine diagnosis of ATL at the IIER
MATERIAL AND METHODS
Patients. The study was conducted on a convenience sample of 128 patients who attended the Instituto de Infectologia Emílio Ribas (IIER), São Paulo, Brazil from March 2007 to January 2012, with clinical signs and symptoms compatible with CL or ML. Cutaneous leishmaniasis was clinically suspected by the presence of ulcer, while mucosal leishmaniasis by the presence of nasal bleeding, drilling nasal septum or infiltrate in mucosal 7 13 24 . The results of the clinical examinations, Montenegro skin test, epidemiologic data, and administered drugs were obtained by reviewing the medical records. Collected samples from 128 lesions were transferred for our lab to be analyzed by Direct Investigation, in vitro culture and kDNA PCR.
The study was approved by the Research Ethics Committee of IIER (Process number 326/2009), and by the Research Ethics Committee of the Instituto de Medicina Tropical de São Paulo (Process CEP-IMT 046/2009).
Montenegro skin test (MST). Montenegro skin test was performed by Instituto Adolfo Lutz, São Paulo - SP. The technique consists of application of 0.5 mL of antigen of L. (L) amazonensis (MHOM/BR/73/PH8), distributed by the Brazilian Ministry of Health. After 72 h the papule was measured using a ruler and the result is expressed in mm. It is considered positive if the test is higher than 5 mm 10 31 . The results of the MST were obtained by reviewing the medical records of the suspected ATL patients.
Direct investigation (DI). Lesion samples were obtained by biopsy after asepsis, and local anesthesia using a 4 mm diameter punch. The excess blood was removed from the samples and lesion imprints were collected on glass slides. After air drying, the slides were fixed in methanol, stained with Giemsa, and microscopically examined by two technicians. The results were based on the following criteria: the presence of typical Leishmania amastigotes indicated a positive result; the absence of amastigotes indicated a negative result; and the presence of atypical forms of parasite was considered to suggest a positive result.
In vitro culture and isolation of the parasites. Parts of the lesion samples were transferred to a plastic tube with a physiological salt solution with 100 U penicillin, 100 mg/mL streptomycin, and 50 µg/mL 5-fluorocytosine, and forwarded to the Instituto de Medicina Tropical (IMT-USP). Each sample was further divided for the in vitro culture of parasites and for the DNA extraction procedures. A fraction of the sample designated for parasite isolation was incubated in Media 199, (SIGMA, USA) with 10% fetal bovine serum at 26 °C in BOD and it was examined weekly for a month.
Reference strains. L. (V.) braziliensis (MHOM/BR/75/M2903), and L. (L.) amazonensis (IFLA/BR/67/PH8) promastigote forms were cultivated in Media 199 with 10% fetal bovine serum at 26 °C in BOD. The promastigotes were collected at the exponential growth phase with approximately 10 7 cells per mL (+/- 96 h).
DNA extraction. DNA extraction from tissue lesion samples from patients and promastigote reference strains was processed using the Wizard Genomic DNA Purification Kit (Promega, USA) following the manufacturer's protocol. The quantification and quality control of the DNA extraction procedures were performed using a nano spectrophotometer (NanoDrop 1000, Thermo Fisher Scientific). All reactions were performed in appropriated places, following the good practice of laboratories to avoid sample contamination.
kDNA-PCR: The technique was performed based on the protocol described previously 41 using the following primers that amplify a 120 bp fragment of the conserved region of a Leishmania kDNA minicircle: kDNA20 forward, 5′- GGG (G/T)AG GGG CGT TCT (G/C)CG AA-3′, and kDNA22 reverse, 5′ (G/C)(G/C)(G/C) (A/T)CT AT(A/T) TTA CAC CAA CCC C- 3′. The reaction mixtures were prepared in a final volume of 20 µL that contained Taq DNA polymerase Buffer with KCl (10 mM Tris-HCl pH8.8, 50 mM KCl, and 0.08% (v/v) Nonidet P-40); 1.0 mM MgCl2; 0.2 mM of each dNTP; 375 pM of each primer; 1 U Taq DNA polymerase (recombinant) (Fermentas); and 4 µL (∼ 200 ng) of DNA. Amplification was conducted using an MWG Biotech Model Primus 96 Plus Thermal Cycler with an initial denaturation step at 94 °C for four min, followed by 35 cycles at 94 °C for one min, 58 °C for one min, 72 °C for 30 s, and a final extension step at 72 °C for five min. The amplicons were visualized by electrophoresis on 2% agarose gels stained with ethidium bromide.
Positive controls that contained the DNA from the reference strains, and a negative control with no DNA were included in each reaction set. In addition, the samples that were negative according to kDNA-PCR protocol were submitted to PCR amplification with primers directed to human β-globin PCR 1 to verify the quality of the DNA extraction procedure.
PCR-RFLP (kDNA- HaeIII): Finally, 10 µL microliters of the positive kDNA-PCR products were digested at 37 °C for three hours with 10 U of HaeIII enzyme (Fermentas), with specific buffer and deionized water, total volume of 15 µL, according to the manufacturer's protocol. The restriction fragments were separated on a 10% polyacrylamide gel, and stained with ethidium bromide.
RESULTS
Analysis of the results of the traditional diagnosis methods. A total of 128 patients with clinically suspected ATL were enrolled in this study: 59 (46.1%) patients with a suspicious mucosal lesion (sML), and 69 (53.9%) patients with suspicious of cutaneous lesions (sCL). As a result of problems encountered during the data collection from medical records, which were often incomplete lacking important information, we were unable to get the results of the three traditional methods for routine diagnosis of all 128 patients. Then, tests were analyzed in the following frequencies: 89.1% by in vitro culture, 60.9% by MST, and 59.4% by DI, only kDNA-PCR was performed on all 128 collected samples (Table 1). As shown in Table 1, we observed that DI test was performed more frequently in sCL patients than for sML patients: 81.2% (56/69), and 33.9% (20/59), respectively. On the contrary, MST was performed on 71.2% (42/59) of the sML patients, and on 52.2% (36/69) of the sCL patients. The results of the DI, MST and in vitro tests were analyzed considering only the number of samples truly performed for each test (76/128 samples for DI, 78/128 for MST and 114/128 for culture (Table 1).
Table 1. Results of the four diagnostic methods performed with samples from patients with suspected ATL who attended the Instituto de Infectologia Emílio Ribas (IIER) in São Paulo, Brazil.
| Clinical manifestation | Cutaneous | Mucosal | Total | |||
|---|---|---|---|---|---|---|
| N | (%) | N | (%) | N | (%) | |
| Patients | 69 | 53.9% | 59 | 46.1% | 128 | |
| Direct investigation (76) | ||||||
| Not performed (52) | ||||||
| Positive | 38 | 67.9% | 9 | 45.0% | 47 | 61.8% |
| Negative | 6 | 10.7% | 4 | 20.0% | 10 | 13.2% |
| Suggestive | 12 | 21.4% | 7 | 35.0% | 19 | 25.0% |
| Total | 56 | 20 | 76 | |||
| Montenegro skin test (78) | ||||||
| Not performed (50) | ||||||
| Positive (> 5 mm) | 18 | 50.0% | 31 | 73.8% | 49 | 62.8% |
| Negative (< 5 mm) | 18 | 50.0% | 11 | 26.2% | 29 | 37.2% |
| Total | 36 | 42 | 78 | |||
| In vitro culture (114) | ||||||
| Not performed (14) | ||||||
| Positive | 14 | 23.7% | 8 | 14.5% | 22 | 19.3% |
| Negative | 34 | 57.6% | 17 | 30.9% | 51 | 44.7% |
| Contamination | 11 | 18.6% | 30 | 54.5% | 41 | 36.0% |
| Total | 59 | 55 | 114 | |||
| kDNA-PCR (128) | ||||||
| Positive | 61 | 88.4% | 51 | 86.4% | 112 | 87.5% |
| Negative | 8 | 11.6% | 8 | 13.6% | 16 | 12.5% |
| Total | 69 | 59 | 128 | |||
Evaluation of the efficiency of the kDNA-PCR method and traditional diagnostic methods. kDNA-PCR was performed on all 128 collected samples, and amplified Leishmania DNA was observed in 112/128 (87.5%) clinically suspected ATL samples (Table 1). The 16 remaining samples (12.5%) were considered negative after confirmation of DNA integrity by human β-globin PCR analysis. In addition, these samples were also negative for ATL according to the traditional methods, excluding one sample (Table 2).
Table 2. Comparison of the kDNA-PCR results with those obtained by the traditional diagnostic methods with samples from patients with suspected of cutaneous and mucosal leishmaniasis*.
| kDNA-PCR | Traditional diagnostic methods | |||||||
|---|---|---|---|---|---|---|---|---|
| In vitro culture | DI test | MST | ||||||
| P | N | Cont. | P | N | Sug. | P | N | |
| Positive | 22 (100%) | 41 (80.4%) | 36 (87.8%) | 47 (100%) | 5 (50%) | 15 (78.9%) | 48 97.9%) | 25 (86.2%) |
| Negative | 0 | 10 (19.6%) | 5 (12.2%) | 0 | 5 (50%) | 4 (21%) | 1 (2%) | 4 (13.8%) |
kDNA-PCR: Polymerase chain reaction using specific primers for Leihmania's kinetoplast DNA, DI test: Direct Investigation test, MST: Montenegro skin test. P: Positive result, N: Negative result; Sug.: Suggestive, Cont.: contamination.
The same sample could be analyzed by more than one diagnostic method.
Thus, we can conclude that kDNA-PCR was the most efficient test, with a positivity of 87.5%, followed by the MST with 62.8%, DI with 61.8%, and in vitro culture with 19.3%. We observed that the efficiency of DI, and MST methods was distinct for each clinical manifestation. DI test was 1.5 times more efficient for samples from sCL patients, than for samples from sML patients. The opposite was observed in the MST results: an efficiency 1.5 times greater for sML patients (73.8% of positivity), than for sCL patients (50.0% of positivity).
For DI test and in vitro culture we respectively observed high frequencies of suggestive results (25.0%) and, contamination (36.0%) indicating the limitations of these techniques. Curiously, in vitro culture contamination was 2.9 times more frequent in sML samples (30/55 or 54.5%) than in sCL samples (11/59 or 18.6%).
As only kDNA-PCR was performed on all collected samples, we can compare the efficiency of the three traditional diagnostic methods (DI test, MST and in vitro culture) with PCR. We can see in Table 2 that kDNA-PCR was able to detect the parasite in all samples that were positive for the DI test47, and for in vitro culture 22 . Surprisingly, from the 49 samples considered positive after the MST analysis, one was negative for kDNA-PCR (it was derived from a patient with chronic ATL presenting a recurrent lesion).
Besides that, we verified that the molecular method detected Leishmania DNA in 77 samples considered negative or contaminated by in vitro culture (41 and 36, respectively), and in the 20 samples considered negative or suggestive by the DI test (5 and 15, respectively). Interestingly, from the 29 samples considered negative by MST, 25 (or 86.2%) were positive after kDNA-PCR amplification (17 samples from sCL patients, and eight from sML patients - data not shown). From the 16 samples negative by kDNA-PCR, 15 present the same result when performed by traditional methods; just one sample presents different results for kDNA-PCR and MST, as discussed above.
The importance of the kDNA-PCR in the clinical practice. We divided the 128 DNA samples enrolled in this study into two groups: the confirmed ATL patients (CATL), that presented at least one positive result for traditional diagnosis tests (DI, MST or in vitro culture); and the non-confirmed ATL patients (NCATL), composed by samples with non positive results following the same traditional techniques (respectively with 83 and 45 DNA samples - Table 3). As expected, kDNA-PCR was able to detect parasite DNA in 98.8% of the DNA samples from the CATL group (82/83), where the diagnosis had been previously confirmed by a combination of the three traditional diagnostic methods. The exception was only the patient with chronic ATL with a recurrent lesion, already discussed. The importance of the kDNA-PCR amplification in the routine diagnosis becomes evident when we analyzed the results of the NCATL group. In this group of samples with negative results by the traditional diagnostic methods, kDNA-PCR was able to detect the parasite in 66.6% (or 30/45).
Table 3. Number of ATL suspected samples presenting none or at least one positive result by traditional methods compared with the kDNA-PCR results.
| Result/group | Traditional methods | |||
|---|---|---|---|---|
| Positive* CATL | Negative NCATL | Total | ||
| kDNA-PCR | Positive | 82 (98.8%) | 30 (66.6%) | 112 |
| Negative | 1 (1.2%) | 15 (33.3%) | 16 | |
| Total | 83 | 45 | 128 | |
Positive result for at least one reference method (direct investigation, Montenegro skin test or in vitro culture).
CATL: ATL DNA samples confirmed by at least one of the traditional method(s). NCATL: ATL DNA samples without confirmation by traditional method(s).
To better verify the acceptance of the kDNA-PCR results, we checked the physician follow-up through the analysis of the medical records. Out of 128 samples analyzed, only 92 records presented correct data on the conduct adopted by the physician. According to the records, most of these patients, 95.7% (88/92), received specific treatment for ATL. Of these 88 treated patients, 18 were positive only by kDNA-PCR analysis, 63 presented a positive result in at least one of the traditional diagnostic methods (and also positive kDNA-PCR positive results), and seven patients were treated based exclusively on the clinical examination (when all diagnostic tests, including PCR, presented negative or inconclusive results). Interestingly, one of the seven treated patients that presented a negative result for kDNA-PCR, was later diagnosed as paracoccidioidomycosis, confirming the previous PCR result. Contrary, two patients from this same group responded clinically well to the ATL drug administration, besides the negative results in all diagnosis tests. No information concerning the response of treatment about the remaining four treated patients was found in the records. Overall, 3/92 patients were not treated even though they presented positive kDNA-PCR results (one patient presented positive results for both kDNA-PCR and MST). The remaining untreated patient was negative by kDNA-PCR analysis.
PCR-RFLP results. To verify the contribution of the PCR-RFLP method for identification of L. (V.) braziliensis, HaeIII restriction digestion was performed in all 112 amplification products of the kDNA-PCR reaction. We verified that 96/112 (or 85.7%) of these samples presented the two expected DNA fragments (80 bp and 40 bp), characteristic of the L. (V.) braziliensis electrophoresis pattern 41 (Fig. 1, lanes 2 to 7). The frequency of L. (V.) braziliensis according to the suspected clinical manifestation was 51/61(83.6%) in sCL samples, and 45/51(88.23%) in sML samples.
Fig. 1. A 10% polyacrylamide gel electrophoresis representing the products of PCR-RFLP (kDNA/HaeIII): 1- 7 samples from LTA suspected patients, Lb- DNA from L. (V.) braziliensis (positive control).
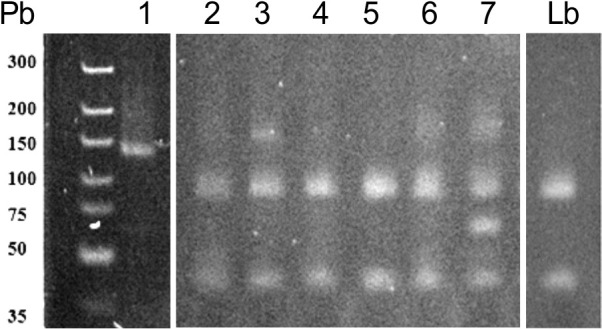
16/112 kDNA-PCR amplification products (10 from sCL samples and six from sML samples) did not present the cleavage of the 120 pb amplification product (Fig. 1, lane 1). Eight samples were from patients that have been in endemic regions of L. (L.) amazonensis: five from Bahia, one from Maranhão, one from Minas Gerais and one from Pernambuco, which can be an evidence of infection caused by this or other Leishmania species. One individual reported to have acquired the disease in Angola where L. (L.) infantum is the main specie that causes leishmaniasis, where specific zymodemes can be related with LC 24 . Four patients have not reported probable local of infection. The remaining three individuals indicated São Paulo and Paraná to be the local of infection, which are states where L. (V.) braziliensis was the only specie causative of human disease. This last result may indicate limitation of the PCR-RFLP or absence of correct information concerning the probable locals of infection.
DISCUSSION
This study evaluated the applicability and efficiency of PCR based on kDNA as a routine diagnostic method for ATL, comparing these results with the results of tests performed routinely for leishmaniasis diagnosis. The data that were obtained indicate that inclusion of the PCR-RFLP (kDNA-HaeIII) technique in the routine diagnosis of ATL would improve the accuracy of the diagnosis, support an appropriate prognosis, and ensure adequate treatment. Moreover, the data indicated that the kDNA-PCR results are in agreement with the clinical practice performance and confirmed the clinical findings (Table 3) that were negative according to the traditional methods (Table 2).
The higher efficiency of the kDNA-PCR method over the traditional methods for ATL diagnosis (Table 1) observed here is in agreement with the literature, which describes sensitivities that range from 75% - 98%, and it is attributed to the naturally amplified DNA in the kinetoplast minicircle 2 4 5 6 14 17 20 30 34 35 40 .
Comparative analysis between the sensitivity levels of the methods tested here and those from previous studies, was difficult to process due to a variety of the techniques and the type of biological samples that were used, the inclusion criteria of the samples and the differences among the sequences of the kDNA primers 34 , and the expertise of the technicians. Nevertheless, we performed a comparison of the efficiency among the kDNA-PCR and traditional methods (Tables 1 and 2) that allowed us to evaluate the limitations of each laboratory method.
The in vitro culture presented a lower percentage of positivity and/or parasite isolation (19.3%, Table 1), than those described by other authors, which ranged from 30.3% to 81.5% 9 17 18 23 . The low sensitivity of the in vitro culture test was evidenced in Table 2 were 77 contaminated or negative samples by this method were positive by kDNA-PCR. On the other hand, these results also indicate that the efficiency of the PCR method was not affected by secondary infections, as pointed by BOGGILD et al. 6 .
Concerning the DI test, several studies have demonstrated that the quality of the prepared slides, the age of the lesions, the presence of secondary microorganisms 6 24 , and pre-treatment of the lesions can lead to atypical forms of the parasite 23 , resulting in a wide variation in the level of sensitivity of the DI tests ranging from 10% to 74.4% 3 4 5 11 15 17 . Corroborating this, we observed that 20 of the 29 samples presenting negative or suggestive results by DI test were positive when evaluated by kDNA-PCR (Table 2). Eleven of these 20 patients received treatment after the positive kDNA-PCR result which confirmed the clinical findings and may indicate the acceptance of the kDNA-PCR for diagnosis of ATL by the physicians. Thus, we suggest that samples with results that are negative or suggestive by DI test require an additional laboratory method, as kDNA-PCR to confirm the final diagnosis.
We verified that MST method was more efficient for sML patients (73.8% of positivity), than for sCL patients (50% of positivity) (Table 1). These low positivity rates are in accordance with the literature, which reports a range from 51.9%-100% for this test 14 16 22 28 , and could be a consequence of immunosuppression 25 or even misinterpretation of the result. It is also reported that the sensitivity of the MST can vary with the presence of secondary infections 6 , or the age of the patient 15 . In our study, we also observed that 25 of the 29 samples presenting negative results by MST were positive when evaluated by kDNA-PCR amplification (Table 2). We noted that 17 of these 25 kDNA-PCR positive samples came from sCL patients, and eight samples from sML patients, which could indicate a recent infection or a weak response from the immune system of the patients 12 25 29 33 . In contrast, one patient that presented positive result for MST was considered negative by kDNA-PCR. This ambiguity may be due to the fact that the patient had a chronic lesion and few or no parasites might be present in the sample subjected to kDNA-PCR reaction. As proposed by DA-CRUZ et al. 12 the inflammatory lesion, in this case, could be due to the activation of T-cells (CD8 +), and not due to the presence of parasites. Detection of false-positive MST results was also reported in 35% (20 of 57) of the patients with reactions up to 11 mm in diameter by GOMES et al. 22 These patients had negative results for in vitro culture, stained tissue smears and PCR for Leishmania's mini exon SL RNA gene. The authors suggested that false-positive results in MST could also be result of an allergic process to the antigen diluent, or an immune response of patients who are not sick, but have already had contact with the parasite in an endemic area 22 36 .
Therefore, we can infer that the limitation factors that have been observed for other laboratory methods, such as contamination by secondary microorganisms 4 33 , the age of infection, cross-reaction to antigens or other reagents that are used in antigen production, the intrinsic characteristics of the parasite, and the pre-treatment of patients 22 does not seem to influence the kDNA-PCR results. In addition, the molecular method has several advantages: complex procedures are not required to collect and to maintain DNA samples for PCR reaction, in contrast to in vitro culture and DI test. Complementary, a wide variety of biological material can be used as sources of DNA, including material obtained by less invasive or non-invasive methods, such as blood 28 34 , lesion impressions on filter paper 4 34 , scraped, and aspirated lesions 6 28 , urine 40 , samples fixed in paraffin 9 and fixed and stained material from glass slides 35 37 . Despite the promising results of the kDNA-PCR method, carry-over contamination must be avoided 14 , and in our experiments, each PCR step was always performed in separate rooms using appropriate equipment.
According to our kDNA-HaeIII PCR-RFLP data, 83.6% of sCL patients could be infected with L. (V.) braziliensis, suggesting that these patients may be at risk of developing the mucosal manifestation or have re-activation of lesions, if they do not receive adequate treatment and clinical assistance. Although kDNA-HaeIII PCR-RFLP reaction can contribute to the identification of L. (V.) braziliensis infected samples, the confirmation of the species of the parasite by other molecular methods is recommended, once similar electrophoretic patterns may occur within related species from Viannia subgenus as reported in other PCR-RFLP studies 2 38 39 .
Studies describing techniques based on PCR for identification of the species from Viannia subgenus using DNA of clinical samples have been reported such as the Polymorphism-Specific PCR (PS-PCR) 26 , G6PD PCR 8 , LBF1-LBR1 PCR 27 . Despite the ability to identify the Leishmania's species, the use of these techniques as routine may not be validated since they require high concentration of total DNA 27 and the employment of several primers for each species 8 26 . On the contrary, the kDNA-HaeIII PCR-RFLP protocol used in our study was simple to execute and presented sensibility higher than the traditional routine methods.
On the other hand, most of the 128 patients analyzed here are from regions where L. (V.) braziliensis is the main causative species responsible for the severe manifestation of the disease, which supports the use of kDNA-HaeIII PCR-RFLP to assist in posterior clinical practices.
The medical reports review has shown that kDNA-PCR results contributed to the treatment of at least 18 patients who had got negative results by the traditional methods. The decision to treat these patients was also based on the clinical symptoms and epidemiological data, confirming that kDNA-PCR is important as a complementary test for the diagnosis and treatment of ATL.
Unfortunately, we failed to collect all results of the traditional diagnostic tests, as well as the response of the patients' treatment, mainly because the medical records were incomplete or lacked important information, which made our comparative analysis difficult (Tables 1, and 3). The difficulty in establishing a gold standard method for the diagnosis of ATL based on a comparison of diagnostic methods has been reported in previous studies 22 . Moreover, we observed that the results of the reference diagnostic methods that were recommended by the National Council of Health of the Brazil Ministry of Health (MST for suspected ML patients, and DI for suspected CL patients) were not registered in the medical records or solicited by the physicians. The same limitations were observed in other studies that were conducted in the following states of Brazil: Pernambuco 36 , Mato Grosso 33 , and Ribeirão Preto, SP 19 . These findings demonstrate that standardization of the tests that are solicited for ATL diagnosis, and improvement or modernization of the medical records systems in public hospitals in Brazil is needed.
Finally, we strongly recommend the use of the kDNA-PCR to be added as routine method for diagnosis of ATL. Patients with suspicion of cutaneous lesions (sCL) after the clinical examination, must collect biopsies of the border lesion, and send for DI test. Patients with suspicion of mucosal lesions (sML) must be submitted for the MST. Patients with positive results according to the DI, and/or MST tests must be treated. kDNA-PCR can be solicited by the physician in addition to DI/MST or performed for disease confirmation when the results of the DI tests, and MST were negative. Patients with positive results for kDNA-PCR can be treated. If suspected leishmaniasis patient present negative results for DI test, MST and kDNA-PCR ATL could be discharged without treatment. The complementary kDNA-HaeIII PCR-RFLP technique can be done to help L. (V.) braziliensis identification of infected samples.
Acknowledgments
We would like to thank Marta Teixeira (Instituto de Ciências Biológicas, Universidade de São Paulo, Brazil) for the reference species of Leishmania, and Regina Maia, and Elisabete Ourique (Instituto de Medicina Tropical de São Paulo) for their technical assistance.
Footnotes
Financial Support
This study was supported by the Fundação de Amparo à Pesquisa do Estado de São Paulo - FAPESP (Process No.: 2010/16963-4), the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), CNPq and the Laboratório de Investigação Médica 38 e 48 (LIM 38 and 48).
REFERENCES
- 1.Al-Jawabreh A, Schnur L, Nasereddin A, Schwenkenbecher J, Abdeen Z, Barghuthy F, et al. The recent emergence of Leishmania tropica in Jericho (A'riha) and its environs, a classical focus of L. major . Trop Med Int Health. 2004;9:812–6. doi: 10.1111/j.1365-3156.2004.01268.x. [DOI] [PubMed] [Google Scholar]
- 2.Andrade RV, Massone C, Lucena MN, Talhari AC, Talhari S, Guerra JAO, et al. The use of polymerase chain reaction to confirm diagnosis in skin biopsies consistent with American tegumentary leishmaniasis at histopathology: a study of 90 cases. An Bras Dermatol. 2011;86:892–6. doi: 10.1590/s0365-05962011000500005. [DOI] [PubMed] [Google Scholar]
- 3.Barrio A, Mora MC, Ramos F, Moreno S, Samson R, Basombrío MA. Short report: use of kDNA-based polymerase chain reaction as a sensitive and differentially diagnostic method of American tegumentary leishmaniasis in disease-endemic areas of northern Argentina. Am J Trop Med Hyg. 2007;77:636–9. [PubMed] [Google Scholar]
- 4.Bensoussan E, Nasereddin A, Jonas F, Schnur LF, Jaffe CL. Comparison of PCR assays for diagnosis of cutaneous leishmaniasis. J Clin Microbiol. 2006;44:1435–9. doi: 10.1128/JCM.44.4.1435-1439.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boggild AK, Ramos AP, Valencia BM, Veland N, Miranda-Verastegui C, et al. Clinical and demographic stratification of test performance: a pooled analysis of five laboratory diagnostic methods for American cutaneous leishmaniasis. Am J Trop Med Hyg. 2010;83:345–50. doi: 10.4269/ajtmh.2010.09-0414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boggild AK, Ramos AP, Valencia BM, Veland N, Calderon F, Arevalo J, et al. Diagnostic performance of filter paper lesion impression PCR for secondarily infected ulcers and nonulcerative lesions caused by cutaneous leishmaniasis. J Clin Microbiol. 2011;49:1097–100. doi: 10.1128/JCM.02457-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brasil . Manual de vigilância da leishmaniose tegumentar americana. In: Gentil K, Pamplona M, Assunção L, Souza F, editors. 2 nd Ed. Brasília: Editora do Ministério da Saúde; 2010. [Google Scholar]
- 8.Castilho T, Shaw J, Floeter-Winter L. New PCR assay using glucose-6-phosphate dehydrogenase for identification of Leishmania species. J Clin Microbiol. 2003;41:540–6. doi: 10.1128/JCM.41.2.540-546.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coelho LI, Paes M, Guerra J, Barbosa M, Coelho C, Lima B, et al. Characterization of Leishmania spp. causing cutaneous leishmaniasis in Manaus, Amazonas, Brazil. Parasitol Res. 2011;108:671–7. doi: 10.1007/s00436-010-2139-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Costa C, de Toledo VP, Genaro O, Williams P, Mayrink W. Montenegro skin test-evaluation of the composition and stability of the antigen preparation. Mem Inst Oswaldo Cruz. 1996;91:193–4. doi: 10.1590/s0074-02761996000200013. [DOI] [PubMed] [Google Scholar]
- 11.Curti MC, Silveira TG, Arraes SM, Bertolini DA, Zanzarini PD, Venazzi E, et al. Epidemiological and clinical characteristics of cutaneous leishmaniasis and their relationship with the laboratory data, south of Brazil. Braz J Infect Dis. 2011;15:12–6. [PubMed] [Google Scholar]
- 12.Da Cruz AM, Bertho AL, Oliveira-Neto MP, Coutinho SG. Flow cytometric analysis of cellular infiltrate from American tegumentary leishmaniasis lesions. Br J Dermatol. 2005;153:537–43. doi: 10.1111/j.1365-2133.2005.06647.x. [DOI] [PubMed] [Google Scholar]
- 13.David CV, Craft N. Cutaneous and mucocutaneous leishmaniasis. Dermatol Ther. 2009;22:491–502. doi: 10.1111/j.1529-8019.2009.01272.x. [DOI] [PubMed] [Google Scholar]
- 14.Degrave W, Fernandes O, Campbell D, Bozza M, Lopez U. Use of molecular probes and PCR for detection and typing of Leishmania - a mini-review. Mem Inst Oswaldo Cruz. 1994;89:463–9. doi: 10.1590/s0074-02761994000300032. [DOI] [PubMed] [Google Scholar]
- 15.Delgado O, Silva S, Coraspe V, Ribas MA, Rodriguez-Morales AJ, Navarro P, et al. American cutaneous leishmaniasis in children and adolescents from northcentral Venezuela. Trop Biomed. 2008;25:178–83. [PubMed] [Google Scholar]
- 16.Diniz J, Costa M, Gonçalves D. Mucocutaneous leishmaniasis: clinical markers in presumptive diagnosis. Braz J Otorhinolaryngol. 2011;77:380–4. doi: 10.1590/S1808-86942011000300018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fagundes A, Schubach A, Paula CC, Bogio A, Antonio L de F, Schiavoni PB, et al. Evaluation of polymerase chain reaction in the routine diagnosis for tegumentary leishmaniasis in a referral centre. Mem Inst Oswaldo Cruz. 2010;105:109–12. doi: 10.1590/s0074-02762010000100018. [DOI] [PubMed] [Google Scholar]
- 18.Farahmand M, Nahrevanian H, Shirazi HA, Naeimi S, Farzanehnejad Z. An overview of a diagnostic and epidemiologic reappraisal of cutaneous leishmaniasis in Iran. Braz J Infect Dis. 2011;15:17–21. [PubMed] [Google Scholar]
- 19.Ferreira M, Roselino A, Nascimento M, Aires J, Figueiredo J. Sensitivity of an immunoenzymatic test for the detection of anti-L. braziliensis antibodies compared to other tests used for the diagnosis of American cutaneous leishmaniasis. Rev Inst Med Trop Sao Paulo. 2006;48:215–7. doi: 10.1590/s0036-46652006000400008. [DOI] [PubMed] [Google Scholar]
- 20.Garcia AL, Kindt A, Quispe-Tintaya K, Bermudez H, Llanos A, Arevalo J, et al. American tegumentary leishmaniasis: antigen-gene polymorphism, taxonomy and clinical pleomorphism. Infect Genet Evol. 2005;5:109–16. doi: 10.1016/j.meegid.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 21.Garcia L, Kindt A, Bermudez H, Llanos-Cuentas A, Doncker SD, Arevalo J, et al. Culture-independent species typing of neotropical Leishmania for clinical validation of a PCR-based assay targeting heat shock protein 70 genes. J Clin Microbiol. 2004;42:2294–97. doi: 10.1128/JCM.42.5.2294-2297.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gomes AHS, Armelin IMI, Menon SSZ, Pereira-Chioccola VL. Leishmania (V.) braziliensis: detection by PCR in biopsies from patients with cutaneous leishmaniasis. Exp Parasitol. 2008;119:319–24. doi: 10.1016/j.exppara.2008.02.014. [DOI] [PubMed] [Google Scholar]
- 23.Gomes AHS, Ferreira IM, Lima ML, Cunha EA, Garcia AS, Araujo MF, et al. PCR identification of Leishmania in diagnosis and control of canine leishmaniasis. Vet Parasitol. 2007;144:234–41. doi: 10.1016/j.vetpar.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 24.Goto H, Lindoso JAL. Current diagnosis and treatment of cutaneous and mucocutaneous leishmaniasis. Expert Rev Anti Infect Ther. 2010;8:419–33. doi: 10.1586/eri.10.19. [DOI] [PubMed] [Google Scholar]
- 25.Lindoso JA, Barbosa RN, Posada-Vergara MP, Duarte MI, Oyafuso LK, Amato VS, et al. Unusual manifestations of tegumentary leishmaniasis in AIDS patients from the New World. Br J Dermatol. 2009;160:311–8. doi: 10.1111/j.1365-2133.2008.08908.x. [DOI] [PubMed] [Google Scholar]
- 26.Marco JD, Barroso PA, Mimori T, Locatelli FN, Tomatani A, Mora MC, et al. Polymorphism-specific PCR enhances the diagnostic performance of American tegumentary leishmaniasis and allows the rapid identification of Leishmania species from Argentina. BMC Infect Dis. 2012;12:191. doi: 10.1186/1471-2334-12-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marcussi VM, Marcussi LM, Barbosa-Tessmann IP, Lonardoni MVC, Silveira TGV. Leishmania (Viannia) braziliensis: new primers for identification using polymerase chain reaction. Exp Parasitol. 2008;120:300–5. doi: 10.1016/j.exppara.2008.08.005. [DOI] [PubMed] [Google Scholar]
- 28.Marques MJ, Volpini AC, Machado-Coelho GL, Machado-Pinto J, Costa CA, Mayrink W, et al. Comparison of polymerase chain reaction with other laboratory methods for the diagnosis of American cutaneous leishmaniasis: diagnosis of cutaneous leishmaniasis. Diagn Microbiol Infect Dis. 2006;54:37–43. doi: 10.1016/j.diagmicrobio.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 29.Martins L, Alexandrino A, Guimarães G. The detection of Leishmania braziliensis DNA in American tegumentary leishmaniasis patients. Rev Saúde Pública. 2010;44:571–4. doi: 10.1590/s0034-89102010005000004. [DOI] [PubMed] [Google Scholar]
- 30.Medeiros A, Rodrigues S, Roselino A. Comparison of the specificity of PCR and the histopathological detection of Leishmania for the diagnosis of American cutaneous leishmaniasis. Braz J Med Biol Res. 2002;35:421–4. doi: 10.1590/s0100-879x2002000400002. [DOI] [PubMed] [Google Scholar]
- 31.Melo MN, Mayrink W, Da Costa CA, Magalhaes PA, Dias M, Williams P, et al. Padronização do antígeno de Montenegro. Rev Inst Med Trop Sao Paulo. 1977;19:161–4. [PubMed] [Google Scholar]
- 32.Mitropoulos P, Konidas P, Durkin-Konidas M. New World cutaneous leishmaniasis: updated review of current and future diagnosis and treatment. J Am Acad Dermatol. 2010;63:309–22. doi: 10.1016/j.jaad.2009.06.088. [DOI] [PubMed] [Google Scholar]
- 33.Murback ND, Hans Filho G, Nascimento RA, Nakazato KR, Dorval ME. American cutaneous leishmaniasis: clinical, epidemiological and laboratory studies conducted at a university teaching hospital in Campo Grande, Mato Grosso do Sul, Brazil. An Bras Dermatol. 2011;86:55–63. doi: 10.1590/s0365-05962011000100007. [DOI] [PubMed] [Google Scholar]
- 34.Oliveira DM, Lonardoni MV, Teodoro U, Silveira TG. Comparison of different primes for PCR-based diagnosis of cutaneous leishmaniasis. Braz J Infect Dis. 2011;15:204–10. doi: 10.1016/s1413-8670(11)70176-3. [DOI] [PubMed] [Google Scholar]
- 35.Oliveira JGS, Novais FO, Oliveira CI, Cruz Junior AC, Campos LF, Rocha AV, et al. Polymerase chain reaction (PCR) is highly sensitive for diagnosis of mucosal leishmaniasis. Acta Trop. 2005;94:55–9. doi: 10.1016/j.actatropica.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 36.Reis LDC, Brito ME, Almeida EL, Félix SM, Medeiros AC, Silva CJ, et al. Clinical, epidemiological and laboratory aspects of patients with American cutaneous leishmaniasis in the State of Pernambuco. Rev Soc Bras Med Trop. 2008;41:439–43. doi: 10.1590/s0037-86822008000500001. [DOI] [PubMed] [Google Scholar]
- 37.Romero GAS, Noronha EF, Pirmez C, Pires FESS, Fernandes O, Nehme NS, et al. Sensitivity and reproducibility of a PCR assay for Leishmania detection using skin biopsy imprints on filter paper. Acta Trop. 2009;109:74–7. doi: 10.1016/j.actatropica.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 38.Schönian G, Nasereddin A, Dinse N, Schweynoch C, Schallig HDFH, Presber W, et al. PCR diagnosis and characterization of Leishmania in local and imported clinical samples. Diagn Microbiol Infect Dis. 2003;47:349–58. doi: 10.1016/s0732-8893(03)00093-2. [DOI] [PubMed] [Google Scholar]
- 39.Silva L, De Sousa C, Da Graça G, Porrozzi R, Cupolillo E. Sequence analysis and PCR-RFLP profiling of the hsp70 gene as a valuable tool for identifying Leishmania species associated with human leishmaniasis in Brazil. Infect Genet Evol. 2010;10:77–83. doi: 10.1016/j.meegid.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 40.Veland N, Espinosa D, Valencia BM, Ramos AP, Calderon F, Arevalo J, et al. Polymerase chain reaction detection of Leishmania kDNA from the urine of Peruvian patients with cutaneous and mucocutaneous leishmaniasis. Am J Trop Med Hyg. 2011;84:556–61. doi: 10.4269/ajtmh.2011.10-0556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Volpini AC, Passos VM, Oliveira GC, Romanha AJ. PCR-RFLP to identify Leishmania (Viannia) braziliensis and L. (Leishmania) amazonensis causing American cutaneous leishmaniasis. Acta Trop. 2004;90:31–7. doi: 10.1016/j.actatropica.2003.10.008. [DOI] [PubMed] [Google Scholar]


