Abstract
Purpose
To study the association of apolipoprotein E (APOE) polymorphisms and primary open-angle glaucoma (POAG).
Methods
After a systematic literature search, all relevant studies evaluating the association between APOE polymorphisms and POAG were included. All statistical tests were calculated with Stata 11.0.
Results
Twelve independent studies on the APOE gene (1,971 cases, 1,756 controls) and POAG were included. A significant association between the APOE gene and POAG was found in the genetic model of ε4/ε4 versus ε3/ε3 (odds ratio [OR] = 2.09, 95% confidence interval [CI] = 1.12–3.88, p = 0.02). However, no association was detected in the models of ε2/ε2 versus ε3/ε3, ε2/ε3 versus ε3/ε3, ε2/ε4 versus ε3/ε3, ε3/ε4 versus ε3/ε3, allele ε2 versus allele ε3, and allele ε4 versus allele ε3. Subgroup analyses showed that a statistically significant association between the APOE gene and the risk of POAG existed in the genetic model of ε4/ε4 versus ε3/ε3 in Asians (OR = 3.55, 95% CI = 1.06–11.87, p = 0.04). No association was identified between the APOE gene and the risk of POAG in Caucasians.
Conclusions
The present meta-analysis indicated that the ε4/ε4 genotype is associated with increased risk of POAG in Asians.
Introduction
Glaucoma, a degenerative group of diseases characterized by visual field loss and optic nerve degenerating changes, is the second major cause of blindness in the world [1]. As a major type of primary glaucoma in most populations, primary open-angle glaucoma (POAG) is defined by an open anterior chamber angle and elevated intraocular pressure (IOP), without other comorbidities [2-4]. However, this disease progresses slowly with concealed symptoms, which are barely detectable until evident and irreversible loss in visual field emerges. Although the pathogenesis of POAG is not fully understood, many previous studies have noted that multiple genes, as well as environmental factors, play vital roles in the development of POAG [5-10]. Thus far, many genetic loci have been predicted to associate with POAG, and among them, only three genes (GLC1A [myocilin, MYOC, OMIM 601652], GLC1E [optineurin, OPTN; OMIM 602432], and GLC1G [WD repeat domain 36, WDR36; OMIM 609669]) have been confirmed [11-15].
Recently, studies have supported the existence of a strong association between Alzheimer disease (AD) and POAG [16,17]. It has also been suggested that loss of retinal ganglion cells and optic nerve degeneration occur in patients with AD [18,19]. The genotype of the apolipoprotein E (APOE;OMIM 107741) gene is one of the major genetic risk factors for AD. Therefore, a new research field has developed to study the associations between POAG and AD by focusing on the APOE gene and its variants. As a player in lipid metabolism, apolipoprotein E (ApoE) plays a vital role in the transportation of cholesterol and triglyceride [20-22]. The human APOE gene is located on chromosome 19q13.2, with three common alleles (ε2, ε3, and ε4) encoding 3 protein isoforms (E2, E3, and E4). As the most common isoform, E3 contains cysteine and arginine at amino acid positions 112 and 158, respectively. In contrast, E2 and E4 contain only cysteine and arginine residues in these positions, respectively. Since each individual inherits one allele from each parent, six possible combinations of genotypes can be generated by three alleles: ε2/ε2, ε2/ε3, ε2/ε4, ε3/ε3, ε3/ε4, and ε4/ε4 [23]. Possible association between the APOE gene and POAG has been investigated in several studies; however, the results were conflicting. To clarify this question, a systematic meta-analysis was performed to ascertain the associations between APOE polymorphisms and the risk of POAG.
Methods
Literature search strategy
The data were obtained from PubMed, Web of Science, Embase, and China National Knowledge Infrastructure (CNKI). The following index terms were used in the search strategy to include all possible studies: (APOE OR apolipoprotein E) AND (primary open-angle glaucoma OR POAG OR high tension glaucoma OR HTG OR normal tension glaucoma OR NTG).
Inclusion criteria and data extraction
The inclusion criteria for the selected articles were as follows: (1) case–control study, (2) reports on the association between APOE polymorphisms and POAG, (3) studies with full text articles, and (4) the number of APOE genotypes/alleles in the case and control groups was calculated. Genotype ε3/ε3 was assigned as the reference group in our study. Thus, seven genetic models were analyzed (ε2/ε2 versus ε3/ε3, ε2/ε3 versus ε3/ε3, ε2/ε4 versus ε3/ε3, ε3/ε4 versus ε3/ε3, ε4/ε4 versus ε3/ε3, allele ε2 versus allele ε3, and allele ε4 versus allele ε3). The articles were reviewed independently by two investigators (Rongfeng Liao and Minjie Ye), who also extracted and evaluated the quality of the data. A third reviewer (Xiping Xu) participated in the investigation and evaluation if there were any disagreements. For each study, the extracted information includes the first author's name, ethnicity (country), publication year, and the number of each allele and genotype in the cases and controls. (See Table 1.)
Table 1. Characteristics of studies included in this meta-analysis.
| First author | Year | Ethnicity (country) | SS(case/control) |
Genotypes distribution (case/control) |
HWE |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ε2/ε2 | ε2/ε3 | ε2/ε4 | ε3/ε3 | ε3/ε4 | ε4/ε4 | ε2 | ε3 | ε4 | P-value (control) | ||||
| Huiping Yuan [26] |
2007 |
Asian(China) |
36/57 |
0/0 |
0/6 |
6/12 |
12/31 |
15/8 |
3/0 |
6/18 |
39/76 |
27/20 |
<0.001 |
| Li Yun Jia [27] |
2009 |
Asian(China) |
176/200 |
2/1 |
25/29 |
5/4 |
112/136 |
29/28 |
3/2 |
34/35 |
280/329 |
38/36 |
0.964 |
| Ching Yan Lam [28] |
2006 |
Asian(China) |
400/300 |
0/0 |
74/42 |
5/8 |
280/203 |
40/47 |
1/0 |
79/50 |
674/495 |
47/55 |
0.124 |
| Fumihiko Mabuchi [29] |
2005 |
Asian(Japan) |
310/179 |
0/0 |
14/18 |
2/0 |
259/123 |
35/38 |
0/0 |
16/18 |
567/302 |
37/38 |
0.188 |
| Yijun Hu [30] |
2007 |
Asian(China) |
142/77 |
1/0 |
11/11 |
4/0 |
95/52 |
28/14 |
3/0 |
17/11 |
229/129 |
38/14 |
0.576 |
| A Jünemann [31] |
2003 |
Caucasian(Germany) |
96/32 |
0/0 |
18/3 |
1/6 |
51/14 |
26/9 |
0/0 |
19/9 |
146/40 |
27/15 |
0.027 |
| E. Saglar [32] |
2009 |
Caucasian(Turkey) |
75/119 |
0/0 |
12/9 |
1/1 |
53/88 |
8/19 |
1/2 |
13/10 |
126/204 |
11/24 |
0.929 |
| Madeleine Zetterberg [33] |
2007 |
Caucasian(Estonia) |
242/187 |
1/2 |
42/34 |
6/4 |
145/110 |
44/35 |
4/2 |
50/42 |
376/289 |
58/43 |
0.976 |
| Al-Dabbagh [34] |
2009 |
Caucasian(Saudi-Arabia) |
60/130 |
0/0 |
0/0 |
0/0 |
50/119 |
7/11 |
3/0 |
0/0 |
107/249 |
13/11 |
NA |
| James C. Vickers [35] |
2002 |
Caucasian(Tasmania) |
142/51 |
6/2 |
8/9 |
7/2 |
78/30 |
42/6 |
1/2 |
27/15 |
208/75 |
51/12 |
0.328 |
| S Lake [36] |
2004 |
Caucasian(America) |
155/349 |
1/3 |
16/37 |
10/13 |
91/208 |
31/81 |
6/7 |
28/56 |
229/534 |
53/108 |
0.472 |
| Thomas Ressiniotis [37] | 2004 | Caucasian(England) | 137/75 | −/− | −/− | −/− | −/− | −/− | −/− | 35/16 | 199/114 | 40/20 | NA |
SS: sample size. −/−: Ressiniotis et al. only provided the frequencies of APOE alleles, the frequencies of APOE genotypes could not be calculated. HWE: Hardy–Weinberg equilibrium; NA: not available.
Statistical analysis
The statistical analyses were performed by using Stata 11.0 (StataCorp, College Station, TX). The association between APOE polymorphisms and risk of POAG was expressed as odds ratio (OR) and 95% confidence interval (CI). The effects of heterogeneity were quantified with the I2 statistic, which detected variations among publications due to heterogeneity rather than chance. All ORs were calculated with the fixed effects model (the Mantel-Haenszel method) or the random effects model (the DerSimonian-Laird method) according to the heterogeneity [24,25]. When there was no heterogeneity among studies, a fixed effects model was applied; otherwise, a random effects model was applied. Subgroup analyses were performed based on ethnicity and POAG subtype. Since genotype distributions of the control group might be important in the studies, a chi-square test was applied to determine if the genotype distributions of the control group reported conformed to Hardy–Weinberg equilibrium (HWE; p≤0.05 was representative of statistical significance). Finally, the funnel plots and Egger’s regression test were used to evaluate publication bias visually.
Results
Characteristics of studies
In Figure 1, the study inclusion process in this meta-analysis is described. Twelve studies (1,971 cases, 1,756 controls) were included [26-37]. Among them, five studies were performed in Asians (1,064 cases and 813 controls) and seven in Caucasians (907 cases and 943 controls). Three studies on Asians and two studies on Caucasians examined the relationships between the APOE gene and high tension POAG (HTG), while three additional studies evaluated the association between the APOE gene and normal tension glaucoma (NTG). The HWE test was performed on the genotype distribution of the controls in all included studies, all of which showed p>0.05 in HWE, except four studies (two [26,31] showed p<0.05; two [34,37] lacked data). Detailed characteristics of each included study are presented in Table 1.
Figure 1.
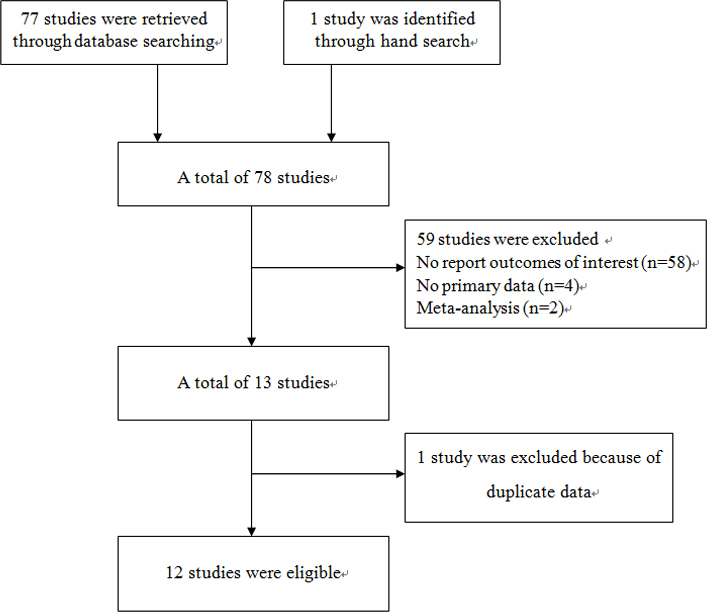
Flow diagram of the study selection process.
Meta-analysis results
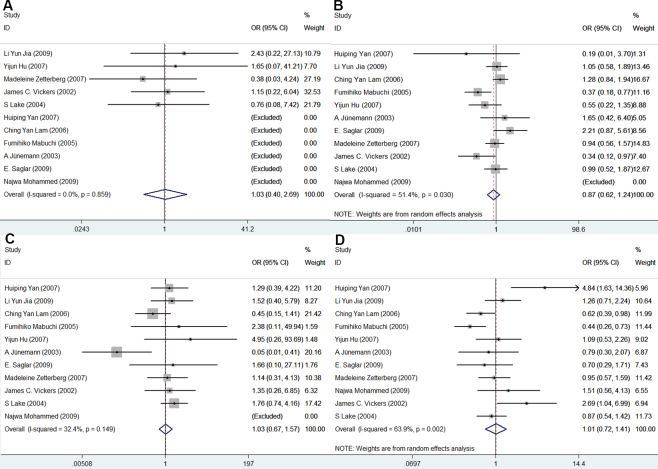
The association between APOE gene polymorphisms and the risk of POAG was statistically significant in the genetic model of ε4/ε4 versus ε3/ε3 (OR = 2.09, 95% CI = 1.12–3.88, p = 0.02; Figure 2). However, compared with the ε3/ε3 genotype, no significant associations were observed in ε2/ε2 (OR = 1.03, 95% CI = 0.40–2.69, p = 0.95; Figure 3A), ε2/ε3 (OR = 0.87, 95% CI = 0.62–1.24, p = 0.44; Figure 3B), ε2/ε4 (OR = 1.03, 95% CI = 0.67–1.57, p = 0.90; Figure 3C), and ε3/ε4 (OR = 1.01, 95% CI = 0.72–1.41, p = 0.97; Figure 3D). There was no significant association between APOE gene polymorphisms and the risk of POAG in the allele ε2 versus allele ε3 (OR = 0.99, 95% CI = 0.83–1.18, p = 0.91; Figure 4A) and the allele ε4 versus allele ε3 (OR = 1.07, 95% CI = 0.81–1.42, p = 0.65; Figure 4B). The meta-analysis results of the association between the APOE gene and the risk of POAG are illustrated in Table 2.
Figure 2.
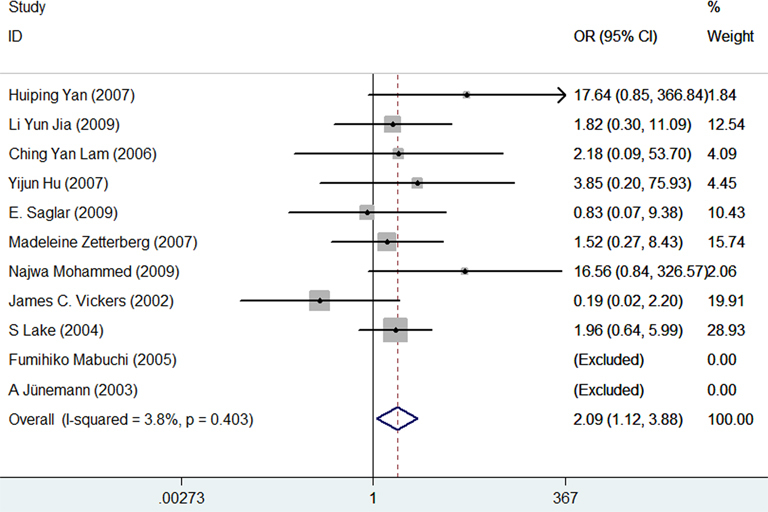
Forest plot for the genetic model of ε4/ε4 vs. ε3/ε3. Every study was represented by a square whose size was proportional to the weight of the study. Diamond indicated summary odds ratios (OR) with its corresponding the pseudo 95% confidence limits (95% CI).
Figure 3.
Forest plots of the association of AOPE polymorphisms with primary open-angle glaucoma. Every study was represented by a square whose size was proportional to the weight of the study. Diamond indicated summary odds ratios (OR) with its corresponding the pseudo 95% confidence limits (95% CI). A: Forest plot for APOE polymorphisms and POAG risk in the genetic model of ε2/ε2 vs. ε3/ε3. B: Forest plot for APOE polymorphisms and POAG risk in the genetic model of ε2/ε3 vs. ε3/ε3. C: Forest plot for APOE polymorphisms and POAG risk in the genetic model of ε2/ε4 vs. ε3/ε3. D: Forest plot for APOE polymorphisms and POAG risk in the genetic model of ε3/ε4 vs. ε3/ε3.
Figure 4.
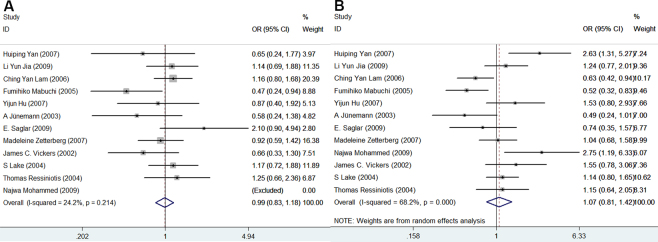
Forest plots for APOE polymorphisms and primary open-angle glaucoma risk in the genetic models of ε2 allele vs. ε3 allele and ε4 allele vs. ε3 allele. Every study was represented by a square whose size was proportional to the weight of the study. Diamond indicated summary odds ratios (OR) with its corresponding the pseudo 95% confidence limits (95% CI). A: Forest plot for APOE polymorphisms and POAG risk in the genetic model of ε2 allele vs. ε3 allele. B: Forest plot for APOE polymorphisms and POAG risk in the genetic model of ε4 allele vs. ε3 allele.
Table 2. Results of meta-analysis for APOE gene polymorphism and risk for primary open-angle glaucoma.
| Category | ε2/ε2 versus ε3/ε3 OR (95%CI) P value (Model*) | ε2/ε3 versus ε3/ε3 OR (95%CI) P value (Model*) | ε2/ε4 versus ε3/ε3 OR (95%CI) P value (Model*) | ε3/ε4 versus ε3/ε3 OR (95%CI) P value (Model*) | ε4/ε4 versus ε3/ε3 OR (95%CI) P value (Model*) | ε2allele versus ε3 allele OR (95%CI) P value (Model*) | ε4 allele versus ε3 allele OR (95%CI) P value (Model*) | Reference |
|---|---|---|---|---|---|---|---|---|
| POAG: |
1.03(0.40–2.69)
0.95F |
0.87(0.62–1.24)
0.44R |
1.03(0.67–1.57)
0.90F |
1.01(0.72–1.41)
0.97R |
2.09(1.12–3.88)
0.02F |
0.99(0.83–1.18)
0.91F |
1.07(0.81–1.42)
0.65R |
26–37 |
| Subgroup: | ||||||||
| Asian |
2.10(0.30–14.63)
0.45F |
0.74(0.42–1.29)
0.29R |
1.09(0.58–2.05)
0.79F |
1.00(0.54–1.87)
0.99R |
3.55(1.06–11.87)
0.04F |
0.96(0.75–1.23)
0.77F |
1.06(0.61–1.82)
0.84R |
26–30 |
| Caucasian |
0.79(0.26–2.44)
0.68F |
1.00(0.72–1.39)
0.99F |
0.87(0.30–2.57)
0.80R |
1.02(0.77–1.34)
0.90F |
1.65(0.79–3.45)
0.19F |
1.02(0.80–1.30)
0.90F |
1.10(0.89–1.35)
0.37F |
31–37 |
| HTG |
1.14(0.28–4.59)
0.85F |
0.91(0.52–1.57)
0.73R |
0.69(0.20–2.36)
0.55R |
0.93(0.69–1.25)
0.63F |
1.19(0.39–3.61)
0.76F |
0.87(0.56–1.36)
0.53R |
0.96(0.66–1.41)
0.84R |
27, 28, 30, 31, 35 |
| NTG | 1.53(0.42–5.59) 0.52F | 0.80(0.48–1.33) 0.38F | 1.86(0.87–4.01) 0.11F | 1.37(0.67–2.81) 0.40R | 1.66(0.65–4.26) 0.29F | 1.06(0.73–1.55) 0.77F | 1.30(0.97–1.76) 0.08F | 30, 35, 36 |
*If the results of the studies were heterogeneous, the random effects model was used for meta-analysis; otherwise, the fixed-effects model was used. F means fixed-effects model; R means random effects model. POAG: primary open-angle glaucoma. HTG: high tension glaucoma. NTG: normal tension glaucoma.
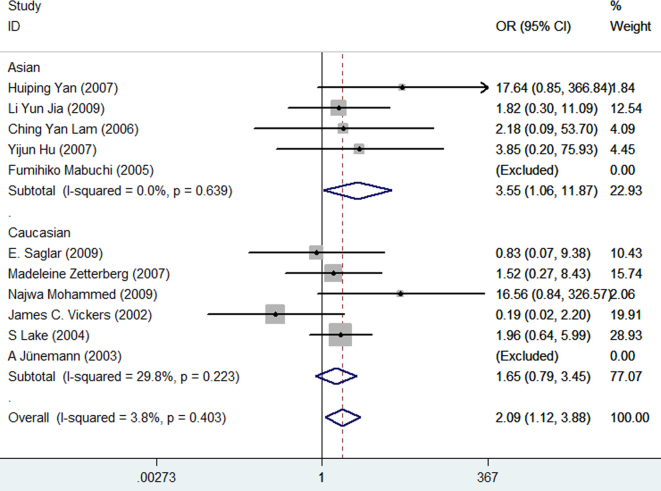
Furthermore, subgroup analyses were conducted on ethnicity and subtypes of POAG (HTG, NTG). The association between the APOE gene and the risk of POAG was statistically significant in Asians in the genetic model of ε4/ε4 versus ε3/ε3 (OR = 3.55, 95% CI = 1.06–11.87, p = 0.04; Figure 5) but not in Caucasians (OR = 1.65, 95% CI = 0.79–3.45, p = 0.19; Figure 5). No results showed a significant association between the APOE gene and POAG in other genetic models in Asians and Caucasians. Similarly, we did not find any correlation between APOE and HTG or NTG. The results of the subgroup analyses are illustrated in Table 2.
Figure 5.
Subgroup analysis stratified by ethnicity in the genetic model of ε4/ε4 vs. ε3/ε3. Every study was represented by a square whose size was proportional to the weight of the study. Diamond indicated summary odds ratios (OR) with its corresponding the pseudo 95% confidence limits (95% CI).
Potential publication bias
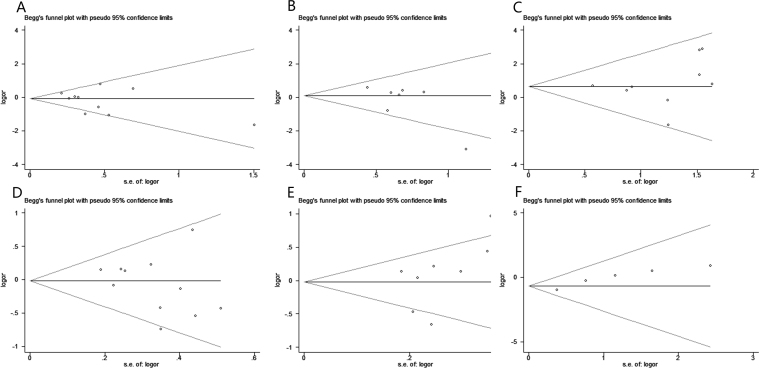
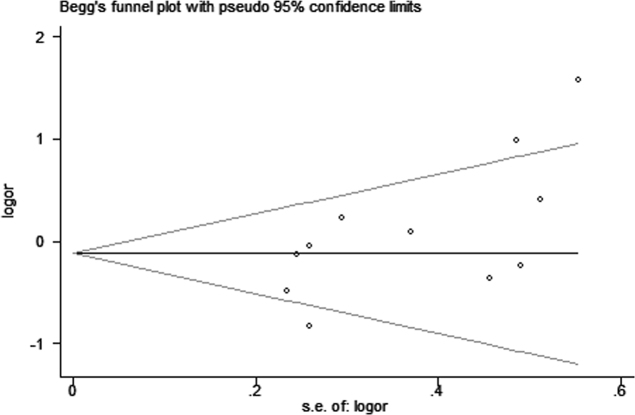
Funnel plots and Egger’s test were applied to assess potential publication bias for APOE. The genetic models of ε2/ε3 versus ε3/ε3 (p = 0.293; Figure 6A), ε2/ε4 versus ε3/ε3 (p = 0.780; Figure 6B), ε4/ε4 versus ε3/ε3 (p = 0.560; Figure 6C), allele ε2 versus allele ε3 (p = 0.267; Figure 6D), and allele ε4 versus allele ε3 (p = 0.255; Figure 6E) revealed no publication bias among the studies. However, publication bias was observed in the genetic models of ε2/ε2 versus ε3/ε3 (p = 0.008; Figure 6F), as well as ε3/ε4 versus ε3/ε3 (p = 0.034; Figure 7).
Figure 6.
Begg’s funnel plots of publication bias analyses. The horizontal line in the figure means the overall estimated log-transformed odds ratio (OR) and the two diagonal lines represent the pseudo 95% confidence limits of the effect estimate(95% CI). A: Funnel plot for the genetic model of ε2/ε3 vs. ε3/ε3. B: Funnel plot for the genetic model of ε2/ε4 vs. ε3/ε3. C: Funnel plot for the genetic model of ε4/ε4 vs. ε3/ε3. D: Funnel plot for the genetic model of ε2 allele vs. ε3 allele. E: Funnel plot for the genetic model of ε4 allele vs. ε3 allele. F: Funnel plot for the genetic model of ε2/ε2 vs. ε3/ε3.
Figure 7.
Begg’s Funnel plot for the meta-analysis of the genetic model of ε3/ε4 vs. ε3/ε3. The horizontal line in the figure means the overall estimated log-transformed odds ratio (OR) and the two diagonal lines represent the pseudo 95% confidence limits of the effect estimate(95% CI).
Discussion
Genetic factors are major factors in the development of POAG. Previously, several studies investigated the association between APOE gene polymorphisms and POAG, but the results were controversial. Recently, two meta-analysis studies were performed, and both indicated no association between the APOE gene and the POAG risk. Song et al. [38] conducted a meta-analysis based on nine case-control studies to evaluate the association between the APOE gene ε2/ε3/ε4 polymorphism and the risk of POAG. However, some issues must be addressed: (1) The study included two “eligible studies” that were published that may have used the same case series [28,39]. (2) The eligible studies of the meta-analysis included a study that evaluated the association between the APOE gene and patients who had POAG and AD [40]. However, since the APOE ε4 allele is regarded as a major risk for AD, the frequencies of the APOE genotypes may be affected by AD in patients who also have POAG. These issues may imply that the results of this meta-analysis were not completely accurate. Wang et al. [41] performed a similar meta-analysis that had the same eligible studies as our study, but they evaluated only the genetic models of the allele ε2 versus allele ε3, allele ε4 versus allele ε3, e2 carriers versus allele ε3, and e4 carriers versus allele ε3, and ignored the functions of the genotypes of the APOE gene. Thus, we performed an updated meta-analysis to better ascertain the role of APOE gene polymorphisms in POAG pathogenesis.
Apolipoprotein E (ApoE) is one of the major apolipoproteins in the central nervous system. Compared with ApoE2 and ApoE3, neurons have a lower cholesterol uptake rate and a less efficient cholesterol efflux when lipids are bound to ApoE4. Expression of ApoE3, but not ApoE4, protects neurons against excitotoxin-induced neuronal damage and age-dependent neurodegeneration [42]. Individuals with APOE ε4 have severe amyloid plaque, neurofibrillary tangle pathology, and increased mitochondrial damage compared to individuals with other APOE polymorphisms [43]. Previous studies have shown that the ε4 allele has been linked to central nervous diseases, such as Parkinson disease, Alzheimer disease, and amyotrophic lateral sclerosis [44-46]. In fact, POAG can be considered a neurodegenerative disease as well [47]. In the retina, retinal ganglion cells and optic nerve axons are vulnerable to degeneration. ApoE proteins are synthesized by Müller cells, absorbed by retinal ganglion cells, and transported to the optic nerve, which may play an important role in retinal ganglion cell metabolism and neuronal survival [48]. Copin et al. reported that the APOE promoter gene polymorphism affected visual field loss and optic nerve damage [49]. Therefore, the pathogenic mechanisms of POAG may also be linked to the ε4 allele.
Our study showed that the risk of development of POAG in ε4/ε4 genotype carriers was 2.09 fold higher than in individuals with the ε3/ε3 genotype. However, there was no significant association between APOE gene polymorphisms and the risk of POAG in the allele ε4 versus allele ε3. Therefore, the ε4/ε4 genotype of APOE is a possible genetic predisposition factor for POAG. To further investigate the association between the allele ε4 and POAG risk, the following settings were used. Similar to other studies, we defined individuals who have the ε2/ε2 and ε2/ε3 genotypes as carriers of the ε2 allele, and individuals with the ε3/ε4 and ε4/ε4 genotypes as carriers of the ε4 allele; we chose the carriers of the ε3/ε3 genotype as the reference group [50]. Although there was no direct evidence of any association between APOE gene polymorphisms and the risk of POAG in the carriers of ε2 allele versus ε3/ε3 (OR = 0.95, 95% CI = 0.76–1.17, p = 0.61) and the carriers of ε4 allele versus ε3/ε3 (OR = 1.07, 95% CI = 0.76–1.52, p = 0.69). Moreover, we investigated the association between the APOE gene and risk of POAG/NTG/HTG in the genetic model of ε4 carrier versus non-ε4 carrier. The results illustrated that there was no association between APOE gene polymorphisms and the risk of POAG in the genetic model of ε4 carrier versus non-ε4 carrier (OR = 1.05, 95% CI = 0.75–1.48, p = 0.77). Similarly, we did not find any correlation between APOE and HTG or NTG in the genetic model of ε4 carrier versus non-ε4 carrier (OR = 0.99, 95% CI = 0.62–1.61, p = 0.98; OR = 1.29, 95% CI = 0.92–1.81, p = 0.14, respectively).
Why was the risk for POAG associated with the ε4/ε4 genotype but not with the ε4 allele? There were several possible explanations for this discrepancy: (1) Compared with ε2/ε4 and ε3/ε4, the ε4/ε4 alleles encode only ApoE4. The homozygote ε4 carriers do not have protection from ApoE2 and ApoE3 proteins. As a result, these carriers are susceptible to glaucoma. (2) Subjects with homozygote ε4 may be more susceptible to POAG than those with only one ε4 allele, which is supported by the study by Corder et al., who claimed that the effects of the ε4 allele dose are associated with increased risk for AD [45]. Similarly, Schmechel et al. also noted that patients with two ε4 alleles exhibited a distinct neuropathological phenotype compared with other patients [51].
The present meta-analysis suggested that the genotype ε4/ε4 of APOE increases the risk of POAG in Asians but not in Caucasians, which may be related to differences in lifestyle, environmental factors, nutrition, and genetic factors. No significant differences were observed between the APOE gene and the risk of HTG and NTG, probably because of the small size of the samples and limited trials.
In our study, we detected heterogeneities in meta-analyses of ε2/ε3 versus ε3/ε3, ε3/ε4 versus ε3/ε3, and ε4 allele versus ε3 allele. The heterogeneities could be due to the sample sizes, diversity in study designs, inclusion criteria, and genotyping methods. Since the subjects came from different populations that perhaps have genetic heterogeneity, subgroup analyses were conducted on ethnicity. The results revealed no heterogeneity in the majority of the genetic models, except the models of ε2/ε3 versus ε3/ε3, ε3/ε4 versus ε3/ε3, and ε4 allele versus ε3 allele among Asians and the model of ε2/ε3 versus ε3/ε3 among Caucasians. The overall analysis involved two subtypes of POAG (NTG and HTG), which have possible differences in etiopathogenesis and genetic risks, and they could be another factor that causes heterogeneities.
Some limitations of our study should be considered. First, our meta-analysis included only studies with accessible full-text articles, in English or Chinese. Therefore, missing some otherwise eligible studies that were reported in other languages could lead to inevitable publication bias in the results. Second, due to the lack of detailed data in the primary articles, subgroup analysis was not conducted according to factors such as age and gender. Third, Asian and Caucasian populations possess a low frequency of the APOE ε4 allele, especially the homozygote ε4 [52,53]. In several included studies, neither the case nor control groups involved ε2/ε2 or ε4/ε4 genotypes. These studies were excluded when we performed analyses in the genetic models of ε2/ε2 versus ε3/ε3 and ε4/ε4 versus ε3/ε3, which reduced the overall sample size. Our results indicated that the ε4/ε4 genotype is associated with increased risk for POAG in Asians. With a small number of cases/controls carried homozygote ε4, more research that supports our results is needed. Last, the included studies lack data about potential gene–gene interactions. Since the roles of several genes in the pathogenesis of POAG have been established, further investigations should be performed in this direction.
In summary, the present meta-analysis suggested that ε4/ε4 is associated with increased risk of POAG in Asian populations but not in Caucasian populations. Further studies are required to further clarify the associations between APOE polymorphisms and genetic predisposition for POAG.
Acknowledgments
We thank Bo Chen, Lei Cao and Jingjing Tong for helpful comments.
References
- 1.Coleman AL, Brigatti L. The glaucomas. Minerva Med. 2001;92:365–79. [PubMed] [Google Scholar]
- 2.Quigley HA, Broman AT. The number of people with glaucoma worldwide in 2010 and 2020. Br J Ophthalmol. 2006;90:262–7. doi: 10.1136/bjo.2005.081224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cedrone C, Mancino R, Cerulli A, Cesareo M, Nucci C. Epidemiology of primary glaucoma: prevalence, incidence, and blinding effects. Prog Brain Res. 2008;173:3–14. doi: 10.1016/S0079-6123(08)01101-1. [DOI] [PubMed] [Google Scholar]
- 4.Rouland JF, Berdeaux G, Lafuma A. The economic burden of glaucoma and ocular hypertension: implications for patient management: a review. Drugs Aging. 2005;22:315–21. doi: 10.2165/00002512-200522040-00004. [DOI] [PubMed] [Google Scholar]
- 5.Bron A, Chaine G, Villain M, Colin J, Nordmann JP, Renard JP, Rouland JF. Risk factors for primary open-angle glaucoma. J Fr Ophtalmol. 2008;31:435–44. doi: 10.1016/s0181-5512(08)71443-8. [DOI] [PubMed] [Google Scholar]
- 6.Charliat G, Jolly D, Blanchard F. Genetic risk factor in primary open-angle glaucoma: a case-control study. Ophthalmic Epidemiol. 1994;1:131–8. doi: 10.3109/09286589409047221. [DOI] [PubMed] [Google Scholar]
- 7.van Koolwijk LM, Despriet DD, van Duijn CM, Pardo Cortes LM, Vingerling JR, Aulchenko YS, Oostra BA, Klaver CC, Lemij HG. Genetic contributions to glaucoma: heritability of intraocular pressure, retinal nerve fiber layer thickness, and optic disc morphology. Invest Ophthalmol Vis Sci. 2007;48:3669–76. doi: 10.1167/iovs.06-1519. [DOI] [PubMed] [Google Scholar]
- 8.Liu Y, Allingham RR. Molecular genetics in glaucoma. Exp Eye Res. 2011;93:331–9. doi: 10.1016/j.exer.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zanon-Moreno V, Garcia-Medina JJ, Zanon-Viguer V, Moreno-Nadal MA, Pinazo-Duran MD. Smoking, an additional risk factor in elder women with primary open-angle glaucoma. Mol Vis. 2009;15:2953–9. [PMC free article] [PubMed] [Google Scholar]
- 10.Ozcura F, Aydin S. Is diabetes mellitus a risk factor or a protector for primary open angle glaucoma? Med Hypotheses. 2007;69:233–4. doi: 10.1016/j.mehy.2006.11.030. [DOI] [PubMed] [Google Scholar]
- 11.Stone EM, Fingert JH, Alward WL, Nguyen TD, Polansky JR, Sunden SL, Nishimura D, Clark AF, Nystuen A, Nichols BE, Mackey DA, Ritch R, Kalenak JW, Craven ER, Sheffield VC. Identification of a gene that causes primary open angle glaucoma. Science. 1997;275:668–70. doi: 10.1126/science.275.5300.668. [DOI] [PubMed] [Google Scholar]
- 12.Budde WM. Heredity in primary open-angle glaucoma. Curr Opin Ophthalmol. 2000;11:101–6. doi: 10.1097/00055735-200004000-00006. [DOI] [PubMed] [Google Scholar]
- 13.Rezaie T, Child A, Hitchings R, Brice G, Miller L, Coca-Prados M, Héon E, Krupin T, Ritch R, Kreutzer D, Crick RP, Sarfarazi M. Adult-onset primary open-angle glaucoma caused by mutations in optineurin. Science. 2002;295:1077–9. doi: 10.1126/science.1066901. [DOI] [PubMed] [Google Scholar]
- 14.Monemi S, Spaeth G, DaSilva A, Popinchalk S, Ilitchev E, Liebmann J, Ritch R, Héon E, Crick RP, Child A, Sarfarazi M. Identification of a novel adult-onset primary open-angle glaucoma (POAG) gene on 5q22.1. Hum Mol Genet. 2005;14:725–33. doi: 10.1093/hmg/ddi068. [DOI] [PubMed] [Google Scholar]
- 15.Fan BJ, Wang DY, Lam DS, Pang CP. Gene mapping for primary open angle glaucoma. Clin Biochem. 2006;39:249–58. doi: 10.1016/j.clinbiochem.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 16.Tsolaki F, Gogaki E, Tiganita S, Skatharoudi C, Lopatatzidi C, Topouzis F, Tsolaki M. Alzheimer's disease and primary open-angle glaucoma: is there a connection? Clin Ophthalmol. 2011;5:887–90. doi: 10.2147/OPTH.S22485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bayer AU, Ferrari F. Severe progression of glaucomatous optic neuropathy in patients with Alzheimer's disease. Eye (Lond) 2002;16:209–12. doi: 10.1038/sj.eye.6700034. [DOI] [PubMed] [Google Scholar]
- 18.Blanks JC, Hinton DR, Sadun AA, Miller CA. Retinal ganglion cell degeneration in Alzheimer's disease. Brain Res. 1989;501:364–72. doi: 10.1016/0006-8993(89)90653-7. [DOI] [PubMed] [Google Scholar]
- 19.Hinton DR, Sadun AA, Blanks JC, Miller CA. Optic-nerve degeneration in Alzheimer's disease. N Engl J Med. 1986;315:485–7. doi: 10.1056/NEJM198608213150804. [DOI] [PubMed] [Google Scholar]
- 20.Ordovas JM, Schaefer EJ. Genetic determinants of plasma lipid response to dietary intervention: the role of the APOA1/C3/A4 gene cluster and the APOE gene. Br J Nutr. 2000;83(Suppl 1):S127–36. doi: 10.1017/s0007114500001069. [DOI] [PubMed] [Google Scholar]
- 21.Eichner JE, Dunn ST, Perveen G, Thompson DM, Stewart KE, Stroehla BC. Apolipoprotein E polymorphism and cardiovascular disease: a HuGE review. Am J Epidemiol. 2002;155:487–95. doi: 10.1093/aje/155.6.487. [DOI] [PubMed] [Google Scholar]
- 22.Dallongeville J, Lussier-Cacan S, Davignon J. Modulation of plasma triglyceride levels by apoE phenotype: a meta-analysis. J Lipid Res. 1992;33:447–54. [PubMed] [Google Scholar]
- 23.Lahiri DK, Sambamurti K, Bennett DA. Apolipoprotein gene and its interaction with the environmentally driven risk factors: molecular, genetic and epidemiological studies of Alzheimer's disease. Neurobiol Aging. 2004;25:651–60. doi: 10.1016/j.neurobiolaging.2003.12.024. [DOI] [PubMed] [Google Scholar]
- 24.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719–48. [PubMed] [Google Scholar]
- 25.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–88. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 26.Yuan HP, Xiao Z, Yang BB. A study on the association of apolipoprotein E genotypes with primary open-angle glaucoma and primary angle-closure glaucoma in northeast of China. Zhonghua Yan Ke Za Zhi. 2007;43:416–20. [PubMed] [Google Scholar]
- 27.Jia LY, Tam PO, Chiang SW, Ding N, Chen LJ, Yam GH, Pang CP, Wang NL. Multiple gene polymorphisms analysis revealed a different profile of genetic polymorphisms of primary open-angle glaucoma in northern Chinese. Mol Vis. 2009;15:89–98. [PMC free article] [PubMed] [Google Scholar]
- 28.Lam CY, Fan BJ, Wang DY, Tam PO, Yung Tham CC, Leung DY, Ping Fan DS, Chiu Lam DS, Pang CP. Association of apolipoprotein E polymorphisms with normal tension glaucoma in a Chinese population. J Glaucoma. 2006;15:218–22. doi: 10.1097/01.ijg.0000212217.19804.a7. [DOI] [PubMed] [Google Scholar]
- 29.Mabuchi F, Tang S, Ando D, Yamakita M, Wang J, Kashiwagi K, Yamagata Z, Iijima H, Tsukahara S. The apolipoprotein E gene polymorphism is associated with open angle glaucoma in the Japanese population. Mol Vis. 2005;11:609–12. [PubMed] [Google Scholar]
- 30.Hu YJ. The APOE gene and its interactions with SNPs of other genes in primary open angle glaucoma and age-related macular degeneration. China. Joint Shantou International Eye Center of Shantou University and Chinese University of Hongkong 2007. [Google Scholar]
- 31.Jünemann A, Bleich S, Reulbach U, Henkel K, Wakili N, Beck G, Rautenstrauss B, Mardin C, Naumann GO, Reis A, Kornhuber J. Prospective case control study on genetic assocation of apolipoprotein epsilon2 with intraocular pressure. Br J Ophthalmol. 2004;88:581–2. doi: 10.1136/bjo.2003.020305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saglar E, Yucel D, Bozkurt B, Ozgul RK, Irkec M, Ogus A. Association of polymorphisms in APOE, p53, and p21 with primary open-angle glaucoma in Turkish patients. Mol Vis. 2009;15:1270–6. [PMC free article] [PubMed] [Google Scholar]
- 33.Zetterberg M, Tasa G, Palmér MS, Juronen E, Teesalu P, Blennow K, Zetterberg H. Apolipoprotein E polymorphisms in patients with primary open-angle glaucoma. Am J Ophthalmol. 2007;143:1059–60. doi: 10.1016/j.ajo.2007.01.031. [DOI] [PubMed] [Google Scholar]
- 34.Al-Dabbagh NM, Al-Dohayan N, Arfin M, Tariq M. Apolipoprotein E polymorphisms and primary glaucoma in Saudis. Mol Vis. 2009;15:912–9. [PMC free article] [PubMed] [Google Scholar]
- 35.Vickers JC, Craig JE, Stankovich J, McCormack GH, West AK, Dickinson JL, McCartney PJ, Coote MA, Healey DL, Mackey DA. The apolipoprotein epsilon4 gene is associated with elevated risk of normal tension glaucoma. Mol Vis. 2002;8:389–93. [PubMed] [Google Scholar]
- 36.Lake S, Liverani E, Desai M, Casson R, James B, Clark A, Salmon JF. Normal tension glaucoma is not associated with the common apolipoprotein E gene polymorphisms. Br J Ophthalmol. 2004;88:491–3. doi: 10.1136/bjo.2003.023366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ressiniotis T, Griffiths PG, Birch M, Keers S, Chinnery PF. The role of apolipoprotein E gene polymorphisms in primary open-angle glaucoma. Arch Ophthalmol. 2004;122:258–61. doi: 10.1001/archopht.122.2.258. [DOI] [PubMed] [Google Scholar]
- 38.Song Q, Chen P, Liu Q. Role of the APOE ε2/ε3/ε4 polymorphism in the development of primary open-angle glaucoma: evidence from a comprehensive meta-analysis. PLoS ONE. 2013;8:e82347. doi: 10.1371/journal.pone.0082347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fan BJ, Wang DY, Fan DS, Tam PO, Lam DS, Tham CC, Lam CY, Lau TC, Pang CP. SNPs and interaction analyses of myocilin, optineurin, and apolipoprotein E in primary open angle glaucoma patients. Mol Vis. 2005;11:625–31. [PubMed] [Google Scholar]
- 40.Tamura H, Kawakami H, Kanamoto T, Kato T, Yokoyama T, Sasaki K, Izumi Y, Matsumoto M, Mishima HK. High frequency of open-angle glaucoma in Japanese patients with Alzheimer's disease. J Neurol Sci. 2006;246:79–83. doi: 10.1016/j.jns.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 41.Wang W, Zhou M, Huang W, Chen S, Zhang X. Lack of association of apolipoprotein E (Apo E) ε2/ε3/ε4 polymorphisms with primary open-angle glaucoma: a meta-analysis from 1916 cases and 1756 controls. PLoS ONE. 2013;8:e72644. doi: 10.1371/journal.pone.0072644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Buttini M, Orth M, Bellosta S, Akeefe H, Pitas RE, Wyss-Coray T, Mucke L, Mahley RW. Expression of human apolipoprotein E3 or E4 in the brains of Apoe−/− mice: isoform-specific effects on neurodegeneration. J Neurosci. 1999;19:4867–80. doi: 10.1523/JNEUROSCI.19-12-04867.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bekris LM, Yu CE, Bird TD, Tsuang DW. Genetics of Alzheimer disease. J Geriatr Psychiatry Neurol. 2010;23:213–27. doi: 10.1177/0891988710383571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Benjamin R, Leake A, Edwardson JA, McKeith IG, Ince PG, Perry RH, Morris CM. Apolipoprotein E genes in Lewy body and Parkinson’s disease. Lancet. 1994;343:1565. doi: 10.1016/s0140-6736(94)92961-0. [DOI] [PubMed] [Google Scholar]
- 45.Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, Roses AD, Haines JL, Pericak-Vance MA. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science. 1993;261:921–3. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- 46.Drory VE, Birnbaum M, Korczyn AD, Chapman J. Association of APOE epsilon4 allele with survival in amyotrophic lateral sclerosis. J Neurol Sci. 2001;190:17–20. doi: 10.1016/s0022-510x(01)00569-x. [DOI] [PubMed] [Google Scholar]
- 47.Schumer RA, Podos SM. The nerve of glaucoma! Arch Ophthalmol. 1994;112:37–44. doi: 10.1001/archopht.1994.01090130047015. [DOI] [PubMed] [Google Scholar]
- 48.Amaratunga A, Abraham CR, Edwards RB, Sandell JH, Schreiber BM, Fine RE. Apolipoprotein E is synthesized in the retina by Muller glial cells, secreted into the vitreous, and rapidly transported into the optic nerve by retinal ganglion cells. J Biol Chem. 1996;271:5628–32. doi: 10.1074/jbc.271.10.5628. [DOI] [PubMed] [Google Scholar]
- 49.Copin B, Brezin AP, Valtot F, Dascotte JC, Bechetoille A, Garchon HJ. Apolipoprotein E-promoter single-nucleotide polymorphisms affect the phenotype of primary open-angle glaucoma and demonstrate interaction with the myocilin gene. Am J Hum Genet. 2002;70:1575–81. doi: 10.1086/340733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Song Y, Stampfer MJ, Liu S. Meta-analysis: apolipoprotein E genotypes and risk for coronary heart disease. Ann Intern Med. 2004;141:137–47. doi: 10.7326/0003-4819-141-2-200407200-00013. [DOI] [PubMed] [Google Scholar]
- 51.Schmechel DE, Saunders AM, Strittmatter WJ, Crain BJ, Hulette CM, Joo SH, Pericak-Vance MA, Goldgaber D, Roses AD. Increased amyloid beta-peptide deposition in cerebral cortex as a consequence of apolipoprotein E genotype in late-onset Alzheimer disease. Proc Natl Acad Sci USA. 1993;90:9649–53. doi: 10.1073/pnas.90.20.9649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van der Flier WM, Pijnenburg YA, Schoonenboom SN, Dik MG, Blankenstein MA, Scheltens P. Distribution of APOE genotypes in a memory clinic cohort. Dement Geriatr Cogn Disord. 2008;25:433–8. doi: 10.1159/000124750. [DOI] [PubMed] [Google Scholar]
- 53.Borenstein AR, Mortimer JA. Ding Ding, Schellenberg GD, DeCarli C, Qianhua Zhao, Copenhaver C, Qihao Guo, Shugang Chu, Galasko D, Salmon DP, Qi Dai, Yougui Wu, Petersen R, Zhen Hong. Effects of apolipoprotein E-epsilon4 and -epsilon2 in amnestic mild cognitive impairment and dementia in Shanghai: SCOBHI-P. Am J Alzheimers Dis Other Demen. 2010;25:233–8. doi: 10.1177/1533317509357736. [DOI] [PMC free article] [PubMed] [Google Scholar]