Summary
The Pseudomonas aeruginosa Wsp signal transduction system produces cyclic-di-GMP (c-di-GMP), an intracellular messenger that stimulates biofilm formation and suppresses motility. The Wsp system is homologous to chemotaxis systems and includes a membrane-bound receptor protein, WspA, and a response regulator GGDEF protein, WspR, that catalyses c-di-GMP synthesis when phosphorylated. We found that the subcellular distributions of fluorescent protein-tagged WspA and WspR differed markedly from their chemotaxis counterparts. WspA–YFP formed patches in cells whereas WspR–YFP was dispersed when unphosphorylated and formed bright cytoplasmic clusters when phosphorylated. WspR formed clusters in cells of a ΔwspF mutant, a genetic background that causes constitutive phosphorylation of WspR, but was dispersed in cells of a wspA mutant, a genetic background necessary for WspR phosphorylation. In addition, WspR mutated at Asp70, its predicted site of phosphorylation, did not form clusters. C-di-GMP synthesis was not required for cluster formation. WspR–YFP was dispersed in liquid-grown wild-type cells, but formed clusters that sometimes appeared and disappeared over the course of a few minutes in cells grown on an agar surface. Our results suggest that the compartmentalized production of c-di-GMP in response to a stimulus associated with growth on a surface is an important functional characteristic of the Wsp system.
Introduction
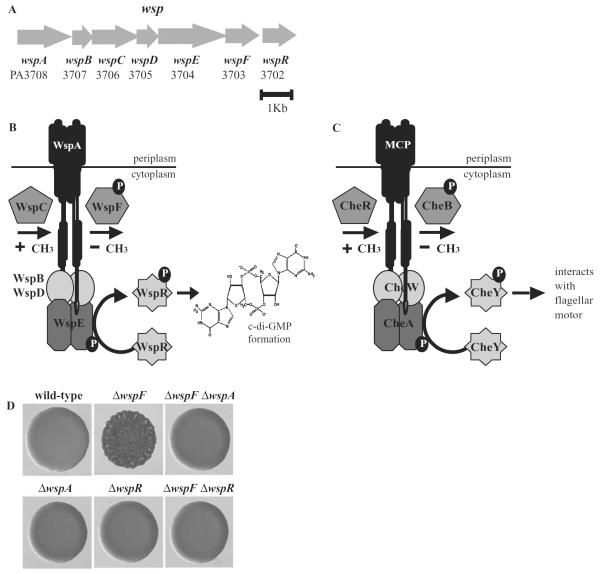
We have been studying a chemosensory-type system called Wsp from Pseudomonas aeruginosa. The wrinkly spreader phenotype (Wsp) was first recognized as a group of adaptive changes that allowed Pseudomonas fluorescens to form thick pellicles in standing liquid cultures and distinctive wrinkled colonies on agar plates (Rainey and Travisano, 1998; Spiers et al., 2002). Subsequent work with P. aeruginosa showed that wrinkly colony formation is an indicator of strains that form thick biofilms, communities of cells encased in a self-produced extracellular matrix (Hickman et al., 2005). The mutations that caused the Wsp phenotype mapped to wspF, a homologue of the chemotaxis gene cheB (D'Argenio et al., 2002). wspF is located in a cluster of genes that are predicted to encode a signal transduction system homologous to a bacterial chemotaxis system (Fig. 1A).
Fig. 1.
Characteristics of the Wsp signal transduction system.
A. Organization of the genes encoding the Wsp system.
B. The Wsp proteins are predicted to be a membrane-bound methyl-accepting chemotaxis protein (WspA), a CheR-like methyltransferase (WspC), a CheB-like methylesterase (WspF), two CheW homologues (WspB and WspD) that are predicted to serve as linkers between WspA and a hybrid histidine kinase response regulator (WspE). The response regulator protein, WspR, has a GGDEF domain and catalyses the synthesis of c-di-GMP when phosphorylated. As described in the text, a wspF mutation is predicted to lock the Wsp system into a configuration where WspR is constantly phosphorylated and thus constantly producing c-di-GMP.
C. The physical organization of homologous Che proteins (encoded by PA1456–1459, PA1464 and PA3349) and (D) colony morphologies of P. aeruginosa PAO1 wild-type and wsp deletion strains. wspF mutants have high levels of intracellular c-di-GMP relative to wild-type cells (Hickman et al., 2005).
The Wsp system is conserved in all Pseudomonas species and includes a predicted membrane-bound [methyl-accepting chemotaxis protein (MCP)]-like receptor (WspA), CheW-like scaffolding proteins (WspB and WspD), a CheA/Y hybrid histidine sensor kinase with a receiver domain (WspE), a methyltransferase CheR homologue (WspC), a methylesterase CheB homologue (WspF) and a two domain response regulator GGDEF protein, WspR, which catalyses the synthesis of cyclic-di-GMP (c-di-GMP) (Fig. 1B). By analogy with chemotaxis, the Wsp signal transduction system is predicted to activate WspR by phosphorylation. As expected from this, purified phosphorylated WspR from P. aeruginosa catalyses c-di-GMP synthesis at faster rates than the unphosphorylated form of the protein (Hickman et al., 2005).
C-di-GMP is an intracellular signalling molecule that has been proposed to control the transition between biofilm and planktonic modes of growth (Römling et al., 2005; Jenal and Malone, 2006; Römling and Amikam, 2006; Ryan et al., 2006; Cotter and Stibitz, 2007; Tamayo et al., 2007). High intracellular c-di-GMP promotes the production of biofilms, adhesive organelles such as pili or stalks, and reduces motility in number of species (Boles and McCarter, 2002; Aldridge et al., 2003; Garcia et al., 2004; Simm et al., 2004; Tischler and Camilli, 2004; Hickman et al., 2005; Kader et al., 2006; Lim et al., 2006). The synthesis and degradation of c-di-GMP is catalysed by diguanylate cyclases with GGDEF domains and phosophodiesterases with EAL domains respectively. Most species of Gram-negative bacteria encode multiple GGDEF and EAL proteins. P. aeruginosa strain PAO1 encodes 17 proteins with GGDEF domains, one of which is WspR, five proteins with an EAL domain and 16 proteins with both domains (Kulasakara et al., 2006).
In chemotaxis, receptors called methyl-accepting chemotaxis proteins bind chemoattractants and then rapidly signal associated CheA proteins to alter the phosphorylation status of the response regulator CheY by phosphotransfer (Fig. 1C). CheY-P interacts with flagella motors to change swimming behaviours and affect chemotactic responses (Stock and Surette, 1996; Falke et al., 1997; Stock et al., 2000). CheA also controls the phosphorylation status of a second response regulator, CheB, with a slower kinetics. CheB-P is active as a methylesterase, which modifies the methylation state of glutamate residues on the receptor proteins in conjunction with a constitutively active CheR methyltransferase. Changes in the methylation state of the receptors resets their signalling activity, thereby allowing them to adapt to the original input stimulus. Receptors are hypermethylated in cheB mutants. This causes CheA to become constitutively active, which results in the generation of continuously phosphorylated CheY. By analogy with chemotaxis, the wspF mutation likely has the effect of locking the Wsp chemosensory system into a configuration where it is constantly signalling, so that the output protein, WspR, is constantly phosphorylated and constantly producing c-di-GMP. A P. aeruginosa wspF mutant produces elevated levels of intracellular c-di-GMP (Hickman et al., 2005). Consistent with its role as a signal transduction system, disruption of the Wsp system by mutation of wspA or wspR causes the wspF wrinkly colony phenotype to convert to smooth (Fig. 1D).
The system of rapid response and slower adaptation that is an inherent feature of chemotaxis systems allows motile cells to continuously sense and swim up concentration gradients of chemoattractants. By extension, we hypothesize that the Wsp signal transduction system allows P. aeruginosa to respond and adapt to an as-yet-unknown, but constantly varying, environmental stimulus to fine tune intracellular levels of c-di-GMP. This may provide a mechanism to allow cells to transition in and out of the biofilm state.
Here we analysed the subcellular localization of Wsp proteins as an avenue to gain information about the functional characteristics of this novel chemotaxis-like signal transduction system and to expand our knowledge of the behaviour of a GGDEF protein and accompanying c-di-GMP production in individual cells. We determined that WspA receptor proteins localize in patches all over cells rather than at cell poles, as is the case with P. aeruginosa chemoreceptors. We also found that WspR dynamically clusters in the cytoplasm of cells when it is phosphorylated and we present evidence that the formation of cytoplasmic clusters of WspR protein is indicative of Wsp signal transduction. From this we concluded that growth on a solid agar surface stimulates Wsp signal transduction and therefore c-di-GMP production. Our data indicate that the Wsp system produces c-di-GMP at discreet subcellular locations, implying that cells may have specific Wsp-associated targets of c-di-GMP action. Furthermore, although chemotaxis and Wsp systems share many features in common, the details of signal transduction by the two systems differ in important ways.
Results
YFP-tagged WspR and WspA proteins are functional
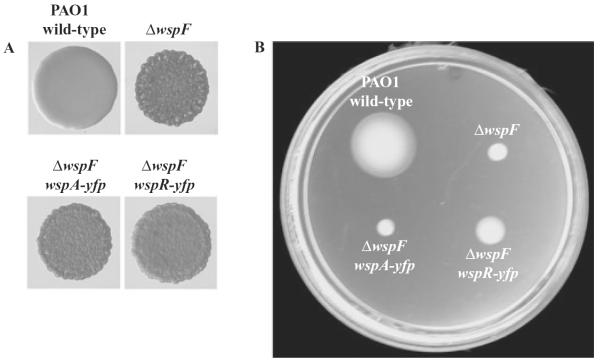
To visualize the subcellular localization of Wsp proteins, we fused a yellow fluorescent protein gene (yfp) to the C-termini of the wspR and wspA genes. We carried out allelic replacements so that yfp gene fusions replaced the wild-type copy of the gene and were present at their native sites on the chromosome, expressed from their original promoters. The phenotypes of ΔwspF strains expressing wspR–yfp or wspA–yfp were similar to those of the ΔwspF strain carrying wild-type wspR and wspA alleles in that all strains had wrinkly colonies and were compromised in swimming motility. This indicates that WspR–YFP and WspA–YFP are functional (Fig. 2).
Fig. 2.
WspR–YFP and WspA–YFP fusion proteins are functional. The introduction of the fusion constructs into the chromosome of a ΔwspF strain by homologous recombination to replace wild-type wspR or wild-type wspA does not alter the (A) colony morphologies or (B) motility in soft agar swim plates, of the strains. This indicates that the Wsp signal transduction pathway has remained intact.
WspA localizes laterally in cells rather than primarily at the poles, as is the case for chemotaxis receptors
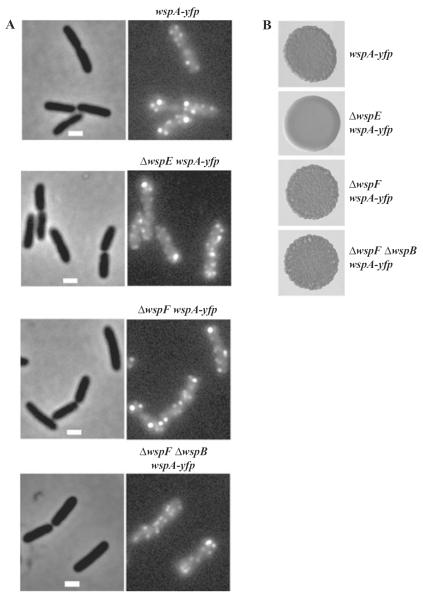
WspA–YFP formed multiple lateral patches in wild-type cells (Fig. 3A). As WspA is a predicted membrane-bound protein, it is likely that the patches are located at the peripheries of cells. This pattern of localization differs from that of MCPs involved in chemotaxis, which localize mainly at cell poles as a large patch (Maddock and Shapiro, 1993; Sourjik and Berg, 2000; Güvener et al., 2006), although a few lateral patches of MCPs are also typically present (Maddock and Shapiro, 1993; Sourjik and Berg, 2000; Thiem et al., 2007). The lateral localization of WspA–YFP occurred throughout growth and the pattern of localization that we observed was very similar in broth-grown and surface-grown cells (data not shown). A wspA–yfp fusion strain in a wild-type background formed wrinkly colonies (Fig. 3B), suggesting that the addition of YFP to the C-terminus of WspA caused the receptor to adopt a conformation such that it stimulated Wsp signal transduction constitutively in a wild-type genetic background. That a ΔwspE wspA–yfp strain formed smooth colonies (Fig. 3B) confirms the prediction that WspE is necessary for WspA-mediated signalling and is an essential part of the Wsp signal transduction system (see Fig. 1B). The deletion of wspB, one of the two cheW homologues in the Wsp operon, did not alter wrinkly colony formation by the ΔwspF mutant strain (Fig. 3B).
Fig. 3.
WspA–YFP localization in cells of wild-type and various mutant strains grown on LB agar.
A. Phase-contrast images are shown on left and fluorescent images on the right. The marker bar is 1 μm.
B. Colony morphologies of strains.
This suggests that WspB does not have an essential role in Wsp signal transduction.
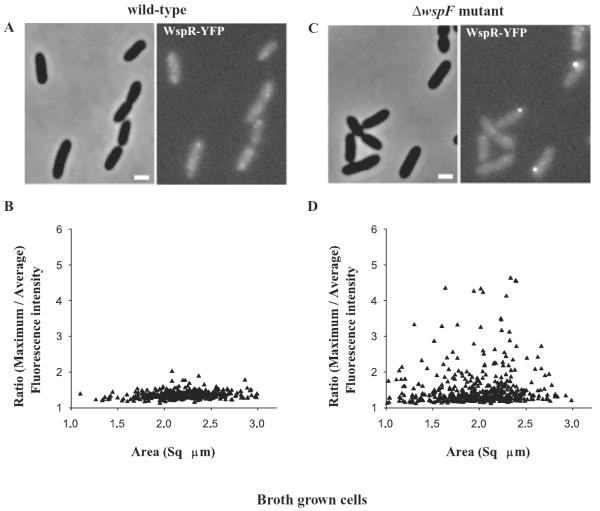
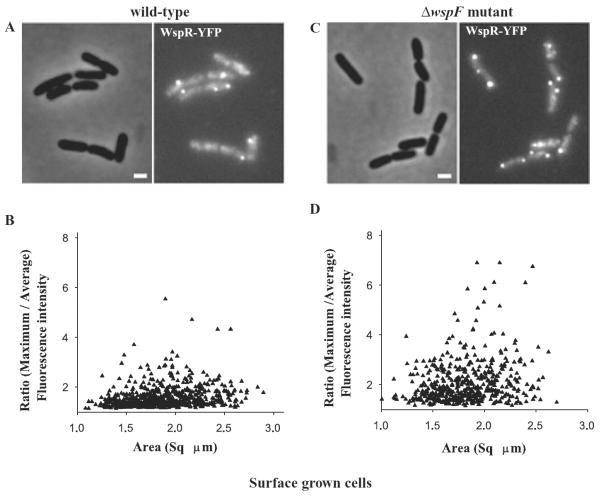
WspR forms cytoplasmic clusters in liquid-grown ΔwspF cells, but not in liquid-grown wild-type cells
We only occasionally saw spots of WspR–YFP in liquid-grown wild-type cells observed by fluorescence microscopy. From this we concluded that WspR was likely dispersed in the cytoplasm rather than present at a particular subcellular location (Fig. 4A). WspR has previously been shown to be a cytoplasmic protein (Hickman et al., 2005). In keeping with this, the measured fluorescence intensities were relatively uniform across individual cells (Fig. 4B). By contrast, WspR–YFP localized in clusters visualized as bright spots in liquid-grown ΔwspF mutant cells (Fig. 4C). This was reflected as a high ratio of maximum to average fluorescence intensities in individual cells (Fig. 4D). Quantification of fluorescence intensities in single cells indicated that about 20% of ΔwspF cell population had WspR–YFP clusters (Table 1). We saw WspR–YFP clustering in 1–2% of the wild-type cell population. Western blot analysis with anti-GFP antibody indicated that the amounts of WspR–YFP were approximately equal in wild-type and ΔwspF cells.
Fig. 4.
WspR–YFP localization in (A) wild-type cells grown in liquid cultures. The phase-contrast (left) and fluorescence (right) images of cells expressing wspR–yfp are shown.
B. Single cell measurements of fluorescence signal intensities expressed as the ratio of maximum to average signal intensities plotted against the area of the respective single cells in wild-type cells expressing WspR–YFP.
C. ΔwspF mutant cells grown in liquid cultures and expressing WspR–YFP.
D. Single cell measurements of fluorescence signal intensities in ΔwspF mutant cells expressing WspR–YFP. Single cell fluorescence data were collected from three independent experiments.
Table 1.
Quantitative analysis of WspR–YFP cluster formation in wild-type and ΔwspF mutant cells grown in liquid cultures.
| Strain | Percentage of cells with WspR–YFP localized in clustersa | Total cells scored |
|---|---|---|
| wspR–yfp | 1.45 (±1.3) | 417 |
| Δ wspF wspR–yfp | 19.73 (±4.8) | 484 |
| Δ wspF ΔwspA wspR–yfp | 2 (±1.67) | 426 |
| Δ wspF wspR[D70N]–yfp | 1.5 (±0.7) | 236 |
| Δ wspF wspR[E253A]–yfp | 35.5 (±13.4) | 115 |
| ΔwspA wspR–yfp/pJN105 | 0.45 (±0.6) | 262 |
| ΔwspA wspR–yfp/pJNspA | 1.58 (±0.9) | 571 |
| ΔwspF ΔwspA wspR–yfp/pJN105 | 1.25 (±0.07) | 151 |
| ΔwspF ΔwspA wspR–yfp/pWspA | 19.4 (±14.9) | 548 |
WspR–YFP clustering was scored based on the presence of a well-defined fluorescent spot or spots in cells. This evaluation was based on the measurement of maximum and average signal intensities in single cells as described in the Experimental procedures. After calculating ratios of maximum to average signal intensities, a ratio value corresponding to presence of a well-defined spot was used as a threshold to sort the cells. At least two independent experiments were performed. Average values are given for the percentage of cells that scored above the ratio threshold value. Standard deviations are indicated in parentheses. Cells were grown in LB broth.
Only phosphorylated WspR forms visible clusters in the cytoplasm
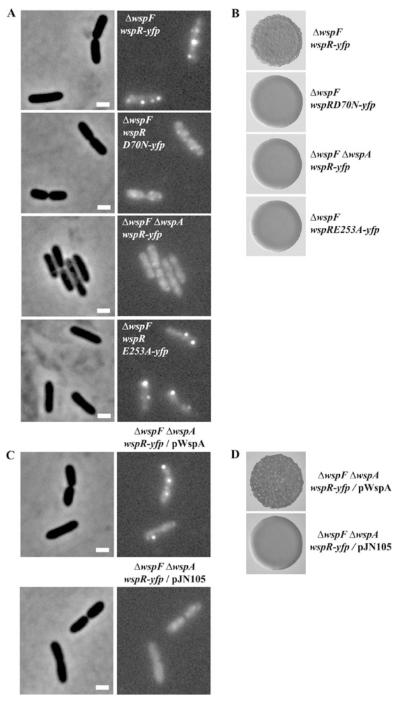
Our observation that WspR–YFP formed significant numbers of bright clusters only in ΔwspF cells (Fig. 5A), a condition where WspR is predicted to be continuously phosphorylated, suggested that clustering of WspR–YFP is an indication of the presence of phosphorylated WspR. To test this, we mutagenized the predicted phosphor-accepting residue, Asp70, critical for the activity of WspR in P. aeruginosa strain PAO1. Mutations in Asp70 disrupt diguanylate cyclase activity, and as a result wrinkly colonies become smooth (J.W. Hickman and C.S. Harwood, unpublished). We created an yfp fusion to a mutant wspR allele carrying an Asn change at the residue Asp70. ΔwspF cells expressing an yfp-tagged Asp70Asn allele of wspR (WspR[D70N]–YFP) formed smooth colonies as expected (Fig. 5B). WspR[D70N]–YFP was dispersed in ΔwspF mutant cells (Fig. 5A) indicating residue Asp70 is also essential for WspR–YFP clustering. Immunoblot assays confirmed that full-length fusion proteins were similarly present in crude cell extracts of strains expressing YFP-tagged wild-type WspR or the YFP-tagged Asp70Asn variant (data not shown). This rules out the possibility that delocalization was due to a cleavage at a junction that would release the Asp70Asn variant and YFP proteins.
Fig. 5.
Phosphorylation is required for WspR–YFP cluster formation.
A. The localization of WspR–YFP in ΔwspF wspR–yfp or ΔwspF ΔwspA wspR–yfp mutant cells and the localization of various WspR–YFP mutant proteins in ΔwspF cells. Phase-contrast images of cells are shown on the left. The marker bar is 1 μm.
B. Colony morphologies of the strains.
C. WspR–YFP cluster formation is restored in a ΔwspF ΔwspA wspR–yfp mutant strain by providing WspA in trans.
D. The colony morphologies of the ΔwspF ΔwspA wspR–yfp strain complemented with empty vector or wspA.
WspR–YFP clusters did not form in ΔwspF ΔwspA mutant cells (Fig. 5A). This indicates that WspA is essential for WspR–YFP clustering probably through WspA-dependent phosphorylation. The expression of wspA in trans from an arabinose-inducible promoter on a low-copy plasmid (pWspA) in ΔwspF ΔwspA mutant cells restored the lack of WspR–YFP clustering and wrinkly colony formation in this strain (Fig. 5C and D). The percentage of cells with WspR–YFP clusters in a WspA-complemented strain was similar to that of the ΔwspF strain (Table 1). A cytoplasmic fragment of WspA failed to restore clustering of WspR–YFP or wrinkly colony formation in a ΔwspF ΔwspA deletion strain (data not shown). We fused an yfp gene to the cytoplasmic fragment of WspA and found that the fusion protein was diffused throughout the cytoplasm (data not shown).
C-di-GMP generation is not important for WspR–YFP clustering. We determined this by examining the subcellular localization of an YFP-tagged Glu253Ala variant of WspR that was unable to catalyse c-di-GMP production due to a mutation in its conserved GGEEF motif (J.W. Hickman and C.S. Harwood, unpublished). This YFP-tagged Glu253Ala variant formed tight clusters similar to the YFP-tagged wild-type WspR in the ΔwspF mutant cells (Fig. 5A).
WspR–YFP clustering and wrinkly colony formation occurred in ΔwspF ΔcheA cells (data not shown), indicating the Wsp signalling pathway does not depend on the chemotaxis-signalling pathway.
WspR–YFP and WspA–CFP colocalize transiently
That WspA and WspE are required for wrinkly colony formation supports the idea that this receptor and sensor kinase act together to phosphorylate WspR by phosphotransfer between a WspA/WspE complex and WspR as is shown in Fig. 1B. When we examined the behaviour of YFP-tagged WspR and CFP-tagged WspA coexpressed in ΔwspF cells, we found that these proteins sometimes appeared to be colocalized, but this was not the general rule (Fig. 6). We reported previously that the P. aeruginosa chemotaxis output regulator CheY is always seen colocalized with membrane-bound receptor-CheA complexes at cell poles (Güvener et al., 2006), as is the case in Escherichia coli (Sourjik and Berg, 2000). The behaviour of the WspR proteins thus differs from its homologous chemotaxis counterparts.
Fig. 6.
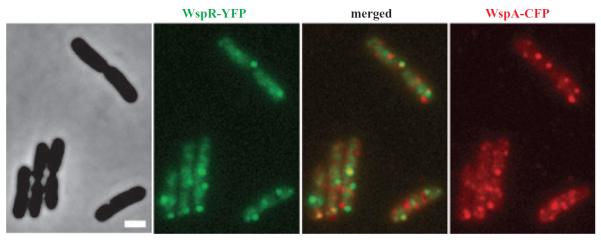
WspR–YFP and WspA–CFP sometimes colocalize in ΔwspF mutant cells. WspR–YFP is coloured green. WspA-CFP is coloured red and the merged image of the two is shown in the middle. A phase-contrast image of the cells is shown on the left. Cells were grown on LB agar. The marker bar is 1 μm.
Surface growth stimulates Wsp signal transduction
That WspR forms visible clusters in cells when it is phosphorylated suggested to us that we could use the subcellular localization characteristics of WspR–YFP to identify conditions that stimulate Wsp signalling. We found that although WspR–YFP was dispersed in wild-type cells growing in Luria–Bertani (LB) broth, it formed fluorescent spots in cells grown to confluence on the surface of LB agar for 20 h (Fig. 7A). Quantification of WspR–YFP fluorescence signals in single cells showed that about 30% of the wild-type cells grown on agar surfaces that we observed had WspR–YFP clusters (Fig. 7B and Table 2). Cells did not form WspR–YFP clusters in the absence of WspA, and wspA expressed in trans (from plasmid pWspA) restored the lack of clustering at levels similar to that of the parent fusion strain (Table 2). The average signal intensity per cell was similar in broth-grown cells (269 ± 14 signal intensity) and surface-grown cells (290 ± 18 signal intensity) expressing WspR–YFP, indicating that the total amount of Wsp protein synthesized was the same under the two conditions. In addition, we found that the amount of wspR mRNA transcript synthesized was the same in broth-grown and surface-grown cells. We also observed WspR–YFP cluster formation in ΔwspF cells grown on agar surfaces (Fig. 7C and D). Interestingly, the percentage of agar-grown ΔwspF cells that had WspR–YFP clusters was higher than the percentage of liquid-grown ΔwspF cells that had clusters (Tables 1 and 2). One interpretation of this is that signal transduction can be further stimulated in the `locked on' ΔwspF mutant.
Fig. 7.
Surface growth stimulates WspR–YFP cluster formation in wild-type cells.
WspR–YFP localization in (A) wild-type cells grown on LB agar. The phase-contrast (left) and fluorescence (right) images of cells expressing wspR–yfp are shown.
B. Single cell measurements of fluorescence signal intensities expressed as the ratio of maximum to average signal intensities plotted against the area of the respective single cells in wild-type cells expressing WspR–YFP.
C. ΔwspF mutant cells grown on agar and expressing WspR–YFP.
D. Single cell measurements of fluorescence signal intensities in ΔwspF mutant cells expressing WspR–YFP. Single cell fluorescence data were collected from three independent experiments.
Table 2.
Quantitative analysis of WspR–YFP cluster formation in various strains grown on agar surfaces.
| Strain | Percentage of cells with WspR–YFP localized in clustersa | Total cells scored |
|---|---|---|
| wspR–yfp | 29.7 (±2.9) | 692 |
| Δ wspF wspR–yfp | 59.8 (±11.8) | 493 |
| Δ wspA wspR–yfp | 0.22 (±0.3) | 253 |
| Δ wspF ΔwspA wspR–yfp | 1.32 (±1.2) | 270 |
| Δ wspF wspR[E253A]–yfp | 74.95 (±1.9) | 290 |
| ΔwspA wspR–yfp/pJN105 | 0.5 (±0.7) | 299 |
| ΔwspA wspR–yfp/pWspA | 37.6 (±4.3) | 280 |
| ΔwspF ΔwspA wspR–yfp/pJN105 | 0.3 (±0.4) | 190 |
| ΔwspF ΔwspA wspR–yfp/pWspA | 58.1 (±4.6) | 414 |
WspR–YFP clustering was scored based on the presence of a well-defined fluorescent spot in cells. This evaluation was based on the measurement of maximum and average signal intensities of single cells as described in the Experimental procedures. After calculating ratios of maximum to average signal intensities, a ratio value corresponding to presence of a well-defined spot was used as a threshold to sort the cells. At least two independent experiments were performed. Average values are given for the percentage of cells that scored above the ratio threshold value. Standard deviations are indicated in parentheses. Cells were grown on LB medium solidified with 2.5% agar as described in Experimental procedures.
The amount of agar used to solidify LB medium was not critical for clustering. WspR–YFP clustering occurred in wild-type cells grown on agar surfaces solidified with 1% or 2.5% agar. Changes in the amount of yeast extract or tryptone or elimination of one of the two from LB medium did not have an effect on clustering of WspR–YFP in wild-type cells. Removal of NaCl from LB agar medium abolished clustering in wild-type but not in ΔwspF cells. We found that 85 mM NaCl, the amount of salt in LB, was needed for the formation of well-defined fluorescent spots. The substitution of 85 mM KCl or 100 mM KH2PO4 for NaCl in LB medium also supported the formation of WspR–YFP clusters. WspR–YFP clusters formed in wild-type cells grown on tryptic soy broth plates. Several defined growth media, including M63, minimal A medium and M9 (Sambrook et al., 1989), solidified with agar, supported the formation of WspR–YFP clustering. Succinate (20 mM), citrate (20 mM) or glucose (17 mM) served as the carbon sources in the defined media tested. WspR–YFP clustering was similarly efficient in cells grown on rich medium or defined minimal medium solidified with Bacto agar, Nobel agar or agarose. However, cells had a slower growth rate on defined medium solidified with agarose. We did not observe WspR–YFP cluster formation in wild-type cells when we made gross changes in viscosity by adding Ficoll to LB broth (not shown). High osmolarity was detrimental to WspR–YFP clustering in wild-type and ΔwspF mutant cells. Cells grown on LB plates supplemented with 300 mM NaCl or 700 mM sucrose did not have clusters of WspR–YFP.
Cells growing on a solid surface tend to be touching and making cell-to-cell contact. P. aeruginosa has a number of different kinds of surface appendages that bind cells together and that could be important for initiating a Wsp signalling event. We visually inspected WspR–YFP cluster formation in several independently grown cultures of mutants lacking flagella (a fliC mutant), type IV pili (a pilA mutant), or exopolysaccharide (EPS) (a ΔpelΔpsl mutant). We found that the quantity and quality of WspR–YFP clusters formed in the cells of each type of mutant observed after about 20 h of growth on LB agar plates were very similar to those of wild-type cells.
Dynamic behaviour of WspR clusters in cells
WspR–YFP clusters were mobile in cells. We used time-lapse microscopy to obtain fluorescent images of individual wild-type and ΔwspF cells grown on agar plates and transferred to an agarose pad. Cells typically had between one and four fluorescent spots. Figure 8 shows representative fluorescence and phase-contrast images of wild-type cells. Images were captured at intervals of 30 s. Several different patterns are evident. Some spots were continuously present but moved inside cells and varied in brightness over the period of observation (Fig. 8A). In some cases spots disappeared and reappeared (Fig. 8B), and we also observed the partitioning of spots from one into two (Fig. 8C). We also observed situations in which spots were continuously present in cells and dynamic without accompanying changes in brightness (not shown). We analysed time lapse images acquired every 30 s over a period of 270 s for 20 ΔwspF cells and 49 wild-type cells. The variety of patterns of WspR fluorescent spots, their movement and the numbers of spots that we saw per cell were approximately the same for ΔwspF and wild-type cells. However, a higher percentage of ΔwspF cells had spots that were continuously present in cells over the interval of observation (65% of cells) compared with wild-type cells (38%) and the spots were brighter. Wild-type cells tended to have a greater proportion of WspR fluorescent clusters that appeared and disappeared or partitioned during the interval of observation.
Fig. 8.
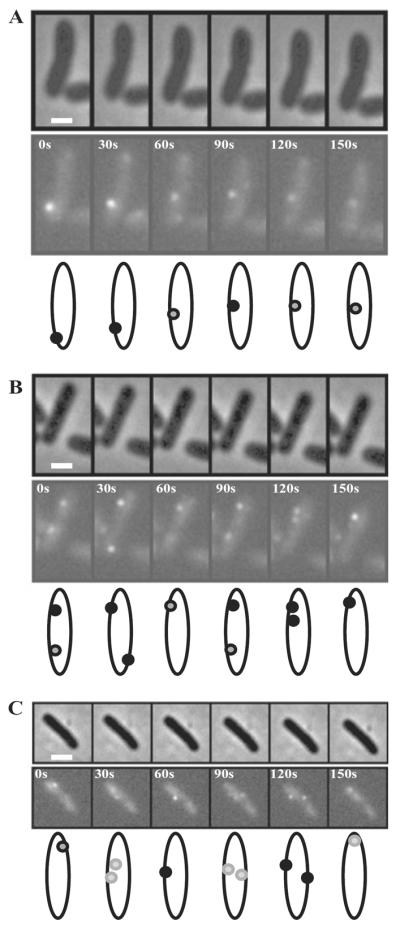
Time-lapse fluorescence and phase-contrast images of surface-grown wild-type cells expressing the WspR–YFP fusion protein. Images were acquired every 30 s. Individual cells were followed over a 150 s time period in (A), (B) and (C). Cartoons depicting the movement and change in fluorescence intensity of the spots in each cell are shown below each panel. The marker bar is 1 μm.
Discussion
Our observations indicate that clusters of WspR–YFP protein become visible in the cytoplasm of cells in which the Wsp system is active. Clusters of fluorescent protein-tagged WspR were dynamic and moved around inside cells. Fluorescent spots of WspR increased and decreased in brightness. This could reflect movement of the complexes of protein in and out of the plane of focus, or it may reflect the formation and dissolution of complexes of active WspR. These observations suggest that that Wsp-mediated c-di-GMP formation occurs at discreet subcellular sites. This could allow the Wsp system to deliver localized high concentrations of c-di-GMP to specific c-di-GMP receptor proteins. Known c-di-GMP binding domains include the I site of some diguanylate cyclases (Christen et al., 2006). First defined for the Caulobacter crescentus PleD protein, the I site binds c-di-GMP to cause feedback inhibition of diguanylate cyclase activity in a non-competitive manner (Chan et al., 2004). The PelD protein from P. aeruginosa binds c-di-GMP at a site that resembles the I site in secondary structure. PelD, which is not a diguanylate cyclase, controls EPS synthesis in response to c-di-GMP by a mechanism that is still to be elucidated (Lee et al., 2007). EAL domain phosphodiesterases that convert c-di-GMP to pGpG also bind c-di-GMP, although the binding site has not been defined. Finally, a c-di-GMP binding domain called PilZ has been described (Amikam and Galperin, 2006; Ryjenkov et al., 2006; Christen et al., 2007; Pratt et al., 2007). P. aeruginosa has eight PilZ domain proteins, all of which have been shown to bind c-di-GMP (Merighi et al., 2007; Ramelot et al., 2007). All are conserved hypothetical proteins of unknown function with the exception of PilZ itself (PA2960), which is required for normal pilus assembly (Alm et al., 1996), and Alg44 (PA3542), an alginate bio-synthesis gene (Remminghorst and Rehm, 2006) which responds to c-di-GMP to modulate the production of this EPS (Merighi et al., 2007). Any of one or a combination of the described P. aeruginosa c-di-GMP receptor proteins could be targets of Wsp-generated c-di-GMP and it is also possible that Wsp-generated c-di-GMP binds to other protein domains that have yet to be described.
Although investigators are beginning to identify c-di-GMP binding proteins as having functions that influence motility or biofilm formation, the mechanism by which any protein exerts its effects once it has bound c-di-GMP is not fully understood. In general, c-di-GMP appears to affect the activities of large protein complexes including cellulose synthases, alginate synthase, and flagella motors and pilus assembly (Huang et al., 2003; Kazmierczak et al., 2006; Ryjenkov et al., 2006; Christen et al., 2007; Merighi et al., 2007; Pratt et al., 2007; Ramelot et al., 2007). C-di-GMP also influences the expression of genes for EPS synthesis, virulence and motility (Hickman et al., 2005; Tischler and Camilli, 2005; Mendez-Ortiz et al., 2006; Kim and McCarter, 2007). Our initial characterization of WspR in cells suggests that its function is not to exclusively influence processes that occur at cell poles because it is not exclusively present at poles when in its activated form. In this sense it differs from the response regulator GGDEF protein PleD from C. crescentus and the GGDEF-EAL phosphodiesterase, FimX, from P. aeruginosa, both of which localize to a cell pole (Paul et al., 2004; Kazmierczak et al., 2006). PleD resembles WspR in that it has a visible subcellular location only when it is active. The localization characteristics of PleD change when it is activated by phosphorylation and dimerization such that it is sequestered to the cell pole destined for stalk biogenesis (Paul et al., 2004; Paul et al., 2007). A pleD mutant is defective in the distinctive flagella ejection and stalk elongation that occur as part of the C. crescentus cell cycle (Aldridge and Jenal, 1999) and a fimX mutant is defective in type IV pilus assembly (Huang et al., 2003).
Non-polar protein complexes that would be logical targets of activation by WspR-generated c-di-GMP are complexes associated with the synthesis of Pel and Psl EPS in P. aeruginosa strain PAO1 (Friedman and Kolter, 2004a,b; Jackson et al., 2004; Matsukawa and Greenberg, 2004; Ma et al., 2007). We know that the pel and psl operons are responsible for the wrinkly colonies and robust biofilms that are formed when WspR-generated c-di-GMP accumulates in wspF strains because we have observed that a ΔwspFΔpelΔpsl strain forms smooth colonies similar in texture to the wild type (J.W. Hickman and C.S. Harwood, unpublished).
Our conclusion that growth on an agar surface stimulates Wsp signal transduction supports the notion that a condition associated with cell-to-cell contact or cell-surface interactions stimulates P. aeruginosa to form biofilms. Growth on a solid surface rather than transient contact with a solid surface seems to be required for the stimulation of Wsp signal transduction because WspR–YFP did not form clusters in broth-grown wild-type cells that were transferred to phosphate-buffered saline (PBS) agarose pads and observed by microscopy over a period of an hour. When we observed individual cells inoculated onto M9-citrate medium agarose pads and allowed to grow over a period of about 18 h, we saw that bright spots of WspR–YFP appeared and disappeared from cells photographed at 30 min intervals once cells grew to form a group of about eight or more cells (not shown). These observations suggest that cells need to be touching or very close to each other to signal through the Wsp system. In many ways the Wsp system seems analogous to the Dif chemosensory pathway in Myxococcus xanthus, which regulates EPS production, though not necessarily by a c-di-GMP based mechanism (Yang et al., 2000; Bellenger et al., 2002; Black and Yang, 2004). Evidence suggests that type IV pili may stimulate Dif signal transduction (Black et al., 2006). We tested several kinds of P. aeruginosa cell surface structures that might bind cells together including flagella, type IV pili and EPS but failed to find any clear evidence for their involvement in Wsp signalling. It is also possible that a physical parameter that is subtly altered during growth on an agar surface stimulates Wsp signal transduction.
A surprising feature of the WspA and WspR proteins is how different their subcellular distributions are from those of their chemotaxis homologues. These differences likely reflect functional differences between the two systems. In chemotaxis, MCPs and associated signal transduction and signal adaptation proteins organize as arrays in large patches at cell poles (Zhang et al., 2007). It has been estimated that on the order of 7500 receptor dimers are packed into E. coli polar arrays (Li and Hazelbauer, 2004). The arrays, which include multiple different receptors, each specific for a different set of chemoeffectors, integrate stimuli for a common output. The structural arrangement of the arrays is also such that input signals are amplified so that environmental stimuli are sensed over a wide dynamic range (Duke and Bray, 1999; Sourjik and Berg, 2004; Studdert and Parkinson, 2004; Wolanin and Stock, 2004). Our observations showing that WspA is distributed at multiple locations around cells and that the WspR protein is transiently associated with WspA suggest that signal integration and signal amplification of the type that occurs in chemotaxis might not be fundamental features of the Wsp system. In the future it will be of interest to determine the structural features of the Wsp proteins that determine their subcellular location and to determine whether the movement of WspR in cells is due to specific associations with the cytoskeleton or other cytoplasmic proteins.
Experimental procedures
Bacterial strains and growth condition
Pseudomonas aeruginosa strains used in this study are listed in Table 3. E. coli strains were DH5α, obtained from Gibco-BRL and S17-1 (Simon et al., 1983), which was the donor strain used to transfer plasmids to P. aeruginosa by conjugation. E. coli and P. aeruginosa strains were grown in LB medium (Sambrook et al., 1989) at 37°C or at room temperature as specified. Strains to be used for microscopy were freshly streaked on LB agar plates from frozen stocks and incubated overnight at 37°C. Cells were inoculated into a 16 mm test tube containing 3 ml LB. Tubes were incubated with shaking until the culture reached an OD600 of 0.2–0.3. These broth-grown cells were then diluted to an OD600 of 0.005. This served as the inoculum for broth-grown or surface-grown cells. To generate surface-grown cells, a 125 ml sample was spread on an LB agar plate solidified with 2.5% Bacto agar. Plates were incubated at room temperature for about 20 h. Cells scraped from plates at the end of this period of incubation typically yielded an OD600 of 0.15–0.3 upon resuspension in 25 ml LB. To generate broth-grown cells, 25 ml diluted cultures were aerated in 125 ml Erlenmeyer flasks at room temperature to mid-log phase in the range of 0.2–0.5 OD600. Antibiotic concentrations were 100 mg ml−1 ampicillin and 10 μg ml−1 gentamicin (Gm) for E. coli and 50 μg ml−1 Gm for P. aeruginosa. L-arabinose at a final concentration of 0.1% was included in growth media to induce gene expression from a low-copy plasmid.
Table 3.
Strains.
| Strain | Relative genotype | Source and/or reference |
|---|---|---|
| PA01 | Wild-type; twitching motility+ | Iglewski (Stover etal., 2000) |
| PAO1100 | Δ wspF | Hickman etal. (2005) |
| PAO1101 | Δ wspA | Hickman etal. (2005) |
| PAO1102 | Δ wspE | Hickman etal. (2005) |
| PAO1103 | Δ wspR | Hickman etal. (2005) |
| PAO1104 | Δ wspF AwspR | Hickman etal. (2005) |
| PAO1251 | wspR–yfp | PAO1 (this study) |
| PAO1252 | Δ wspA wspR–yfp | PAO1101 (this study) |
| PAO1254 | Δ wspF wspR–yfp | PAO1100 (this study) |
| PAO1258 | Δ wspF wspR–yfp wspA-cfp | PAO1254 (this study) |
| PAO1260 | Δ wspF AwspA wspR–yfp | PAO1254 (this study) |
| PAO1262 | Δ wspF AwspA | PAO1100 (this study) |
| PAO1265 | Δ wspF Ache A wspR–yfp | PAO1254 (Ferrández etal., 2002; this study) |
| PAO1268 | Δ wspF wspR[D70N]–yfp | PAO1100 (this study) |
| PAO1275 | wspA–yfp | PAO1 (this study) |
| PAO1276 | Δ wspE wspA–yfp | PAO1102 (this study) |
| PAO1287 | Δ wspF wspR[E253A]–yfp | PAO1100 (this study) |
| PAO1290 | Δ wspF Apel Apsl wspR–yfp | PAO1543 (this study) |
| PAO1501 | ΔwspF ΔwspA/pJN105 | PAO1262 (this study) |
| PAO1512 | ΔwspF ΔwspA wspR–yfp/pJN105 | PAO1260 (this study) |
| PAO1514 | A wspA wspR–yfp/pJ N105 | PAO1252 (this study) |
| PAO1516 | ΔwspF ΔwspA/pWspA(truncated) | PAO1262 (this study) |
| PAO1526 | fliC∷Tn5 wspR–yfp | PAO1540 (this study) |
| PAO1529 | Δ pilA wspR–yfp | PAO1541 (this study) |
| PAO1530 | Δ pel Δpsl wspR–yfp | PAO1542 (this study) |
| PAO1534 | Δ wspF ΔwspA/pWspA | PAO1262 (this study) |
| PAO1535 | Δ wspA wspR–yfp/pWspA | PAO1252 (this study) |
| PAO1536 | Δ wspF ΔwspA wspR–yfp/pWspA | PAO1260 (this study) |
| PAO1537 | Δ wspF wspA–yfp | PAO1100 (this study) |
| PAO1539 | Δ wspF ΔwspB wspA–yfp | PAO1537 (this study) |
| PAO1540 | fliC∷Tn5 | L. L. McCarter |
| PAO1541 | Δ pilA | M. Parsek |
| PAO1542 | Δ pel Δpsl | M. Parsek |
| PAO1543 | Δ wspF Δpel Δpsl | PAO1542 (this study) |
Colony morphologies were visualized by spotting 2 μl of LB broth-grown cells that had been diluted to an OD600 of 0.005 onto Congo Red plates (Friedman and Kolter, 2004a) and allowing growth to occur at room temperature for 2 days.
DNA manipulations
Cloning, gel electrophoresis, and related molecular biology work were carried out according to standard protocols (Sambrook et al., 1989). Herculase Enhanced DNA polymerase (Stratagene) was used to amplify DNA. DNA was isolated with Qiagen DNA extraction kits. Oligonucleotides were synthesized by Integrated DNA Technologies. DNA sequencing was performed at the DNA sequencing facility of the University of Washington. wspR mRNA transcript levels were determined by real-time PCR as described (Bagge et al., 2004).
Generation of chromosomal fusions of yfp or cfp to P. aeruginosa genes
yfp or cfp translational fusions at the C-terminus of wspA or wspR were generated as previously described by using suicide vectors pEX19EYFP or pEX19ECFP (Güvener et al., 2006). wspA was amplified from the P. aeruginosa chromosome with a primer pair in which both the 5′ and 3′ ends had an engineered KpnI sites. The 3′ end excluded the stop codon and was followed by two bases prior to the KpnI site. This KpnI fragment was cloned into either pEX19EYFP or pEX19ECFP just upstream of the yfp or cfp start codon and a clone with the correct orientation of the KpnI fragment was determined. Flanking DNA downstream of wspA was amplified from the P. aeruginosa chromosome to give a PCR fragment with XbaI and HindIII engineered sites. This XbaI-HindIII fragment was cloned into the previous construct immediately downstream of the yfp or cfp gene to yield a wspA–yfp or wspA–cfp fusion with downstream flanking DNA. PCR amplified fragments containing wspR sequences had engineered EcoRI sites at the 5′ ends and KpnI sites at the 3′ ends. Site-directed mutagenesis to create specific mutations in wspR is described below. All final constructs were sequenced to make sure that they were correct and that no PCR mistakes had been introduced during the cloning steps. Gene replacement was used to move the fusion constructs into the P. aeruginosa chromosome by homologous recombination (Ferrández et al., 2002). Candidates were screened by PCR. For the allelic replacements in which site-specific mutations were moved to the chromosome, a region of the chromosome was PCR-amplified and sequenced to verify that the correct site-specific mutations were present. Western blot analysis with antiserum to GFP (Clontech) showed that full-length fusion proteins were expressed to the same levels in P. aeruginosa cells in all genetic backgrounds tested.
Site-directed mutagenesis to change WspR residue Asp70 to Asn was carried out by overlap extension PCR (Horton et al., 1993). Intergenic primers, which carried mismatched bases to change the codon Asp70 to Asn, were 5′-GTCGACGCCGGGCATCACCAGGTTCTG GAGG ATC-3′ (WspRD70N-R) and 5′-GATCCTCCAGAACCTGGTGATGCCCGGCGTCGAC-3′ (WspRD70N-F). Mismatched bases are underlined. Outer primer sequences were 5′-GCCATGGAATTCCCGGGTTCGACTCCGGCCAGGCGCCAG-3′ (WspR-F) and 5′-GCTATCGGTACCGGGCCCGCCGGGGCCGGCGGCACCGGCTGTTCCATCAGGCCCACCTGG-3′ (WspR-R). EcoRI and KpnI sites are shown in bold letters. A stop codon was omitted in the WspR-R primer but it contained two extra bases and an engineered KpnI site to allow creation of an in-frame fusion to yfp in pEX19EYFP. An Ala change in the WspR residue Glu253 was accomplished by a similar strategy. The wspRE253A allele was amplified with WspR-F and WspR-R primers to make an in-frame fusion to yfp when cloned into pEX19EYFP.
Construction of an in-frame deletion of wspB
To make a ΔwspB deletion in a wspA–yfp fusion strain, we created a ΔwspB in-frame deletion construct in pEX19Gm (Hoang et al., 1998). The deletion construct was generated by using PCR to amplify two regions of chromosome. One was a 0.757 kb fragment immediately upstream of wspB, corresponding to the junction between wspA–yfp and wspB genes. The other was a 1.239 kb fragment immediately downstream of wspB. Primers for the region upstream of wspB were 5′-AGTCCCGGTACCGGTCGCCACCATGGTGAGCAAG-3′ (WspB–YFPupF) and 5′-GCAGGCGTTCGAAGCGATCGTTCATGCGGTCACAGCCTCTA GAG TCGCG-3′ (WspB–YFPupR), and for the region downstream of wspB were 5′-CGCGACTCTAGAGGCTGTGACCGCATGAACGATCGCTTCGAACGCCTGC-3′ (WspBdownF) and 5′-GC CACAAAGCTTCTTTGTTCACTCCACGGGCCGCCGTTG-3′ (WspBdownR). KpnI and HindIII sites are shown in bold letters and regions complementary to downstream of wspB (in WspB–YFPupR) or upstream of wspB (in WspBdownF) are underlined. A mixture of the two PCR products served as templates for a final PCR reaction, which generated a fused product of the two templates. This was then crossed into wspA–yfp strain PAO1275 (Table 3) by conjugation, and homologous recombinants were selected as described by Ferrández et al. (2002). Mutant candidates were screened by PCR.
Generation of arabinose-inducible gene expression constructs
Plasmid pGFP-C was used to generate an arabinose-inducible wspA expression construct. pGFP-C is a derivative of pJN105, a broad host-range vector that carries an l-arabinose-inducible promoter and the araC regulator gene (Newman and Fuqua, 1999). We originally constructed pGFP-C to make in-frame translational fusions to green fluorescence protein gene (gfp). However, here we used this plasmid because it contained an optimized ribosome-binding site for gene expression. To construct pGFP-C, an NheI-PvuI fragment was obtained from plasmid pDSW208 (Weiss et al., 1999). This fragment contained an optimized ribosome-binding site, a methionine initiation codon (ATG) and multiple restriction enzyme sites followed by the gfp gene. The NheI-PvuI fragment was ligated just 3′ of the arabinose-inducible promoter of pJN105, to give pGFP-C. The wspA-coding region starting with the second codon, and including its stop codon, was amplified from the chromosome of P. aeruginosa with a primer set that had engineered XbaI sites. The 3′ primer also included an engineered HindIII site prior to the XbaI site. The amplified PCR product was digested with XbaI and cloned into XbaI-digested pGFP-C. A clone carrying the correct orientation of the XbaI fragment was determined. A HindIII digestion released the gfp gene from this construct. HindIII-digested and gel-purified vector was self-ligated creating the final construct, pWspA. The cloning procedure introduced 11 codons for amino acids Glu-Phe-Glu-Leu-Gly-Thr-Arg-Gly-Ser-Ser-Arg between the first and second codons of wspA in the pWspA construct.
An expression construct of a truncated version of wspA, which consisted of only the predicted cytoplasmic domain, was constructed similarly to the pWspA construct. The truncated wspA excluded the N-terminal 286 codons predicted to encode its periplasmic sensing and transmembrane domains. A primer pair, of which the 5′ primer had an engineered EcoRI site and the 3′ primer had a HindIII site, was used to amplify the C-terminus of wspA, a region of 256 amino acids with a stop codon. The PCR fragment was digested with EcoRI and HindIII and ligated into EcoRI-HindIII-digested and gel-purified pGFP-C. This procedure also eliminated the gfp gene sequence from pGFP-C. The construct encoding the truncated wspA had a methionine start codon followed by two non-native codons (Glu and Phe) before the first residue 287Gln of the WspA C-terminus.
Fluorescence microscopy
Samples were prepared for microscopy from broth-grown or surface-grown cells. Broth-grown cultures were appropriately diluted in PBS (pH 7.4) and 3 μl of cell suspension was spotted on the surface of agarose pads that were prepared with 1.0% agarose made in PBS. An agarose gel had previously been prepared in a standard gel casting system. Blocks of agarose were cut out and transferred on microscope slides and samples were applied and then covered with coverslips. Surface-grown cells were transferred to agarose pads with cotton swabs. Images were acquired with a Nicon 80i microscope equipped with 100× PlanApochromat objective (numerical aperture, 1.4), and were recorded with a Cool-Snap HQ camera (Photometrics). The imaging system was operated with Metamorph 6.3r2 software. Filter sets were purchased from Chroma Technology Corp. The filter sets were ET-EYFP (Chroma # 49003) for YFP excitation and ET-CFP (Chroma # 49001) for CFP excitation. All images shown are fluorescence and phase-contrast images. All images that were compared with each other in the text and figures were acquired in the same day with the same microscope settings. Images were minimally processed with Adobe Photoshop 7.0 and ImageJNIH software. All images that were compared with each other were manipulated (brightness and contrast adjustments and resizing of files) identically. At least three independent experiments were performed, and at least five different fields of view from each experiment were analysed. Figures show representative images.
Single cell fluorescence intensity measurements
Phase-contrast images were used as a reference point to select well-isolated single cells in the fields of view. The corresponding fluorescence images were then analysed with Metamorph software to quantify greyscale intensity and pixel brightness. Average grey values were extracted that indicated the average of the pixel greyscale values in selected cells. Integrated grey values were extracted that indicated the sum of the greyscale values for all pixels in individual cells. Maximum grey values were extracted that indicated maximum areas of pixel intensity within cells. These values were used to assess WspR clustering. Integrated grey values were plotted against the area of selected individual cells. Data points that deviated from a linear curve were not considered for analysis. Average grey values for the wspR–yfp strain were low but were above that seen for a nonfluorescent wild-type strain. By determining ratios of maximum to average grey values, it was possible to combine data obtained from different days. With the microscope settings used, a ratio of 1.64 was determined to be the threshold value for a well-defined fluorescent spot as judged by visual inspection of images of wild-type cells grown on surfaces. Data shown in graphs are the ratios that were plotted against the areas of selected cells. These analyses were done on images acquired from at least three independent experiments from about 10 different fields of view.
Acknowledgements
Public Health Service Grant GM56665 from the National Institute of General Medical Sciences supported this work. We thank Linda McCarter, Matthew Parsek and Jason Hickman for providing strains. We thank Andrew Hawkins for making the pGFP-C construct, and Karine Gibbs for technical help with image quantification by Metamorph software and discussions.
References
- Aldridge P, Jenal U. Cell cycle-dependent degradation of a flagellar motor component requires a novel-type response regulator. Mol Microbiol. 1999;32:379–391. doi: 10.1046/j.1365-2958.1999.01358.x. [DOI] [PubMed] [Google Scholar]
- Aldridge P, Paul R, Goymer P, Rainey P, Jenal U. Role of the GGDEF regulator PleD in polar development of Caulobacter crescentus. Mol Microbiol. 2003;47:1695–1708. doi: 10.1046/j.1365-2958.2003.03401.x. [DOI] [PubMed] [Google Scholar]
- Alm RA, Bodero AJ, Free PD, Mattick JS. Identification of a novel gene, pilZ, essential for type 4 fimbrial biogenesis in Pseudomonas aeruginosa. J Bacteriol. 1996;178:46–53. doi: 10.1128/jb.178.1.46-53.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amikam D, Galperin MY. PilZ domain is part of the bacterial c-di-GMP binding protein. Bioinformatics. 2006;22:3–6. doi: 10.1093/bioinformatics/bti739. [DOI] [PubMed] [Google Scholar]
- Bagge N, Schuster M, Hentzer M, Ciofu O, Givskov M, Greenberg EP, Hoiby N. Pseudomonas aeruginosa biofilms exposed to imipenem exhibit changes in global gene expression and beta-lactamase and alginate production. Antimicrob Agents Chemother. 2004;48:1175–1187. doi: 10.1128/AAC.48.4.1175-1187.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellenger K, Ma X, Shi W, Yang Z. A CheW homologue is required for Myxococcus xanthus fruiting body development, social gliding motility, and fibril biogenesis. J Bacteriol. 2002;184:5654–5660. doi: 10.1128/JB.184.20.5654-5660.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black WP, Yang Z. Myxococcus xanthus chemotaxis homologs DifD and DifG negatively regulate fibril polysaccharide production. J Bacteriol. 2004;186:1001–1008. doi: 10.1128/JB.186.4.1001-1008.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black WP, Xu Q, Yang Z. Type IV pili function upstream of the Dif chemotaxis pathway in Myxococcus xanthus EPS regulation. Mol Microbiol. 2006;61:447–456. doi: 10.1111/j.1365-2958.2006.05230.x. [DOI] [PubMed] [Google Scholar]
- Boles BR, McCarter LL. Vibrio parahaemolyticus scrABC, a novel operon affecting swarming and capsular polysaccharide regulation. J Bacteriol. 2002;184:5946–5954. doi: 10.1128/JB.184.21.5946-5954.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C, Paul R, Samoray D, Amiot NC, Giese B, Jenal U, Schirmer T. Structural basis of activity and allosteric control of diguanylate cyclase. Proc Natl Acad Sci USA. 2004;101:17084–17089. doi: 10.1073/pnas.0406134101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christen B, Christen M, Paul R, Schmid F, Folcher M, Jenoe P, et al. Allosteric control of cyclic di-GMP signaling. J Biol Chem. 2006;281:32015–32024. doi: 10.1074/jbc.M603589200. [DOI] [PubMed] [Google Scholar]
- Christen M, Christen B, Allan MG, Folcher M, Jeno P, Grzesiek S, Jenal U. DgrA is a member of a new family of cyclic diguanosine monophosphate receptors and controls flagellar motor function in Caulobacter crescentus. Proc Natl Acad Sci USA. 2007;104:4112–4117. doi: 10.1073/pnas.0607738104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotter PA, Stibitz S. c-di-GMP-mediated regulation of virulence and biofilm formation. Curr Opin Microbiol. 2007;10:17–23. doi: 10.1016/j.mib.2006.12.006. [DOI] [PubMed] [Google Scholar]
- D'Argenio DA, Calfee MW, Rainey PB, Pesci EC. Autolysis and autoaggregation in Pseudomonas aeruginosa colony morphology mutants. J Bacteriol. 2002;184:6481–6489. doi: 10.1128/JB.184.23.6481-6489.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duke TA, Bray D. Heightened sensitivity of a lattice of membrane receptors. Proc Natl Acad Sci USA. 1999;96:10104–10108. doi: 10.1073/pnas.96.18.10104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falke JJ, Bass RB, Butler SL, Chervitz SA, Danielson MA. The two-component signaling pathway of bacterial chemotaxis: a molecular view of signal transduction by receptors, kinases, and adaptation enzymes. Annu Rev Cell Dev Biol. 1997;13:457–512. doi: 10.1146/annurev.cellbio.13.1.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrández A, Hawkins AC, Summerfield DT, Harwood CS. Cluster II che genes from Pseudomonas aeruginosa are required for an optimal chemotactic response. J Bacteriol. 2002;184:4374–4383. doi: 10.1128/JB.184.16.4374-4383.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman L, Kolter R. Two genetic loci produce distinct carbohydrate-rich structural components of the Pseudomonas aeruginosa biofilm matrix. J Bacteriol. 2004a;186:4457–4465. doi: 10.1128/JB.186.14.4457-4465.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman L, Kolter R. Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Mol Microbiol. 2004b;51:675–690. doi: 10.1046/j.1365-2958.2003.03877.x. [DOI] [PubMed] [Google Scholar]
- Garcia B, Latasa C, Solano C, Garcia-del Portillo F, Gamazo C, Lasa I. Role of the GGDEF protein family in Salmonella cellulose biosynthesis and biofilm formation. Mol Microbiol. 2004;54:264–277. doi: 10.1111/j.1365-2958.2004.04269.x. [DOI] [PubMed] [Google Scholar]
- Güvener ZT, Tifrea DF, Harwood CS. Two different Pseudomonas aeruginosa chemosensory signal transduction complexes localize to cell poles and form and remould in stationary phase. Mol Microbiol. 2006;61:106–118. doi: 10.1111/j.1365-2958.2006.05218.x. [DOI] [PubMed] [Google Scholar]
- Hickman JW, Tifrea DF, Harwood CS. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc Natl Acad Sci USA. 2005;102:14422–14427. doi: 10.1073/pnas.0507170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene. 1998;212:77–86. doi: 10.1016/s0378-1119(98)00130-9. [DOI] [PubMed] [Google Scholar]
- Horton RM, Ho SN, Pullen JK, Hunt HD, Cai Z, Pease LR. Gene splicing by overlap extension. Methods Enzymol. 1993;217:270–279. doi: 10.1016/0076-6879(93)17067-f. [DOI] [PubMed] [Google Scholar]
- Huang B, Whitchurch CB, Mattick JS. FimX, a multidomain protein connecting environmental signals to twitching motility in Pseudomonas aeruginosa. J Bacteriol. 2003;185:7068–7076. doi: 10.1128/JB.185.24.7068-7076.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson KD, Starkey M, Kremer S, Parsek MR, Wozniak DJ. Identification of psl, a locus encoding a potential exopolysaccharide that is essential for Pseudomonas aeruginosa PAO1 biofilm formation. J Bacteriol. 2004;186:4466–4475. doi: 10.1128/JB.186.14.4466-4475.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenal U, Malone J. Mechanisms of cyclic-di-GMP signaling in bacteria. Annu Rev Genet. 2006;40:385–407. doi: 10.1146/annurev.genet.40.110405.090423. [DOI] [PubMed] [Google Scholar]
- Kader A, Simm R, Gerstel U, Morr M, Römling U. Hierarchical involvement of various GGDEF domain proteins in rdar morphotype development of Salmonella enterica serovar Typhimurium. Mol Microbiol. 2006;60:602–616. doi: 10.1111/j.1365-2958.2006.05123.x. [DOI] [PubMed] [Google Scholar]
- Kazmierczak BI, Lebron MB, Murray TS. Analysis of FimX, a phosphodiesterase that governs twitching motility in Pseudomonas aeruginosa. Mol Microbiol. 2006;60:1026–1043. doi: 10.1111/j.1365-2958.2006.05156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YK, McCarter LL. ScrG, a GGDEF-EAL protein, participates in regulating swarming and sticking in Vibrio parahaemolyticus. J Bacteriol. 2007;189:4094–4107. doi: 10.1128/JB.01510-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulasakara H, Lee V, Brencic A, Liberati N, Urbach J, Miyata S, et al. Analysis of Pseudomonas aeruginosa diguanylate cyclases and phosphodiesterases reveals a role for bis-(3′-5′)-cyclic-GMP in virulence. Proc Natl Acad Sci USA. 2006;103:2839–2844. doi: 10.1073/pnas.0511090103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee VT, Matewish JM, Kessler JL, Hyodo M, Hayakawa Y, Lory S. A cyclic-di-GMP receptor required for bacterial exopolysaccharide production. Mol Microbiol. 2007;65:1474–1484. doi: 10.1111/j.1365-2958.2007.05879.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Hazelbauer GL. Cellular stoichiometry of the components of the chemotaxis signaling complex. J Bacteriol. 2004;186:3687–3694. doi: 10.1128/JB.186.12.3687-3694.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim B, Beyhan S, Meir J, Yildiz FH. Cyclic-diGMP signal transduction systems in Vibrio cholerae: modulation of rugosity and biofilm formation. Mol Microbiol. 2006;60:331–348. doi: 10.1111/j.1365-2958.2006.05106.x. [DOI] [PubMed] [Google Scholar]
- Ma L, Lu H, Sprinkle A, Parsek MR, Wozniak D. Pseudomonas aeruginosa Psl is a galactose- and mannose-rich exopolysaccharide. J Bacteriol. 2007;189:8353–8356. doi: 10.1128/JB.00620-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddock JR, Shapiro L. Polar location of the chemoreceptor complex in the Escherichia coli cell. Science. 1993;259:1717–1723. doi: 10.1126/science.8456299. [DOI] [PubMed] [Google Scholar]
- Matsukawa M, Greenberg EP. Putative exopolysaccharide synthesis genes influence Pseudomonas aeruginosa biofilm development. J Bacteriol. 2004;186:4449–4456. doi: 10.1128/JB.186.14.4449-4456.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez-Ortiz MM, Hyodo M, Hayakawa Y, Membrillo-Hernandez J. Genome-wide transcriptional profile of Escherichia coli in response to high levels of the second messenger 3′,5′-cyclic diguanylic acid. J Biol Chem. 2006;281:8090–8099. doi: 10.1074/jbc.M510701200. [DOI] [PubMed] [Google Scholar]
- Merighi M, Lee VT, Hyodo M, Hayakawa Y, Lory S. The second messenger bis-(3′-5′)-cyclic-GMP and its PilZ domain-containing receptor Alg44 are required for alginate biosynthesis in Pseudomonas aeruginosa. Mol Microbiol. 2007;65:876–895. doi: 10.1111/j.1365-2958.2007.05817.x. [DOI] [PubMed] [Google Scholar]
- Newman JR, Fuqua C. Broad-host-range expression vectors that carry the 1-arabinose-inducible Escherichia coli araBAD promoter and the araC regulator. Gene. 1999;227:197–203. doi: 10.1016/s0378-1119(98)00601-5. [DOI] [PubMed] [Google Scholar]
- Paul R, Weiser S, Amiot NC, Chan C, Schirmer T, Giese B, Jenal U. Cell cycle-dependent dynamic localization of a bacterial response regulator with a novel di-guanylate cyclase output domain. Genes Dev. 2004;18:715–727. doi: 10.1101/gad.289504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul R, Abel S, Wassmann P, Beck A, Heerklotz H, Jenal U. Activation of the diguanylate cyclase PleD by phosphorylation-mediated dimerization. J Biol Chem. 2007;282:29170–29177. doi: 10.1074/jbc.M704702200. [DOI] [PubMed] [Google Scholar]
- Pratt JT, Tamayo R, Tischler AD, Camilli A. PilZ domain proteins bind cyclic diguanylate and regulate diverse processes in Vibrio cholerae. J Biol Chem. 2007;282:12860–12870. doi: 10.1074/jbc.M611593200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainey PB, Travisano M. Adaptive radiation in a heterogeneous environment. Nature. 1998;394:69–72. doi: 10.1038/27900. [DOI] [PubMed] [Google Scholar]
- Ramelot TA, Yee A, Cort JR, Semesi A, Arrowsmith CH, Kennedy MA. NMR structure and binding studies confirm that PA4608 from Pseudomonas aeruginosa is a PilZ domain and a c-di-GMP binding protein. Proteins. 2007;66:266–271. doi: 10.1002/prot.21199. [DOI] [PubMed] [Google Scholar]
- Remminghorst U, Rehm BH. Alg44, a unique protein required for alginate biosynthesis in Pseudomonas aeruginosa. FEBS Lett. 2006;580:3883–3888. doi: 10.1016/j.febslet.2006.05.077. [DOI] [PubMed] [Google Scholar]
- Römling U, Amikam D. Cyclic di-GMP as a second messenger. Curr Opin Microbiol. 2006;9:218–228. doi: 10.1016/j.mib.2006.02.010. [DOI] [PubMed] [Google Scholar]
- Römling U, Gomelsky M, Galperin MY. C-di-GMP: the dawning of a novel bacterial signalling system. Mol Microbiol. 2005;57:629–639. doi: 10.1111/j.1365-2958.2005.04697.x. [DOI] [PubMed] [Google Scholar]
- Ryan RP, Fouhy Y, Lucey JF, Dow JM. Cyclic di-GMP signaling in bacteria: recent advances and new puzzles. J Bacteriol. 2006;188:8327–8334. doi: 10.1128/JB.01079-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryjenkov DA, Simm R, Römling U, Gomelsky M. The PilZ domain is a receptor for the second messenger c-di-GMP: the PilZ domain protein YcgR controls motility in enterobacteria. J Biol Chem. 2006;281:30310–30314. doi: 10.1074/jbc.C600179200. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1989. [Google Scholar]
- Simm R, Morr M, Kader A, Nimtz M, Römling U. GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition from sessility to motility. Mol Microbiol. 2004;53:1123–1134. doi: 10.1111/j.1365-2958.2004.04206.x. [DOI] [PubMed] [Google Scholar]
- Simon R, Priefer U, Puhler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology. 1983;1:784–789. [Google Scholar]
- Sourjik V, Berg HC. Localization of components of the chemotaxis machinery of Escherichia coli using fluorescent protein fusions. Mol Microbiol. 2000;37:740–751. doi: 10.1046/j.1365-2958.2000.02044.x. [DOI] [PubMed] [Google Scholar]
- Sourjik V, Berg HC. Functional interactions between receptors in bacterial chemotaxis. Nature. 2004;428:437–441. doi: 10.1038/nature02406. [DOI] [PubMed] [Google Scholar]
- Spiers AJ, Kahn SG, Bohannon J, Travisano M, Rainey PB. Adaptive divergence in experimental populations of Pseudomonas fluorescens. I. Genetic and phenotypic bases of wrinkly spreader fitness. Genetics. 2002;161:33–46. doi: 10.1093/genetics/161.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu Rev Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- Stock JB, Surette MG. Chemotaxis. In: Neidhardt FC, Curtis R III, Ingraham JL, Lin ECC, Low KB, Magasanik B, et al., editors. Escherichia coli and Salmonella: Cellular and Molecular Biology. American Society for Microbiology; Washington, DC: 1996. pp. 1103–1129. [Google Scholar]
- Stover CK, Pham XQ, Erwin AL, Mizoguchi SD, Warrener P, Hickey MJ, et al. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature. 2000;406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- Studdert CA, Parkinson JS. Crosslinking snapshots of bacterial chemoreceptor squads. Proc Natl Acad Sci USA. 2004;101:2117–2122. doi: 10.1073/pnas.0308622100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamayo R, Pratt JT, Camilli A. Roles of cyclic diguanylate in the regulation of bacterial pathogenesis. Annu Rev Microbiol. 2007;61:131–148. doi: 10.1146/annurev.micro.61.080706.093426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiem S, Kentner D, Sourjik V. Positioning of chemosensory clusters in E. coli and its relation to cell division. EMBO J. 2007;26:1615–1623. doi: 10.1038/sj.emboj.7601610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tischler AD, Camilli A. Cyclic diguanylate (c-di-GMP) regulates Vibrio cholerae biofilm formation. Mol Microbiol. 2004;53:857–869. doi: 10.1111/j.1365-2958.2004.04155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tischler AD, Camilli A. Cyclic diguanylate regulates Vibrio cholerae virulence gene expression. Infect Immun. 2005;73:5873–5882. doi: 10.1128/IAI.73.9.5873-5882.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss DS, Chen JC, Ghigo JM, Boyd D, Beckwith J. Localization of FtsI (PBP3) to the septal ring requires its membrane anchor, the Z ring, FtsA, FtsQ, and FtsL. J Bacteriol. 1999;181:508–520. doi: 10.1128/jb.181.2.508-520.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolanin PM, Stock JB. Bacterial chemosensing: cooperative molecular logic. Curr Biol. 2004;14:R486–R487. doi: 10.1016/j.cub.2004.06.018. [DOI] [PubMed] [Google Scholar]
- Yang Z, Ma X, Tong L, Kaplan HB, Shimkets LJ, Shi W. Myxococcus xanthus dif genes are required for biogenesis of cell surface fibrils essential for social gliding motility. J Bacteriol. 2000;182:5793–5798. doi: 10.1128/jb.182.20.5793-5798.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Khursigara CM, Hartnell LM, Subramaniam S. Direct visualization of Escherichia coli chemotaxis receptor arrays using cryo-electron microscopy. Proc Natl Acad Sci USA. 2007;104:3777–3781. doi: 10.1073/pnas.0610106104. [DOI] [PMC free article] [PubMed] [Google Scholar]