Summary
Reversible posttranslational modifications are emerging as critical regulators of mitochondrial proteins and metabolism. Here, we use a label-free quantitative proteomic approach to characterize the lysine succinylome in liver mitochondria and its regulation by the desuccinylase SIRT5. A total of 1190 unique sites were identified as succinylated, and 386 sites across 140 proteins representing several metabolic pathways including β-oxidation and ketogenesis were significantly hypersuccinylated in Sirt5−/− animals. Loss of SIRT5 leads to accumulation of medium- and long-chain acylcarnitines and decreased β-hydroxybutyrate production in vivo. In addition, we demonstrate that SIRT5 regulates succinylation of the rate-limiting ketogenic enzyme 3-hydroxy-3-methylglutaryl-CoA synthase 2 (HMGCS2) both in vivo and in vitro. Finally, mutation of hypersuccinylated residues K83 and K310 on HMGCS2 to glutamic acid strongly inhibits enzymatic activity. Taken together, these findings establish SIRT5 as a global regulator of lysine succinylation in mitochondria and present a mechanism for inhibition of ketogenesis through HMGCS2.
Introduction
Proteins undergo posttranslational modifications that modulate their structural conformation, activity, stability, or subcellular localization. Small acyl groups, such as acetyl, modify lysine side chains, resulting in lysine acetylation and neutralization of its positive charge. Succinylation was recently identified as another significant modification of lysine residues (Zhang et al., 2011). The substrate for succinylation is presumably the succinyl group derived from succinyl-CoA. Since the succinyl group is more bulky and causes a greater change in charge than an acetyl group, one could expect that succinylation will affect protein function in ways that are different from acetylation. Acetyl-CoA and succinyl-CoA are important metabolites at central nodes of intermediary metabolism, including oxidative and anaplerotic fluxes through the tricarboxylic acid (TCA) cycle. During catabolic conditions, acyl-CoAs of different lengths can be generated from the breakdown of diverse nutrient substrates, and their levels may fluctuate substantially under different metabolic conditions (He et al., 2012; Newman et al., 2012). The different acyl-CoAs and their modification of proteins might therefore constitute an important signaling mechanism for cells to sense and respond to changes in metabolic status (He et al., 2012; Newman et al., 2012).
Accumulating evidence suggests that protein acetylation dynamically interacts with proteins involved in energy metabolism. In prokaryotes and eukaryotes, acetylation regulates the functions of many metabolic enzymes (Wang et al., 2010; Zhao et al., 2010). Lysine acetylation is regulated by the competing activities of acetyltransferases, such as p300, and deacetylases, such as the nicotinamide adenine dinucleotide (NAD+)-dependent class III protein deacetylases sirtuins (Glozak et al., 2005). Three of the 7 mammalian sirtuins, SIRT3, 4 and 5, are located in mitochondria. SIRT3 is the major deacetylase in mitochondria (Lombard et al., 2007). Our recent label-free quantitative proteomic study of the lysine acetylome reveals that SIRT3 targets about 13% of all acetylated lysines identified on 136 proteins in mitochondria, many of which are critical metabolic enzymes (Rardin et al., 2013). Mitochondrial protein hyperacetylation due to SIRT3 deficiency has severe consequences and may lead to metabolic disorders in mice and humans (He et al., 2012; Hirschey et al., 2011).
The regulation of lysine succinylation is less well understood. SIRT5, a mitochondrial sirtuin with very weak deacetylase activity, is a potent desuccinylase (Du et al., 2011; Peng et al., 2011). The catalytic reaction involves the removal of a succinyl group from the lysine side chain of protein substrates, consumes NAD+ as a co-substrate, and generates nicotinamide (NAM) and 2’-O-succinyl-ADP-ribose (Figure 1A). Two amino acids—tyrosine (Y102) and arginine (R105), located within the catalytic pocket of SIRT5—are required for desuccinylase activity (Du et al., 2011). Their positive charges may explain the preference of SIRT5 for negatively charged acyl groups, such as succinyllysine. Studies that address the biological role of lysine succinylation and its regulation by SIRT5 have been hampered by a lack of knowledge of the proteins and lysine residues that contain succinyl modifications. Only two studies have characterized SIRT5 function. These reported a role for SIRT5 in regulating the urea cycle enzyme carbamoyl phosphate synthetase 1 (CPS1) by desuccinylation (Du et al., 2011) or possibly deacetylation (Nakagawa et al., 2009). Therefore, characterization of the lysine succinylome across the entire mitochondrial proteome will shed light on the biological function of succinylation and SIRT5.
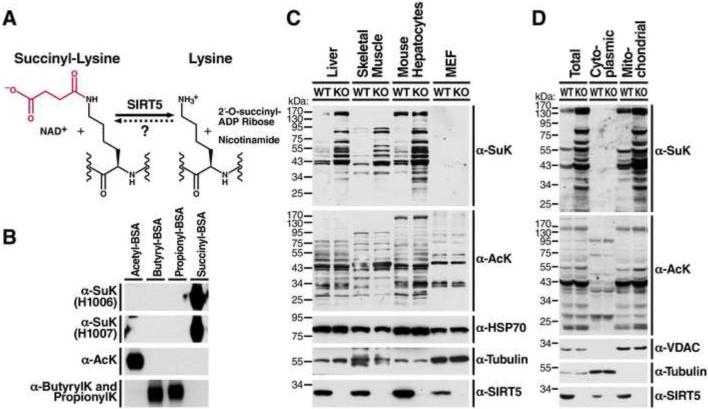
Figure 1. Generation of succinyl-lysine-specific antibodies and characterization of succinylation distribution in mouse tissues, cultured cells and subcellular compartments.
(A) The structure of lysine succinylation and the catalytic reaction of desuccinylation by SIRT5. The succinyl group on the lysine residue is indicated in red. (B) Succinyl-lysine antibodies specifically detected succinyl-BSA, but not acetyl-BSA or butyryl- or propionyl-BSA. (C) Western blot using succinyl-lysine-specific antibodies was performed to assess succinylation and acetylation levels in mouse liver, skeletal muscle, primary cultured mouse hepatocytes and MEFs derived from wild-type (WT) or Sirt5−/− mice (See also Figure S1A). Equal amount of proteins were loaded into each lane. Loading controls, α-tubulin and mitochondrial protein HSP70, were also examined. Western blot for SIRT5 confirmed the absence of SIRT5 protein in Sirt5−/− mouse tissues or cells. (D) Protein succinylation and acetylation in whole cell lysates, cytoplasmic or mitochondrial fractions derived from WT or Sirt5−/− mouse livers. Western blot for cytoplasmic protein α-tubulin and mitochondrial protein voltage dependent anion channel (VDAC) confirmed the purity of subcellular fractionation. Western blot for SIRT5 showed the presence of SIRT5 in both cytoplasm and mitochondria of WT livers, but not in Sirt5−/− livers.
In the present study, we generated antibodies for the specific enrichment of succinyl-lysine containing peptides and applied a label-free quantitative proteomic method called MS1 Filtering (Schilling et al., 2012) to characterize the mitochondrial succinylome in a metabolically active and highly aerobic organ, liver of wild-type (WT) and Sirt5−/− mice. Pathway analysis of SIRT5 targeted proteins followed by both in vitro and in vivo investigation further revealed an important role for SIRT5 in regulating fatty acid β-oxidation and ketone body synthesis.
Results
Lysine succinylation is strongly enriched in liver mitochondria
To detect pan-lysine succinylation, we generated polyclonal antibodies by immunizing a rabbit with succinylated keyhole limpet hemocyanin. The specificity of newly developed antibodies was tested by western blot analysis with BSA carrying several distinct acylation modifications. Two rabbit sera containing succinyl-lysine antibodies, H1006 and H1007, showed strong and specific reaction with succinyl-BSA, but not with acetyl-BSA, butyryl-BSA, or propionyl-BSA (Figure 1B). In contrast, the acetyl-lysine antibody strongly recognized acetyl-BSA and the pan-butyryl- and propionyl-lysine antibody recognized butyryl-BSA and propionyl-BSA (Figure 1B).
Next, we screened global protein succinylation in various organs and primary cell lines of WT or Sirt5−/− mice with these antibodies. In agreement with published data showing a potent desuccinylase activity of SIRT5 (Du et al., 2011; Peng et al., 2011), protein succinylation increased in Sirt5−/− mouse tissues, including liver, skeletal muscle, and primary hepatocytes (Figures 1C and S1A). In contrast, protein acetylation levels were unchanged in both Sirt5−/− mouse tissues and primary cell lines (Figures 1C and S1A). Importantly, levels of succinyl-CoA and succinate remain unchanged in the Sirt5−/− animals under both the fed and fasted state implicating SIRT5 as the major regulator of lysine succinylation (Figure S1B and S1C). Surprisingly, lysine succinylation was least abundant in mouse embryonic fibroblasts (MEFs) when compared with equal amounts of protein (Figure 1C). As SIRT5 is localized to both the cytoplasmic and mitochondrial compartments, we examined the amount of total protein succinylation in these two subcellular fractions. Mouse liver mitochondria were strongly enriched in lysine succinylated proteins while a mitochondrial depleted cytoplasmic fraction showed minimal staining when equal amounts of protein were compared (Figure 1D). These results confirm the broad desuccinylase activity of SIRT5 across tissues in vivo and indicate succinylated proteins are strongly enriched in liver mitochondria.
Defining and quantifying the lysine succinylome in liver mitochondria
To identify proteins and specific sites of lysine succinylation in mitochondria, we developed a workflow to enrich succinylated peptides for identification by mass spectrometry (MS) (Figure 2A). Mouse liver mitochondria were isolated from five WT and five Sirt5−/− mice by differential centrifugation. Equal amounts of individual protein samples were digested with trypsin, and 15 μg was removed to quantify protein expression levels in WT and knock out (KO) mice. A heavily labeled succinylated lysine (SuK) peptide, ADIAESQVNsuKLR[13C 156N4], was spiked into the remaining mitochondrial protein digest as a process-loading control and normalization factor for subsequent label-free quantification. As we showed that multiple antibodies increase the diversity of enrichment (Schilling et al., 2012), succinylated peptides were immunoprecipitated with equal amounts of two polyclonal antibodies (Figure 1B). Samples were analyzed in duplicate by liquid chromatography (LC)-MS/MS on a TripleTOF 5600 MS, and data were searched against the mouse proteome. We identified 2183 SuK peptides with a false discovery rate (FDR) of ≤1% (Dataset S1) that correspond to 1190 lysine succinylation sites across 252 proteins (Figure 2B). Of the 1190 sites identified, 64% were identified only in the KO, 10% only in the WT, and 26% in both (Figure S2). When the data were searched against other lysine modifications, such as acetylation and malonylation, no additional sites were identified, demonstrating the specificity of the antibodies for lysine succinylation.
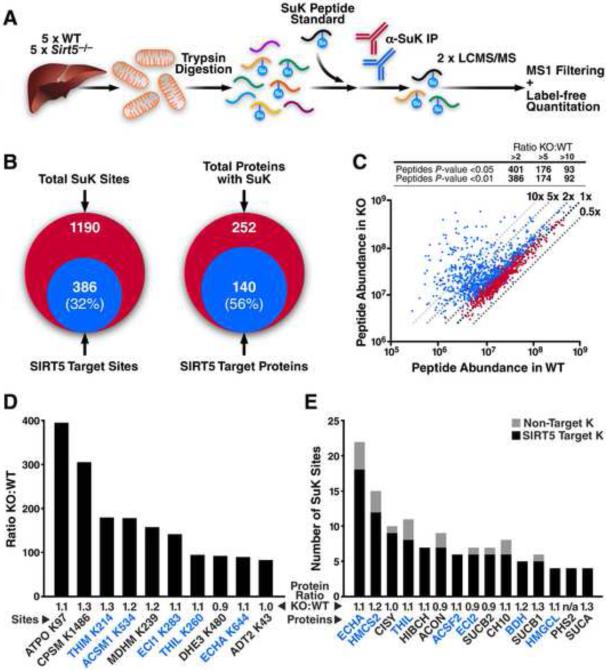
Figure 2. Enrichment and identification of liver mitochondrial lysine succinylome by label-free quantitation.
(A) Liver mitochondria were isolated from five individual WT and Sirt5−/− mice. Mitochondrial protein from each of the 10 samples was digested separately with trypsin, desalted, and 150 fmol of a heavy isotope-labeled succinyl-lysine peptide standard was added. Succinyl-lysine-containing peptides were immunoprecipitated and analyzed in duplicate by LC-MS/MS. Precursor ion intensity chromatograms were integrated using MS1 Filtering in Skyline and for label-free quantitation. Venn diagrams of lysine succinylated proteins and peptides identified in WT and Sirt5−/− are shown in Figure S2. (B) Overlap of the number of succinylated peptides and proteins identified with the number that were targeted by SIRT5 (> twofold increase and p <0.01). For peptide information and quantitation of individual sites see Dataset S1 and S2. (C) Scatter plot of the SuK peptides quantitated by MS1 filtering. Dashed lines indicate the fold change of abundance in KO samples in comparison to WT samples. Peptides with a significant change are shown in blue (p <0.05), and the others are in red. Inset summarizes the number of SuK peptides that show significant change in KO samples. (D) Largest fold-changes (KO:WT) for individual SuK sites with the average protein expression ratio displayed. (E) Highly regulated proteins enriched with SIRT5 target sites with the average protein expression ratio displayed. Number of SIRT5 target sites in black, number of non-target sites in grey. See Dataset S3 and S4 for mass spec details of identified peptides used for protein quantification.
Abbreviations include ATP synthase subunit O (ATPO), carbamoyl-phosphate synthetase 1 (CPSM), 3-ketoacyl-CoA thiolase (THIM), acyl-coenzyme A synthetase medium-chain family member 1 (ACSM1), malate dehydrogenase (MDHM), enoyl-CoA delta isomerase (ECI1), acetyl-CoA acetyltransferase (THIL), glutamate dehydrogenase (DHE3), trifunctional enzyme α subunit (ECHA), ADP/ATP translocase 2 (ADT2), 3-hydroxy-3-methylglutaryl-CoA synthase 2 (HMCS2), citrate synthase (CISY), 3-hydroxyisobutyryl-CoA hydrolase (HIBCH), aconitase (ACON), acyl-CoA synthetase family member 2 (ACSF2), enoyl-CoA delta isomerase 2 (ECI2), GDP-specific succinyl-CoA synthetase β subunit (SUCB2), HSP10 (CH10), 3-hydroxybutyrate dehydrogenase (BDH), ADP-specific succinyl-CoA synthetase β subunit (SUCB1), 3-hydroxy-3-methylglutaryl-CoA lyase (HMGCL), prostaglandin G/H synthase 2 (PHS2), and succinyl-CoA synthetase α subunit (SUCA).
Within the mouse liver succinylome, we sought to identify substrates of SIRT5 with a label-free quantitative method, MS1 Filtering (Schilling et al., 2012). With this method, we can measure intact precursor ion abundance for succinylated peptides across WT and KO samples. The peptide standard had a coefficient of variation (CV) of 24% across all samples indicating strong reproducibility between enrichments. We used published criteria to select representative peptides for quantitation, including charge state abundance, tryptic cleavage, and lack of secondary modifications (Rardin et al., 2009). Precursor ion intensities were normalized to the peptide standard before calculating peptide ratios (KO:WT). Quantitative data generated on 992 sites from 221 proteins revealed 32% of SuK sites (386) across 56% of the identified proteins (140) were increased by more than twofold (p <0.01) (Figure 2B and Dataset S2). Sites of increased succinylation in the KO samples showed a wide distribution of changes with 386 sites increased by more than twofold and 92 sites increased by more than 10-fold (Figure 2C). To control for possible changes in protein expression levels in response to the loss of SIRT5, we used MS1 Filtering to quantitatively analyze peptides from the total mitochondrial protein digest before enrichment. Samples from each mouse were analyzed in duplicate by LC-MS/MS, and peptides quantified from 203 of the 252 succinylated proteins identified in our SuK enrichment showed no changes in expression and average peptide KO:WT ratios near 1.0. These results indicate the observed changes in SuK peptide abundances were due to change in protein succinylation in the absence of SIRT5 and not to altered protein expression levels (Dataset S3). Peptides quantified from an additional 33 non-succinylated mitochondrial proteins were also unchanged in the KO mice (Dataset S3), suggesting limited remodeling of the mitochondrial proteome in response to SIRT5 deletion.
Loss of SIRT5 leads to hyper-succinylation of mitochondrial proteins
MS1 Filtering analysis revealed large-scale increases in lysine succinylation at specific sites from a variety of mitochondrial proteins (Figure 2D). We identified 12 SuK sites on the electron transport chain (ETC) complex V subunit, ATPO, that showed the most robust increase in succinylation at K97 with a > 300-fold increase. Several other sites on this protein were unchanged (Dataset S2). Other proteins showed strong increases in SuK, including the urea cycle enzyme carbamoyl phosphate synthetase 1 at K1486, the TCA cycle enzyme malate dehydrogenase at K239, and several enzymes involved in fatty acid metabolism, including 3-ketoacyl-CoA thiolase, acyl-coenzyme A synthetase medium-chain family member 1, enoyl-CoA delta isomerase 1, and the trifunctional enzyme α subunit. Therefore, loss of SIRT5 leads to a dramatic increase in site-specific SuK of mitochondrial proteins across several metabolic pathways.
We calculated the enrichment of SIRT5 target SuK on proteins by using a binomial distribution analysis. Fifteen proteins are significantly more abundant with SIRT5 target sites than expected from the total number of SuK sites on each protein (Figure 2E, p <0.05). For instance, SIRT5 targets 18 out of 22 SuK sites on the trifunctional enzyme α subunit, 12 out of 15 sites on 3-hydroxy-3-methylglutaryl-CoA synthase 2, and 9 out of 10 sites on citrate synthase (Figure 2E) (Dataset S2). Importantly, multiple enzymes involved in fatty acid metabolism and ketone body metabolism show up on the lists of highly regulated sites (Figure 2D) and highly regulated proteins (Figure 2E), which are highlighted in blue on both figures.
Succinylation does not appear to be evenly distributed across the mitochondrial proteome. More than 80 proteins have a single SuK site, and 12 proteins have at least 15 SuK sites (Figure 3A). Almost half of succinylated proteins contain a single SIRT5 target site, while four proteins have more than 10 SIRT5 target sites (Figure 3B).
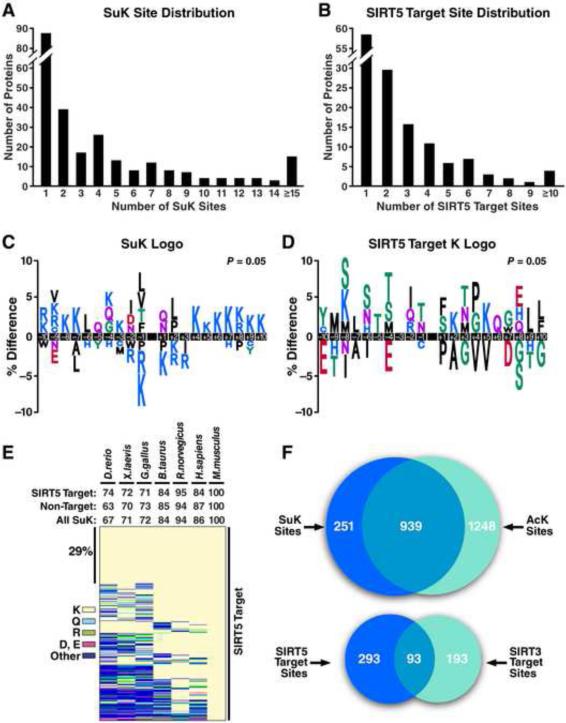
Figure 3. Site distribution, conservation, and sequence logo analysis.
(A) Distribution of the number of succinylation sites per protein. (B) Distribution of the number of SIRT5 target sites per protein. For peptide information and quantitation of individual sites see Dataset S1 and S2. (C) Consensus sequence logo plot for succinylation sites ± 10 amino acids from the lysine of all succinylated sites identified, (D) from the lysine of all SIRT5 target sites (> twofold and p <0.01). SIRT5 target sequence context heatmap is also shown in Figure S3A. (E) Heatmap depicting the conservation index of the SIRT5 target sites across seven vertebrate species. Percent conservation calculated for all succinylated sites, SIRT5 target sites or non-target sites is shown above the heatmap. Lysine (K), glutamine (Q), arginine (R), aspartic acid (D) or glutamic acid (E), and other amino acids are presented in different colors. Heatmap depicting the conservation index of non-target sites is available at Figure S3B. See Dataset S5 for conservation index analysis details. (F) Venn diagrams showing the overlap of succinylated and acetylated lysine residues identified in mouse liver mitochondria and the overlap of sites that are targeted by SIRT5 and SIRT3.
Succinylation and SIRT5 target sites with distinct sequence motif features
As succinylation and SIRT5 appear to selectively target specific sites, we were intrigued to determine if there is a common sequence motif for succinylation or SIRT5 regulation. We compared the amino acid sequences surrounding all succinylated sites to non-succinylated sites (Figure 3C) or all SIRT5 target sites to non-target succinylated sites (Figure 3D) by iceLogo (https://code.google.com/p/icelogo/). Notably, positively charged amino acids (lysine or arginine) were strongly excluded from positions −1 and +1 of the succinylation logo (Figure 3C). Positions close to the modified site (−2, −1 and +2) had modest enrichment for small nonpolar hydrophobic amino acids, such as leucine, valine, or isoleucine, while positions farther away (−10 to −7 and +4 to +10) were enriched with positively charged lysine or arginine (Figure 3C). The SIRT5 target logo is distinct from the succinylation logo. Notably, serine or threonine was enriched at multiple positions (−8, −6, −5, −4, −1, +1 and +3) of the SIRT5 target logo (Figure 3D). The average KO/WT fold-change of all SuK peptides with a given amino acid at a given position was calculated and illustrated in a sequence context heatmap (Figure S3A). Interestingly, serine and threonine were preferred at positions −6, −4, −1, +1, +3 and +5, which coincide with the SIRT5 target sequence logo generated using iceLogo.
Conservation analysis of succinylation and SIRT5 target lysine sites
Lysine succinylation is found from yeast to mammal (Zhang et al., 2011). Sirtuins are a highly conserved family of proteins (Greiss and Gartner, 2009). We therefore determined if succinylated sites or SIRT5 target sites are evolutionarily conserved by generating a conservation index across vertebrate proteomes with AL2CO (Pei and Grishin, 2001). Of the SuK sites identified in mouse liver mitochondria, 86% were conserved in human (Homo sapiens) and 67% were conserved in zebrafish (Danio rerio) (Figure 3E). The conservation of SuK sites or SIRT5 target sites in any of the other six species (human, rat, cattle, bird, frog, zebrafish) across vertebrates was above 60%, when compared against mouse. About 29% of SIRT5 target sites were 100% conserved in all seven species. The conservation index was however not apparently different between SIRT5 target sites and non-target sites, except in zebrafish (p <0.001, chi squared test) (Figures 3E and S3B).
Lysine acetylome and succinylome overlap significantly in the mitochondria
To determine the extent of the overlap of lysine modifications in liver mitochondria with succinylation, we compared our succinylation data to our recently published dataset of SIRT3-targeted mitochondrial lysine acetylation in mouse liver that employed similar enrichment and label-free quantification methods (Rardin et al., 2013). Of 1190 sites of lysine succinylation across 252 proteins identified in the present study, we find that 939 sites (~79%) were also acetylated in the SIRT3 study (Figure 3F). This indicates a strong overlap of succinylation and acetylation on lysine residues in mitochondrial proteins. However, of all identified sites, only 93 were targeted by both SIRT5 and SIRT3 when we used a twofold increase cutoff (p <0.01) in their respective knockout models (Figure 3F).
Pathway analysis reveals fatty acid β-oxidation and ketone body production as highly targeted by SIRT5
To gain more insight into how succinylation and SIRT5 regulate mitochondrial metabolic networks, we performed pathway enrichment analysis with Reactome (www.reactome.org). Many metabolic pathways are significantly enriched with succinylation target proteins (Figure 4A) or SIRT5 target proteins (Figure 4B). The top pathways enriched with SIRT5 targets are β-oxidation, branched-chain amino acid catabolism, TCA cycle, ATP-synthesis, ketone body synthesis, and propionyl-CoA catabolism, which are all critical nodes in energy metabolic networks. Strikingly, 100% of proteins (4 out of 4) in ketogenesis and 93% of proteins (14 out of 15) in β-oxidation are targeted by SIRT5, as compared to 58% (11 out of 19) in branched-chain amino acid catabolism, 43% (10 out of 23) in TCA cycle, or 43% (9 out of 21) in ATP-synthesis.
Figure 4. Fatty acid β-oxidation and ketone body synthesis are highly targeted by SIRT5.
(A, B) Pathway analysis of succinylation (A) and SIRT5 targets (B) with the number of proteins identified per pathway. Fatty acid β-oxidation and ketone body synthesis are highlighted in blue. (C) Schematic depicting the core machinery of fatty acid β-oxidation and ketone body synthesis with SIRT5 target sites indicated on each protein. (D) Succinylation profiles of enzymes involved in fatty acid β-oxidation and ketone body synthesis. KO:WT ratio of each SuK site is shown in scatter plots. Red horizontal bar represents the median KO:WT ratio of all SuK sites on each protein. Dotted line indicates a KO:WT ratio of 2.
Abbreviations include very long chain acyl-CoA dehydrogenase (ACADV), long chain acyl-CoA dehydrogenase (ACADL), medium chain acyl-CoA dehydrogenase (ACADM), short chain acyl-CoA dehydrogenase (ACADS), trifunctional enzyme α subunit (ECHA) and β subunit (ECHB), 2,4-dienoyl-CoA reductase (DECR), enoyl-CoA delta isomerase 1 (ECI1), short chain enoyl-CoA hydratase (ECHM), short chain 3-hydroxyacyl-CoA dehydrogenase (HCDH), acetoacetyl-CoA thiolase (THIL), 3-hydroxy-3-methylglutaryl coenzyme A synthase 2 (HMCS2), 3-hydroxy-3-methylglutarate-CoA lyase (HMGCL), and 3-hydroxybutyrate dehydrogenase (BDH).
β-Oxidation is the process of fatty acid catabolism into acetyl-CoA in the mitochondria. Four core steps are carried out sequentially by acyl-CoA dehydrogenase family proteins (ACAD) (dehydrogenation) and the trifunctional enzyme (hydration→oxidation→thiolysis) (Figure 4C). During each cycling, a two-carbon acetyl group is liberated and activated to form acetyl-CoA. The remaining acyl-CoA shortened by two carbons continues cycling through the four steps until complete oxidation. Three ACAD family proteins of various chain-length preferences, including very long chain, long chain, and medium chain, and the trifunctional enzyme α and β subunits are all targeted by SIRT5, with some proteins targeted at multiple sites (Figures 4C and 4D). SIRT5 targeted sites are exceptionally abundant on the trifunctional enzyme α subunit which is also a target of SIRT3 (Rardin et al., 2013). The two catalytic domains enoyl-CoA hydratase and 3-hydroxyacyl-CoA dehydrogenase responsible for two sequential steps of β-oxidation-hydration and oxidation are heavily targeted by SIRT5.
Acetyl-CoA produced during β-oxidation can enter the ketone body synthesis pathway in the liver to generate ketone bodies such as acetoacetate and β-hydroxybutyrate for energy use in extrahepatic tissues such as the heart and brain when carbohydrates become limiting (Figure 4C). All four enzymes in ketone body synthesis pathway are targeted by SIRT5 at multiple sites (Figures 4C and 4D). Notably, 3-hydroxy-3-methylglutaryl-CoA synthase 2 (HMGCS2), which controls the rate-limiting conversion of acetoacetyl-CoA and acetyl-CoA into 3-hydroxy-3-methylglutaryl-CoA (HMG-CoA), contains the most succinyllysine sites and the most SIRT5 target sites (Figure 4C and 4D).
Decreased fatty acid oxidation and increased accumulation of acylcarnitines in the absence of SIRT5
The striking enrichment of SIRT5 desuccinylation targets in the fatty acid β-oxidation pathway prompted us to determine if hypersuccinylation due to loss of SIRT5 alters fatty acid metabolism. Oxidation of deuterium-labeled palmitate was measured in primary cultures of hepatocytes isolated from WT or Sirt5−/− mice. After incubating the cells with deuterium-labeled palmitate-BSA conjugates, deuterium in C-2H bonds was released from palmitate and incorporated into water. The rate of palmitate oxidation was calculated from the production of deuterium-labeled water in the culture medium. Oxidation of palmitate increased over time in both WT and Sirt5−/− cells (Figure 5A). However, Sirt5−/− cells oxidized significantly less palmitate at both time points. When cells were cultured in glucose-deprived medium for 24 hours, oxidation of palmitate significantly increased (Figure 5B). Sirt5−/− cells still oxidized significantly less palmitate even under glucose-deprived conditions. Etomoxir, an irreversible carnitine palmitoyltransferase 1 inhibitor, blocks long-chain fatty acid transport into the mitochondria and therefore inhibits fatty acid β-oxidation. Etomoxir co-incubation drastically attenuated palmitate oxidation in both WT and Sirt5−/− cells, supporting the sensitivity and specificity of this assay (Figure 5B). These data suggest a potential deficiency of β-oxidation in Sirt5−/− hepatocytes.
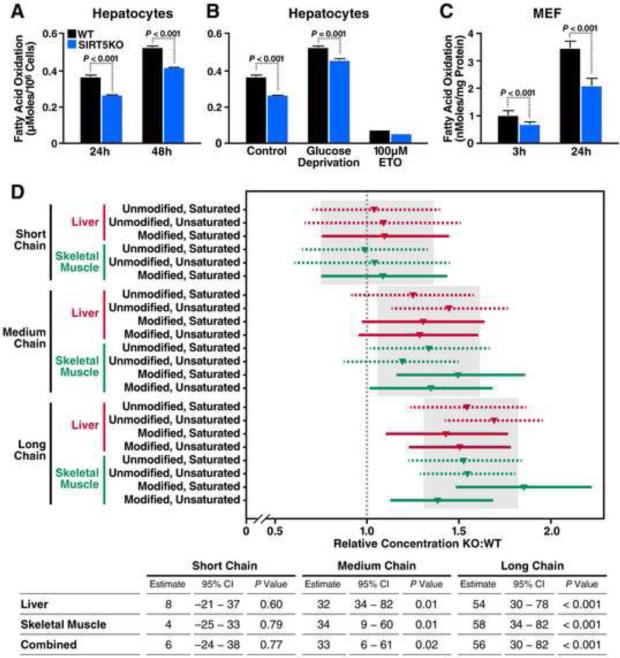
Figure 5. Lack of SIRT5 is associated with impaired fatty acid β-oxidation.
(A) Oxidation of deuterium-labeled palmitate in primary cultured hepatocytes isolated from WT (black bars) or Sirt5−/− (blue bars) mice measured after a 24- or 48-hour incubation. Y axis represents μmoles of deuterium-labeled palmitate oxidized per 106 cells (n=3). Results are shown as the mean ± standard deviation. (B) Oxidation of deuterium-labeled palmitate, when cultured in control medium, or glucose-deprived medium, or treated with 100 μM Etomoxir (ETO), measured after 24-hour incubation (n=3). Results are shown as the mean ± standard deviation. (C) Oxidation of tritium labeled palmitate in mouse embryonic fibroblasts (MEF) derived from WT (black bars) or Sirt5−/− (blue bars) mice after a 3- or 24-hour incubation. Y axis represents nmoles of tritium-labeled palmitate oxidized per mg of proteins (3 h: n=8; 24 h: n=7). Results are shown as the mean ± standard deviation. (D) Concentrations of acylcarnitines in liver or skeletal muscle of Sirt5−/− mice are shown as relative values compared to WT. The grey rectangles and their corresponding vertical grey lines show the chain-length-level inference (posterior mean and 95% CI). The horizontal lines and shapes show stratum-specific inference. Table below shows estimated percent increase, 95% CI, and P-value calculated for relative acylcarnitine concentrations in KO vs. WT in liver, skeletal muscle, or combined. Relative concentrations of individual acylcarnitines in KO vs. WT are also shown in Figure S4.
To further confirm these results, we used mouse embryonic fibroblasts (MEFs) derived from WT or Sirt5−/− mice and applied another tracer approach with tritium labeling. In agreement with the findings in primary hepatocytes, oxidation of tritium-labeled palmitate was also significantly reduced in Sirt5−/− MEFs after a 3- or 24-hour incubation (Figure 5C).
Next, to explore the role of lysine succinylation in fatty acid metabolism in vivo, we examined the levels of acylcarnitines, which are intermediates, of β-oxidation, in WT and Sirt5−/− mouse tissues. In both liver and skeletal muscle tissues from KO animals there was a broad accumulation of medium-and long-chain acylcarnitines including saturated acylcarnitine species, poly- and mono-unsaturated acylcarnitines, hydroxyacylcarnitines, and dicarboxylates (Figure 5D) consistent with the global protein hypersuccinylation observed in these tissues from Sirt5−/− mice (Figure 1C and Figure S1A). Statistical analysis of both liver and skeletal muscle acylcarnitines in a combined model demonstrated a significant increase in medium- and long-chain acylcarnitines in KO tissues (p <0.001 for long-chain; p =0.02 for medium-chain) when compared to WT tissues, while no significant difference was observed in short-chain acylcarnitines (Figure 5D). Taken together, this abnormal accumulation of β-oxidation intermediates supports in vitro flux assays and is consistent with a deficient oxidation of endogenous fatty acids in the absence of SIRT5 in both liver and skeletal muscle.
Lack of SIRT5 is associated with decreased ketone body production and hypersuccinylation of HMGCS2
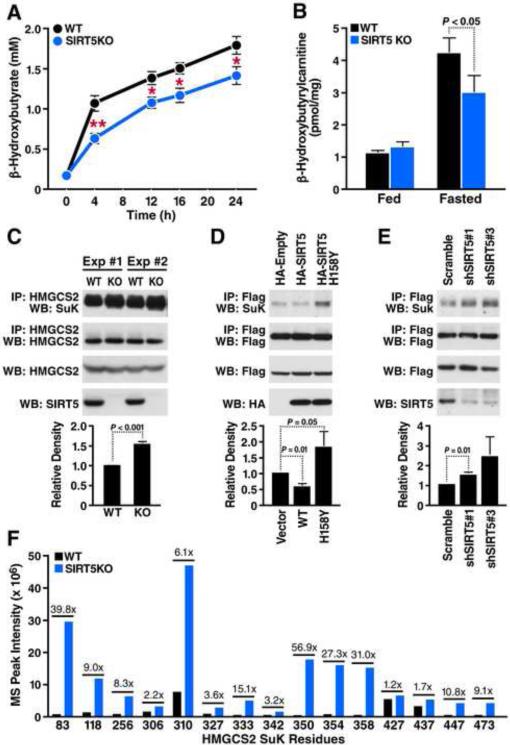
Given that all four key enzymes in ketone body synthesis are highly succinylated and targeted by SIRT5, we further tested whether ketone body production was altered in the absence of SIRT5 by measuring plasma β-hydroxybutyrate levels in WT or Sirt5−/− mice. No difference was observed between WT and Sirt5−/− mice under basal condition (Figure 6A). However, under fasting condition, Sirt5−/− mice showed significantly decreased β-hydroxybutyrate production at all time points measured starting from 4h until 24h fasting (Figure 6A). β-hydroxybutyrylcarnitine levels can reflect the pool of β-hydroxybutyrate in cells, as it is a mitochondrial intermediate derived from β-hydroxybutyryl-CoA. Levels of β-hydroxybutyrylcarnitine in Sirt5−/− mouse liver were significantly lower than in WT mouse liver under 24h fasted condition (Figure 6B), which further supports a deficient ketone body production in Sirt5−/− mice.
Figure 6. Lack of SIRT5 leads to decreased β-hydroxybutyrate production and hypersuccinylation of HMGCS2.
(A) Plasma β-hydroxybutyrate levels in WT and Sirt5−/− mice at 0, 4, 12, 16 and 24 hour following fasting (n=5, **p<0.001, *p<0.01). Results are shown as the mean ± standard error. (B) Liver β-hydroxybutyrylcarnitine levels in WT and Sirt5−/− mice under fed or 24 hour fasted condition (n=5). Results are shown as the mean ± standard error. (C) Western blot showing succinylation of endogenous HMGCS2 immunoprecipitated from WT or Sirt5−/− mouse liver (n=3). Integrated density values were calculated and are shown relative to WT mice. (D) Expression plasmids for WT HMGCS2 were transfected into HEK293 cells with an empty vector control, SIRT5 WT, or SIRT5 H158Y (catalytically inactive mutant). Immunoprecipitation and western blot were performed to examine succinylation levels of HMGCS2 (n=3). Integrated density values were calculated and are shown relative to empty vector control. (E) HEK293 cells over-expressing WT HMGCS2 were infected with lentivirus carrying scrambled shRNA or two different shRNAs for specifically knocking down SIRT5. Immunoprecipitation and western blot were performed to examine succinylation levels of HMGCS2 (n=5). Integrated density values were calculated and are shown relative to scrambled shRNA treatment. (F) Mass spectrum peak intensity of all identified succinyllysine-containing peptides measured in WT (black bars) and Sirt5−/− mice (blue bars). Fold change of each site (KO:WT) is indicated above the bars.
To investigate how succinylation and SIRT5 may regulate ketone body production, we focused on HMGCS2, the rate-limiting step of ketone body synthesis. HMGCS2 is hypersuccinylated at 12 of 15 lysine residues in the absence of SIRT5 (Figures 4C and 4D). To examine SIRT5 regulation of HMGCS2 succinylation in vivo, we immunoprecipitated endogenous HMGCS2 from WT or Sirt5−/− mouse livers and examined its succinylation by western blot (Figure 6C). Quantification of three independent experiments confirmed that succinylation of HMGCS2 is significantly higher in Sirt5−/− mice when compared to WT mice (Figure 6C). In contrast, overexpression of SIRT5 reduced succinylation of HMGCS2 in HEK293 cells (Figure 6D). Surprisingly, overexpression of a catalytically inactive mutant SIRT5-H158Y led to hypersuccinylation of HMGCS2, suggesting it may act as a dominant negative protein. Finally, knockdown of SIRT5 via shRNA led to hypersuccinylation of over-expressed HMGCS2 in HEK293 cells (Figure 6E).
Succinylation of lysine residues within the substrate binding pocket inhibits HMGCS2 activity
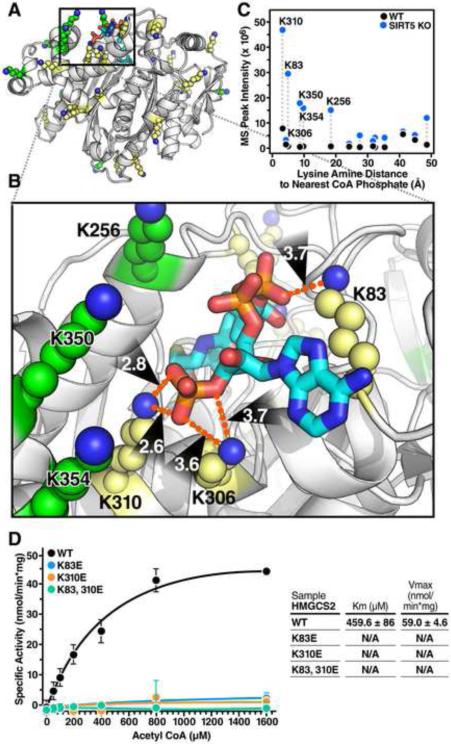
Among the 15 succinylated lysine residues identified on HMGCS2, several sites appear to be highly targeted by SIRT5 including K83, K310, K350, K354 and K358 (Figure 6F). To further investigate the possible effects of succinylation at these residues, we examined the crystal structure of human HMGCS2 bound to HMG-CoA (Figure 7A) (Shafqat et al., 2010). The structure reveals a cluster of lysine residues targeted by SIRT5 adjacent to the substrate-binding pocket of HMGCS2 with lysine amine nitrogens for residues 83, 306 and 310 within 4Å of the phosphate oxygen atoms of the substrate acetyl-CoA (Figure 7B). The observed peak intensity of specific succinyl lysine residue-containing peptides is plotted with respect to their distance to the nearest CoA phosphate group (Figure 7C). Strikingly, lysine residues K83 and K310, which are among the closest to the CoA phosphate group, are also most intensely succinylated. This observation suggests the negative charge of added succinyl groups would likely disrupt these lysines’ interactions with the phosphate groups of acetyl-CoA if not removed by SIRT5.
Figure 7. HMGCS2 mutations at sites of succinylation within the substrate binding region regulate its enzymatic activity.
(A) Ribbon diagram of the crystal structure of human HMGCS2 (PDB entry 2WYA) bound with HMG-CoA. Lysines fully observed in the crystal structure are shown in yellow, and lysines without fully defined conformations in the crystal structure are modeled in PyMol using low-energy all-trans rotamers as allowed by local backbone and steric interactions and displayed as green atomic spheres at 50% scale. (B) The interactions between several lysine side chains and the negatively charged CoA phosphate groups in the substrate binding pocket are shown in detail. Lysine amine nitrogens for residues 83, 306 and 310 are within 4Å of the phosphate oxygen atoms. (C) The observed mass spectrum peak intensity of succinyl-lysine-containing peptides plotted with respect to their distance to the nearest CoA phosphate. Dotted lines connect the same peptide identified in WT and Sirt5−/− mice. (D) Steady state kinetic analysis of WT HMGCS2 and succinyllysine mimetics HMGCS2-K83E, HMGCS2-K310E, and HMGCS2-K83,310E enzymatic activity as measured by DTNB detection of CoA-SH released from increasing concentrations of acetyl-CoA at 412 nM. Graph is representative of two independent experiments, n=3 measurements/sample, mean ± SD. Inset contains values for average Km and Vmax ± SEM. The purity and equal amounts of immunoprecipitated HMGCS2 WT or mutants are shown in Figure S5.
To test the possible effects of succinylation at these sites on HMGCS2 activity, we generated FLAG-tagged constructs of WT HMGCS2, or mutants to mimic the negative charge state of the succinyl group including K83E, K310E, and the double mutant (K83/310E). Individual constructs were transfected into HEK293 cells and proteins were isolated by immunoprecipitation (Figure S5). Steady state kinetic analysis was carried out using acetyl-CoA and acetoacetyl-CoA as substrates and activity was measured by release of CoA-SH at increasing concentrations of acetyl-CoA (Figure 7D). While Km and Vmax for WT HMGCS2 kinetic activity was reasonably similar to previous reports (Andrew Skaff and Miziorko, 2010; Shimazu et al., 2010) the individual mutants (K83E, K310E) and the double mutant (K83/310E) exhibited a complete loss of enzymatic activity (Figure 7D). Given the robust changes in succinylation at K83 and K310 in Sirt5−/− mice, their close proximity to the substrate binding pocket, and the loss of function when replaced with a negatively charged amino acid, our data support the model that lysine succinylation and desuccinylation by SIRT5 can regulate HMGCS2 activity through disrupting and restoring the binding pocket for phosphate groups of acetyl-CoA respectively.
Discussion
The reversible succinylation of lysines is a recently identified posttranslational modification with largely unknown prevalence or biological function. The present study identified 1190 unique succinyllysine sites on 252 proteins in mouse liver mitochondria in a robust affinity-enrichment and rigorous label-free quantification approach. Furthermore, SIRT5 deficiency in the liver leads to the selective hypersuccinylation (at least twofold) at 386 sites on 140 proteins, indicating 32% of identified SuK sites are SIRT5 target sites and about 56% of succinylated proteins are SIRT5 target proteins. Proteins in major metabolic pathways are targeted by succinylation and SIRT5. These pathways include ketogenesis, fatty acid β-oxidation, TCA cycle, and ATP-synthesis, which are crucial components of a cell's energy metabolic networks. In addition, we show that hypersuccinylation occurs on almost all proteins involved in fatty acid β-oxidation and ketogenesis in the absence of SIRT5, and that this switch in the succinylation state is associated with accumulation of both medium and long chain acylcarnitines as well as decreased ketone body production during fasting. Finally, we propose a mechanism by which lysine succinylation can negatively regulate ketogenesis through modification of the substrate binding pocket of HMGCS2.
After completion of our study, data emerged on lysine succinylation in liver tissue and SIRT5 targeted succinylation in mouse embryonic fibroblasts (MEFs) from WT and Sirt5−/− animals (Park et al., 2013). Interestingly of the ~1200 sites they identified in MEF's, only 294 were also identified in liver tissue, suggesting a dramatic rearrangement of the succinylome when moving into a cell culture based system. As sirtuins are thought to play a major role in metabolism, this may be reflective of the transition from in vivo to in vitro conditions. We observed a similar dramatic decrease in global protein succinylation when comparing mouse tissues with MEFs by western blot (Figure 1C). Of the 997 quantifiable sites in MEFs only 18% overlapped with our liver mitochondrial analysis, indicating that Park et al. were able to generate a broader cellular overview of lysine succinylation while our approach gained much deeper coverage of SIRT5-targeted mitochondrial proteins.
The biological function of SIRT5 remains largely obscure. Global Sirt5−/− mice develop normally and appear normal under basal, unchallenged conditions (Lombard et al., 2007). The regulation of CPS1 by SIRT5 has a role in ammonia detoxification and disposal during conditions when protein catabolism increases, such as fasting, long-term calorie restriction or a high protein diet resulting in abnormally elevated ammonia level (Nakagawa et al., 2009). The present study has demonstrated a role for SIRT5 in regulating ketogenesis through hypersuccinylation of HMGCS2. Interestingly, succinyl-CoA was previously proposed to inhibit HMGCS2 by auto-succinylation of the active site cysteine residue (Lowe and Tubbs, 1985). Follow up studies suggested that increased levels of succinyl-CoA in vitro could affect HMGCS2 activity presumably through succinylation (Quant et al., 1989). However, succinyl-CoA levels in our studies appeared unchanged in both the fed and fasted state indicating that increased succinylation was due to loss of SIRT5 and not to an overall increase in succinyl-CoA levels. Based on the crystal structure, our data indicate that lysines adjacent to the substrate binding site of HMGCS2 are intensely succinylated (Figure 7C), which suggests that succinyl-CoA may interact with lysine residues around the catalytic pocket leading to non-enzymatic modifications of these lysines. Importantly, a negative charge at residues K83 and K310 strongly inhibits HMGCS2 activity, probably through disrupting the binding pocket for phosphate groups of the substrate acetyl-CoA. While other robustly succinylated residues on HMGCS2 (K256, K350, and K354) near the binding pocket would not directly interact with the bound CoA group in the crystal structure, these sites may contribute to initial steps of acetyl-CoA binding through electrostatic interactions.
Succinylation is an abundant lysine modification in mitochondria. The 252 identified succinylated proteins have 1190 SuK sites and a total of 6579 lysines. Since our affinity enrichment approach was unlikely to cover all possible succinylated peptides, we can conclude that at least 18% of lysines on these proteins are modified by succinylation. Succinylation however is not evenly distributed among all proteins as some have more succinylated sites than others despite containing a similar number of total lysines, suggesting site-specificity of succinylation. For instance, the trifunctional enzyme α subunit has 66 lysines, of which 22 are succinylated (33%). In contrast, nicotinamide nucleotide transhydrogenase, an enzyme involved in mitochondrial reactive oxygen species detoxification, has 61 lysines, of which only three are succinylated (5%) (Dataset S2). This difference is not due to protein abundance as their expression levels in KO livers are not significantly altered from WT livers (Dataset S3).
To date, no protein with lysine acetyl- or succinyl-transferase activity has been identified within mitochondria. This raises a question as to what mechanism is responsible for the widespread, but specific, lysine acylation of mitochondrial proteins that we and others have identified (Hebert et al., 2013; Kim et al., 2006; Rardin et al., 2013). In the absence of evidence for such a transferase, chemical modification due to the abundance of acyl-CoAs (e.g., acetyl-CoA, succinyl-CoA) and the relative high pH in mitochondria is a possibility (Wagner and Payne, 2013). If chemical, nonenzymatic reactions are producing these modifications, protein biophysical parameters, such as specific lysine's pKa, solvent accessibility, and tertiary structure, as well as physiological parameters, such as micro-environmental acyl-CoA levels, protein super complex-compartmentalization in mitochondria, would be expected to determine the specificity of these acylation reactions.
Our quantitative analysis by MS1 Filtering measured the relative precursor ion abundance of succinylated peptides. In contrast to the Park et al. study we employed a label-free quantitative approach to control for biological variability by comparing lysine succinylation directly from mouse liver mitochondria using 5 WT and 5 KO animals as opposed to a single biological replicate using MEFs. This approach allowed us to demonstrate statistically that over 90 sites were significantly increased in the KO by more than 10-fold as compared to only 28 observed in MEFs from the Park et al. study. This difference is likely due to the greater overall abundance of lysine succinylation in liver tissue as compared to MEFs, as well as lysine succinylation being more abundant in mitochondria as opposed to other cellular compartments. In addition our previous analysis of lysine acetylation in liver mitochondria from WT and Sirt3−/− mice, where only 25 sites had greater than a 10-fold increase (Rardin et al., 2013), indicates a greater relative response to the lack of SIRT5 desuccinylase activity in the Sirt5−/− mouse than the difference seen in the Sirt3−/− mouse model. However, it remains to be determined if differences exist in the relative site occupancy of succinylation versus acetylation as methods for directly determining precise stoichiometries have not been successfully applied to lysine acyl modifications. Moreover, our two-fold cutoff does not preclude the possibility that lower fold changes may have functional significance. All quantified peptides with statistical significance are provided as a resource for interrogation of individual sites and proteins (Dataset S2). Finally, caution should also be taken in over-interpreting some of these very large changes (>20-fold) as in many cases the WT signal was near the lower limit of detection.
Our previous SIRT3 study has also allowed us to examine the overlap between acetylation and succinylation at specific sites. Remarkably, nearly 80% overlap between sites of lysine succinylation and acetylation is observed, with 24% of SIRT5 target sites also targeted by SIRT3. SIRT3 and SIRT5 target proteins are enriched in overlapping pathways including fatty acid β-oxidation, ketogenesis, and TCA cycle. Surprisingly, Park et al.'s comparison in MEFs identified only 24% overlap between the two acyl modifications indicating that within mitochondria there is much greater likelihood for co-regulation of mitochondrial proteins than in other subcellular compartments. Crosstalk between post-translational modifications has been widely observed across histones and signaling proteins such as p53 (Latham and Dent, 2007; Olsson et al., 2007). The differential regulation of proteins in common pathways by two mitochondrial sirtuins with distinct enzymatic activities may suggest a crosstalk or synergy between acetylation and succinylation in regulating mitochondrial metabolic networks. These important questions will be the focus of future investigations on the role of protein acylation in mitochondrial biology and metabolic regulation.
Experimental Procedures
Mass spectrometry (MS) and chromatographic parameters
All samples used for MS1 Filtering experiments were analyzed by reverse-phase LC-ESI-MS/MS with an Eksigent Ultra Plus nano-LC 2D HPLC system (Dublin, CA) connected to a quadrupole time-of-flight TripleTOF 5600 mass spectrometer (AB SCIEX) in direct injection mode. See supplementary methods for technique details.
Bioinformatic database searches
MS data sets were analyzed and searched using Mascot server version 2.3.02 (Matrix Sciences, Boston, MA) and ProteinPilot (AB SCIEX 4.5 B) with the Paragon algorithm (4.5, 1656). All data files were searched using the SwissProt 2013_01 database with a total of 538,849 sequences, but restricted to Mus musculus (16,580 protein sequences). See supplementary methods for details.
Generation of sequence logos
To generate sequence logos, we first determined the 21–amino acid sequence context for each modified peptide. We aligned the modified sequence to the full protein sequence downloaded from UniProt (mouse proteome, accessed 5/10/2013) and extracted the 21–amino acid interval from the UniProt sequence. These context sequences were analyzed with IceLogo (Colaert et al., 2009) by comparison to context sequences from an appropriate, non-redundant set of control lysines. As one such control set, we extracted the sequence context for all lysines on all proteins that appear in our data set from the UniProt sequences. All IceLogo analyses were performed on non-redundant sets (e.g., we subtracted SuK context sequences from the control set of all lysines before logo generation).
Conservation index of lysine succinylation sites
Succinylated peptides that are non-redundant, with distinct peaks, were mapped to orthologous proteins for conservation analysis (Dataset S5). We downloaded the full sequence from UniProt and aligned it to the nr database using blastpgp (BLAST suite 2.2.18) (Altschul et al., 1997). To ensure high-quality multiple alignment, we required (i) sequence identity of 30–94% to the query mouse protein, and (ii) >10 such hits could be found. We generated the multiple alignment with CLUSTALW (2.0.12) using default settings (Larkin et al., 2007). We computed conservation indices for each modified lysine by: (i) counting the number of conserved lysines across the seven queried species and (ii) using AL2CO on the alignment (Pei and Grishin, 2001). When calculating rates of mutation to various amino acids, we excluded sites that are absent from the multiple alignment in a given species.
Pathway analysis
Pathway analysis was performed with Reactome (Vastrik et al., 2007), which uses all mouse proteins as the comparison set. For other over-representation analyses, such as over-representation of SIRT5 target sites on particular proteins, we calculated a binomial probability based on the overall distribution of a feature in the dataset (e.g., fraction of SuK sites that are SIRT5 targets in the overall data set).
Sample preparation for mouse liver mitochondrial protein lysate
Livers were collected from five wild-type and five Sirt5−/− male mice at 16 weeks of age in the presence of deacetylase inhibitors (20 mM nicotinamide and 1 M trichostatin A), and mitochondria were isolated by differential centrifugation as described previously (Hirschey et al., 2009). For additional details see Supplementary Information.
Supplementary Material
Highlights.
-Liver mitochondrial succinylome is quantified in wild type and Sirt5−/− mice
-386 lysine sites on 140 proteins are hypersuccinylated in Sirt5−/− mouse liver
-SIRT5 targets enzymes in fatty acid oxidation and ketone body production
-Inhibition of HMGCS2 activity by lysine succinylation at K83 and K310
Acknowledgements
We thank F. W. Alt for generously providing the original Sirt5−/− mouse strain, C. Her for primary hepatocyte preparation, M. Finucane for statistical analysis, J. Carroll and T. Roberts for figure preparation, G. Howard for editorial assistance, and K. E. Dittenhafer-Reed for experimental advice. This work was supported by National Institutes of Health Grants T32AG000266 (M.J.R.), RO1 DK090242 (E.S.G.), PL1 AG032118 (B.W.G.), R24 DK085610 (E.V.), and institutional support from the J. David Gladstone Institutes (E.V.). This work was also supported in part by the Shared Instrumentation Grant S10 RR024615 (B.W.G.) and the generous access of a TripleTOF 5600 by AB SCIEX at the Buck Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Deuterium-palmitate oxidation assay and Tritium-palmitate oxidation assay. See Supplementary Methods for detailed information.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrew Skaff D, Miziorko HM. A visible wavelength spectrophotometric assay suitable for high-throughput screening of 3-hydroxy-3-methylglutaryl-CoA synthase. Anal Biochem. 2010;396:96–102. doi: 10.1016/j.ab.2009.08.030. [DOI] [PubMed] [Google Scholar]
- Colaert N, Helsens K, Martens L, Vandekerckhove J, Gevaert K. Improved visualization of protein consensus sequences by iceLogo. Nat Methods. 2009;6:786–787. doi: 10.1038/nmeth1109-786. [DOI] [PubMed] [Google Scholar]
- Du J, Zhou Y, Su X, Yu JJ, Khan S, Jiang H, Kim J, Woo J, Kim JH, Choi BH, et al. Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase. Science. 2011;334:806–809. doi: 10.1126/science.1207861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glozak MA, Sengupta N, Zhang X, Seto E. Acetylation and deacetylation of non-histone proteins. Gene. 2005;363:15–23. doi: 10.1016/j.gene.2005.09.010. [DOI] [PubMed] [Google Scholar]
- Greiss S, Gartner A. Sirtuin/Sir2 phylogeny, evolutionary considerations and structural conservation. Mol Cells. 2009;28:407–415. doi: 10.1007/s10059-009-0169-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W, Newman JC, Wang MZ, Ho L, Verdin E. Mitochondrial sirtuins: regulators of protein acylation and metabolism. Trends Endocrinol Metab. 2012;23:467–476. doi: 10.1016/j.tem.2012.07.004. [DOI] [PubMed] [Google Scholar]
- Hebert AS, Dittenhafer-Reed KE, Yu W, Bailey DJ, Selen ES, Boersma MD, Carson JJ, Tonelli M, Balloon AJ, Higbee AJ, et al. Calorie restriction and SIRT3 trigger global reprogramming of the mitochondrial protein acetylome. Mol Cell. 2013;49:186–199. doi: 10.1016/j.molcel.2012.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschey MD, Shimazu T, Huang JY, Schwer B, Verdin E. SIRT3 regulates mitochondrial protein acetylation and intermediary metabolism. Cold Spring Harb Symp Quant Biol. 2011;76:267–277. doi: 10.1101/sqb.2011.76.010850. [DOI] [PubMed] [Google Scholar]
- Hirschey MD, Shimazu T, Huang JY, Verdin E. Acetylation of mitochondrial proteins. Methods Enzymol. 2009;457:137–147. doi: 10.1016/S0076-6879(09)05008-3. [DOI] [PubMed] [Google Scholar]
- Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, Cheng T, Kho Y, Xiao H, Xiao L, et al. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol Cell. 2006;23:607–618. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Latham JA, Dent SY. Cross-regulation of histone modifications. Nat Struct Mol Biol. 2007;14:1017–1024. doi: 10.1038/nsmb1307. [DOI] [PubMed] [Google Scholar]
- Lombard DB, Alt FW, Cheng HL, Bunkenborg J, Streeper RS, Mostoslavsky R, Kim J, Yancopoulos G, Valenzuela D, Murphy A, et al. Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Mol Cell Biol. 2007;27:8807–8814. doi: 10.1128/MCB.01636-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe DM, Tubbs PK. Succinylation and inactivation of 3-hydroxy-3-methylglutaryl-CoA synthase by succinyl-CoA and its possible relevance to the control of ketogenesis. Biochem J. 1985;232:37–42. doi: 10.1042/bj2320037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Lomb DJ, Haigis MC, Guarente L. SIRT5 Deacetylates carbamoyl phosphate synthetase 1 and regulates the urea cycle. Cell. 2009;137:560–570. doi: 10.1016/j.cell.2009.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman JC, He W, Verdin E. Mitochondrial protein acylation and intermediary metabolism: regulation by sirtuins and implications for metabolic disease. J Biol Chem. 2012;287:42436–42443. doi: 10.1074/jbc.R112.404863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson A, Manzl C, Strasser A, Villunger A. How important are post-translational modifications in p53 for selectivity in target-gene transcription and tumour suppression? Cell Death Differ. 2007;14:1561–1575. doi: 10.1038/sj.cdd.4402196. [DOI] [PubMed] [Google Scholar]
- Park J, Chen Y, Tishkoff DX, Peng C, Tan M, Dai L, Xie Z, Zhang Y, Zwaans BM, Skinner ME, et al. SIRT5-mediated lysine desuccinylation impacts diverse metabolic pathways. Mol Cell. 2013;50:919–930. doi: 10.1016/j.molcel.2013.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei J, Grishin NV. AL2CO: calculation of positional conservation in a protein sequence alignment. Bioinformatics. 2001;17:700–712. doi: 10.1093/bioinformatics/17.8.700. [DOI] [PubMed] [Google Scholar]
- Peng C, Lu Z, Xie Z, Cheng Z, Chen Y, Tan M, Luo H, Zhang Y, He W, Yang K, et al. The first identification of lysine malonylation substrates and its regulatory enzyme. Mol Cell Proteomics. 2011;10:M111 012658. doi: 10.1074/mcp.M111.012658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quant PA, Tubbs PK, Brand MD. Treatment of rats with glucagon or mannoheptulose increases mitochondrial 3-hydroxy-3-methylglutaryl-CoA synthase activity and decreases succinyl-CoA content in liver. Biochem J. 1989;262:159–164. doi: 10.1042/bj2620159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rardin MJ, Newman JC, Held JM, Cusack MP, Sorensen DJ, Li B, Schilling B, Mooney SD, Kahn CR, Verdin E, et al. Label-free quantitative proteomics of the lysine acetylome in mitochondria identifies substrates of SIRT3 in metabolic pathways. Proc Natl Acad Sci U S A. 2013;110:6601–6606. doi: 10.1073/pnas.1302961110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling B, Rardin MJ, MacLean BX, Zawadzka AM, Frewen BE, Cusack MP, Sorensen DJ, Bereman MS, Jing E, Wu CC, et al. Platform-independent and label-free quantitation of proteomic data using MS1 extracted ion chromatograms in skyline: application to protein acetylation and phosphorylation. Mol Cell Proteomics. 2012;11:202–214. doi: 10.1074/mcp.M112.017707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafqat N, Turnbull A, Zschocke J, Oppermann U, Yue WW. Crystal structures of human HMG-CoA synthase isoforms provide insights into inherited ketogenesis disorders and inhibitor design. J Mol Biol. 2010;398:497–506. doi: 10.1016/j.jmb.2010.03.034. [DOI] [PubMed] [Google Scholar]
- Shimazu T, Hirschey MD, Hua L, Dittenhafer-Reed KE, Schwer B, Lombard DB, Li Y, Bunkenborg J, Alt FW, Denu JM, et al. SIRT3 deacetylates mitochondrial 3-hydroxy-3-methylglutaryl CoA synthase 2 and regulates ketone body production. Cell Metab. 2010;12:654–661. doi: 10.1016/j.cmet.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vastrik I, D'Eustachio P, Schmidt E, Gopinath G, Croft D, de Bono B, Gillespie M, Jassal B, Lewis S, Matthews L, et al. Reactome: a knowledge base of biologic pathways and processes. Genome Biol. 2007;8:R39. doi: 10.1186/gb-2007-8-3-r39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner GR, Payne RM. Widespread and Enzyme-independent N[epsilon]-Acetylation and N[epsilon]-Succinylation of Proteins in the Chemical Conditions of the Mitochondrial Matrix. J Biol Chem. 2013;288:29036–29045. doi: 10.1074/jbc.M113.486753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Zhang Y, Yang C, Xiong H, Lin Y, Yao J, Li H, Xie L, Zhao W, Yao Y, et al. Acetylation of metabolic enzymes coordinates carbon source utilization and metabolic flux. Science. 2010;327:1004–1007. doi: 10.1126/science.1179687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Tan M, Xie Z, Dai L, Chen Y, Zhao Y. Identification of lysine succinylation as a new post-translational modification. Nat Chem Biol. 2011;7:58–63. doi: 10.1038/nchembio.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T, Yao J, Zhou L, Zeng Y, Li H, et al. Regulation of cellular metabolism by protein lysine acetylation. Science. 2010;327:1000–1004. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.