Summary
The anatomical and functional architecture of the human brain is largely determined by prenatal transcriptional processes. We describe an anatomically comprehensive atlas of mid-gestational human brain, including de novo reference atlases, in situ hybridization, ultra-high resolution magnetic resonance imaging (MRI) and microarray analysis on highly discrete laser microdissected brain regions. In developing cerebral cortex, transcriptional differences are found between different proliferative and postmitotic layers, wherein laminar signatures reflect cellular composition and developmental processes. Cytoarchitectural differences between human and mouse have molecular correlates, including species differences in gene expression in subplate, although surprisingly we find minimal differences between the inner and human-expanded outer subventricular zones. Both germinal and postmitotic cortical layers exhibit fronto-temporal gradients, with particular enrichment in frontal lobe. Finally, many neurodevelopmental disorder and human evolution-related genes show patterned expression, potentially underlying unique features of human cortical formation. These data provide a rich, freely-accessible resource for understanding human brain development.
Keywords: Human brain, Transcriptome, Microarray, Development, Gene expression, Evolution
The human brain develops following a complex, highly stereotyped series of histogenic events that depend on regulated differential gene expression, and acquired or inherited disruption can lead to devastating consequences. Largely due to limitations in access to human prenatal tissue, most developmental studies are performed in mouse or non-human primate (but see1–5). However, significant species differences exist, necessitating the study of human brain. For example, the human neocortex has undergone massive evolutionary expansion, particularly in superficial layers, likely due to differences in rates of progenitor pool expansion during neurogenesis compared to other species6. A secondary progenitor zone, the subventricular zone (SZ) is present in all mammals, but is split into an outer and inner region in primates7. The transient subplate zone (SP) is greatly expanded in human8, as is the subpial granular zone (SG), a transient compartment at the pial surface composed primarily of tangentially migrating neurons9. Furthermore, there is evidence for species differences in the developmental origin of cortical GABAergic interneurons. In mouse, nearly all originate from the striatal ganglionic eminences (GEs) of the ventral telencephalon10; however, the origin of human cortical interneurons remains controversial7,11–14. Finally, understanding the emergence of cortical specialization for language can only be studied in humans.
Recent studies have begun to analyze the developing brain and neocortical transcriptome. Profiling of mid-gestational human brain3,5 identified many genes differentially expressed between major regions, including genes associated with human-accelerated conserved noncoding sequences (haCNSs)15. Expression varies between cell populations, and more detailed analysis of layers of fetal mouse neocortex found >2500 genes differentially expressed between ventricular zone (VZ), SZ, intermediate zone (IZ), and cortical plate (CP)16. Species differences in distinct fetal transient zones, including the SZ2, SP17,18, CP6, and SG9, have also been described.
The goal of the current project was to create resources for studying prenatal human brain development and the early roots of neurodevelopmental and psychiatric disorders19. These include anatomical reference atlases similar to those for model organisms20–22, and an anatomically comprehensive, detailed transcriptional profiling of normal mid-gestational brain modeled on atlases of adult mouse and human brain23,24 and using methods for selective analysis of discrete structural nuclei and layers25. These data are freely accessible as part of the BrainSpan Atlas of the Developing Human Brain (http://brainspan.org) via the Allen Brain Atlas data portal (http://www.brain-map.org).
Comprehensive transcriptome analysis of prenatal brain
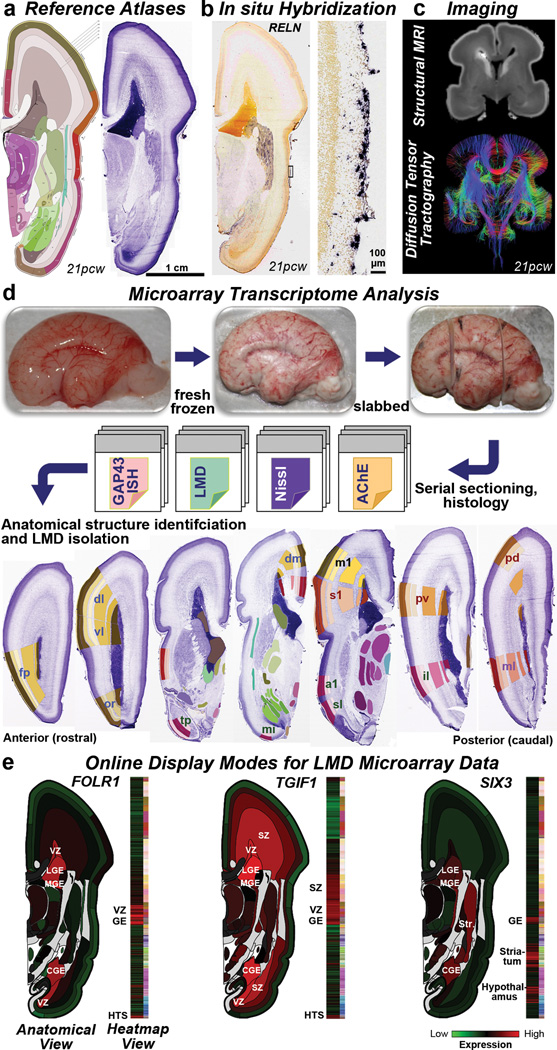
Four intact high quality mid-gestational brains, two from 15–16 post-conceptual weeks (pcw) and two from 21pcw (Suppl. Table 1), were used to create detailed de novo reference atlases and transcriptome datasets (Fig. 1). The entire left hemisphere of each specimen was coronally, serially cryosectioned onto polyethylene naphthalate (PEN) membrane slides for laser microdissection (LMD), with interleaved slides for histological staining (Nissl, acetylcholinesterase (AChE), and in situ hybridization (ISH) for GAP43) for detailed structure identification. Approximately 300 anatomical regions per specimen were isolated for RNA isolation, amplification and microarray analysis on custom 64K Agilent microarrays23 (Fig. 1d; Suppl. Table 2; Extended Data Fig. 1). For one 16pcw and one 21pcw specimen, the right hemisphere was processed similarly but used for ISH (Fig. 1b) and Nissl staining. These data were anatomically delineated to make digital reference atlases (Fig. 1a), which allow the representation of transcriptome data in native anatomical coordinates. Figure 1e illustrates the specificity of anatomical sampling using this representation. For example, the folate receptor FOLR1 is selectively expressed in VZ and GE. Sufficient folate intake is essential for proper neuronal development,26 and mutations in FOLR1 cause severe neurological sequelae due to cerebral folate transport deficiency. Similarly, two genes associated with abnormal cortical development in holoprosencephaly, TGIF1 and SIX327, are also enriched in cortical germinal zones. Finally, structural magnetic resonance imaging (MRI) and diffusion weighted MRI (DWI) data from 3 approximately age-matched brains (Fig. 1c; Extended Data Fig. 2), as well as reconstruction of fetal white matter tracts for three additional brains (Extended Data Fig. 3; see28), are included for anatomical reference in the online resource.
Figure 1. Prenatal human brain atlas components.
a. Nissl stained (right) and corresponding annotated reference atlas (left) plate, color coded by structure. b. ISH for RELN showing expression in Cajal-Retzius cells at low (left) and high (right) magnification in MZ. c. High resolution MRI and tract DWI of fixed ex cranio brain. d. Experimental strategy for systematic histology, anatomical delineation and LMD-based isolation of discrete anatomical structures for microarray analysis. Nissl, acetylcholinesterase (AChE) and GAP43 ISH were used to identify structures. e. Quantitative representation of microarray data for FOLR1, TGIF1, and SIX3. GE: ganglionic eminence. See Supplemental Table 2 for other anatomical abbreviations.
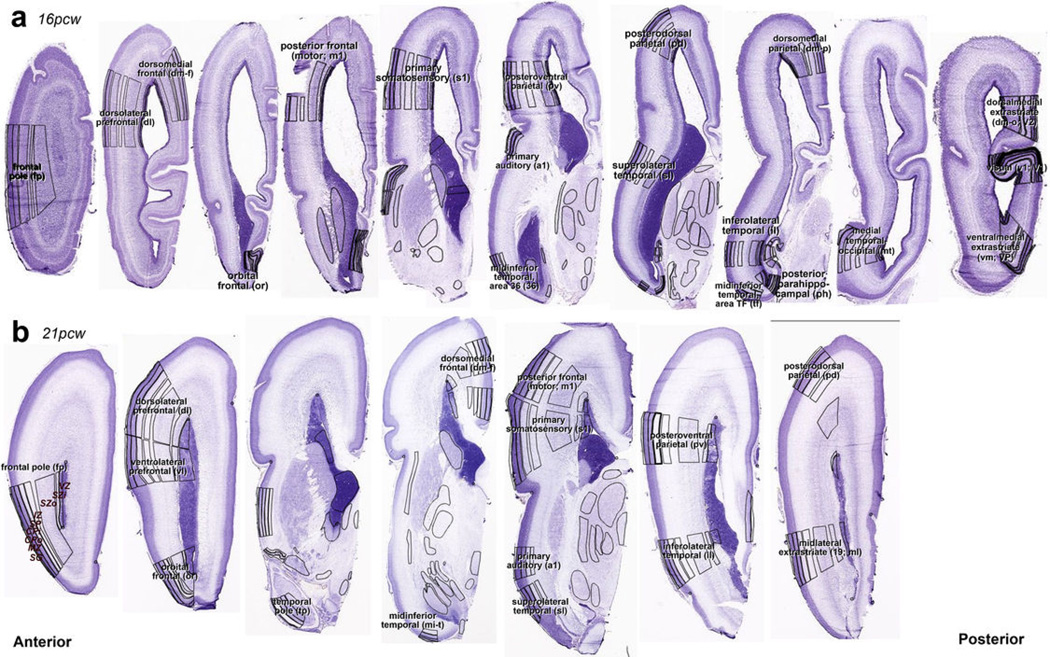
Extended Data Figure 1. Representative Nissl sections for laser microdissection (LMD) of 16 pcw and 21 pcw brains.
Nissl-stained sections were annotated and used to determine LMD region boundaries for 16 pcw (a) and 21 pcw (b) brains. Regions from adjacent sections on PEN membrane slides were captured using these annotations as guidelines. Labels show full name and abbreviation for representative planes of section through presumptive neocortical regions. (b) is a higher resolution modified version of the bottom row in Figure 1c of the main manuscript.
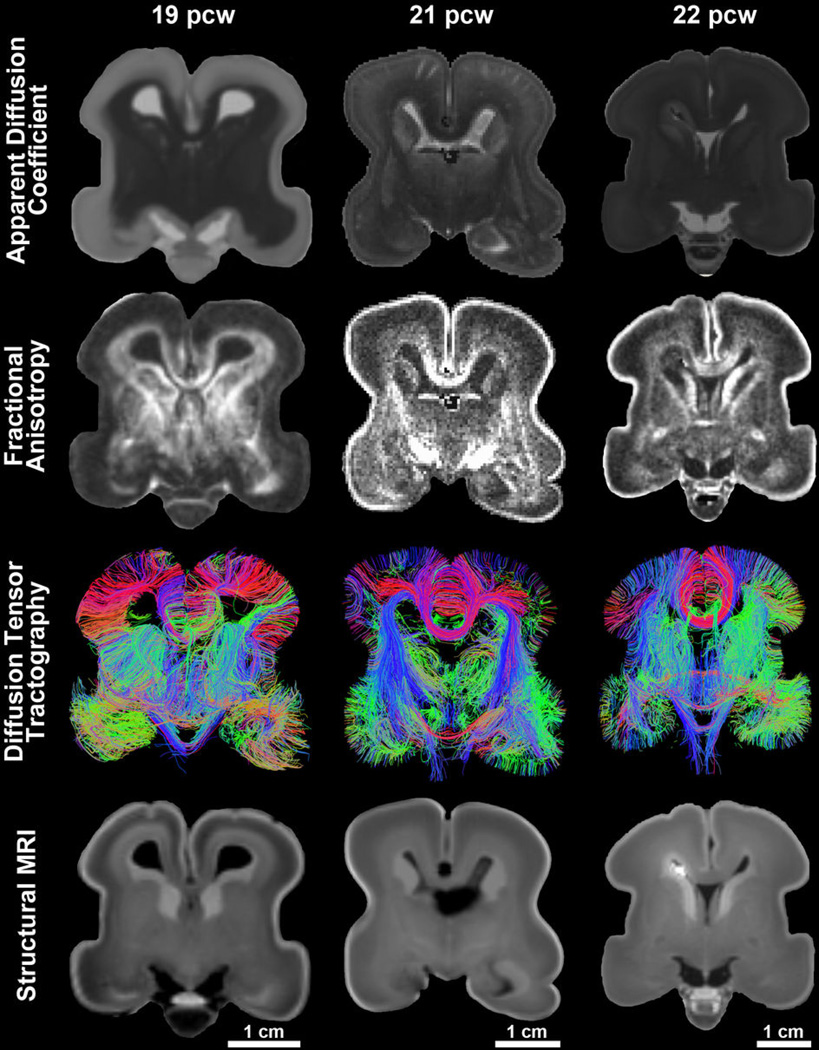
Extended Data Figure 2. Overview of magnetic resonance imaging data acquired from the post-mortem, formalin-fixed human fetal brain samples.
Diffusion-weighted MRI were acquired for each sample using a steady state free precession sequence (b= 730s/mm^2; 44 directions), yielding maps of apparent diffusion coefficient (ADC) (1st row) and of fractional anisotropy (FA) (2nd row). Whole-brain deterministic tractography results (3rd row) represent visualization of tractography output data filtered by a coronal slice filter. Structural data were acquired for each sample using a multi-echo flash sequence with images acquired at alpha = 40 providing optimal contrast to identify cortical and subcortical structures of interest (4th row).
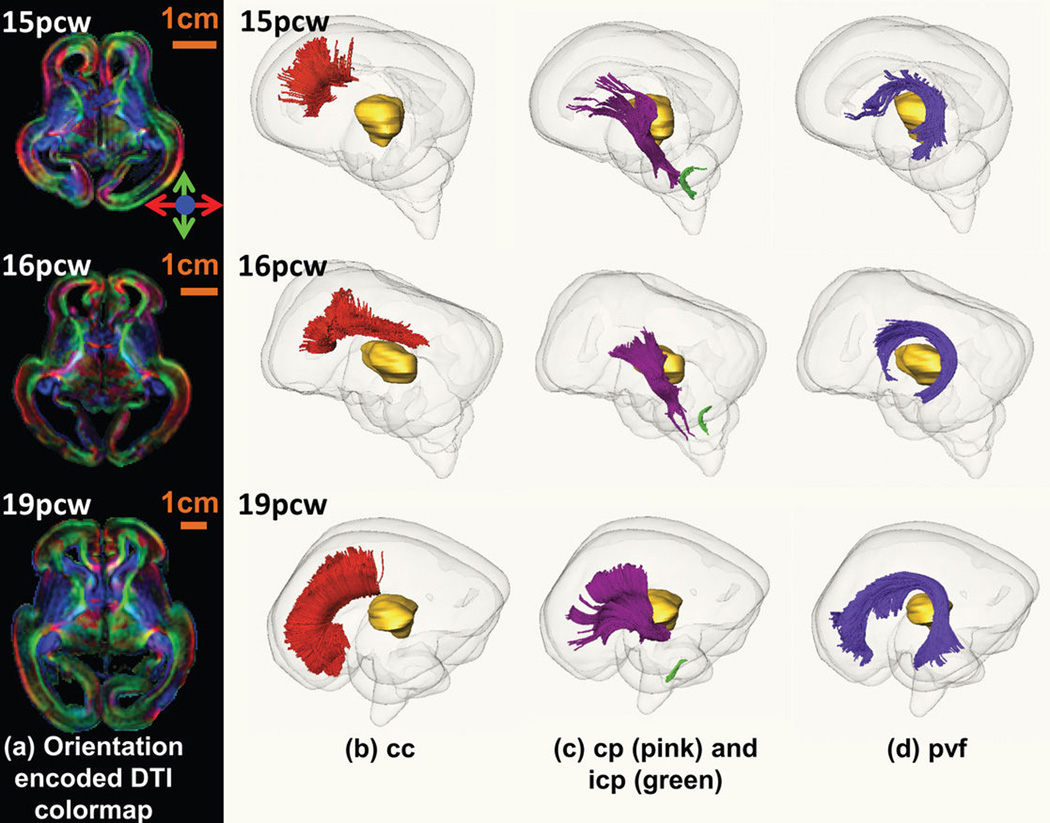
Extended Data Figure 3. White matter fiber tracts in fetal human brain.
Orientation-encoded diffusion tensor imaging (DTI) colormaps in the left panel (a) and the three-dimensionally reconstructed fetal white matter fibers in the right panels (b, c and d) for fetal brains at 15pcw (upper row), 16pcw (middle row) and 19pcw (lower row). The orientation-encoded DTI colormaps are in axial planes at the anterior commissure level. The red, pink, green and purple fibers in the right panels are cc in (b), cp and icp in (c), and pvf in (d), respectively. The transparent whole brain and yellow thalamus are also shown as anatomical guidance in (b), (c) and (d). The scale bars are shown in the left panel (a). The fiber name abbreviations are as follows. cc: corpus callosum; cp: cerebral peduncle; icp: inferior cerebellar peduncle; pvf: periventricular fibers (transient fibers coursing around the germinal matrix and only existing in the prenatal fetal brain).
Laminar transcriptional patterning
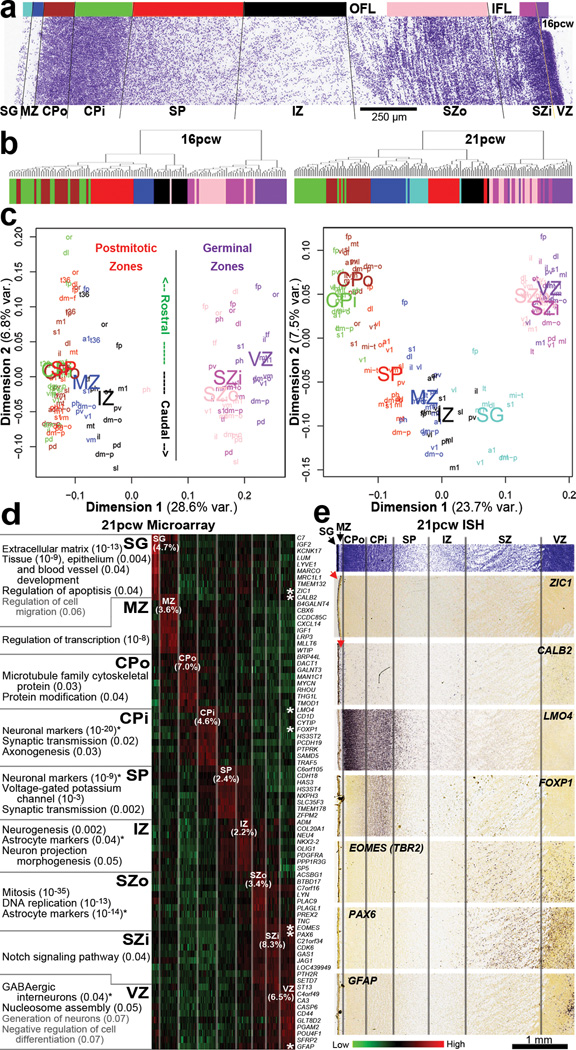
We assayed ~25 areas of the developing neocortex, delineating nine layers per area (here referring to fetal mitotic and postmitotic zones and not layers 1–6 of mature neocortex)29: SG, marginal zone (MZ), outer and inner CP (CPo; CPi), SP, IZ (or inner SP), outer and inner SZ (SZo; SZi), and VZ (Fig. 2a). Approximately 95% of RefSeq genes are expressed in developing neocortex, compared with 84% identified using these arrays in adult23. Different layers show robust and unique molecular signatures, and samples group by layer using hierarchical clustering (using differential genes from ANOVA, p<10−29) at both timepoints (Fig. 2b). Samples also cluster with multidimensional scaling (MDS), where dimension one separates samples by layer, with germinal zones (VZ, SZi, SZo) distinct from layers containing primarily postmitotic cells, and dimension two roughly reflects rostrocaudal position (Fig. 2c).
Figure 2. Laminar gene expression mirrors developmental processes in prenatal neocortex.
a. Nissl section from 16pcw cortex showing layers dissected for analysis (represented by color bar). The outer and inner fiber layers (OFL, IFL) were omitted from the dissection. b. Sample clustering based on 100 most significant genes differentiating layers by ANOVA (p<10−29) groups samples by layer at both timepoints. c. MDS using all genes demonstrates clustering of postmitotic versus germinal zones (Dimension 1) and to a lesser extent by rostrocaudal position (Dimension 2). d. Layer-enriched gene expression, based on correlation to binary templates at 21pcw (Methods). Enriched genes in each layer relate to cellular makeup and developmental maturity of those cells. e. Validation of laminar enrichment by ISH for genes with asterisks in (d). See Supplemental Table 2 for anatomical abbreviations.
To identify laminar signatures at 21pcw, we correlated each gene with a binary vector (1 in tested layer versus 0 elsewhere), which identified ~2000 layer-enriched genes with R>0.5 in both brains (Suppl. Table 3). Each layer included genes with high laminar specificity (Fig. 2d), although the SZi profiles tended to overlap with the neighboring SZo and VZ. ISH validated the specificity of layer enriched genes (Fig. 2e). For example, the Cajal-Retzius cell marker CALB230 showed enrichment in MZ and SG as expected. The cortical progenitor markers TBR2 and PAX6 are enriched in germinal layers as in mouse, although PAX6 in mouse is restricted to VZ whereas it is also highly expressed in human SZ31. VZ-restricted GFAP expression likely marks radial glia (RG)32. Finally, expression of ZIC1, associated with Dandy-Walker congenital brain malformation33, was restricted to the pia mater overlying the cortex, therefore indicating that SG samples captured pial cells in addition to granule cells. However, while mouse Zic1 is expressed by virtually all Cajal-Retzius neurons34, our results indicate that this is not true in human as ZIC1 and CALB2 expression do not overlap in MZ (Figs. 1b and 2e).
These laminar expression patterns mirror cellular composition and developmental processes, shown by enrichment analysis (Fig. 2d; Suppl. Table 4; Methods). SZo-enriched categories primarily related to cell division and contained many astrocytic markers likely expressed in outer radial glia (ORG)7. Functional ontology of postmitotic layers reflected developmental maturity. SP, which contains the earliest-generated neurons, showed enrichment for mature neuronal markers and synaptic transmission, reflecting early thalamic afferent input by midgestation35. The next oldest neurons in CPi are additionally enriched for genes involved in forming connections, whereas the youngest neurons in CPo are primarily enriched for terms related to metabolism rather than mature neuronal function.
Gene networks discriminate fetal cell types
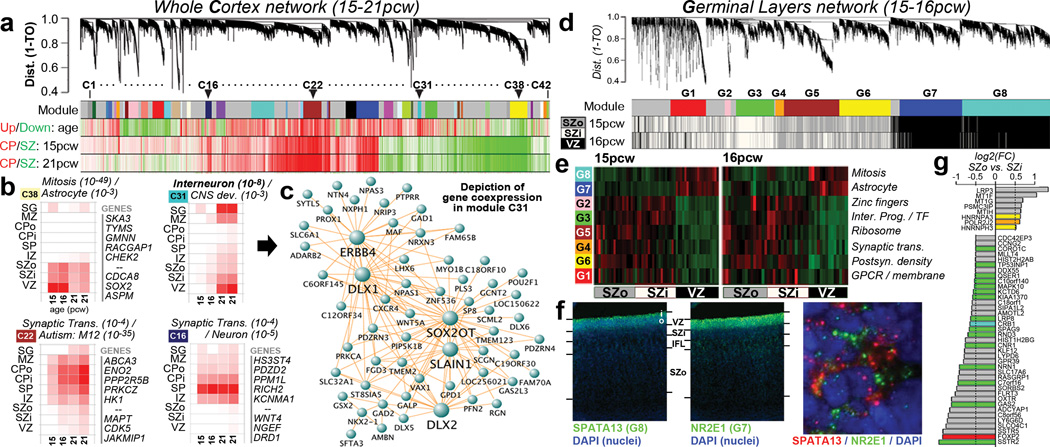
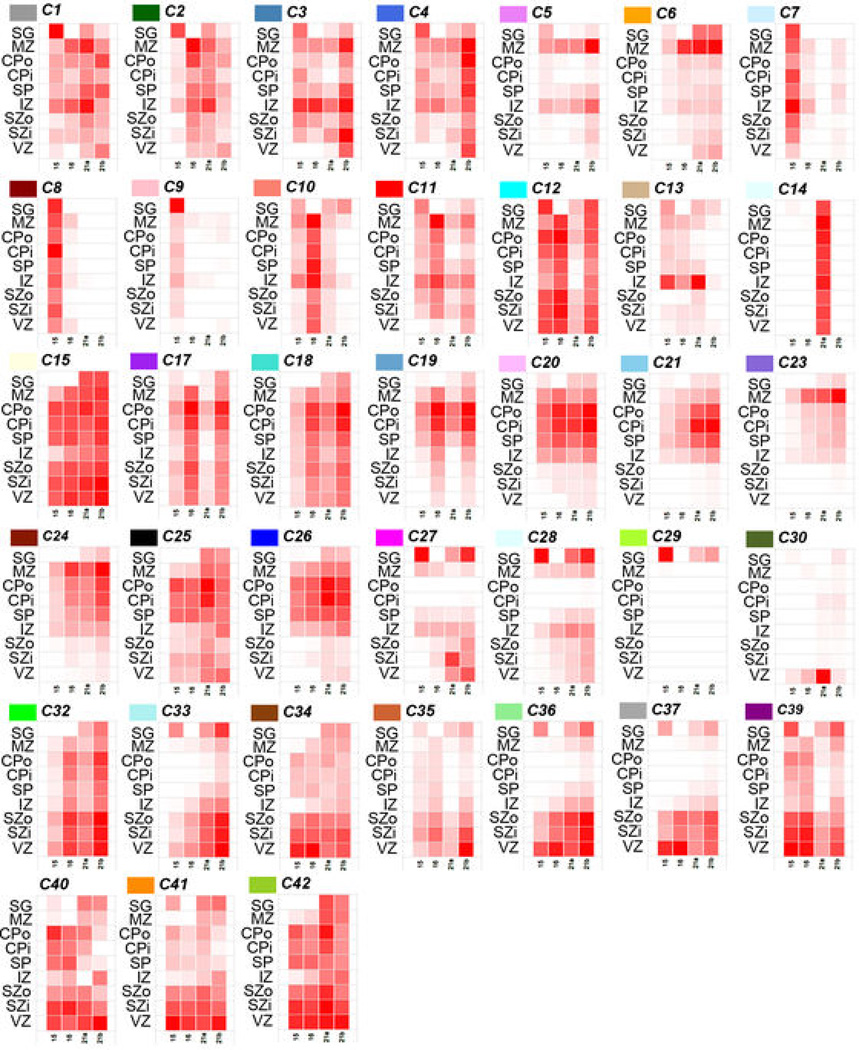
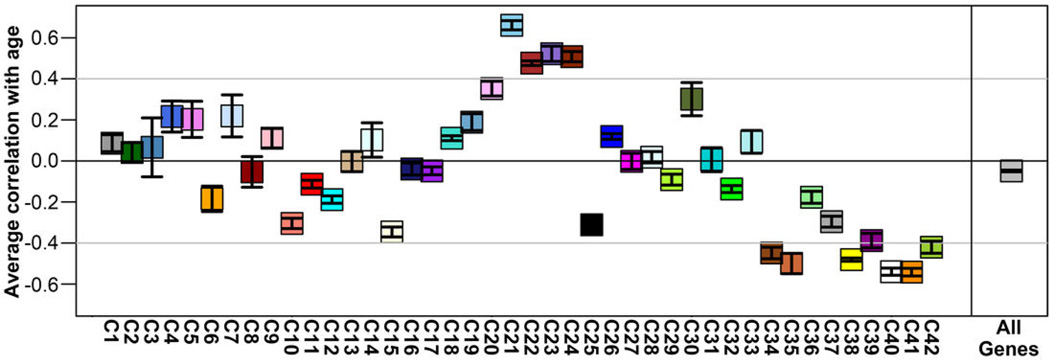
To identify principal features of the developing cortical transcriptome, we performed weighted gene co-expression network analysis (WGCNA)36 on all 526 neocortical samples, and identified 42 modules of co-expressed genes (Fig. 3a; Suppl. Tables 5–6; Suppl. Methods). WGCNA clusters genes with similar expression patterns in an unbiased manner, allowing a biological interpretation of transcriptional patterns (layer, cell type, biological process, disease, etc.)23,36–38. Here, most gene clusters ("modules") corresponded to layers and/or changes with age, (Fig. 3a-b; Extended Data Fig. 4) while areal patterning appeared to be a more subtle transcriptional feature. For example, module C16 is enriched in SP (Fig. 3b, lower right), and shows hallmarks of mature neuronal function. Module C38 contains genes enriched in germinal layers, and also decreased expression with age (Fig. 3b, upper left). This module has a large signature of glia and cell division, suggesting that these genes reflect decreasing progenitor cell division. Conversely, module C22 is enriched in newly generated postmitotic neurons of the CP, and increases with age (Fig. 3b, lower left). Importantly given the small sample size, this temporal patterning in C38 and C22 is corroborated by RNA-seq data from a larger timeseries of cortical development contained in the BrainSpan resource (Extended Data Fig. 5). Interestingly, genes in module C22 significantly overlap genes showing altered expression in postnatal human brains of patients affected by autism38. This suggests involvement of autism risk factors in early development of excitatory cortical neurons, consistent with other recent studies39,40.
Figure 3. Co-expression analyses of prenatal cortex.
a. WGCNA cluster dendrogram on all 526 neocortical samples groups genes into 42 distinct modules (First row). Rows 2–4: Strong differential expression relationships are seen between module genes and age or cortical layer. b. Module eigengene (ME) expression of four notable modules in (a), averaged across brain and layer. Modules are biologically characterized with significant category enrichments and representative genes. Many top gene-gene connections for module C31 are shown in (c), including several known GABAergic interneuron genes. d. Cluster dendrogram for consensus network focused on germinal layers identifies modules (Row 1) enriched in each layer (Rows 2,3). e. ME heatmap shows differential VZ/SZ expression for 8 modules, along with enriched gene sets. f. FISH on 15pcw frontal cortex shows enrichment of SPATA13 and NR2E1 in mutually exclusive subcellular localization in VZ. g. Genes enriched in SZ, and differentially expressed between SZo and SZi. Genes color-coded by module (gray = unassigned).
Extended Data Figure 4. Module eigengene expression of remaining modules in the cortical network.
Module eigengene expression of remaining 38 modules averaged across brain and layer. Each box corresponds to average module eigengene expression of all samples in that layer (rows) and brain (columns). Red = higher expression.
Extended Data Figure 5. Temporal patterning of whole cortex WGCNA modules across early to mid-gestational periods in BrainSpan RNA-seq cortical data.
RNA-seq RPKM values for 8–22pcw specimens in the BrainSpan database for genes assigned to WGCNA modules (Figure 3 in main manuscript) were correlated with age. For each module (Fig. 3a–c; x-axis), the average correlation (+/− standard error of the mean) between expression of genes in that module and age (y-axis) is plotted. Many of the modules show increases (positive correlation) or decreases (negative correlation) with age. In particular, modules C38 (increasing with age) and C22 (decreasing with age) presented in the main manuscript (see Fig. 3b, left column) show consistent trends with age in both datasets.
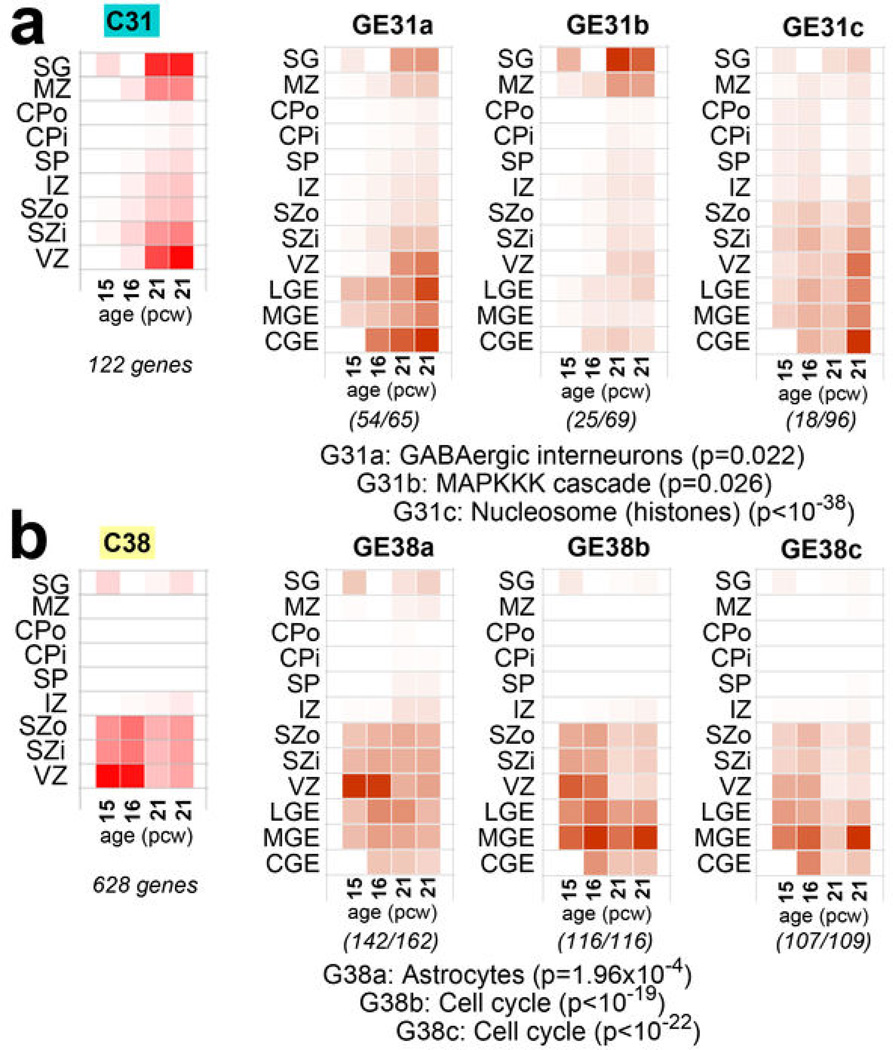
Finally, we identified a module (C31) with particular enrichment in SG and VZ (Fig. 3b, upper right), containing many interneuron-associated genes (Fig. 3c). DLX1 and DLX2, homeobox transcription factors essential for interneuron migration and survival10, were central ("hub") genes therein. There is controversy regarding the origin of cortical interneurons in primates, where the argument has been made that a substantial proportion of interneurons are generated locally in the VZ12,13. However, several recent studies have shown strong evidence that, as with rodents, primate interneurons are generated extracortically in the GEs11,14. To address this issue, we generated a new network using neocortical as well as GE samples, and examined the distribution of C31 genes therein. Most genes were assigned to a module showing common enrichment in both the GEs and VZ (Extended Data Fig. 6). While this finding does not resolve the origin of cortical interneurons, it shows that transcriptional programs associated with these cells in both structures are highly similar.
Extended Data Figure 6. Gene sets corresponding to GABAergic interneurons and proliferating layers also are highly expressed in the ganglionic eminences.
To examine the relationship between genes enriched in the cortical VZ, including gene modules associated with GABAergic interneurons and mitotically active proliferative cells, WGCNA was performed on the combined cortical and GE samples (referred to as the "GE network"). a. Genes from module C31 in the whole cortex WGNCA (GABA neurons) are assigned primarily to three modules in the GE network. GE31a has a similar pattern in cortex as C31, is highly expressed in GE and is enriched in genes associated with GABAergic interneurons. Other genes from C31 were assigned to modules with other cortical patterns and functional ontological associations (GE31b, GE31c). b. Genes from module C38 in the whole cortex WGNCA also divide primarily into three GE modules that are enriched in both the cortical germinal layers and the GE. These modules are enriched for genes expressed in astrocytes, potentially reflecting expression in radial glia, and are associated with cell cycle. For all plots, module eigengene (ME) expression is averaged across brain and layer (as in Fig. 3b of the main manuscript), also including LGE, MGE, and CGE (referred to here collectively as GE). Numbers in parentheses below each plot show the number of genes from module C31 in a, or C38 in b, out of the total number of module genes in the newly-generated network. One representative enrichment category for each module is shown with enrichment p-value.
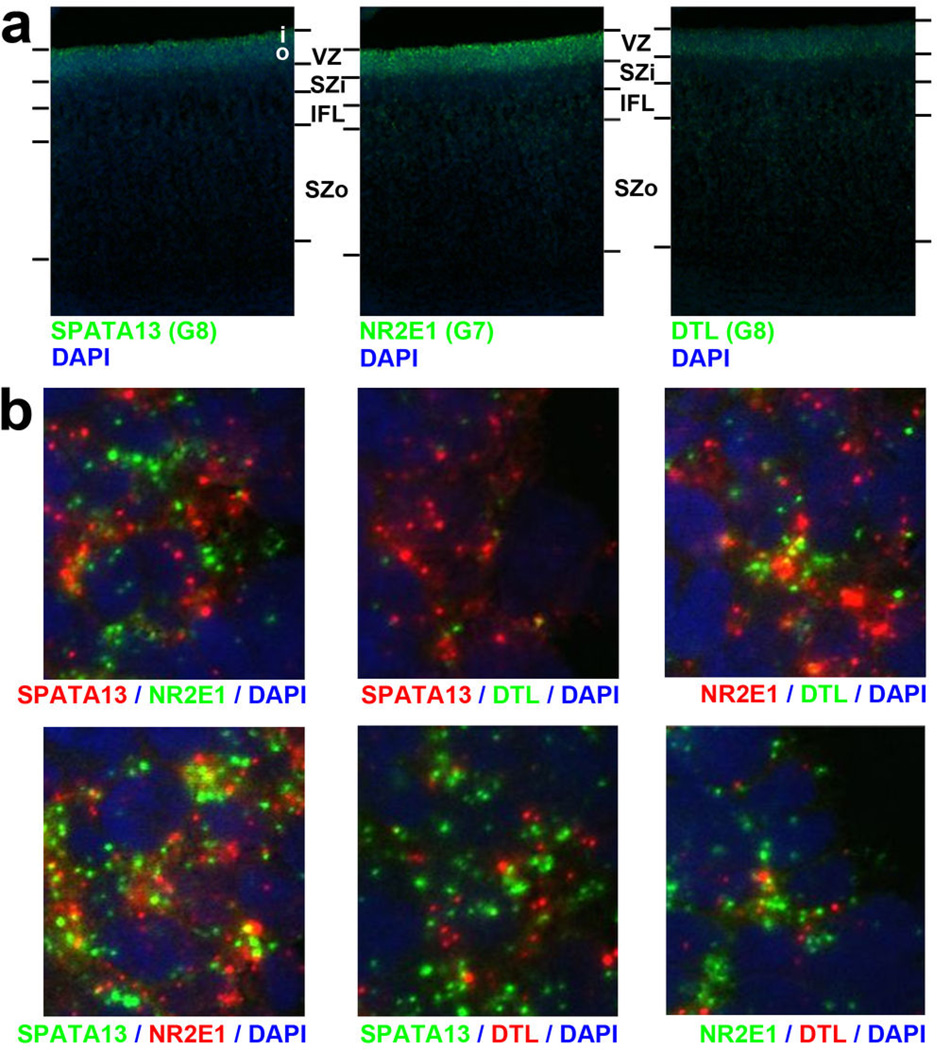
Germinal layers contain various cortical progenitors including RG in the VZ, intermediate progenitors (IP) in the SZi, and ORG in the SZo (reviewed in41), and these RG may be quite diverse42. To search for coherent expression profiles marking putative progenitor populations, we created a consensus co-expression network using samples from VZ, SZi, and SZo in the 15/16 pcw brains, only including genes differentially expressed between these layers (Fig. 3d; Suppl. Table 7; Suppl. Methods). This network will only identify intergenic relationships common to both brains, and should therefore produce robust co-expression relationships that exclude specimen-specific features or changes with age. We found eight co-expression modules with selective enrichment in either the SZ or the VZ (Fig. 3d,e), highly conserved between brains (p<10−50), and highly distinct. Both VZ-enriched modules (G7, G8) contained cell cycle genes and many astrocyte markers, suggesting that these modules may represent different RG populations. Fluorescent ISH (FISH) confirmed that representative genes in both G7 (NR2E1) and G8 (SPATA13) are enriched in VZ versus SZ (Fig. 3f; Extended Data Fig. 7). Surprisingly, rather than labeling distinct cell populations in VZ, these genes labeled mutually exclusive subcellular locations within the same VZ cells (note non-overlapping cytoplasmic localization of SPATA13 and NR2E1), suggesting that these modules represent differentially regulated biological processes within RG cells.
Extended Data Figure 7. FISH of hub genes in VZ-enriched modules shows expected laminar enrichment and largely non-overlapping subcellular distributions.
a. Fluorescent in situ hybridization (FISH) in proliferative layers of 15 post-conceptual week human cortex for three genes in modules G7 and G8 in the germinal layers network shown in Figure 3 of the manuscript (see Fig. 3d–f)—SPATA13, NR2E1, and DTL. All three genes show enrichment in the VZ compared to the SZ as expected based on microarray data. Nuclei are labeled with DAPI (blue). b. High magnification images in the VZ show double labeling for each pair of genes (with fluor reversal, lower row) and show complex subcellular distributions. SPATA13, NR2E1, and (to a lesser extent) DTL appear to be expressed in most cells in the VZ, but these genes are typically expressed in non-overlapping punctate cytoplasmic locations (excluded from DAPI-stained nuclei in blue). b is at 50× magnification relative to a.
We were particularly interested in identifying differences between the SZi and the SZo, which is absent in mice. Modules G2 and G3 are enriched in the SZi (Fig. 3d, white bars in bottom two rows), and G3 is enriched for genes marking Svet1+ IPs in E14 mouse SZ43, including ELAVL4, LRP8, NEUROD1, NRN1, SLC17A6, SSTR2, and TP53INP1. Modules G4-G6 are enriched in the SZo (Fig. 3d, gray bars); however, these modules are enriched for neuron-associated categories likely expressed in postmitotic cells during their radial migration. To identify expression specific to SZo or SZi using a more targeted approach, we searched for genes both maximally expressed in the SZ and differentially expressed between SZo and SZi (Fig. 3g; t>4, p<0.01, log2(FC)>0.5). Remarkably, few genes met both criteria at 15–16 pcw: 39 genes were enriched in SZi (including several in module G3), while only eight were specifically enriched in SZo. These results are consistent with a previous study of laminar enrichment in 13–16pcw human prenatal cortex, which also found few genes specifically enriched in SZo (55) or SZi (61)2.
Species differences in subplate
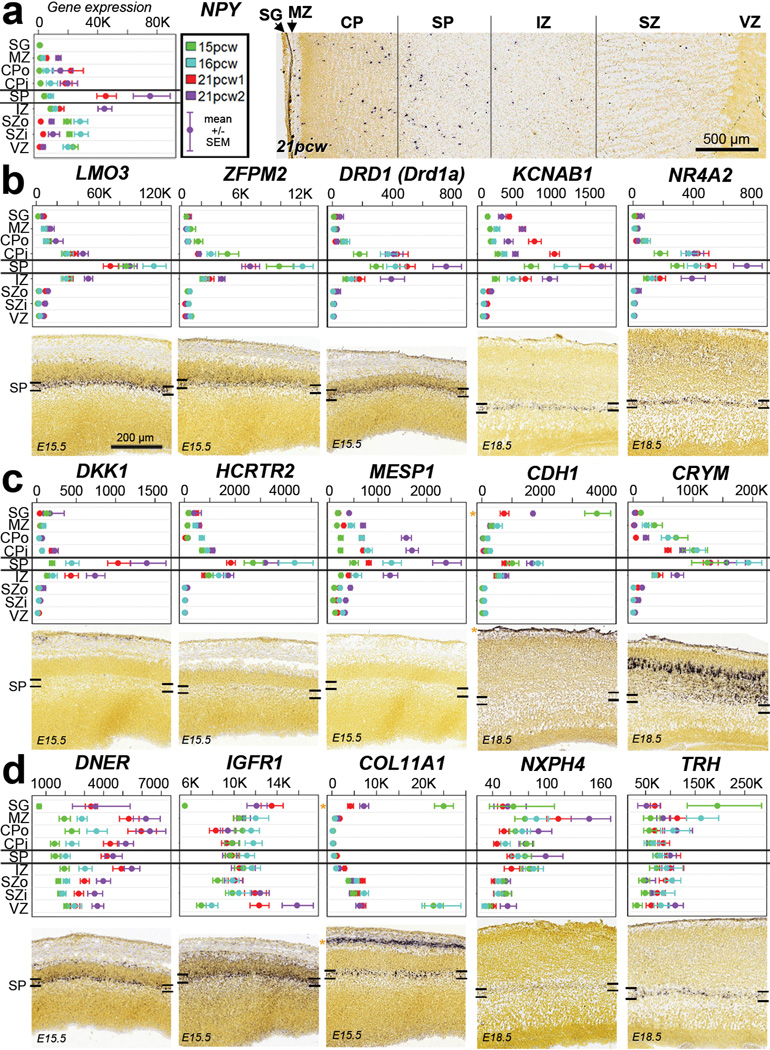
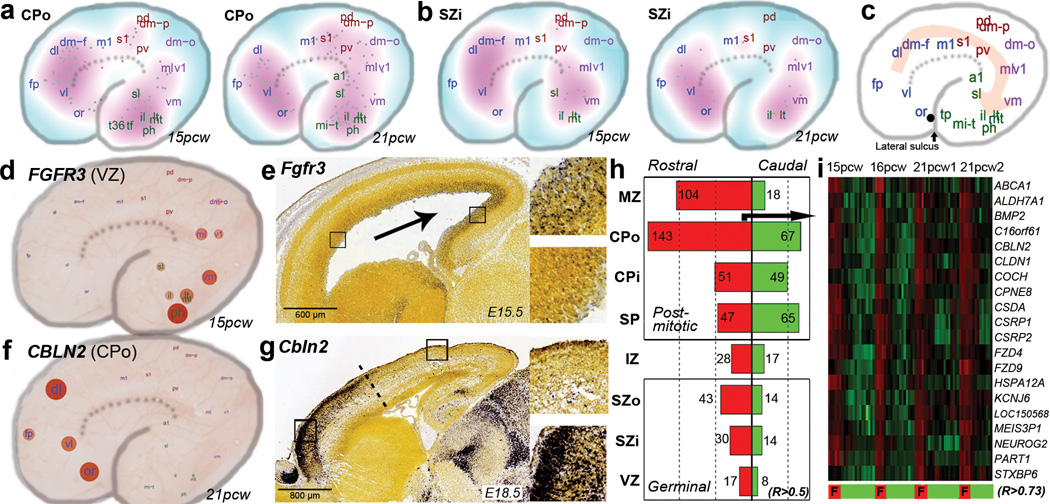
The SP is a largely transient zone beneath the CP that plays an important role in establishment of thalamocortical connectivity (see35 for review). SP generation is protracted in primates8, and its thickness particularly expanded in human8. In mouse and other species18 this layer is molecularly distinct, and our laminar profiling also identified many SP-enriched genes in human (Fig. 2). For example, NPY is enriched in SP at 21pcw (but not 15–16pcw) as shown both by microarray and ISH (Fig. 4a). To facilitate a comparative analysis, we identified a high confidence set of 150 SP-enriched genes in human and mouse (Suppl. Table 8). Many genes showed similar enrichment in the developing mouse and human SP, including the known SP markers KCNAB1 and NR4A217,18,44 (Fig. 4b). Several genes showed enrichment in developing human but not mouse SP, including the hypocretin (orexin) receptor HCRTR2 (Fig. 4c; Extended Data Fig. 8a), which is thought to regulate sleep-wakefulness and is highly expressed in mouse hypothalamus45. Conversely, Trh and Nxph4 show enriched expression in mouse but not human SP (Fig. 4d; Extended Data Fig. 8b). Interestingly, Trh is also not expressed in the rat SP18, suggesting this pattern is specific to mouse. These results indicate that the evolutionary elaboration of SP in primates is associated with altered gene expression.
Figure 4. Common and distinct subplate markers in human and mouse.
a. NPY is enriched in SP at 21pcw but not 15–16pcw based on microarray (left) and ISH (right). Microarray data is plotted as the average +/− standard error of the mean (SEM) for each layer in each brain. b. Genes with SP enrichment in both species, based on microarray data in human (upper row) and ISH data at E15.5 or E18.5 mouse (lower row). c. Genes with SP enrichment in human but either no expression (DKK1, CRTR2 and MESP1) or no SP enrichment (CHD1 and CRYM) in mouse. d. Genes with SP enrichment in developing mouse, but not human, SP. Asterisks indicate common expression between human and mouse in other layers (i.e., SG/pia mater). Mouse ISH images taken from the Allen Developing Mouse Brain Atlas.
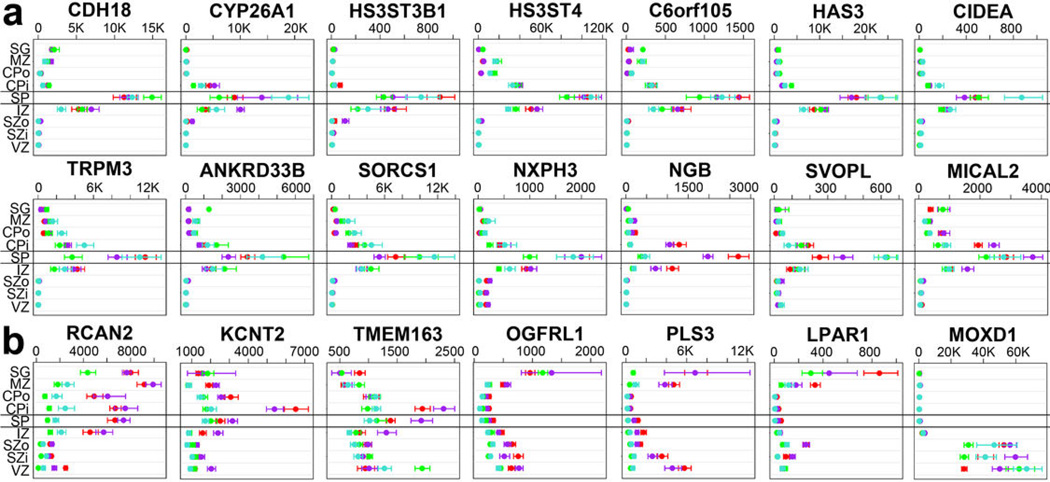
Extended Data Figure 8. Laminar gene expression of putative SP markers for human and mouse in prenatal human cortex.
(a) Novel human subplate-enriched genes showing at least 8-fold enrichment in SP in all four prenatal human brains. CDH18, a known SP marker in mouse, is presented as a positive control. (b) Genes with differences in subplate expression between mouse and human. These genes have been reported as subplate-enriched in mouse studies but do not show human subplate enrichment. Labeling as in Figure 4a of the main manuscript. Microarray data is plotted as the average +/− standard error of the mean (SEM) for each layer in each of the four brains analyzed (colors).
Developmental gradients in neocortex
Cortical patterning is likely a result of intrinsic signaling, controlled in part by graded expression of transcription factors during early cortical development, followed by extrinsic signaling from thalamic afferents after the start of corticogenesis46–49. We sought to identify putative patterning centers, defined as regions where many genes show peak expression tapering off with distance, for each layer of the human prenatal cortex using an unbiased approach. To do so, we first assigned 3D coordinates to each cortical sample, and then identified the location of maximum expression of the most graded genes in all four brains. In several layers, including CPo and SZi (Fig. 5a–b), the majority of these genes peaked in the frontal or temporal lobes. Rather than peaks in presumptive functional areas, this suggests a generally rostro-caudal organization axis that is better characterized as fronto-temporal, following the contour of the developing cortex. To identify such fronto-temporal patterning genes directly, we correlated gene expression in each cortical layer against the angular position of each neocortical region, as illustrated schematically in Figure 5c. All layers contained gradient genes conserved across all four brains (p~0, permutation analysis; FDR<2.4%; Suppl. Methods), and each layer contained distinct sets of gradient genes, particularly when comparing germinal with postmitotic layers (Suppl. Table 9). Gradient genes in VZ likely reflect intrinsic areal specification as VZ does not receive thalamic innervation. Interestingly, rostral and caudal genes that can be identified in grossly dissected cortex3 largely represent gradients in postmitotic cells, as they show significant overlap with gradient genes in MZ, CP, and IZ (Extended Data Fig. 9).
Figure 5. Areal patterning in developing neocortex.
a–b. Density plot showing the location of highest expression for genes with gradient-like expression in CPo (a) or SZi (b) in each brain. c. Schematic illustrating the predominant direction of gene gradients, which follow a fronto-temporal trajectory. d. FGFR3 shows caudal enrichment in germinal zones of developing human cortex. Samples are plotted on a schematic of the prenatal cortex, with expression level indicated by circle size and color. e. Similar enrichment is seen in developing mouse cortex. f–g. CBLN2 shows rostral enrichment in CP of both human (f) and mouse (g). h. Barplot showing common rostrally- (red) and caudally- (green) enriched genes across brains for each layer. i. Heatmap representation of the top 20 rostral-enriched in the CPo shows selective enrichment in frontal lobe (F).
Extended Data Figure 9. Areal gradients are consistent with patterns in BrainSpan RNA-seq cortical data, particularly for postmitotic layers.
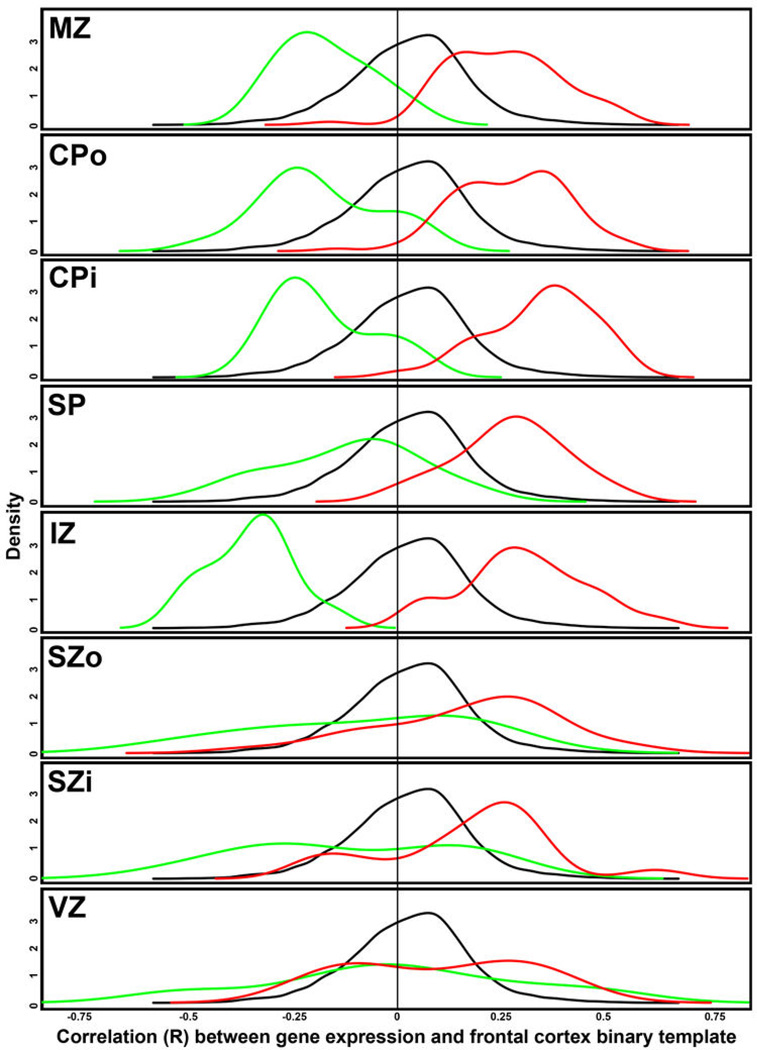
RNA-seq RPKM values for 8–22pcw specimens in the BrainSpan database were used to assess rostral caudal patterning for all genes in prenatal development. Specifically, gene expression was correlated with a template of frontal cortex samples (1) vs. samples from other cortical layers (0), such that positive correlations correspond to rostral enrichment. The same density plot of the resulting correlations is plotted for each layer in black. For each layer (except SG), density plots for the subset of rostral (red) and caudal (green) genes identified in this study (Fig. 5h) are shown. Note the significant offset of density curves for rostral and caudal genes in MZ, CPo, CPi, and IZ (and other layers to a lesser extent), indicating good agreement in areal gradient genes between studies.
Some features of areal patterning appear to be preserved between mouse and human. For example, FGFR3, which is known to cause defects in human temporal cortex when mutated50, shows significant caudal enrichment in all germinal layers in both species (Fig. 5d–e). Conversely, CBLN2 shows rostral enrichment in human CPo (Fig. 5f) and in mouse (Fig. 5g), with an abrupt expression cutoff, implicating CBLN2 in early rostral patterning. Comparing these data to a microarray analysis of rostral versus caudal cortex in E14 mouse51 identified 20 additional genes with consistent rostrocaudal gradation between species (Suppl. Table 9; Suppl. Methods). Interestingly, we find more rostrally- than caudally-enriched genes in nearly every cortical layer (Fig. 5h), whereas two studies of gradient expression in prenatal mouse brain identified more49 or comparable51 caudally-enriched genes, indicating potential species differences related to areal patterning. This human frontal asymmetry is most apparent in the outer postmitotic layers MZ and CPo (Fig. 5i), and in SZo, which generates most superficial CP neurons in primate7,8, suggesting that these genes may play a part in the expansion and reorganization of human PFC8. Alternatively, this asymmetry could reflect temporal differences, as peak generation of excitatory neurons in visual cortex is delayed relative to frontal cortex52.
Patterned expression of genes near haCNSs
Conserved non-coding sequences (CNSs) are genomic regions with exceptionally high similarity across divergent organisms, and therefore thought to be important for organism viability. CNSs are typically located by genes important for developmental regulation, and many show significant enhancer activity in brain5. Genes near CNSs with significantly accelerated rates of substitution in the human lineage (haCNSs)15 are particularly likely to show differential expression between regions of developing human neocortex3,5, suggesting transcriptional regulation by haCNSs may be important in human-specific neurodevelopment. Our results confirm and extend these findings (Table 1; Suppl. Table 9). Rostrally-enriched genes include significantly more haCNSs than caudally-enriched genes, consistent with the expanded frontal cortex in primates and the developmental role of haCNSs. We also find more haCNSs with areal expression patterns in postmitotic than germinal layers, even after accounting for the larger number of areal genes in postmitotic layers. Interestingly, nearly 25% of regional genes in IZ (11/45) were haCNSs. This result, which cannot be explained by over-representation of neural adhesion genes in IZ, suggests that IZ is of particular importance in areal cortical identity during human development53.
Table 1.
Human-specific neurodevelopmental processes enriched for genes near haCNSs
| Category | Subcategory | O | O/E | % total | P-value |
|---|---|---|---|---|---|
| Areal | All | 46 | 3.54 | 9.9% | 2.56E-14 |
| Rostral (R) | 34 | 4.24 | 11.8% | 2.60E-13 | |
| Caudal (C) | 14 | 2.77 | 7.7% | 1.78E-04 | |
| Postmitotic (P) | 35 | 3.37 | 9.4% | 1.01E-10 | |
| IZ | 11 | 8.80 | 24.4% | 2.34E-09 | |
| Germinal (G) | 7 | 2.68 | 7.4% | 4.65E-03 | |
| Network | C31 (interneuron) | 18 | 5.31 | 14.8% | 1.15E-09 |
| C16 (subplate) | 25 | 3.98 | 11.1% | 9.80E-10 |
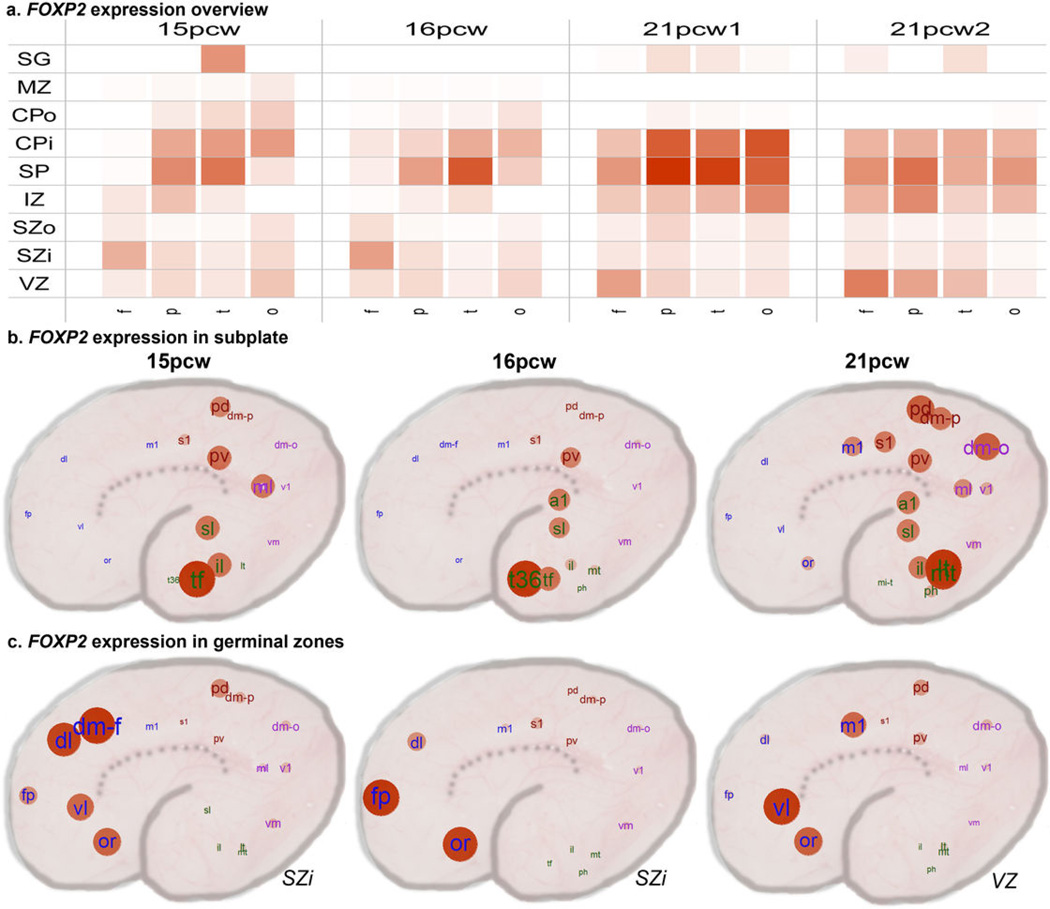
Finally, we assessed the distribution of haCNSs in modules from our whole cortex network (Fig 3a–c). Only two of the 42 tested modules were enriched for haCNSs: the interneuron-related (C31) and SP-enriched (C16) modules, which both mark processes with features potentially distinct to the human lineage. Interestingly, FOXP2, implicated in the specialization of language areas54, is included in the module C16, and shows enrichment in parietal and temporal lobes including presumptive language areas (Extended Data Fig. 10). No layers other than SP were significantly enriched for haCNSs (data not shown). Together these results support the hypothesis that transcriptional networks underlying the evolution of human neocortex can at least partly be traced to haCNSs.
Extended Data Figure 10. Areal and laminar expression patterning of FOXP2.
a. Summarized expression levels of FOXP2 across each lobe, layer, and brain. b. FOXP2 shows enrichment in parietal and temporal regions overlapping Wernicke's area in SP at all three time points. c. FOXP2 shows enrichment in frontal cortex in germinal zones. Red = higher expression.
Discussion
Studies of the developing human brain are essential for elucidating the details of human brain formation, function, and evolutionary differences, and for understanding developmental mechanisms underlying neurodevelopmental disorders such as autism39,40 and schizophrenia19. The atlas of the mid-gestational human brain described here, part of the BrainSpan Atlas of the Developing Human Brain, builds on digital molecular brain atlasing efforts in mouse20,22,24 and adult human23 by providing transcriptome resources on prenatal specimens typically inaccessible for research. Several recent studies have assayed a limited set of brain structures1,3–5 and layers2 from prenatal human brain. In contrast, the current project aimed for anatomical comprehensiveness at a fine nuclear/laminar level, albeit with a small number of specimens. This degree of specificity necessitated using available methods for small sample amplification and DNA microarrays (the same platform recently used for adult human23), but newer techniques may soon allow moving to the resolution of single cells using RNA sequencing for complete transcriptome coverage55.
Many differences in cortical development between human, non-human primate and rodent have been documented, including an expanded SP8 and SG9, expansion of association areas particularly in frontal lobe8, expansion of superficial layers that greatly increase the extent of cortico-cortical connections, and the appearance of a secondary proliferative zone, the SZo, that likely allow the massive expansion of human cortex6,7. We find transcriptional features related to each of these anatomical features, although we were able to identify only minimal molecular differences between the SZi and SZo, leaving open the question of what distinguishes this primate-specific zone of cortical precursors. These data also provide a powerful map to pin an anatomical and developmental locus on genes related to neurodevelopmental disease origins and human-specific brain function and evolution. Although the current analyses only scratch the surface, these data will be extremely useful for generating and testing new hypotheses about molecular substrates for specific features of human brain development and function.
Methods (online version)
Post-mortem tissue acquisition and screening
Tissue was provided by the Birth Defects Research Laboratory (BDRL) at the University of Washington and Advanced Bioscience Resources Incorporated (ABR; Alameda, CA). All work was performed according to guidelines for the research use of human brain tissue (ABR) or the UAGA and NOTA guidelines for the acquisition and distribution of human tissue for bio-medical research purposes (BDRL) and with approval by the Human Investigation Committees and Institutional Ethics Committees of each institute from which samples were obtained. Appropriate written informed consent was obtained and all available non-identifying information was recorded for each sample. Specimens for microarray profiling consisted of two 21 pcw females, one 15 pcw male and one 16 pcw female (Suppl. Table 1).
Laser microdissection and RNA isolation
Slabs from the frozen brains were serially cryosectioned at 14 µm onto PEN slides for LMD (Leica Microsystems, Inc., Bannockburn, IL) and a 1:10 Nissl series was generated for neuroanatomical reference. After drying for 30 min at room temperature, PEN slides were frozen at −80°C. Slides were later rapidly fixed in ice cold 70% ethanol, lightly stained with cresyl violet to allow cytoarchitectural visualization, dehydrated, and frozen at −80°C. LMD was performed on a Leica LMD6000 (Leica Microsystems, Inc.) using the cresyl violet stain to identify target brain regions. Samples captured include cortical and subcortical regions and are listed for each brain in the ontological sample map (Suppl. Table 2).
Microdissected tissue was collected directly into RLT buffer from the RNeasy Micro kit (QIAGEN Inc., Valencia, CA) supplemented with β-mercaptoethanol. Samples were volume adjusted with RLT Buffer to 75µl, vortexed, centrifuged, and frozen at −80°C. RNA was isolated for each brain region following the manufacturer’s directions. RNA samples were eluted in 14µl, and 1µl was run on the Agilent 2100 Bioanalyzer (Agilent Technologies, Inc., Santa Clara, CA) using the Pico 6000 assay kit. Samples were quantitated using the Bioanalyzer concentration output. The average RNA Integrity Number (RIN) of all 1,202 passed experimental samples was 6.3.
mRNA profiling
Sample amplification, labeling, and microarray processing were performed by the Covance Genomics Laboratory (Seattle, WA). Briefly, samples were amplified using a custom two-cycle RT-IVT amplification protocol. For each sample, 5ng of total RNA was mixed with 250ng of pBR322 (Life Technologies) to act as a carrier. The MessageAmp II aRNA Amplification Kit was utilized for the first round of amplification and the Amino Allyl MessageAmp II aRNA Amplification Kit for the second round of amplification (Life Technologies). Following amplification, 5µg of cRNA was labeled with Cy3 mono-Reactive Dye (GE Healthcare). Each labeled aRNA was resolved using a Bioanalyzer with RNA 6000 Nano kit reagents (Agilent Technologies) before hybridization. Samples were evaluated for yield and size distribution, then normalized to 600ng input, fragmented, and hybridized to Agilent Human 8×60K Arrays. Gene expression data quality was assessed using standard Agilent quality control metrics. To control for batch effects, common RNA pool control samples were amplified and hybridized in each batch. A total of 1,225 samples passed sample quality control (QC), including 1,202 experimental samples and 23 control samples. The data discussed in this publication are accessible through the Allen Brain Atlas data portal (http://www.brain-map.org) or directly at http://www.brainspan.org.
Microarray data analysis
All microarray data was subjected to QC and ERCC spike-in assessments, and any failing samples were omitted from the analysis. Biological outliers were identified by comparing samples from related structures using hierarchical clustering and inter-array correlation measures. Data for samples passing QC were normalized in three steps: 1) “within-batch” normalization to the 75th percentile expression values; 2) “cross-batch” bias reduction using ComBat57; and 3) "cross-brain" normalization as in step 1. Differential expression assessments were done using template vector correlation, where 1="in group" and 0="not in group", or by measuring the fold change, defined as mean expression in category divided by mean expression elsewhere. False discovery rates were estimated using permutation tests (Suppl. Methods). WGCNA was performed on all neocortical samples using the standard method36,58, and on germinal layers by defining a consensus module in the 15 and 16pcw brains59, only including genes differentially expressed across these layers (5494 genes; ANOVA p<0.01, Benjamini-Hochberg adjusted). Gene list characterizations were made using a combination of module eigengene / representative gene expression, gene ontology enrichment using DAVID60, and enrichment for known brain-related categories (i.e.,61,62) using userListEnrichment63. Module C31 is depicted using VisANT64: the top 250 gene-gene connections based on topological overlap are shown, with histone genes removed for clarity. Rostral-caudal areal gradient genes were identified as follows: first, the center of each neocortical region was identified at 21pcw in Euclidean coordinates; second, the rostral/caudal region position was estimated as an angle along the lateral face of the brain centered at the temporal/frontal lobe juncture (ordering lobes roughly as frontal, parietal, occipital, temporal; Fig. 5c); third, for each brain gene expression in each layer was (Pearson) correlated with this region position; and finally, genes with R>0.5 in all four brains were identified. A similar strategy was used to identify unbiased areal gradient genes (Suppl. Methods). Enrichment of haCNSs was determined using hypergeometric tests. Samples in all plots are ordered in an anatomically relevant manner. Unless otherwise noted, all p-values are Bonferroni corrected for multiple comparisons.
In situ hybridization
Nonisotopic colorimetric ISH was performed as described previously24 with some modifications such as a reduction in proteinase K concentration. Briefly, following cryosectioning of fresh-frozen samples at 20µm, tissue sections were fixed, acetylated, and subsequently dehydrated. Digoxigenin-based riboprobe labeling coupled with TSA amplification and alkaline-phosphatase-based colorimetric detection was used to label target mRNAs in expressing cells. Riboprobes were designed to overlap probe designs for homologous genes in mouse in the Allen Developing Mouse Brain Atlas (http://www.developingmouse.brain-map.org/). FISH was run as previously dd that high resolution images were captured on an Olympus Fluoview 1000 Confocal Microscope at 60× magnification.
Supplementary Material
Acknowledgments
We wish to thank the Allen Institute founders, P. G. Allen and J. Allen, for their vision, encouragement, and support. We express our gratitude to past and present Allen Institute staff members Rachel Adams, Andreas Alpisa, Andrew Boe, Emi Byrnes, Mike Chapin, Jefferey Chen, Catherine Copeland, Nadezhda Dotson, Korrin Fotheringham, Erich Fulfs, Mary Gasparrini, Terri Gilbert, Zeb Haradon, Nika Hejazinia, Nishi Ivanov, John Kinnunen, Allison Kriedberg, Jacob Laoenkue, Samuel Levine, Vilas Menon, Erika Mott, Nathanael Motz, Julie Pendergraft, Lydia Potekhina, Joshuah Redmayne-Titley, David Rosen, Cecilli Simpson, Shu Shi, Lissette Velasquez, Udipta Wagley, Natalie Wong, and Brian Youngstrom for their technical assistance. We would also like to thank Jean Augustinack, Thomas Benner, Azma Mayaram, Michelle Roy, Andre van der Kouwe, and Larry Wald from Dr. Fischl's lab. Also, we wish to acknowledge Covance Genomics Laboratory (Seattle, WA) for microarray probe generation, hybridization and scanning. In addition, we express our gratitude to Advanced Bioscience Resources Inc., for providing tissue used for expression profiling and reference atlas generation as well as to the Laboratory of Developmental Biology, University of Washington, for providing tissue used for expression profiling and reference atlas generation. The Laboratory of Developmental Biology work was supported by NIH Award Number 5R24HD0008836 from the Eunice Kennedy Shriver National Institute of Child Health & Human Development. The BrainSpan project was supported by Award Number RC2MH089921 (PIs: Ed Lein & Michael Hawrylycz, Allen Institute for Brain Science) from the National Institute of Mental Health. The content is solely the responsibility of the respective authors and does not necessarily represent the official views of the National Institute of Mental Health or the National Institutes of Health.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributors:
E.S.L, S.-L.D., K.A.S. and S.M.S. contributed significantly to the overall project design. S.M.S, K.A.S., A.E., A.B., and P.W. managed the tissue and sample processing in the laboratory. K.A., Ja.A., C.B., D.B., K.B., S.B., S.C., A.C., C.C., R.A.D., G.Ge., J.G., L.G., B.W.G., R.E.H., T.A.L., Na.M., N.F.M., N-K N., A.O., E.O., J.Pa., P.D.P., S.E.P., M.P ., Me.R., J.J.R., K.R., D.S., Me.S., S.S., N.V.S., and Mi.S. contributed to tissue and sample processing. E.H.S., Z.L.R., T.N.-C., and I.A.G. contributed to establishing the tissue acquisition pipeline. N.D., J.N. and A.B. contributed to protocol development. A.S.P., L.Z., B.F., and H.H. contributed to MR and DWI imaging and analysis. J.M.J., C.R.S., and D.W. provided engineering support. S.-L.D., R.A.D., P.D.P., D.S., and J.G.H. contributed to the neuroanatomical design and implementation. S.-L.D., B.A.C.F., Ph.L., B.M., J.J.R., R.He., N.Se. and J.G.H. contributed to the reference atlas design, quality control and implementation. L.N., A.S., and C.D. managed the creation of the data pipeline, visualization and mining tools. L.N., A.S., T.A.D., D.F., T.P.F., G.Gu, C.L.K., C.La., F.L., N.Sj., and A.J.S. contributed to the creation of the data pipeline, visualization and mining tools. J.A.M., S.-L.D., R.F.H., C.-K.L., M.J.H., S.M.S, and E.S.L. contributed to data analysis and interpretation. M.B.G., D.H.G., J.A.K., Pa.L., J.W.P., N.Se, and A.R.J. contributed to overall project design and consortium management. E.S.L. and M.J.H. conceived the project, and the manuscript was written by J.A.M. and E.S.L. with input from all other authors.
These data are freely accessible as part of the BrainSpan Atlas of the Developing Human Brain (http://brainspan.org), also available via the Allen Brain Atlas data portal (http://www.brain-map.org).
The authors declare no competing financial interests.
References
- 1.Colantuoni C, et al. Temporal dynamics and genetic control of transcription in the human prefrontal cortex. Nature. 2011;478:519–523. doi: 10.1038/nature10524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fietz S, et al. Transcriptomes of germinal zones of human and mouse fetal neocortex suggest a role of extracellular matrix in progenitor self-renewal. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:11836–11841. doi: 10.1073/pnas.1209647109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johnson M, et al. Functional and evolutionary insights into human brain development through global transcriptome analysis. Neuron. 2009;62:494–509. doi: 10.1016/j.neuron.2009.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kang HJ, et al. Spatio-temporal transcriptome of the human brain. Nature. 2011;478:483–489. doi: 10.1038/nature10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lambert N, et al. Genes expressed in specific areas of the human fetal cerebral cortex display distinct patterns of evolution. PLoS ONE. 2011;6:e17753. doi: 10.1371/journal.pone.0017753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kriegstein A, Noctor S, Martínez-Cerdeño V. Patterns of neural stem and progenitor cell division may underlie evolutionary cortical expansion. Nature reviews. Neuroscience. 2006;7:883–890. doi: 10.1038/nrn2008. [DOI] [PubMed] [Google Scholar]
- 7.Hansen D, Lui J, Parker P, Kriegstein A. Neurogenic radial glia in the outer subventricular zone of human neocortex. Nature. 2010;464:554–561. doi: 10.1038/nature08845. [DOI] [PubMed] [Google Scholar]
- 8.Smart I, Dehay C, Giroud P, Berland M, Kennedy H. Unique morphological features of the proliferative zones and postmitotic compartments of the neural epithelium giving rise to striate and extrastriate cortex in the monkey. Cerebral cortex (New York, N.Y. : 1991) 2002;12:37–53. doi: 10.1093/cercor/12.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brun A. The subpial granular layer of the foetal cerebral cortex in man. Its ontogeny and significance in congenital cortical malformations. Acta pathologica et microbiologica Scandinavica. 1965;179(Suppl):173–198. [PubMed] [Google Scholar]
- 10.Anderson SA, Eisenstat DD, Shi L, Rubenstein JL. Interneuron migration from basal forebrain to neocortex: dependence on Dlx genes. Science. 1997;278:474–476. doi: 10.1126/science.278.5337.474. [DOI] [PubMed] [Google Scholar]
- 11.Ma T, et al. Subcortical origins of human and monkey neocortical interneurons. Nat Neurosci. 2013;16:1588–1597. doi: 10.1038/nn.3536. [DOI] [PubMed] [Google Scholar]
- 12.Yu X, Zecevic N. Dorsal radial glial cells have the potential to generate cortical interneurons in human but not in mouse brain. J Neurosci. 2011;31:2413–2420. doi: 10.1523/JNEUROSCI.5249-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Letinic K, Zoncu R, Rakic P. Origin of GABAergic neurons in the human neocortex. Nature. 2002;417:645–649. doi: 10.1038/nature00779. [DOI] [PubMed] [Google Scholar]
- 14.Hansen DV, et al. Non-epithelial stem cells and cortical interneuron production in the human ganglionic eminences. Nat Neurosci. 2013;16:1576–1587. doi: 10.1038/nn.3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prabhakar S, Noonan J, Pääbo S, Rubin E. Accelerated evolution of conserved noncoding sequences in humans. Science (New York, N.Y.) 2006;314:786. doi: 10.1126/science.1130738. [DOI] [PubMed] [Google Scholar]
- 16.Ayoub A, et al. Transcriptional programs in transient embryonic zones of the cerebral cortex defined by high-resolution mRNA sequencing. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:14950–14955. doi: 10.1073/pnas.1112213108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Montiel J, et al. Hypothesis on the dual origin of the Mammalian subplate. Frontiers in neuroanatomy. 2011;5 doi: 10.3389/fnana.2011.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang WZ, et al. Comparative aspects of subplate zone studied with gene expression in sauropsids and mammals. Cerebral cortex (New York, N.Y. : 1991) 2011;21:2187–2203. doi: 10.1093/cercor/bhq278. [DOI] [PubMed] [Google Scholar]
- 19.Insel TR. Rethinking schizophrenia. Nature. 2010;468:187–193. doi: 10.1038/nature09552. [DOI] [PubMed] [Google Scholar]
- 20.Paxinos G, Franklin K. Paxinos and Franklin's the Mouse Brain in Stereotaxic Coordinates, Fourth Edition. Academic Press; 2012. [Google Scholar]
- 21.Saleem K, Logothetis N. A Combined MRI and Histology Atlas of the Rhesus Monkey Brain in Stereotaxic Coordinates, Second Edition. Academic Press; 2012. [Google Scholar]
- 22.Dong HW. Allen reference atlas : a digital color brain atlas of the C57Black/6J male mouse. Wiley; 2008. [Google Scholar]
- 23.Hawrylycz M, et al. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature. 2012;489:391–399. doi: 10.1038/nature11405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lein ES, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- 25.Bernard A, et al. Transcriptional architecture of the primate neocortex. Neuron. 2012;73:1083–1099. doi: 10.1016/j.neuron.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Steinfeld R, et al. Folate receptor alpha defect causes cerebral folate transport deficiency: a treatable neurodegenerative disorder associated with disturbed myelin metabolism. American journal of human genetics. 2009;85:354–363. doi: 10.1016/j.ajhg.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dubourg C, et al. Molecular screening of SHH ZIC2, SIX3, and TGIF genes in patients with features of holoprosencephaly spectrum: Mutation review and genotype-phenotype correlations. Human mutation. 2004;24:43–51. doi: 10.1002/humu.20056. [DOI] [PubMed] [Google Scholar]
- 28.Huang H, et al. Anatomical characterization of human fetal brain development with diffusion tensor magnetic resonance imaging. J Neurosci. 2009;29:4263–4273. doi: 10.1523/JNEUROSCI.2769-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bystron I, Blakemore C, Rakic P. Development of the human cerebral cortex: Boulder Committee revisited. Nature reviews. Neuroscience. 2008;9:110–122. doi: 10.1038/nrn2252. [DOI] [PubMed] [Google Scholar]
- 30.Weisenhorn DM, Prieto EW, Celio MR. Localization of calretinin in cells of layer I (Cajal-Retzius cells) of the developing cortex of the rat. Brain research. Developmental brain research. 1994;82:293–297. doi: 10.1016/0165-3806(94)90171-6. [DOI] [PubMed] [Google Scholar]
- 31.Englund C, et al. Pax6, Tbr2, and Tbr1 are expressed sequentially by radial glia, intermediate progenitor cells, and postmitotic neurons in developing neocortex. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2005;25:247–251. doi: 10.1523/JNEUROSCI.2899-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Levitt P, Rakic P. Immunoperoxidase localization of glial fibrillary acidic protein in radial glial cells and astrocytes of the developing rhesus monkey brain. The Journal of comparative neurology. 1980;193:815–840. doi: 10.1002/cne.901930316. [DOI] [PubMed] [Google Scholar]
- 33.Grinberg I, et al. Heterozygous deletion of the linked genes ZIC1 and ZIC4 is involved in Dandy-Walker malformation. Nat Genet. 2004;36:1053–1055. doi: 10.1038/ng1420. [DOI] [PubMed] [Google Scholar]
- 34.Inoue T, Ogawa M, Mikoshiba K, Aruga J. Zic deficiency in the cortical marginal zone and meninges results in cortical lamination defects resembling those in type II lissencephaly. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2008;28:4712–4725. doi: 10.1523/JNEUROSCI.5735-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kostovic I, Judas M. The development of the subplate and thalamocortical connections in the human foetal brain. Acta Paediatr. 2010;99:1119–1127. doi: 10.1111/j.1651-2227.2010.01811.x. [DOI] [PubMed] [Google Scholar]
- 36.Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Statistical applications in genetics and molecular biology. 2005;4 doi: 10.2202/1544-6115.1128. [DOI] [PubMed] [Google Scholar]
- 37.Oldham M, et al. Functional organization of the transcriptome in human brain. Nature neuroscience. 2008;11:1271–1282. doi: 10.1038/nn.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Voineagu I, et al. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature. 2011;474:380–384. doi: 10.1038/nature10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parikshak NN, et al. Integrative functional genomic analyses implicate specific molecular pathways and circuits in autism. Cell. 2013;155:1008–1021. doi: 10.1016/j.cell.2013.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Willsey AJ, et al. Coexpression networks implicate human midfetal deep cortical projection neurons in the pathogenesis of autism. Cell. 2013;155:997–1007. doi: 10.1016/j.cell.2013.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gaspard N, Vanderhaeghen P. Laminar fate specification in the cerebral cortex. F1000 biology reports. 2011;3 doi: 10.3410/B3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Betizeau M, et al. Precursor diversity and complexity of lineage relationships in the outer subventricular zone of the primate. Neuron. 2013;80:442–457. doi: 10.1016/j.neuron.2013.09.032. [DOI] [PubMed] [Google Scholar]
- 43.Kawaguchi A, et al. Single-cell gene profiling defines differential progenitor subclasses in mammalian neurogenesis. Development (Cambridge, England) 2008;135:3113–3124. doi: 10.1242/dev.022616. [DOI] [PubMed] [Google Scholar]
- 44.Butler DM, Ono JK, Chang T, McCaman RE, Barish ME. Mouse brain potassium channel beta1 subunit mRNA: cloning and distribution during development. Journal of neurobiology. 1998;34:135–150. [PubMed] [Google Scholar]
- 45.Gerashchenko D, et al. Hypocretin-2-saporin lesions of the lateral hypothalamus produce narcoleptic-like sleep behavior in the rat. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2001;21:7273–7283. doi: 10.1523/JNEUROSCI.21-18-07273.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mallamaci A, Stoykova A. Gene networks controlling early cerebral cortex arealization. The European journal of neuroscience. 2006;23:847–856. doi: 10.1111/j.1460-9568.2006.04634.x. [DOI] [PubMed] [Google Scholar]
- 47.O'Leary DD. Do cortical areas emerge from a protocortex? Trends in Neurosciences. 1989;12:400–406. doi: 10.1016/0166-2236(89)90080-5. [DOI] [PubMed] [Google Scholar]
- 48.Rakic P. Specification of cerebral cortical areas. Science (New York, N.Y.) 1988;241:170–176. doi: 10.1126/science.3291116. [DOI] [PubMed] [Google Scholar]
- 49.Sansom S, Livesey F. Gradients in the brain: the control of the development of form and function in the cerebral cortex. Cold Spring Harbor perspectives in biology. 2009;1 doi: 10.1101/cshperspect.a002519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hevner R. The cerebral cortex malformation in thanatophoric dysplasia: neuropathology and pathogenesis. Acta neuropathologica. 2005;110:208–221. doi: 10.1007/s00401-005-1059-8. [DOI] [PubMed] [Google Scholar]
- 51.Pinto L, et al. AP2gamma regulates basal progenitor fate in a region- and layer-specific manner in the developing cortex. Nat Neurosci. 2009;12:1229–1237. doi: 10.1038/nn.2399. [DOI] [PubMed] [Google Scholar]
- 52.Rakic P. Neurogenesis in adult primates. Progress in brain research. 2002;138:3–14. doi: 10.1016/S0079-6123(02)38067-1. [DOI] [PubMed] [Google Scholar]
- 53.Allendoerfer KL, Shatz CJ. The subplate, a transient neocortical structure: its role in the development of connections between thalamus and cortex. Annual review of neuroscience. 1994;17:185–218. doi: 10.1146/annurev.ne.17.030194.001153. [DOI] [PubMed] [Google Scholar]
- 54.Enard W, et al. Molecular evolution of FOXP2, a gene involved in speech and language. Nature. 2002;418:869–872. doi: 10.1038/nature01025. [DOI] [PubMed] [Google Scholar]
- 55.Qiu S, et al. Single-neuron RNA-Seq: technical feasibility and reproducibility. Front Genet. 2012;3:124. doi: 10.3389/fgene.2012.00124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sorensen SA, et al. Correlated Gene Expression and Target Specificity Demonstrate Excitatory Projection Neuron Diversity. Cereb Cortex. 2013 doi: 10.1093/cercor/bht243. [DOI] [PubMed] [Google Scholar]
References (for online methods)
- 57.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics (Oxford, England) 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 58.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559–559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Langfelder P, Horvath S. Eigengene networks for studying the relationships between co-expression modules. BMC Systems Biology. 2007;1:54–54. doi: 10.1186/1752-0509-1-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huang DW, Sherman B, Lempicki R. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protocols. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 61.Cahoy J, et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2008;28:264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sugino K, et al. Molecular taxonomy of major neuronal classes in the adult mouse forebrain. Nat Neurosci. 2006;9:99–107. doi: 10.1038/nn1618. [DOI] [PubMed] [Google Scholar]
- 63.Miller J, et al. Strategies for aggregating gene expression data: the collapseRows R function. BMC bioinformatics. 2011;12:322–322. doi: 10.1186/1471-2105-12-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hu Z, Mellor J, Wu J, DeLisi C. VisANT: an online visualization and analysis tool for biological interaction data. BMC Bioinformatics. 2004;5:17–17. doi: 10.1186/1471-2105-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.