SUMMARY
In female mice, two forms of X chromosome inactivation (XCI) ensure the selective silencing of female sex chromosomes during mouse embryogenesis. Beginning at the four cell stage, imprinted XCI (iXCI) exclusively silences the paternal X (Xp). Later, around implantation, epiblast cells of the ICM that give rise to the embryo reactivate the Xp and undergo a random form of XCI (rXCI)1,2. Xist, a long non-coding RNA crucial for both forms of XCI, is activated by the ubiquitin ligase Rnf12/RLIM3-5. While Rnf12/RLIM is required for triggering iXCI in mice, its importance for rXCI has been controversial. Here, we show that Rnf12/RLIM levels are downregulated in embryonic cells undergoing rXCI. Using mouse genetics we demonstrate that female cells lacking Rnf12/RLIM from pre-implantation stages onwards display hallmarks of XCI including Xist clouds and H3K27me3 foci and display full embryogenic potential. These results provide evidence that Rnf12/RLIM is dispensable for rXCI, indicating that in mice an Rnf12/RLIM-independent mechanism activates Xist in the embryo proper.
The RING finger ubiquitin ligase Rnf12/RLIM is encoded by the X-linked gene Rnf126,7 and functions as a sex-specific epigenetic regulator of nurturing tissues in female mice. In mammary glands of pregnant and lactating females, Rnf12/RLIM expressed from the Xp serves survival factor for milk-producing alveolar cells8,9. In contrast, female embryos with a maternal deletion of Rnf12 in the germline (Δm) display defective Xist cloud formation and subsequent Xp silencing during iXCI, and die between embryonic day (E) 5.5 and E10.5 due to a lack of extraembryonic trophoblast tissues in the placenta5. Using an embryonic stem cell (ESC) model, evidence has been provided that Rnf12/RLIM can serve as a dose-dependent activator of XCI. Indeed, the overexpression of Rnf12/RLIM induces ectopic Xist clouds in male and female ESCs4, and, in this system, homozygous disruption of Rnf12 results in a failure to initiate Xist transcription during rXCI3,10. Thus, current models of rXCI depict RLIM/Rnf12 as a crucial activator of Xist11,12, although some evidence suggests a less important role in mice5.
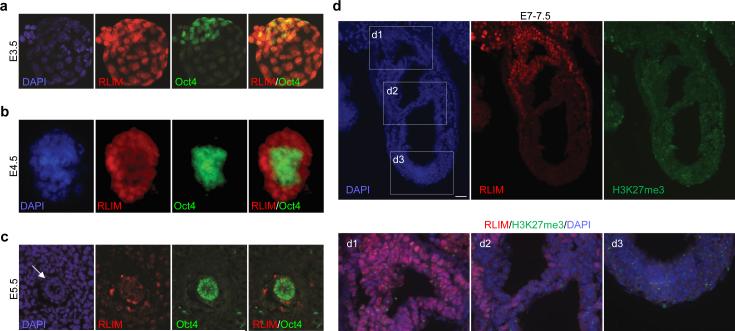
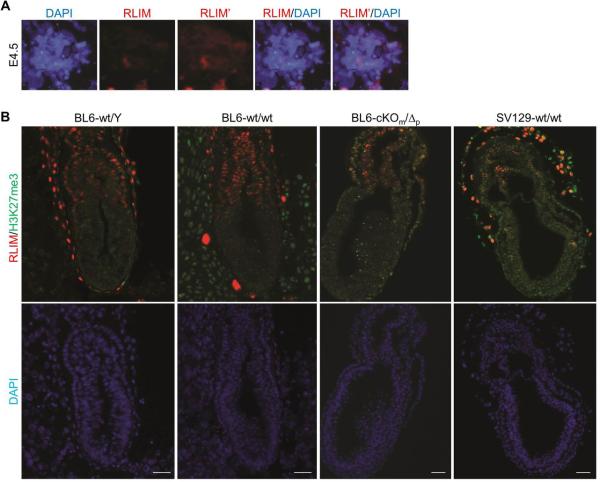
During mouse development RLIM-encoding mRNA is ubiquitously expressed while its protein expression profile appears more restricted6,13. Thus, to investigate in vivo functions of Rnf12/RLIM for rXCI, we first analyzed its protein expression and co-stained early embryos with antibodies against RLIM and the pluripotency factor Oct4 that identifies undifferentiated cells in the inner cell mass (ICM) undergoing rXCI. While RLIM appeared uniformly expressed in cells of E3.5 embryos (Fig. 1a), at E4.5 and E5.5 only low levels were detected in Oct4-positive cells in contrast to extraembryonic cell types (Fig. 1b, c; Extended Data Fig. 1a; not shown). Likewise, at E6.5 and E7-7.5 we detected low levels of RLIM in embryonic epiblast tissues and amnion tissues, whereas levels were high in many extraembryonic tissues, including cells of the ectoplacental cone, extraembryonic ectoderm, and visceral endoderm (Fig. 1d, Extended Data Fig. 1b; not shown). Indeed, due to low levels of RLIM, it was difficult to distinguish wt from embryos lacking RLIM in embryonic tissues by immunostaining (Fig. S1b). Although we did not genotype E5.5 embryos for technical reasons, in all ten wt embryos analyzed we detected no/very low levels of RLIM in Oct4-positive nuclei. Moreover, we observed similarly low RLIM levels in genotyped males and female embryos at E4.5 and 7-7.5 of various genetic backgrounds (Fig. 1; Extended Data Fig. 1; not shown). Since rXCI occurs around E5-E5.5 our data suggest that levels of RLIM/Rnf12 are downregulated in mouse epiblast cells before rXCI.
Fig. 1.
Down-regulation of RLIM levels in early embryonic tissues before and after implantation. a, b) Co-stainings of whole wt/wt pre-implantation embryos at E3.5 (a) and E4.5 (b) (n=5 and 8, respectively) using antibodies against RLIM (red) and Oct4 (green). Embryos were first photographed and then processed for genotyping. c) Co-stainings of sections of wt E5.5 post-implantation embryos (arrow; n=10) within placental tissues. Note lack of RLIM staining (red) in nuclei of Oct4-positive cells (green). d) Co-stainings of sections of wt/wt E7-7.5 embryos (n=5) within placental tissues using antibodies against RLIM (red) and H3K27me3 (green). Boxed regions are shown in lower panels. d1: ectoplacental cone region, d2: amnion region and d3: embryonic epiblast region. Note low RLIM staining intensity in cells in the amnion and epiblast regions. Scale bars = 30μm (c) and 75μm (d).
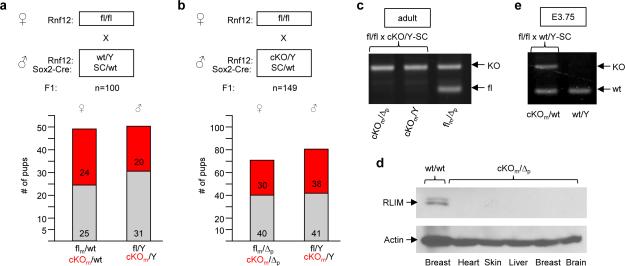
To induce the conditional knockout of Rnf12 (cKO) in female embryos after the occurrence of iXCI but before induction of rXCI, we used Sox2-Cre (SC) transgenic mice 14,15, as Sox2 is specifically expressed in embryonic epiblast cells of E3.5 blastocysts 16, and a paternally transmitted SC transgene is robustly transcribed in ICM cells of preimplantation blastocysts 14. Indeed, from a fl/fl x wt/Y-SC cross, female and male pups with a maternal cKO (cKOm) are born in Mendelian sex ratios, and the percentages of male and female pups carrying a cKOm were similar (Fig. 2a), confirming normal iXCI. Importantly, female cKOm/Δp pups generated via a fl/fl x cKO/Y-SC cross are born at Mendelian ratios (Fig. 2b), and, except for a mammary phenotype, adult cKOm/Δp females appear normal and are fertile (not shown). In genotyping these animals via PCR we detected only the KO allele but no longer the fl allele (Fig. 2c), and RLIM protein in was undetectable in somatic tissues (Fig. 2d). Moreover, in matings using cKO/Y-SC males we never observed the transmission of an unrecombined fl allele (not shown). Consistent with published data14,15 these results indicate complete penetrance of the SC-mediated cKO. Analyzing embryos from fl/fl x wt/Y-SC crosses the recombination of the fl allele was robustly detectable via PCR in blastocysts as early as E3.75 (Fig. 2e), well before initiation of rXCI.
Fig. 2.
Rnf12 is dispensable at post-implantation stages in female mice. a, b) Schematic diagram showing parental genotypes, the total number (n) of born and genotyped F1 offsprings. Number, gender and genotypes of offsprings are indicated. m (maternal) and p (paternal) indicate the origin of the floxed Rnf12 allele. Maternally transmitted cKO alleles are indicated in red. c) High penetrance of the SC-mediated cKO in somatic tissues. PCR analysis of genomic DNA isolated from tail tissue samples using primer combinations that detect floxed and KO Rnf12 alleles. Parental genotypes are indicated. d) Lack of RLIM in somatic tissues of adult cKOm/Δp females in Western blots hybridized with RLIM antibodies. e) Recombination via paternally transmitted Sox2-Cre occurs in pre-implantation embryos. PCR genotyping of E3.75 blastocysts using primers that detect wt and KO Rnf12 alleles. Parental genotypes are indicated.
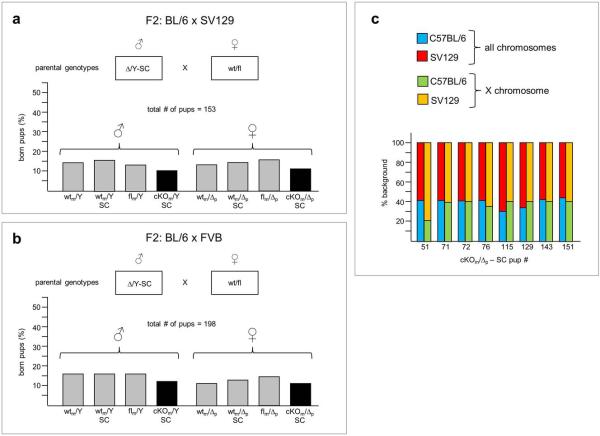
To test whether an essential function of RLIM/Rnf12 in rXCI might be masked by the C57BL/6 mouse background, we generated F1 parents in mixed C57BL/6-SV129 and C57BL/6-FVB F2 hybrid backgrounds suitable for generating SC-mediated cKOm offspring. While numbers of cKOm F2 pups were slightly underrepresented compared to wtm pups, there was no difference between the numbers of male and female cKOm pups (Extended Data Fig. 2a, b), indicating that this effect is XCI-independent. Sequencing analyses of cKOm/Δp pups in mixed C57BL/6-SV129 backgrounds using 156 strain-specific SNPs distributed among all chromosomes revealed no general bias towards the C57BL/6 background (Extended Data Fig. 2c). Together with strain-independent low RLIM/Rnf12 protein levels in the embryonic epiblast (Fig. 1; Extended Data Fig. 1) these data indicate that the genetic background has little or no influence on pup numbers.
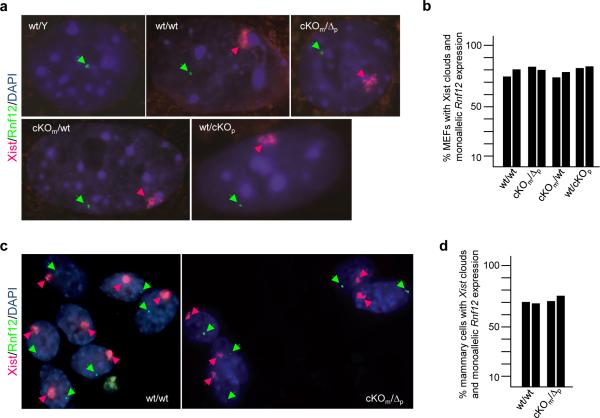
We next examined XCI via RNA fluorescence in situ hybridization (RNA-FISH) in mouse embryonic fibroblasts (MEFs) isolated from E12.5embryos, co-hybridizing with probes specific for Xist 17 and Rnf12 5. As expected, male wt/Y MEFs only displayed Rnf12 transcription foci but no Xist clouds (Fig. 3a). In contrast, the majority of female wt/wt, cKOm/wt, wt/cKOp and cKOm/Δp MEFs displayed monoallelic expression of both genes with Xist painting the Xi and foci of Rnf12 transcription marking the Xa (Figs. 3a, b). XCI in cKOm/Δp MEFs was verified by co-staining cells with antibodies directed against RLIM and H3K27me3, an XCI marker downstream of Xist 18 (Extended Data Fig. 3). We observed a similar pattern for primary mammary epithelial cells isolated from 3 month old adult females (Figs. 3c, d, data not shown). We also examined X skewing in MEFs isolated from female embryos with either a maternal or paternal cKO allele, using transgenic mice containing an X-linked GFP transgene (XGFP) 19 on the wild type X chromosome. MEFs of both cKOm/XGFPp and XGFPm/cKOp embryos displayed approximately 1:1 ratios of GFP-positive:GFP-negative cells (Extended Data Fig. 4). These results indicate normal rXCI in somatic tissues.
Fig. 3.
Detection of Xist clouds in somatic tissues of cKOm/Δp females. RNA FISH analysis using Xist (red) and Rnf12 (green) as probes. a) RNA FISH analysis on mouse embryonic fibroblasts (MEFs) isolated from wt/Y, wt/wt-SC, cKOm/wt, wt/cKOp and cKOm/Δp embryos at E12.5. Representative images are shown. b) Summary graph of RNA FISH experiments on MEFs. Numbers of counted cells from independent biological duplicates are 51/53 (wt/wt-SC); 79/88 (cKOm/Δp); 101/83 (cKOm/wt,); 75/77 (wt/cKOp). c) RNA FISH analysis on primary mammary epithelial cells isolated from virgin cKOm/Δp adult females and wt/wt, as control. d) Summary graph of RNA FISH experiments. Numbers of counted cells from independent biological duplicates are 103/108 (wt/wt-SC); 102/121 (cKOm/Δp).
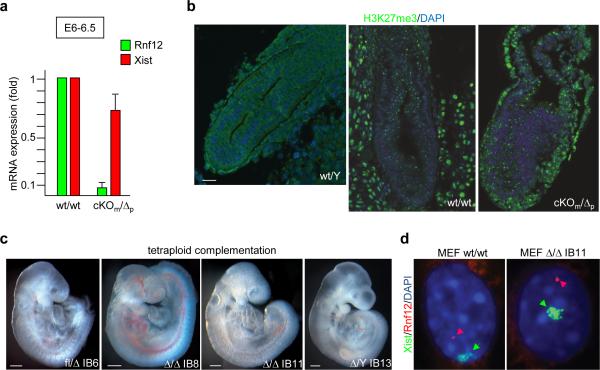
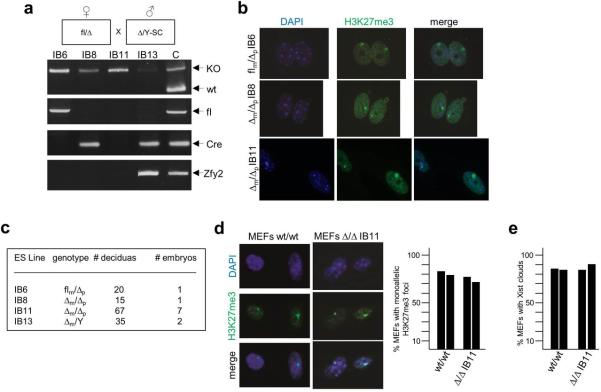
Focusing our analyses on embryonic stages E5.5-E7.5, when rXCI occurs in vivo 1,2, we dissected epiblast tissue of E6-6.5 wt/wt and cKOm/Δp embryos and measured mRNA levels of Rnf12 and Xist by RT-qPCR. While levels of Rnf12 were greatly diminished in cKOm/Δp embryos, those of Xist were only slightly reduced (Fig. 4a). The residual levels of Rnf12 in cKOm/Δp embryos are likely due to minor amounts of contaminating extraembryonic tissue in epiblast dissections. Consistent with the kinetics of developing H3K27me3 marks on the Xi 18, we first detected H3K27me3 foci in wt/wt embryonic tissues at stages E7-E7.5 but not earlier post-implantation embryos (Fig. 4b; not shown). These signals were indistinguishable from those of E7-7.5 cKOm/Δp embryos, indicating similar XCI kinetics in the absence of RLIM. Moreover, cKOm/Δp and wt/wt embryos were similar in size, and in immunostainings using antibodies directed against cleaved caspase 3, no increase in cells undergoing apoptosis were detected in cKOm/Δp embryos (data not shown). To obtain definitive evidence that rXCI does not require the presence of Rnf12/RLIM in cells, we performed tetraploid complementation assays in which tetraploid wt/wt embryos were aggregated with low passage Rnf12 Δ/Δ (IB8 or IB11), and control flm/Δp (IB6) or male Δm/Y ES cells (and IB13), freshly isolated from blastocysts. These ESC lines were generated by a flm/Δp x Δ/Y-SC cross, and are capable of developing H3K27me3 foci upon differentiation in culture indicating XCI (Extended Data Fig. 5a, b). In tetraploid injections the development of embryos derived from Δ/Δ ES cells was indistinguishable from control ESCs at E9-9.5 (Fig. 4c) and no significant differences in complementation efficiencies between these ES cell lines were detected (Fig. S5c). Examining rXCI in MEFS isolated from E10.5 embryos generated from Δ/Δ IB11 cells revealed that more than 75% of Δ/Δ MEFS developed H3K27me3 foci and Xist clouds (Fig. 4d, Extended Data Figs. 5d, e). Combined with the presence of a single Rnf12 transcription focus in cells indicated that ES cells lacking RLIM are capable of undergoing XCI in vivo. Together, our results provide strong evidence that Rnf12/RLIM is dispensable for rXCI in mice. Moreover, because maternal Rnf12 is crucial for iXCI and the Δ/Δ ES line IB11 is SC-negative (Extended Data Fig. 5a) therefore containing maternal and paternal Δ alleles, these data suggest that the process of iXCI is not required in pre-implantation embryos for the epiblast precursor cell lineage that will give rise to the embryo proper.
Fig. 4.
Normal kinetics of XCI in female mouse embryos lacking RLIM/Rnf12. a) RT-qPCR analysis on RNA preparations prepared from epiblast disections of E6-6.5 cKOm/Δp embryos examining expression of Rnf12 and Xist. Values from 3 independent experiments were normalized against actin and are shown relative to wt/wt control. Error bars represent s.d. b) Immunohistochemical analysis of embryos sections at E7-7.5 using antibodies directed against H3K27me3. Sections of wt/Y, wt/wt and cKOm/Δp embryos within placental tissues are shown. Scale bar = 75μm. Images show a representative embryo of at least 3 obtained from independent matings. c) Tetraploid complementation assay using wt/wt blastocysts complemented with low passage Δ/Δ and, as control, with Δ/Y and fl/Δ ES cells. Embryos were harvested at E9-9.5 and image was recorded prior to genotyping. Scale bars = 0.3 mm. d)Δ/Δ ES cells are able to undergo XCI in vivo. RNA FISH experiments on MEFs isolated from E10.5 embryos generated via tetraploid complementation using Δ/Δ ES line IB11 reveals monoallelic expression of both Xist (green) and Rnf12 (red).
Clearly, Rnf12/RLIM can serve as an activator of Xist in some systems 3-5,10. The fact that in cKOm/Δp embryos, Xist levels are only mildly affected and H3K27me3 signals are established with kinetics similar to wt/wt (Figs. 4a, b) combined with the finding that RLIM levels are down-regulated in wt embryos (Fig. 1, Extended Data Fig. 1), suggests that in female mouse embryos, an RLIM-independent mechanism is used to activate/upregulate Xist transcription during rXCI. This is further underscored by our results that E4.5 blastocyst stage female embryos with a maternal and paternal germline Rnf12 KO (Δ/Δ) can develop Xist clouds 5. Because rXCI occurred with low frequency in the Rnf12 KO ESC system 3, it appears likely that two independent mechanisms for Xist activation exist in mouse: one that is independent of RLIM and may be inactive/downregulated in certain ESC systems in culture, and another that is RLIM-dependent. Such a model is consistent with the presence of Xist activators other than RLIM/Rnf12, such as Jpx RNA 20,21, and would explain why RLIM is necessary for XCI in specific ESCs but not in embryos. A similar case, where ESC systems do not accurately reflect the in vivo situation, has been reported for Nanog: while Nanog is required for maintenance of pluripotency in the mouse epiblast 22, it is dispensable for somatic pluripotency in ESCs 23. Moreover, a specific down-regulation of Rnf12/RLIM in mice may also provide an explanation for reported differences of its embryonic functions, as phenotypes in morphogenesis upon Rnf12 mutation have been observed in zebrafish 24 but not in mice 5.
Our results suggest important functions of parental germline imprinting on RLIM/Rnf12 expression and the rXCI process. In addition, differences in Rnf12 mutant alleles may exert some effect on the discrepancies reported as from the Rnf12 KO locus a protein containing 83 amino acids is predicted to be expressed in mice 5 compared to 340 amino acids in the ESC system3,4. Thus, we cannot exclude the possibility that truncated RLIM produced from the respective Rnf12 KO loci may differentially influence the rXCI process. In summary, while there is no doubt about the crucial function of Rnf12 for Xist function during iXCI, we propose that Rnf12 is dispensable for rXCI in mice.
Methods Summary
To investigate functions of RLIM/Rnf12 during rXCI, the mouse conditional Rnf12 KO 5 was targeted to the inner cell mass in preimplantation blastocysts using a paternally transmitted Sox2-Cre transgene 14,15 as driver. Analyses of cells and embryos at pre- and post-implantation stages was carried out as described previously using RNA FISH 25, immunological staining methods 5,26, Western blotting 27 and RT-qPCR 5. Rnf12 Δ/Δ ES cell lines were newly generated 28 and used for tetraploid injections. MEFs were prepared as described 29.
Methods
Mice
Mice used in this study and genotyping have been described Rnf12 fl/fl 5, Sox2-Cre 14, GFPX 19, Sox2-Cre, GFPX, wt SV129, and FVB mice were purchased from the Jackson Laboratories. Rnf12 mice were generally bred in a congenic C57BL/6 background. However, to examine background effects on XCI, the floxed Rnf12 and Sox2-Cre alleles were introduced into C57BL6/SV129 and C57BL6/FVB hybrid backgrounds. The extent of C57BL/6 vs SV129 background of hybrid F2 cKOm/Δp newborn females was determined on genomic DNA using the Genome Scanning Services provided by The Jackson Laboratories. All mice were housed in the animal facility of UMMS according to NIH guidelines, established by the Institute of Animal Care and Usage Committee.
Generation of ES cells and Tetraploid Injection experiments
ES cell lines Rnf12 Δ/Δ (IB8 and IB11), flm/Δp (IB6), and Δ/Y (IB13) were generated as described 28. Briefly, E3.5 blastocyst-stage embryos from a flm/Δp x Δ/Y-Sox2-Cre cross were recovered and cultured onto mitotically-inactivated SNL-76 feeder cells in DMEM plus antibiotics and 15% FBS supplemented with β–mercaptoethanol, non-essential amino acids, and nucleosides. Five days after hatching and expansion, each blastocyst culture was rinsed with phosphate-buffered saline and the inner cell mass collected by manual pipetting and placed into trypsin. Following disaggregation, the cells were plated into an individual well of a 96-well feeder dish and expanded appropriately for genotyping and freezing. ES cells were scored by genotyping and further expanded for mycoplasma testing, ability to undergo XCI and for use in tetraploid injection experiments.
For tetraploid complementation, ES cells were microinjected into 8-cell stage embryos that had undergone tetraploid fusion at the two-cell stage using a CF-150B Electrofusion Instrument (BLS Laboratory, Budapest Hungary). ES cells were harvested from feeder cell layers, rinsed, and placed into DMEM plus 20% fetal bovine serum. Approximately 8-10 ES cells were microinjected into the tetraploid-fused embryos, and the injected embryos then surgically placed into the oviducts of pseudo-pregnant female (E0.5) mice. The recipient female mice were sacrificed nine days later, and embryos were recovered and assessed for proper embryonic development and formation of structures derived from the three germ cell layers prior to genotyping. Mouse embryonic fibroblasts were prepared as described 29.
Preparation of embryos and RT-qPCR
Apart from embryos generated via tetraploid injection, all blastocysts and embryos were generated by naturally mating 8-14-week-old female mice with males. Embryos were collected at stages E3.5 to E12.5 for further processing. Genotyping of embryos was carried out as described 5. Analyses of dissected epiblast tissues of E6.5 embryos via RT-qPCR including primer sequences have previously been described 5. Values obtained from 3 independent biological samples were normalized against actin and Standard Deviation was calculated.
Blastocyst outgrowths and culturing of primary mammary cells
Blastocysts were obtained at E4.5 and cultured for 24h prior to immunocytochemical stainings as described 5. Embryos were genotyped after image recording. Primary mammary epithelial cells were isolated from adult virgin female mice (12 weeks), cultured as described 8 and then processed for RNA FISH or immunostainings.
RNA fluorescence in situ hybridization (RNA FISH)
RNA FISH experiments on MEFs and primary mammary epithelial cells were performed as described previously 5,8. For the synthesis of specific Xist probes, we used plasmids containing mouse Xist exon 1 and 6 that recognize Xist and Tsix 17. For the Rnf12 probe, we used a plasmid containing genomic Rnf12 sequences upstream of the KO site that detects specific Rnf12 mRNAs transcribed from both wild type and KO alleles 5.
Antibodies and Western blots
Primary antibodies used for immunostainings were rabbit RLIM 7, guinea pig RLIM 13, Oct4 (Santa Cruz, sc5279, Abcam, ab27985), H3K27me3 (Abcam, ab6002, Millipore, 07-447), GFP (Rockland, 600-101-215), cleaved Caspase 3 (Cell signaling, 5A1E) and Actin (Sigma, A 4700). Secondary antibodies were Alexa Fluor® 488 Donkey Anti-Rabbit IgG (Invitrogen, A21206), Alexa Fluor® 488 Goat Anti-mouse IgG (Invitrogen, A11029), Alexa Fluor® 546 Goat Anti-Guinea Pig IgG (Invitrogen A11074), Alexa Fluor® 568 Goat Anti-Rabbit IgG (Invitrogen, A11011). Western blots were carried out as reported 27.
Immunohistochemistry
Immunohistochemical staining of embryonic sections was carried out essentially as described 26. After dissection and removal of the ectoplacental cone for genotyping, embryos were gently placed into a small section of uterus, in order to facilitate the embedding protocol and maintain orientation for sectioning. Embryos/uteri were fixed for histology in 4% paraformaldehyde (PFA) for 2 hours at room temp or overnight at 4C. Embryos in uteri were dehydrated in methanol washes; 20 minutes at 25%, 50%, 75% methanol in phosphate buffered saline/0.01% tween20 (PBT), followed by two 100% methanol washes. After overnight incubation in 100% Xylenes, embryos were incubated 2 hours in molten paraffin, prior to embedding and sectioning 26. Sections were mounted and dried on superfrost plus slides. Wax was removed via three 10-minute xylene washes and rehydrated with three 5-minute washes in 100% ethanol, followed successive washes in 90%, 80%, 70% ethanol and finally water (1 min each). Antigen retrieval consisted of boiling for 5 minutes in 0.01M Tris Base pH 10.0 with 0.05% Tween20. After cooling to room temperature slides were washed twice in PBT for 2 minutes prior to blocking with 0.5% milk in PBT (2h at room temperature) in a humidified chamber. Primary antibodies were incubated in 0.05% milk/PBT overnight at 4C. Three 15-minute PBT washes were done before 1-hour secondary antibody incubation (also in 0.05% milk/PBT) in a humid chamber at room temperature. Slides were washed twice in PBT for 15 minutes and once in PBS. Nuclei were stained using DAPI (Roche or Molecular Probes) 1:10,000 in PBS for 2 minutes and rinsed once with PBS. Prolong Gold (Invitrogen) was used to seal and coverslip the slides. Images of sectioned embryos were taken with a Nikon Eclipse TE2000-S inverted fluorescence microscope and QImaging Retiga Exi Fast 1394 camera using NIS-Elements BR Software.
Extended Data
Extended Data Fig. 1.
Strain-independent downregulation of RLIM in the mouse embryonic epiblast. a) Test of specificity of the RLIM antibody (compare with Fig. 1b). Shown is an E4.5 Δ/Y blastocyst outgrowth stained with RLIM (red). Increasing the general signal levels (RLIM’) reveals augmented unspecific staining mainly in the cytoplasm of cells. b) Co-stainings of sections of E7-7.5 embryos within placental tissues using antibodies directed against RLIM (red) and H3K27me3 (green). Representative C57BL/6 and SV129 embryos are shown (n = out of at least 3 that were stained. Scale bars = 75μm.
Extended Data Fig. 2.
Little or no influence of the genetic background on Rnf12/RLIM dispensability during rXCI. a, b) C57BL/6-SV129 (A) and C57BL/6-FVB (B) hybrid F1 parents were generated by crossings of fl/Y (C57BL/6) males with wt/wt (SV129 or FVB) females and wt/Y (SV129 or FVB) males with cKO/Δ-SC (C57BL/6) females. F1 wt/fl females and cKO/Y-SC males were then backcrossed to generate F2 cKOm offsprings. Percentages of offsprings (grouped in female and male) and their genotypes with respect to Rnf12 and SC are indicated in the abscissa and ordinate, respectively, and the total number (n) of born and genotyped F2 pups is shown. Maternally transmitted cKO alleles are indicated in black. c) No discrimination against SV129 in born cKOm/Δp pups with a mixed C57BL/6-SV129 background. Sequencing analyses of genomic DNA isolated from 8 hybrid cKOm/Δp pups using 156 strain-specific SNPs distributed among all chromosomes (blue/red) or 10 SNPs distributed on the X chromosome (green/orange). Note SV129 contribution in born cKOm/Δp pups of up to 70% (total) and 80% (on the X).
Extended Data Fig. 3.
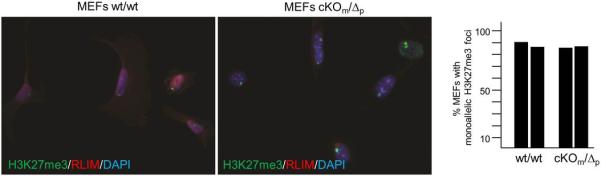
Co-stainings of MEFs isolated from cKOm/Δp and wt/wt embryos using antibodies directed against H3K27me3 (green) and RLIM (red). a) Representative images are shown. b) Summary graph of cells displaying H3K27me3 foci is shown on the right. Numbers of counted cells from independent biological duplicates are 111/112 (wt/wt) and 110/104 (cKOm/Δp).
Extended Data Fig. 4.
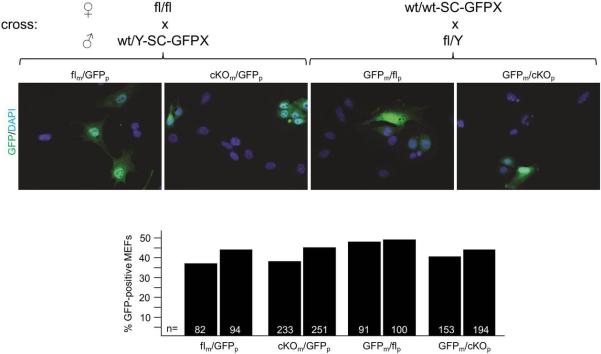
Random XCI in female mice heterozygous for the Sox2-Cre-mediated deletion of Rnf12. Female mice were generated carrying either a paternal or maternal Rnf12 deletion and a GFP transgene on the other X chromosome (XGFP). Mating schemes to generate these females are indicated. Female littermates without the Sox2-Cre transgene were used as controls. Numbers of cells counted from independent biological duplicates are indicated.
Extended Data Fig. 5.
Female ES cells lacking RLIM/Rnf12 are able to undergo rXCI in vivo. a) Parental cross used for the generation of ES cell lines is indicated on top. Newly generated ES cell lines were genotyped via PCR for the presence of wt, floxed or KO Rnf12 alleles, as well as Cre driver (SC) and Y chromosome (Zfy2). b) Newly isolated female ES cells were differentiated for 5 days in culture and stained with the H3K27me3 antibody. c) Summary of the tetraploid complementation assays showing the injected ES cell lines, genotypes, number of deciduas and embryos obtained (see also Fig. 4c). All embryos generated via tetraploid complementation were genotyped for the presence of wt, fl and KO Rnf12 alleles as well as Zfy2. d) Left panel: MEFs isolated from an E10.5 embryo generated via tetraploid injection of Δ/Δ line IB11 ES cells were cultured for 24h prior to staining with antibodies against H3K27me3. Right panel: Summary of H3K27me3 stainings Numbers of counted cells from independent biological duplicates are 60/62 (wt/wt) and 78/75 (cKOm/Δp). e) Summary of RNA FISH experiments on MEFs isolated from embryos generated via tetraploid injection of Δ/Δ line IB11 ES cells (n=109) (see Fig. 4d). MEFs isolated from wt/wt embryos (n=106) served as control.
Acknowledgments
We are grateful to H. Ma for help in the mouse facility, M. Keeler for culturing ES cells, T. Fazzio, N. Lawson and Y. Yoon for advice, and T. Fazzio for helpful discussion and reading of the manuscript. I.B. is a member of the University of Massachusetts DERC (DK32520). This work was supported from NIH grants CA131158 to I.B., CA077735 to S.N.J. and GM053234 to J.B.L.
Footnotes
Author Contributions
J.S. and I.B. conceived and designed the experiments. M.Bo. generated the floxed Rnf12 mice. J.G. and S.N.J. generated the ES cell lines and performed the tetraploid injections. M.By. and J.B.L. carried out RNA FISH experiments and M.C.W., C.M. and J.M. performed IHC on early embryos. All authors analyzed the data. I.B. wrote the manuscript with input from J.S.
The authors declare no competing financial interests.
References
- 1.Augui S, Nora EP, Heard E. Regulation of X-chromosome inactivation by the X-inactivation centre. Nature reviews. Genetics. 2011;12:429–442. doi: 10.1038/nrg2987. doi:10.1038/nrg2987. [DOI] [PubMed] [Google Scholar]
- 2.Lee JT. Gracefully ageing at 50, X-chromosome inactivation becomes a paradigm for RNA and chromatin control. Nature reviews. Molecular cell biology. 2011;12:815–826. doi: 10.1038/nrm3231. doi:10.1038/nrm3231. [DOI] [PubMed] [Google Scholar]
- 3.Barakat TS, et al. RNF12 activates Xist and is essential for X chromosome inactivation. PLoS genetics. 2011;7:e1002001. doi: 10.1371/journal.pgen.1002001. doi:10.1371/journal.pgen.1002001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jonkers I, et al. RNF12 is an X-Encoded dose-dependent activator of X chromosome inactivation. Cell. 2009;139:999–1011. doi: 10.1016/j.cell.2009.10.034. doi:10.1016/j.cell.2009.10.034. [DOI] [PubMed] [Google Scholar]
- 5.Shin J, et al. Maternal Rnf12/RLIM is required for imprinted X-chromosome inactivation in mice. Nature. 2010;467:977–981. doi: 10.1038/nature09457. doi:10.1038/nature09457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bach I, et al. RLIM inhibits functional activity of LIM homeodomain transcription factors via recruitment of the histone deacetylase complex. Nature genetics. 1999;22:394–399. doi: 10.1038/11970. doi:10.1038/11970. [DOI] [PubMed] [Google Scholar]
- 7.Ostendorff HP, et al. Ubiquitination-dependent cofactor exchange on LIM homeodomain transcription factors. Nature. 2002;416:99–103. doi: 10.1038/416099a. doi:10.1038/416099a. [DOI] [PubMed] [Google Scholar]
- 8.Jiao B, et al. Paternal RLIM/Rnf12 is a survival factor for milk-producing alveolar cells. Cell. 2012;149:630–641. doi: 10.1016/j.cell.2012.02.056. doi:10.1016/j.cell.2012.02.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiao B, et al. Functional activity of RLIM/Rnf12 is regulated by phosphorylation-dependent nucleocytoplasmic shuttling. Molecular biology of the cell. 2013;24:3085–3096. doi: 10.1091/mbc.E13-05-0239. doi:10.1091/mbc.E13-05-0239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gontan C, et al. RNF12 initiates X-chromosome inactivation by targeting REX1 for degradation. Nature. 2012;485:386–390. doi: 10.1038/nature11070. doi:10.1038/nature11070. [DOI] [PubMed] [Google Scholar]
- 11.Dupont C, Gribnau J. Different flavors of X-chromosome inactivation in mammals. Current opinion in cell biology. 2013;25:314–321. doi: 10.1016/j.ceb.2013.03.001. doi:10.1016/j.ceb.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 12.Schulz EG, Heard E. Role and control of X chromosome dosage in mammalian development. Current opinion in genetics & development. 2013;23:109–115. doi: 10.1016/j.gde.2013.01.008. doi:10.1016/j.gde.2013.01.008. [DOI] [PubMed] [Google Scholar]
- 13.Ostendorff HP, et al. Dynamic expression of LIM cofactors in the developing mouse neural tube. Developmental dynamics : an official publication of the American Association of Anatomists. 2006;235:786–791. doi: 10.1002/dvdy.20669. doi:10.1002/dvdy.20669. [DOI] [PubMed] [Google Scholar]
- 14.Hayashi S, Lewis P, Pevny L, McMahon AP. Efficient gene modulation in mouse epiblast using a Sox2Cre transgenic mouse strain. Mechanisms of development. 2002;119(Suppl 1):S97–S101. doi: 10.1016/s0925-4773(03)00099-6. [DOI] [PubMed] [Google Scholar]
- 15.Hayashi S, Tenzen T, McMahon AP. Maternal inheritance of Cre activity in a Sox2Cre deleter strain. Genesis. 2003;37:51–53. doi: 10.1002/gene.10225. doi:10.1002/gene.10225. [DOI] [PubMed] [Google Scholar]
- 16.Avilion AA, et al. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes & development. 2003;17:126–140. doi: 10.1101/gad.224503. doi:10.1101/gad.224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Panning B. X inactivation in mouse ES cells: histone modifications and FISH. Methods in enzymology. 2004;376:419–428. doi: 10.1016/S0076-6879(03)76028-5. doi:10.1016/S0076-6879(03)76028-5. [DOI] [PubMed] [Google Scholar]
- 18.Plath K, et al. Role of histone H3 lysine 27 methylation in X inactivation. Science. 2003;300:131–135. doi: 10.1126/science.1084274. doi:10.1126/science.1084274. [DOI] [PubMed] [Google Scholar]
- 19.Hadjantonakis AK, Cox LL, Tam PP, Nagy A. An X-linked GFP transgene reveals unexpected paternal X-chromosome activity in trophoblastic giant cells of the mouse placenta. Genesis. 2001;29:133–140. doi: 10.1002/gene.1016. [DOI] [PubMed] [Google Scholar]
- 20.Tian D, Sun S, Lee JT. The long noncoding RNA, Jpx, is a molecular switch for X chromosome inactivation. Cell. 2010;143:390–403. doi: 10.1016/j.cell.2010.09.049. doi:10.1016/j.cell.2010.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun S, et al. Jpx RNA activates Xist by evicting CTCF. Cell. 2013;153:1537–1551. doi: 10.1016/j.cell.2013.05.028. doi:10.1016/j.cell.2013.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mitsui K, et al. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113:631–642. doi: 10.1016/s0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- 23.Chambers I, et al. Nanog safeguards pluripotency and mediates germline development. Nature. 2007;450:1230–1234. doi: 10.1038/nature06403. doi:10.1038/nature06403. [DOI] [PubMed] [Google Scholar]
- 24.Zhang L, et al. RNF12 controls embryonic stem cell fate and morphogenesis in zebrafish embryos by targeting Smad7 for degradation. Molecular cell. 2012;46:650–661. doi: 10.1016/j.molcel.2012.04.003. doi:10.1016/j.molcel.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 25.Hall LL, et al. An ectopic human XIST gene can induce chromosome inactivation in postdifferentiation human HT-1080 cells. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:8677–8682. doi: 10.1073/pnas.132468999. doi:10.1073/pnas.132468999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Griffith GJ, et al. Yin-yang1 is required in the mammalian oocyte for follicle expansion. Biology of reproduction. 2011;84:654–663. doi: 10.1095/biolreprod.110.087213. doi:10.1095/biolreprod.110.087213. [DOI] [PubMed] [Google Scholar]
- 27.Tursun B, et al. The ubiquitin ligase Rnf6 regulates local LIM kinase 1 levels in axonal growth cones. Genes & development. 2005;19:2307–2319. doi: 10.1101/gad.1340605. doi:10.1101/gad.1340605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robertson EJ. Embryo-derived stem cell lines. In: Roberston EJ, editor. Teratocarcinomas and Embryonic Stem Cells - a Practical Approach. IRL Press Ltd; Oxford, England: 1987. pp. 71–112. Chapter 4. [Google Scholar]
- 29.Nagy A, et al. Manipulating the mouse embryo- a laboratory manual. 3rd ed. CSH press; Cold Spring Harbor, New York: 2003. Isolation and Culture of blastocyst-derived stem cell lines. pp. 359–397. Chapter 8. [Google Scholar]