SUMMARY
The generation of Aβ, the main component of senile plaques in Alzheimer’s disease (AD), is precluded by α-secretase cleavage within the Aβ domain of the amyloid precursor protein (APP). We identified two rare mutations (Q170H and R181G) in the prodomain of the metalloprotease, ADAM10, that co-segregate with late-onset AD (LOAD). Here, we addressed the pathogenicity of these mutations in transgenic mice expressing human ADAM10 in brain. In Tg2576 AD mice, both mutations attenuated α-secretase activity of ADAM10 and shifted APP processing toward β-secretase-mediated cleavage, while enhancing Aβ plaque load and reactive gliosis. We also demonstrated ADAM10 expression potentiates adult hippocampal neurogenesis, which is reduced by the LOAD mutations. Mechanistically, both LOAD mutations impaired the molecular chaperone activity of ADAM10 prodomain. Collectively, these findings suggest that diminished α-secretase activity, owing to LOAD ADAM10 prodomain mutations, leads to AD-related pathology, strongly supporting ADAM10 as a promising therapeutic target for this devastating disease.
Keywords: ADAM10, Alzheimer’s disease, APP, Aβ, prodomain, chaperone
INTRODUCTION
Alzheimer’s disease (AD) is the most common form of dementia in the elderly, with more than five million patients in the US alone. The greatest known risk factor for AD is advanced age with incidence doubling every decade after 60 years of age. The second greatest risk factor for AD is family history. Heritability for AD is estimated to be as high as 80% (Gatz et al., 2006). Early-onset familial AD (EO-FAD) can be caused by fully penetrant mutations in three genes, APP and the two presenilins (PSEN1 and PSEN2). The most well-established late-onset AD (LOAD) gene is apolipoprotein E (APOE), in which the ε4 variant increases risk by 3.7-fold (one copy) to >10-fold (two copies) (Bertram et al., 2010). AD is characterized by the cerebral neuronal loss and deposition of amyloid β-protein (Aβ) in senile plaques. Vast amounts of clinical and biochemical data, in addition to the four established AD genes, support the hypothesis that abnormal processing of APP and the accumulation of its metabolite, Aβ, play key roles in the etiology and pathogenesis of AD (Hardy and Selkoe, 2002).
APP is a type one transmembrane protein that can be processed into a variety of proteolytic fragments. Aβ, a 4 kDa-sized fragment, is generated via serial cleavage of APP by β-secretase (BACE1) at ectodomain and γ-secretase at intra-membranous sites. In contrast, cleavage of APP at the juxtamembrane by α-secretase precludes Aβ generation. α- versus β-secretase cleavage of APP may also lead to different functional consequences. The secreted APP ectodomain generated by α-secretase, sAPPα, has neurotrophic and neuroprotective properties in vivo and in vitro (Mattson et al., 1993; Ring et al., 2007). In contrast, the β-secretase-derived product, sAPPβ, is not as neuroprotective, and upon further processing, can render pro-apoptotic and neurodegenerative effects on neuronal cells (Nikolaev et al., 2009). Several members of the ADAM (a disintegrin and metalloprotease) family proteases, including ADAM9, ADAM10, and ADAM 17 have been reported to possess α-secretase activity at least under in vitro conditions. However, recent evidence indicates that ADAM10 is the major α-secretase responsible for the ectodomain shedding of APP in the mouse brain (Jorissen et al., 2010; Kuhn et al., 2010; Postina et al., 2004).
Nascent ADAM10 is produced as a zymogen and matures into an active protease only after the liberation of its prodomain by furin or proconvertase 7 in the trans-Golgi secretory pathway (Lammich et al., 1999). Following maturation, the majority of the ADAM10 is transported to the plasma membrane, where it acts as a sheddase for cell surface proteins, such as APP. Recent studies have shown that ADAM10, itself, is also subject to shedding in cultured cells (Parkin and Harris, 2009; Tousseyn et al., 2009). Numerous studies of metalloprotease family proteins suggest that the prodomain of ADAM10 plays critical roles in the maturation of the enzyme. The primary function of the prodomain is to keep the ADAM10 zymogen in an inactive status via direct interaction with the catalytic site (Moss et al., 2007), thereby preventing autocatalysis during biosynthesis. Another function of the prodomain is to serve as an intra-molecular chaperone, facilitating the proper folding of other domains of the protein (Cao et al., 2000; Shinde et al., 1997). While recombinant ADAM10 lacking the prodomain upon initial synthesis is catalytically inactive, co-expression of the prodomain in trans has been shown to restore the α-secretase activity of the enzyme (Anders et al., 2001).
In addition to function as a major α-secretase for APP, ADAM10 plays an essential role in embryonic neurogenesis and brain development (Jorissen et al., 2010; Pan and Rubin, 1997) . Both ADAM10 knockout (KO) and conditional KO mice are lethal in early developmental stages (Hartmann et al., 2002; Jorissen et al., 2010), potentially due to the lack of ectodomain shedding of Notch and its ligands by ADAM10. Recent studies have shown that neurogenesis in adult hippocampus plays an essential role in learning and memory (Zhao et al., 2008). Whether ADAM10 plays a role in adult hippocampal neurogenesis has yet to be addressed. To assess the candidacy of ADAM10 as a LOAD susceptibility gene, we previously genotyped 30 SNPs that span the gene to test for genetic association with AD. These analyses, followed by targeted re-sequencing of ADAM10, ultimately led to the identification of two non-synonymous mutations, Q170H and R181G in seven LOAD families (Kim et al., 2009). Over-expression of both mutant forms of ADAM10 in cell cultures significantly increased Aβ levels as compared to wild-type (WT) ADAM10 controls (Kim et al., 2009). Here, we set out to demonstrate the pathogenicity of the two ADAM10 mutations in transgenic mouse models. For this purpose, we generated transgenic mice that express either WT or LOAD mutant forms of human ADAM10, and compared the differences in APP processing among the multiple lines of WT and mutant transgenic mice expressing similar levels of ADAM10 in brain. The impact of the mutations on Aβ accumulation and plaque deposition was further assessed by crossing these ADAM10 transgenic mice with Tg2576 mice, a well-characterized AD mouse model that overexpresses human APP Swedish mutation (APPswe) and is known to cause an APP ectodomain cleavage shift that favors the β- over the α-site. We also assessed the effect of the LOAD mutations on adult hippocampal neurogenesis, and finally, we explored the underlying molecular mechanisms by which the LOAD mutations in the ADAM10 prodomain attenuate α-secretase activity.
RESULTS
The ADAM10 prodomain mutations attenuate ectodomain shedding of ADAM10
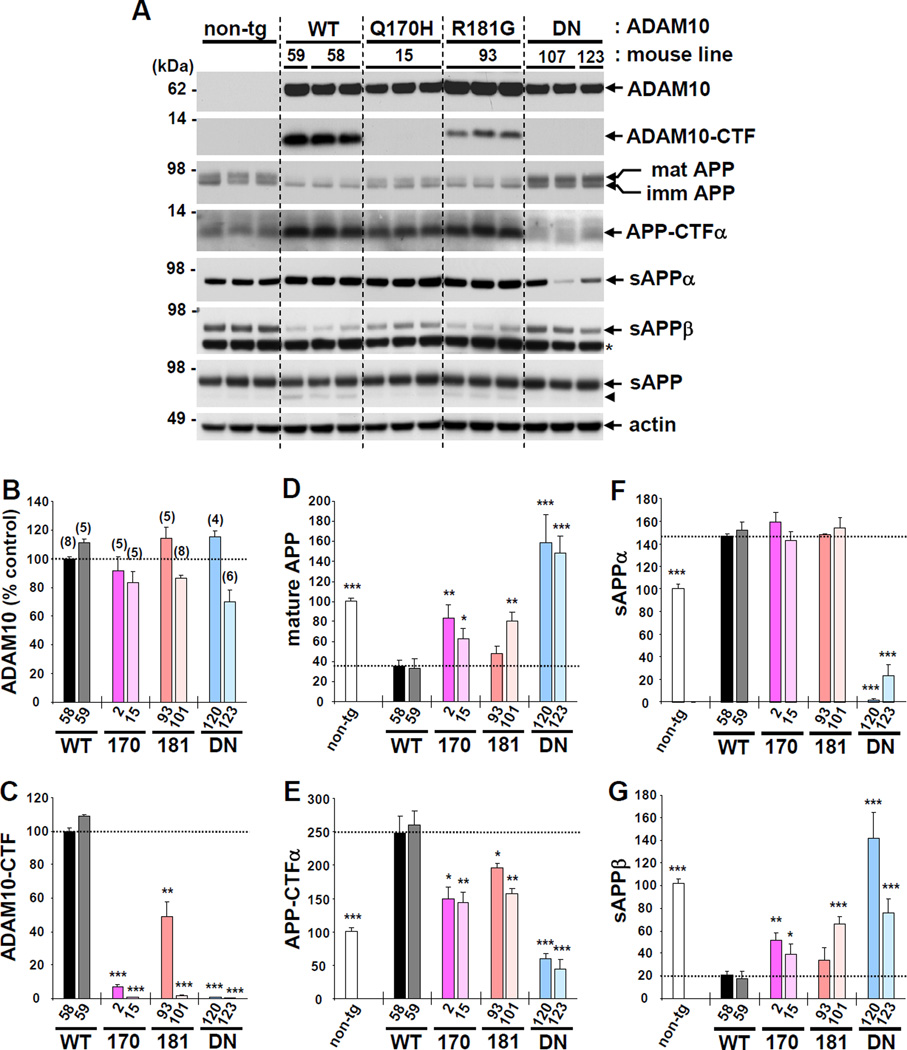
To test the in vivo effects of the two LOAD-associated ADAM10 mutations on α-secretase activity, we generated transgenic mice overexpressing human WT ADAM10, ADAM10 harboring the LOAD-associated mutations Q170H, or R181G, and an artificial dominant-negative (DN) mutation, E384A. All transgenes were driven by the mouse prion protein promoter (MoPrP) and tagged with hemagglutinin (HA) at the C-terminus of the protein (Figures S1A and S1B). For each ADAM10 genotype, we obtained three WT, three Q170H, eight R181G and three DN F1 transgenic mice, which were bred with non-transgenic littermates to maintain mouse lines. The brains from F2 and F3 progenies of each line were analyzed for ADAM10 expression and APP processing. Western blot analysis of 12-week-old mouse brains revealed that a WT transgenic line (WT-58) expressed ~2.5 fold higher level of mature ADAM10 in brain than non-transgenic control (Figure S1C). In addition to the pro and mature forms of ADAM10, high levels of ADAM10-CTF (~10 kDa) were detected in the membrane fraction (Figure 1A–1C and S1D). Previous studies have shown that these ADAM10-CTFs are generated by ectodomain shedding of ADAM10 mature forms (Parkin and Harris, 2009; Tousseyn et al., 2009). Interestingly, as compared to the ADAM10-WT transgenic mice, the levels of ADAM10-CTF were significantly reduced in mice expressing either of the two LOAD mutations, and were undetectable in mice expressing DN mutation (Figure 1A and 1C). This pattern of reduced ADAM10-CTF was consistently observed in all the ADAM10 LOAD and DN mutant lines as compared to WT transgenic lines (two WT, three Q170H, six R181G and three DN ADAM10 transgenic lines) and in the three single F1 mice (one WT and two R181G, which failed to produce progeny). The decrease in ADAM10-CTF generation was also detected in primary cortical neurons derived from the LOAD mutant mice (Figure S1E). These results indicate that both the LOAD and DN ADAM10 mutations decreased ectodomain shedding of the metalloprotease. Consistent with our findings, previous studies have shown that artificial mutations at the prodomain cleavage or catalytic sites, which block enzyme activity of the corresponding ADAM proteases (ADAM 13 and ADAM 19), also result in the prevention of its own ectodomain shedding at their cysteine-rich domains (Gaultier et al., 2002; Kang et al., 2002).
Figure 1. Effect of ADAM10 LOAD prodomain mutations on ADAM10 ectodomain shedding and APP processing in brain.
(A) Representative western blot images for ADAM10 expression and APP processing in 12-week-old mouse brains. Anti-HA and anti-APP C-terminal antibody (A8717) were used to detect full-length and CTF forms of ADAM10 and APP, respectively in RIPA buffer-soluble total brain lysates. TBS-soluble fraction was used to detect sAPP (22C11, anti-APP N-terminus), sAPPα and sAPPβ. Arrow head indicates C-terminal truncated sAPP. *, non-specific band. (B-G) Densitometric quantification of western blot results of two mouse lines each from the four different genotypes. Number in parenthesis denotes the number of mouse analyzed. Relative value of 100 is given for WT-58 mouse line for mature ADAM10 (B), ADAM10-CTF (C). Relative value of 100 is given for non-transgenic mouse for the analysis of APP processing product, mature APP (D), APP-CTFα (E), sAPPα (F), and sAPPβ (G). Values are mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001 versus WT-58 mouse line (t-test). See also Figure S1.
In vitro studies have previously suggested that ectodomain shedding of ADAM10 depends on the activity of ADAM family proteins, including ADAM9 and ADAM17 (Parkin and Harris, 2009; Tousseyn et al., 2009). Conversely, ADAM10 activity was found to be essential for the ADAM9 function (Taylor et al., 2009). To further investigate the ectodomain shedding of ADAM10, we assessed the expression and processing of endogenous ADAM9 in each genotype of ADAM10 transgenic mice. Levels of pro- and mature ADAM9, and ADAM9-CTF were unaffected by the expression of WT or mutant forms of ADAM10 (Figure S1F). In addition, overexpression of ADAM10-DN did not interfere with the generation of ADAM10-CTF from either transgenic or endogenous ADAM10 proteins (Figure S1G). Taken together, these results suggest that the decrease in ADAM10-CTF levels observed in mice expressing LOAD mutations is due to the reduced auto-proteolytic activity of the mutant ADAM10. A reduced ratio of pro- vs. mature ADAM10 in the Q170H mutant lines suggested that the mutation might also affect the liberation of its prodomain (Figure S1H). However, the marked variability of the ratio of pro- vs. mature ADAM10 in mice expressing the other ADAM10 mutations, R181G and DN, indicates that ADAM10 prodomain cleavage does not depend on the enzyme activity of the metalloprotease.
Expression of ADAM10 LOAD prodomain mutations attenuates non-amyloidogenic processing of endogenous mouse APP
To examine the effect of the LOAD ADAM10 mutations on endogenous APP processing, we selected two mouse lines from each of the four genotypes, expressing comparable levels of mature ADAM10 (Figure 1A and 1B), and analyzed the levels of APP and its cleavage products in the brain. Compared to non-transgenic control, ADAM10 WT transgenic mice exhibited lower levels of mature APP and sAPPβ, and higher levels of APP-CTFα and sAPPα (Figure 1A and 1D–1G). Mature APP is cleaved primarily by α-secretase at the cell surface into APP-CTFα and sAPPα, and accumulating evidence supports that APP is cleaved competitively by α- and β-secretase in neural cells (Colombo et al., 2012; Lee et al., 2005; Postina et al., 2004). Thus, overexpression of ADAM10-WT increased α-secretase cleavage while decreasing β-secretase cleavage of endogenous APP. In contrast, expression of ADAM10-DN had an opposite effect on APP processing. Compared to the WT transgenic controls, both Q170H and R181G mutant transgenic mice exhibited significant attenuation of APP processing, i.e. less of an increase in APP-CTFα levels and less of a decrease in mature APP and sAPPβ levels. Interestingly, however, the level of sAPPα in the two LOAD mutant mice was not reduced when compared to that of ADAM10-WT mice (Figure 1A and 1F). This is likely due to the enhanced degradation of sAPPα in the brains expressing ADAM10-WT over the other mutant forms. In support of this hypothesis, we observed higher levels of ~70 kDa sAPP degradation products in the brains of ADAM10 WT as compared to the two LOAD mutant mice (Figure 1A). Moreover, compared to the non-transgenic control, the amount of total sAPP was barely different in ADAM10 transgenic mice expressing either WT or any mutant forms. These data suggest a presence of a potential compensatory mechanism that maintains the total level of sAPP in the brain regardless of ADAM10 expression levels. We also measured the levels of secreted APP (sAPPα, sAPPβ and total sAPP) in cerebrospinal fluid (CSF) and found similar patterns depending on the ADAM10 genotypes (Figure S2A). Levels of endogenous Aβ40 and Aβ42 (measured by ELISA) were significantly lower in ADAM10-WT mice compared to the non-transgenic controls, while the levels were dramatically higher in ADAM10-DN mice (Figure S2B). For mice expressing either Q170H or R181G mutations, a trend was observed in which endogenous Aβ levels were increased compared to ADAM10-WT mice. Taken together, these data reveal that both ADAM10 LOAD prodomain mutations attenuate, but do not entirely abolish α-secretase activity of ADAM10 on APP.
Next, we asked whether the prodomain mutations affect the cleavage of other ADAM10 substrates besides APP (Pruessmeyer and Ludwig, 2009). We examined the processing of candidate ADAM10 substrates in the brain including APLP2, Notch-1 and N-cadherin. The processing of APLP2 showed very similar patterns to those of APP in that ADAM10-WT overexpression resulted in the increased cleavage of mature APLP2 while this was attenuated in mice expressing either of the two LOAD mutations (Figure S2C). Interestingly, ~57 kDa-sized C-terminus-truncated soluble APLP2, analogous to the soluble APP cleavage product, was detected more abundantly in ADAM10-WT than the LOAD mutant mouse brains. In contrast, western blot analysis of adult brains and primary neurons derived from ADAM10 transgenic mice showed ADAM10 overexpression barely affected N-cadherin level or N-cadherin-CTF generation (Figure S2D). In contrast to embryonic brains, most of Notch-1 protein in adult brains is cleaved and present as extracellular truncated forms in the membrane. We found that neither WT nor mutant ADAM10 overexpression changed the level of full-length or truncated forms of Notch-1 (Figure S2E). Together, these data suggest that the processing of APP family proteins is particularly vulnerable to loss-of-function mutations in ADAM10.
LOAD ADAM10 mutations increase amyloidogenic processing of APP and Aβ levels in Tg2576/ADAM10 double transgenic mouse brains
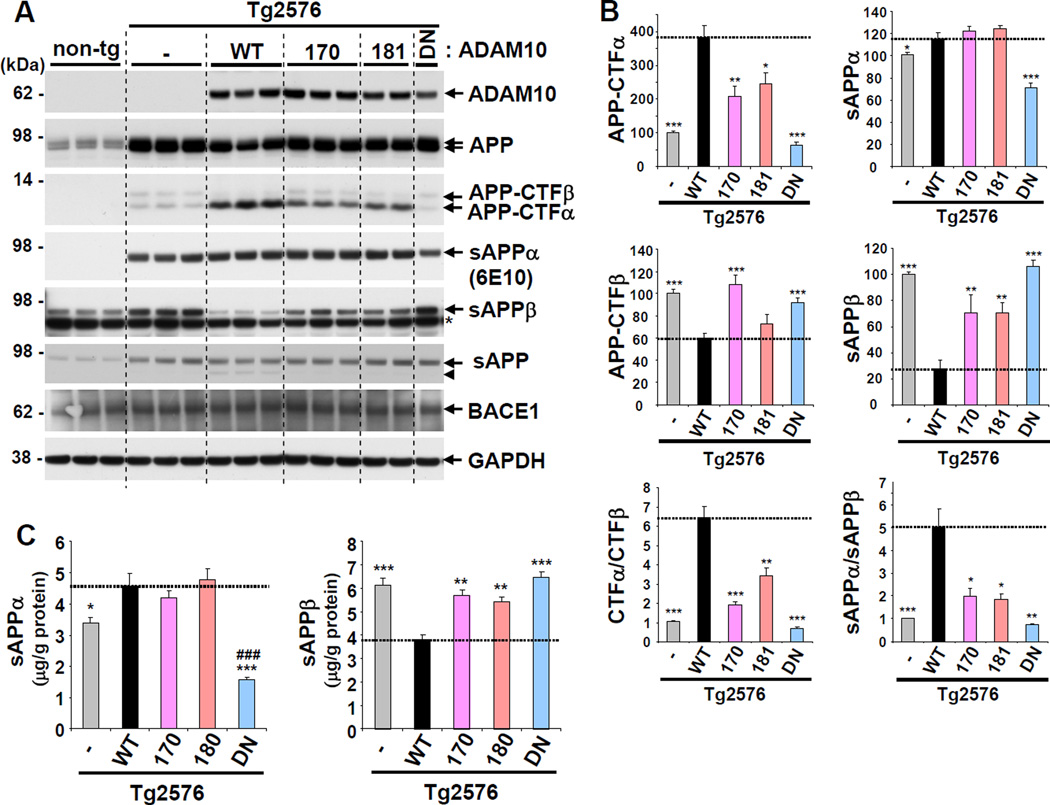
To evaluate the effects of the two LOAD ADAM10 mutations on AD pathogenesis, we crossed transgenic mice expressing WT or mutant forms of ADAM10 with Tg2576 AD mice, and assessed APP processing, Aβ levels, and amyloid plaque load. As in the ADAM10 transgenic mice, the prion protein promoter was utilized in the Tg2576 mice to express human APPswe. Western blot analysis of mouse brain lysates revealed that APP expression was several-fold higher in Tg2576 mice than in non-transgenic control. However, ADAM10 expression in Tg2576/ADAM10 double transgenic mice was maintained at similar levels to those in parental ADAM10 single transgenic mice (Figures 2A and S3A). Attenuation of α-secretase activity by the LOAD mutations was clearly evident in the mutant Tg2576/ADAM10 double transgenic mice. Compared to the double transgenic mice expressing ADAM10-WT (Tg2576/WT), the decrease of mature APP and increase of APP-CTFα were significantly reduced in 3 month-old brains expressing either Q170H (Tg2576/Q170H) or R181G (Tg2576/R181G) ADAM10 mutations (Figure 2A and 2B). Moreover, the levels of sAPPβ and APP-CTFβ were elevated by both LOAD mutations in comparison to Tg2576/WT mice. Quantitative analysis of brain sAPPα and sAPPβ by ELISA revealed similar patterns as compared to the results from western blots (Figure 2C). The ratios of both APP-CTFα:APP-CTFβ and sAPPα:sAPPβ indicate that both the LOAD mutations shifted more than 50% of the APP processing from the α-secretase to β-secretase pathway. While the ratio of α- vs. β-cleavage was still higher in Tg2576/Q170H and Tg2576/R181G mice than Tg2576, the DN mutation modestly shifted APP processing toward β-cleavage. However, the increase in β-secretase cleavage of APP by mutant ADAM10 expression was not caused by altered BACE1 expression (Figure 2A). Notably, as observed in the ADAM10 single transgenic mice, no differences were found in sAPPα levels among Tg2576/WT, Tg2576/Q170H and Tg2576/R181G double transgenic mice (Figure 2A and 2B). Instead, C-terminal truncated sAPP were detected more abundantly in mice expressing the WT form (Figures 2A, S3B and Figure S3C). Given the robust increase of APP-CTFα and concurrent decrease of APP-CTFβ by ADAM10-WT expression, the C-terminus truncated sAPP are likely generated from sAPPα.
Figure 2. Impact of ADAM10 LOAD prodomain mutations on non-amyloidogenic processing of APP in Tg2576/ADAM10 double transgenic mice.
(A) APP processing in the brains of three-month-old Tg2576/ADAM10 double transgenic mice. Endogenous mouse sAPPα was barely detected by the 6E10 antibody, the epitope for which is based on human APP. Arrow head indicates C-terminal truncated sAPP. *, non-specific band. (B) Densitometric analysis of APP-CTFα, APP-CTFβ, sAPPα and sAPPβ, and ratios of CTFα:CTFβ and sAPPα:sAPPβ. Analyzed mouse number: Tg2576 (n=6), Tg2576/WT (5), Tg2576/170 (5), Tg2576/181 (4) Tg2576/DN (5). *p < 0.05, **p < 0.01, ***p < 0.001 versus Tg2576/ WT (t-test). (C) ELISA for sAPPα and sAPPβ levels in TBS-soluble brain lysates. Analyzed mouse number: Tg2576 (n=10), Tg2576/WT (5), Tg2576/170 (5), Tg2576/181 (5) Tg2576/DN (5). *p < 0.05, **p < 0.01, ***p < 0.001 versus Tg2576/ WT; ###p < 0.001 versus Tg2576 (one-way ANOVA and Tukey’s test). See also Figure S2.
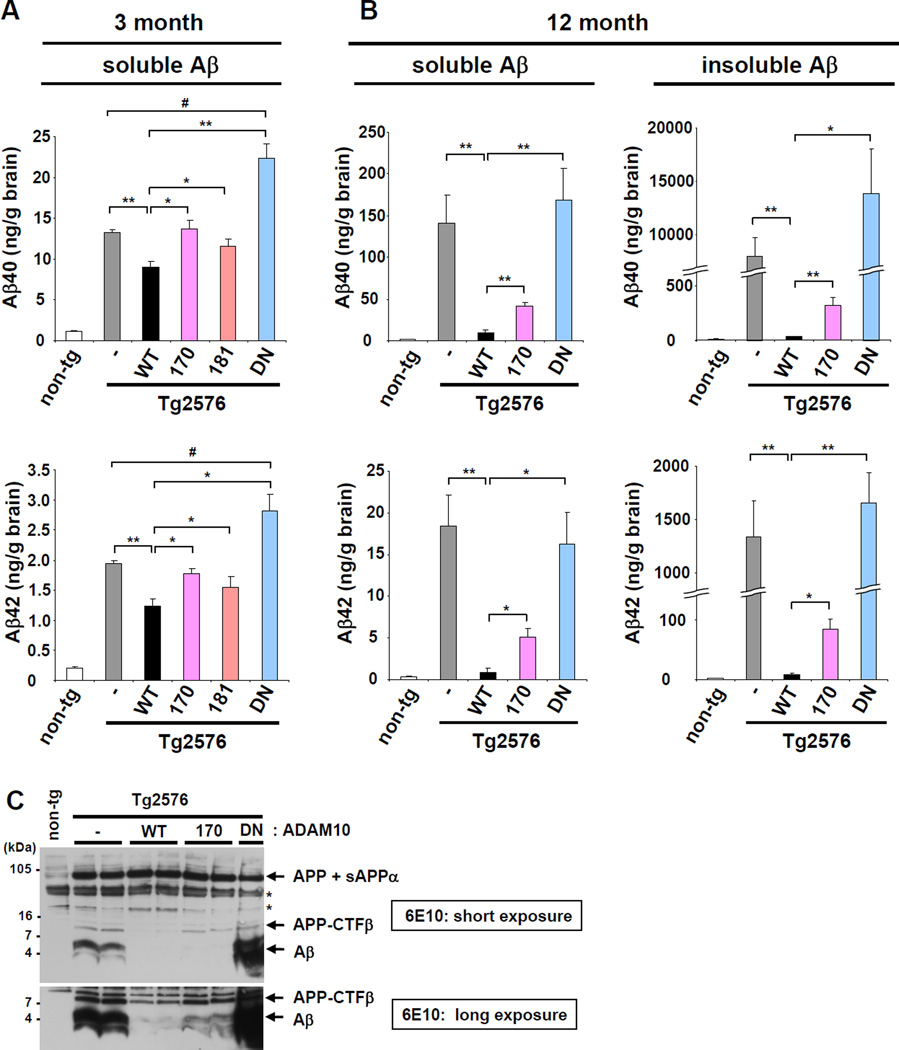
Next, we examined Aβ levels in the Tg2576/ADAM10 double transgenic mice. In the brains of 3-month-old Tg2576/WT mice, both TBS-soluble Aβ40 and Aβ42 levels were reduced ~35% compared to Tg2576 control (Figure 3A). However, the ADAM10-mediated decrease in Aβ40 and Aβ42 was significantly attenuated in both Tg2576/Q170H and Tg2576/R181G mice. Tg2576/DN mice exhibited higher Aβ levels than Tg2576 alone, which indicates decreased non-amyloidogenic processing of APP in the presence of the DN form. In 3 month-old brains, TBS-insoluble Aβ was barely detectable in Tg2576 or Tg2576/ADAM10 mice (data not shown). As the deposition of insoluble Aβ occurs at 7–8 month in the brains of Tg2576 mice (Kawarabayashi et al., 2001), the total Aβ levels at 12 months were hundreds-fold higher than those at 3 months (Figure 3B). Correspondingly, in 12-month-old mice, the reduction of Aβ levels in Tg2576/WT was dramatically amplified in both TBS-soluble (>90%) and insoluble (>99%) Aβ fractions (Figure 3B). Compared to the Tg2576/WT, there was much less of a decrease in Aβ levels in Tg2576/Q170H mice. However, Tg2576/Q170H mice also showed a robust decrease in brain Aβ levels as compared to Tg2576. This decrease was most likely due to partial, but not complete loss of α-secretase activity by the LOAD mutation. Combined with endogenous ADAM10, transgenic mice overexpressing the LOAD mutation still had overall higher α-secretase activity than ADAM10 non-transgenic control (Figures 1 and 2). The decrease in Aβ levels by ADAM10-WT or -Q170H overexpression was also confirmed by western blot analysis of 12 month-old AD mouse brain (Figure 3C). The Tg2576/R181G mice were examined only in small numbers (n<3) at 12 month- old age, due to the similarities noted in the patterns of APP processing and Aβ generation with Tg2576/Q170H mice at 3 months of age (Figure 2A and 2B). These mice also exhibited an increase in Aβ levels as compared to the Tg2576/WT (data not shown). Other APP cleavage fragments remained at similar levels between 3- and 12-months old (Figures 2A and S3D). Thus, the enhanced impact of ADAM10 on cerebral Aβ amount at 12 month, compared to 3 month, might be the result of accumulated effect of APP processing change (amyloidogenic vs. non-amyloidogenic) and Aβ deposition over the several months time period.
Figure 3. Increased Aβ40 and Aβ42 levels in AD mice expressing the ADAM10 LOAD prodomain mutations.
TBS buffer-soluble and insoluble Aβ40 and Aβ42 levels were measured for 3- (A) and12-month-old (B) Tg2576 single or Tg2576/ADAM10 double transgenic mice by sandwich ELISA. Analyzed mouse number −3 months old groups: Tg2576 (n= 4 Male and 4 Female), Tg2576/WT (3M, 2F), Tg2576/170 (2M, 3F), Tg2576/181 (3M, 2F), Tg2576/DN (2M, 3F); −12 months old groups: Tg2576 (4M, 4F), Tg2576/WT (3M, 2F), Tg2576/170 (3M, 3F), Tg2576/DN (3M, 3F). *p < 0.05, **p < 0.01 versus Tg2576/WT; #p < 0.05 versus Tg2576 (t-test). Breaks are inserted to adjust different scales in graphs for Aβ level in 12-month-old brains. (C) WB analysis of detergent (RIPA)-soluble total brain lysates from 12-month-old mice using 16% Tricine gel with 6E10 antibody. The human APP, sAPPα, APP-CTFβ, and Aβ were detected in the immunoblot by 6E10. *, nonspecific band. WB analysis for TBS-buffer insoluble, but formic acid-soluble, fraction produced similar results (data not shown). See also Figure S3.
We did not detect changes in the ratio of Aβ40 and Aβ42 in brains expressing either WT or mutant forms of ADAM10 (Figure 3A, 3B and S2B). This suggests that the change in ADAM10 activity did not affect the preference of γ-secretase cleavage site in APP-CTFβ. Most early onset familial AD mutations in APP, PSEN1, and PSEN2 increase the Aβ42/Aβ40 ratio, with a few exceptions (e.g. APP Swedish and APP duplication mutations) which increase total Aβ levels. Our results indicate that the ADAM10 LOAD mutations elevate both Aβ40 and Aβ42 levels in the brain, without affecting the Aβ42/Aββ40 ratio.
ADAM10 activity regulates Aβ plaque load and morphology, and reactive gliosis in AD mouse brain
Immunofluorescence staining of 12-month-old brain sections with anti-Aβ antibodies revealed notable amounts of Aβ plaques in Tg2576 mice (Figure 4A and 4B). However, virtually no plaque was detected in Tg2576/WT brain sections. Compared to Tg2576, reduced Aβ plaque counts in Tg2576/Q170H mouse brains indicate the prodomain mutation diminished, but did not abolish the α-secretase (anti-amyloidogenic) activity of ADAM10. In contrast, both Aβ levels and plaque counts were elevated in Tg2576/DN (compared to Tg2576 mice), suggesting that ADAM10-DN expression down-regulates α-secretase activity of endogenous mouse ADAM10. In 18–20 month-old brains, Aβ plaque loads were markedly increased in Tg2576 and Tg2576/DN, whereas small numbers of only tiny plaques were observed in Tg2576/WT mice (Figure 4A and 4C). The differences in plaque load among the four groups remained consistent up to the 18–20 month-old time point (Figure 4B and 4C).
Figure 4. Effect of WT and mutant forms of ADAM10 expression on Aβ plaque load in AD mice.
(A) Aβ plaques were detected by anti-Aβ antibodies (red) in 12 month-old (first row, cortex) and 20 month-old (second and fourth rows, cortex/hippocampus) in Tg2576 single or Tg2576/ADAM10 double transgenic mice. Dense neuritic plaques were stained by Thioflavin S (green, third and fourth rows). Large and small arrows indicate neuritic and diffuse plaques, respectively. DAPI (blue) was used for nuclei counter staining. Bar= 100 µm. (B) Counts of total Aβ plaque numbers in 12 month-old mice (6–12 sections/mouse). Red: plaque size > 50 µm, blue: < 50 µm. Analyzed mouse number: Tg2576 (n=4M, 4F), Tg2576/WT (3M, 2F), Tg2576/170 (3M, 3F), Tg2576/DN (3M, 3F). *p < 0.05, **p < 0.01 (t -test). (C) Aβ plaque load in the cortex and hippocampus of 18–20 month-old Tg2576/ADAM10 mice. Analyzed mouse number: Tg2576 (n=5M, 5F), Tg2576/WT (2M, 2F), Tg2576/170 (2M, 3F), Tg2576/DN (3M, 3F). *p < 0.05, **p < 0.01, ***p < 0.001 (t-test). (D) Brain sections of Tg2576/170 (upper) and Tg2576/DN (lower) mice were immunolabeled with 3D6 Aβ antibody followed by staining with Thioflavin S. (E) Counts of Thioflavin S-stained plaque in 18–20 month old mice. ***p < 0.001 (t-test).
Interestingly, in addition to this dramatic difference in Aβ plaque load, the morphology of plaque was also affected by ADAM10 activity. Tg2576 mice at this age have both neuritic and diffuse plaques (Figure 4A) (Kawarabayashi et al., 2001). Co-staining with Thioflavin S and anti-Aβ antibody (3D6) showed that core-containing neuritic plaques (Thioflavin S-positive) were more predominant in Tg2576/DN than in Tg2576 brains (Figure 4A and 4E). On the contrary, most Aβ aggregates in Tg2576/Q170H or Tg2576/WT mice were diffuse plaques (Thioflavin S-negative). Overall, the ratio of neuritic vs. diffuse plaque is inversely correlated with ADAM10 activity in Tg2576/ADAM10 mice. This relationship is more evident when plaque morphology is compared between 12 month-old Tg2576/DN and 20 month-old Tg2576/Q170H mice, which harbor comparable plaque loads (Figure 4D). Collectively, these findings suggest that ADAM10 activity affects not only the plaque load, but plaque morphology as well.
In conjunction with senile neuritic plaques, reactive gliosis is one of the key pathological markers in AD brains (Simpson et al., 2010). Compared to non-transgenic control, 18–20 month-old Tg2576 mice showed robust increases in microglia and astrocytes activation. The number of Iba1-positive microglia was dramatically increased in association with Congo red-stained neuritic plaques (Figure 5A and 5B), and GFAP-expressing hypertrophic astrocytes became widespread in the cortex (Figure 5C and 5D). Glial cell activation was enhanced in Tg2576/DN mouse brains, but barely detectable in Tg2576/WT mice. Tg2576/Q170H showed higher levels of gliosis than Tg2576/WT. Overall, the levels of gliosis in Tg2576/ADAM10 brains were correlated well with those of neuritic plaques. In ADAM10 single transgenic mice, we found no notable change in glial cell activation (Figure S4A and S4B). Taken together, these results suggest that non-amyloidogenic processing of APP by ADAM10 reduces reactive gliosis in AD pathogenesis.
Figure 5. Effect of ADAM10 on gliosis in AD mouse brains.
(A) Photomicrographs of 20 month-old Tg2576/ADAM10 mouse cortex immunolabeled with Iba1 (brown), a microglial marker, and Congo red (orange). Arrows indicate Congo-red stained neuritic plaques. High magnification (bottom) of Iba1 staining showed the high density of activated microglia around neuritic plaques. Bar= 50 µm. (B) Percentile of Iba1-immunoreactive area in cortex. Analyzed mouse number for Iba1 and GFAP immunostaining: Tg2576 (n=5M, 5F), Tg2576/WT (2M, 2F), Tg2576/170 (2M, 3F), Tg2576/DN (3M, 3F). *p < 0.05, **p < 0.01, ***p < 0.001 (t-test). (C) Photomicrographs of Tg2576/ADAM10 mouse cortex stained with an astrocyte marker, GFAP. Arrows indicate Congo-red stained neuritic plaques. High magnification (bottom) of GFAP staining revealed hypertrophy of astrocyte around neuritic plaques. Bar= 100 µm. (D) Percentile of GFAP-immunoreactive area in cortex. *p < 0.05, **p < 0.01, ***p < 0.001 (t -test). See also Figure S4.
Decreased ADAM10 activity significantly reduces adult hippocampal neurogenesis
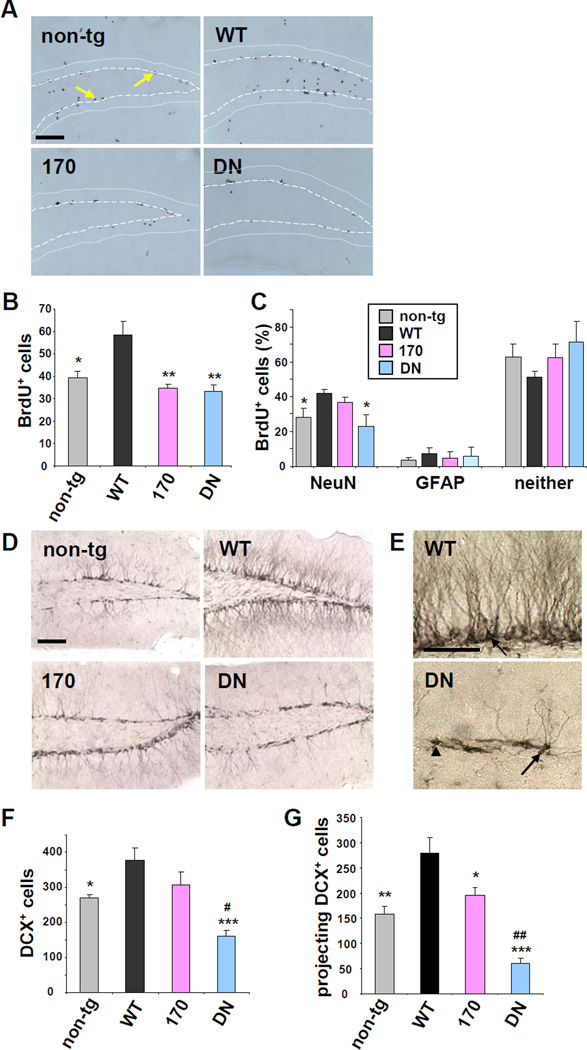
To further examine the physiological consequences of reduced ADAM10 activity caused by the two LOAD mutations, we tested for effects of WT versus mutant ADAM10 on adult hippocampal neurogenesis. Recent studies have revealed that application of sAPPα increases the proliferation of neural progenitor cells (NPCs) in vitro and in vivo (Caille et al., 2004; Demars et al., 2011) and impairments in hippocampal neurogenesis have been linked to cognitive deficits in AD (Zhao et al., 2008). Proliferation of NPCs was analyzed by the incorporation of BrdU, a nucleotide analog, into newly born cells in the hippocampus. One day after intraperitoneal injection of BrdU, the number of BrdU-positive cells was significantly increased (48%) in the dendate gyrus of 4 month-old ADAM10-WT transgenic mice as compared to non-transgenic controls (Figure 6A and 6B). In contrast, in the Q170H and DN ADAM10 mice, the proliferation of NPCs remained at levels similar to those of the control non-transgenic mice. The level of NPC proliferation in ADAM10-R181G mice was similar compared to that of Q170H mice (data not shown). The effect of ADAM10 on differentiation of the NPCs was measured at 2 weeks after BrdU injection. Triple labeling of brain sections with anti-BrdU antibody combined with antibodies for neuronal (NeuN) and glial lineages (GFAP) revealed a 49% increase in NPC differentiation into neurons in the ADAM10-WT mice as compared to non-transgenic controls (Figures 6C and S5A). The fraction of BrdU+:NeuN+ cells was modestly increased in ADAM10-Q170H mice, however, the degree of increase was lower than that of ADAM10-WT mice. Only a minor fraction (<10%) of BrdU+ cells were co-labeled with GFAP, and no significant differences were detected among the different mouse groups for the glial differentiation. Together, these results indicate that ADAM10 activity regulates the proliferation of NPCs and their differentiation into neurons in adult hippocampus. Moreover, the ADAM10 Q170H and DN mutations inhibited the proliferation and differentiation of NPCs in adult brains.
Figure 6. ADAM10 regulates proliferation and differentiation of neural progenitor cells (NPCs) and dendritic development of immature neurons in hippocampus.
(A) Immunohistochemical staining for BrdU in the dentate gyrus of non-transgenic, WT, Q170H, and DN ADAM10 transgenic mice. Area between dots and dashed lines is granular cell layer in hippocampus. Black dots (arrows) indicate BrdU-positive NPCs. Bar= 100 µm. (B) Counts of BrdU-positive proliferating cells in dentate gyrus at 1 day after BrdU injection. Analyzed mouse number: n=6–7. *p < 0.05, **p < 0.01 versus ADAM10-WT (one-way ANOVA and Tukey’s test). (C) Percentile of BrdU-positive cells co-stained with either NeuN or GFAP, or neither of the two cell markers in each mouse group at 2-weeks after BrdU injection. n=6–7. *p < 0.05 versus ADAM10-WT (one-way ANOVA and Tukey’s test). (D) Photographs of dentate gyrus stained with doublecortin (DCX). Most cell bodies of the DCX-positive immature neurons were located in the subgranular cell layer. (E) High magnification of DCX-stained neurons in dentate gyrus. Arrow, dendrite-projecting; arrow head, tangential immature neurons. (F) Counts of DCX-positive cells in dentate gyrus. n=6–7. (G) Counts of dendrite-projecting DCX-positive cells. *p < 0.05, **p < 0.01, ***p < 0.001 versus ADAM10-WT; #p < 0.05, ##p < 0.01 versus non-transgenic control (one- way ANOVA and Tukey’s test). See also Figure S5.
The impact of ADAM10 expression was further examined for potential effects on the dendritic development of the new-born neurons in hippocampus. Immunostaining of brain sections with doublecortin (DCX), a marker for immature neurons, revealed that most of DCX-positive neurons in subgranular cell layer of non-transgenic, ADAM10-WT, and -Q170H transgenic mice project dendrites into or beyond the granular cell layer (projecting DCX+ neurons; Figures 6D and 6E). However, in ADAM10-DN mice, the length of dendrite in DCX-positive neuron was markedly decreased and many of the immature neurons were found without dendrites in the subgranular cell layer (tangential DCX+ neurons). Both total DCX-positive and projecting DCX-positive immature neurons are significantly elevated in ADAM10-WT, but reduced in ADAM10-DN mice (Figure 6F and 6G). In the LOAD ADAM10-Q170H mice, which exhibit attenuated α-secretase activity, the number of dendrite projecting immature neurons was significantly lower than that of ADAM10-WT.
To test for effects of APP processing on the regulation of hippocampal neurogenesis, levels of sAPPα and sAPPβ were measured in the TBS-soluble fraction of hippocampal lysates. Similar to the whole brain lysates, WB analysis revealed an elevated ratio of sAPPα/sAPPβ in the hippocampus of ADAM10-WT mice compared to that of non-transgenic or the LOAD mutant ADAM10 mice (Figure S5B). However, no difference in the expression level and pattern of Notch-1, a major ADAM10 substrate contributing to embryonic neurogenesis, was observed in the adult hippocampus among non-transgenic, WT and mutant ADAM10 transgenic mice (Figure S5C and S5D). Taken together, these findings support that ADAM10 activity is tightly linked to the regulation of hippocampal neurogenesis, and that the LOAD and DN mutations impair the neurogenic activity of ADAM10 in adult brain.
The LOAD mutations do not alter the cellular trafficking of ADAM10, but impair the molecular chaperone function of the prodomain
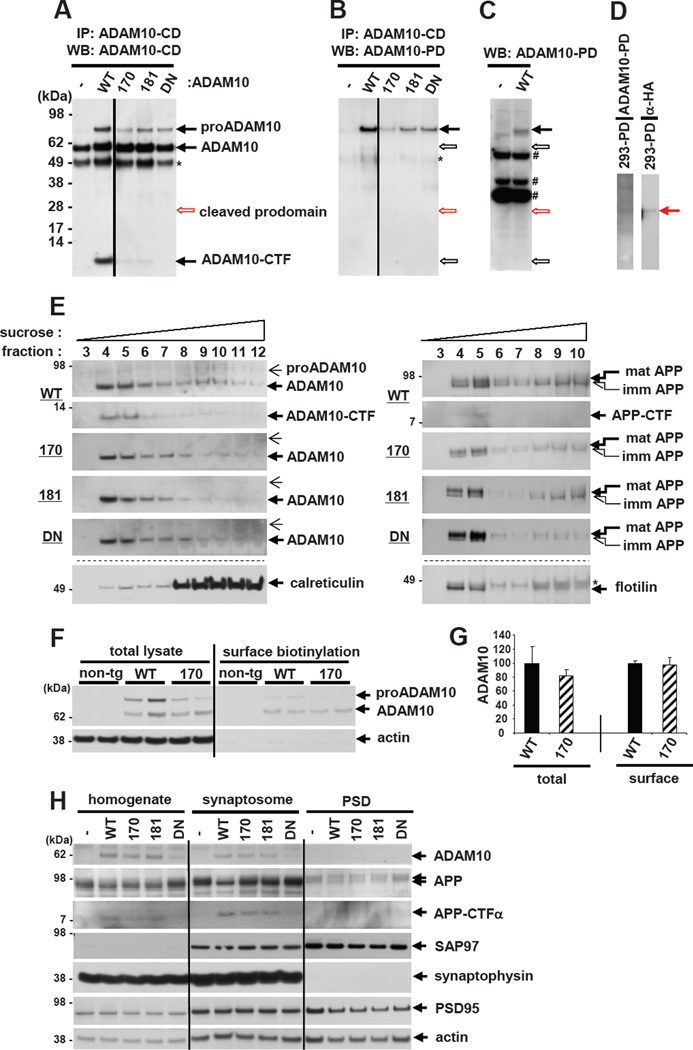
The prodomain in the ADAM family proteins serves to ensure correct protein folding and maintain the enzyme in a latent form (Anders et al., 2001). While the majority of cleaved prodomain is readily degraded after liberation, under certain conditions, the released prodomain remains and binds to the mature enzyme (Moss et al., 2007), suggesting that it may have a biological function following the cleavage. To determine the underlying mechanism responsible for the attenuated α-secretase activity by the prodomain mutations, we first attempted to measure the level of liberated ADAM10 prodomain in mouse brains. However, immunoprecipitation followed by WB analysis using a combination of antibodies targeting the ADAM10 prodomain and cytosolic domain revealed that the cleaved ADAM10 prodomain is undetectable (Figures 7A–7D). These results suggest that the liberated ADAM10 prodomain is rapidly degraded following cleavage in brain, and that the impact of the liberated prodomain on ADAM10 enzyme activity is likely to be minimal. This also implies that the ADAM10 LOAD mutations may affect the prodomain function prior to its liberation.
Figure 7. Effects of ADAM10 LOAD prodomain mutations on cellular trafficking of ADAM10.
(A,B) Total brain lysates from each genotype of ADAM10 transgenic mice were immunoprecipitated with an antibody targeting ADAM10 cytoplasmic domain (CD), followed by WB with anti-ADAM10-CD (A) or anti-ADAM10-prodomain (PD) (B) antibodies. Similar results with (B) were obtained by immunoprecipitation with anti-ADAM10-PD antibody followed by WB with either anti-ADAM10-CD or anti-ADAM10-PD antibodies. Empty arrows indicate the position of corresponding ADAM10 or ADAM10 cleavage products. *IgG heavy chain. Straight vertical lines are inserted to mark two separate parts in the same blot. (C,D) WB analysis of brain lysates from ADAM10-WT or non-transgenic mice with anti-ADAM10 prodomain (~25 kDa) antibody (C) and HEK293 cell lysates from HA-tagged ADAM10 prodomain transfected cultures with anti-ADAM10-prodomain or anti-HA antibodies (D). # non-specific band. (E) Mouse brain homogenates were separated by ultracentrifugation in discontinuous sucrose gradients. Mature forms of ADAM10 (large arrow, left panel), APP (large arrow, right panel) and their CTFs are enriched in lipid raft fractions (fraction 4 and 5). Both proADAM10 (small arrow, left panel) and immature APP (small arrow, right panel) are enriched in ER. Calreticullin and flotillin were used as cellular markers for ER and lipid raft, respectively. * non-specific band. (F) Primary cortical neurons (DIV13) derived from non-transgenic, ADAM10-WT or -Q170H transgenic mouse embryos were biotinylated to measure ADAM10 levels in plasma membrane (right panel). Actin is as a negative control for surface biotinylation. (G) Densitometric quantification of total and surface ADAM10 levels in primary neurons. n=3. (H) Synaptosome and post synaptic density (PSD) were isolated by sequential centrifugations of mouse brain homogenates. 15 µg of proteins were loaded for brain homogenates or synaptosome fraction and 5 µg for PSD fraction. Synaptophysin and PSD95 were probed as pre- and post-synaptic markers, respectively.
Next, we asked whether the prodomain mutations interfere with the cellular trafficking of ADAM10. Previously, it has been shown that the introduction of an artificial mutation (Leu73Pro) in the ADAM12 prodomain results in the complete retention of the enzyme in ER (Cao et al., 2002). Sucrose gradient fractionation of brain lysates revealed that the mature forms of ADAM10 and APP are enriched in lipid raft fractions, where ectodomain shedding of ADAM10 itself and α-secretase cleavage of APP mainly occur (Figure 7E). However, neither the prodomain LOAD mutations nor DN mutations altered the cellular trafficking of ADAM10 and APP to ER and lipid rafts. Surface biotinylation of primary cortical neurons derived from ADAM10 transgenic mouse embryos also indicated that the Q170H prodomain mutation did not change the trafficking of the enzyme to the plasma membrane (Figure 7F and 7G), a major location responsible for APP cleavage by α-secretase. We also examined whether the prodomain mutations affect ADAM10 trafficking to the synapse, in which the activity of the metalloprotease is regulated by synapse-associated protein-97 (SAP-97) (Marcello et al., 2007). As shown in Figure 7H, compared to the whole brain homogenates, the levels of both APP and APP-CTFα were elevated in synaptosomal fraction and LOAD mutations decreased APP-CTFα levels. However, ADAM10 levels at the synapse were not changed by the prodomain mutations. Together, these results suggest that the attenuated α-secretase cleavage of APP by the LOAD mutations is not caused by altered ADAM10 trafficking.
We next tested whether the LOAD ADAM10 mutations affects the prodomain chaperone function. Previous studies have shown that addition of a prodomain in trans to a prodomain-deleted enzyme enables the completion of protein folding and restores the enzyme activity for many types of proteases, including ADAM10 (Anders et al., 2001; Cao et al., 2000). Thus, we asked whether the ADAM10 prodomain in trans affects the activity of prodomain-deleted ADAM10 (ADAM10Δpro). We also tested if the two prodomain mutations affect the chaperone activity of ADAM10 prodomain as compared to WT. To this end, neuroblastoma H4 cells stably overexpressing APP were transfected with either ADAM10Δpro, alone, or in combination with ADAM10 prodomain constructs expressing either WT, Q170H or R181G forms of ADAM10. Forty-eight hours after transfection, the cells were harvested and analyzed for APP and ADAM10 processing. While WT prodomain in trans dramatically increased the activity of prodomain-deleted ADAM10 (as evidenced by increased levels of APP-CTFα), the ADAM10 prodomain harboring either Q170H or R181G mutations failed to restore the enzyme activity of ADAM10 (Figure 8A and 8B). These results suggest that the LOAD mutations impair the chaperone function of ADAM10 prodomain.
Figure 8. Molecular chaperone function of ADAM10 prodomain is compromised by ADAM10 LOAD prodomain mutations.
(A) H4 cells were transfected with prodomain deletion cDNA construct (ADAM10 ΔPro)
alone or in combination with ADAM10 prodomain cDNA (WT, Q170H, or R181G form). GFP and empty pCDNA3.1 (mock) vectors were used to adjust total transfected DNA amounts (800 ng). Cells were harvested and analyzed for APP (A8717) and ADAM10 expression (anti-HA) 48 hr after transfection. (B) Quantification of APP-CTFα level in transfected H4 cells. Results are combined from 3 independent transfection experiments. n=5. *p < 0.05, **p < 0.01, ***p < 0.001 versus ADAM10ΔPro + WT prodomain group (one-way ANOVA and Tukey’s test). (C) Schematic model for attenuated α-secretase activity by the ADAM10 prodomain mutations. Green and yellow represent prodomain and the rest of ADAM10 domain, respectively. Red depicts LOAD mutation within the prodomain.
DISCUSSION
Although up-regulation of α-secretase activity has been proposed previously as a potential therapeutic strategy for AD by precluding the generation of Aβ (Donmez et al., 2010; Fahrenholz and Postina, 2006), no genetic variants supporting this premise had been reported until our recent finding of the two ADAM10 prodomain mutations, Q170H and R181G, in several LOAD families (Kim et al., 2009). To investigate the potential pathogenic effects of these LOAD mutations in vivo, in the current study, we generated transgenic mice expressing human ADAM10: WT, each prodomain LOAD mutation, and an artificial dominant-negative mutation. The impact of the mutations on AD pathology was assessed by crossing these ADAM10 mice with the Tg2576 AD mouse model.
Several novel insights have emerged from these efforts (Figure 8C). First, we found that the two LOAD mutations diminished α-secretase activity of ADAM10 and shifted APP processing toward β-secretase-mediated cleavage. The ectodomain shedding of ADAM10 itself was also dramatically attenuated by the prodomain mutations. Second, we showed that the ADAM10 mutations elevate Aβ levels, plaque load and reactive gliosis in Tg2576 AD mice. Plaque morphology (diffuse vs. neuritic) was also affected by ADAM10 activity. Third, we demonstrated that ADAM10 plays critical roles in adult hippocampal neurogenesis and the LOAD and DN mutations impair this activity. Finally, with regard to the pathogenic mechanism, we showed that both LOAD ADAM10 mutations impair molecular chaperone function of the ADAM10 prodomain. Beyond the four established AD genes (APP, PSEN1, PSEN2, and APOE), this is the first report documenting additional AD-associated pathogenic gene mutations in vivo. The evidence presented here support that ADAM10 is a bona fide AD susceptibility gene that can harbor rare mutations causing LOAD.
ADAM10 transgenic mice and ADAM10 processing
To test for the in vivo effects of the LOAD ADAM10 prodomain mutations, we compared transgenic mice expressing either WT or mutant forms of human ADAM10 in the brain. To reduce potential bias stemming from individual mouse line dependent variables (e.g. different expression level), we analyzed all F1 mice and their progeny from different ADAM10 genotypes (minimum 3 mouse lines per genotype). Two lines from each genotype, all of which possess ADAM10 expression level comparable to a control line (WT-58), were selected for further analysis of APP processing. Immunoblot analysis of the transgenic mouse brains confirmed that the pattern of ADAM10 and APP processing are dependent on the ADAM10 genotype, not individual mouse lines (Figure 1).
To address the impact of the ADAM10 mutations on brain pathology, we chose Tg2576 as an AD mouse model. These mice overexpress human APP harboring the Swedish mutation at the β-secretase cleavage site, under the expression control of prion protein promoter sequence, in B6/SJL hybrid background. Thus, to avoid the influence on phenotype expression in Tg2576/ADAM10 double transgenic mice, particularly with regard to Aβ generation and deposition, we generated ADAM10 transgenic mice by employing the same expression promoter and genetic background as in the Tg2576 mice.
To account for potential incomplete penetrance of the prodomain mutations, akin to effects on AD pathology reported for transgenic mice expressing the ε4 risk allele of APOE, we included an artificial DN mutation of ADAM10 (E384A), as a positive control to simulate a fully penetrant mutation. The E384A DN mutation was originally reported in Drosophila at the zinc-binding catalytic site of the enzyme, making the protease as an inactive form (Pan and Rubin, 1997). However, in vivo overexpression of the defective protease resulted in dominant negative signaling pattern related to the enzyme activity, probably by competing with endogenous ADAM10 for its substrates and auxiliary factors essential for the enzyme activity (Lammich et al., 1999; Pan and Rubin, 1997). In this study, we also observed dramatic effects of the artificial dominant negative mutant form of ADAM10 on APP processing, Aβ accumulation and hippocampal neurogenesis. Meanwhile, the two LOAD mutations in the ADAM10 prodomain exerted significant, but less dramatic effects on ADAM10 activity in brain. Although complete ablation of endogenous mouse ADAM10 results in lethal developmental defects in brain (Hartmann et al., 2002; Jorissen et al., 2010), partial reduction of the endogenous metalloprotease activity by the overexpression of ADAM10-DN form did not produce any notable abnormality in brain morphology up to 24 months old (data not shown). All the ADAM10 transgenic mouse lines used in this study maintain endogenous mouse ADAM10. Therefore, the impact of different ADAM10 genotypes (WT, Q170H, R181G, and DN) on substrates (e.g. APP) processing would likely be affected by the presence of the wild-type form of endogenous mouse ADAM10. However, we deemed it necessary to retain the endogenous ADAM10 to prevent potential developmental defects that might have occurred in its absence.
The first evidence that the two ADAM10 LOAD mutations attenuate enzyme activity was derived from our observation of reduced ectodomain shedding of ADAM10, itself. In agreement with the recent findings from in vitro studies of ADAM10 and other ADAM protease processing (Gaultier et al., 2002; Kang et al., 2002; Taylor et al., 2009; Tousseyn et al., 2009), the complete absence of ADAM10-CTF in all the DN mouse lines (Figure 1 and S1) suggest that ADAM10 activity regulates its own ectodomain shedding at the cysteine-rich domain. Structurally, the C-shaped conformation of the metalloprotease family proteins enables the protease domain to come in close proximity to the cysteine-rich domain (Takeda et al., 2006). This unique structural characteristic also supports the possibility of an auto-catalytic mechanism in ADAM10 ectodomain shedding. The ADAM10 LOAD mutations in the prodomain may interfere with the ectodomain shedding by decreasing either the enzyme activity (protease domain) or substrate accessibility (cysteine-rich domain) of ADAM10.
Competition between α- and β- secretases on APP processing and Aβ generation
In the ADAM10 transgenic mice, the prodomain and catalytic-site mutations decrease α-site cleavage of APP (less APP-CTFα). Notably, reduced α-secretase activity was accompanied by an increase in β-secretase processing of APP (higher levels of APP-CTFβ, sAPPβ, and Aβ). Concordantly, a missense mutation, which was recently found in an early onset dementia family precisely at the APP α-secretase cleavage site (K16N), led to a decrease in APP-CTFα coupled with increases in levels of APP-CTFβ and Aβ (Kaden et al., 2012). Inverse effects have been reported in mice with altered β-secretase gene expression. BACE1 KO mice produced elevated APP-CTFα (Luo et al., 2001), and BACE1 transgenic mice revealed reduced APP-CTFα with increased APP-CTFβ and sAPPβ (Lee et al., 2005) . Although several cell-based studies produced inconsistent results with regard to these alternative cleavages (Colombo et al., 2012), studies using genetically modified mice have consistently shown the presence of competition between α- and β-secretases on APP processing in the brain (Lee et al., 2005; Luo et al., 2001; Postina et al., 2004).
Remarkably, while ADAM10-WT overexpression in Tg2576 mice decreased ~35% of Aβ levels at 3 months old, the impact was dramatically magnified at 12 months, at which point, Aβ40 and Aβ42 levels were decreased by more than 99% in the ADAM10-WT mice (Figure 3). LOAD mutant forms of ADAM10, which possess attenuated α-secretase activity (60–70% of WT), did not produce notable decreases in Aβ levels in 3 month-old double transgenic mice. However, Aβ levels were dramatically down-regulated (~95%) in the brains of 12 month-old double transgenics, as compared to Tg2576 control. The robust decrease in Aβ plaque load was maintained up to 18–20 months old (Figure 4). This profound impact on plaque load by ADAM10 in older brains is consistent with a previous report that employed transgenic mice overexpressing bovine ADAM10 and human APP London mutation (Postina et al., 2004). In addition to the potential direct cleavage of Aβ by ADAM10 (Lammich et al., 1999), this increased effect in older mice might be the result of accumulated production and deposition of excess Aβ in brains. As the half life of Aβ in brains is only ~ 2 hr (Cirrito et al., 2003), changes in Aβ generation rate would greatly affect the accumulation and deposition of Aβ over several months in the brains of APPswe overexpressing Tg2576 mice. Aging-enhanced Aβ accumulation was also reported in Bace1+/+ mice compared to Bace1+/− mice (McConlogue et al., 2007). Collectively, these findings suggest that modest modulation of α- or β-secretase activity for extended time period can have a profound impact on Aβ pathology in aged brain.
Beyond the level of plaque load, ADAM10 activity also affected the morphology of Aβ plaque. While Tg2576/DN double transgenic mice had more neuritic plaques with compact cores (versus Tg2576), most plaques found in the double transgenic mice overexpressing WT or Q170H displayed irregular diffuse morphology. Neuritic plaques are known to be more tightly associated with AD pathogenesis than diffuse plaque. For example, fibrillar-core containing neuritic plaques are predominant in AD brains, whereas diffuse plaques are more frequent in non-demented elderly (Selkoe, 2001). Furthermore, neuritic, but not diffuse plaques are associated with pathological phenotypes of the disease, including dystrophic neurites, activated microglia and reactive astrocytes (Figure 5). While further studies are warranted to delineate the mechanism underlying the observed differences in plaque morphology, our findings suggest that enhanced ADAM10 activity may lessen Aβ pathology not only by decreasing plaque load, but also by affecting plaque morphology.
Currently, it is unclear how the two secreted APP ectodomains, sAPPα and sAPPβ, engender different effects - neurotrophic vs. neurodegenerative - on neurons. Interestingly, a 35 kDa fragment derived from sAPPβ has been demonstrated to bind the cell surface death receptor DR6 and trigger axonal degeneration in neurons (Nikolaev et al., 2009). In addition to the extra 16 amino acids at the C-terminus of sAPPα, the difference in where these ectodomains are generated, cell surface for sAPPα and endosome for sAPPβ, may play a key role in determining their distinct biological functions. At the cell surface, APP can be present as a dimer in cis or trans formation (Wang and Ha, 2004). Structural and imaging studies have shown that liberated sAPPα can bind as a ligand to APP at cell surface and disrupt APP dimer complex to exert its neuroprotective effect (Gralle et al., 2009; Wang and Ha, 2004). Therefore, it is interesting to speculate that ADAM10 cleavage of APP may shift the complex formation toward neurotrophic APP-sAPPα (or its cleavage derivatives) versus APP-APP dimerization at the cell surface.
Hippocampal neurogenesis and molecular chaperone function of ADAM10 prodomain
Accumulating evidence shows that elevated hippocampal neurogenesis improves memory function (Zhao et al., 2008), and that down-regulation of hippocampal neurogenesis is associated with cognitive impairments in AD (Choi et al., 2008). Notably, adult neurogenesis has been reported to be affected by all three early-onset familial AD genes, APP, PSEN1 and PSEN2, and by Aβ in AD mouse models (Mu and Gage, 2011), suggesting its tight link to the etiology and pathogenesis of the disease. With regard to ADAM10, Bell and colleagues reported that moderate expression of bovine ADAM10 or exogenous infusion of sAPPα in the cortex increases presynaptic bouton density, indicative of neurotrophic effects of ADAM10 on cortical synaptogenesis (Bell et al., 2008). Additionally, lateral ventricular injection of sAPPα increased the proliferation of NPCs in the subventricular zone, another neurogenic niche in mouse brain (Caille et al., 2004).
All ADAM family proteins contain an N-terminal prodomain, which acts to chaperone and ensure the proper folding of this family of metalloproteases. Our results show that the cleaved ADAM10 prodomain appears to be quickly degraded in the brain and that cellular trafficking of ADAM10 is not affected by the prodomain mutations. Interestingly, while the prodomain may be degraded, it may still have the ability to affect the mature enzyme via its intramolecular chaperone function. This phenomenon, dubbed ‘protein memory’, was reported in mutant subtilysin, a serine protease harboring a mutation in its prodomain (Shinde et al., 1997). The point mutations yielded mature subtilysin that had a different structure and activity via ‘structural imprinting’ during protein folding (Shinde et al., 1997). Improperly chaperoned, mis-folded proteases can be restructured and become active by ectopic expression of WT prodomain (Cao et al., 2000). In our mammalian cell-based studies, WT but not mutant ADAM10 prodomain rescued the α-secretase activity of inactive ADAM10 expressed from a prodomain-deleted cDNA construct. This indicates impairment of the chaperone function of the prodomain by the LOAD mutations (Figure 8C). In further support of this conclusion, secondary structure predictions showed that the only α-helix in the prodomain can be terminated by the R181G mutation (McGuffin et al., 2000). To date, a few pathogenic amino acid substitutions, which are not present in the mature forms, have been associated with diseases. But to our knowledge, the two LOAD ADAM10 mutations are the first to be associated with the etiology of any disease by impairing the intramolecular chaperone function of a prodomain.
Increased ADAM10 α-secretase activity could potentially be achieved by multiple different mechanisms, including the activation of ADAM10 gene transcription by retinoic acid, the inhibition of natural ADAM10 inhibitors (e.g., TIMPs, tissue inhibitor of metalloproteases), and the modulation of ADAM10 cellular trafficking (Lichtenthaler, 2011). While dozens of proteins have been reported as ADAM10 substrates (Pruessmeyer and Ludwig, 2009), only a handful are related to brain and neuronal function. Moreover, in contrast to the ADAM10 knockdown, our data and a previous report by Postina et al. (2004) support that modest elevation of ADAM10 is relatively well tolerated and does not affect Notch-1 signaling in adult brain. Therefore, while a careful evaluation will be essential to assess the potential unwanted side effects elicited by chronic ADAM10 activation, our results suggest that brain specific α-secretase-based therapy could be a useful alternative or combination therapy with current AD therapeutics, e.g. targeting of β- or γ-secretases.
For past two decades, more than 1400 genetic studies have been carried out to elucidate the genetic loci influencing risk for AD (Bertram et al., 2010). Most recently, a high priority has been placed on identifying rare functional variants with high penetrance on disease risk by re-sequencing for inherited diseases, including AD (Pottier et al., 2012). In this study, we have presented in vivo functional analyses of two highly penetrant LOAD mutations in ADAM10, which we originally found by re-sequencing this gene in follow up to the observation of genetic association of several ADAM10 SNPs with AD (Kim et al., 2009). The multiple in vivo functional effects of these ADAM10 prodomain LOAD mutations presented here suggest that up-regulation of ADAM10 α-secretase activity may be beneficial for AD by two distinct, but functionally closely related biological mechanisms: 1. Decrease of neurotoxic Aβ accumulation by non-amyloidogenic cleavage of APP in brain, and 2. Up-regulation of neurogenesis in hippocampus. A tractable ADAM10-specific activator possessing these two neuroprotective properties could potentially be used as a potent therapeutic intervention for treating and preventing AD.
EXPERIMENTAL PROCEDURES
Procedures are described in detail in Supplemental Experimental Procedures. Detailed methods for Mouse Brain lysate Preparation, Western Blotting and Immunoprecipitation, sAPPα and sAPPβ ELISA, Primary Cortical Neurons and Surface Biotinylation, Analysis of Reactive Gliosis, ADAM10 Prodomain Chaperone Activity Assay, Cerebrospinal Fluid (CSF) Collection, Sucrose Gradient Fractionation, and Synaptosome and Postsynaptic Density Isolation are described in Supplemental Experimental Procedures.
ADAM10 Transgenic Mice
ADAM10 transgenic mice were generated by the injection of human ADAM10 cDNA (2.23 kb) of WT, Q170H, R181G and E384A mutant forms into embryos derived from B6SJLF1 female mice. Except one ADAM10-DN mouse line (DN-120), which was ~30% smaller in size from birth, all other ADAM10 transgenic mice were inconspicuous in morphology, breeding, and daily handling, compared to non-transgenic control mice. Immunohistochemical analysis using anti-HA antibodies showed that human ADAM10 is highly expressed in the cortex and hippocampus of transgenic mice. All animal generation, husbandry, and experimental procedures were approved by the MGH Subcommittee on Research Animal Care (SRAC). See Supplemental Experimental Procedures for details.
Aβ ELISA and Plaque Load Analysis
TBS brain lysates were centrifuged at 100,000 g for 1 hr and the supernatants were saved for ‘soluble Aβ’ measurements. Pellets were resuspended and further homogenized in 70% formic acid, followed by centrifugation at 100,000 g. Formic acid supernatants are neutralized with 1 M Tris for ‘insoluble Aβ’ analysis. The amounts of soluble and insoluble Aβ40 and Aβ42 were determined by sandwich ELISA using commercially available kits (Wako). For Aβ plaque load analysis, 4% paraformaldehyde-fixed brain sections were probed with anti-Aβ antibody and DAPI. For 12 months old brain sections, the numbers of Aβ plaque in cortex and hippocampus were counted without the genotype information. For 18–20 month old mice, the area covered by Aβ were analyzed by ImageJ. The numbers of Thioflavin S-stained neuritic plaques were also analyzed by ImageJ. See Supplemental Experimental Procedures for details.
NPCs Proliferation and Differentiation Analysis
Mice were injected once daily with BrdU (100 mg/kg, i.p.) for three consecutive days. Half of the mice in each group were sacrificed 1 day after the final injection of BrdU to identify proliferating neural progenitor cells (NPCs). The remaining mice were killed 2 weeks after the final injection of BrdU to determine survival and neuronal differentiation of the newborn cells. See Supplemental Experimental Procedures for details.
Statistical Analyses
Data are presented as the mean ± standard error of the mean (SEM). Comparisons between two groups were performed with the independent-samples t test, and those among more than two groups were performed with analysis of variance (ANOVA) followed by Tukey’s test. All analyses were performed with SPSS version 12.0. P value of < 0.05 was considered statistically significant.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. Lin Wu for her scientific expertise and technical assistance in generating ADAM10 transgenic mice. We also thank Dr. Jorg Bartsch for providing ADAM10 prodomain antibody; Dr. Sam Sisodia for providing MoPrP.XhoI plasmid; Dr. Matthew Frosch for providing Tg2576 mice; Drs. Basavaraj Hooli and Can Zhang for helpful discussions and advice; William Wisdom for mouse tail genotyping. This study was supported by the Cure Alzheimer’s Fund, grants from the NIA, and NIMH (RET), and the American Health Assistance Foundation (JS).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Anders A, Gilbert S, Garten W, Postina R, Fahrenholz F. Regulation of the alpha-secretase ADAM10 by its prodomain and proprotein convertases. FASEB J. 2001;15:1837–1839. doi: 10.1096/fj.01-0007fje. [DOI] [PubMed] [Google Scholar]
- Bell KF, Zheng L, Fahrenholz F, Cuello AC. ADAM-10 over-expression increases cortical synaptogenesis. Neurobiol Aging. 2008;29:554–565. doi: 10.1016/j.neurobiolaging.2006.11.004. [DOI] [PubMed] [Google Scholar]
- Bertram L, Lill CM, Tanzi RE. The genetics of Alzheimer disease: back to the future. Neuron. 2010;68:270–281. doi: 10.1016/j.neuron.2010.10.013. [DOI] [PubMed] [Google Scholar]
- Caille I, Allinquant B, Dupont E, Bouillot C, Langer A, Muller U, Prochiantz A. Soluble form of amyloid precursor protein regulates proliferation of progenitors in the adult subventricular zone. Development. 2004;131:2173–2181. doi: 10.1242/dev.01103. [DOI] [PubMed] [Google Scholar]
- Cao J, Hymowitz M, Conner C, Bahou WF, Zucker S. The propeptide domain of membrane type 1-matrix metalloproteinase acts as an intramolecular chaperone when expressed in trans with the mature sequence in COS-1 cells. J Biol Chem. 2000;275:29648–29653. doi: 10.1074/jbc.M001920200. [DOI] [PubMed] [Google Scholar]
- Cao Y, Kang Q, Zhao Z, Zolkiewska A. Intracellular processing of metalloprotease disintegrin ADAM12. J Biol Chem. 2002;277:26403–26411. doi: 10.1074/jbc.M110814200. [DOI] [PubMed] [Google Scholar]
- Choi SH, Veeraraghavalu K, Lazarov O, Marler S, Ransohoff RM, Ramirez JM, Sisodia SS. Non-cell-autonomous effects of presenilin 1 variants on enrichment-mediated hippocampal progenitor cell proliferation and differentiation. Neuron. 2008;59:568–580. doi: 10.1016/j.neuron.2008.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirrito JR, May PC, O’Dell MA, Taylor JW, Parsadanian M, Cramer JW, Audia JE, Nissen JS, Bales KR, Paul SM, et al. In vivo assessment of brain interstitial fluid with microdialysis reveals plaque-associated changes in amyloid-beta metabolism and half-life. J Neurosci. 2003;23:8844–8853. doi: 10.1523/JNEUROSCI.23-26-08844.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo A, Wang H, Kuhn PH, Page R, Kremmer E, Dempsey PJ, Crawford HC, Lichtenthaler SF. Constitutive alpha- and beta-secretase cleavages of the amyloid precursor protein are partially coupled in neurons, but not in frequently used cell lines. Neurobiol Dis. 2012;49C:137–147. doi: 10.1016/j.nbd.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demars MP, Bartholomew A, Strakova Z, Lazarov O. Soluble amyloid precursor protein: a novel proliferation factor of adult progenitor cells of ectodermal and mesodermal origin. Stem Cell Res Ther. 2011;2:36. doi: 10.1186/scrt77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donmez G, Wang D, Cohen DE, Guarente L. SIRT1 suppresses beta-amyloid production by activating the alpha-secretase gene ADAM10. Cell. 2010;142:320–332. doi: 10.1016/j.cell.2010.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Fahrenholz F, Postina R. Alpha-secretase activation--an approach to Alzheimer’s disease therapy. Neurodegener Dis. 2006;3:255–261. doi: 10.1159/000095264. [DOI] [PubMed] [Google Scholar]
- Gatz M, Reynolds CA, Fratiglioni L, Johansson B, Mortimer JA, Berg S, Fiske A, Pedersen NL. Role of genes and environments for explaining Alzheimer disease. Arch Gen Psychiatry. 2006;63:168–174. doi: 10.1001/archpsyc.63.2.168. [DOI] [PubMed] [Google Scholar]
- Gaultier A, Cousin H, Darribere T, Alfandari D. ADAM13 disintegrin and cysteine-rich domains bind to the second heparin-binding domain of fibronectin. J Biol Chem. 2002;277:23336–23344. doi: 10.1074/jbc.M201792200. [DOI] [PubMed] [Google Scholar]
- Gralle M, Botelho MG, Wouters FS. Neuroprotective secreted amyloid precursor protein acts by disrupting amyloid precursor protein dimers. J Biol Chem. 2009;284:15016–15025. doi: 10.1074/jbc.M808755200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer’s disease: progress and problems on the road to therapeutics. Science. 2002;297:353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- Hartmann D, de Strooper B, Serneels L, Craessaerts K, Herreman A, Annaert W, Umans L, Lubke T, Lena Illert A, von Figura K, et al. The disintegrin/metalloprotease ADAM10 is essential for Notch signalling but not for alpha-secretase activity in fibroblasts. Hum Mol Genet. 2002;11:2615–2624. doi: 10.1093/hmg/11.21.2615. [DOI] [PubMed] [Google Scholar]
- Jorissen E, Prox J, Bernreuther C, Weber S, Schwanbeck R, Serneels L, Snellinx A, Craessaerts K, Thathiah A, Tesseur I, et al. The disintegrin/metalloproteinase ADAM10 is essential for the establishment of the brain cortex. J Neurosci. 2010;30:4833–4844. doi: 10.1523/JNEUROSCI.5221-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaden D, Harmeier A, Weise C, Munter LM, Althoff V, Rost BR, Hildebrand PW, Schmitz D, Schaefer M, Lurz R, et al. Novel APP/Abeta mutation K16N produces highly toxic heteromeric Abeta oligomers. EMBO Mol Med. 2012;4:647–659. doi: 10.1002/emmm.201200239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang T, Park HI, Suh Y, Zhao YG, Tschesche H, Sang QX. Autolytic processing at Glu586-Ser587 within the cysteine-rich domain of human adamalysin 19/disintegrin-metalloproteinase 19 is necessary for its proteolytic activity. J Biol Chem. 2002;277:48514–48522. doi: 10.1074/jbc.M208961200. [DOI] [PubMed] [Google Scholar]
- Kawarabayashi T, Younkin LH, Saido TC, Shoji M, Ashe KH, Younkin SG. Age-dependent changes in brain, CSF, and plasma amyloid (beta) protein in the Tg2576 transgenic mouse model of Alzheimer’s disease. J Neurosci. 2001;21:372–381. doi: 10.1523/JNEUROSCI.21-02-00372.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Suh J, Romano D, Truong MH, Mullin K, Hooli B, Norton D, Tesco G, Elliott K, Wagner SL, et al. Potential late-onset Alzheimer’s disease-associated mutations in the ADAM10 gene attenuate {alpha}-secretase activity. Hum Mol Genet. 2009;18:3987–3996. doi: 10.1093/hmg/ddp323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn PH, Wang H, Dislich B, Colombo A, Zeitschel U, Ellwart JW, Kremmer E, Rossner S, Lichtenthaler SF. ADAM10 is the physiologically relevant, constitutive alpha-secretase of the amyloid precursor protein in primary neurons. EMBO J. 2010;29:3020–3032. doi: 10.1038/emboj.2010.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammich S, Kojro E, Postina R, Gilbert S, Pfeiffer R, Jasionowski M, Haass C, Fahrenholz F. Constitutive and regulated alpha-secretase cleavage of Alzheimer’s amyloid precursor protein by a disintegrin metalloprotease. Proc Natl Acad Sci U S A. 1999;96:3922–3927. doi: 10.1073/pnas.96.7.3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee EB, Zhang B, Liu K, Greenbaum EA, Doms RW, Trojanowski JQ, Lee VM. BACE overexpression alters the subcellular processing of APP and inhibits Abeta deposition in vivo. J Cell Biol. 2005;168:291–302. doi: 10.1083/jcb.200407070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenthaler SF. Alpha-secretase in Alzheimer’s disease: molecular identity, regulation and therapeutic potential. J Neurochem. 2011;116:10–21. doi: 10.1111/j.1471-4159.2010.07081.x. [DOI] [PubMed] [Google Scholar]
- Luo Y, Bolon B, Kahn S, Bennett BD, Babu-Khan S, Denis P, Fan W, Kha H, Zhang J, Gong Y, et al. Mice deficient in BACE1, the Alzheimer’s beta-secretase, have normal phenotype and abolished beta-amyloid generation. Nat Neurosci. 2001;4:231–232. doi: 10.1038/85059. [DOI] [PubMed] [Google Scholar]
- Marcello E, Gardoni F, Mauceri D, Romorini S, Jeromin A, Epis R, Borroni B, Cattabeni F, Sala C, Padovani A, et al. Synapse-associated protein-97 mediates alpha-secretase ADAM10 trafficking and promotes its activity. J Neurosci. 2007;27:1682–1691. doi: 10.1523/JNEUROSCI.3439-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattson MP, Cheng B, Culwell AR, Esch FS, Lieberburg I, Rydel RE. Evidence for excitoprotective and intraneuronal calcium-regulating roles for secreted forms of the beta-amyloid precursor protein. Neuron. 1993;10:243–254. doi: 10.1016/0896-6273(93)90315-i. [DOI] [PubMed] [Google Scholar]
- McConlogue L, Buttini M, Anderson JP, Brigham EF, Chen KS, Freedman SB, Games D, Johnson-Wood K, Lee M, Zeller M, et al. Partial reduction of BACE1 has dramatic effects on Alzheimer plaque and synaptic pathology in APP Transgenic Mice. J Biol Chem. 2007;282:26326–26334. doi: 10.1074/jbc.M611687200. [DOI] [PubMed] [Google Scholar]
- McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- Moss ML, Bomar M, Liu Q, Sage H, Dempsey P, Lenhart PM, Gillispie PA, Stoeck A, Wildeboer D, Bartsch JW, et al. The ADAM10 prodomain is a specific inhibitor of ADAM10 proteolytic activity and inhibits cellular shedding events. J Biol Chem. 2007;282:35712–35721. doi: 10.1074/jbc.M703231200. [DOI] [PubMed] [Google Scholar]
- Mu Y, Gage FH. Adult hippocampal neurogenesis and its role in Alzheimer’s disease. Mol Neurodegener. 2011;6:85. doi: 10.1186/1750-1326-6-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolaev A, McLaughlin T, O’Leary DD, Tessier-Lavigne M. APP binds DR6 to trigger axon pruning and neuron death via distinct caspases. Nature. 2009;457:981–989. doi: 10.1038/nature07767. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Pan D, Rubin GM. Kuzbanian controls proteolytic processing of Notch and mediates lateral inhibition during Drosophila and vertebrate neurogenesis. Cell. 1997;90:271–280. doi: 10.1016/s0092-8674(00)80335-9. [DOI] [PubMed] [Google Scholar]
- Parkin E, Harris B. A disintegrin and metalloproteinase (ADAM)-mediated ectodomain shedding of ADAM10. J Neurochem. 2009;108:1464–1479. doi: 10.1111/j.1471-4159.2009.05907.x. [DOI] [PubMed] [Google Scholar]
- Postina R, Schroeder A, Dewachter I, Bohl J, Schmitt U, Kojro E, Prinzen C, Endres K, Hiemke C, Blessing M, et al. A disintegrin-metalloproteinase prevents amyloid plaque formation and hippocampal defects in an Alzheimer disease mouse model. J Clin Invest. 2004;113:1456–1464. doi: 10.1172/JCI20864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pottier C, Hannequin D, Coutant S, Rovelet-Lecrux A, Wallon D, Rousseau S, Legallic S, Paquet C, Bombois S, Pariente J, et al. High frequency of potentially pathogenic SORL1 mutations in autosomal dominant early-onset Alzheimer disease. Mol Psychiatry. 2012 doi: 10.1038/mp.2012.15. [DOI] [PubMed] [Google Scholar]
- Pruessmeyer J, Ludwig A. The good, the bad and the ugly substrates for ADAM10 and ADAM17 in brain pathology, inflammation and cancer. Semin Cell Dev Biol. 2009;20:164–174. doi: 10.1016/j.semcdb.2008.09.005. [DOI] [PubMed] [Google Scholar]
- Ring S, Weyer SW, Kilian SB, Waldron E, Pietrzik CU, Filippov MA, Herms J, Buchholz C, Eckman CB, Korte M, et al. The secreted beta-amyloid precursor protein ectodomain APPs alpha is sufficient to rescue the anatomical, behavioral, and electrophysiological abnormalities of APP-deficient mice. J Neurosci. 2007;27:7817–7826. doi: 10.1523/JNEUROSCI.1026-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selkoe DJ. Alzheimer’s disease: genes, proteins, and therapy. Physiol Rev. 2001;81:741–766. doi: 10.1152/physrev.2001.81.2.741. [DOI] [PubMed] [Google Scholar]
- Shinde UP, Liu JJ, Inouye M. Protein memory through altered folding mediated by intramolecular chaperones. Nature. 1997;389:520–522. doi: 10.1038/39097. [DOI] [PubMed] [Google Scholar]
- Simpson JE, Ince PG, Lace G, Forster G, Shaw PJ, Matthews F, Savva G, Brayne C, Wharton SB. Astrocyte phenotype in relation to Alzheimer-type pathology in the ageing brain. Neurobiol Aging. 2010;31:578–590. doi: 10.1016/j.neurobiolaging.2008.05.015. [DOI] [PubMed] [Google Scholar]
- Takeda S, Igarashi T, Mori H, Araki S. Crystal structures of VAP1 reveal ADAMs’ MDC domain architecture and its unique C-shaped scaffold. EMBO J. 2006;25:2388–2396. doi: 10.1038/sj.emboj.7601131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor DR, Parkin ET, Cocklin SL, Ault JR, Ashcroft AE, Turner AJ, Hooper NM. Role of ADAMs in the ectodomain shedding and conformational conversion of the prion protein. J Biol Chem. 2009;284:22590–22600. doi: 10.1074/jbc.M109.032599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tousseyn T, Thathiah A, Jorissen E, Raemaekers T, Konietzko U, Reiss K, Maes E, Snellinx A, Serneels L, Nyabi O, et al. ADAM10, the rate-limiting protease of regulated intramembrane proteolysis of Notch and other proteins, is processed by ADAMS-9, ADAMS-15, and the gamma-secretase. J Biol Chem. 2009;284:11738–11747. doi: 10.1074/jbc.M805894200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Ha Y. The X-ray structure of an antiparallel dimer of the human amyloid precursor protein E2 domain. Mol Cell. 2004;15:343–353. doi: 10.1016/j.molcel.2004.06.037. [DOI] [PubMed] [Google Scholar]
- Zhao C, Deng W, Gage FH. Mechanisms and functional implications of adult neurogenesis. Cell. 2008;132:645–660. doi: 10.1016/j.cell.2008.01.033. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.