Summary
Chromosomal rearrangements often occur at genomic loci with DNA secondary structures, such as common fragile sites (CFSs) and palindromic repeats. We developed assays in mammalian cells that revealed CFS-derived AT-rich sequences and Alu inverted repeats (Alu-IRs) are mitotic recombination hotspots, requiring the repair functions of CtIP and the Mre11/Rad50/Nbs1 complex (MRN). We also identified an endonuclease activity of CtIP which is dispensable for end resection and homologous recombination (HR) at I-SceI-generated "clean" double-strand breaks (DSBs), but is required for repair of DSBs occurring at CFS-derived AT-rich sequences. In addition, CtIP nuclease defective mutants are impaired in Alu-IRs-induced mitotic recombination. These studies suggest that an end resection-independent CtIP function is important for processing DSB ends with secondary structures to promote HR. Furthermore, our studies uncover an important role of MRN, CtIP and their associated nuclease activities in protecting CFSs in mammalian cells.
Keywords: CtIP, MRN, common fragile sites, inverted repeats, DNA double-strand break repair, end resection
Introduction
Gross chromosomal rearrangement (GCR) is a hallmark of cancer cells, and DNA double strand breaks (DSBs) are one major source to cause such genome instability (Kasparek and Humphrey, 2011). Common fragile sites (CFSs) are part of normal chromosomal structures, extend over hundreds of kilobases, and upon replication stress, exhibit gaps and breaks on metaphase chromosomes, which are termed as CFS expression (Glover et al., 2005). Repetitive DNA sequences comprise a large portion of the human genome (Lander et al., 2001), and inverted repeats (IRs) can form hairpin or cruciform structures (Lilley, 1980; Panayotatos and Wells, 1981) to trigger gene amplification and chromosomal translocation (Kato et al., 2012; Tanaka et al., 2005). CFSs and IRs are often associated with pathogenic breakpoint junctions in tumors (Abeysinghe et al., 2004; Arlt et al., 2006; Tanaka et al., 2005), but the detailed mechanisms underlying the prevention of genome instability at these sites are not well understood.
The Mre11/Rad50/Nbs1 complex (MRN) is well-conserved and critical for DSB repair (D'Amours and Jackson, 2002). CtIP, a homologue of Sae2 in budding yeast (Sartori et al., 2007), binds to MRN and BRCA1 (Chen et al., 2008; Yu and Chen, 2004) and promotes end resection to initiate homologous recombination (HR)-mediated DSB repair (Jazayeri et al., 2006; Sartori et al., 2007). In budding yeast, the Mre11/Rad50/Xrs2 complex (MRX) and Sae2 are required for cleaving covalent Spo11-DNA intermediates in meiosis (Keeney, 2001) and processing hairpin structures formed by IRs at DSB ends (Lobachev et al., 2002; Rattray et al., 2005). Consistently, Mre11 and Sae2 exhibit endonuclease activities to cleave hairpin DNA and single-strand DNA (ssDNA) adjacent to hairpins, respectively (Lengsfeld et al., 2007; Paull and Gellert, 1999).
In this study, we demonstrated that AT-rich CFS sequences and Alu-IRs induce mitotic recombination, especially under replication stress in mammalian cells, a process which strongly depends on MRN, CtIP, and the nuclease activity of Mre11. We also identified a CtIP-associated endonuclease activity that is dispensable for end resection and HR at I-SceI-generated "clean" DSBs, but is required when AT-rich CFS sequences are present in proximity to DSBs. This CtIP endonuclease activity is also important for Alu-IRs-induced mitotic recombination. Significantly, MRN and CtIP, and their associated nuclease activities, are important for maintaining CFS stability in mammalian cells.
Results
AT-rich sequences derived from CFS FRA16D induce mitotic recombination in mammalian cells
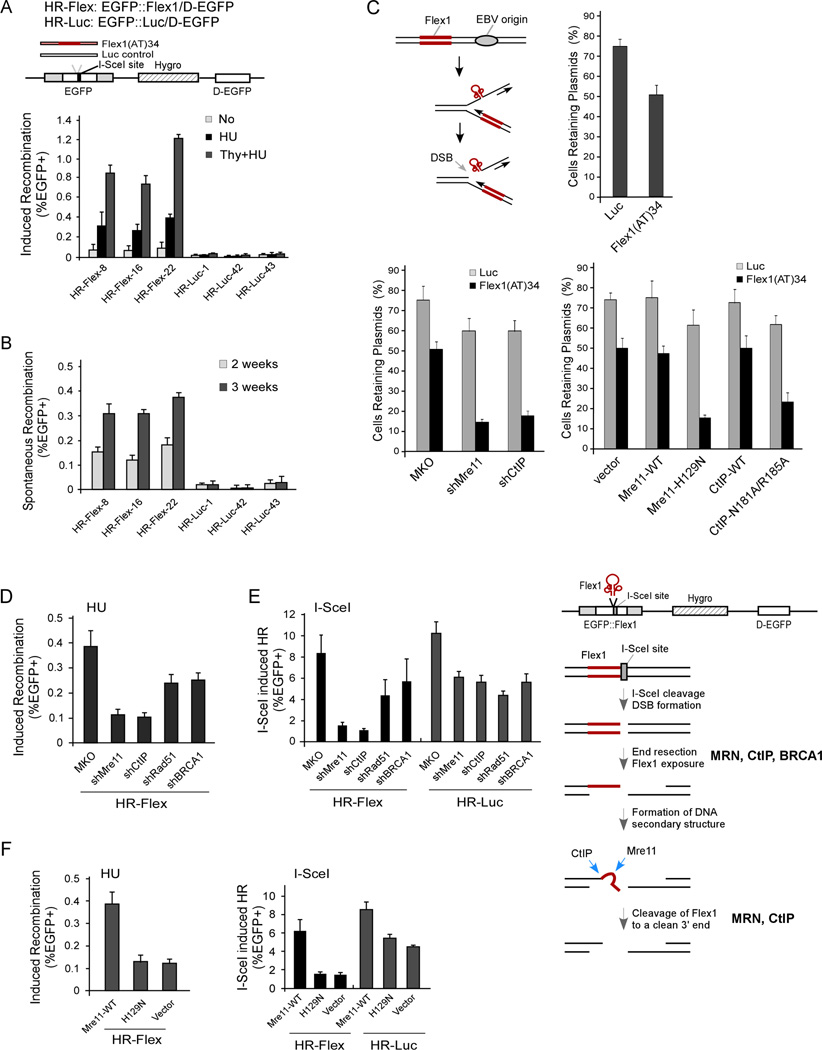
To study CFS-induced genome instability in mammalian cells, we generated EGFP-based HR substrates, in which a small AT-rich and hairpin-forming DNA sequence, Flex1(AT)34 (0.33 kb) with 34 copies of AT-dinucleotide repeats in the middle, derived from CFS FRA16D, or a comparable-sized luciferase fragment, was inserted into the EGFP cassette (Figure 1A, top, HRFlex and HR-Luc). Flex1 was shown to stall DNA replication and cause DSB formation in yeast (Zhang and Freudenreich, 2007). Multiple U2OS cell lines carrying HR-Flex or HR-Luc stably integrated at different genomic loci were obtained. Hydroxyurea (HU)- and aphidicolin (Aph)-induced HR in HR-Flex was significantly higher than in the control HR-Luc, and enrichment of S-phase cells by double thymidine block further increased HU-induced HR frequency in HRFlex (Figures 1A, and S1A). Flex1, but not Luc, also induced spontaneous mitotic recombination (Figure 1B). Four Flex1 sequences, identified from human population with varying numbers of the AT repeats due to polymorphism (Finnis et al., 2005), all induced HR after HU treatment, with longer AT repeats showing stronger effects (Figure S1B). Flex1 also induced plasmid instability of the Epstein-Barr virus (EBV) replication origin-containing plasmids (Figures 1C, top), propagated in mammalian cells as episomes (Reeves et al., 1985; Shen et al., 2009). These results suggest that CFS-derived AT-rich sequences are genetically unstable, supporting the notion that these AT-rich sequences contribute to CFS fragility.
Figure 1. The AT-rich Flex1 sequences derived from CFS FRA16D are mitotically unstable.
A. The HR-Flex and HR-Luc substrates with I-SceI cleavage site at one side (top, D-EGFP: donor EGFP fragment). Three U2OS clones with single-integration of HR-Flex or HR-Luc were mock treated (No), treated with 2 mM hydroxyurea (HU) for 24 hr, or subjected to double-thymidine block then HU-treated for 24 hr (Thy + HU), and assayed for EGFP-positive events 72 hr after HU treatment (bottom).
B. U2OS clones carrying HR-Flex or HR-Luc were sorted for EGFP-negative cells, cultured for 2 or 3 weeks, and assayed for spontaneous mitotic recombination.
C. A model for DSB formation on stalled replication forks induced by the Flex1 sequence close to an EBV origin (top left). Plasmid instability assay of U2OS cells carrying pCEP4-Flex1(AT)34 or pCEP4-Luc after culturing without hygromycin for 1 week (top right), or in cell lines expressing shRNAs against Mre11, CtIP or control MKO (bottom left), or expressing Mre11 (WT or H129N) or CtIP (WT or N181A/R185A), with endogenous Mre11 or CtIP silenced by shRNAs.
D–F. U2OS cells carrying HR-Flex or HR-Luc, expressing indicated shRNAs or control MKO, or in F, expressing Myc-Mre11 (WT or H129N) or vector, with endogenous Mre11 silenced, were induced with HU (2 mM) for 24 hr (left) or with I-SceI (right) and assayed 72 hr later. In E, right, a model for end resection and processing of structure-forming sequences (such as Flex1) at DSB ends to generate clean 3’ ends for strand invasion.
See also Figure S1.)
In all experiments, error bars represent standard deviation (s.d.) of three independent experiments.
The Mre11 complex and CtIP are specifically required for repairing DSBs with AT-rich Flex1 sequences at DSB ends
When Mre11 or CtIP was inactivated by shRNAs, HU-induced HR at Flex1 was reduced to a greater extent than inactivating BRCA1 or Rad51 (Figures 1D and S1D). Yet, HU-induced γH2AX foci were increased in cells expressing Mre11 or CtIP shRNAs compared to control cells (Figure S1E), showing that Mre11 or CtIP inactivation causes DSB accumulation. Meanwhile, when DSBs were induced by I-SceI cleavage at the side of Flex1 or Luc (Figure 1A), inactivation of Mre11, CtIP, BRCA1, BRCA2 or Rad51 led to a similar reduction at HR-Luc (Figures 1E, S1D and S1F), but interestingly, Mre11 or CtIP inactivation reduced HR at Flex1 more so than BRCA1, BRCA2 or Rad51 inactivation. The Mre11 nuclease mutant Mre11-H129N also exhibited a similar reduction of HU-induced mitotic recombination as Mre11 knock-down cells, and was more defective in HR after I-SceI cleavage at Flex1 than the Luc control (Figures 1F and S1G). These data suggest that besides their general roles in the BRCA1- and Rad51-dependent HR pathway, MRN and CtIP carry additional functions requiring Mre11 nuclease activity to repair DSBs with structure-forming DNA sequences at ends.
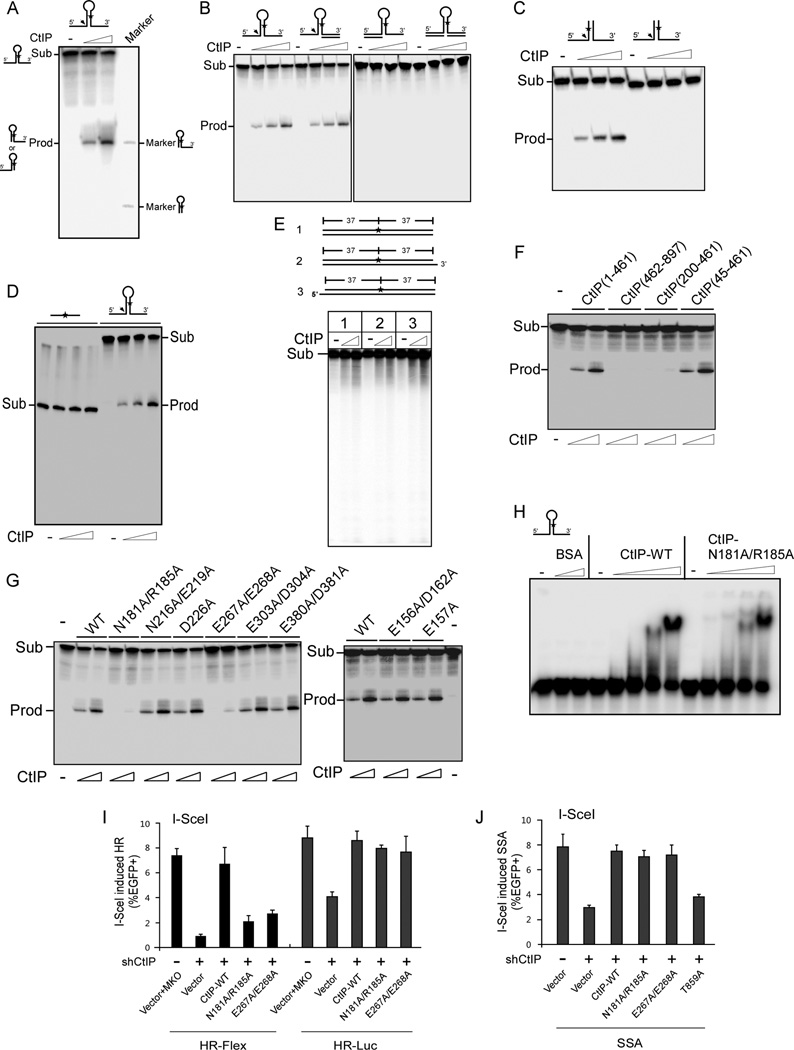
CtIP has an endonuclease activity
Sae2 possesses endonuclease activities to process hairpin structures at DSB ends (Lengsfeld et al., 2007). We also detected an endonuclease activity of human CtIP to cleave one ssDNA tail of a hairpin substrate (Figures 2A and S2A). Apparently, CtIP exhibits specificity for 5’ ssDNA tails and does not cleave 3’ ssDNA nor double-strand DNA (dsDNA) tails of the substrate (Figures 2B and S2B). To test whether the hairpin loop is required for CtIP activity as with Sae2 (Lengsfeld et al., 2007), we removed the loop and found that CtIP cleaves Y-shaped branched DNA substrates with similar activity as hairpin loop substrates at the base of 5’ ssDNA tails (Figure 2C). No endonuclease and exonuclease activities of CtIP were detected on linear ssDNA and dsDNA (Figures 2D and 2E), and CtIP has higher endonuclease activity in the presence of manganese than magnesium (Figure S2C).
Figure 2. CtIP is associated with an endonuclease activity.
A–C. Internally 32P-labeled (asterisk) DNA hairpin substrates with two 30 bp ssDNA tails (A), ssDNA and/or dsDNA tails (B), or Y-shaped branched substrates with ssDNA tails (C) were used for nuclease assays with CtIP at concentrations 20 and 60 nM (A), or 5, 20 and 60 nM (B and C) or without at 37°C for 30 min. Sub: substrates, Prod: products. DNA hairpin substrates with one 3’ tail or no tail were run on the gel as markers in A.
D. Internally 32P-labeled 66-bp ssDNA or a hairpin substrate with ssDNA tails were incubated with 5, 20, 60 nM CtIP or without at 37°C for 30 min.
E. dsDNA with blunt, or 3’ or 5’ overhangs were internally labeled with 32P and used as substrates for nuclease assays with CtIP concentrations of 70 and 200 nM or without at 37°C for 1 hr.
F and G. CtIP fragments of indicated lengths (in F) and CtIP fragment 45–461 containing various point mutations (in G) were assayed for nuclease activity at CtIP concentrations of 20 and 60 nM or without using the hairpin substrate from A. Coomassie blue staining of these purified CtIP fragments is shown in Figure S2D.
H. Gel mobility shift was performed with CtIP (45-461) WT or N181A/R185A mutant at 2, 5, 20 and 60 nM or without and using 3 nM hairpin DNA substrate from A, with BSA (100 and 200 nM) as control.
I and J. U2OS cells carrying HR-Flex or HR-Luc (in I) or EGFP-SSA (in J, Figure S2N) and expressing HA-CtIP (WT, N181A/R185A or E267A/E268A) or vector, with endogenous CtIP silenced, were induced with I-SceI and assayed.
See also Figure S2.)
In all experiments, error bars represent standard deviation (s.d.) of three independent experiments.
Deletion analysis revealed that the CtIP N-terminal fragment, CtIP (1-461), but not the C-terminal fragment, CtIP (462-897), exhibited endonuclease activities (Figures 2F and S2D). Deleting the N-terminal 199 residues [CtIP (200–461)], but not the N-terminal 44 residues [CtIP (45–461)], abolished the nuclease activity. Since CtIP enzymatic activity depends on metal ions, and glutamic acid (E) and aspartic acid (D) residues often contribute to metal ion binding (Shen et al., 1997), we mutated a series of E and D residues to alanine (A) in CtIP (45-461). While mutating E156, E157, D162, N216, E219, D226, E303, D304, E380 and D381 did not show an effect, both CtIP (45-461)-E267A/E268A and full-length CtIP-E267A/E268A significantly reduced CtIP-associated endonuclease activity (Figures 2G, S2D and S2E). In addition, alignment of human CtIP N-terminus with that of other species revealed a putative conserved NxxxR/K motif (Figure 3F), and mutating conserved residues N181 and/or R185 in CtIP (45–461) and full-length CtIP reduced endonuclease activity (Figures 2G and S2D–G). Both CtIP N181A/R185A and E267A/E268A mutants exhibited similar affinity to bind the hairpin DNA substrate and MRN (Figures 2H, S2H and data not shown), and were recruited to DSBs in live cells with similar kinetics as CtIP-WT (wild-type, Figure S2I). Quantitative PCR (qPCR)-based in vitro assays also revealed that CtIP-WT but not the N181A/R185A and E267A/E268A endonuclease mutants removed Flex1 ssDNA much more efficiently than Luc ssDNA (Figure S2J, top and bottom left). Presence of ssDNA tail 3' to Flex1 (substrate 3) did not block CtIP-mediated cleavage of Flex1, and the 3' ssDNA tail was largely removed as an intact piece (Figure S2J, top and bottom right), consistent with an endonuclease activity of CtIP. Therefore, CtIP possesses an endonuclease activity that is associated with its N-terminus and is sufficient to process DNA ends with secondary structures.
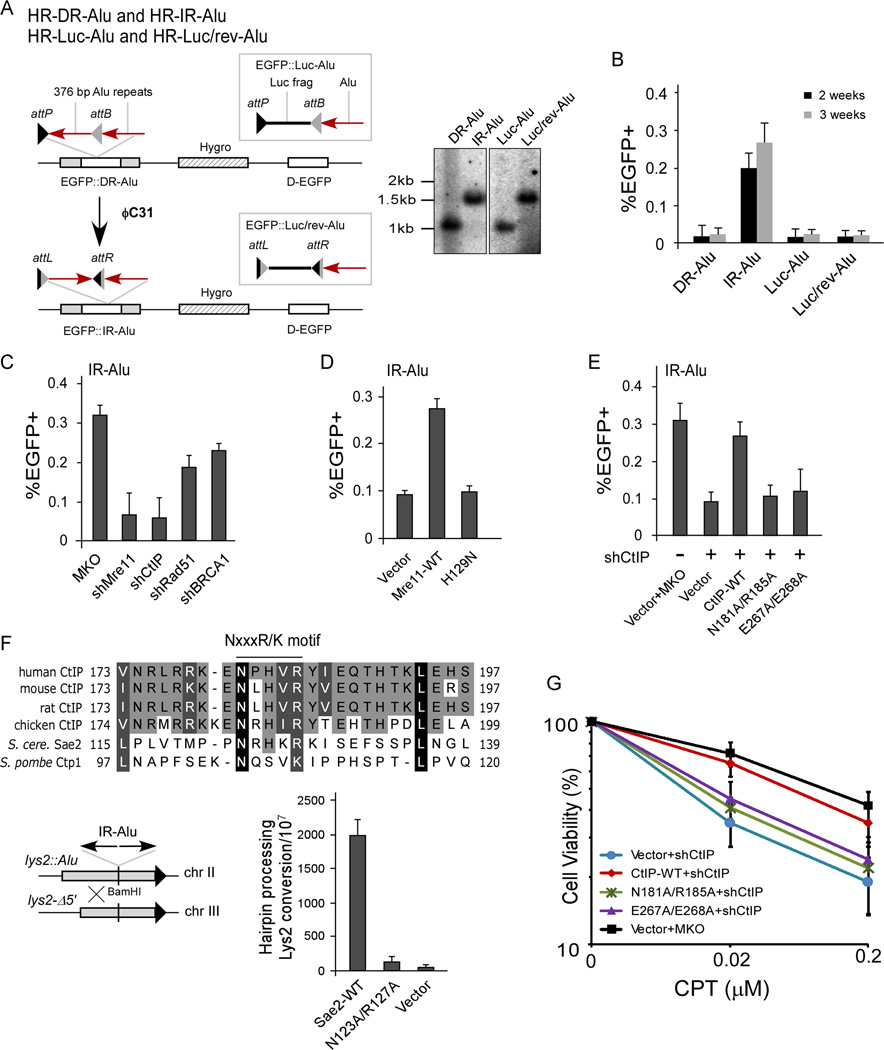
Figure 3. CtIP exhibits a conserved function required for IRs-induced mitotic recombination.
A. The HR-Alu substrates (left). Southern blot analysis of HR-Alu substrates (see also Figure S3B), to distinguish DR-Alu and Luc-Alu from IR-Alu and Luc/rev-Alu, respectively (right).
B. U2OS cells carrying HR-DR-Alu or HR-IR-Alu or control HR-Luc-Alu and HR-Luc/rev-Alu were assayed for spontaneous HR of non-green cells after sorting and 2- or 3-week culturing.
C–E. U2OS (HR-IR-Alu) cells were expressed with indicated shRNAs or control MKO (in C) or expressed with HA-Mre11 (WT or H129N), with endogenous Mre11 silenced (in D), or with HA-CtIP (WT or indicated mutants), with endogenous CtIP silenced (in E). Spontaneous HR was assayed after 3-week culturing.
F. Top: Alignment of human CtIP and its homologues from indicated species, with the conserved NxxxR/K motif shown. Bottom: Alu-IRs-induced recombination between two lys2 alleles as described (Lobachev et al., 2002) was assayed in S. cerevisiae Δsae2 yeast strain expressing Sae2 (WT or N123A/R127A) or vector.
G. Clonogenic survival assay was performed in U2OS cells expressing HA-CtIP (WT or indicated mutants), with endogenous CtIP silenced, after CPT treatment with indicated concentrations for 1hr. (See also Figure S3.)
In all experiments, error bars represent standard deviation (s.d.) of three independent experiments.
CtIP-associated endonuclease activity is important for repairing DSBs at CFSs but is dispensable for end resection and HR at “clean” I-SceI-induced DSBs
We observed that EBV-Flex1 plasmids become more unstable than EBV-Luc plasmids in CtIP- and CtIP nuclease-deficient cells, and similarly in Mre11- or Mre11 nuclease-deficient cells (Figures 1C, bottom and S1C). To more directly examine the role of CtIP-associated endonuclease activity for DSB repair, we assayed for I-SceI-induced HR using the CtIPN181A/ R185A and CtIP-E267A/E268A mutants. Interestingly, these mutants did not show defects in HR-mediated DSB repair using HR-Luc, but even with N181A and R185A single mutations, a significant reduction of HR was observed when Flex1 is present at DSBs (HR-Flex) after I-SceI cleavage (Figures 2I, S2K and S2L). Furthermore, combining the N181A/R185A mutant with the end resection defective CDK mutant CtIP-T847A (Huertas and Jackson, 2009) reduced HR in HR-Luc to the level of T847A single mutant, and further decreased HR in HRFlex (Figure S2M). These data suggest that CtIP endonuclease activity is dispensable for end resection required for HR at general DSBs, but is specifically required for processing DSBs with secondary structures formed at ends. In agreement, while the end resection defective mutant CtIP-T859A (Wang et al., 2013) was impaired in single-strand annealing (SSA), CtIP-N181A/ R185A and CtIP-E267A/E268A mutants were not (Figures 2J and S2N). In addition, CtIP-dependent RPA binding to DSB surrounding regions due to ssDNA accumulation was at similar levels in CtIP N181A/R185A and E267A/E268A mutant and CtIP-WT cell lines (Figure S2O). These data support that the CtIP-associated endonuclease activity is not required for end resection at general DSBs.
Inverted Alu repeats induce mitotic recombination in mammalian cells
In budding yeast, Mre11 and Sae2 are critical for IRs-induced mitotic recombination with no significant contribution to general mitotic recombination (Lobachev et al., 2002). Like CFS-derived AT-rich sequences (Zhang and Freudenreich, 2007), IRs also stall replication forks, possibly due to hairpin formation at the lagging strand during DNA replication [(Voineagu et al., 2008), Figure S3A]. To study IRs-induced genome instability in mammalian cells, we developed a novel EGFP-based repair assay, (Figure 3A, left). Two identical Alu sequences were placed in a direct orientation (DR-Alu) in the EGFP ORF, with the upstream Alu flanked by attP and attB recombination sites for the phage integrase ϕC31 in a reversed orientation (Belteki et al., 2003). As ϕC31-mediated recombination would generate hybrid sites, attL and attR sites that cannot recombine further (Thorpe et al., 2000), a stable inverted Alu repeat (IR-Alu) would form at the same genomic locus where DR-Alu is located. U2OS cell lines with a single chromosomal integration of the EGFP::DR-Alu cassette were generated, and the corresponding cell lines with inverted Alu sequences (EGFP::IR-Alu) were isolated after ϕC31 expression and confirmed by Southern blot analysis (Figures 3A, right, and S3B). Mitotic recombination frequency in the cell lines with IR-Alu was significantly higher compared to those carrying DR-Alu at the same genomic locus (Figure 3B). Inverting the Luc in the control cell line (Luc/rev-Alu) did not show such an effect. These studies suggest that Alu-IRs are mitotic hotspots in mammalian cells.
Inactivation of Mre11 or CtIP by shRNAs dramatically reduced IRs-induced mitotic recombination, while inactivation of Rad51 or BRCA1 had a much less effect (Figures 3C and S3C). The Mre11-H129N nuclease mutant and CtIP N181A/R185A and E267A/E268A endonuclease mutants were severely defective in Alu-IRs-induced mitotic recombination (Figures 3D, 3E, S3D and S3E). These studies suggest that the Mre11 and CtIP nuclease activities are important for IRs-induced mitotic recombination.
To see whether the CtIP endonuclease activity has a conserved role for DSB repair, we mutated the corresponding sites of CtIP N181 and R185 in the yeast homologue Sae2, and expressed them from a low-copy CEN plasmid in Δsae2 S. cerevisiae strains. We found that Sae2-N123A/R127A, N123A and R127A mutants were defective in Alu-IRs-induced mitotic recombination (Figures 3F and S3F). Furthermore, like the human CtIP N181A/R185A and E267A/E268A endonuclease mutants (Figures 3G and S3G), yeast Sae2-N123A/R127A also showed sensitivity to camptothecin (CPT, Figure S3H). Thus, the CtIP N181 and R185 sites required for human CtIP endonuclease activity are functionally conserved in budding yeast.
MRN and CtIP are important for protecting CFSs
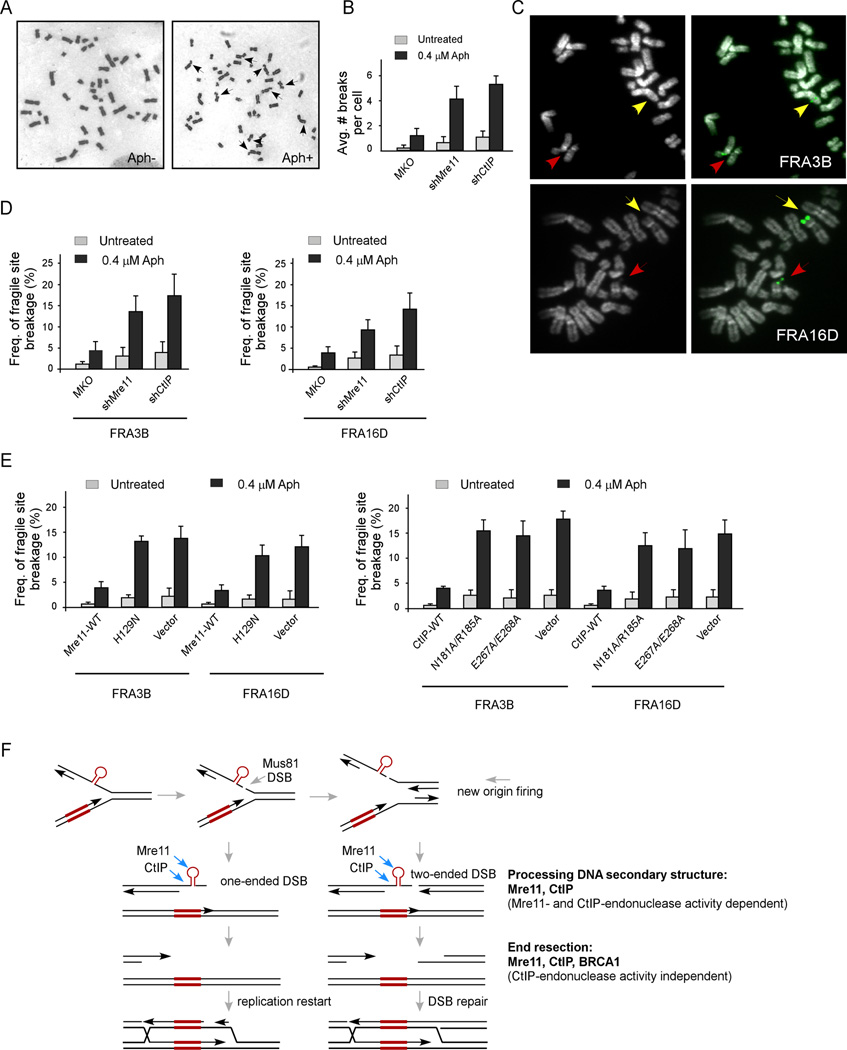
Since MRN and CtIP are specifically required for repairing DSBs at CFS-derived sequences, we asked whether they are also important for protecting CFSs in mammalian cells. Chromosomal gaps and breakages per cell were elevated when Mre11 or CtIP was inactivated by shRNAs in HCT116 and HeLa cells, which were further increased after Aph treatment (0.4 µM, 24 hr), the condition that induces CFS expression (Figures 4A, 4B, S4A and data not shown). More specifically, the CFS expression at FRA16D and FRA3B was significantly increased when Mre11 or CtIP was deficient, as revealed by FISH analysis (Figures 4C and 4D), suggesting that MRN and CtIP are indeed important for CFS protection. Furthermore, the Mre11-H129N nuclease mutant and CtIP N181A/R185A and E267A/E268A endonuclease mutants were impaired in protecting FRA16D and FRA3B stability (Figures 4E, S4B and S4C). Thus, the Mre11 nuclease activity and the CtIP endonuclease activity are both required for CFS protection.
Figure 4. Mre11 and CtIP are important for CFS protection.
A. Metaphase spread of HCT116 cells before (Aph−) and after aphidicolin (Aph+) treatment (Aph, 0.4 µM, 24 hr), with arrows indicating chromosomal breakages.
B. Overall chromosome gaps and breaks per cell in HCT116 cells expressing shRNAs against Mre11, CtIP or control MKO, before and after Aph treatment.
C. FISH analysis of HCT116 cells using probes against FRA3B (top panels) or FRA16D (bottom panels), with DAPI staining (left) and FISH probe hybridization (right). Red and yellow arrows indicate broken and normal chromosomes, respectively.
D–E. Frequency of CFS expression at FRA3B or FRA16D in HCT116 cells expressing Mre11, CtIP or control MKO shRNAs (in D) or expressing HA-Mre11 (WT or H129N), with endogenous Mre11 silenced (in E, left), or HA-CtIP (WT or indicated mutants), with endogenous CtIP silenced (in E, right), before and after Aph treatment.
F. Proposed model for HR-mediated DSB repair and replication restart at collapsed replication forks due to structure-forming DNA (such as hairpins). See Discussion.
See also Figure S4.)
In all experiments, error bars represent standard deviation (s.d.) of three independent experiments.
Discussion
DNA sequences with unusual secondary structures are associated with GCRs leading to cancer and other human diseases (Chen et al., 2010). We established novel EGFP-based HR repair systems in mammalian cells, allowing for a mechanistic study of the genome instability caused by structure-forming DNA sequences derived from CFSs and Alu-IRs in the chromosomal context. We demonstrated that CtIP possesses an endonuclease activity which is dispensable for end resection and HR at general DSBs, but is specifically required for repairing DSBs containing DNA secondary structures at the ends.
Structure-forming DNA sequences contribute to CFS fragility
CFSs expand over large chromosomal regions and are prone to breakage upon replication stress (Glover, 2006; Glover et al., 2005). Recent studies suggest that paucity of replication initiation is one cause for fragility of CFSs (Debatisse et al., 2012; Letessier et al., 2011; Palumbo et al., 2010). In this study, we demonstrated that the small AT-rich structure-forming DNA sequence fragments (0.33kb) derived from CFS FRA16D, when inserted into a new chromosomal locus, sufficiently induces mitotic recombination both spontaneously and under replication stress, and these AT-rich CFS-associated sequences cause plasmid instability in mammalian cells. These data support the notion that CFSs contain core sequences which tend to form DNA secondary structures during replication, inducing DSBs and DSB-associated repair (Figure 4F). Thus, fragility of CFSs is likely caused by the combination of perturbation of replication fork progression by unusual DNA sequences and inherent origin paucity in CFSs. Indeed, slower replication rates at AT-rich sequences and insufficient origin firing were both observed at FRA16C (Ozeri-Galai et al., 2011).
CtIP possesses an endonuclease activity with a conserved repair function
We showed that human CtIP exhibits an endonuclease activity to cleave 5’ ssDNA at the base of dsDNA of hairpins or Y-shaped branched structures, reminiscent of a common feature of the DNA structures that stall replication (Figure 4F). Since inactivation of CtIP or Mre11 leads to more rather than fewer DSBs compared to control cells, Mre11 and CtIP, unlike Mus81, are probably not involved in generating DSBs at stalled replication forks (Hanada et al., 2007). Upon fork collapse, replication can be reassumed by DSB repair-coupled restart (Lambert et al., 2007), or be completed by new origin firing from adjacent origins, which generates two-ended DSBs when new forks encounter collapsed forks, followed by HR-mediated DSB repair (Petermann et al., 2010). CtIP and Mre11 may use their nuclease activities to process DNA secondary structures after DSB formation, so that a clean 3’ end can be generated for strand invasion (Figure 4F). The specificity of CtIP endonuclease for 5’ ssDNA of hairpins is congruent with its potential role in removing structured DNA at 3’ ends to facilitate HR.
By analyzing different CtIP fragments, we found that the N-terminal part of CtIP is important for its endonuclease activity, which is consistent with the finding from Paull’s group using oxidative cleavage analysis [(Makharashvili et al., 2014), accompanying paper]. The N181 and R185 residues of a conserved NxxxR/K motif at the N-terminus of CtIP and the putative metal binding residues E267 and E268, conserved in higher eukaryotes (Figure S4D), are important for CtIP endonuclease activities. Mutating these residues in human CtIP and the CtIP N181/R185-corresponding residues N123 and R127 in Sae2 resulted in sensitivity to CPT and defects in Alu-IRs-induced mitotic recombination, supporting a conserved role of CtIP/Sae2 endonuclease activity in DSB repair. Furthermore, deleting the N-terminal 44 residues, which are essential for CtIP dimerization (Wang et al., 2012), does not alter CtIP biochemical activity, thus forming a CtIP dimer is not required for its nuclease function.
The endonuclease activity of CtIP is independent of its end resection function
By using the CtIP endonuclease mutants N181A/R185A and E267A/E268A, we demonstrated that CtIP endonuclease activity is not required for end resection and HR at “clean” DSBs generated by I-SceI, but is needed for processing DSB ends at CFS-derived sequences with secondary structures. This suggests that CtIP possesses two separate functions: to promote general end resection and to carry out end processing of structure-forming DSB ends.
To remove DNA secondary structures at DSB ends, CtIP utilizes its endonuclease activity and may function cooperatively with Mre11. It was described that Mre11 enlarges the ssDNA gap at hairpin-capped ends to generate substrates favorable for Sae2 cleavage (Lengsfeld et al., 2007). It is also possible that Mre11 and CtIP exhibit different substrate and cleavage specificities, and thus both are required to generate a “clean” 3’ ssDNA tail for strand invasion. Biochemical studies show that Mre11 is more favored to cleave at the tip of hairpin loops, while CtIP/Sae2 cleaves at ssDNA adjacent to hairpins [(Lengsfeld et al., 2007; Paull and Gellert, 1999), this study and (Makharashvili et al., 2014), accompanying paper]. In fission yeast, Rad32 (Mre11 homologue) and Ctp1 (CtIP homologue) have distinct requirements to remove covalently bound Top1 and Top2 from DNA (Hartsuiker et al., 2009). Of note, CFS-formed secondary structures often contain multiple hairpin loops with complex secondary structures (Zlotorynski et al., 2003), and different enzymatic activities may be required for efficient removal of these structures. In support of CtIP and Mre11 acting at the same genetic step to process structure-forming DNA at DSB ends, inactivation of CtIP endonuclease activity does not further reduce HR function at Flex1 in Mre11-deficient cells (Figure S4E). As for end resection, CtIP itself may not directly cleave DNA, but may play a role in facilitating the MRN biochemical activity and promoting BLM/Exo1 recruitment to DSBs (Eid et al., 2010; Nicolette et al., 2010; Wang et al., 2013).
The Role of Mre11 and CtIP in CFS protection
It was described that the BRCA1/Rad51-dependent HR pathway is involved in CFS protection (Arlt et al., 2006). In this study, we showed that not only the HR activity of MRN and CtIP, but also HR-dispensable enzymatic activities required for processing DNA secondary structures are important for CFS protection. Thus, these studies reveal new roles and the underlying mechanisms of MRN and CtIP in the maintenance of CFS stability, which will help to understand the biology of CFS protection, and also the maintenance of genome stability at other structure-forming DNA sequences associated with GCRs in mammalian cells.
Experimental Procedures
Detailed descriptions of the HR-Flex, HR-Luc, HR-DR-Alu and HR-IR-Alu reporter substrates, the SSA reporter substrate, analysis of DSB repair assays, protein purification, biochemical nuclease reaction assays, plasmid stability assay, gel shift assay, and in vitro cleavage assay are provided in the Supplemental Experimental Procedures. Plasmid construction, cell lysis, immunoblotting, immunostaining, metaphase chromosomal analysis, fluorescence in situ hybridization (FISH), yeast recombination assays, chromatin immunoprecipitation (ChIP), and laser microirradiation were described previously (Casper et al., 2002; Chen et al., 2008; Lengsfeld et al., 2007; Wang et al., 2012; Wilke et al., 1996), with additional details in the Supplemental Experimental Procedures.
Supplementary Material
Highlights.
AT-rich CFS sequences and inverted Alu repeats induce mitotic recombination.
Human CtIP exhibits an endonuclease activity that is dispensable for end resection.
The CtIP endonuclease activity is required for repairing structure-forming DSBs.
The nuclease activities of CtIP and Mre11 are important for CFS protection.
Acknowledgements
We thank Drs. Catherine Freudenreich (Tufts University), Kirill Lobachev (Georgia Tech), Sang Eun Lee (University of Texas Health Science Center at San Antonio), Tanya Paull (University of Texas at Austin), Andras Nagy (University of Toronto), Michele Calos (Stanford University School of Medicine), Thomas W. Glover (University of Michigan Medical School), Bing Xia (Rutgers University) and Eros Lazzerini Denchi (The Scripps Research Institute) for kindly providing valuable reagents. This work was supported by NIH Grants CA102361, GM080677, CA140972, CA102361-07S1 and CA140972-03S1 to X.W.; an NIH grant R01CA073764 to B.H.S.; and the Beckman Laser Institute Foundation to M.W.B.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no conflicts of interest.
References
- Abeysinghe SS, Stenson PD, Krawczak M, Cooper DN. Gross Rearrangement Breakpoint Database (GRaBD) Human mutation. 2004;23:219–221. doi: 10.1002/humu.20006. [DOI] [PubMed] [Google Scholar]
- Arlt MF, Durkin SG, Ragland RL, Glover TW. Common fragile sites as targets for chromosome rearrangements. DNA Repair (Amst) 2006;5:1126–1135. doi: 10.1016/j.dnarep.2006.05.010. [DOI] [PubMed] [Google Scholar]
- Belteki G, Gertsenstein M, Ow DW, Nagy A. Site-specific cassette exchange and germline transmission with mouse ES cells expressing phiC31 integrase. Nature biotechnology. 2003;21:321–324. doi: 10.1038/nbt787. [DOI] [PubMed] [Google Scholar]
- Casper AM, Nghiem P, Arlt MF, Glover TW. ATR regulates fragile site stability. Cell. 2002;111:779–789. doi: 10.1016/s0092-8674(02)01113-3. [DOI] [PubMed] [Google Scholar]
- Chen JM, Cooper DN, Ferec C, Kehrer-Sawatzki H, Patrinos GP. Genomic rearrangements in inherited disease and cancer. Seminars in cancer biology. 2010;20:222–233. doi: 10.1016/j.semcancer.2010.05.007. [DOI] [PubMed] [Google Scholar]
- Chen L, Nievera CJ, Lee AY, Wu S. Cell cycle-dependent complex formation of BRCA1.CtIP.MRN is important for DNA double-strand break repair. J Biol Chem. 2008;283:7713–7720. doi: 10.1074/jbc.M710245200. [DOI] [PubMed] [Google Scholar]
- D'Amours D, Jackson SP. The Mre11 complex: at the crossroads of dna repair and checkpoint signalling. Nat Rev Mol Cell Biol. 2002;3:317–327. doi: 10.1038/nrm805. [DOI] [PubMed] [Google Scholar]
- Debatisse M, Le Tallec B, Letessier A, Dutrillaux B, Brison O. Common fragile sites: mechanisms of instability revisited. Trends Genet. 2012;28:22–32. doi: 10.1016/j.tig.2011.10.003. [DOI] [PubMed] [Google Scholar]
- Eid W, Steger M, El-Shemerly M, Ferretti LP, Pena-Diaz J, Konig C, Valtorta E, Sartori AA, Ferrari S. DNA end resection by CtIP and exonuclease 1 prevents genomic instability. EMBO Rep. 2010;11:962–968. doi: 10.1038/embor.2010.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnis M, Dayan S, Hobson L, Chenevix-Trench G, Friend K, Ried K, Venter D, Woollatt E, Baker E, Richards RI. Common chromosomal fragile site FRA16D mutation in cancer cells. Human molecular genetics. 2005;14:1341–1349. doi: 10.1093/hmg/ddi144. [DOI] [PubMed] [Google Scholar]
- Glover TW. Common fragile sites. Cancer Lett. 2006;232:4–12. doi: 10.1016/j.canlet.2005.08.032. [DOI] [PubMed] [Google Scholar]
- Glover TW, Arlt MF, Casper AM, Durkin SG. Mechanisms of common fragile site instability. Human molecular genetics. 2005;14(2):R197–R205. doi: 10.1093/hmg/ddi265. [DOI] [PubMed] [Google Scholar]
- Hanada K, Budzowska M, Davies SL, van Drunen E, Onizawa H, Beverloo HB, Maas A, Essers J, Hickson ID, Kanaar R. The structure-specific endonuclease Mus81 contributes to replication restart by generating double-strand DNA breaks. Nat Struct Mol Biol. 2007;14:1096–1104. doi: 10.1038/nsmb1313. [DOI] [PubMed] [Google Scholar]
- Hartsuiker E, Neale MJ, Carr AM. Distinct requirements for the Rad32(Mre11) nuclease and Ctp1(CtIP) in the removal of covalently bound topoisomerase I and II from DNA. Mol Cell. 2009;33:117–123. doi: 10.1016/j.molcel.2008.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huertas P, Jackson SP. Human CtIP mediates cell cycle control of DNA end resection and double strand break repair. J Biol Chem. 2009;284:9558–9565. doi: 10.1074/jbc.M808906200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jazayeri A, Falck J, Lukas C, Bartek J, Smith GC, Lukas J, Jackson SP. ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat Cell Biol. 2006;8:37–45. doi: 10.1038/ncb1337. [DOI] [PubMed] [Google Scholar]
- Kasparek TR, Humphrey TC. DNA double-strand break repair pathways, chromosomal rearrangements and cancer. Seminars in cell & developmental biology. 2011;22:886–897. doi: 10.1016/j.semcdb.2011.10.007. [DOI] [PubMed] [Google Scholar]
- Kato T, Kurahashi H, Emanuel BS. Chromosomal translocations and palindromic AT-rich repeats. Curr Opin Genet Dev. 2012;22:221–228. doi: 10.1016/j.gde.2012.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney S. Mechanism and control of meiotic recombination initiation. Current topics in developmental biology. 2001;52:1–53. doi: 10.1016/s0070-2153(01)52008-6. [DOI] [PubMed] [Google Scholar]
- Lambert S, Froget B, Carr AM. Arrested replication fork processing: interplay between checkpoints and recombination. DNA Repair (Amst) 2007;6:1042–1061. doi: 10.1016/j.dnarep.2007.02.024. [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Lengsfeld BM, Rattray AJ, Bhaskara V, Ghirlando R, Paull TT. Sae2 is an endonuclease that processes hairpin DNA cooperatively with the Mre11/Rad50/Xrs2 complex. Mol Cell. 2007;28:638–651. doi: 10.1016/j.molcel.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letessier A, Millot GA, Koundrioukoff S, Lachages AM, Vogt N, Hansen RS, Malfoy B, Brison O, Debatisse M. Cell-type-specific replication initiation programs set fragility of the FRA3B fragile site. Nature. 2011;470:120–123. doi: 10.1038/nature09745. [DOI] [PubMed] [Google Scholar]
- Lilley DM. The inverted repeat as a recognizable structural feature in supercoiled DNA molecules. Proc Natl Acad Sci U S A. 1980;77:6468–6472. doi: 10.1073/pnas.77.11.6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobachev KS, Gordenin DA, Resnick MA. The Mre11 complex is required for repair of hairpin-capped double-strand breaks and prevention of chromosome rearrangements. Cell. 2002;108:183–193. doi: 10.1016/s0092-8674(02)00614-1. [DOI] [PubMed] [Google Scholar]
- Makharashvili N, Tubbs AT, Yang S, Wang H, Barton O, Zhou Y, Deshpande RA, Lee J, Lobrich M, Sleckman BP, et al. Catalytic and non-catalytic roles of the CtIP endonuclease in double-strand break end resection. Mol Cell. 2014 doi: 10.1016/j.molcel.2014.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolette ML, Lee K, Guo Z, Rani M, Chow JM, Lee SE, Paull TT. Mre11-Rad50-Xrs2 and Sae2 promote 5' strand resection of DNA double-strand breaks. Nat Struct Mol Biol. 2010;17:1478–1485. doi: 10.1038/nsmb.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozeri-Galai E, Lebofsky R, Rahat A, Bester AC, Bensimon A, Kerem B. Failure of origin activation in response to fork stalling leads to chromosomal instability at fragile sites. Mol Cell. 2011;43:122–131. doi: 10.1016/j.molcel.2011.05.019. [DOI] [PubMed] [Google Scholar]
- Palumbo E, Matricardi L, Tosoni E, Bensimon A, Russo A. Replication dynamics at common fragile site FRA6E. Chromosoma. 2010;119:575–587. doi: 10.1007/s00412-010-0279-4. [DOI] [PubMed] [Google Scholar]
- Panayotatos N, Wells RD. Cruciform structures in supercoiled DNA. Nature. 1981;289:466–470. doi: 10.1038/289466a0. [DOI] [PubMed] [Google Scholar]
- Paull TT, Gellert M. Nbs1 potentiates ATP-driven DNA unwinding and endonuclease cleavage by the Mre11/Rad50 complex. Genes Dev. 1999;13:1276–1288. doi: 10.1101/gad.13.10.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petermann E, Orta ML, Issaeva N, Schultz N, Helleday T. Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Mol Cell. 2010;37:492–502. doi: 10.1016/j.molcel.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattray AJ, Shafer BK, Neelam B, Strathern JN. A mechanism of palindromic gene amplification in Saccharomyces cerevisiae. Genes Dev. 2005;19:1390–1399. doi: 10.1101/gad.1315805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves R, Gorman CM, Howard B. Minichromosome assembly of nonintegrated plasmid DNA transfected into mammalian cells. Nucleic Acids Res. 1985;13:3599–3615. doi: 10.1093/nar/13.10.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartori AA, Lukas C, Coates J, Mistrik M, Fu S, Bartek J, Baer R, Lukas J, Jackson SP. Human CtIP promotes DNA end resection. Nature. 2007;450:509–514. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B, Nolan JP, Sklar LA, Park MS. Functional analysis of point mutations in human flap endonuclease-1 active site. Nucleic Acids Res. 1997;25:3332–3338. doi: 10.1093/nar/25.16.3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Do H, Li Y, Chung WH, Tomasz M, de Winter JP, Xia B, Elledge SJ, Wang W, Li L. Recruitment of fanconi anemia and breast cancer proteins to DNA damage sites is differentially governed by replication. Mol Cell. 2009;35:716–723. doi: 10.1016/j.molcel.2009.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H, Bergstrom DA, Yao MC, Tapscott SJ. Widespread and nonrandom distribution of DNA palindromes in cancer cells provides a structural platform for subsequent gene amplification. Nat Genet. 2005;37:320–327. doi: 10.1038/ng1515. [DOI] [PubMed] [Google Scholar]
- Thorpe HM, Wilson SE, Smith MC. Control of directionality in the site-specific recombination system of the Streptomyces phage phiC31. Mol Microbiol. 2000;38:232–241. doi: 10.1046/j.1365-2958.2000.02142.x. [DOI] [PubMed] [Google Scholar]
- Voineagu I, Narayanan V, Lobachev KS, Mirkin SM. Replication stalling at unstable inverted repeats: interplay between DNA hairpins and fork stabilizing proteins. Proc Natl Acad Sci U S A. 2008;105:9936–9941. doi: 10.1073/pnas.0804510105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Shao Z, Shi LZ, Hwang PY, Truong LN, Berns MW, Chen DJ, Wu X. CtIP Protein Dimerization Is Critical for Its Recruitment to Chromosomal DNA Double-stranded Breaks. J Biol Chem. 2012;287:21471–21480. doi: 10.1074/jbc.M112.355354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Shi LZ, Wong CC, Han X, Hwang PY, Truong LN, Zhu Q, Shao Z, Chen DJ, Berns MW, et al. The interaction of CtIP and Nbs1 connects CDK and ATM to regulate HR-mediated double-strand break repair. PLoS Genet. 2013;9:e1003277. doi: 10.1371/journal.pgen.1003277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilke CM, Hall BK, Hoge A, Paradee W, Smith DI, Glover TW. FRA3B extends over a broad region and contains a spontaneous HPV16 integration site: direct evidence for the coincidence of viral integration sites and fragile sites. Human molecular genetics. 1996;5:187–195. doi: 10.1093/hmg/5.2.187. [DOI] [PubMed] [Google Scholar]
- Yu X, Chen J. DNA damage-induced cell cycle checkpoint control requires CtIP, a phosphorylation-dependent binding partner of BRCA1 C-terminal domains. Mol Cell Biol. 2004;24:9478–9486. doi: 10.1128/MCB.24.21.9478-9486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Freudenreich CH. An AT-rich sequence in human common fragile site FRA16D causes fork stalling and chromosome breakage in S. cerevisiae. Mol Cell. 2007;27:367–379. doi: 10.1016/j.molcel.2007.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlotorynski E, Rahat A, Skaug J, Ben-Porat N, Ozeri E, Hershberg R, Levi A, Scherer SW, Margalit H, Kerem B. Molecular basis for expression of common and rare fragile sites. Mol Cell Biol. 2003;23:7143–7151. doi: 10.1128/MCB.23.20.7143-7151.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.