Abstract
Objective
Endothelial cells are central to the initiation of atherosclerosis, yet there has been limited success in studying their gene expression in the mouse aorta. To address this, we developed a method for determining the global transcriptional changes that occur in the mouse endothelium in response to atherogenic conditions and applied it to investigate inflammatory stimuli.
Approach and Results
We characterized a method for the isolation of endothelial cell RNA with high purity directly from mouse aortas and adapted this method to allow for the treatment of aortas ex vivo before RNA collection. Expression array analysis was performed on endothelial cell RNA isolated from control and hyperlipidemic prelesion mouse aortas, and 797 differentially expressed genes were identified. We also examined the effect of additional atherogenic conditions on endothelial gene expression, including ex vivo treatment with inflammatory stimuli, acute hyperlipidemia, and age. Of the 14 most highly differentially expressed genes in endothelium from prelesion aortas, 8 were also perturbed significantly by ≥1 atherogenic conditions: 2610019E17Rik, Abca1, H2-Ab1, H2-D1, Pf4, Ppbp, Pvrl2, and Tnnt2.
Conclusions
We demonstrated that RNA can be isolated from mouse aortic endothelial cells after in vivo and ex vivo treatments of the murine vessel wall. We applied these methods to identify a group of genes, many of which have not been described previously as having a direct role in atherosclerosis, that were highly regulated by atherogenic stimuli and may play a role in early atherogenesis.
Keywords: atherosclerosis, endothelium, genetics
The cells that compose a healthy artery, particularly the vascular endothelial cells (ECs) that line the lumen, are critical to the development of atherosclerotic plaques.1,2 The activation of ECs leads to the recruitment of leukocytes, such as monocytes, and the increased permeability of the endothelial layer permits the entry of low-density lipoproteins (LDL) to the intima, where they are taken up by macrophages to form foam cells.3 As atherosclerosis progresses, smooth muscle cells (SMCs) that reside in the media of the artery migrate to the developing lesion, where they proliferate and join the accumulating foam cells and cellular debris in the growing atheroma. Although the general events that occur in the vessel wall during the transition of a healthy artery to an atherogenic vessel have been established, many of the specific molecular changes that take place during disease progression remain unclear. Determining the precise genetic perturbations that occur in the endothelium at the initiation of atherogenesis, before lipid accumulation and leukocyte infiltration, would be vital in understanding the events that underlie atherosclerosis initiation.
Microarray analysis of ECs from the carotid arteries of human atherosclerosis samples provides valuable insight into the gene expression differences present in early and advanced lesions.4 However, these ECs are dissected from vessels that have already developed atherosclerosis, making it difficult to separate the transcriptional profile of the initial response to atherogenic conditions from downstream perturbations. Genome-wide transcriptional studies of in vivo ECs have been performed in healthy swine, comparing atherosusceptible disturbed flow regions of the vessel wall with atheroprotective undisturbed laminar flow regions to identify genetic pathways that differ between these conditions.5,6 These studies have shown that the atherosusceptible endothelium exhibits elevated levels of proinflammatory and endoplasmic reticulum stress genes, whereas the atheroprotective endothelium has elevated levels of antioxidant genes. Similar studies on ECs isolated from swine maintained on a brief hypercholesterolemic diet supported these findings and reported an upregulation of ABCA1 in response to diet across all regions tested.7 Although swine is a useful model of human atherosclerosis and its size allows for successful in vivo endothelial expression studies, there are significant limitations for their use in genetic studies. Although the small size of the mouse aorta does not allow for the spatial resolution achieved in swine, inbred mouse strains allow for genetic manipulation, biological replicates, and more high-throughput approaches.8,9 Furthermore, the atherosclerotic disease pathology of an inbred mouse strain has been studied in detail,10,11 and the healthy and prelesional disease states can be clearly identified, predicted, and studied.
The mouse is the most commonly used animal model of human atherosclerosis,9,12 yet no studies have been published that examine the global transcriptional changes in the endothelium during atherosclerosis. This may be partially because of the difficulty in obtaining pure, differentiated cell cultures of mouse aortic ECs (MAECs), as cell culture conditions require growing cells away from their native environment and may lead to rapid dedifferentiation.13 The study of cells isolated directly from the aorta as opposed to cell culture offers significant advantages: the data gathered would closely represent the in vivo physical state of these cells because the cell–cell interactions remain intact up until the moment of cell isolation, and it would be possible to assess the transcriptional response of the cells to in vivo conditions, such as hyperlipidemia. However, commonly used cell-isolation techniques, such as fluorescence-activated cell sorting and laser capture microdissection, cannot be used to successfully isolate MAECs in a high-throughput manner because of the tendency of ECs to adhere to one another, the irregular shape of this cell type, and the curvature of the mouse aorta.14 Previous studies on preparations described as mouse aortic intimal cells have explored differential gene expression in disturbed compared with laminar flow regions.15,16 Studies have also examined the effect of hyperlipidemia on the expression of previously identified candidate genes in MAECs,17–20 but a whole-genome hypothesis-generating study has not yet been reported for this cell type.
We characterized cells obtained by a method to isolate the intimal layer from disease-free mouse aortas15 as ECs, and adapted this method to allow for the treatment of these cells with proinflammatory agents ex vivo. These methods were then used to compare the gene expression profiles of MAECs isolated from aortas predisposed to atherosclerosis as compared with those from healthy vessels to determine the major transcriptionally perturbed genes. We used the inbred strain C57BL/6, or BL6, fed a chow diet for these studies; as BL6 is both the most commonly used inbred strain and one of the most atherosusceptible, 9,12 it is valuable to analyze the genetic perturbations that occur in the endothelium during atherosclerosis initiation of this widely studied and useful strain. Wild-type BL6 mice fed a normal chow diet do not develop atherosclerosis, whereas BL6 mice with an apolipoprotein-deficient (ApoE−/−) mutation on a chow diet are hyperlipidemic and do develop atherosclerosis. By comparing the gene expression profiles between the healthy wild-type and atherosclerotic ApoE−/− mice, we were able to assess the transcriptional differences that occur during the earliest stages of atherosclerosis initiation. We further characterized the top differentially expressed genes in the endothelium of the prelesion aorta by observing their transcriptional activity in the following atherogenic conditions: proinflammatory stimuli, age, and acute hyperlipidemia. In addition, as the roles of inflammatory mediators in atherosclerosis initiation remain unclear, we applied our methods to provide a global picture of the genetic perturbations caused by 3 inflammatory agents: 1-palmitoyl-2-arachidonoyl-sn-glycero-3-phosphocholine (oxPAPC), oxidized LDL (oxLDL), and the more general inflammatory molecule lipopolysaccharide (LPS). We present a comprehensive study of genes that are differentially regulated between healthy and atherogenic mice in discrete MAEC cell preparations and provide valuable insight into the genetics of atherosclerosis initiation.
Materials and Methods
Materials and Methods are available in the online-only Supplement.
Results
Cells Isolated From the Intimal Layer of Healthy Mouse Aortas Are Highly Enriched ECs
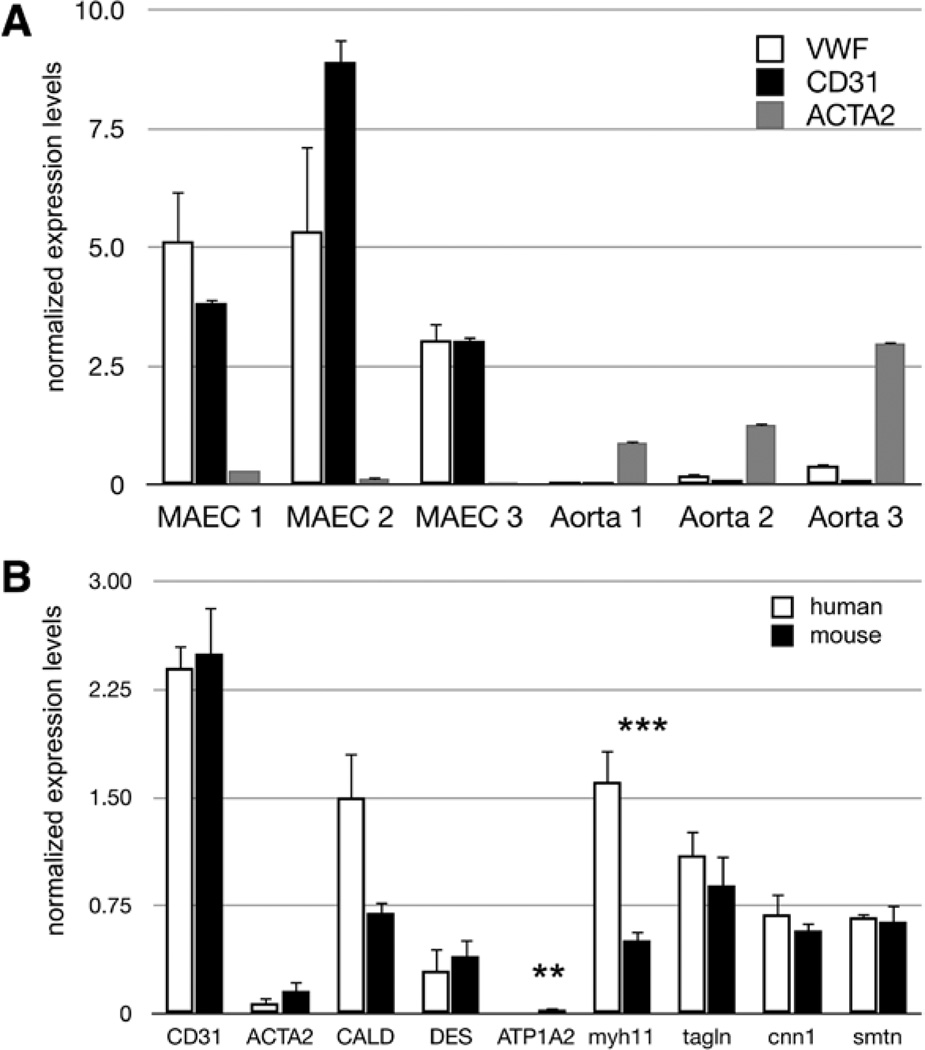
Transcript levels of the EC-specific markers von Willebrand factor (vWF), platelet/endothelial cell adhesion molecule 1 (CD31), cadherin 5 (VE Cad), and endothelial nitric oxide synthase (eNOS) were enriched significantly in healthy aortic intimal cell preparations from wild-type BL6 mice compared with RNA extracted from whole mouse aortas (Figure 1A; Table I in the online-only Data Supplement), indicating that the intimal preparations are predominantly ECs. Expression levels of macrophage, leukocyte, and adipose markers were below detection thresholds, indicating the lack of these cell types in our preparations. Previous studies have reported ≈215 CD68+ and 8 CD3+ cells in the entire ascending aorta of the BL6 mouse strain at the ages of 3 to 6 months and an EC density of ≈2500 ECs per millimeter squared in the murine aorta.15 As the aortic arch has a surface area ≥9 mm,2 leukocytes would represent <1% of >22 500 cells in this region; therefore, we conclude that our preparations are >99% pure ECs. Studies in swine report 0.72% presence of leukocytes in preparations taken from the porcine aortic arch endothelium using a scraping method for cell isolation, further indicating that the presence of leukocytes adherent to the EC layer is <1%.6
Figure 1.
Cells isolated from the intima of healthy mouse aortas are endothelial cells (ECs). A, Graph represents relative expression of RNA normalized to the housekeeping gene B2M. RNA was isolated from mouse aortic endothelial cell (MAEC) preparations and whole aortas, from 3 animals each, and expression values are shown for individual mice. Normalized gene expression for the 2 EC markers von Willebrand factor (vWF) and platelet/endothelial cell adhesion molecule 1 (CD31) show significant enrichment in the intimal preparations and are nearly undetectable in the whole aorta samples, whereas the smooth muscle cell (SMC) protein marker ACTA2 is expressed to a much higher level in the whole aorta samples. Error bars show SD for 2 technical replicates. B, Relative transcription levels of 8 different smooth muscle cell protein markers and the EC marker CD31 in MAECs and human aortic endothelial cell (HAEC) cultures as determined by reverse transcription quantitative polymerase chain reaction, normalized to the housekeeping gene B2M. mRNA transcript levels for all SMC protein markers are present in the HAEC cultures, which have been confirmed as SMC free using protein markers and serve as a positive control for our purposes. SMC protein transcript levels for the mouse homologs of these genes show similar expression levels in our intimal preparations, indicating a confidence level of EC purity equal to that for HAEC cultures. Error bars show SE for 6 biological replicates for each sample set. t test: **P <0.01; and ***P <0.005.
RNA expression levels of the SMC protein marker, Acta2, were significantly lower in the intimal preparations compared with whole aortas (Figure 1A). Because the amount of cells obtained does not yield sufficient amounts of protein for Western blots, we were unable to assess directly protein expression of the cell preparations. However, we were able to determine indirectly the protein purity levels through comparison with human aortic EC (HAEC) cultures (Figure 1B). These HAEC cultures have been characterized previously as at ≥99% pure as judged by endothelial protein markers and do not show expression of smooth muscle proteins.21 We compared the mRNA expression levels of the EC marker CD31 and 8 different SMC protein markers in our MAEC preparations with those present in HAECs. We found that CD31 levels were equivalent between mouse and human samples, and all SMC protein markers tested are expressed as mRNA transcripts in HAEC cultures as well. Thus, ECs express several transcripts characteristic of SMCs, but these are not translated into proteins, a finding that has been reported in previous studies on ECs.22 These data indicate that cells isolated from the intimal layer of healthy mouse aortas are highly enriched for ECs and can be used as such with the same level of confidence as HAEC cultures.
The RNA obtained consistently represents cells from both high and low areas of shear stress in the aortic arch. We chose to include the entire aortic arch region primarily for ease of reproducibility across the mice used for these studies and to collect enough RNA of high quality. Although separating different spatial regions on a larger animal such as swine is feasible, this is more challenging in a smaller animal such as the mouse, and day-to-day variability could result in identification of false-positive differential expression based on aortic region and not the environmental effects studied.
Amplification of RNA Isolated From MAECs Does Not Affect Relative Abundance of Transcripts
As the MAEC isolation process yielded ≈3 ng of RNA per mouse, we wanted to identify a RNA amplification procedure that would not alter the relative expression levels of transcript in the samples. Although the manufacturer and additional independent research laboratories have demonstrated that the single primer isothermal amplification method used by NuGEN (San Carlos, CA) yields expression data that are concordant with native RNA expression,23,24 we confirmed this in our specific data set using individual genes relevant to our studies. We tested 4 samples: 2 that had been treated with oxPAPC before isolation and 2 control samples incubated in media only. After isolation, half the RNA was converted to cDNA and the other half was amplified using the NuGEN RNA amplification kit. Both the cDNA and the amplified RNA product were assayed for transcript levels of vWF, heme oxygenase 1 (Hmox1), vascular cell adhesion molecule 1 (Vcam1). Both the raw and the normalized reverse transcription quantitative polymerase chain reaction (PCR) values of the native cDNA and amplified RNA products correspond closely, indicating that the expression data acquired from amplified RNA will provide relative expression levels consistent with the native RNA sample (Figure II in the online-only Data Supplement).
MAECs From Prelesion Aortas Show a Differential Expression Pattern Compared With MAECs From Healthy Aortas
To understand the changes in EC gene expression in the initiation and later stages of atherosclerosis, we used 4-week-old BL6 wild-type mice as on a chow diet as healthy controls and 4-week-old hyperlipidemic BL6 ApoE−/− mice on a chow diet as prelesioned mice. BL6 ApoE−/− mice at 24 weeks on a chow diet develop lesions, whereas BL6 wild-type mice at 24 weeks on a chow diet do not develop lesions (Figure III in the online-only Data Supplement).25 At 4 weeks, BL6 ApoE−/− mice on a chow diet have not yet developed lipid deposits observable by oil-red-O staining (Figure IIIA in the online-only Data Supplement), which is why we term these ApoE−/− mice pre-lesioned aortas as opposed to the healthy BL6 wild-type mice that will not develop lesions under these experimental conditions. High-resolution, freeze-etch electron microscopy indicates that monocytes have not yet adhered to the endothelium in a 4-week-old BL6 ApoE−/− chow-fed mouse because these cells are not observable until 5 weeks.26 We did not detect macrophage transcripts in unamplified RNA collected from wildtype mice (Table I in the online-only Data Supplement) and do not see a difference in the transcription levels of the macrophage markers CD68 and Msr1, which are barely detectable even in amplified RNA, in BL6 wild-type compared with BL6 ApoE−/− 4-week-old mice (Figure IV in the online-only Data Supplement). We wanted to study the intima at a time point before lipid accumulation and monocyte adhesion and determined the 4-week time point to be ideal based on our observations and previous reports.
MAEC RNA was isolated, amplified, and subjected to gene expression microarray analysis. A total of 797 genes were differentially expressed between ECs isolated from the 4-week-old healthy and prelesion aortas at a false-discovery rate of 10% (Figure 2; Table 1), and 32 genes were differentially expressed at the stringent standards of 2-fold change at 5% false-discovery rate (Table 1). Functional annotation analysis of the 797 differentially expressed genes (Table II in the online-only Data Supplement) identified the plasma membrane and immune response–related categories as the most enriched.
Figure 2.
Treatment of whole aortas with proinflammatory factors before mouse aortic endothelial cell (MAEC) isolation effectively perturbs gene expression of inflammatory markers without altering endothelial cell (EC) marker levels. Relative transcription levels of EC marker genes and select stress–response genes in MAECs isolated after treatment with Dulbecco’s Modified Eagle’s Medium (DMEM) cell culture medium containing various additives, as determined by reverse transcription quantitative polymerase chain reaction and normalized to B2M. A, Expression levels of the EC markers von Willebrand factor (vWF), platelet/endothelial cell adhesion molecule 1 (CD31) in native MAEC preparations compared with MAECs isolated from aortas incubated in DMEM for 4 hours. No significant expression differences in EC cell marker expression after incubation in cell culture media were observed as determined by a t test. Error bars show SE from 4 biological replicates for each condition. B, EC markers and heme oxygenase 1 (Hmox1) in MAECs isolated from aortas incubated in DMEM compared with DMEM containing 1-palmitoyl-2-arachidonoyl-sn-glycero-3-phosphocholine (oxPAPC) for 4 hours. Although the transcription levels of EC cell markers remained the same, there was a strong induction of Hmox1 in response to treatment with oxidized phospholipid. Error bars show SE from 3 biological replicates. t test: ***P <0.001. C, Expression levels of EC markers Hmox1 in MAECs isolated from aortas incubated in DMEM vs DMEM containing oxidized low-density lipoprotein (oxLDL) for 4 hours. We see an induction of Hmox1 but not EC markers in response to treatment. Error bars show SE from 3 biological replicates. t test: *P <0.05. D, Transcription levels of EC markers, Hmox1, and vascular cell adhesion molecule 1 (Vcam1) in MAECs isolated from aortas incubated in DMEM vs DMEM containing lipopolysaccharide (LPS) for 4 hours. No response was observed in Hmox1 expression, but Vcam1 transcript levels are induced as expected. Error bars show SE from 2 biological replicates. t test: *P <0.05.
Table 1.
Number of Genes Differentially Expressed in Endothelial Cells Isolated From Healthy Compared With Prelesion Aortas
| FDR 5% | FDR 10% | |
|---|---|---|
| >1.2-Fold change | 316 | 797 |
| >1.5-Fold change | 175 | 341 |
| >2-Fold change | 32 | 51 |
Fold changes of 1.2, 1.5, and >2 and FDR cut-offs of 5% and 10% are shown. FDR indicates false-discovery rate.
MAECs Isolated From Treated Whole Aortas Respond to Activation With Proinflammatory Agents
To study the response of MAECs to proatherogenic stimuli in the context of the vessel wall, we applied a protocol used for treating HAEC cultures to whole mouse aortas27 (Figure I in the online-only Data Supplement). Neither incubation of the aortas in media before EC isolation affects significantly the expression levels of EC markers (Figure 3A) nor did treatment with the oxidized phospholipid oxPAPC, oxLDL, or LPS (Figure 3B–3D). We next determined whether endothelium isolated from treated vessels displayed gene regulation previously observed in HAEC cell cultures. We found that expression of Hmox-1, a gene induced during oxidative stress and the inflammatory response in HAECs, is upregulated ≈10-fold in response to oxPAPC (Figure 3B), a response also seen in HAEC cultures.28 A similar response was seen after treatment with oxLDL (Figure 3C). LPS treatment (Figure 3D) showed increased expression of proinflammatory marker Vcam1 but no induction of Hmox-1, also consistent with prior observations in HAECs.29 Because gene regulation was consistent with past studies, we proceeded with the expression array analysis.
Figure 3.
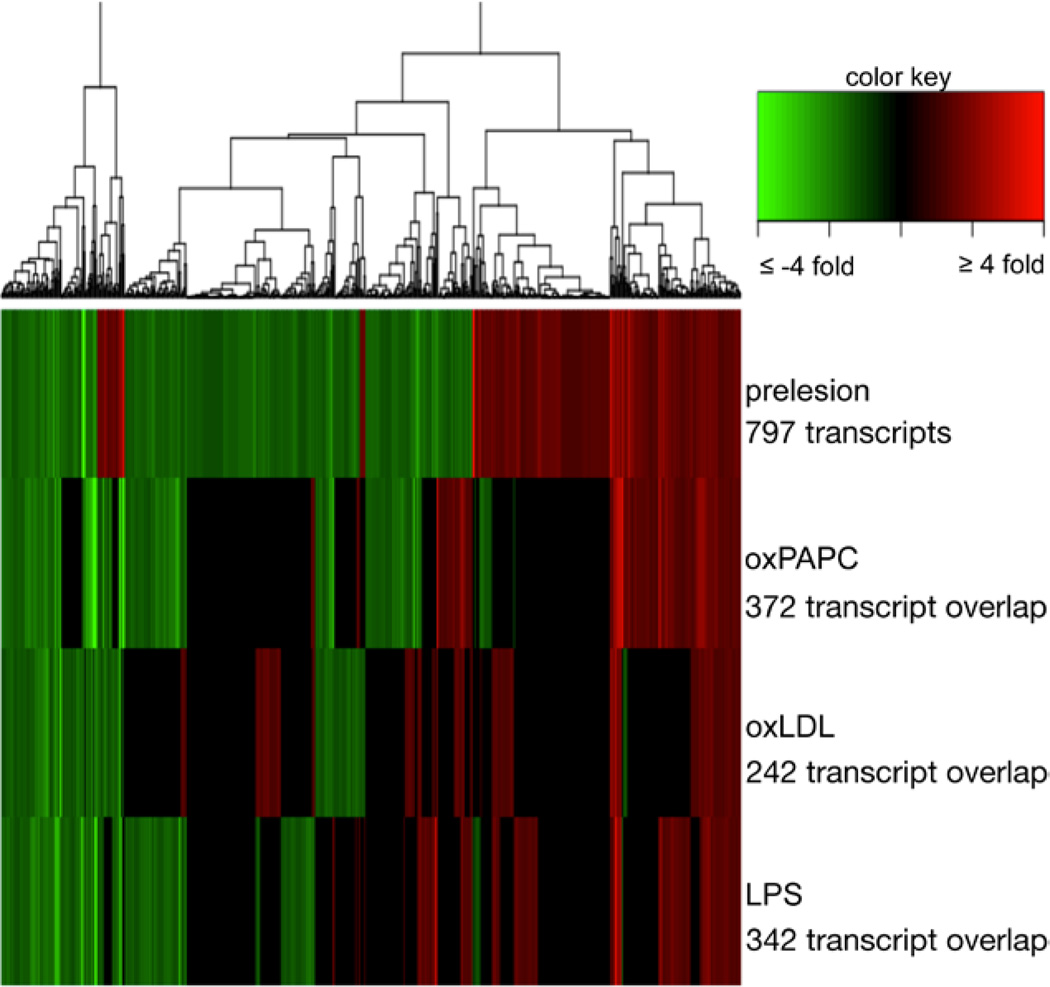
Differential gene expression of mouse aortic endothelial cells (MAECs) isolated from prelesion compared with healthy aortas and the overlap with treated MAECs. Heatmap displaying differential expression of gene expression transcripts in MAECs. The top bar displays differential expression of MAECs isolated from healthy to prelesion aortas with 10% false-discovery rate. The red bars indicate upregulation of expression from healthy to prediseased, whereas green bars indicate downregulation. The next 3 rows show differential expression data of the same 797 differentially expressed prelesion transcripts in MAECs treated with 1-palmitoyl-2-arachidonoyl-sn-glycero-3-phosphocholine (oxPAPC), oxidized low-density lipoprotein (oxLDL), or lipopolysaccharide (LPS), respectively. Gene expression cut-off for treatment is 1.2-fold and is maximized at an absolute value of 4 for purposes of graph visualization. Numbers below treatments indicate the number of transcripts that are regulated in the same direction as prelesion samples.
Patterns of Differential Expression Elicited by oxPAPC Exhibits the Highest Similarity With ECs From Prelesion Aortas
Because many of the early EC changes in atherogenesis are thought to be affected by activation of the inflammatory response, we compared the endothelial gene expression profile after treatment with proinflammatory agents to the gene expression of MAECs in the early stages of atherosclerosis as determined in the previous section. An additional set of MAEC expression data was generated by treating aortas for 4 hours with medium alone or media containing oxPAPC, oxLDL, or LPS before cell isolation and then amplifying the RNA and performing microarray analysis. Differentially expressed genes were determined using MAECs collected from aortas incubated in control media as the baseline. Of the 797 differentially expressed transcripts in ECs from healthy to prelesion aortas, nearly half are regulated in the same direction after treatment with oxPAPC, followed closely with treatment of LPS and oxLDL (Figure 2). Although there is overlap among the 3 different proinflammatory treatments, there are also distinct sets of transcripts that differ. There are also smaller sets of transcripts that are differentially expressed in opposite direction in the prelesion samples compared with the treated samples and a subset of transcripts in ECs from prelesion aortas whose regulation is not seen in the treatments tested, represented by the black areas on the heat map presented in Figure 2.
Differential Expression Analysis Identifies New Candidates for a Role in Atherosclerosis
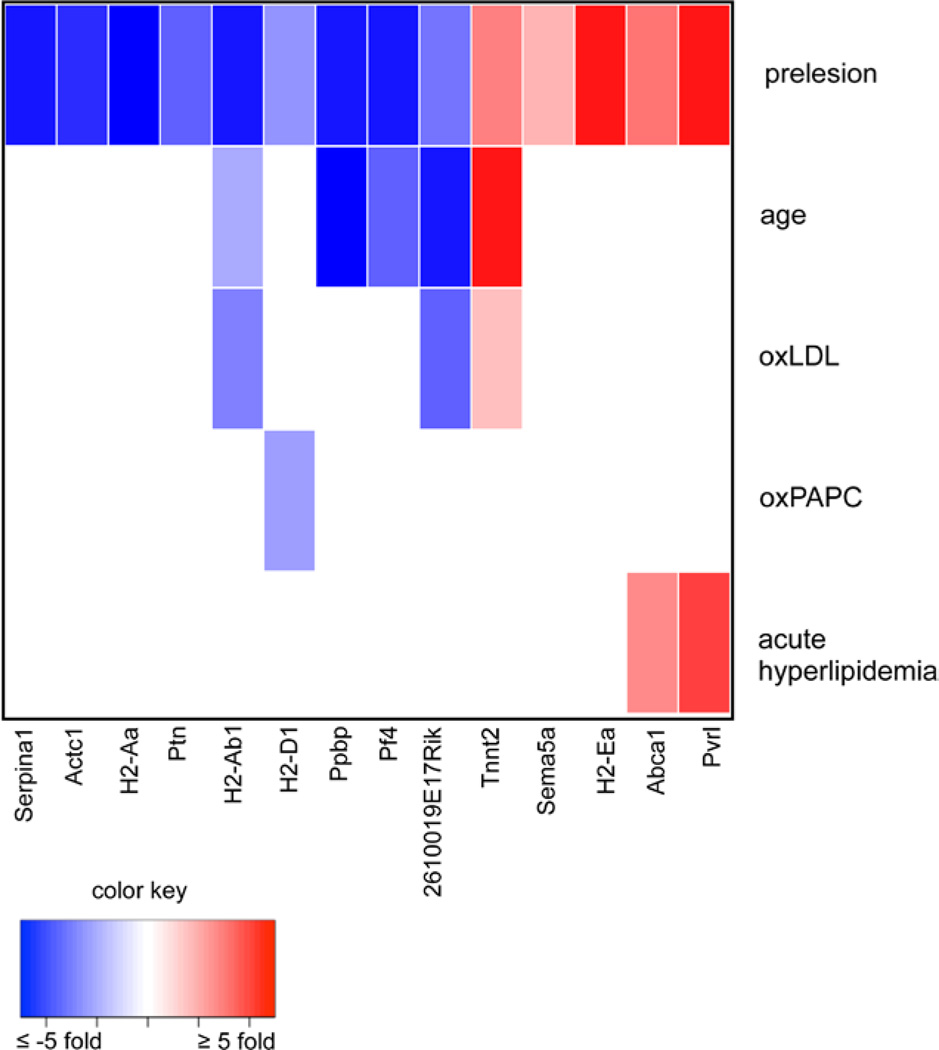
To identify genes that may play a critical role in EC-mediated atherogenesis, we used a more stringent threshold to identify the top differentially expressed gene transcripts from healthy to MAECs from prelesion aortas as determined by fold change in the microarray data. The expression levels of the genes represented by these microarray probe sets were tested by reverse transcription quantitative PCR. Genes that were determined to have significant differential expression in both microarray and reverse transcription quantitative PCR data have a correlation coefficient of 0.91 and are shown in Table 2. The ApoE−/− mutation commonly used to obtain hyperlipidemic mice could affect gene expression separate from the perturbations that result from the proatherogenic environment. To gain additional evidence that the most regulated genes in the endothelium of prelesion aortas resulted from an atherogenic stimulus, we determined their transcriptional response to additional atherogenic stimuli. These experimental comparisons were all performed using BL6 wild-type mice and did not include the ApoE−/− mutation as a variable. In addition to the oxPAPC, oxLDL, and LPS treatments previously described, we examined transcriptional differences of these highly regulated genes in MAECs isolated from aged mice and mice with acute hyperlipidemia. We compared MAEC expression from BL6 wild-type mice at 4 to 24 weeks to represent aging because BL6 wild-type mice on a chow diet >1 year begin to show lipid accumulation in the aortic root, but not in the aortic arch (Figure IIID and IIIE in the online-only Data Supplement). Although these mice are not hyperlipidemic, as atherosclerosis increases with age in humans,30 they represent the aspects of atherosclerosis that may be regulated by advanced age. We also collected MAEC RNA from mice that had been injected intravenously with human LDL twice during a 5-day time period to represent acute hyperlipidemia (Table III in the online-only Data Supplement). Cell preparations from these treatments were compared with MAECs of prelesion aortas from BL6 ApoE−/− mice at 4 weeks. Eight of the top 14 differentially expressed genes in the prelesioned condition were also perturbed by these separate atherogenic treatments: 2610019E17Rik, H2-Ab1, H2-D1, Pf4, Ppbp, Pvrl2, and Tnnt2 (Figure 4; Table 2). None of the top differentially expressed genes were responsive to LPS treatment, the most nonspecific of the 3 proinflammatory substances tested.
Table 2.
Top Differentially Expressed Genes as Determined by Microarray Analysis Identify 14 Highly Regulated Transcripts
| Fold Change From Healthy to Diseased |
RT-qPCR Fold Change Under Additional Conditions |
|||||
|---|---|---|---|---|---|---|
| Gene | RT-qPCR | Microarray | oxPAPC | oxLDL | LDL Injection | Wild-Type Age |
| 2610019E17Rik | −2.97* | −2.64* | … | −3.49** | … | −13.49** |
| Abca1 | 3.13** | 2.91** | −3.06* | … | 2.53** | … |
| Actc1 | −4.68 | −2.80* | … | … | … | … |
| H2-Aa | −5.60** | −3.04* | … | … | … | … |
| H2-Ab1 | −10.20* | −3.45** | … | −2.71* | −1.95* | |
| H2-D1 | −2.29** | −2.63 | −2.02* | … | … | … |
| H2-Ea | 442^ | 4.48 | … | … | … | … |
| Pf4 | −9.59* | −2.53* | … | … | … | −3.37* |
| Ppbp | −9.63** | −7.48** | … | … | … | −5.51* |
| Ptn | −3.37* | −2.75 | … | … | … | … |
| Pvrl2 | 5.88* | 4.06** | … | … | 4.08* | … |
| Sema5a | 1.76* | 2.75* | … | … | … | … |
| Serpina1 | −14.25* | −6.23* | … | … | … | … |
| Tnnt2 | 2.87* | 3.50* | … | 1.35* | … | 13.40* |
These genes were selected using a threshold of 10% FDR and fold change of ≥2.5 in microarray data and confirmed using RT-qPCR analysis. P values for fold changes from healthy to diseased MAECs as measured by qPCR are determined by t test; P values for fold changes determined by microarrays are adjusted using Benjamini–Hochberg. Gene expression for these 14 genes was also assessed using qPCR in MAECs from: aortas treated with oxPAPC and oxLDL, mice intravenously injected with human LDL, and wild-type BL6 mice at 4 vs 24 weeks (wild-type age). FDR indicates false-discovery rate; LDL, low-density lipoprotein; MAEC, mouse aortic endothelial cell; oxLDL, oxidized LDL; oxPAPC, 1-palmitoyl-2-arachidonoyl-sn-glycero-3-phosphocholine; and RT-qPCR, reverse transcription quantitative polymerase chain reaction.
P ≤0.05;
P ≤0.01; and
extreme differential expression from very low expression at base level; P=0.112.
Figure 4.
Differential expression overlaps in atherogenic conditions and top prelesion genes. Differential expression of the top differentially expressed genes in mouse aortic endothelial cells from 4-week healthy vs prelesioned aortas was determined in 5 additional conditions relevant to atherosclerotic disease: aortas treated with 1-palmitoyl-2-arachidonoyl-sn-glycero-3-phosphocholine (oxPAPC) and oxidized low-density lipoprotein (oxLDL), mice intravenously injected with human LDL, and wild-type BL6 mice at 4 vs 24 weeks (wild-type age).
Discussion
We characterized fully and adapted a method15 to isolate ECs from mouse aortas that allowed us to profile gene expression in ECs during the early stages of atherosclerosis. This method provides a quick and efficient way to collect MAEC RNA, generally yielding RNA from a mouse in <1 hour after harvesting, and has several advantages instead of cell culture methods. Not only do we avoid the passage and dedifferentiation issues that occur during cell culture procedures but we are able to directly assay the transcriptional state of the cells that closely represents their in vivo status. In these experiments, we were able to obtain the transcriptional state of ECs in prelesion aortas, and through the use of 1 RNA amplification, we were able to perform microarray analysis on MAECs taken from a single mouse. Moreover, we demonstrate that the whole aorta, with its endothelium intact, can be treated in cell culture medium similar to conventional cell culture with the added advantage of not disrupting the natural environment and cell–cell communication. This method does not allow for the isolation of protein for downstream applications such as Western blots, and because the cells die after the hematoxylin staining step, this procedure would not be appropriate for downstream cell-sorting techniques. We also found that the small size of the mouse aorta made it difficult to consistently gather cells from discrete flow regions. A limitation of the resulting data is that cells from both atherosusceptible and atheroresistant laminar flow regions have been collected, resulting in a heterogeneous RNA sample representing both types of cells. As some of the aortic regions from which we have collected RNA will never develop atherosclerosis31 and are, therefore, not representative of prelesion cells, the resulting differential expression profile may contain false-positives. Further experiments that compare ECs that are isolated exclusively from atherosusceptible aortic regions of the mouse would refine these results. However, we envision that our characterization of this method and its potential applications for various biological treatments will be of use to vascular biologists who require MAEC preparations that are highly enriched and provide a reliable representation of in vivo transcriptional activity.
This method was used to assay the transcriptional profile of MAECs from prelesion aortas for the first time. We found that ≈800 genes are perturbed in the in vivo endothelium before lipid accumulation, a significant alteration to the transcriptional profile of healthy ECs. The most highly regulated genes were enriched for immune response–related categories (Table II in the online-only Data Supplement), a finding that is consistent with prior studies in swine,5 and plasma membrane/cell surface categories, perhaps indicative of the endothelial activation and dysfunction that occur during atherosclerosis initiation.32 We also assessed the role of biologically relevant proinflammatory substances in atherogenic transcriptional regulation through ex vivo treatment. We found that the oxidized phospholipid oxPAPC has the most overlap with ECs from prelesion aortas, and that none of the 14 top differentially expressed genes in these ECs are regulated by LPS treatment, whereas some are regulated by oxPAPC and oxLDL. We also observed that there are sets of genes that are both up- and downregulated in endothelium of prelesion aortas that are unaffected by inflammatory mediators. These findings indicate that although oxidized lipids may be more biologically relevant than LPS in the inflammatory response leading to the initiation of atherosclerosis, these inflammatory mediators are not likely to be solely responsible for the transcriptional changes seen in the early hyperlipidemic endothelium.
We identified 14 highly differentially expressed genes in the prelesion endothelium. We were able to further characterize the transcriptional activity of these novel genes in response to relevant proinflammatory agents using our ex vivo treatment method. Eight of the top differentially expressed genes in pre-lesion MAECs were also differentially expressed in response to additional disease-relevant stimuli and represent top candidates for further study (Figure 4; Table 2). Among these genes are some that have been described previously as playing a role in the atherosclerotic endothelium, particularly Abca1. This gene has been identified as upregulated in the swine endothelium in response to a brief hypercholesterolemic diet,7 a finding that is consistent with its regulation in our ApoE−/− hyperlipidemic and acute hyperlipidemic mice. The protein encoded by Abca1 has been shown to modulate the transport of phospholipids and cholesterol to apo AI through the endothelial layer and is atheroprotective in vascular ECs because of this role in cholesterol efflux.33,34 The upregulation of this gene that we observe in the hyperlipidemic prelesion intima and the acute hyperlipidemic mice may reflect the response to the influx of LDL in these mice. Pvrl2, a gene expressed by ECs and involved with the immune response,35 shows significant differential expression as well. Its closely related cell surface ligand, Pvr, has been shown to regulate the movement of leukocytes across the endothelium36,37 and interact with vascular endothelial growth factor 2 (VEGFR2),38 suggesting that Pvrl2 may also be involved with endothelial transmigration and VEGF-induced angiogenesis. This gene may also affect EC permeability because it has been shown to control cell–cell adhesion.39 The strong upregulation we observe in both the hyperlipidemic prelesion and the acute hyperlipidemic endothelium, a similar pattern to Abca1, suggests that this gene is strongly responsive to LDL levels.
Our study also identified several genes that have not been linked previously to the endothelial role in atherogenesis, such as the sarcomere protein gene Tnnt2.40 The Tnnt2 transcript, which is homologous to human TNNT2, encodes the protein cardiac muscle troponin T, also known as cTnT. Circulating cardiac troponin T is the most widely used biomarker for the detection of myocardial injury,41 and the extent of coronary atherosclerosis is associated with increasing circulating levels of high-sensitive cardiac troponin T.42,43 We observe elevation of this transcript in mice susceptible for atherosclerosis before developing the disease, as they advance in age, and after treatment with oxLDL. It is possible that this biomarker or its mRNA transcript could indicate endothelial activation during atherogenesis at a much earlier stage of coronary artery disease than myocardial injury. A pair of related transcripts, the platelet genes Pf4 and Ppbp,44 is downregulated in both the prelesion and the aged mouse endothelium. Pf4 has been shown to inhibit EC proliferation and angiogenesis in vitro and in vivo.45 Because atherosclerosis has high levels of angiogenesis, 46 downregulation of an inhibitory gene, such as Pf4, could be a key step in allowing the progression of atherosclerosis to proceed once initiation has occurred. Ppbp may also be involved in angiogenesis47 although less is known about its specific role in the process.
We also observe the perturbation of 4 histocompatibility genes: H2-Aa, H2-Ab1, H2-D1, and H2-Ea. These genes are all located in the same cluster in both mouse and human, and a Genome Wide Association Study (GWAS) meta-analysis identified a single-nucleotide polymorphism within this region that is associated with coronary artery disease48 (Table 3). Transcripts for the mouse homolog of HLA-B, H2-D1, were downregulated in response to treatment with the oxidized phospholipid oxPAPC and in the prelesion endothelium. When transfected into ECs, a pulmonary hypertension risk allele of HLA-B influences the production of endothelin -1 (Edn1) and endothelial nitric oxide synthase (eNOS) and induces both ER stress and the unfolded protein response,49 all processes that are central to endothelial activation during atherosclerosis.50 The human homologs to mouse H2-Aa, H2-Ab1, and H2-Ea, HLA-DR and HLA-DQ, are major histocompatibility complex class II members that regulate T-cell–dependent immune responses. Vascular SMCs in atherosclerotic lesions express HLA-DR,51 and as studies have shown that hypoxia induces both HLA-DR expression and secretion in EC cultures,52 it is likely that MAEC expresses this transcript as part of the inflammatory response in atherogenesis as we have seen here. Although the HLA-DQ transcripts are also expressed in ECs,53 little is known about the HLA-DQ subtypes in the context of atherosclerosis and why they would be downregulated in atherosusceptible conditions as we show here. The previously unstudied gene 2610019E17RIK is downregulated in the prelesion endothelium, by oxLDL treatment, and in older mice. It is a noncoding RNA of 494 base pairs located on mouse chromosome 17 and may merit further characterization as noncoding RNAs have been shown to play central roles in disease.54
Table 3.
Differential Expression of Mouse Genes Homologous to Human 6p21.3 Region
| Gene Expression Fold Change in MAECs |
|||||||
|---|---|---|---|---|---|---|---|
| Human Gene | Mouse Gene | MHC Class | From rs3869109, bp | Prelesioned | oxPAPC | oxLDL | Wild-Type Age |
| HLA-B | H2-D1 | I | 137 453 | −2.29** | −2.02* | … | … |
| HLA-DRA | H2-Ea | II | 1 223 423 | 442^ | … | … | … |
| HLA-DQA1 | H2-Aa | II | 1 420 987 | −5.60** | … | … | … |
| HLA-DQB1 | H2-Ab1 | II | 1 443 045 | −10.20* | … | −2.71* | −1.95* |
The human homologs of differentially expressed mouse genes are listed, along with their MHC class, distance from the previously published SNP associated with CAD in Genome Wide Association Study (GWAS) meta-analysis, and fold changes of the transcript in MAECs from prelesion aortas, oxPAPC-, or oxLDL-treated aortas, and 24-week BL6 wild-type aortas compared with 4-week endothelium. CAD indicates coronary artery disease; MAEC, mouse aortic endothelial cell; MHC, major histocompatibility complex; oxLDL, oxidized low-density lipoprotein; oxPAPC, 1-palmitoyl-2-arachidonoyl-sn-glycero-3-phosphocholine; and RT-qPCR, reverse transcription quantitative polymerase chain reaction.
P ≤0.05;
P≤0.01; and
extreme differential expression from very low expression at base level.
In summary, our study provides several valuable leads for the study of the genes mediating atherosclerosis in the vessel wall. Our characterization of a vascular endothelium isolation method and demonstration that it is applicable to treatment of cells in the aorta provide new tools to researchers performing individual gene follow-up studies in the mouse vasculature.55 Future studies could apply these methods to observe differences in discrete flow regions and compare them from atheroresistant and atherosusceptible mouse strains to refine these data and identify additional genetic factors. The application of this method identifies several genes that are differentially expressed in the endothelium during the early stages of atherogenesis. Although most of these genes have been described previously as having roles in inflammation, ECs, or heart disease, this is the first time the majority of these transcripts have been correlated directly to vascular ECs in atherosclerosis and provide exciting leads for further study.
Materials and Methods
Animals
C57BL/6J (BL6) male mice fed a chow diet were used for experiments. BL6 mice were obtained from the Jackson Laboratory or bred in our colony; BL6 ApoEtm1Unc null mice (ApoE -/-) were maintained in a colony in the UCLA vivarium. Wild-type BL6 mice were used in samples representing healthy vascular cells, and ApoE -/- BL6 mice were used for pre-lesioned vascular cells. The aortas from BL6 ApoE -/- mice fed a chow diet at four weeks of age were considered prelesioned, and BL6 ApoE -/- mice at 24-weeks of age exhibited clear atherosclerosis (Supp Fig 3). Mice were euthanized using isofluorane in accordance with Animal Research Committee policies.
Microscopy/oil red O staining
Sections of aortic arch were fixed onto Superfrost slides and stained with oil red O dye. Microscopy photos were taken at a magnification of 20x.
Aorta treatment procedure (Supp Fig 1)
DMEM with 1% FBS was used as the cell culture medium; vessels were isolated, connective tissue removed, and were incubated for 4 hours in a cell culture incubator at 37° C, 5% CO2. 1-palmitoyl-2-arachidonoyl-sn-glycero-3-phosphocholine 1 (OxPAPC) and oxLDL 2 were prepared as previously described. Lipopolysaccharide (LPS) was purchased from List Biological Laboratories. OxPAPC and oxLDL were added to DMEM for a final concentration of 50 μg/mL, LPS was formulated for a final concentration of 4 ng/mL. After aortas were opened en face, each was placed in one well of a 12-well cell culture plate along with 1 mL of the appropriate DMEM formulation pre-warmed to 37° C. After the 4 hour treatment, aortas were removed from media and placed lumen side up on a glass slide; isolation continued as described below, beginning with hematoxylin treatment.
Intravenous injection of LDL into mice
Human LDL was isolated as previously described 3. LDL in buffer (0.15 M NaCl, 1.0 mM EDTA in PBS pH 7.2) was injected into the tail vein of 10-week old BL6 WT mice in order to induce acute hyperlipidemia 3. Two doses of 3.2 mg LDL in 240 μL were injected two days apart and MAECs were isolated five days after the first injection. Control mice were injected with buffer only. Plasma lipid levels of these mice are shown in Supp Table 1.
MAEC cell isolation procedure
The cell isolation protocol was adapted from a previously published method for intimal cell isolation 4 (Supp Fig 1). After euthanization of the mouse, the chest cavity was opened, lungs, trachea, and esophagus removed, and the aortic arch excised under a dissecting microscope. Care was taken to maintain consistency in dissection of the arch region. No perfusion of vasculature was performed. After rinsing in cold PBS, the vessel was placed on a glass slide, the surrounding connective tissue was removed, and the aorta was opened en face. In order to visualize the endothelial layer, the opened aorta was stained with 30 μL hematoxylin for 3 minutes. The stain was rinsed off with cold PBS. The collagenase liberase blendzyme 2 (Roche) was diluted 1:100 with PBS, and 25 μL was added to the top of the aorta on the slide and incubated at 37° C for 8 minutes. The slide with collagenase-treated aorta was then placed under a dissecting microscope, and the endothelial cells were gently pried off using a 26-gauge needle. This process continued until all endothelial cells were removed, determined by the lack of hematoxylin-dyed nuclei on the surface of the sample. The liquid containing the endothelial cells was then pipetted with a thin pipet tip into RNA extraction buffer.
MAEC RNA extraction
RNA was extracted using the RNAqueous®-Micro Kit by Ambion. The buffer and collagenase solution containing scraped endothelial cells was pipetted directly into 100 μL of Lysis buffer in a 200 μL PCR tube, vortexed, then incubated at 42° C for 30 minutes on a PCR thermalcycler. RNA isolation continued following the manufacturer’s protocol, including suggestions to pre-wet the filter assembly with 30 μL L Lysis buffer prior to isolation and to preheat elution solution to 95° C. RNA was stored at -80° C after isolation. RNA quality was tested using the Agilent 2100 Bioanalyzer and the Agilent RNA 6000 Pico Kit, and only RNA samples with RNA integrity numbers (RIN) of 7.0 or higher were used for data collection. RNA expression profiles from three to six mice meeting these conditions were analyzed individually for each condition tested.
RNA amplification
The NuGEN WT-Ovation One-Direct RNA amplification system was used to amplify RNA isolated from MAECs. The procedure followed manufacturer’s protocol; pre-amplification work was conducted in a sterile hood treated with RNase ZAP and DNA-OFF and using pipetmen and other lab materials exclusive to pre-amplification in order to eliminate contamination.
cDNA synthesis and RT-qPCR
As the output of the NuGEN WT-Ovation One-Direct RNA amplification system is cDNA, no separate cDNA synthesis was required for amplified RNA. Roche and KAPA Biosystems SYBR green reagents were used for RT-qPCR, and reactions were processed on 384-well plates on the Roche LightCycler 480. Primer sequences are listed in Supp Table 4 and Cp value ranges are shown in Supp Table 5.
Expression microarrays
The products of RNA amplification were treated with the NuGEN Encore Biotin Module prior to hybridization according to manufacturer’s protocol. Affymetrix HT Mouse Genome 430 array plates were used for microarray analysis.
Statistical analyses
Statistical analyses were executed in the R programming environment. T-tests were calculated for RT-qPCR results using the “t.test” command. Microarray results were normalized using RMA in the “affy” package, and differential expression p-values and Venn diagrams were calculated using the “limma” package. Heat maps were constructed using the “heatmap.2” package. Microarray p-values were adjusted for multiple comparisons using Benjamini-Hochberg. Pearson’s coefficient between microarray and RT-qPCR values was calculated using the “cor.test” command; H2-Ea values were removed from this calculation due to absence of expression in control mice. Expression data can be obtained from Gene Expression Omnibus (GEO) databases (accession no. GSE39264). Functional annotation of microarray gene lists was determined using the DAVID Bioinformatics resource 5, 6.
Supplementary Material
Significance.
Vascular endothelial cells are central to the initiation and progression of atherosclerosis, but there have been few studies that directly assess the genetic changes that occur in vivo during these events. As the mouse is the most commonly used animal model of human atherosclerosis, isolating and studying the endothelial cells from the mouse aorta would provide valuable information on their role in disease. This study characterizes cells isolated from the intima of the mouse aorta as endothelial cells and applies this method to identify a set of genes that are highly differentially expressed in endothelial cells of mice susceptible to atherosclerosis compared with mice that are resistant. The expression of these genes is also studied in response to several atherogenic stimuli. On the basis of these data, we identify 8 candidate genes for a role in the endothelium during atherosclerosis initiation, many of which have not been described previously in this context.
Acknowledgments
We thank Jan Danciger for her advice on endothelial cells, Ladan Vakili for the oxidized low-density lipoprotein, and Xuping Wang and Judy Wu for sectioning and staining the aortic arch samples.
Sources of Funding
This research was funded by National Institutes of Health (NIH) grant PO1-HL030568 (A.J. Lusis, J. Berliner), NIH grant HL-064731 (J. Berliner), American Heart Association predoctoral fellowship (A. Erbilgin), and Ruth L. Kirschstein National Research Service Award GM007185 (A. Erbilgin).
Nonstandard Abbreviations and Acronyms
- ApoE−/−
apolipoprotein deficient
- EC
endothelial cell
- HAEC
human aortic endothelial cell
- LDL
low-density lipoprotein
- LPS
lipopolysaccharide
- MAEC
mouse aortic endothelial cell
- oxLDL
oxidized low-density lipoprotein
- oxPAPC
1-palmitoyl-2-arachidonoyl-sn-glycero-3-phosphocholine
- SMC
smooth muscle cell
Footnotes
The online-only Data Supplement is available with this article at http://atvb.ahajournals.org/lookup/suppl/doi:10.1161/ATVBAHA.113.301989/-/DC1.
Disclosures
N. Siemers, P. Kayne, and W.-p. Yang are employees of Bristol-Myers Squibb. The other authors report no conflicts.
References
- 1.Kathiresan S, Srivastava D. Genetics of human cardiovascular disease. Cell. 2012;148:1242–1257. doi: 10.1016/j.cell.2012.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Libby P, Ridker PM, Hansson GK. Progress and challenges in translating the biology of atherosclerosis. Nature. 2011;473:317–325. doi: 10.1038/nature10146. [DOI] [PubMed] [Google Scholar]
- 3.Hansson GK, Hermansson A. The immune system in atherosclerosis. Nat Immunol. 2011;12:204–212. doi: 10.1038/ni.2001. [DOI] [PubMed] [Google Scholar]
- 4.Volger OL, Fledderus JO, Kisters N, Fontijn RD, Moerland PD, Kuiper J, van Berkel TJ, Bijnens AP, Daemen MJ, Pannekoek H, Horrevoets AJ. Distinctive expression of chemokines and transforming growth factorbeta signaling in human arterial endothelium during atherosclerosis. Am J Pathol. 2007;171:326–337. doi: 10.2353/ajpath.2007.061196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Passerini AG, Polacek DC, Shi C, Francesco NM, Manduchi E, Grant GR, Pritchard WF, Powell S, Chang GY, Stoeckert CJ, Jr, Davies PF. Coexisting proinflammatory and antioxidative endothelial transcription profiles in a disturbed flow region of the adult porcine aorta. Proc Natl Acad Sci USA. 2004;101:2482–2487. doi: 10.1073/pnas.0305938101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Civelek M, Manduchi E, Riley RJ, Stoeckert CJ, Jr, Davies PF. Chronic endoplasmic reticulum stress activates unfolded protein response in arterial endothelium in regions of susceptibility to atherosclerosis. Circ Res. 2009;105:453–461. doi: 10.1161/CIRCRESAHA.109.203711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Civelek M, Grant GR, Irolla CR, Shi C, Riley RJ, Chiesa OA, Stoeckert CJ, Jr, Karanian JW, Pritchard WF, Davies PF. Prelesional arterial endothelial phenotypes in hypercholesterolemia: universal ABCA1 upregulation contrasts with region-specific gene expression in vivo. Am J Physiol Heart Circ Physiol. 2010;298:H163–H170. doi: 10.1152/ajpheart.00652.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nguyen D, Xu T. The expanding role of mouse genetics for understanding human biology and disease. Dis Model Mech. 2008;1:56–66. doi: 10.1242/dmm.000232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eppig JT, Blake JA, Bult CJ, Kadin JA, Richardson JE. Mouse Genome Database Group. The Mouse Genome Database (MGD): comprehensive resource for genetics and genomics of the laboratory mouse. Nucleic Acids Res. 2012;40(Database issue):D881–D886. doi: 10.1093/nar/gkr974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stylianou IM, Bauer RC, Reilly MP, Rader DJ. Genetic basis of atherosclerosis: insights from mice and humans. Circ Res. 2012;110:337–355. doi: 10.1161/CIRCRESAHA.110.230854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daugherty A. Mouse models of atherosclerosis. Am J Med Sci. 2002;323:3–10. doi: 10.1097/00000441-200201000-00002. [DOI] [PubMed] [Google Scholar]
- 12.Breslow JL. Mouse models of atherosclerosis. Science. 1996;272:685–688. doi: 10.1126/science.272.5262.685. [DOI] [PubMed] [Google Scholar]
- 13.Frid MG, Kale VA, Stenmark KR. Mature vascular endothelium can give rise to smooth muscle cells via endothelial-mesenchymal trans-differentiation: in vitro analysis. Circ Res. 2002;90:1189–1196. doi: 10.1161/01.res.0000021432.70309.28. [DOI] [PubMed] [Google Scholar]
- 14.Beer M, Doepping S, Hildner M, Weber G, Grabner R, Hu D, Mohanta SK, Srikakulapu P, Weih F, Habenicht AJ. Laser-capture microdissection of hyperlipidemic/ApoE−/− mouse aorta atherosclerosis. Methods Mol Biol. 2011;755:417–428. doi: 10.1007/978-1-61779-163-5_35. [DOI] [PubMed] [Google Scholar]
- 15.Jongstra-Bilen J, Haidari M, Zhu SN, Chen M, Guha D, Cybulsky MI. Low-grade chronic inflammation in regions of the normal mouse arterial intima predisposed to atherosclerosis. J Exp Med. 2006;203:2073–2083. doi: 10.1084/jem.20060245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nam D, Ni CW, Rezvan A, Suo J, Budzyn K, Llanos A, Harrison D, Giddens D, Jo H. Partial carotid ligation is a model of acutely induced disturbed flow, leading to rapid endothelial dysfunction and atherosclerosis. Am J Physiol Heart Circ Physiol. 2009;297:H1535–H1543. doi: 10.1152/ajpheart.00510.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang XQ, Nigro P, World C, Fujiwara K, Yan C, Berk BC. Thioredoxin interacting protein promotes endothelial cell inflammation in response to disturbed flow by increasing leukocyte adhesion and repressing Kruppellike factor 2. Circ Res. 2012;110:560–568. doi: 10.1161/CIRCRESAHA.111.256362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao J, Ishigaki Y, Yamada T, Kondo K, Yamaguchi S, Imai J, Uno K, Hasegawa Y, Sawada S, Ishihara H, Oyadomari S, Mori M, Oka Y, Katagiri H. Involvement of endoplasmic stress protein C/EBP homologous protein in arteriosclerosis acceleration with augmented biological stress responses. Circulation. 2011;124:830–839. doi: 10.1161/CIRCULATIONAHA.110.014050. [DOI] [PubMed] [Google Scholar]
- 19.Cheng C, Tempel D, Den Dekker WK, Haasdijk R, Chrifi I, Bos FL, Wagtmans K, van de Kamp EH, Blonden L, Biessen EA, Moll F, Pasterkamp G, Serruys PW, Schulte-Merker S, Duckers HJ. Ets2 determines the inflammatory state of endothelial cells in advanced atherosclerotic lesions. Circ Res. 2011;109:382–395. doi: 10.1161/CIRCRESAHA.111.243444. [DOI] [PubMed] [Google Scholar]
- 20.Simeone SM, Li MW, Paradis P, Schiffrin EL. Vascular gene expression in mice overexpressing human endothelin-1 targeted to the endothelium. Physiol Genomics. 2011;43:148–160. doi: 10.1152/physiolgenomics.00218.2009. [DOI] [PubMed] [Google Scholar]
- 21.Navab M, Liao F, Hough GP, Ross LA, Van Lenten BJ, Rajavashisth TB, Lusis AJ, Laks H, Drinkwater DC, Fogelman AM. Interaction of monocytes with cocultures of human aortic wall cells involves interleukins 1 and 6 with marked increases in connexin43 message. J Clin Invest. 1991;87:1763–1772. doi: 10.1172/JCI115195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krenek P, Hamaide MC, Morel N, Wibo M. A simple method for rapid separation of endothelial and smooth muscle mRNA reveals Na/K+-ATPase alpha-subunit distribution in rat arteries. J Vasc Res. 2006;43:502–510. doi: 10.1159/000095963. [DOI] [PubMed] [Google Scholar]
- 23.Clément-Ziza M, Gentien D, Lyonnet S, Thiery JP, Besmond C, Decraene C. Evaluation of methods for amplification of picogram amounts of total RNA for whole genome expression profiling. BMC Genomics. 2009;10:246. doi: 10.1186/1471-2164-10-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Singh R, Maganti RJ, Jabba SV, Wang M, Deng G, Heath JD, Kurn N, Wangemann P. Microarray-based comparison of three amplification methods for nanogram amounts of total RNA. Am J Physiol, Cell Physiol. 2005;288:C1179–C1189. doi: 10.1152/ajpcell.00258.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakashima Y, Plump AS, Raines EW, Breslow JL, Ross R. ApoE-deficient mice develop lesions of all phases of atherosclerosis throughout the arterial tree. Arterioscler Thromb. 1994;14:133–140. doi: 10.1161/01.atv.14.1.133. [DOI] [PubMed] [Google Scholar]
- 26.Tamminen M, Mottino G, Qiao JH, Breslow JL, Frank JS. Ultrastructure of early lipid accumulation in ApoE-deficient mice. Arterioscler Thromb Vasc Biol. 1999;19:847–853. doi: 10.1161/01.atv.19.4.847. [DOI] [PubMed] [Google Scholar]
- 27.Romanoski CE, Lee S, Kim MJ, Ingram-Drake L, Plaisier CL, Yordanova R, Tilford C, Guan B, He A, Gargalovic PS, Kirchgessner TG, Berliner JA, Lusis AJ. Systems genetics analysis of gene-by-environment interactions in human cells. Am J Hum Genet. 2010;86:399–410. doi: 10.1016/j.ajhg.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Romanoski CE, Che N, Yin F, Mai N, Pouldar D, Civelek M, Pan C, Lee S, Vakili L, Yang WP, Kayne P, Mungrue IN, Araujo JA, Berliner JA, Lusis AJ. Network for activation of human endothelial cells by oxidized phospholipids: a critical role of heme oxygenase 1. Circ Res. 2011;109:e27–e41. doi: 10.1161/CIRCRESAHA.111.241869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walton KA, Gugiu BG, Thomas M, Basseri RJ, Eliav DR, Salomon RG, Berliner JA. A role for neutral sphingomyelinase activation in the inhibition of LPS action by phospholipid oxidation products. J Lipid Res. 2006;47:1967–1974. doi: 10.1194/jlr.M600060-JLR200. [DOI] [PubMed] [Google Scholar]
- 30.Go AS, Mozaffarian D, Roger VL, et al. American Heart Association Statistics Committee and Stroke Statistics Subcommittee. Heart disease and stroke statistics-2013 update: a report from the American Heart Association. Circulation. 2013;127:e6–e245. doi: 10.1161/CIR.0b013e31828124ad. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Davies PF, Civelek M, Fang Y, Fleming I. The atherosusceptible endothelium: endothelial phenotypes in complex haemodynamic shear stress regions in vivo. Cardiovasc Res. 2013;99:315–327. doi: 10.1093/cvr/cvt101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pober JS, Sessa WC. Evolving functions of endothelial cells in inflammation. Nat Rev Immunol. 2007;7:803–815. doi: 10.1038/nri2171. [DOI] [PubMed] [Google Scholar]
- 33.Cavelier C, Rohrer L, von Eckardstein A. ATP-Binding cassette transporter A1 modulates apolipoprotein A-I transcytosis through aortic endothelial cells. Circ Res. 2006;99:1060–1066. doi: 10.1161/01.RES.0000250567.17569.b3. [DOI] [PubMed] [Google Scholar]
- 34.Prosser HC, Ng MK, Bursill CA. The role of cholesterol efflux in mechanisms of endothelial protection by HDL. Curr Opin Lipidol. 2012;23:182–189. doi: 10.1097/MOL.0b013e328352c4dd. [DOI] [PubMed] [Google Scholar]
- 35.Tahara-Hanaoka S, Shibuya K, Onoda Y, Zhang H, Yamazaki S, Miyamoto A, Honda S, Lanier LL, Shibuya A. Functional characterization of DNAM-1 (CD226) interaction with its ligands PVR (CD155) and nectin-2 (PRR-2/CD112) Int Immunol. 2004;16:533–538. doi: 10.1093/intimm/dxh059. [DOI] [PubMed] [Google Scholar]
- 36.Sullivan DP, Seidman MA, Muller WA. Poliovirus receptor (CD155) regulates a step in transendothelial migration between PECAM and CD99. Am J Pathol. 2013;182:1031–1042. doi: 10.1016/j.ajpath.2012.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bottino C, Castriconi R, Pende D, Rivera P, Nanni M, Carnemolla B, Cantoni C, Grassi J, Marcenaro S, Reymond N, Vitale M, Moretta L, Lopez M, Moretta A. Identification of PVR (CD155) and Nectin-2 (CD112) as cell surface ligands for the human DNAM-1 (CD226) activating molecule. J Exp Med. 2003;198:557–567. doi: 10.1084/jem.20030788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kinugasa M, Amano H, Satomi-Kobayashi S, Nakayama K, Miyata M, Kubo Y, Nagamatsu Y, Kurogane Y, Kureha F, Yamana S, Hirata K, Miyoshi J, Takai Y, Rikitake Y. Necl-5/poliovirus receptor interacts with VEGFR2 and regulates VEGF-induced angiogenesis. Circ Res. 2012;110:716–726. doi: 10.1161/CIRCRESAHA.111.256834. [DOI] [PubMed] [Google Scholar]
- 39.Samanta D, Ramagopal UA, Rubinstein R, Vigdorovich V, Nathenson SG, Almo SC. Structure of Nectin-2 reveals determinants of homophilic and heterophilic interactions that control cell-cell adhesion. Proc Natl Acad Sci USA. 2012;109:14836–14840. doi: 10.1073/pnas.1212912109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Frey N, Luedde M, Katus HA. Mechanisms of disease: hypertrophic cardiomyopathy. Nat Rev Cardiol. 2012;9:91–100. doi: 10.1038/nrcardio.2011.159. [DOI] [PubMed] [Google Scholar]
- 41.Newby LK, Rodriguez I, Finkle J, Becker RC, Hicks KA, Hausner E, Chesler R, Harper C, Targum S, Berridge BR, Lewis E, Walker DB, Dollery C, Turner JR, Krucoff MW. Troponin measurements during drug development-considerations for monitoring and management of potential cardiotoxicity: an educational collaboration among the Cardiac Safety Research Consortium, the Duke Clinical Research Institute, and the US Food and Drug Administration. Am Heart J. 2011;162:64–73. doi: 10.1016/j.ahj.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 42.Saunders JT, Nambi V, de Lemos JA, Chambless LE, Virani SS, Boerwinkle E, Hoogeveen RC, Liu X, Astor BC, Mosley TH, Folsom AR, Heiss G, Coresh J, Ballantyne CM. Cardiac troponin T measured by a highly sensitive assay predicts coronary heart disease, heart failure, and mortality in the Atherosclerosis Risk in Communities Study. Circulation. 2011;123:1367–1376. doi: 10.1161/CIRCULATIONAHA.110.005264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu B, Barbalic M, Brautbar A, et al. CARDIoGRAM Consortium. Association of genome-wide variation with highly sensitive cardiac troponin- T levels in European Americans and Blacks: a meta-analysis from atherosclerosis risk in communities and cardiovascular health studies. Circ Cardiovasc Genet. 2013;6:82–88. doi: 10.1161/CIRCGENETICS.112.963058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pitsilos S, Hunt J, Mohler ER, Prabhakar AM, Poncz M, Dawicki J, Khalapyan TZ, Wolfe ML, Fairman R, Mitchell M, Carpenter J, Golden MA, Cines DB, Sachais BS. Platelet factor 4 localization in carotid atherosclerotic plaques: correlation with clinical parameters. Thromb Haemost. 2003;90:1112–1120. doi: 10.1160/TH03-02-0069. [DOI] [PubMed] [Google Scholar]
- 45.Bikfalvi A. Platelet factor 4: an inhibitor of angiogenesis. Semin Thromb Hemost. 2004;30:379–385. doi: 10.1055/s-2004-831051. [DOI] [PubMed] [Google Scholar]
- 46.Aidoudi S, Bikfalvi A. Interaction of PF4 (CXCL4) with the vasculature: a role in atherosclerosis and angiogenesis. Thromb Haemost. 2010;104:941–948. doi: 10.1160/TH10-03-0193. [DOI] [PubMed] [Google Scholar]
- 47.Powell JA, Jr, Mousa SA. Neutrophil-activating protein-2- and interleukin- 8-mediated angiogenesis. J Cell Biochem. 2007;102:412–420. doi: 10.1002/jcb.21302. [DOI] [PubMed] [Google Scholar]
- 48.Davies RW, Wells GA, Stewart AF, et al. A genome-wide association study for coronary artery disease identifies a novel susceptibility locus in the major histocompatibility complex. Circ Cardiovasc Genet. 2012;5:217–225. doi: 10.1161/CIRCGENETICS.111.961243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lenna S, Townsend DM, Tan FK, Kapanadze B, Markiewicz M, Trojanowska M, Scorza R. HLA-B35 upregulates endothelin-1 and downregulates endothelial nitric oxide synthase via endoplasmic reticulum stress response in endothelial cells. J Immunol. 2010;184:4654–4661. doi: 10.4049/jimmunol.0903188. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 50.Tabas I. The role of endoplasmic reticulum stress in the progression of atherosclerosis. Circ Res. 2010;107:839–850. doi: 10.1161/CIRCRESAHA.110.224766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hansson GK, Jonasson L. The discovery of cellular immunity in the atherosclerotic plaque. Arterioscler Thromb Vasc Biol. 2009;29:1714–1717. doi: 10.1161/ATVBAHA.108.179713. [DOI] [PubMed] [Google Scholar]
- 52.Lahat N, Bitterman H, Weiss-Cerem L, Rahat MA. Hypoxia increases membranal and secreted HLA-DR in endothelial cells, rendering them T-cell activators. Transpl Int. 2011;24:1018–1026. doi: 10.1111/j.1432-2277.2011.01304.x. [DOI] [PubMed] [Google Scholar]
- 53.Mannam VK, Lewis RE, Cruse JM. The fate of renal allografts hinges on responses of the microvascular endothelium. Exp Mol Pathol. 2013;94:398–411. doi: 10.1016/j.yexmp.2012.06.002. [DOI] [PubMed] [Google Scholar]
- 54.Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152:1298–1307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Erbilgin A, Civelek M, Romanoski CE, Pan C, Hagopian R, Berliner JA, Lusis AJ. Identification of CAD candidate genes in GWAS loci and their expression in vascular cells. J Lipid Res. 2013;54:1894–1905. doi: 10.1194/jlr.M037085. [DOI] [PMC free article] [PubMed] [Google Scholar]
References
- 1.Watson AD, Leitinger N, Navab M, Faull KF, Horkko S, Witztum JL, Palinski W, Schwenke D, Salomon RG, Sha W, Subbanagounder G, Fogelman AM, Berliner JA. Structural identification by mass spectrometry of oxidized phospholipids in minimally oxidized low density lipoprotein that induce monocyte/endothelial interactions and evidence for their presence in vivo. J Biol Chem. 1997;272:13597–13607. doi: 10.1074/jbc.272.21.13597. [DOI] [PubMed] [Google Scholar]
- 2.Subbanagounder G, Leitinger N, Schwenke DC, Wong JW, Lee H, Rizza C, Watson AD, Faull KF, Fogelman AM, Berliner JA. Determinants of bioactivity of oxidized phospholipids. Specific oxidized fatty acyl groups at the sn-2 position. Arteriosclerosis, Thrombosis, and Vascular Biology. 2000;20:2248–2254. doi: 10.1161/01.atv.20.10.2248. [DOI] [PubMed] [Google Scholar]
- 3.Nievelstein PF, Fogelman AM, Mottino G, Frank JS. Lipid accumulation in rabbit aortic intima 2 hours after bolus infusion of low density lipoprotein. A deep-etch and immunolocalization study of ultrarapidly frozen tissue. Arterioscler Thromb. 1991;11:1795–1805. doi: 10.1161/01.atv.11.6.1795. [DOI] [PubMed] [Google Scholar]
- 4.Jongstra-Bilen J, Haidari M, Zhu SN, Chen M, Guha D, Cybulsky MI. Lowgrade chronic inflammation in regions of the normal mouse arterial intima predisposed to atherosclerosis. J Exp Med. 2006;203:2073–2083. doi: 10.1084/jem.20060245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang da WSB, Lempicki RA. Systematic and integrative analysis of large gene lists using david bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 6.Huang da WSB, Lempicki RA. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.