Abstract
TRPML3 is a member of the mucolipin branch of the transient receptor potential cation channel family. A dominant missense mutation in Trpml3 (also known as Mcoln3) causes deafness and vestibular impairment characterized by stereocilia disorganization, hair cell loss, and endocochlear potential reduction. Both marginal cells of the stria vascularis and hair cells express Trpml3 mRNA. Here we used in situ hybridization, quantitative RT-qPCR, and immunohistochemistry with several antisera raised against TRPML3 to determine the expression and subcellular distribution of TRPML3 in the inner ear as well as in other sensory organs. We also use Trpml3 knockout tissues to distinguish TRPML3-specific from nonspecific immunoreactivities.
We find that TRPML3 localizes to vesicles of hair cells and strial marginal cells but not to stereociliary ankle links or pillar cells, which nonspecifically react with two antisera raised against TRPML3. Upon cochlear maturation, TRPML3 protein is redistributed to perinuclear vesicles of strial marginal cells and is augmented in inner hair cells vs. outer hair cells. Mouse somato-sensory neurons, retinal neurons, and taste receptor cells do not appear to express physiologically relevant levels of TRPML3. Finally, we found that vomeronasal and olfactory sensory receptor cells do express TRPML3 mRNA and protein, which localizes to vesicles in their somas and dendrites as well as at apical den dritic knobs.
Keywords: mucolipin, VNO, taste receptor, circumvallate, CVP, retina, lysosome, endosome, transient receptor potential, TRP, ion channel, Mcoln3
Most types of sensory receptor cells form part of a specialized neuroepithelia devoted to detecting a form of environmental stimulation. Rod and cone photoreceptors of retina in the eye detect light. Taste receptors of taste buds in the tongue detect tastants. Olfactory receptor neurons (ORNs) of the olfactory epithelium (OE) in the nose detect odorants. Vomeronasal sensory neurons (VSNs) of the vomeronasal organ (VNO) in the nose detect pheromones. Hair cells (HCs) of the vestibular (saccule, utricle, and crista ampullaris) and auditory (organ of Corti) sensory epithelia in the inner ear detect displacements caused by head movements or sound. Despite the diverse nature of the stimuli they detect, sensory receptor cells display substantial anatomical and functional similarities and express many common genes.
For example, hair cells, vomeronasal sensory neurons, and taste receptor cells concentrate their macromolecular transduction complexes on actin-rich microvilli, and all contain the filamentous actin-bundling protein espin (Sekerkova et al., 2004); Usher syndrome, the most common cause of combined deafness and blindness, is caused by mutations in several genes used by both hair cells and photoreceptors (Kremer et al., 2006).
Varitint-waddler (Va and VaJ) mice suffer deafness and vestibular impairment characterized by an initial disorganization, fusion, and clumping of hair cell stereocilia, followed by loss of hair cells and reduction or elimination of the endocochlear potential (Cable and Steel, 1998; Deol, 1954; Di Palma et al., 2002). In situ hybridization with nonoverlapping probes on sections of neonatal inner ear revealed the cells that express Trpml3 mRNA (Nagata et al., 2008). These include marginal cells of the stria vascularis, the secretory epithelium that contributes to generating the endocochlear potential, and hair cells. A mutation shared by the Va and VaJ alleles causes an A419P substitution in TRPML3 (Di Palma et al., 2002) that favors its open state, generating a lethal cationic inward current (Grimm et al., 2007; Kim et al., 2007; Nagata et al., 2007, 2008; van Aken et al., 2008; Xu et al., 2007). Cultured epithelial cells heterologously expressing TRPML3(A419P) protrude from the epithelium and die in a manner similar to dying hair cells in varitint-waddler mice (Nagata et al., 2008). These observations explained the hair cell loss of varitint-waddler mice. They also revealed the existence in the inner ear of novel TRPML3-containing channels of unknown function.
The mucolipin branch of the transient receptor potential (TRP) family of cation channels (Venkatachalam et al., 2006) includes TRPML1 and TRPML2 in vertebrates, plus TRPML3 in amphibians, reptiles, birds, and mammals, but not in bony fishes (Flores and García-Añoveros, 2010). All mucolipins appear to localize to vesicles of the endocytic pathway (Cheng et al., 2010). TRPML1 localizes to lysosomes, and loss-of-function mutations in Trpml1 (a.k.a. Mcoln1) cause mucolipidosis type IV, a neurodegenerative lipid-storage disorder (Cheng et al., 2010; Puertollano and Kiselyov, 2009; Zeevi et al., 2007). Evidence for the presence of TRPML3 in late endosomes and lysosomes comes from localization of fluorescently tagged, heterologously expressed TRPML3 and from immunoreactivities on cells thought to express TRPML3 endogenously (Kim et al., 2009; Martina et al., 2009; Nagata et al., 2008; Venkatachalam et al., 2006; Zeevi et al., 2009). However, some evidence also points to localization of TRPML3 on hair cell stereocilia. In espin-expressing CL4 cells, a culture model of hair cells (Zheng et al., 2010), heterologously expressed TRPML3::GFP localized to late endosomes/lysosomes as well as to the espin-enlarged micro-villi that resemble stereocilia (Nagata et al., 2008). Furthermore, several antisera raised against TRPML3 immunoreacted with vesicles as well as stereocilia of neonatal hair cells (Di Palma et al., 2002). Particularly, two antisera raised against different domains of TRPML3 (the cytosolic carboxyl-terminus and the extracellular third loop) both labeled the ankle links (van Aken et al., 2008), structures that connect postnatal stereocilia (from P2 to P12) at their base (Goodyear et al., 2005; van Aken et al., 2008). Mutations in several genes that contribute ankle link components cause Usher syndrome, the most common form of combined deafness and blindness (Adato et al., 2005; Delprat et al., 2005; Kremer et al., 2006; Kussel-Andermann et al., 2000; McGee et al., 2006; Senften et al., 2006). Hence, these results suggest that TRPML3 may contribute to ankle link function and that deafness in varitint-waddler mice may relate to dysfunction of these stereociliary connectors.
The above-mentioned localization studies were not conclusive for several reasons. 1) Heterologously expressed tagged protein may mislocalize as a result of tagging or overexpression. 2) Antisera often immunoreact with proteins other than the one against which they were raised. To determine the distribution of TRPML3 in the inner ear and other sensory epithelia unambiguously, we used several antisera raised against separate domains of TRPML3 to identify cells and organelles that might contain TRPML3 protein. We also used in situ hybridization with nonoverlap-ping probes to confirm that cells expressing TRPML3 protein also express its mRNA and Trpml3 knockout tissues to rule out nonspecific immunoreactivities. We found that the ankle link immunoreactivities do not represent TRPML3 protein, which localizes to vesicles of hair cells as well as of marginal cells of the stria vascularis. We also found that vomeronasal and olfactory chemosensory neurons of the nose, but not taste receptor cells of tongue, retinal neurons, or somatosensory neurons of dorsal root and trigeminal ganglia, express TRPML3.
MATERIALS AND METHODS
Animals
All animal care and procedures were in strict accordance with the Guide for the care and use of laboratory animals published by the National Institutes of Health and were approved by the Northwestern University Institutional Animal Care and Use Committee. Mice were housed in the barrier rooms of Northwestern University's animal facility. We obtained tissues from either Swiss Webster or CD1 mice (Charles River) or from Trpml3−/− and Trpml3+/+ littermates with a genetic background ~75% C57BL/6 and ~25% Sv129/Ola. The Trpml3 knockout (Trpml3− ) allele was generated by homologous recombination in HM1/129Ola ES cells, which are derived from Sv129/Ola mice. Chimeras obtained from these mice were crossed to C57BL/6, and, after germline transmission, their Trpml3−/+ heterozygous progeny were mated once more to C57BL/6. The resulting Trpml3−/+ males and females were mated, and their wild-type and knockout progeny were used to obtain the various tissues used in this study.
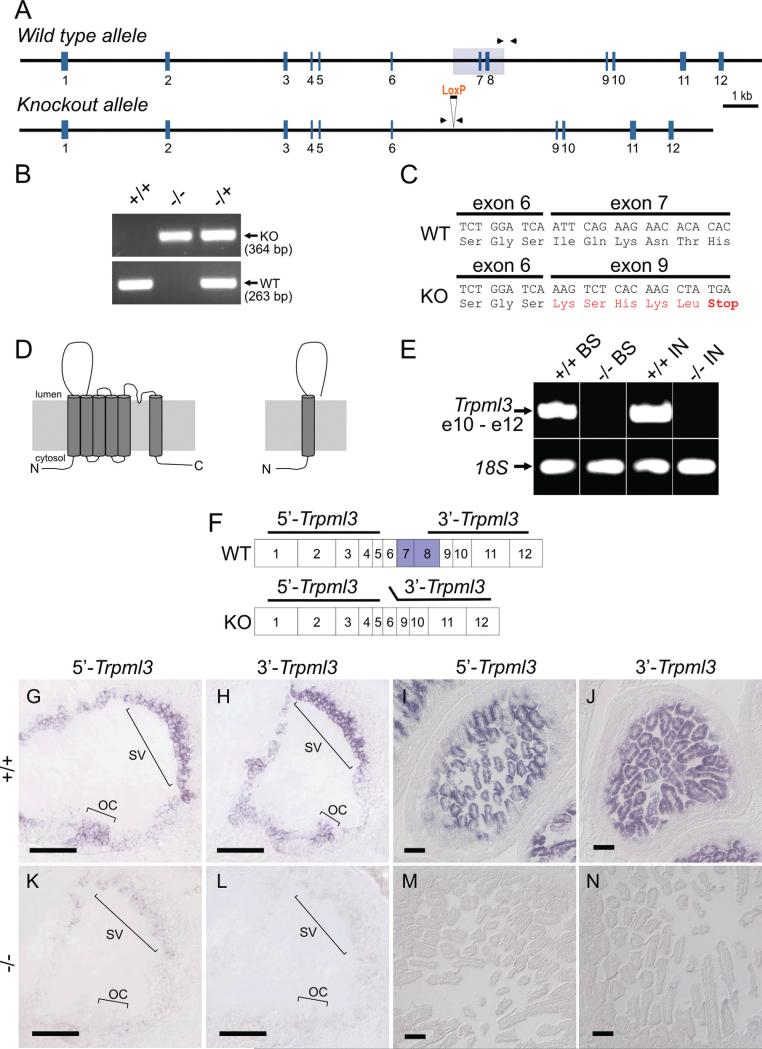
Tissues were genotyped by PCR from genomic DNA (Fig. 1A,B). A forward primer located within the deleted portion of intron 8 (WTgntyp1; 5’-GTGGAGCCTTGACTGTCTAG) and a reverse primer located within the nondeleted portion of intron 8 (WTgntyp4; 5’-CTGTGAGACCTCTTAACAACTCT) generate a 263-base-pair (bp) product from the wild-type (Trpml3+) allele and no product from the knockout (Trpml3− ) allele. Another forward primer located in intron 6, upstream of the deletion (ML3KO-fInt6; 5’-GGCAAGAGCTGAGGATATCTT), and a reverse primer located in intron 8, downstream of the deletion (ML3KOrInt8; 5’-GAACTCTCTCGATCTAACCACTC), generate a 364-bp product from the knockout (Trpml3− ) allele and no product from the wild-type (Trpml3+) allele, unless the PCR extension time is lengthened to permit amplification of a 1,697-bp product containing exons 7 and 8.
Figure 1.
Deletion of Trpml3 exons 7 and 8 truncates mRNA translation and greatly reduces mRNA levels. A: Schematic representation of Trpml3 wild-type and knockout alleles. Exons (vertical blue lines) and introns (horizontal black line) shown to scale. The region deleted is shown as a light gray box over exons 7 and 8. LoxP site remaining after excision also shown. Arrowheads represent primers (located in introns) used for PCR genotyping. B: Representative PCR genotyping results from Trpml3 wild-type (+/+), knockout (−/−), and heterozygous (−/+) mice. The wild-type primers generate a product of 263 bp, and the knockout primers generate a product of 364 bp. C: Sequence of RT-PCR products obtained using RNA extracted from Trpml3−/− (KO) inner ear and melanocyte-containing back skin (P13) as template. Primers located upstream (exon 4) and downstream (exon 9) of the deletion. In the mutants, splicing joins exon 6 with exon 9, generating five out-of-frame codons (in red), followed by a premature stop codon. The wild-type (WT) sequence is included for comparison. D: Schematic representation of the wild-type TRPML3 protein and of the truncated protein fragment that could theoretically be translated from the KO mRNA, should this be sufficiently stable for protein production. The N and C termini are predicted to be cytoplasmic in wild-type TRPML3. E: RT-PCR from mRNA of back skin (BS, from a P13 pup, which is rich in melanocytes) and intestine (IN) with primers from exons 10–12 reveals a great reduction of Trpml3 mRNA in Trpml3−/− mice compared with Trpml3+/+ mice. RT-PCR on the same templates with primers for 18S rRNA demonstrates the presence of integral RNA in the Trpml3−/− samples. F: Location of Trpml3 in situ hybridization probes. Horizontal lines over the exons depict the cDNA fragments used to generate the nonoverlapping 5’ and 3’ probes for in situ hybridization. Exons 7 and 8, deleted in the KO, are labeled blue in the wild type. G-N: In situ hybridization with probes that detect fragments of Trpml3 mRNA upstream (5’) or downstream (3’) of the deleted exons reveals great reduction or elimination of mRNA in neonatal (P2) inner ear and intestine of Trpml3−/− mice compared with Trpml3+/+ mice. OC, organ of Corti; SV, stria vascularis. Scale bars ¼ 100 μm. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Antisera characterization
The present study used triple-controlled immunohisto-chemistry to determine the tissue and subcellular expression pattern of TRPML3 protein. We employ antisera raised against distinct regions of TRPML3 (see Fig. 2M). We also compare TRPML3 immunoreactivities with available in situ hybridization (ISH) analyses. Finally, we determine which immunoreactivities are absent from the tissues of a Trpml3−/− mouse. Below is a summary of knowledge prior to our study. See Table 1 for additional details, including epitope information.
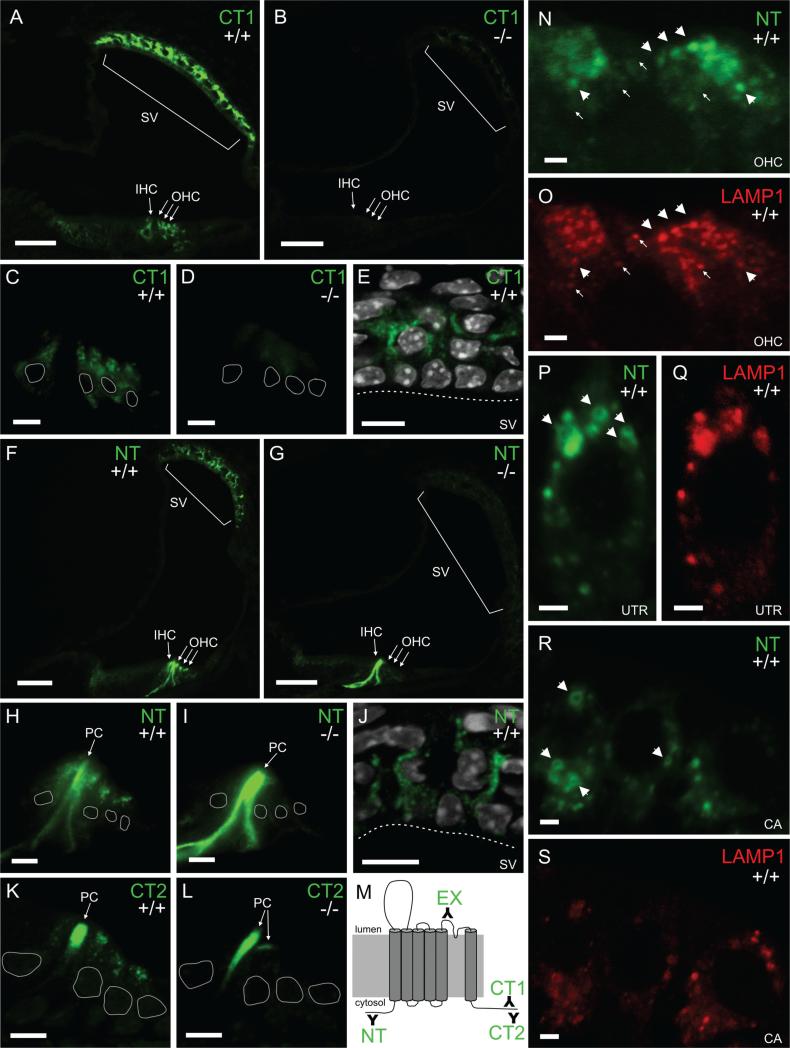
Figure 2.
TRPML3 protein localizes to stria vascularis and hair cell vesicles of neonatal cochleae. Immunohistochemistry on sections of mouse inner ear with three different antibodies raised against different parts of TRPML3 (schematically shown in M). A-E: The carboxyl-terminal CT1 labels inner and outer hair cells (IHC and OHC) as well as cells of the stria vascularis (SV) in Trpml3+/+ (A,C,E) but not Trpml3−/− (B,D) cochleae. White perimeters indicate the position of hair cell nuclei. White dashed lines delineate the apical edge of marginal cells in the stria vascularis. E: DAPI (in white) labels the nuclei of SV cells. F-J: The amino-terminal NT also labels IHC, OHC, and SV cells in addition to Pillar cells (PC) in Trpml3+/+ (F,H,J) but labels only PCs in Trpml3−/− (G,I) cochleae, indicating that the PC immunore-activity is nonspecific. J: DAPI (in white) labels the nuclei of SV cells. K,L: The carboxyl-terminal CT2 also labels IHC, OHC, and PC Trpml3+/+ (K) but labels only PCs in Trpml3−/− (L) cochleae, indicating that the PC immunoreactivity is nonspecific. M: Schematic representation of the TRPML3 protein (membrane-spanning domains depicted as cylinders) and of the antibodies raised against it (represented with Y signs and termed NT, EX, CT1, and CT2). The N and C termini are predicted to be cytoplasmic. N,O: Double immunohistochemistry with antibodies to NT (N) and the late endosome and lysosome marker LAMP1 (O) reveals colocalization of both proteins in vesicles of three outer hair cells. Approximately 66% of TRPML3-positive vesicles were also LAMP1 positive. Arrowheads point to vesicles that contain TRPML3 and LAMP1, whereas arrows point to vesicles that contain LAMP1 but not TRPML3. P-S: Double immunohistochemistry with antibodies to NT (P,R) and LAMP1 (Q,S) also reveals colocalization of both proteins in vesicles of hair cells from utricle (UTR) and crista ampullaris (CA). Arrowheads point to TRPML3-positive vesicles that are large enough to be visible as rings. All pairs of Trpml3+/+ and Trpml3−/− images were acquired with the same exposure, laser, and gain settings. The brightness and contrast settings were adjusted identically for each pair of images. A-L and P-S are from postnatal day 4 (P4) cochleae. N,O are from P7 cochleae. A magenta-green version of this figure is supplied as Supporting Information Figure 1. Scale bars = 50 μm in A,B,F,G; 10 μm in C-E,H–L; 2 μm in N-S.
TABLE 1.
Antibodies Used
| Name | Immunogen | Origin, species, type |
|---|---|---|
| TRPML3-NT | Synthetic peptide equivalent to aa 26–43 of human TRPML3 (TSPSEELLLEDQMRRKLK) conjugated to KLH | Sigma No. M7570 (Lot 067K4822), rabbit, polyclonal, affinity purified with unconjugated immunizing peptide |
| TRPML3-EX | Synthetic peptide equivalent to aa 446–462 of mouse TRPML3 (RVSECLFSLINGDDMFS) | Original name: PB221 (Di Palma et al., 2002; van Aken et al., 2008), rabbit, polyclonal, affinity purified with immunizing peptide |
| TRPML3-CT1 | Synthetic peptide equivalent to aa 528–547 of mouse TRPML3 (CKDLPNSGKYRLEDDPPGSL) | Xu et al., 2007; rabbit, polyclonal, affinity purified with immunizing peptide |
| TRPML3-CT2 | Synthetic peptide equivalent to aa 529–548 of mouse TRPML3 (KDLPNSGKYRLEDDPPGSLL) | Original name: HL4460 (Di Palma et al., 2002; van Aken et al., 2008), rabbit, polyclonal, affinity purified with immunizing peptide |
| LAMP1 | Antibodies generated by immunizing rats with NIH3T3 cell membrane fraction (Hughes et al., 1981); subsequent analysis by 2D gel and tryptic mapping revealed that clone 1D4B recognizes the N-terminus of LAMP1 (Chen et al., 1985) | Developmental Studies Hybridoma Bank, clone 1D4B, rat, monoclonal |
| TuJ1 | Antibody raised against microtubules derived from rat brain (manufacturer's technical information) | Covance No. MMS-435P, mouse IgG2a |
| Espin | Purified recombinant, full-length rat espin 2B (Genbank AA050330) with His tag | Rabbit polyclonal, affinity purified with same recombinant His-tagged rat Espin 2B (Sekerkova et al., 2008) |
TRPML3-NT
The NT antiserum (Sigma; M7570) recognized a single band (~75 kDa) in a Western blot of total protein extracts from HEK cells transfected with human Trpml3 cDNA but not in extracts from untransfected HEK cells. Preincubation of antiserum with antigenic peptide removed detection of this band (product information sheet; Sigma, St. Louis, MO). In addition, shRNA mediated knockdown of human Trpml3 mRNA and preincubation with the antigenic peptide reduced endogenous NT immunoreactivity in HEK cells (Zeevi et al., 2009).
TRPML3-CT1
The CT1 antiserum recognized a single band of ~60 kDa (close to the 59 kDa predicted for TRPML3) in a Western blot of protein obtained from HEK cells heterologously expressing mouse HA-TRPML3 fusion protein after immunoprecipitation with the same antisera. The presence of antigenic peptide during the immunoprecipitation abolished detection of this band. Signal was also absent from untransfected HEK cells. The CT1 antiserum also recognized a single band (~60 kDa) in a Western blot of cultured mouse melanocyte (melan-a2) lysate. In the same Western, the antigenic peptide successfully competed the endogenous protein, eliminating the CT1 band (Xu et al., 2007). In situ hybridization confirmed that melanocytes express Trpml3 mRNA (Xu et al., 2007; and our unpublished results).
TRPML3-CT2
The CT2 antiserum (originally termed HL4460) recognized heterologously expressed mouse TRPML3 protein tagged with GFP on Western blots and colocalized with GFP-tagged TRPML3 by immunohistochemistry (van Aken et al., 2008). The CT2 antiserum also labeled a single band (~68 kDa) in a Western blot of wild-type mouse cochlear and vestibular lysates (van Aken et al., 2008). These are tissues previously shown to express Trpml3 mRNA by ISH (Nagata et al., 2008). CT2 also recognized structures in the ankle link region of stereocilia similar to those recognized by independent antiserum EX.
TRPML3-EX
The EX antiserum (originally termed PB221) labeled hair cell soma in a punctate pattern. This label was absent after incubation with preimmune serum or excess specific peptide (Di Palma et al., 2002). This affinity-purified antiserum also recognized structures in the ankle link region of stereocilia similar to those recognized by independent antiserum CT2 (van Aken et al., 2008).
LAMP1-1D4B
Immunogold labeling consistently showed 1D4B location immediately inside lysosomal membranes (Chen et al., 1985). 1D4B also colabeled with lysosome marker acridine orange. In Western blots of tissue lysates obtained from wild-type and LAMP1 knockout mice, LAMP1 recognized a band (~90 kDa) in the predicted size range in wild-type tissues only (Andrejewski et al., 1999).
TuJ1
Mouse monoclonal antibody was raised against micro-tubules derived from rat brain (manufacturer's technical information). It specifically recognizes rat brain bIII -tubulin as a 50-kD band in Western blots of rat and chicken proteins. In developing neural epithelia, it reacts primarily with postmitotic cells of the marginal zone (Lee et al., 1990).
Espin
The espin antiserum has numerous examples demonstrating its specificity for rat and mouse espins (Sekerkova et al., 2008). In Western blots, bands labeled by this antibody on protein extracts from native tissues coincide with bands seen in protein extracts from transfected cells heterologously expressing espin cDNAs (Sekerkova et al., 2004). Furthermore, the espin antibody does not react in Western blotting or immunohistochemistry with protein or tissues prepared from homozygous jerker mice, which lack espin proteins because of a mutation in their espin gene (Sekerkova et al., 2006; Zheng et al., 2000).
Tissue processing
We used unfixed tissue for ISH (P2 VNO) and some immunohistochemistry (adult CVP). These tissues were dissected, embedded in OCT, and immediately snap frozen in dry ice isopentane. For fixed adult tissues (inner ear and VNO), we cardially perfused the animal with 2% paraformaldehyde, dissected out the bone-encased organs, postfixed for 1 hour in 2% paraformaldehyde, and rinsed three times in 1 × PBS. We decalcified the bone in RDO rapid decalcifier (Electron Microscopy Sciences, Fort Washington, PA) for 10–15 minutes and rinsed three times in 1 × PBS. We then took the tissue through a sucrose gradient (1 hour each 5%, 10%, 20%), ending with an overnight incubation in 20% sucrose and 50% OCT (Tissue-Tek; Sakura). We mounted the tissue in OCT and froze it on dry ice. For neonatal inner ears, we did not perfuse but rather punctured several small holes in the cochlea to facilitate the entry of fixative prior to the 1 hour of fixation. The overnight embedding in 20% sucrose and 50% OCT was critical to preserving anatomy during cryosectioning of adult inner ears. We also prepared our own paraformaldehyde fresh from powder. Consistency in fixation was very important, because we often saw background autofluorescence that we could ascribe to over- or inconsistent fixation.
Immunohistochemistry
With noted exceptions, we performed all TRPML3 immunohistochemistry using a tyramide signal amplification system (TSA; Alexa488 tyramide; Invitrogen, Carlsbad, CA). When noted, we used the ABC/DAB (avidin-biotin complex with diaminobenzidine reaction) signal amplification system (Vector, Burlingame, CA). We also performed immunohistochemistry (shown in Fig. 4) with CT2 and EX antisera exactly as described previously for these antisera (van Aken et al., 2008). LAMP1, TuJ1, and espin were detected by using conventional Texas red (Jackson Immunoresearch, West Grove, PA)- or Alexa568 (Invitrogen)-labeled secondary antibodies. The following paragraphs provide brief protocols for our immunohisto-chemistry techniques.
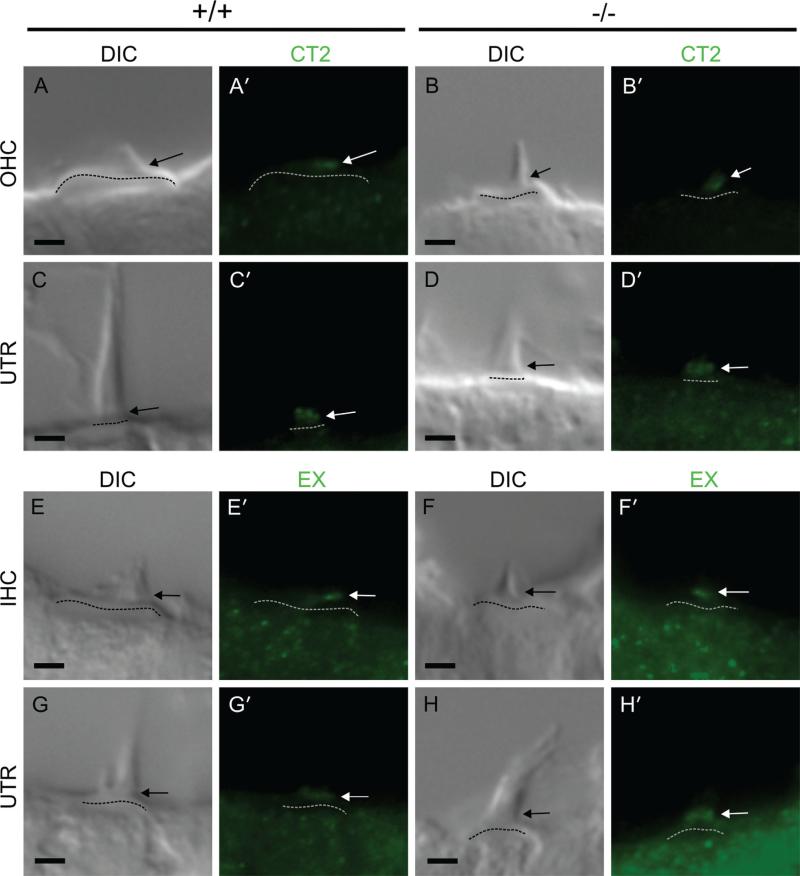
Figure 4.
Ankle link immunoreactivity on hair cells does not represent TRPML3 protein. A-H: Differential Interference Contrast (DIC) images of hair cell stereociliary bundles. A’-H’: Fluorescent images of the same stereociliary bundles immunostained in the absence of antigen retrieval with anti-TRPML3 antibodies CT2 (A’-D’) and EX (E’-H’; shown in Fig. 2M). Arrows point to immunoreactivities at the base of auditory and vestibular stereociliary bundles in both Trpml3+/+ and Trpml3−/− tissues. All pairs of Trpml3+/+ and Trpml3−/− images were acquired using same exposure settings. The brightness and contrast settings were adjusted identically for each bundle type pair of images. Dashed lines indicate cuticular plates. OHC, outer hair cell; IHC, inner hair cell; UTR, utricular hair cell. All images are from postnatal day 2 (P2) mice. Scale bars = 2 μm. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Tyramide signal amplification
Air dry fixed sections for 15 minutes and vacuum dry for 15 additional minutes. Rinse for 3 × 5 minutes in 1 × PBS (Lonza). Thaw unfixed sections briefly, then postfix for 10 minutes in freshly prepared 2% paraformaldehyde. Rinse 3 × 5 minutes in 1 × PBS. Retrieve antigens by incubating in 10 mM sodium citrate, pH 6, with 0.25% Triton for 20 minutes at 92°C. After cooling for 30 minutes at room temperature, rinse 3 × 5 minutes in 1 × PBS. Quench endogenous peroxidase by incubating in 1% H2O2, 10% methanol, in 1 × PBS for 30 minutes. Rinse 3 × 5 minutes in 1 × PBS. Block for 1 hour in 1% TSA block solution (Invitrogen). Incubate with primary antibody (NT, 1:5,000–1:10,000; CT1, 1:1,000–1:2,000; CT2, 1:1,000) in 1% TSA block solution (no azide) overnight at 4°C. On the next day, rinse 4 × 10 minutes in 1 × PBS. Incubate with secondary antibody (1:100, goat anti-rabbit; Invitrogen) in 1% TSA block solution for 1 hour in the dark. Rinse 4 × 10 minutes in 1 × PBS. Proceed with tyramide labeling reaction in groups of five or six slides. Apply tyramide (1:150; Invitrogen) working solution to slide. Incubate for 10 minutes at room temperature in the dark. Rinse 3 × 10 minutes in 1 × PBS, in the dark. Add DAPI (1 μM) for 10 minutes. Rinse 2 × 10 minutes in 1 × PBS. Rinse with ddH2O. Mount with Prolong Gold (Invitrogen).
With adult inner ears (Fig. 3), we retrieved antigens using milder conditions (10 mM sodium citrate, pH 6, with 0.025% Triton for 10 minutes at 92°C) to facilitate tissue adhesion to the slide. For double immunohisto-chemistry using TSA to detect one of the TRPML3 antibodies, we obtained our best results when performing TSA detection of the TRPML3 antibody, followed by the conventional detection of the second primary antibody.
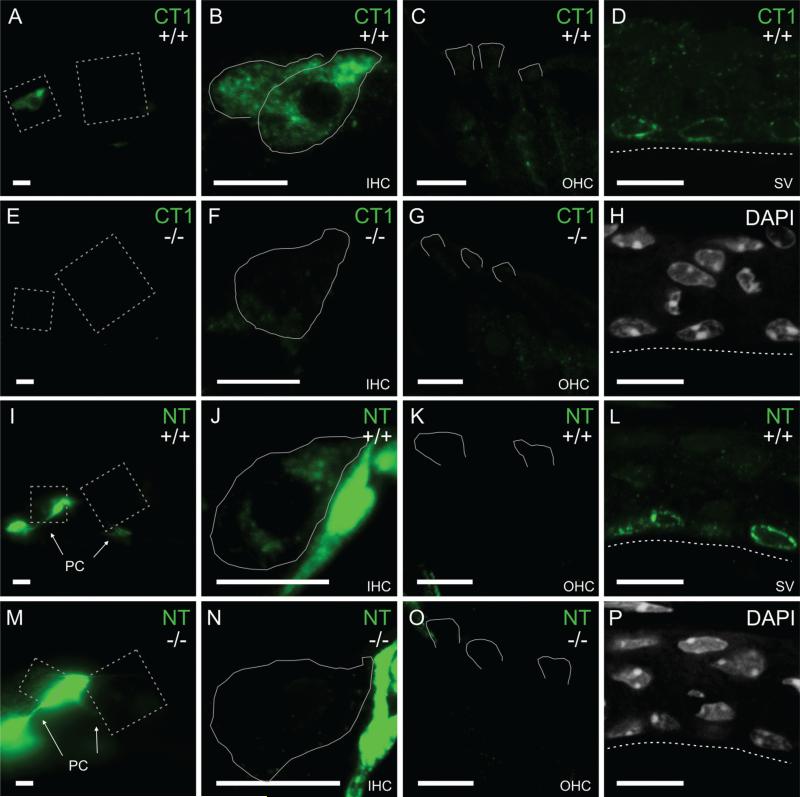
Figure 3.
TRPML3 redistribution to perinuclear vesicles of marginal strial cells and to inner hair cells of mature cochleae. A-C,E-G,I-K,M-O: Immunoreactivities on sections of the organ of Corti from juvenile (P29) mice with an antibody raised against a carboxyl-terminal domain of TRPML3 (CT1; A-C,E-G) and an antibody raised against an amino-terminal domain of TRPML3 (NT; I-K,M-O). Immunoreactivities were used with the same conditions as for neonatal inner ears (Fig. 2) or, to facilitate tissue adhesion to the slide, with milder conditions (shown here). Both antibodies label inner hair cells (IHCs; A,B,I,J), but not outer hair cells (OHCs; C,K) from Trpml3+/+ mice. These immunoreactivities likely represent TRPML3 protein, because they are not detected on sections from Trpml3−/− mice (E-G,M-O). As with the neonatal organ of Corti, the NT antibody labels pillar cells (PC) nonspecifically in both Trpml3+/+ (I,J) and Trpml3−/− (M,N) inner ears. D,H,L,P: Immunoreactivities on sections of adult (P147) stria vascularis with the same two antibodies (CT1 in D and NT in L) reveal that TRPML3 redistributed to vesicles surrounding the nuclei (labeled with DAPI in H and P) of marginal cells. Tissue from Trpml3−/− mice lacked these immunoreactivities. All pairs of Trpml3+/+ and Trpml3−/− images were acquired with same exposure, laser, and gain settings. The brightness and contrast settings were adjusted identically for each pair of images. Dashed boxes in A,E,I,and M indicate areas magnified to show IHCs and OHCs. White lines delineate full IHCs or apical regions of OHCs as determined by LAMP1 immunoreactivity (not shown). Dashed lines indicate apical side of marginal cells in stria vascularis. Scale bars = 10 μm. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
ABC/DAB signal amplification
Air dry fixed sections for 15 minutes and vacuum dry for 15 additional minutes. Rinse 3 × 5 minutes in 1 × PBS (Lonza). Thaw unfixed sections briefly, then postfix for 10 minutes in freshly prepared 2% paraformaldehyde. Rinse 3 × 5 minutes in 1 × PBS. Retrieve antigens by incubating in 10 mM sodium citrate, pH 6, with 0.25% Triton for 20 minutes at 92°C. After cooling for 30 minutes at room temperature, rinse 3 × 5 minutes in 1 × PBS. Quench endogenous peroxidase by incubating in 1% H2O2,10% methanol, in 1 × PBS for 30 minutes. Rinse 3 × 5 minutes in 1 × PBS. Block for 2 hours in 10% normal goat serum, in 1 × PBS. Incubate primary antibody (NT, 1:2,000; CT1, 1:1,000; CT2, 1:1,000) in 10% normal goat serum block + 0.1% Triton (no azide) overnight at 4°C. On the next day, rinse 4 × 10 minutes in 1 × PBS 0.1% Triton. Incubate with biotinylated secondary antibody (1:200, goat anti-rabbit; Vector) in 10% normal goat serum block + 0.1% Triton for 1 hour. Rinse 4 10 minutes in 1 × PBS + 0.1% Triton, 50 rpm. ABC solution (Vector) Prepare 30 minutes prior to use. Incubate in ABC solution for 1 hour. Rinse 3 × 5 minutes in 1 × PBS. Incubate with DAB solution (Sigma) for at least 7 minutes until tissue turns light brown. Rinse 3 × 5 minutes in 1 × PBS. Add DAPI (1 μM) for 10 minutes. Rinse 2 × 10 minutes in 1 × PBS. Rinse with ddH2O. Mount using Prolong Gold (Invitrogen).
In situ hybridization
We performed in situ hybridization on cryostat sections of snap frozen, unfixed tissues from CD1, Trpml3+/+, and Trpml3−/− mice using protocols previously described (Duggan et al., 2008; Nagata et al., 2008; Schaeren-Wiemers and Gerfin-Moser, 1993). Freshly dissected and unfixed tissues were immediately snap frozen by dipping in isopentane cooled to −30°C with dry ice and sectioned (10-12 μm). For ISH, we used two nonoverlapping cRNA probes for mouse Trpml3 mRNA (Genbank ID NM_134160). These are a 5’ probe, which corresponds to nucleotides 179-723 (from codon 60 at the end of exon 1 to codon 240 at the end of exon 5) and a 3’ probe, which corresponds to nucleotides 1005-1594 (from codon 335 in the middle of exon 8 to codon 531 in the middle of exon 12, the last exon). The probe for mouse Pkd1l3 corresponds to nucleotides 5803-6241 of its mRNA (Genbank ID NM_181544).
We PCR amplified these cDNA fragments from mouse inner ear or CVP mRNA and TA cloned them into vector pCRII. We generated digoxigenin-labeled antisense and sense (control) cRNA probes using the DIG-RNA labeling kit (Roche, Indianapolis, IN) according to the manufacturer's instructions. Sections were hybridized with antisense or sense probes as previously described (Schaeren-Wiemers and Gerfin-Moser, 1993). Sections were mounted for observation. Only cell types that labeled with both the 5’ and the 3’ Trpml3 probes were considered positive for Trpml3 mRNA.
Image acquisition and analysis
We acquired images using either a Nikon E600 pan fluorescence microscope (×20 0.75 N.A., ×60 1.4 N.A., or ×100 1.4 N.A. objectives) equipped with a CCD camera (Spot RC-Slider) or a Zeiss LSM 510 confocal microscope (×63 1.4 N.A. or ×100 1.46 N.A. objectives). When comparing wild-type and knockout immunoreactivities, we captured images under identical conditions. In practice, this meant capturing images with identical exposure settings (pan fluorescence) or identical laser and gain settings (confocal). For even illumination, we flat field corrected and white balanced the color (Spot RC-Slider) camera prior to acquiring DIC images.
After acquisition, we identically processed image pairs of wild-type tissues and their corresponding knockout controls. This included deconvolution of pan fluorescence images as well as adjustment for brightness and contrast of all images. Confocal images shown in this paper represent a maximum-intensity projection of either two or three Z-stack image slices. We used ImageJ for all post-acquisition processing, with the exception of deconvolution, for which we used MetaMorph.
RT-PCR
We isolated total RNA of P13 back skin, inner ears, and intestine from both Trpml3+/+ and Trpml3−/− mice using Trizol reagent (Invitrogen). Two micrograms of total RNA from each tissue was reverse transcribed with Superscript III (Invitrogen) and used for standard RT-PCR (Invitrogen) with primers for the Trpml3 allele: Trpml3ex4f, 5’TAAACTCAACCTGAGCCTGG; Trpml3ex9r, 5’ATAGTTG ACGTCCCGAGAAG; Trpml3ex10f, 5’TGCTGCGCTGCT ATGATCTA; Trpml3ex12r, 5’GAGGTCTTTGCATTCCGCTA. PCR was performed with a Mastercycler (Eppendorf) programmed as follows: 1 cycle of 95°C for 10 minutes; followed by 35 cycles of 95°C for 30 seconds, 59°C for 30 seconds, 72°C for 1 minute; followed by a final extension at 72°C. RT-PCR products were run on 1.5% agarose gels prestained with ethidium bromide and imaged on a BioDoc-it UV imaging system. We cloned and sequenced PCR products to confirm their molecular identity.
Quantification of mRNA copy number by RT-qPCR
For RT-qPCR (reverse transcription of RNA followed by quantitative polymerase chain reaction), we used tissues from animals cardially perfused with 1 × PBS (to remove blood) prior to decapitation and dissection. All tissues were dissected as quickly and as cleanly as possible and immediately snap frozen on dry ice, until homogenized in Trizol (Invitrogen). We homogenized all tissues in the same volume of Trizol using a microcentrifuge tube and microcentrifuge pestle, followed by five passages through a 20-G needle, and performed RNA isolation with Trizol according to the manufacturer's instructions. RNA concentration was determined by UV absorption (OD260). This value helped determine the volume of RNA used per RT reaction, with the goal of reverse transcribing 1 μg total RNA per reaction. Prior to RT, we subjected the 1 μg total RNA to DNaseI treatment (Invitrogen) to eliminate genomic DNA according to the manufacturer's protocol. This DNaseI-treated 1 μg total RNA was then subjected to first-strand cDNA synthesis with Superscript III reverse transcriptase (Invitrogen) according to the manufacturer's protocol.
We performed RT-qPCR with a Mastercycler Realplex2 machine (Eppendorf) on ~100 ng (2 μl of a 20-μl RT reaction) of first-strand cDNA with SYBR Green PCR Master-mix (Applied Biosystems, Foster City, CA) in triplicate, according to the manufacturer's instructions. The following primers (IDT) were designed on mouse sequence and used in qPCR: Trpml3ex8f, 5’-ATGGAGTTCATCAACGGGTG; Trpml3ex9r, 5’-ATAGTTGACGTCCCGAGAAG; 18Sf, 5’TTGACGGAAGGGCACCACCAG; 18Sr, 5’-GCACCACCACC CACGGAATCG; Trpa1ex12f, 5’-GGCGCATCAATACATGTC AG; Trpa1ex13r, 5’-CGTGATGCAAAGCAGTCCAG; Pkd1l3ex2f, 5’-GTCACACACTCTAATGCTGTCTT; Pkd1l3ex3r, 5’-CAGGAC CAAACTCATTTCTCCAA. Melting curve analysis and gel electrophoresis of PCR products indicated single products of the correct size for each primer pair used. In addition, we performed three amplification attempts from each Trpml3−/− tissue, and all failed to detect any product. Insofar as the Trpml3e8f primer falls within exon 8, which is deleted in Trpml3−/− , these negative results confirm that the RT-qPCR represents mRNA from Trpml3 and not from another gene.
To estimate the number of mRNA molecules per tissue sample, we first generated a standard curve for Trpml3 using an amplicon with a full-length Trpml3 cDNA insert. We quantified this template DNA by gel densitometry in comparison with DNA of known amounts. We then serially diluted the template DNA in nuclease-free water from 107 to 102 copies/2 μl. We measured threshold cycle (Ct) values for each dilution in duplicate and plotted them on the y-axis against the logarithm of their initial template copy number on the x-axis. We generated the standard curve by linear regression analysis and calculated PCR amplification efficiency (E) with the equation: 10(−1/slope)-1.
We quantified the absolute concentration of Trpml3 mRNA in each tissue by relating the Ct values obtained with 100 ng total RNA as template to the standard curve. We then normalized all tissues with ribosomal 18S rRNA as the reference gene. Among all the tissues harvested, the VNO had the lowest 18S Ct value, indicating the highest amount (i.e., least degradation) of RNA. By using the equation: 2 −ΔCt, where ΔCt = 18S Ct (tissue) - 18S Ct (VNO), we normalized amounts of RNA in all tissues to the VNO, which had the highest amounts of 18S. We adjusted Trpml3 mRNA copy numbers per 100 ng total RNA for each tissue based on this calibration.
We used the genes Pkd1l3 and Trpa1 as positive controls for CVP and DRG. We normalized and calibrated the data with the 2 −ΔΔCt method, where ΔΔCt = (Cttarget – Ctreference)positive control – (Cttarget – Ctreference)TRPML3, with 18S rRNA as the reference gene. This method provided us with the relative amount of Pkd1l3 and Trpa1 compared with Trpml3, which was then used to estimate the number of Pkd1l3 and Trpa1 mRNAs per 100 ng total RNA for each tissue. We finally estimated the total number of Trpml3, Pkd1l3, and Trpa1 mRNA molecules per organ by adjusting their concentration (per 100 ng of total RNA) to the complete total RNA mass obtained from each organ.
RESULTS
To control for nonspecific immunoreactivities, we used KO mice lacking exons 7 and 8 of Trpml3 (Fig. 1A). These exons were deleted by homologous recombination in HM1/129Ola ES cells, which derive from Sv129/Ola mice. After two generations of crosses to C57BL/6, mice had a genetic background of ~75% C57BL/6 and ~25% Sv129/Ola. PCR amplification from genomic DNA using a forward primer located upstream of exon 7 and a reverse primer located downstream of exon 8 confirmed the deletion of these two exons in mice with one or two KO alleles (Fig. 1B). Lack of PCR amplification from genomic DNA with at least one of the primers located within the deleted area confirmed the absence of the deleted DNA in homo-zygous KOs (Fig. 1B). Amplification from reverse-transcribed RNA with primers located in exon 7 or exon 8 also confirmed the absence of the deleted exons in the mRNA from Trpml3−/− mice (negative data not shown) and their presence in the mRNA from Trpml3+/+ (see Fig. 6). The generation and characterization of these KO mice will be described elsewhere (N.N.R., J.G.-A., in preparation). Here, we used tissues from these animals to test the specificity of antisera raised against TRPML3.
Figure 6.
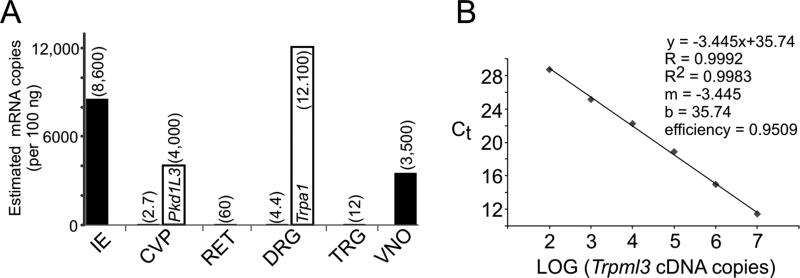
Levels of Trpml3 mRNA in different sensory organs estimated by RT-qPCR. A,B: One microgram of total RNA from each sensory organ was reverse transcribed, and one-tenth of the resulting cDNA served as template for quantitative PCR. The values per sensory organ are the average of three separate reactions, except for Trpml3 in retina and in circumvallate papillae, for which only one of the three reactions detected any mRNA, so the number given should be considered an upper estimate. Only results from Trpml3+/+ tissues are shown, because no amplification product was obtained from Trpml3−/− tissues, confirming the specificity of the product. 18S rRNA was used as the reference gene, and VNO, which had the highest amounts of 18S rRNA and thus probably the least degraded RNA, was used as the tissue calibrator by means of 2−ΔΔCt. By comparing with qPCR values from known copy numbers (A; over six orders of magnitude) of Trpml3 cDNA template, we estimated the number of mRNAs in each tissue sample (A; indicated in parenthesis over each bar). Sensory organs or tissues examined were inner ear (IE), circumvallate papillae with adjacent tongue tissue (CVP), retina without pigmented epithelium and choroid (RET), L4 dorsal root ganglion (DRG), trigeminal ganglion (TRG), and vomeronasal organ (VNO). Open bar for CVP indicates estimated copy number of Pkd1l3 mRNA, and open bar for DRG indicates estimated copy number of Trpa1 mRNA.
By RT-PCR, we found that the mRNA produced in the KO results from splicing exon 6 to exon 9, which ends up out of frame and contains multiple stop codons (Fig. 1C). Hence, the mutant mRNA cannot synthesize the C-terminal half of the TRPML3 protein (residues 278 to the C-terminal 554), which form transmembrane domains 2-6 and the pore of the channel. The mutated out-of-frame mRNA could theoretically produce an N-terminal fragment of TRPML3 (residues 1-277) containing the first trans-membrane domain and part of the first extracellular loop (Fig. 1D), although this mRNA is expected to be degraded by nonsense-mediated mRNA decay. By performing in situ hybridization with probes both upstream (exons 1-5) and downstream (exons 8-12) of the deletion, we confirmed that the Trpml3 KO mRNA is greatly reduced in inner ear as well as in other tissues that express this gene, such as neonatal intestine (Fig. 1F-N). We obtained similar results by RT-PCR analysis of mRNA from neonatal intestine and neonatal back skin rich in melanocytes (Fig. 1E). We conclude that the Trpml3 KO mice have little Trpml3 mRNA, which is anyway incapable of producing most of the TRPML3 protein domains.
Neonatal stria vascularis and hair cells, but not pillar cells, express TRPML3
To determine the expression and localization of TRPML3 in the inner ear, we used four polyclonal antisera raised against three different regions of TRPML3: the amino-terminus (NT) and the carboxyl-terminus (CT1 and CT2), both predicted to be cytosolic, and the third extracellular loop (EX; these regions and the antibodies are shown schematically in Fig. 2M). Although raised to virtually identical peptide sequences (Table 1), CT1 and CT2 were generated independently by separate groups of investigators. Because prior in situ hybridization demonstrated Trpml3 mRNA in neonatal (P2, P5, and P6) hair cells and stria vascularis (Nagata et al., 2008; and Fig. 1G,H,K,L), we performed immunohistochemistry on sections of neonatal inner ears first. Three antisera to TRPML3 (NT, CT1, and CT2) labeled inner and outer hair cells as well as cells of the stria vascularis in sections obtained from Trpml3+/+ pups (Fig. 2). These immunoreactivities appear to represent specific TRPML3 protein recognition, because the antisera failed to immunoreact (NT and CT2) or did so very weakly (CT1) on sections from Trpml3−/− pups. The weak immunoreactivity detected with CT1 on knockout hair cells (Fig. 2D) probably represents nonspecific labeling of another protein, because, as indicated above, the codons of Trpml3 encoding the carboxyl-terminus of TRPML3 are out of frame in these mutants. Notably, the immunoreactivity with NT was abolished in the knockouts (Fig. 2G,I), indicating that they do not produce detectable levels of the truncated amino-terminal fragment of TRPML3 that is encoded in the mutated mRNAs.
Two of these three antisera (NT and CT2) also labeled pillar cells in wild-type inner ears, and this label was enhanced by antigen retrieval. Because these antisera were raised against, and affinity-purified with, two different domains of TRPML3 (its amino- and carboxyl-terminal tails), these immunoreactivities seem to suggest TRPML3 expression in pillar cells. However, these immunoreactivities were not affected by deletion of Trpml3 (Fig. 2F- I,K,L), and therefore do not represent expression of TRPML3 in pillar cells. These results underscore the importance of using knockout tissues as controls for determining the localization of endogenous proteins unambiguously, even when multiple antisera to non-overlapping epitopes label the same cells.
In the stria vascularis, TRPML3 antisera labeled cytoplasmic vesicular structures located between the nuclei of marginal (apical, facing the scala media), intermediate, and basal cells (Fig. 2E,J). These probably correspond to the basal processes of marginal cells, which interdigitate with those of intermediate cells, because in situ hybridization previously showed that only marginal cells express Trpml3 mRNA (Nagata et al., 2008). In addition, at later stages, the vesicles containing TRPML3 clearly localize around the nuclei of marginal cells (Fig. 3D,H,L,P; see below).
TRPML3 localizes to LAMP-1-containing vesicles
Prior subcellular localization studies, with either heterologously expressed tagged TRPML3 or antisera to this protein on cultured cells, revealed TRPML3 localization to vesicles of the endocytic pathway that include late endosomes and lysosomes (Kim et al., 2009; Martina et al., 2009; Nagata et al., 2008; Zeevi et al., 2009). For example, 45% of TRPML3-containing vesicles in transfected HeLa cells (Kim et al., 2009) and 50% in transfected ARPE19 cells (Martina et al., 2009) also express LAMP1, a marker for late endosomes and lysosomes. Similarly, by using our controlled immunoreactivities in neonatal hair cells, we found that 66% of vesicles containing endogenous TRPML3 also immunoreact with anti-LAMP1 antibodies and that 43% of all LAMP1-containing vesicles also contain TRPML3 (n = 112 vesicles from three postnatal outer hair cells imaged by confocal microscopy; Fig. 2N- O). When imaged in confocal optical sections, TRPML3 appeared in the periphery of some of the larger vesicles, consistent with its expected localization at their membrane (Fig. 2P,R). We conclude that in hair cells TRPML3 localizes to vesicles that include late endosomes and lysosomes.
Cochlea maturation results in enrichment of TRPML3 in IHCs, decrease in OHCs, and perinuclear redistribution within marginal cells of SV
The mouse cochlea matures during the first 2-3 weeks of life. This maturation involves changes in the stria vascularis (which contributes to forming the endolymphatic fluid composition and the endocochlear potential) as well as the hair cells. Hence, we examined the distribution of TRPML3 in mature cochleae. With two of the antisera to TRPML3 described above (NT and CT1), we observed that TRPML3-specific immunoreactivities (that is, present in Trpml3+/+ but not in Trpml3−/− ears) were much stronger in inner hair cells (IHCs) than in the adjacent outer hair cells (OHCs; Fig. 3A-C,E-G,I-K,M-O). These observations indicate that TRPML3 protein remains or increases in adult IHCs but decreases in adult OHCs. With the same two antisera (CT1 and NT), we also revealed that TRPML3 in the adult stria vascularis (SV) is concentrated in vesicles surrounding the nuclei of the marginal cells (Fig. 3D,H,L,P).
TRPML3 does not localize to ankle links of postnatal HCs
Prior studies suggested that TRPML3 localizes to stereocilia of postnatal hair cells, and particularly to the ankle links at the base of the stereocilia (Di Palma et al., 2002; van Aken et al., 2008). These studies used two antisera, each raised against, and affinity-purified with, a different domain of TRPML3: the above-mentioned CT2, raised against a portion of the carboxyl-tail, and an antiserum that we term EX, raised against the third extracellular loop, close to the pore domain (Fig. 2M). With these same antibodies, we also detected immunoreactivity to the base of stereociliary bundles in inner, outer, and vestibular (from utricle, saccule, and crista ampullaris) hair cells (Fig. 4). However, these immunoreactivities were identical in Trpml3+/+ and Trpml3−/− tissues, even though the Trpml3−/− animals cannot produce either the third extracellular loop or the carboxyl-tail of TRPML3. In addition, the other two antibodies raised against TRPML3 (NT and CT1) failed to label ankle links, whereas they (as well as CT2) specifically labeled hair cell vesicles. We conclude that the ankle link immunoreactivities do not represent TRPML3 protein localization.
TRPML3 has close homology with TRPML1 and TRPML2, and the TRPML3 epitope recognized by the EX antibody is highly conserved (of its 17 residues, 15 are identical in TRPML1 and 13 in TRPML2). Thus, we wondered whether the ankle link immunoreactivities might represent TRPML1 or TRPML2 instead of TRPML3. However, EX immunoreactivities are also present in Trpml1−/− tissues (unpublished data). In addition, we detected no Trpml2 mRNA in inner ears by performing in situ hybridization with two nonoverlapping probes, both of which strongly labeled other cell types (unpublished data). Finally, the CT2 epitope is poorly conserved among mucolipins (of its 20 residues, only 7 are identical in TRPML1 and 4 in TRPML2), yet the CT2 antiserum also labels ankle links. We conclude that the ankle link immunoreactivities of EX and CT2 are nonspecific and do not reveal the presence of mucolipin channels at ankle links.
In the case of CT2, antigen retrieval revealed the TRPML3-specific immunoreactivity on hair cell vesicles. In the case of EX, however, the pattern of immunoreactivity in the cochlea differs from the pattern shared by NT, CT1, and CT2, and it was not altered in cochleae from Trpml3−/− mice. Furthermore, EX did not label other cells that express TRPML3, such as vomeronasal sensory and olfactory receptor neurons (see below). We conclude that the EX antisera does not label endogenous TRPML3 under any of the conditions we attempted (with or without antigen retrieval and/or tyramide amplification), whereas CT2 displays both specific (vesicles with antigen retrieval) and nonspecific (ankle links) immunoreactivities.
Mouse tastebuds do not express histochemically detectable levels of Trpml3 mRNA or protein
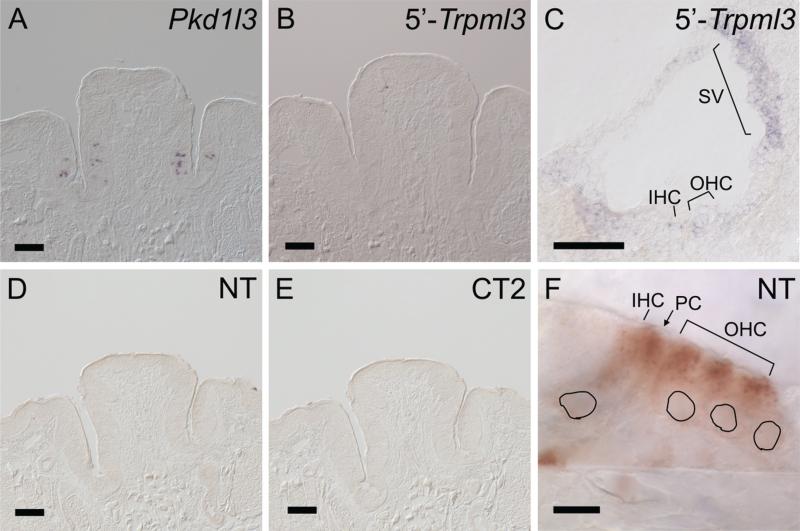
Given that both secretory and sensory epithelia of inner ear express TRPML3, we wondered whether other sensory organs also express this channel. A recent report in the form of a U.S. patent application (public application No. 20090210953) indicated that a subset of taste-receptor cells expressed Trpml3 mRNA and protein. These cells were not labeled with markers of sweet, bitter, umami, or sour receptors and, by default, were considered to be salt taste receptors. Based on this and other considerations, TRPML3 was proposed to mediate gustatory salt-sensing. We performed in situ hybridization in sections of mouse circumvallate papillae (CVP; Fig. 5). These sections contain taste buds with all types of taste receptor cells, as attested by the expression of Pkd1l3 mRNA (a marker of sour taste receptor cells; Ishimaru et al., 2006; Fig. 5A) and of inositol 1,4,5-triphosphate receptor 3 (ITPR3; a marker of bitter, sweet, and umami taste receptors; Clapp et al., 2004). With the same two nonoverlapping probes to Trpml3 previously described (Fig. 1F; Nagata et al., 2008), which detect Trpml3 in inner ear and other tissues (Figs. 1G-N, 5C, 7A,B), we did not detect Trpml3 mRNA in CVP taste receptor cells (Fig. 5B). Furthermore, with two antibodies (NT and CT2) that detect TRPML3 in cochlear hair and strial cells (Figs. 2, 3, 5F), we did not detect TRPML3 protein in CVP or fungiform taste receptor cells (Fig. 5D,E, and data not shown). These negative but controlled results suggest that mouse taste receptor cells do not express TRPML3.
Figure 5.
Taste buds of mouse circumvallate papillae do not express detectable levels of Trpml3 mRNA or protein. A-C: In situ hybridization on sections of the same adult circumvallate papillae (CVP; A,B) and of neonatal (P0) cochlea (C) with antisense probes to the sour taste receptor marker Pkd1l3 mRNA (A) or Trpml3 mRNA (B,C). A: Detection of Pkd1l3 in CVP of CD1 mice confirms the presence of taste buds and controls for the integrity and detection of their mRNA. C: Detection of Trpml3 in cochlea of CD1 mice demonstrates the ability of this probe to detect this mRNA. B: However, this same probe, as well as a nonoverlapping cRNA probe (not shown), failed to detect Trpml3 mRNA in CVP of mice with CD1 or C57BL/6 + Sv129/Ola genetic backgrounds. D-F: (D,E) Immunohistochemistry (ABC/DAB reaction) on sections of the same circumvallate papillae (CVP) obtained from Trpml3+/+ mice and of neonatal (P2; F) organ of Corti with antibodies to the N-terminus of TRPML3 (NT; D,F) or its C-terminus (CT2; E). Although these antibodies recognized TRPML3 protein in hair cells (shown only for NT), neither NT nor CT2 detected TRPML3 in taste buds. Black perimeters indicate the positions of hair cell nuclei. Scale bars = 100 μm in A-E; 10 μm in F. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Figure 7.
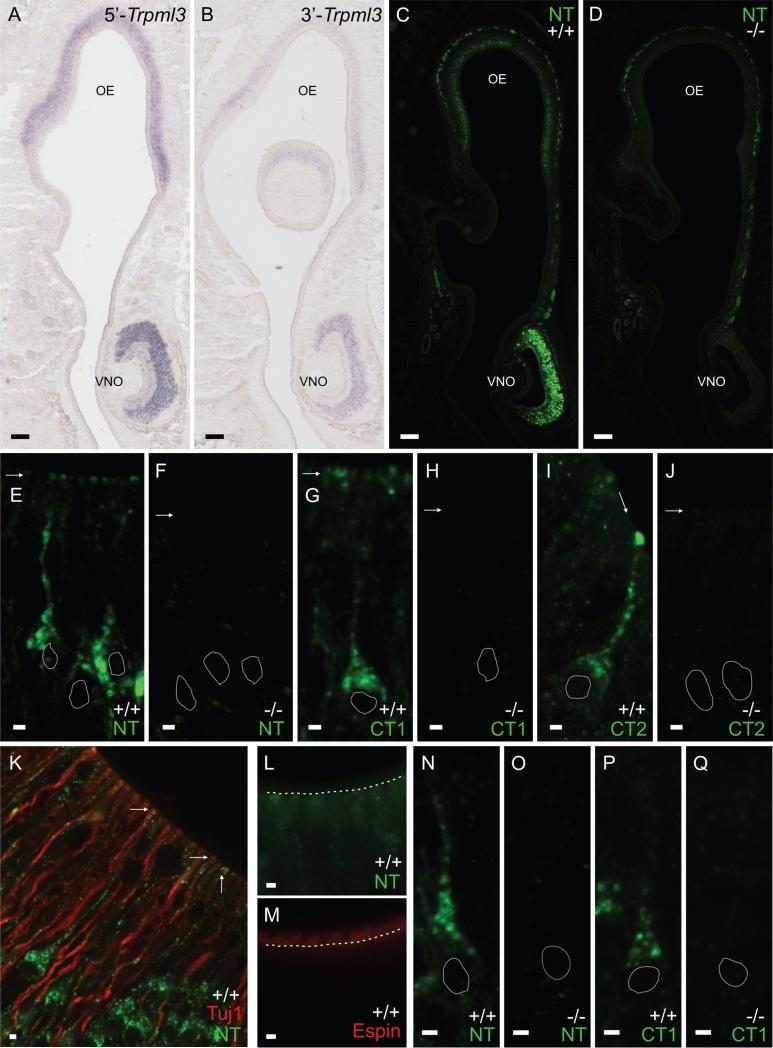
Vomeronasal (VNO) and olfactory (OE) sensory neurons also express Trpml3 mRNA and protein. A,B: In situ hybridization on sections of postnatal (P2) snout from Swiss Webster mice with the Trpml3 antisense 5’ or 3’ probe (5’-Trpml3 and 3’-Trpml3) demonstrates that VNO and OE sensory epithelia express Trpml3 mRNA (lack of signal with control sense probe not shown). Expression was weaker in OE than in VNO. C,D: Immunohistochemistry with TRPML3 antisera NT on sections of postnatal (P4) snout from Trpml3+/+ (C) and Trpml3−/− (D) mice. Expression was weaker in OE than in VNO. E-J: Magnified, high-resolution images of individual VNO sensory neurons from Trpml3+/+ and Trpml3−/− mice using TRPML3 antisera NT (E,F), CT1 (G,H), and CT2 (I,J). Arrows point to apical dendritic knobs. White perimeters indicate the position of neuronal nuclei. Antibodies to TRPML3 labeled vesicles in the soma, along the dendrites, and at the apical dendritic knobs of VNO sensory neurons in Trpml3+/+ but not Trpml3−/− tissues. K: Double immunohistochemistry of adult (P123) VNO with antibodies NT and Tuj1 showing that TRPML3 immunoreactivity remains in the dendrites of VNO sensory organs. L,M: Immunohistochemistry on adjacent sections of adult (P123) VNO using antibodies against NT and espin, showing that TRPML3 immunore-activity is not detected in apical microvilli of VNO sensory neurons. The dotted white line demarcates the boundary between the microvilli and the dendritic knobs, which can be discerned with Nomarski optics. N-Q: Magnified, high-resolution images of individual OE receptor neurons from Trpml3+/+ and Trpml3−/− mice using TRPML3 antisera NT (N,O) and CT1 (P,Q). White perimeters indicate the position of neuronal nuclei. Antibodies to TRPML3 labeled vesicles in OE receptor neurons. A magenta-green version of this figure may be seen as Supporting Information Figure 2. Scale bars = 100 μm in A-D; 2 μm in E-Q.
One potential caveat is that we cannot rule out expression at levels undetectable by in situ hybridization and immunohistochemistry with enzymatic amplification. Hence we tested for the presence of Trpml3 in RNA obtained from several sensory organs by a more sensitive detection method, RT-qPCR (reverse transcription followed by quantitative polymerase chain reaction). As expected, we found abundant levels of Trpml3 mRNA in inner ear (Fig. 6). By comparison with amplification results from fixed numbers of Trpml3 cDNA copies (102, 103, 104, 105, 106, and 107), we estimate 8,600 Trpml3 mRNAs per 100 ng inner ear RNA and ~645,000 mRNAs per whole inner ear. By comparison, we failed to detect any Trpml3 mRNA from a piece of mouse tongue with CVP in two of three amplification trials, and, in the third trial, we barely detected 3,200 times less Trpml3 mRNA than in inner ear. This would represent an estimated 2.7 Trpml3 mRNA molecules per 100 ng total RNA or only 234 molecules per entire CVP (Fig. 6). To confirm the presence of taste receptor cell-specific mRNA in our total RNA sample from CVP, we also performed RT-qPCR with primers for Pkd1l3 and detected 4,000 mRNAs per 100 ng total RNA (~350,000 mRNAs per entire CVP), 1,500 times more than for Trpml3. Given that the CVP contains about 30,000 taste receptor cells (about 300 taste buds, each with about 100 receptor cells) and that 20% of these express PKD1L3 (Ishimaru et al., 2006), our results estimate that each sour taste receptor cell contains 58 Pkd1l3 mRNAs (Table 2). Following the same logic and assuming that salt taste receptor cells also constitute 20% of the all the taste receptors (Chandrashekar et al., 2006, 2010; Chaudhari and Roper, 2010), we estimate that each salt taste receptor cell could contain a maximum of only 0.039 Trpml3 mRNAs (assuming also that all these mRNAs were in taste receptor cells and not in other cells of the tongue; Table 2). This extremely low abundance does not support a physiological role for mouse TRPML3 in taste and explains our inability to detect its mRNA by ISH.
TABLE 2.
Estimated mRNA Copy Numbers in Mouse Circumvallate Papillae (CVP), Retina (RET), and L4 Dorsal Root Ganglion (DRG)
| Tissue/gene | Est. mRNA copies per 100 ng total RNA | Est. mRNA copies per whole organ | Est. number of cells per whole organ | Est. number of cells per whole organ that might express mRNA | Est. mRNA copy number per expressing cell |
|---|---|---|---|---|---|
| CVP/Trpml3 | 0–2.7 | 234 | 300,0001 | 6,000 (20%)1 | 0–0.039 |
| CVP/Pkd1l3 | 4,000 | 350,000 | 300,0001 | 6,000 (20%)1 | 58 |
| RET/Trpml3 | 00–60 | 5,500 | 8,000,0002 | 8,000,000 (100%) | 0–0.00069 |
| DRG/Trpml3 | 4.4 | 243 | 12,0003 | 12,000 (100%) | 0.02 |
| DRG/Trpa1 | 12,100 | 666,000 | 12,0003 | 6720 (56%)4 | 99 |
Neural retina does not express histochemically detectable levels of Trpml3 mRNA or protein
With the same in situ probes and antibodies to TRPML3, we examined sections of neonatal and mature mouse retina and found no evidence of expression (not shown). By RT-qPCR, we also failed to detect any Trpml3 mRNA from neural retina (i.e., excluding choroid and retinal pigmented epithelium) in two of three amplification trials, and in the third trial we barely detected 140 times less than from ear, which yields an estimated 60 mRNA molecules per 100 ng total RNA (Fig. 6). This would amount to ~5,500 copies of Trpml3 mRNA per entire mouse retina (Table 2), a likely insignificant number given the nearly 8 million retinal cells (Jeon et al., 1998).
Somatosensory ganglia do not express histochemically detectable levels of Trpml3 mRNA or protein
Others have previously reported that Trpml3 mRNA is present in DRG neurons at very low levels (detectable by qPCR but not by ISH) but that, upon nerve damage, these levels rise by an average of 7.5 times in most neurons (Staaf et al., 2009). With the in situ probes and antibodies to TRPML3 used here, we examined sections of naïve mouse dorsal root and trigeminal ganglia and found no evidence of expression (not shown). By RT-qPCR, we barely detected an estimated 12 Trpml3 mRNAs per 100 ng total RNA from trigeminal ganglia (700 times less than from ear) and 4.4 Trpml3 mRNAs per 100 ng total RNA from L4 dorsal root ganglion (2,000 times less than from ear; Fig. 6). To confirm the quality of our RNA extraction, we performed RT-qPCR with primers for the nociceptive transducer TRPA1, whose mRNA we previously detected in 56% of DRG neurons by ISH (Garcia-Añoveros and Nagata, 2007; Nagata et al., 2005), and found 12,100 Trpa1 mRNAs per 100 ng total RNA from the same ganglion (~666,000 mRNAs per entire ganglion; Fig. 6). Given that each L4 ganglion contains ~12,000 sensory neurons (Shi et al., 2001), we estimate that each TRPA1-expressing nociceptor contains 99 Trpa1 mRNAs (Table 2). By contrast, because we detected only ~243 Trpml3 mRNAs per whole L4 dorsal root ganglion, we can similarly calculate an average of ~0.02 Trpml3 mRNAs per sensory neuron in naïve ganglia and only ~0.15 Trpml3 mRNAs per sensory neuron in damaged ganglia (assuming the reported increase of 7.5 times; Table 2). The extremely low concentration of Trpml3 mRNA in dorsal root and trigeminal ganglia compared with inner ear or with TRPA1 levels on DRG (2,700 times less) does not support a physiological role for TRPML3 in somato-sensory neurons.
Vomeronasal and olfactory sensory receptor neurons also express TRPML3
The above-mentioned RT-qPCR experiments also revealed a high abundance of Trpml3 mRNA in the vomeronasal organ (Fig. 6). Hence, we performed in situ hybridizations on coronal sections of neonatal (P2) snout containing both vomeronasal and olfactory sensory neuroepithelia. With both nonoverlapping antisense probes to Trpml3 mRNA, we detected strong signal in the neuronal crescent of the VNO and comparatively weaker signal in the neuronal layer of olfactory epithelium (Fig. 7A,B). We did not detect this signal with the control, sense probe (not shown). Hence, these in situ hybridization signals most likely represent Trpml3 mRNA expression. We also performed immunohistochemistry on P4 sections of snout. One of the antisera (NT) labeled the olfactory nerve along the basal lamina, but this immunoreactivity was not specific to TRPML3; it was present in both Trpml3+/+ and Trpml3−/− mice (Fig. 7C,D). However, this and the other two antibodies previously used to detect TRPML3 protein in inner ear (NT, CT1, and CT2) specifically labeled vomeronasal and olfactory receptor neurons of Trpml3+/+, but not Trpml3−/− , mice (Fig. 7C-Q). In OE, labeled cells were those at more apical locations, where mature ORNs reside. Staining of adjacent sections with antibodies to the olfactory marker protein (OMP; Farbman and Margo-lis, 1980; Graziadei et al., 1980) confirmed that these apically located, TRPML3-expressing cells are the mature ORNs (not shown). Subcellularly, TRPML3-specific immunoreactivities labeled vesicular structures strongly at the cell bodies and more weakly along their dendrites, which extend toward the airway. In vomeronasal sensory neurons, all three TRPML3-specific immunoreactivities (NT, CT1, and CT2) were also prominent at the apical, dendritic knobs from which the sensory microvilli emanate. We did not detect TRPML3 in the sensory microvilli, which we could visualize with antibodies to the transduction channel TRPC2 (Liman et al., 1999) and the actin-bundling protein espin (Sekerkova et al., 2004; Fig 7L,M; and data not shown). We could also distinguish TRPML3-containing dendritic knobs from TRPML3-lacking sensory microvilli by colabeling with antibodies to neuronal βIII -tubulin, which immunoreact with dendrites but not microvilli (Fig. 7K). Finally, because the VNO is used largely after sexual maturity, we wondered whether the expression of TRPML3 changes in adults (as it did in inner ear). However, by immunohistochemistry, we detected expression of TRPML3 in adult VNO neurons (Fig. 7K) similar to our analyses in neonatal neurons.
DISCUSSION
This study has elucidated the expression of TRPML3 in sensory organs and clarified its localization in hair cells by using antibodies raised against different domains of TRPML3; nonoverlapping in situ probes to confirm expression of Trpml3 mRNA; and, most critical of all, Trpml3−/− tissues to distinguish immunoreactivities that represent TRPML3 protein expression from nonspecific immunoreactivities. Crucial to our controls is assurance that the TRPML3 protein is absent from Trpml3−/− mice or, if truncated, present at substantially lower levels than in Trpml3+/+ mice. Although some immunoreactivities remained in tissues from Trpml3−/− mice (nerve fibers with CT1, ankle links with EX and CT2, and pillar cells with NT and CT2), these are unlikely to represent TRPML3 protein for several reasons. First, by RT-PCR, we demonstrated that the KO mRNA is out of frame after exon 6 and could not produce the protein domains recognized by all but one of the antibodies (NT). Hence, the patterns obtained by EX, CT1, and CT2 on tissues from Trpml3−/− mice should not represent TRPML3 protein. Second, we also demonstrated a severe reduction of the mutant mRNA levels, so that, even if an amino-terminal fragment was produced, it would be produced at greatly reduced levels. However, the NT immunoreactivity on pillar cells did not vary between Trpml3−/− and Trpml3+/+ mice. Third, other immunoreactivities obtained with NT, CT1, and CT2 antisera (vesicles of hair cells, stria vascularis, vomeronasal and olfactory receptor neurons) did disappear in Trpml3−/− mice. Hence we conclude that the Trpml3 KO we used lacks any TRPML3 full-length protein and most if not all of the truncated amino-terminal TRPML3 fragment.
Prior evidence suggesting that TRPML3 might localize to stereocilia included immunoreactivities with antisera to TRPML3 (Di Palma et al., 2002; van Aken et al., 2008). By far the most convincing evidence was the ankle link immunoreactivities obtained with two antibodies raised against, and affinity-purified to, separate domains of TRPML3. One of these antibodies (HL4460, here called CT2) recognized heterologously expressed GFP-tagged TRPML3 as well as a single band of 68 kDa (near the expected molecular weight of TRPML3) in Western blots from inner ear. Furthermore, there was ample evidence that hair cells did express Trpml3 mRNA (Nagata et al., 2008). The simplest and most reasonable interpretation of this evidence at the time was that TRPML3 localized to ankle links. However, we now find that these ankle link immunoreactivities do not represent TRPML3. It seems highly coincidental that two antisera raised against two separate protein domains would nonspecifically label the same structure, either pillar cells or ankle links. Our experience is that many antibodies nonspecifically immuno-react with pillar cells, perhaps because they are “immunophilic,” but this is not the case with ankle links. Both of the ankle link-labeling antisera (CT2 and EX) were raised simultaneously by the same research group. Although it might appear improbable, we wonder whether the immunized rabbits shared the same environment and thus exposure to common immunogens (such as infectious agents), one of which contained an ankle link-like epitope. Another potential explanation is that both of these antibodies coincidentally cross-react with similar epitopes in other proteins that localize to ankle links, even though CT1 (raised to an epitope nearly identical to that for CT2) does not label ankle links). Whatever the explanation, our observations stress the importance of testing the specificities of antisera with KO tissues before inferring protein expression and localization definitively.
Although we did not detect specific immunoreactivities to ankle links or any other part of hair cell bundles, these negative results cannot rule out the possibility that undetectable levels of TRPML3 exist in stereocilia. What our controlled immunoreactivities demonstrate is that TRPML3 localizes to vesicular structures in hair cells, a localization suggested by previous immunoreactivities as well as by heterologous expression of GFP-tagged TRPML3 (Di Palma et al., 2002; Nagata et al., 2008; van Aken et al., 2008). In addition, we also demonstrated vesicular localization in marginal cells of the stria vascularis.
Although in neonates both IHC and OHC contain vesicles with TRPML3, as the ear matures TRPML3-containing vesicles become much more abundant in IHCs. Compared with OHCs, adult IHCs have many more pre-synaptic vesicles, and these are released at very high rates. To maintain or replenish the synaptic vesicle pool, IHCs constantly endocytose vesicles at their apical membranes and transport them basolaterally to presynaptic sites (Griesinger et al., 2002, 2005). Overexpression and down-regulation experiments in cultured cells suggest that TRPML3 regulates trafficking along the endosomal pathway (Martina et al., 2009). Hence, TRPML3 may contribute to the traffic of vesicles in IHCs. Alternatively, the high levels of TRPML3 in mature inner hair cells may simply reflect their high content of endocytic vesicles. Another feature distinguishing IHCs from OHCs is the high concentration of proteinaceous calcium buffers in the latter (110 times more than in IHCs; Hackney et al., 2005). Given that calcium constantly enters hair cells (apically via transduction channels and basolaterally via voltage-gated channels), one wonders how IHCs avoid reaching lethal concentrations of cytosolic calcium. One possibility is that intracellular stores sequester calcium. Interestingly, lysosomes and endosomes contain high levels of calcium and constitute the recently discovered acidic calcium stores (Patel and Docampo, 2010). Hence, another possibility is that the TRPML3-containing vesicles in IHCs function as calcium stores, perhaps playing a cytoprotective role. The specific role of TRPML3 in vesicles is unknown, but possibilities include release of calcium to empty the acidic stores and/or to trigger vesicle fission, fusion, or transport.
We also have provided conclusive evidence that other types of sensory receptor cells, the olfactory and vomero-nasal receptor neurons, express TRPML3. First, two antisense probes that hybridize to nonoverlapping parts of Trpml3 mRNA labeled the areas of VNO and OE containing the somas of mature receptor neurons, whereas control sense probe did not. Hence, the in situ signals most likely reveal specific hybridization to Trpml3 mRNA and not cross-hybridization to another mRNA or nonspecific hybridization resulting from low stringency in the hybridization conditions. Second, three antibodies (NT, CT1, and CT2) raised to two separate domains of TRPML3 immunoreact with VNO and OE neurons of Trpml3+/+ but not Trpml3−/− mice. These specific immunoreactivities labeled vesicular structures at neuronal soma and along the dendrites, including the dendritic knobs of vomero-nasal sensory neurons, but not the chemosensory micro-villi that emanate from these knobs. This distribution resembles that of mechanosensory hair cells, in which TRPML3 localizes to vesicles distributed along the cell body and at the cuticular plate from which the stereocilia emanate but is not detected in the sensory stereocilia.
Supplementary Material
ACKNOWLEDGMENT
We thank Markus Delling and David Clapham for the generous gift of TRPML3 antiserum here referred to as CT1, Konrad Noben-Trauth for the generous gifts of TRPML3 antisera PB221 and HL4460 (here referred to as EX and CT2, respectively), Janitza Montalvo-Ortiz and Callie Pollock for experimental support during their rotations in our laboratory, Steve DeVries for the use of his confocal microscope, and James Bartles for critical review and advice.
Grant sponsor: National Institutes of Health; Grant number: RO1 NS044363; Grant sponsors: The Hugh Knowles Foundation (to J.G.-A.); National Research Service Award; Grant number: NS041234 (to A.J.C., E.N.F.); Grant number: DC010529 (to N.N.R.).
Additional Supporting Information may be found in the online version of this article.
LITERATURE CITED
- Adato A, Lefevre G, Delprat B, Michel V, Michalski N, Chardenoux S, Weil D, El-Amraoui A, Petit C. Usherin, the defective protein in Usher syndrome type IIA, is likely to be a component of interstereocilia ankle links in the inner ear sensory cells. Hum Mol Genet. 2005;14:3921–3932. doi: 10.1093/hmg/ddi416. [DOI] [PubMed] [Google Scholar]
- Andrejewski N, Punnonen EL, Guhde G, Tanaka Y, Lullmann-Rauch R, Hartmann D, von Figura K, Saftig P. Normal lysosomal morphology and function in LAMP-1-deficient mice. J Biol Chem. 1999;274:12692–12701. doi: 10.1074/jbc.274.18.12692. [DOI] [PubMed] [Google Scholar]
- Cable J, Steel KP. Combined cochleo-saccular and neuroepithelial abnormalities in the varitint-waddler-J (VaJ) mouse. Hear Res. 1998;123:125–136. doi: 10.1016/s0378-5955(98)00107-5. [DOI] [PubMed] [Google Scholar]
- Chandrashekar J, Hoon MA, Ryba NJ, Zuker CS. The receptors and cells for mammalian taste. Nature. 2006;444:288–294. doi: 10.1038/nature05401. [DOI] [PubMed] [Google Scholar]
- Chandrashekar J, Kuhn C, Oka Y, Yarmolinsky DA, Hummler E, Ryba NJ, Zuker CS. The cells and peripheral representation of sodium taste in mice. Nature. 2010;464:297–301. doi: 10.1038/nature08783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhari N, Roper SD. The cell biology of taste. J Cell Biol. 2010;190:285–296. doi: 10.1083/jcb.201003144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JW, Murphy TL, Willingham MC, Pastan I, August JT. Identification of two lysosomal membrane glycoproteins. J Cell Biol. 1985;101:85–95. doi: 10.1083/jcb.101.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X, Shen D, Samie M, Xu H. Mucolipins: intracellular TRPML1–3 channels. FEBS Lett. 2010;584:2013–2021. doi: 10.1016/j.febslet.2009.12.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapp TR, Yang R, Stoick CL, Kinnamon SC, Kinnamon JC. Morphologic characterization of rat taste receptor cells that express components of the phospholipase C signaling pathway. J Comp Neurol. 2004;468:311–321. doi: 10.1002/cne.10963. [DOI] [PubMed] [Google Scholar]
- Delprat B, Michel V, Goodyear R, Yamasaki Y, Michalski N, El-Amraoui A, Perfettini I, Legrain P, Richardson G, Hardelin JP, Petit C. Myosin XVa and whirlin, two deafness gene products required for hair bundle growth, are located at the stereocilia tips and interact directly. Hum Mol Genet. 2005;14:401–410. doi: 10.1093/hmg/ddi036. [DOI] [PubMed] [Google Scholar]
- Deol MS. The anomalies of the labyrinth of the mutants varitint-waddler, shaker-2 and jerker in the mouse. J Genet. 1954;52:562–588. [Google Scholar]
- Di Palma F, Belyantseva IA, Kim HJ, Vogt TF, Kachar B, Noben-Trauth K. Mutations in Mcoln3 associated with deafness and pigmentation defects in varitint-waddler (Va) mice. Proc Natl Acad Sci U S A. 2002;99:14994–14999. doi: 10.1073/pnas.222425399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duggan A, Madathany T, de Castro SC, Gerrelli D, Guddati K, Garcia-Anoveros J. Transient expression of the conserved zinc finger gene INSM1 in progenitors and nascent neurons throughout embryonic and adult neurogenesis. J Comp Neurol. 2008;507:1497–1520. doi: 10.1002/cne.21629. [DOI] [PubMed] [Google Scholar]
- Farbman AI, Margolis FL. Olfactory marker protein during ontogeny: immunohistochemical localization. Dev Biol. 1980;74:205–215. doi: 10.1016/0012-1606(80)90062-7. [DOI] [PubMed] [Google Scholar]
- Flores EN, García-Añoveros J. Trpml2 and the evolution of mucolipins. Adv Exp Med Biol. 2010 doi: 10.1007/978-94-007-0265-3_12. (in press) [DOI] [PubMed] [Google Scholar]
- Garcia-Añoveros J, Nagata K. Trpa1. Hdbk Exp Pharmacol. 2007;179:347–362. doi: 10.1007/978-3-540-34891-7_21. [DOI] [PubMed] [Google Scholar]
- Goodyear RJ, Marcotti W, Kros CJ, Richardson GP. Development and properties of stereociliary link types in hair cells of the mouse cochlea. J Comp Neurol. 2005;485:75–85. doi: 10.1002/cne.20513. [DOI] [PubMed] [Google Scholar]
- Graziadei GA, Stanley RS, Graziadei PP. The olfactory marker protein in the olfactory system of the mouse during development. Neuroscience. 1980;5:1239–1252. doi: 10.1016/0306-4522(80)90197-9. [DOI] [PubMed] [Google Scholar]
- Griesinger CB, Richards CD, Ashmore JF. Fm1–43 reveals membrane recycling in adult inner hair cells of the mammalian cochlea. J Neurosci. 2002;22:3939–3952. doi: 10.1523/JNEUROSCI.22-10-03939.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griesinger CB, Richards CD, Ashmore JF. Fast vesicle replenishment allows indefatigable signalling at the first auditory synapse. Nature. 2005;435:212–215. doi: 10.1038/nature03567. [DOI] [PubMed] [Google Scholar]
- Grimm C, Cuajungco MP, van Aken AF, Schnee M, Jors S, Kros CJ, Ricci AJ, Heller S. A helix-breaking mutation in TRPML3 leads to constitutive activity underlying deafness in the varitint-waddler mouse. Proc Natl Acad Sci U S A. 2007;104:19583–19588. doi: 10.1073/pnas.0709846104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackney CM, Mahendrasingam S, Penn A, Fettiplace R. The concentrations of calcium buffering proteins in mammalian cochlear hair cells. J Neurosci. 2005;25:7867–7875. doi: 10.1523/JNEUROSCI.1196-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimaru Y, Inada H, Kubota M, Zhuang H, Tominaga M, Matsunami H. Transient receptor potential family members PKD1L3 and PKD2L1 form a candidate sour taste receptor. Proc Natl Acad Sci U S A. 2006;103:12569–12574. doi: 10.1073/pnas.0602702103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon CJ, Strettoi E, Masland RH. The major cell populations of the mouse retina. J Neurosci. 1998;18:8936–8946. doi: 10.1523/JNEUROSCI.18-21-08936.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Li Q, Tjon-Kon-Sang S, So I, Kiselyov K, Muallem S. Gain-of-function mutation in TRPML3 causes the mouse Varitint-Waddler phenotype. J Biol Chem. 2007;282:36138–36142. doi: 10.1074/jbc.C700190200. [DOI] [PubMed] [Google Scholar]
- Kim HJ, Soyombo AA, Tjon-Kon-Sang S, So I, Muallem S. The Ca2+ channel TRPML3 regulates membrane trafficking and autophagy. Traffic. 2009;10:1157–1167. doi: 10.1111/j.1600-0854.2009.00924.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer H, van Wijk E, Marker T, Wolfrum U, Roepman R. Usher syndrome: molecular links of pathogenesis, proteins and pathways. Hum Mol Genet. 2006;15:R262–R270. doi: 10.1093/hmg/ddl205. (Spec No. 2) [DOI] [PubMed] [Google Scholar]
- Kussel-Andermann P, El-Amraoui A, Safieddine S, Nouaille S, Perfettini I, Lecuit M, Cossart P, Wolfrum U, Petit C. Vezatin, a novel transmembrane protein, bridges myosin VIIA to the cadherin-catenins complex. EMBO J. 2000;19:6020–6029. doi: 10.1093/emboj/19.22.6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MK, Tuttle JB, Rebhun LI, Cleveland DW, Frankfurter A. The expression and posttranslational modification of a neuron-specific beta-tubulin isotype during chick embryo-genesis. Cell Motil Cytoskeleton. 1990;17:118–132. doi: 10.1002/cm.970170207. [DOI] [PubMed] [Google Scholar]
- Liman ER, Corey DP, Dulac C. TRP2: a candidate transduction channel for mammalian pheromone sensory signaling. Proc Natl Acad Sci U S A. 1999;96:5791–5796. doi: 10.1073/pnas.96.10.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martina JA, Lelouvier B, Puertollano R. The calcium channel mucolipin-3 is a novel regulator of trafficking along the endosomal pathway. Traffic. 2009;10:1143–1156. doi: 10.1111/j.1600-0854.2009.00935.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGee J, Goodyear RJ, McMillan DR, Stauffer EA, Holt JR, Locke KG, Birch DG, Legan PK, White PC, Walsh EJ, Richardson GP. The very large G-protein-coupled receptor VLGR1: a component of the ankle link complex required for the normal development of auditory hair bundles. J Neurosci. 2006;26:6543–6553. doi: 10.1523/JNEUROSCI.0693-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata K, Duggan A, Kumar G, Garcia-Añoveros J. Nociceptor and hair cell transducer properties of TRPA1, a channel for pain and hearing. J Neurosci. 2005;25:4052–4061. doi: 10.1523/JNEUROSCI.0013-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata K, Zheng L, Madathany T, Castiglioni AJ, Bartles JR, Garcia-Añoveros J. The varitint-waddler (Va) deafness mutation in TRPML3 generates constitutive, inward rectifying current and causes cellular degeneration. Society for Neuroscience; San Diego: 2007. p. 699.612/S612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata K, Zheng L, Madathany T, Castiglioni AJ, Bartles JR, Garcia-Añoveros J. The varitint-waddler (Va) deafness mutation in TRPML3 generates constitutive, inward rectifying currents and causes cell degeneration. Proc Natl Acad Sci U S A. 2008;105:353–358. doi: 10.1073/pnas.0707963105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel S, Docampo R. Acidic calcium stores open for business: expanding the potential for intracellular Ca2+ signaling. Trends Cell Biol. 2010;20:277–286. doi: 10.1016/j.tcb.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puertollano R, Kiselyov K. TRPMLs: in sickness and in health. Am J Physiol Renal Physiol. 2009;296:F1245–F1254. doi: 10.1152/ajprenal.90522.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeren-Wiemers N, Gerfin-Moser A. A single protocol to detect transcripts of various types and expression levels in neural tissue and cultured cells: in situ hybridization using digoxigenin-labelled cRNA probes. Histochemistry. 1993;100:431–440. doi: 10.1007/BF00267823. [DOI] [PubMed] [Google Scholar]
- Sekerkova G, Zheng L, Loomis PA, Changyaleket B, Whitlon DS, Mugnaini E, Bartles JR. Espins are multifunctional actin cytoskeletal regulatory proteins in the microvilli of chemosensory and mechanosensory cells. J Neurosci. 2004;24:5445–5456. doi: 10.1523/JNEUROSCI.1279-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekerkova G, Zheng L, Mugnaini E, Bartles JR. Differential expression of espin isoforms during epithelial morpho-genesis, stereociliogenesis and postnatal maturation in the developing inner ear. Dev Biol. 2006;291:83–95. doi: 10.1016/j.ydbio.2005.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekerkova G, Zheng L, Mugnaini E, Bartles JR. Espin actin-cytoskeletal proteins are in rat type I spiral ganglion neurons and include splice-isoforms with a functional nuclear localization signal. J Comp Neurol. 2008;509:661–676. doi: 10.1002/cne.21755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senften M, Schwander M, Kazmierczak P, Lillo C, Shin JB, Hasson T, Geleoc GS, Gillespie PG, Williams D, Holt JR, Muller U. Physical and functional interaction between protocadherin 15 and myosin VIIa in mechano-sensory hair cells. J Neurosci. 2006;26:2060–2071. doi: 10.1523/JNEUROSCI.4251-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi TJ, Tandrup T, Bergman E, Xu ZQ, Ulfhake B, Hokfelt T. Effect of peripheral nerve injury on dorsal root ganglion neurons in the C57 BL/6J mouse: marked changes both in cell numbers and neuropeptide expression. Neuro-science. 2001;105:249–263. doi: 10.1016/s0306-4522(01)00148-8. [DOI] [PubMed] [Google Scholar]
- Staaf S, Oerther S, Lucas G, Mattsson JP, Ernfors P. Differential regulation of TRP channels in a rat model of neuropathic pain. Pain. 2009;144:187–199. doi: 10.1016/j.pain.2009.04.013. [DOI] [PubMed] [Google Scholar]
- van Aken AF, Atiba-Davies M, Marcotti W, Goodyear RJ, Bryant JE, Richardson GP, Noben-Trauth K, Kros CJ. TRPML3 mutations cause impaired mechano-electrical transduction and depolarization by an inward-rectifier cation current in auditory hair cells of varitint-waddler mice. J Physiol. 2008;586:5403–5418. doi: 10.1113/jphysiol.2008.156992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatachalam K, Hofmann T, Montell C. Lysosomal localization of TRPML3 depends on TRPML2 and the mucolipidosis-associated protein TRPML1. J Biol Chem. 2006;281:17517–17527. doi: 10.1074/jbc.M600807200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Delling M, Li L, Dong X, Clapham DE. Activating mutation in a mucolipin transient receptor potential channel leads to melanocyte loss in varitint-waddler mice. Proc Natl Acad Sci U S A. 2007;104:18321–18326. doi: 10.1073/pnas.0709096104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeevi DA, Frumkin A, Bach G. TRPML and lysosomal function. Biochim Biophys Acta. 2007;1772:851–858. doi: 10.1016/j.bbadis.2007.01.004. [DOI] [PubMed] [Google Scholar]
- Zeevi DA, Frumkin A, Offen-Glasner V, Kogot-Levin A, Bach G. A potentially dynamic lysosomal role for the endogenous TRPML proteins. J Pathol. 2009;219:153–162. doi: 10.1002/path.2587. [DOI] [PubMed] [Google Scholar]
- Zheng L, Sekerkova G, Vranich K, Tilney LG, Mugnaini E, Bartles JR. The deaf jerker mouse has a mutation in the gene encoding the espin actin-bundling proteins of hair cell stereocilia and lacks espins. Cell. 2000;102:377–385. doi: 10.1016/s0092-8674(00)00042-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, Zheng J, Whitlon DS, Garcia-Anoveros J, Bartles JR. Targeting of the hair cell proteins cadherin 23, harmonin, myosin XVa, espin, and prestin in an epithelial cell model. J Neurosci. 2010;30:7187–7201. doi: 10.1523/JNEUROSCI.0852-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.