Abstract
Background
The RUNX family, which is composed of RUNX1, RUNX2, and RUNX3, is a sequence-specific transcription factor family and is closely involved in a variety of cellular processes including development, differentiation, participation in the regulation of p53-dependent DNA damage response and/or tumorigenesis. Emerging evidence indicates that RUNX3 is a candidate tumor suppressor in several types of human tumors including colorectal cancer (CRC). However, the correlation of RUNX3 inactivation with CRC remains unclear. In the study reported here, we conducted a systematic review and meta-analysis to quantitatively evaluate the effects of RUNX3 hypermethylation/expression on the incidence of CRC.
Methods
A detailed search of the literature was made using Medline® and Web of Science for related research publications written in English. The methodological quality of the studies was also evaluated. The data were extracted and assessed by two reviewers independently. Analyses of the pooled data were performed. Odds ratios (ORs) and hazard ratios were calculated and summarized, respectively.
Results
A final analysis of 1,427 CRC patients from eleven eligible studies was performed. We observed that RUNX3 hypermethylation was significantly higher in CRC than in normal colorectal mucosa. The pooled OR from six studies comprising 289 CRC and 188 normal colorectal mucosa was OR =0.07 (confidence interval [CI] =0.03–0.18, P<0.00001). Aberrant RUNX3 hypermethylation/expression was significantly higher in advanced CRC than in early staged CRC (OR =0.54, CI =0.41–0.71, P<0.0001). Aberrant RUNX3 hypermethylation/expression was also significantly higher in microsatellite instability (MSI)-positive CRC than in MSI-negative CRC (OR =0.44, CI =0.3–0.66, P<0.0001). In addition, CRC patients with RUNX3 hypermethylation or lacking RUNX3 protein expression had a lower survival rate than those without RUNX3 hypermethylation or those who did not express RUNX3 protein.
Conclusion
The results of this meta-analysis suggest that RUNX3 hypermethylation is associated with an increased risk of CRC, increased risk of progression of CRC, and a poorer CRC survival rate. RUNX3 hypermethylation, which induces the inactivation of RUNX3 gene, plays an important role in colorectal carcinogenesis, high levels of MSI, as well as CRC progression and development.
Keywords: methylation, tumor-suppressor gene, odds ratio, hazard ratio, microsatellite instability
Introduction
Colorectal cancer (CRC) is one of the most malignant types of cancers. In the USA and Europe, CRC is the second most frequent cancer after lung cancer that leads to death.1 The incidence of CRC in the People’s Republic of China has been reported to increase annually and will continue to rise in the next few years.2 Currently, there are approximately 1.25 million patients diagnosed with CRC, and more than 600,000 patients will die from this disease every year worldwide.3 Surgical resection can be performed to remove the tumor if no lymph-node or distant metastasis is present, but the recurrence rate after surgery remains high.4,5 Therefore, investigation of the mechanism of initiation and progression and identification of prognostic markers are still needed in the treatment of CRC.
A proper DNA damage response has been considered a critical barrier to genetic alterations, preventing tumor initiation and progression.6,7 The runt-domain-related (RUNX) family of genes, RUNX1, RUNX2, and RUNX3, have recently come to prominence because of their protein roles as essential regulators of cell fate in development and their paradoxical effects in cancer, as well as in the regulation of p53-dependent DNA damage response and/or tumorigenesis.8–10 The RUNX3 gene is one of the most critical members of the runt-domain family and has a crucial role in the regulation of cell proliferation and cell death by apoptosis, and in angiogenesis, cell adhesion, and invasion.11,12 The RUNX3 gene, localized to chromosome 1p36, a region that exhibits frequent loss-of-heterozygosity events in colon, gastric, breast, and ovarian cancers, is considered a tumor-suppressor gene involved in the transforming growth factor beta (TGF-β) signaling pathway.13 Its precise function has been intensively studied in several tumors, with upregulation of the induction of cell-cycle arrest, apoptosis, and downregulation of cyclin D1 expression.14–18 Inactivation of RUNX3 by promoter methylation (hypermethylation) has been found to play an important role in colorectal epithelial tumorigenesis.19–21 However, its role in CRC and its clinical significance have not been thoroughly investigated. In this study, we review and update the published clinical investigations regarding the effect of RUNX3 on patients with CRC.
Methods
Search strategy and selection criteria
We searched PubMed, Embase, and ISI Web of Knowledge to identify studies from January 1, 1998 to December 2013 using the search terms “colorectal” and “cancer” or “tumor” or “neoplasm” or “carcinoma”, “methylation”, and “RUNX3”. We also manually searched the reference lists of the retrieved articles and reviews for additional articles. Although our search did not initially have language limits, for the full-text reading and final evaluation, we reviewed only studies published in English. Conference abstracts were not selected for our analysis due to their containing insufficient data. After the exclusion of irrelevant and/or redundant publications from the different databases, the full-text versions of the remaining papers were evaluated against inclusion and exclusion criteria (see the following paragraph), their reference lists, and/or bibliographies searched for additional relevant articles. All searched data were retrieved. To avoid duplication, if the same patient population was reported in several publications, the most complete study was chosen for review.
The three inclusion criteria were as follows: 1) RUNX3 methylation and/or expression evaluated in the circulation and/or primary CRC tissues, 2) research revealed the relationship between RUNX3 methylation and/or expression and CRC clinicopathological parameters and prognosis, 3) RUNX3 methylation and/or expression examined by polymerase chain reaction (PCR), 4) sufficient information provided to estimate hazard ratio (HR) concerning overall survival (OS) and 95% confidence interval (CI). The two exclusion criteria were: 1) letters, reviews, case reports, conference abstracts, editorials, expert opinion; and 2) all in vitro/ex vivo studies. Studies on cell lines and human xenografts were also excluded.
Data extraction and methodological assessment
Two authors (WM, JW) independently reviewed and extracted the data from eligible studies. Disagreements were resolved by discussion and consensus. Two authors (QN, NS) reviewed all of articles that fitted the inclusion criteria. The following information was recorded for each study: the first author’s name, year of publication, sample source, number of cases, clinicopathological parameters, cancer tumor-node-metastasis stage, methylation detection method, methylation rate and/or expression, and follow-up. Data for study characteristics and clinical responses were summarized and transferred into table format. The heterogeneity of investigation was evaluated to determine whether the data from the various studies could be analyzed in a meta-analysis.
For the methodological evaluation of the studies, three investigators (WM, JW, and HL) read each publication independently, and assessed and scored them according to Reporting Recommendations for Tumor Marker Prognostic Studies (REMARK) guidelines and the European Lung Cancer Working Party quality scale.22,23 The three readers provided the quality scores and compared them, then reached a consensus value for each item.
Statistical analysis
Analysis was conducted using STATA (v 12.0; StataCorp LP, TX, USA) and Review Manager (v 5.2; The Cochrane Collaboration, Oxford, UK) software. The pooled frequency of RUNX3 hypermethylation and 95% CI were estimated. The frequency of RUNX3 hypermethylation in different tumor characteristics was compared. Heterogeneity among studies was evaluated with Cochran’s Q test24 and the I2 statistic.25,26 When heterogeneity was not an issue (I2 values <50%), a fixed-effects model was used to calculate parameters. If there was substantial heterogeneity (I2 values ≥50%), a random-effects model was used to pool data and attempt to identify potential sources of heterogeneity based on subgroup analyses. The pooled OR was estimated for the association between RUNX3 hypermethylation and clinicopathological features. P-values less than 0.05 were considered statistically significant.
Publication bias was assessed using a method reported by Egger et al.27 We also explored reasons for statistical heterogeneity using meta-regression, subgroup analysis, and sensitivity analysis. The analysis of meta-regression and publication bias was performed using STATA (v 10.0; StataCorp LP).
Results
Identification of relevant studies
Sixty-seven publications were identified using the described search method. Of these, 56 were excluded due to being laboratory studies, non-original articles (ie, reviews), or irrelevant to the current analysis. Eventually, eleven studies were included in the final meta-analysis,19–21,28–35 as shown in Figure 1.
Figure 1.
Flowchart of study selection.
Study characteristics
The eleven studies eligible for meta-analysis were published between 2004 and 2013, and between them reported on a total of 1,427 CRC patients from Japan, Singapore, Brazil, Norway, and the USA. The basic characteristics of these patients are summarized in Table 1.
Table 1.
Basic characteristics of the included studies
| Study | Patients, N | Method(s) | Primary aim | Methylation site | RUNX3 expression |
|---|---|---|---|---|---|
| Imamura et al19 | 92 | MSP | Determination of the methylation status of the RUNX3 gene and correlation of the results with the clinicopathological features of affected patients | Promoter, CpG islands | − |
| Goel et al20 | 91 | MSP, RT-PCR | Determination of RUNX3 expression and the frequency of RUNX3 promoter hypermethylation | Promoter, CpG islands | + |
| Ku al21 | 87 | RT-PCR, MSP | Determination of whether there is promoter methylation of the RUNX3 gene in CRC | Promoter, CpG islands | + |
| Ahlquist et al28 | 25 | MSP | Pinpointing of the epigenetic markers that can discriminate between nonmalignant and malignant tissue from the large bowel | Promoter, CpG islands | − |
| Soong et al29 | 849 | Immunohistochemistry | Determination of the clinical significance of RUNX3 expression in a large series of CRC patients | + | |
| Ito et al30 | 48 | MSP, RT-PCR | Investigation of the possible roles of RUNX3 in regulating the Wnt pathway and in intestinal tumorigenesis | Promoter, CpG islands | + |
| Hibi and Nakao31 | 59 | MSP | Determination of whether RUNX3 methylation status is correlated with the clinicopathological features of the affected patients | Promoter, CpG islands | − |
| Ogino et al32 | 30 | MethyLight | Determination of whether CpG island methylation may predict poor survival in CRC | Promoter, CpG islands | + |
| Tan et al33 | 17 | MSP | Determination of the promoter status of RUNX3 in CRC | Promoter, CpG islands | − |
| Silva et al34 | 10 | Methyl-Profiler™ PCR Array* | Identification of useful epigenetic markers for noninvasive CRC screening | Promoter, CpG islands | − |
| Nishio et al35 | 119 | MSP | Analysis of RUNX3 promoter hypermethylation as a novel tumor marker | Promoter, CpG islands | − |
Note:
Qiagen NV, Venlo, the Netherlands.
Abbreviations: CRC, colorectal cancer; MSP, methylation-specific polymerase chain reaction; PCR, polymerase chain reaction; RT-PCR, reverse transcription followed by conventional polymerase chain reaction.
Correlation of RUNX3 hypermethylation and expression with clinicopathological features
Inactivation of RUNX3 through hypermethylation in CRC and adenoma
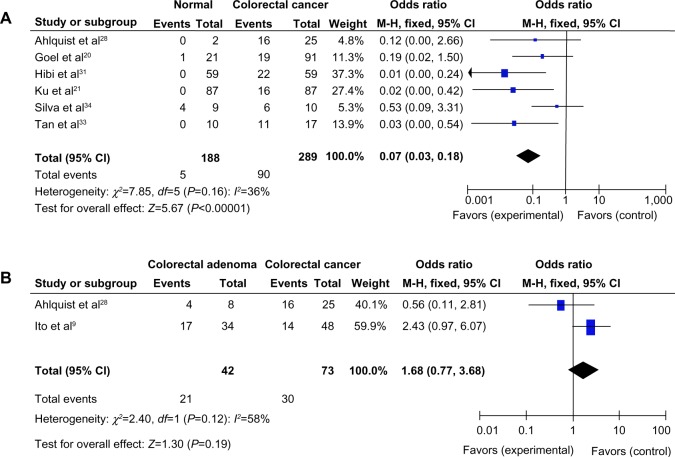
The loss of RUNX3 messenger RNA (mRNA) and protein expression was found to be statistically correlated with the promoter hypermethylation in several types of cancer including CRC.20,30,36–39 RUNX3 hypermethylation was significantly higher in CRC than in normal colorectal mucosa. The pooled OR from six studies of a total of 289 CRC and 188 normal colorectal mucosa, as shown in Figure 2A (OR =0.07, CI =0.03–0.18, P<0.00001) indicates that RUNX3 inactivation through hypermethylation plays an important role in the pathogenesis of CRC. In addition, RUNX3 hypermethylation was found to occur in colorectal adenoma but slightly less so than in CRC, as shown in Figure 2B.
Figure 2.
(A) Pooled odds ratios (ORs) from six studies of 289 colorectal cancer (CRC) and 188 normal colorectal mucosa.a (B) The pooled OR from two studies including 73 CRC and 42 colorectal adenoma.b
Notes: aOR =0.07, CI =0.00–0.54, P<0.00001; bOR =1.68, CI =0.77–3.68, P=0.19.
Abbreviations: CI, confidence interval; M-H, Mantel-Haenszel; OR, odds ratio; df, degrees of freedom.
Role of RUNX3 hypermethylation in CRC development
We analyzed 1,059 CRC patients pooled from three studies to assess whether the aberrant RUNX3 hypermethylation/expression in CRC was associated with advanced stage. As shown in Figure 3, aberrant RUNX3 hypermethylation/expression was significantly higher in advanced CRC compared with in early staged CRC (OR =0.54, CI =0.41–0.71, P<0.0001). These results suggest that epigenetic silencing of RUNX3 gene expression by promoter hypermethylation may play an important role in CRC progression and development.
Figure 3.
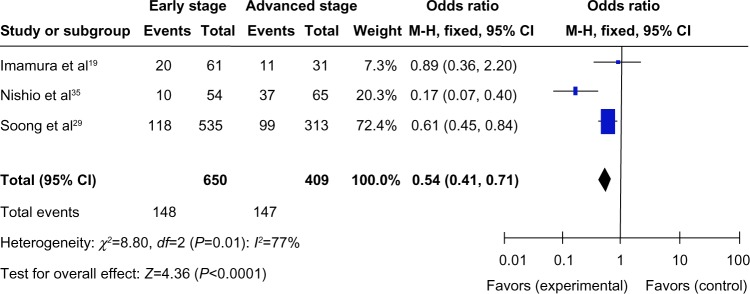
A total of 1,059 colorectal cancer (CRC) patients pooled in three studies. Aberrant RUNX3 hypermethylation/expression was significantly higher in advanced CRC than in early staged CRC.*
Notes: *OR =0.54, CI =0.41–0.71, P<0.0001.
Abbreviations: CI, confidence interval; M-H, Mantel-Haenszel; OR, odds ratio; df, degrees of freedom.
Correlation of RUNX3 hypermethylation/expression with microsatellite instability (MSI) in CRC
We also examined the data for 1,039 CRC patients pooled from four studies to assess whether the aberrant RUNX3 hypermethylation/expression in CRC was associated with MSI. As shown in Figure 4, aberrant RUNX3 hypermethylation/expression was significantly higher in MSI-positive CRC compared with in MSI-negative CRC (OR =0.44, CI =0.3–0.66, P<0.0001). These results suggest that epigenetic silencing of RUNX3 gene expression by promoter hypermethylation may play an important role in high levels of MSI.
Figure 4.
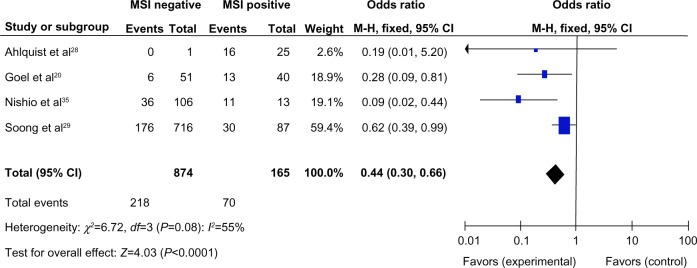
A total of 1,039 colorectal cancer (CRC) patients pooled in four studies. Aberrant RUNX3 hypermethylation/expression was significantly higher in microsatellite instability (MSI)-positive CRC than in MSI-negative CRC.*
Notes: *OR =0.44, CI =0.30–0.66, P<0.0001.
Abbreviations: CI, confidence interval; M-H, Mantel-Haenszel; OR, odds ratio; df, degrees of freedom.
RUNX3 hypermethylation/expression as a prognostic factor for CRC
Only two studies estimated the relationship between OS and RUNX3 hypermethylation/expression in CRC. Ogino et al showed that CRC patients with RUNX3 hypermethylation had a lower 1-year survival than those without RUNX3 hypermethylation (P<0.001).32 Soong et al observed that nuclear RUNX3 expression was associated with significantly better patient survival than no nuclear RUNX3 expression (P=0.025).29 The pooled HR for OS showed that RUNX3 hypermethylation or decreased RUNX3 expression was associated with worse survival in CRC patients, as shown in Figure 5 (HR =0.44, 95% CI =0.27–0.70, P=0.0006).
Figure 5.
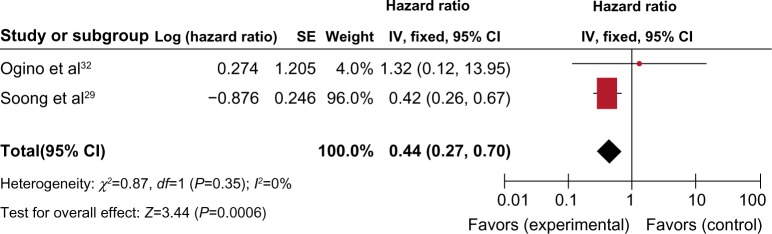
Two studies estimated the relationship between overall survival (OS) and RUNX3 hypermethylation/expression in colorectal cancer (CRC). The pooled hazard ratios (HRs) for OS showed that RUNX3 hypermethylation or decreased RUNX3 expression was associated with worse survival in CRC patients.*
Notes: *HR =0.44, 95% CI =0.27–0.70, P=0.0006.
Abbreviations: CI, confidence interval; M-H, Mantel-Haenszel; HR, hazards ratio; df, degrees of freedom.
Sensitivity analyses and publication bias
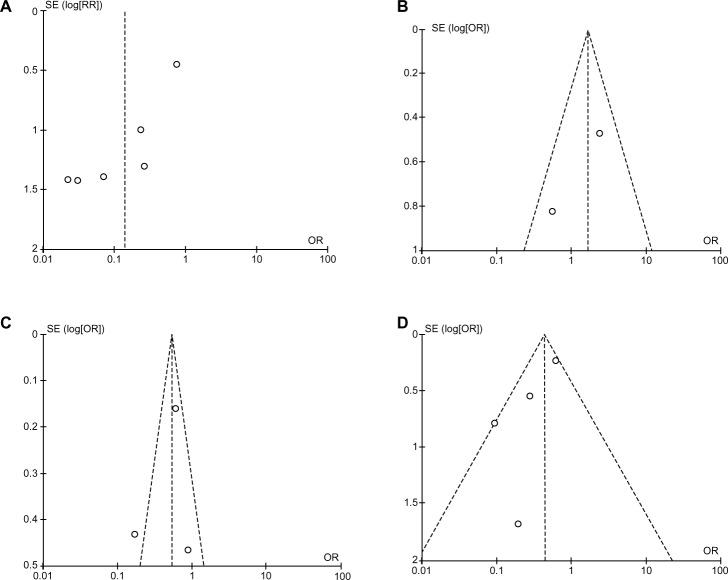
A “sensitivity analysis”, in which one study was removed at a time, was conducted to assess the stability of the results. The pooled ORs and HRs were not significantly changed, indicating the stability of our analyses. The funnel plots were largely symmetrical (Figure 6), suggesting there were no publication biases in our meta-analysis of RUNX3 methylation/expression and clinicopathological features.
Figure 6.
Funnel plots, which are largely symmetrical, suggesting there were no publication biases in the meta-analysis of RUNX3 hypermethylation/expression and clinicopathological features. (A) Funnel plot from six studies comparing colorectal cancer (CRC) with normal colorectal mucosa. (B) Funnel plot from two studies comparing CRC and colorectal adenoma. (C) Funnel plot from three studies comparing different stages of CRC. (D) Funnel plot from four studies comparing microsatellite instability (MSI)-positive and MSI-negative CRC.
Notes: X axis, value of OR; Y axis, SEs multiplied by log scale of OR.
Abbreviations: OR, odds ratio; SE, standard error.
Discussion
“Epigenetics” is the study of heritable and age-related modifications of the genome that occur without a change in the primary DNA sequence. Epigenetic alterations, particularly aberrant DNA methylation, one of the best-characterized epigenetic modifications, contribute to tumor initiation and progression.40,41 RUNX3 belongs to the runt-domain family of transcriptional factors, and its inactivation by promoter hypermethylation plays an important role in the normal development of, and tumorigenesis in, several types of tumors including CRC.14,42–53 To date, there have been some studies describing the precise expression, prognostic impact, and methylation status of RUNX3 in CRC; however, the role of RUNX3 inactivation in CRC and RUNX3 inactivation have not been thoroughly investigated. We conducted the meta-analysis to evaluate the correlation of RUNX3 hypermethylation/low expression with CRC. Analysis of the pooled data showed that: CRC had a higher hypermethylation OR than normal colorectal mucosa; aberrant RUNX3 hypermethylation/expression was significantly higher in advanced CRC compared with in early staged CRC, indicating that epigenetic silencing of RUNX3 gene expression by promoter hypermethylation may play an important role in CRC progression and development; and aberrant RUNX3 hypermethylation/expression was significantly higher in MSI-positive CRC compared with in MSI-negative CRC. In addition, CRC patients with RUNX3 hypermethylation or lacking RUNX3-protein expression had a lower survival rate than those without RUNX3 hypermethylation or with RUNX3-protein expression. Progression from colorectal adenoma to CRC appears to mirror the accumulation of genetic abnormalities, suggesting a stepwise progression of genetic changes. In fact, the hypermethylation of several tumor suppressors has been reported in colorectal adenoma.54–56 Combining RUNX3 with other genes to develop several gene markers will be a good strategy for risk stratification to predict initial carcinogenesis and neoplastic progression, including in CRC.54
RUNX3 exerts pleiotropic effects during tumor suppression. RUNX3 inhibits the oncogenic Wnt signaling pathway via the formation of a complex with the TCF4-β-catenin complex and hampers it from binding to target genes such as c-myc and cyclin D1.42,57 RUNX3 interacts with SMAD3/SMAD4 to activate TGF-β-dependent proliferation inhibition and apoptosis by activation of p21 and Bim. Although only two studies estimated the relationship between OS and RUNX3 hypermethylation/expression in CRC, both showed very similar results. Ogino et al showed that CRC patients with RUNX3 hypermethylation had lower 1-year survival than those without RUNX3 hypermethylation (P<0.001).32 Soong et al observed that nuclear RUNX3 expression was associated with significantly better patient survival than no nuclear RUNX3 expression (P=0.025).29 In addition, detection of RUNX3 methylation status may predict treatment effects among CRC patients who receive fluorouracil-based and irinotecan-based chemotherapy.32
Consistent results were shown in sensitivity analyses, and no evidence of publication bias was found. This study has several potential limitations. First, the possibility of information and selection biases and unidentified confounders could not be completely excluded, because all of the included studies were observational. Second, the search strategy was restricted to articles published in English. Articles with potentially high-quality data published in other languages were not included because of anticipated difficulties in obtaining accurate medical translation. Hence, caution should be taken in interpreting our findings among the general population.
Footnotes
Disclosure
The authors declare no conflicts of interest in this work.
References
- 1.Jemal A, Center MM, DeSantis C, Ward EM. Global patterns of cancer incidence and mortality rates and trends. Cancer Epidemiol Biomarkers Prev. 2010;19(8):1893–1907. doi: 10.1158/1055-9965.EPI-10-0437. [DOI] [PubMed] [Google Scholar]
- 2.Dai Z, Zheng RS, Zou XN, et al. Analysis and prediction of colorectal cancer incidence trend in China. Zhonghua Yu Fang Yi Xue Za Zhi. 2012;46(7):598–603. Chinese. [PubMed] [Google Scholar]
- 3.International Agency for Research on Cancer (IARC) Globocan 2008: Colorectal Cancer Fact Sheet 2008. Lyon: IARC; 2008. Available from: http://globocaniarcfr/factsheets/cancers/colorectalasp. [Google Scholar]
- 4.Weitz J, Koch M, Debus J, Höhler T, Galle PR, Büchler MW. Colorectal cancer. Lancet. 2005;365(9454):153–165. doi: 10.1016/S0140-6736(05)17706-X. [DOI] [PubMed] [Google Scholar]
- 5.McKeown E, Nelson DW, Johnson EK, et al. Current Approaches and Challenges for Monitoring Treatment Response in Colon and Rectal Cancer. J Cancer. 2014;5(1):31–43. doi: 10.7150/jca.7987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Solier S, Zhang YW, Ballestrero A, Pommier Y, Zoppoli G. DNA damage response pathways and cell cycle checkpoints in colorectal cancer: current concepts and future perspectives for targeted treatment. Curr Cancer Drug Targets. 2012;12(4):356–371. doi: 10.2174/156800912800190901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duthie SJ. Folate and cancer: how DNA damage, repair and methylation impact on colon carcinogenesis. J Inherit Metab Dis. 2011;34(1):101–109. doi: 10.1007/s10545-010-9128-0. [DOI] [PubMed] [Google Scholar]
- 8.Ozaki T, Nakagawara A, Nagase H. RUNX Family Participates in the Regulation of p53-Dependent DNA Damage Response. Int J Genomics. 2013;2013:271347. doi: 10.1155/2013/271347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ito Y. RUNX genes in development and cancer: regulation of viral gene expression and the discovery of RUNX family genes. Adv Cancer Res. 2008;99:33–76. doi: 10.1016/S0065-230X(07)99002-8. [DOI] [PubMed] [Google Scholar]
- 10.Blyth K, Cameron ER, Neil JC. The RUNX genes: gain or loss of function in cancer. Nat Rev Cancer. 2005;5(5):376–387. doi: 10.1038/nrc1607. [DOI] [PubMed] [Google Scholar]
- 11.Subramaniam MM, Chan JY, Yeoh KG, Quek T, Ito K, Salto-Tellez M. Molecular pathology of RUNX3 in human carcinogenesis. Biochim Biophys Acta. 2009;1796(2):315–331. doi: 10.1016/j.bbcan.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 12.Lund AH, van Lohuizen M. RUNX: a trilogy of cancer genes. Cancer Cell. 2002;1(3):213–215. doi: 10.1016/s1535-6108(02)00049-1. [DOI] [PubMed] [Google Scholar]
- 13.Levanon D, Negreanu V, Bernstein Y, Bar-Am I, Avivi L, Groner Y. AML1, AML2, and AML3, the human members of the runt domain gene-family: cDNA structure, expression, and chromosomal localization. Genomics. 1994;23(2):425–432. doi: 10.1006/geno.1994.1519. [DOI] [PubMed] [Google Scholar]
- 14.Shiraha H, Nishina S, Yamamoto K. Loss of runt-related transcription factor 3 causes development and progression of hepatocellular carcinoma. J Cell Biochem. 2011;112(3):745–749. doi: 10.1002/jcb.22973. [DOI] [PubMed] [Google Scholar]
- 15.Li QL, Ito K, Sakakura C, et al. Causal relationship between the loss of RUNX3 expression and gastric cancer. Cell. 2002;109(1):113–124. doi: 10.1016/s0092-8674(02)00690-6. [DOI] [PubMed] [Google Scholar]
- 16.Chi XZ, Yang JO, Lee KY, et al. RUNX3 suppresses gastric epithelial cell growth by inducing p21(WAF1/Cip1) expression in cooperation with transforming growth factor {beta}-activated SMAD. Mol Cell Biol. 2005;25(18):8097–8107. doi: 10.1128/MCB.25.18.8097-8107.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wei D, Gong W, Oh SC, et al. Loss of RUNX3 expression significantly affects the clinical outcome of gastric cancer patients and its restoration causes drastic suppression of tumor growth and metastasis. Cancer Res. 2005;65(11):4809–4816. doi: 10.1158/0008-5472.CAN-04-3741. [DOI] [PubMed] [Google Scholar]
- 18.Chen LF. Tumor suppressor function of RUNX3 in breast cancer. J Cell Biochem. 2012;113(5):1470–1477. doi: 10.1002/jcb.24074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Imamura Y, Hibi K, Koike M, et al. RUNX3 promoter region is specifically methylated in poorly-differentiated colorectal cancer. Anticancer Res. 2005;25(4):2627–2630. [PubMed] [Google Scholar]
- 20.Goel A, Arnold CN, Tassone P, et al. Epigenetic inactivation of RUNX3 in microsatellite unstable sporadic colon cancers. Int J Cancer. 2004;112(5):754–759. doi: 10.1002/ijc.20472. [DOI] [PubMed] [Google Scholar]
- 21.Ku JL, Kang SB, Shin YK, et al. Promoter hypermethylation downregulates RUNX3 gene expression in colorectal cancer cell lines. Oncogene. 2004;23(40):6736–6742. doi: 10.1038/sj.onc.1207731. [DOI] [PubMed] [Google Scholar]
- 22.McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM, Statistics Subcommittee of the NCI-EORTC Working Group on Cancer Diagnostics Reporting recommendations for tumor marker prognostic studies (REMARK) J Natl Cancer Inst. 2005;97(16):1180–1184. doi: 10.1093/jnci/dji237. [DOI] [PubMed] [Google Scholar]
- 23.Steels E, Paesmans M, Berghmans T, et al. Role of p53 as a prognostic factor for survival in lung cancer: a systematic review of the literature with a meta-analysis. Eur Respir J. 2001;18(4):705–719. doi: 10.1183/09031936.01.00062201. [DOI] [PubMed] [Google Scholar]
- 24.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7(3):177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 25.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327(7414):557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.DerSimonian R. Meta-analysis in the design and monitoring of clinical trials. Stat Med. 1996;15(12):1237–1248. doi: 10.1002/(SICI)1097-0258(19960630)15:12<1237::AID-SIM301>3.0.CO;2-N. discussion 1249–1252. [DOI] [PubMed] [Google Scholar]
- 27.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315(7109):629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ahlquist T, Lind GE, Costa VL, et al. Gene methylation profiles of normal mucosa, and benign and malignant colorectal tumors identify early onset markers. Mol Cancer. 2008;7:94. doi: 10.1186/1476-4598-7-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Soong R, Shah N, Peh BK, et al. The expression of RUNX3 in colorectal cancer is associated with disease stage and patient outcome. Br J Cancer. 2009;100(5):676–679. doi: 10.1038/sj.bjc.6604899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ito K, Lim AC, Salto-Tellez M, et al. RUNX3 attenuates beta-catenin/T cell factors in intestinal tumorigenesis. Cancer Cell. 2008;14(3):226–237. doi: 10.1016/j.ccr.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 31.Hibi K, Nakao A. Lymph node metastasis is infrequent in patients with highly-methylated colorectal cancer. Anticancer Res. 2006;26(1A):55–58. [PubMed] [Google Scholar]
- 32.Ogino S, Meyerhardt JA, Kawasaki T, et al. CpG island methylation, response to combination chemotherapy, and patient survival in advanced microsatellite stable colorectal carcinoma. Virchows Arch. 2007;450(5):529–537. doi: 10.1007/s00428-007-0398-3. [DOI] [PubMed] [Google Scholar]
- 33.Tan SH, Ida H, Lau QC, et al. Detection of promoter hypermethylation in serum samples of cancer patients by methylation-specific polymerase chain reaction for tumour suppressor genes including RUNX3. Oncol Rep. 2007;18(5):1225–1230. [PubMed] [Google Scholar]
- 34.Silva TD, Vidigal VM, Felipe AV, et al. DNA methylation as an epigenetic biomarker in colorectal cancer. Oncol Lett. 2013;6(6):1687–1692. doi: 10.3892/ol.2013.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nishio M, Sakakura C, Nagata T, et al. RUNX3 promoter methylation in colorectal cancer: its relationship with microsatellite instability and its suitability as a novel serum tumor marker. Anticancer Res. 2010;30(7):2673–2682. [PubMed] [Google Scholar]
- 36.Long C, Yin B, Lu Q, et al. Promoter hypermethylation of the RUNX3 gene in esophageal squamous cell carcinoma. Cancer Invest. 2007;25(8):685–690. doi: 10.1080/07357900701561131. [DOI] [PubMed] [Google Scholar]
- 37.Tonomoto Y, Tachibana M, Dhar DK, et al. Differential expression of RUNX genes in human esophageal squamous cell carcinoma: downregulation of RUNX3 worsens patient prognosis. Oncology. 2007;73(5–6):346–356. doi: 10.1159/000135350. [DOI] [PubMed] [Google Scholar]
- 38.Watanabe T, Kobunai T, Ikeuchi H, et al. RUNX3 copy number predicts the development of UC-associated colorectal cancer. Int J Oncol. 2011;38(1):201–207. [PubMed] [Google Scholar]
- 39.Chen W, Gao N, Shen Y, Cen JN. Hypermethylation downregulates Runx3 gene expression and its restoration suppresses gastric epithelial cell growth by inducing p27 and caspase3 in human gastric cancer. J Gastroenterol Hepatol. 2010;25(4):823–831. doi: 10.1111/j.1440-1746.2009.06191.x. [DOI] [PubMed] [Google Scholar]
- 40.Delpu Y, Cordelier P, Cho WC, Torrisani J. DNA methylation and cancer diagnosis. Int J Mol Sci. 2013;14(7):15029–15058. doi: 10.3390/ijms140715029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ma X, Wang YW, Zhang MQ, Gazdar AF. DNA methylation data analysis and its application to cancer research. Epigenomics. 2013;5(3):301–316. doi: 10.2217/epi.13.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chuang LS, Ito Y. RUNX3 is multifunctional in carcinogenesis of multiple solid tumors. Oncogene. 2010;29(18):2605–2615. doi: 10.1038/onc.2010.88. [DOI] [PubMed] [Google Scholar]
- 43.Jang BG, Kim WH. Molecular pathology of gastric carcinoma. Pathobiology. 2011;78(6):302–310. doi: 10.1159/000321703. [DOI] [PubMed] [Google Scholar]
- 44.Kohya N, Koga Y, Kitajima Y, Miyazaki K. Aberrant promoter hypermethylation in biliary tract carcinoma. J Hepatobiliary Pancreat Surg. 2006;13(4):296–305. doi: 10.1007/s00534-005-1058-0. [DOI] [PubMed] [Google Scholar]
- 45.Lee JH, Pyon JK, Kim DW, et al. Expression of RUNX3 in skin cancers. Clin Exp Dermatol. 2011;36(7):769–774. doi: 10.1111/j.1365-2230.2011.04069.x. [DOI] [PubMed] [Google Scholar]
- 46.Lee JM, Shin JO, Cho KW, et al. Runx3 is a crucial regulator of alveolar differentiation and lung tumorigenesis in mice. Differentiation. 2011;81(4):261–268. doi: 10.1016/j.diff.2011.02.001. [DOI] [PubMed] [Google Scholar]
- 47.Lee YM. Control of RUNX3 by histone methyltransferases. J Cell Biochem. 2011;112(2):394–400. doi: 10.1002/jcb.22969. [DOI] [PubMed] [Google Scholar]
- 48.Levanon D, Brenner O, Otto F, Groner Y. Runx3 knockouts and stomach cancer. EMBO Rep. 2003;4(6):560–564. doi: 10.1038/sj.embor.embor868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shima K, Nosho K, Baba Y, et al. Prognostic significance of CDKN2A (p16) promoter methylation and loss of expression in 902 colorectal cancers: Cohort study and literature review. Int J Cancer. 2011;128(5):1080–1094. doi: 10.1002/ijc.25432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tamura G. Promoter methylation status of tumor suppressor and tumor-related genes in neoplastic and non-neoplastic gastric epithelia. Histol Histopathol. 2004;19(1):221–228. doi: 10.14670/HH-19.221. [DOI] [PubMed] [Google Scholar]
- 51.Tamura G. Alterations of tumor suppressor and tumor-related genes in the development and progression of gastric cancer. World J Gastroenterol. 2006;12(2):192–198. doi: 10.3748/wjg.v12.i2.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhao C, Bu X. Promoter methylation of tumor-related genes in gastric carcinogenesis. Histol Histopathol. 2012;27(10):1271–1282. doi: 10.14670/HH-27.1271. [DOI] [PubMed] [Google Scholar]
- 53.Suzuki M, Shigematsu H, Shames DS, et al. DNA methylation-associated inactivation of TGFβ-related genes, DRM/Gremlin, RUNX3, and HPP1 in human cancers. Br J Cancer. 2013;109(12):3132. doi: 10.1038/bjc.2013.776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cassinotti E, Melson J, Liggett T, et al. DNA methylation patterns in blood of patients with colorectal cancer and adenomatous colorectal polyps. Int J Cancer. 2012;131(5):1153–1157. doi: 10.1002/ijc.26484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Abouzeid HE, Kassem AM, Abdel Wahab AH, El-mezayen HA, Sharad H, Abdel Rahman S. Promoter hypermethylation of RASSF1A, MGMT, and HIC-1 genes in benign and malignant colorectal tumors. Tumour Biol. 2011;32(5):845–852. doi: 10.1007/s13277-011-0156-7. [DOI] [PubMed] [Google Scholar]
- 56.Psofaki V, Kalogera C, Tzambouras N, et al. Promoter methylation status of hMLH1, MGMT, and CDKN2A/p16 in colorectal adenomas. World J Gastroenterol. 2010;16(28):3553–3560. doi: 10.3748/wjg.v16.i28.3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Voon DC, Wang H, Koo JK, et al. Runx3 protects gastric epithelial cells against epithelial-mesenchymal transition-induced cellular plasticity and tumorigenicity. Stem Cells. 2012;30(10):2088–2099. doi: 10.1002/stem.1183. [DOI] [PubMed] [Google Scholar]