Abstract
• Premise of the study: We developed and tested primers for 218 nuclear loci for studying population genetics, phylogeography, and genome evolution in bryophytes.
• Methods and Results: We aligned expressed sequence tags (ESTs) from Ceratodon purpureus to the Physcomitrella patens genome sequence, and designed primers that are homologous to conserved exons but span introns in the P. patens genome. We tested these primers on four isolates from New York, USA; Otavalo, Ecuador; and two laboratory isolates from Austria (WT4 and GG1). The median genome-wide nucleotide diversity was 0.008 substitutions/site, but the range was large (0–0.14), illustrating the among-locus heterogeneity in the species.
• Conclusions: These loci provide a valuable resource for finely resolved, genome-wide population genetic and species-level phylogenetic analyses of C. purpureus and its relatives.
Keywords: chromosomal inversion, expressed sequence tag (EST), phylogeography, Physcomitrella patens, sex-linked loci
Over the past 15 yr, our understanding of the microevolutionary processes that shape variation within bryophyte populations has been revolutionized by the use of DNA sequence variation. Most of these inferences have been drawn from variation in a small number of loci, principally from the chloroplast and nuclear ribosomal regions (Stech and Quandt, 2010). However, these loci may be difficult to align, they may lack sufficient variation to answer many questions, and they may not reflect the full complexity of the organismal history (McDaniel et al., 2010; Vanderpoorten and Shaw, 2010).
To develop new loci for phylogeographic and population genetic inference in Ceratodon purpureus (Hedw.) Brid., we have generated primers for exon-primed intron-spanning loci, based on an alignment of expressed sequence tags (ESTs) from C. purpureus to the Physcomitrella patens (Hedw.) Bruch & Schimp. genome. The common ancestor of P. patens and C. purpureus represents the common ancestor of nearly all of the arthrodontous mosses, comprising ∼95% of moss species (Cox et al., 2010). Thus, although we designed these primers specifically for use in C. purpureus and its relatives, by choosing conserved priming sites we have maximized the chance that these loci will amplify homologous regions in other bryophyte species.
METHODS AND RESULTS
To develop primers for nuclear loci in C. purpureus, we screened the 1677 ESTs available on GenBank at the time. We first clustered the ESTs into 850 unigenes, and aligned them to the P. patens genome using the software BLAT (Kent, 2002; http://genome.ucsc.edu/goldenPath/help/blatSpec.html). This resulted in 450 aligned unigenes, or 1050 aligned ESTs. Using the software Primer3 (Rozen and Skaletsky, 2000), we designed pairs of primers that were homologous to the C. purpureus sequence and that spanned a single intron in the P. patens genome. We designed a set of primers with their 3′ end at least 25 bp from the beginning of the intron. This resulted in primers for 212 nuclear loci. On the intron-spanning unigenes that failed the primer design process, we also designed a set of primers with their 3′ ends at least 5 bp from the beginning of the intron. This resulted in primers for an additional 33 nuclear loci (all primer details are in Table 1). In some cases, the unigene spanned multiple introns, and we designed separate pairs of primers for each intron. Where possible, we also designed alternate primers for each intron in the complete unigene set.
Table 1.
Ceratodon purpureus EST intron primer sequences.
| EST–intron start position | 5′ Primer | 3′ Primer | Forward Tma | Reverse Tmb | Prod. sizec |
| Primer >25 nucleotides from intron | |||||
| AW098097–137 | CCAATAGCAAAAGAATACAATCAAAGC | CGCGTTGTTCCTGGAATGG | 62 | 64 | 318 |
| AF233229–229 | CCGAACTTGAACAGGACAGTTATGG | TGATCACGATTCCGACTGAGG | 65 | 63 | 458 |
| AF233229–433 | GCTCCCAACTCAGGAACACCC | GGAGGCCTGGAGATGGTAACG | 65 | 65 | 510 |
| AW098318–384 | AATCGTCACCGAATGAGAGGG | CAATGTTGGCATGCTCCC | 63 | 61 | 467 |
| AW098631–273 | ATGTGTTGTGCTTTCCCAGG | TGGACAGTCACAACTCCTCTCCC | 60 | 65 | 396 |
| AW086794–139 | TATCGGCGTGTGCAAGGG | GCTGGCTTCTCGAATGTGGG | 64 | 65 | 342 |
| AW086770–115 | TCCGCGAGCTCTGAGTGG | AACAACTTCACCACATCTGCACG | 63 | 64 | 308 |
| AW086590–267 | AACTGTGCACAAACAGCGGC | TGTACGACCATCCAGACTAAGAAGG | 64 | 62 | 593 |
| AW098359–150 | GCGAAGTCCAAGAACCATACCG | AGCCTCGACCTCATCGGC | 65 | 63 | 407 |
| AW087018–146 | CGACATTGCAGTACGAACTTTGC | CAAGTGGAGCCTGTCATCTTTGC | 64 | 65 | 375 |
| AW087018–260 | CTGGTCATGGATCTGGTCGC | CAGTCCTCTCAGCATCCAGG | 64 | 60 | 298 |
| AW086837–165 | CTTGGACGTGCATGGAGGC | GTGTTGAACCGCATGTTGTCG | 65 | 64 | 316 |
| AW086837–243 | ATCAGCTTCGGACCCTTCACC | CTATGTGTGGGGCCAGCG | 64 | 63 | 307 |
| AW086675–86 | CCGCATGTAGAAGAGCTACGCC | TCTCGTCGCAATGCAGGC | 65 | 64 | 277 |
| AW086645–117 | ATGCTTGCAACGCTTGATGG | GTCAGGTTTCGGTGGTCAGTCC | 64 | 65 | 466 |
| AW098393–199 | AAGGCTGAGGCTGAGGGC | AGTGCGGGAAGCCACACG | 63 | 65 | 284 |
| AW098679–454 | CCTTGATGTGGTAGTAATCTGCG | CGAAATTGGTGGTGTGAGGG | 60 | 63 | 438 |
| AW098679–532 | CATCGGGATTGAAAGCAAGAGC | ATTGGTGGTGGTCAAGAGGC | 65 | 62 | 426 |
| AW098515–203 | CTGAAGAATTGGAACCAGTCCG | TTAAGCTTCGCGCAGTCCG | 63 | 64 | 452 |
| AW086579–194 | ATCGGACGACTTCCTGCC | TTCAGGAATACGGTCGGCG | 61 | 64 | 223 |
| AW086579–332 | TGCAACTCCCAAAGCTGCC | TGTTTTGGCACGAGACATGG | 64 | 63 | 412 |
| AW086736–262 | GAGGTCCGAAGAGAGCCACC | TCTTGCATGTGGCAGACACG | 63 | 64 | 404 |
| AW097953–298 | GCCCCTTCAAGAGACTTTGGC | GGCAAGAACATCTTGTCTTTGATGC | 64 | 65 | 354 |
| AW098418–107 | GAAGAAGGAGACGCCCGC | AGACGGTCGGACAACACGG | 63 | 64 | 320 |
| AW098974–394 | TTGGAAGCCAATTTCTCCTTGG | CAGGCACAGGAAGACTGTGAACC | 64 | 65 | 510 |
| AW086877–265 | TTGATCCTCGGCTAGTTGCTCC | TCCAGTGCTTCCAGCAGCC | 64 | 65 | 511 |
| AW086877–378 | ATGGAGATGCGGCTGCTGG | ACCAAGTGAATGCGGTCATATCC | 67 | 64 | 339 |
| AW086549–119 | ACAAGGAAAGACGCAATTATTTATAGG | TGTGTCCTCCGGCTCACG | 61 | 64 | 394 |
| AW086549–238 | AGAGCACAGGCATGACGGC | GCAGAATTGTATGCGCGACG | 64 | 64 | 323 |
| AW086868–74 | CGATGTGGTTTACAACGCGTCC | ACCAGGTACGGAAGGGGGC | 66 | 65 | 377 |
| AW086868–247 | TCGATGCTGCCCCCTTCC | CAGAACGGCCGCATTGG | 66 | 64 | 392 |
| AW086594–170 | TGCTCCAACAAGTCACCAATGC | TTCCCATTGACTTGCGCC | 65 | 63 | 337 |
| AW086594–327 | GCTTGGTCAACCTCCTGCG | TGAGAGTTGCCAAGGTCACTGG | 64 | 64 | 479 |
| AW098340–132 | GCCCAGCAACAGCAAGGG | CACCACCCTGGCCAACTCC | 65 | 66 | 441 |
| AW098700–545 | CCACTTCTGCCATGGTGGG | GCTTCGGTGTGGTTCTGTGG | 64 | 64 | 350 |
| AW086759–94 | CCGTAGCTTCTGCAATATGTTTGAGG | TGGCAAGGCGACCAAAGC | 65 | 65 | 486 |
| AW098624–132 | GACAAGGCCAGTTTGTACGCC | TGGTCTTAGCCTTATTGTGAAAGACG | 64 | 63 | 269 |
| AW098020–216 | TCCGTTCAATTCAGCTCGG | TGACGCAGCTTAGCGGGC | 62 | 65 | 262 |
| AW098462–157 | TGACGATAGAAGGTGTGCGCC | AACAGGAAGGCCAGCCTATGG | 65 | 64 | 576 |
| AW086686–370 | CACTCCCTTTTTGAGATCTTCAGGG | ATGGGGTGCAGATTGGGG | 65 | 64 | 396 |
| AW098568–216 | ACCACATCCATTTCGGACGC | TTCCCAGCTCGACAACATGG | 65 | 64 | 327 |
| AW086975–261 | ACTTATTTCGCCAGAGGATCTGC | TCACTTTGACTCACAGACTGAATGACC | 63 | 65 | 391 |
| AW086975–331 | TTGCGCAAGTTTGACAGTGG | GCAGGAGCCATTCCCAAGG | 63 | 65 | 362 |
| AW098234–164 | TGGAGCTTGCTCACATCGC | CAGCAATACCCACTGCACGG | 63 | 64 | 396 |
| AW098407–97 | GGCGAAGGCTGTGATGGG | TTTTTGCAAATGCAACCTGGG | 65 | 65 | 392 |
| AW086555–375 | GCCATAGTCGCATCAAAATTGG | GGTTGTGGTCAGCAGCCG | 63 | 63 | 438 |
| AW097994–187 | ACGAAGCTGGAGGCTGTGG | CAATGTCTATGGAACCAATCTTTGC | 63 | 62 | 396 |
| AW086517–380 | TTGGTCTTCCTCCGCAGC | TCCGTGGATGATGATCTGGC | 62 | 64 | 473 |
| AW086517–457 | CATGTCTTGCTTCAAGGATGC | TCTGCATTGGAGGCAGAATCCG | 60 | 68 | 267 |
| AW098225–85 | AGGCCAGAGTTGCTGAACCG | ACTCACCCGCCAGCAAGG | 65 | 64 | 531 |
| AW098839–86 | TGACTAGATGCTTTTATTGCTGAAAGG | ATCACATCGAACGGGCAGC | 63 | 64 | 493 |
| AW098839–177 | CAACCTGGTCCAGATCACGC | TACGGTGGCCGCAAGAGC | 64 | 65 | 428 |
| AW086694–137 | TCTTCATGACCTTAGCCACAGCC | AGATGATCTTGATGCGAGGG | 64 | 60 | 304 |
| AW086845–149 | AGGGGAAAGGCACCAGGG | AGTTCACGGTTCCGGTGAGG | 64 | 64 | 679 |
| AW086858–91 | CATCAAGGGAGTGGGGCG | TCAGCTTCATGTCCAAGGCG | 64 | 64 | 451 |
| AW086858–280 | TGGTGATCGTTGCCAACCC | CACCGACGGTTCTTCCACG | 65 | 64 | 406 |
| BI894288–61 | TGGAAGAAGCCTTTTGCTGG | TTCAAACAACCATCGTTGGGG | 62 | 65 | 285 |
| BI894288–188 | GGCTAAGCAGCCCAACTCTGC | TCTCCCACGGCATGTCCC | 65 | 65 | 353 |
| AW087023–114 | GGATCCACAAGGCCAAGTTGC | CATTTCGACGAACGGTGGC | 66 | 64 | 433 |
| AW087023–239 | AGCCATAAACGCAATTCGGG | TGACTGAACCCGTCCGGC | 64 | 65 | 399 |
| AW097931–294 | AGAAGTTCCAAGTCGGGTGGC | TCCGTTGGCGTTCTTCAGC | 64 | 64 | 500 |
| AW098613–99 | CTTTGGCTTGAGAAACAAGGGG | GCCGGCGATACAAATGAAACC | 64 | 65 | 251 |
| AW086659–247 | TTCCAGGTGGGTGGGAGC | AGGGTTTGACCCCGTCGC | 64 | 65 | 494 |
| AW086999–142 | AGCTATTAGGGCGAGTGAAAGCG | GGCTGGAGTGAATCATTTTGGG | 65 | 64 | 368 |
| AW086548–172 | AAATTCGGCAAATAAGAGGGG | CCGGGCAAATCGTTCAGG | 61 | 64 | 428 |
| AW098157–126 | CCTGCGAGGAAGATGACGC | AAGTTTCCACATTCAGAAGAGCCG | 64 | 64 | 514 |
| AJ250735–923 | TCAGCCAGTTTGATGGGC | GGTTTCCATCCGGGGAGC | 60 | 65 | 723 |
| AJ250735–1460 | TCAGGCCCGAGTTGACCC | AAGTGCCCGAAGCCATGC | 64 | 64 | 747 |
| AW097975–117 | GAAGGGAAAGCCTGATTTGCG | GATGGGTCCGGTGATGGC | 65 | 64 | 564 |
| AW098252–97 | CAAGGAGGTGGCGACTGC | CCGGCGAACTGCAAAACC | 63 | 64 | 624 |
| AW086519–461 | CAGCGAAAATCAGACGCTGC | CCGCAAGGACCATCACCC | 64 | 64 | 1025 |
| AW098432–169 | ATGAGTCGGCCGCATGG | TGCAGAGCCTTACACGACGC | 64 | 64 | 759 |
| AW087005–110 | GCTATGTGGTGGCGTCCG | GAACAGCAAAATATTGCCTCTCGG | 63 | 65 | 385 |
| AW098158–174 | TCCCTTGCCCTTCTTGTCTCC | TGTGCTGAGGTTGCAGTGCC | 64 | 65 | 216 |
| AW098158–249 | TGCTGTGTCACGTGCTTGC | GGAAGCAGTCAGGATACGGAGG | 63 | 64 | 443 |
| AW098158–317 | CGGGCTTGGTCTGTCCTCC | TCTTCTGCCCTGGGAAGGC | 65 | 65 | 471 |
| AW098372–71 | TCCATTTGATCGCTGTTCCG | GAACTCAGGCAATCAAAGCAAGC | 64 | 64 | 257 |
| AW097984–288 | GTGGGCTGAAGCGCAAGC | TGATCATGTTGCGTAGGTGGG | 65 | 64 | 237 |
| AW098512–413 | TCGCAACTCAGCCCTCACG | TTGTTCCCGCAAGTGGGC | 66 | 65 | 319 |
| AW098442–86 | AGCTCTGCACTGGGCTGG | TGGAACCGTGAAAGCCCG | 62 | 65 | 279 |
| AW098442–198 | GGATAAGCAGACGCGCCG | CGAAAATGCAACCCCAGGC | 65 | 65 | 271 |
| AW098442–292 | CCCAAAGTTCTTCTTGGGGC | GAAAGATGTGAAGAAGCTGTTGAAGC | 63 | 63 | 252 |
| AW098349–75 | AGATGATGCTGAGATGGAAGCG | TCAAGAGAGGCACCCTCGG | 63 | 63 | 400 |
| AW098572–84 | TGGGCTCTTGATGTTCTGGG | ACCGATCCTCAGCCTGCC | 63 | 63 | 381 |
| AW098572–303 | CCACAAATGGATCGGCAGG | TTGCCTTCAAGCACAGCAGC | 64 | 65 | 316 |
| AW086786–104 | ACGTGCTTGTTGCAAGGGC | AACAGCACGTGTCGGTCAGC | 64 | 64 | 291 |
| AW086989–141 | AAGTGGCGCGGTTTGAGG | GTTTGACAGGAACGGGCAGG | 64 | 65 | 759 |
| AW098304–85 | ATGGAAGCATGTGGTTTTGGG | CGACGCATGTCCTTCGGC | 64 | 66 | 560 |
| AW086668–76 | CAACAGGCCTCTAAATCTTGGCG | TGGAGATTCTCACAGCCCCC | 66 | 64 | 307 |
| AW098058–71 | CCAAGCGGGGCTTGTATGC | GTCCAGCTTGCGTTCGGG | 66 | 65 | 414 |
| AW098449–310 | CTGCCAAGACCATTTTGAGTGG | AAAAAGTCACGACTAATGCTGTGC | 64 | 61 | 369 |
| AW097976–116 | CATGTGCTGGTCTCACATTCCC | GCATTTTGTCTCAAGCCTTCG | 65 | 62 | 485 |
| AW098621–172 | TCGTGGTTGAGTTGAACGGG | TCCCAAAACCTTGCCTCCG | 64 | 65 | 407 |
| AW097924–87 | GCGTGCGTATGTTGTGATTAAGG | GCTCTCTGCTACAGCAGCCTCG | 63 | 66 | 347 |
| AW097924–222 | CATGTTGCGCGAATCAGAGG | AACACAGAGTCTTGAATCTCCAGGG | 65 | 64 | 323 |
| AW086546–344 | TCCTTCTCGGCAGCCTCG | CAACCGTCTTAACACCTACAGGGC | 65 | 65 | 504 |
| AW086789–334 | GACCCCCAGAGCATCTTCG | CCTTGCGCTCCTTGATCTGC | 63 | 65 | 358 |
| AW098343–365 | CCAACGACGCTTCTTCGC | GAGGACGGGATTTGTTCATGC | 63 | 63 | 674 |
| AW098256–171 | TGATGACGAGCTGCTCAGAGG | GCCTTCTCAGCTTTGGCTGC | 64 | 64 | 310 |
| AW098256–290 | AAGTGGCCGTGGAGATACAAGC | GCTGCTCTCTCCTTCTCACGC | 64 | 64 | 393 |
| AW097983–255 | TGCAAGCCCAGCTCTCTGC | TCAAATTCCATTTGCTCGTCC | 65 | 62 | 455 |
| AW097983–358 | GTGGCAATATTAAAGAGGCACCG | GGAATACATGCTGCTCCTTTCTCC | 63 | 64 | 307 |
| AW087043–267 | CCCTTGCCCTTCTTCTTCTGG | TTCTCACTTGGAGGATCAGTTTGC | 64 | 63 | 764 |
| AW086539–362 | GTTCCTGCAGCGCAGTCG | ACTTGCGCCAAGGAGAATGG | 64 | 64 | 425 |
| AW098322–137 | CTTGGGAAGACGTGGCCC | AAGCGCTTCACCCAGGC | 63 | 62 | 339 |
| AW098322–275 | CGTCATTGATGCTGAGGCG | CAAGCCGACGCCAGTTGC | 63 | 66 | 398 |
| AW097946–284 | CGACGCCGACATTCATGC | AGGTCTGCATTCAGCAGATTACCC | 64 | 64 | 370 |
| AW098391–123 | AGCCGAGGATTTAGGGCAGG | AGACCGGTGCCTCATCCG | 64 | 64 | 287 |
| AW098391–191 | AGGTGCTTGGACTGCTCAGG | GGAATGCCAAAGGCGTCG | 62 | 64 | 297 |
| AW097948–171 | GGCCCAAATCACCTACGCC | GTTCTTCAGATGACGACGAGCC | 64 | 63 | 408 |
| AW097948–251 | TGGTCTTCTTTTGGCTCGTCG | CGTCCAACAGAGCCCTAAATGG | 64 | 65 | 244 |
| AW098786–108 | GCACCGGTGGAAAGACCG | ACGTATCACGAGGGCCACC | 65 | 63 | 349 |
| AW098786–315 | ATGGGAAGACTTGCGCGG | TTTGTGACCCATTCGCCC | 64 | 62 | 324 |
| AW098048–269 | TCCCAGGTATCATTGCCCG | AATGAAAGTGGCTGCTATCCAAGC | 64 | 64 | 606 |
| AW098048–348 | ACAGATGTGGCTTGGATAGCAGC | CCGATGTAAGTGTCTCTGCTGATGG | 64 | 66 | 333 |
| AW098804–195 | GGCGGAGACAGGCACATACC | GAGAATTCAGGCCGCTCCG | 65 | 65 | 405 |
| AW086917–156 | GAATGGCCTCTTCCGGGC | TGCAATCTGCACCGACTTGG | 65 | 65 | 333 |
| AW086712–250 | TTACTGCCTTGCTGGGGTCC | AACCAGCCCGCTAAGCTGC | 64 | 64 | 378 |
| AW087002–282 | TTATCGCCAGAGGACAGCG | GCCCATATTTGACAAGGCCG | 62 | 64 | 402 |
| AW086924–329 | GCAGCACCTTGACGTCGC | AGCTTCAAGGCCTGAGAAATGC | 64 | 64 | 600 |
| AW098761–71 | CATGGCGCAGACCAATACG | AAGTATGGGATCAAAGAGTTGTAGAGC | 63 | 61 | 294 |
| AW086619–136 | CACACCTGCCTGTGGATTTGG | CAAGCTCGACAACCCGGC | 65 | 65 | 602 |
| AW087029–297 | CATGGGGTTCTGAGGCCG | GGTCGGGATGAAATGCAAGC | 64 | 64 | 486 |
| AW086641–183 | GGTTGCCTCCCTCAATCGC | GAAGGCTGTCGGCATCTGG | 65 | 64 | 248 |
| AW086641–266 | AAGGTGTAAGCACCTCCGGC | GAAAGCCGGAATCGTCGG | 63 | 63 | 501 |
| AW086618–269 | TGCCTCCTTCGCCTGATCC | AGACATCGGAAAAGAAGTCGAACG | 66 | 64 | 513 |
| AW098004–106 | GGATTGGGCGAAAGAAGCG | AGAGAAGCACAACAGGGCCG | 65 | 65 | 363 |
| AW098004–215 | CCATGGCTCGGAAGGAGG | CCTCCGTAAGGCTGACTGTCC | 64 | 63 | 373 |
| AW098782–68 | GCTTGCTCCTGCTTTGACTGC | CTGAAGGGCTCCCAAATAGCC | 64 | 64 | 422 |
| AW098479–502 | CTGGACAGCCAGTCAAGGC | CCGATGCAATGAATGCCG | 62 | 64 | 281 |
| AW086833–142 | AAGGATCCGCGACAAGTACCC | TTGATGCGCTTCCTTATGACG | 64 | 63 | 298 |
| AW098949–71 | GATTTCAAGACGCATCAGTTGCC | CGAAGATGTCGCCCCTGG | 65 | 65 | 466 |
| AW098949–266 | TGGCGCTATTACCAGGGGC | CTGTCCAGGCATGGTGGG | 65 | 63 | 406 |
| AW098098–311 | CCCCTGATCTTCTTGGCACG | GACCCCCAGAGCATCTTCG | 65 | 63 | 349 |
| AW086738–110 | CATGCAGAACGAGGAGGGC | CCTGAGCGCGAACGAAGC | 64 | 65 | 528 |
| AW086979–128 | TACACATTGCAGCGCATGG | CCATAACAATGAAGGCGCACG | 62 | 65 | 342 |
| AW086878–120 | TCTACCCCGCCGATGACG | GAGAAGGCCAGACTCCAACTGC | 65 | 64 | 654 |
| AW087022–123 | CGCCTTCACAGGCACAGG | CTTGGAAGCCAATTTCTCCTTGG | 64 | 65 | 502 |
| AW086647–346 | AGATGATGGGGGTGGTGTAAGC | GCAACCCCATTGAGAAAGCG | 64 | 65 | 395 |
| AW098140–138 | CCTAGTCACATGCCATGACCACC | CGCTCAGGGAGATACTGACAGC | 65 | 63 | 412 |
| AW097944–86 | AAGGCCGTGTCCAGCTCC | TCGGGGCTAACAATGCACC | 63 | 64 | 236 |
| AW097944–242 | CGTAACGGTGAGCGCAGG | TTGCCTGTGGCAGACACG | 63 | 63 | 538 |
| AW087030–122 | ACTGTTGGAGGATGGTCGTCG | CCTCCATCAACAGCCGAGG | 64 | 64 | 536 |
| AW086635–160 | TTGACTGCGAACTTCCTCATGG | GGCTTTCAATCGCCCAGG | 64 | 63 | 380 |
| AW098279–292 | TTTCGCGACATGGATGGG | AAGTCTCTACCTCTATTGCATCAAGCC | 64 | 63 | 260 |
| AW086909–334 | ACGTCCTCTGCGTCCTCCC | TCCGCTCGTGTCACTGGG | 65 | 64 | 412 |
| AW086827–136 | CTCGGATCCGCGTGTTGC | CGTACGCGGACTCTGGTGG | 66 | 65 | 790 |
| AW086674–69 | CTGCTCTTGCCAGTCTTGAACC | GAACACTGTGACTGCTGAGAAGTTGG | 63 | 65 | 352 |
| AW098248–320 | TTGGACGCCATCTTCGGC | CTGCCTCTCCTGTGACAAAAGC | 65 | 63 | 571 |
| AW098328–149 | TCAAGATGGAGGTGGGAATCG | GCCCCGTCCATGATTTCG | 64 | 64 | 335 |
| AW097937–210 | TCAAATGCGATGAATGTCGC | CTGGACAGCCAGTCAAGGC | 63 | 62 | 291 |
| AW086669–414 | AACGACCAGCGTAGGTGCC | GCTGAGAAGGGTGAAGATGCG | 63 | 64 | 290 |
| AW098185–232 | TATTGCGACTGCCCCACG | GGGGTTATGCTACGGCACG | 64 | 63 | 166 |
| AW087017–243 | CGATCCTGAAACAGGCCACC | GAACTTTGCCTCAAACTTTCCAGC | 65 | 64 | 445 |
| AW087017–331 | TTGGTGGTGGTCAAGAGGC | AAAGCAAGAGCATTGATAGGTCCG | 62 | 64 | 382 |
| AW086830–254 | TTTGGGTGGCCTTCTTCAGC | GGGAGACACCCAACCACTTCG | 64 | 66 | 533 |
| AW086566–259 | TGGGTAGCTCCAATACCGGG | TTCCAGCGTGGGTTGAAGC | 64 | 64 | 431 |
| AW086566–452 | CGATGATCATGTTGCGGAGG | TCCGGGGAGTGGATCTCG | 64 | 64 | 606 |
| AW098560–101 | ATCCGCAAGTGCAAGCCC | TCATCCGTGGTGATTTTCGG | 64 | 64 | 420 |
| AW098281–150 | GTACTTCGAAGACAATGCGGG | CATTCGCAGCACTAGCAATCC | 61 | 63 | 291 |
| AW098587–180 | AGGAAGTTCGTGGTCGTGGG | TTGTGGCGTCCACATCGG | 64 | 65 | 436 |
| AW086962–158 | TGGACTATTCCTTGGGCTTCTCG | CCCCGATTGCGCAGTATCC | 65 | 65 | 287 |
| AW086962–273 | TCGAGCAAGGCAAGTCATAAGG | TCCGATGTTTCCTTCAGCGG | 63 | 65 | 310 |
| AW086962–333 | TTCTCTACATGCTTGCCGC | TTTGATAGCACAGCAGGGTAATGC | 60 | 64 | 272 |
| AW098337–225 | GAACCTGCACCACGACAAAGC | AAGCGTCGGCATTGCTCC | 65 | 64 | 304 |
| AW098364–121 | TGGATATGGGTTGGCGGG | AGCTGCTTGATGTCGGCTCC | 64 | 65 | 518 |
| AW098799–185 | TCAGGCTTTGCCTTGGTGG | AAGGTTGCTGGAGAAAGTAGCCC | 64 | 64 | 354 |
| AW098651–169 | GCAATCCTGCTCTACCTTGCG | CGCTCCAACGGGTAAGGG | 64 | 63 | 307 |
| AW098361–258 | AAATGCAGGCTGTGCGAGC | GCCATGACGACCTCCACG | 64 | 63 | 834 |
| AW086544–267 | TTTCTTCGCAGAGCCCACG | GTGTTGCATCCTGGTCGTGC | 64 | 65 | 331 |
| AW086990–246 | CCCAGACGGCATTCGAGG | TCATCCATTCGCATCTTCGC | 65 | 64 | 499 |
| AW086531–354 | TTCAGCTGTCATGGCCTTCG | ACGCAAGCTTTGGCACCC | 64 | 64 | 373 |
| AW086934–65 | TTTTTCAACGAAAATAGCGAGCC | CGAAGCTTTTGCAGGAACTCCC | 63 | 66 | 265 |
| AW086934–136 | TCGAAAGGGACAAACGGGG | AAGGCTCTCTAATATGGGGGTCG | 65 | 63 | 314 |
| AW087013–133 | CTTCGGAGCCACCATCCC | TTGTTGCCGAATGGGTCG | 63 | 64 | 465 |
| AW098823–115 | AGCGAGCTTGCCTGCACC | GCTTGGCAAAGAGACCAGGC | 65 | 64 | 393 |
| AW098823–199 | GCAACTTTGGTGAAGGCCG | CGGTCGCGGCAGTAGTAGC | 63 | 64 | 308 |
| AW098056–70 | AAGAATGCAGTGTTTGGTGACAGC | CCACATCCGCCTTGAGAGC | 64 | 64 | 542 |
| AW097956–276 | CCGCGACCAATGAGACCC | TGATCTGGATGCACGGGG | 65 | 64 | 710 |
| AW098620–189 | GATGAACAACGCTCGCGG | TGGTGTCGGAGATGTGGGG | 63 | 65 | 326 |
| AW097987–87 | GTCTTAACCTTGGATGGCCGC | GAGTGCTTTGAGTCATCGCTTCC | 65 | 64 | 268 |
| AW097987–167 | TGTGGAGCCAACAGAGATTGAGG | TGCATGCCTGCAAATCAGC | 65 | 64 | 328 |
| AW097987–313 | CATGCAGCCGTTAAGGAAAGG | TCAGCAATCCATTTTCAACCG | 64 | 63 | 279 |
| AW087075–240 | TTGGTGATGCATACTCAAAGGTGG | GGTCATCGAGCTCTCCTGGC | 65 | 64 | 452 |
| AW087075–332 | TCAACTGGCGATCAGAAGCC | CTGGCTGCACTAACACTGCC | 63 | 61 | 232 |
| AW098367–359 | CAATTATCTCCAACGGCGGC | AGTGGTAATCAGCCGTCATCTCG | 64 | 64 | 421 |
| AW086525–450 | ACTCCTGGTTGGTGGCCG | CTTCTTGAGGAAGTTGCAGAAGG | 64 | 61 | 674 |
| AW086525–514 | TGATGGCAAGAAGGTGTCCG | ACCGCATGAAGTTGTGGGC | 64 | 64 | 318 |
| AW098078–239 | CCTCAACACCAGGCTCAATGG | TGATCAAGGGCGCCAAGG | 65 | 65 | 482 |
| AW086765–100 | TGACCCATGTGTTGATGATGAGG | ATTTCAATGCCAGATTCCAGC | 65 | 61 | 484 |
| AF309562–421 | TCCCCTTCGAGGAAAACCC | CTGGCGACCAAAGCTCCG | 63 | 65 | 425 |
| AF309562–538 | TGCTGGCGTTACAGACGACC | GAGCAGCAGCCCTCTGAGC | 64 | 63 | 323 |
| AW087021–183 | GGTTGAAGCTGTGAAGTTGTTCG | GAGGACCTCTCTGGATGGGG | 63 | 63 | 310 |
| AW087021–312 | AGCTTCTGCCATCCTCACTGC | AGTTGCGACGACGAGACGG | 64 | 64 | 315 |
| AW098247–82 | ACGCGTCCATCTGCCAGG | AAGGTCGCCTCCACCTCG | 65 | 63 | 461 |
| AW086944–155 | CTGCCATCCTTCCCGTCC | AAGCAGTTGGTGTTGGCGG | 63 | 64 | 353 |
| AW098672–254 | TTCCTTGGGGGCCTCAGC | ACTGGCCTGGTCTCTTTGCC | 65 | 63 | 313 |
| AW087053–85 | CGTCGTGAGCGTGAGGAGG | AGGTATCCAAGCTTCTCATTGTAGTCC | 65 | 63 | 474 |
| AW098317–317 | TGATATGGGGTCTTCCAGGTCC | CGTTTTAATAAGAGGTCGACAGTGGC | 64 | 65 | 498 |
| AW086622–280 | GCGAAGAGTGGGTAGCTCCG | AAGCCCGAGCCTGTGAGG | 64 | 63 | 346 |
| AW086841–85 | TTCGGAAGCACAAAACTGACG | TCTCATCGCCGTTTAGCCG | 63 | 64 | 279 |
| AW098024–169 | GGAAAGCTGTGACTGCACTTACCC | AATCTGGGCCTGGCCTTACC | 65 | 64 | 578 |
| AW086649–182 | TGCATGAATCACAATGAAGCCC | ACGCATGCGCCATCTGC | 65 | 65 | 463 |
| AW098284–284 | GCAGGACTGAGGAGTCGTCG | TTTTCCTAGTCCCGCACGC | 63 | 63 | 309 |
| AW098191–439 | GACCCGCCACATGAATGC | CTTCCAGCTTGTGGACGGC | 63 | 64 | 572 |
| AW098191–543 | TTGGAAAGGTTGTCAGCTTCTGC | TTCGAAGCATTTGGACCAGG | 64 | 63 | 478 |
| AW086824–186 | GAACGAGGGCAAAACAACACG | TTGTTCGTATCATGAGTCCTTATTTGC | 64 | 63 | 282 |
| AW086824–253 | GTGTACGTCATGGTGTCGATTGG | TGCTGGGTGGAGATGGTCC | 64 | 64 | 456 |
| AW086779–195 | TGGTGCCAGTTTGAGGAAGC | AGCACCTCCGGCTTTGACC | 63 | 64 | 431 |
| AW098026–111 | TGGCAGTGGAAAGCTGCG | CTCATTGGGCATGTGGATGG | 64 | 64 | 395 |
| AW086929–81 | ATGCACACTGCATCCCTTTGC | AAATATGAAAGGAGGGGTCGC | 65 | 61 | 407 |
| AW086753–293 | GGCAGCCCAATTTCATGAGG | ATTGCTTGGAGCCTCTCAATGG | 64 | 64 | 458 |
| AW086737–66 | AACCTACTGGTCGACAAGAACTGG | CGAAGCACTTCCGGTGCC | 62 | 65 | 464 |
| AW098283–167 | TGATGGATGCGCTTGTGG | TCTGCAAGAGAGCCTACCTTGACC | 62 | 65 | 426 |
| AW098019–120 | ATCGGGATTAGGACCAAGGC | GATTCTGCCAGCGCATCC | 62 | 62 | 271 |
| AW098019–199 | TGGAAAGGATGCGCTGGC | ATTGCGCATCATCCATACCG | 65 | 63 | 265 |
| AW098019–297 | CTTGCGTGAGGACTATTTGGC | TTCTGCCCCTCAGAACCAGC | 62 | 65 | 325 |
| AW098233–116 | TTATGCGCAGGAGCTTGGC | CCTCCTCCCGCTACCAACG | 64 | 65 | 453 |
| AW098233–233 | GCAAATCCTGATGGCCGC | CCCGCATATTTGCCAATCC | 65 | 63 | 281 |
| AW086758–136 | AAGGCTTTTGCACTGCACTCG | TGCTGCTCGCACTGGAGG | 64 | 65 | 299 |
| AW087065–177 | CGGGAGCACTTAACGACGC | TCACACCTTCTGCTGTCTGG | 64 | 60 | 575 |
| AW098770–113 | CCGTGAAGGACTGGGACAGG | TTGCACGCCTTGTATCCTCG | 65 | 64 | 282 |
| AW098258–180 | TTTGCACCCATTGCCAACC | GCCGGGTTGTAAGCGAAGC | 65 | 65 | 390 |
| AW098797–116 | AGGATCGGACTCCCTGCC | TTCCCAACTTGTCAACTGCCC | 62 | 64 | 490 |
| AW098643–123 | GGAGGCTTTTGAGGCGAGG | CAATCAGCTGGCAATGAGCG | 64 | 65 | 508 |
| AW086790–163 | TGGCAGAGTTTGATCGAGGC | TGAAGGTGCTCTGGAGCGG | 63 | 65 | 591 |
| AW098780–75 | AGGCGTCAGTCTACGGAATTGG | TGCTCGCAAATCTTGCCC | 64 | 63 | 379 |
| AW098780–211 | CCCACATCCCGATCCACG | ACAATTTTCGCTTCAACAAGATCTCC | 65 | 64 | 324 |
| AW086556–413 | AAGGCAGACTCTCGGGCG | GCGTTCAGAAGGCCAATGC | 64 | 64 | 458 |
| AW098831–338 | TTCAGTGGACGCGCTACCC | TCCACAGTTGAGTTCCGGTGC | 64 | 65 | 456 |
| AW086710–390 | TTTCTCCGGGCTTCCATCC | AGGGCATTGCTCTCAGGGG | 64 | 64 | 531 |
| AW086768–261 | CCTCCTCCCCATACGCTCC | CACAACAACAGCACAAAGCTGC | 64 | 64 | 427 |
| AW087072–205 | CCGCGACCAATGAGACCC | TTGCGAGAAGTTGACCGTGG | 65 | 64 | 387 |
| AW087072–269 | GCAGAAGGTCAAGAAGGCCC | CCGTTGAGAGTGAGGTCACGG | 63 | 65 | 373 |
| AW086870–57 | AACCCCGCCTGCTTCACC | GGTGCATTTGGCTGTGCC | 65 | 63 | 318 |
| AW098197–197 | ACGTGGATTTCAGGCGGC | CGTGCTTCTTCTTCAGTAGCAGGG | 64 | 65 | 489 |
| AW098074–139 | CAGGGGAAAGGCACCTGG | CGGTTCCAGTGAGGATGCG | 63 | 65 | 520 |
| AW098521–456 | ACTTCTCAGACGAGTTGGGGC | CGTGCCGAAAAGGTGCG | 63 | 64 | 288 |
| AW098301–160 | GCTGTGCAGGCGTTGTGG | AGAAAGATGACGCAGATGGCG | 65 | 64 | 599 |
| AW087027–88 | TGGGACTTCTACAAGCAAAAGTTGG | CAGCGCTTGATGCTTCG | 64 | 60 | 268 |
| AW087027–159 | AGGTGCCAGAGTACAAGGATGACC | CTTCTGTTCCTGCACCCTCG | 64 | 63 | 374 |
| AW087027–246 | GCTAAGGCCTCAGAGCAAGAGG | CTTCACTTGTGGGTGCTTTGC | 63 | 63 | 291 |
| AW086848–172 | CAACCCTTTCATGCACGCC | CGCAACTCAGCCCTCACG | 64 | 64 | 513 |
| AW098187–139 | AGTACAATCAGGCTGCCACGC | TTGAGTTGAACTTCATTTACCTGCC | 64 | 62 | 306 |
| AW086969–51 | TCGCATTTATGGCAGAGCAGG | AGATTCTTCAGACGAACAGCCG | 65 | 63 | 280 |
| AW086692–252 | CCGAATCATCAGATGCCAGG | GTCTCGCAGCCGAGTTGG | 63 | 63 | 420 |
| BI894286–170 | TGGACGAGCTGAGCGAGG | TTACGCCATGTCCTTCGCC | 63 | 64 | 494 |
| AW086973–252 | GGATGATTCCGCCAAACAGG | TCTACGACGGCATCAGGGC | 64 | 64 | 309 |
| AW098153–431 | CGAAGACGGCCTTGCACC | CAAATTGGAGCAGCTGTGGC | 65 | 64 | 448 |
| AW098812–178 | ACTGGCCCAGCCTTTCCG | AGAAGTAGCCCCACTGCATCG | 65 | 63 | 430 |
| AW098812–326 | TGAGATTGGGTTGTTCGATGG | CCGCCGTCCACAATCTCG | 63 | 66 | 457 |
| AW086545–249 | TTCCTTGGGGGCCTCAGC | GCCGGTGTGCAAATTGAGG | 65 | 64 | 330 |
| AW086575–297 | CATCTAGGTATTGTCGAGTCCCG | TGAAAGTGGCAAGATGACCAAGG | 62 | 65 | 459 |
| AW086575–420 | AGCAACTGCATCAATAAATTCCTCG | TGGACCCGTGGTCTTAGCC | 64 | 63 | 317 |
| AW086781–77 | CTCGCGTTGCTGGTGTCG | TTCTGCGCATCTTCTTTTTGC | 65 | 62 | 421 |
| AW087048–137 | GAAGGCTGCAATTCAGGAAACG | TGAAAGAGGTTTCTTTGGTTTGTTGG | 65 | 65 | 257 |
| AW087048–215 | GAGAAGAAGAAGGCCAGGGACC | GTGGGCATTCGTTTCCGC | 64 | 64 | 515 |
| AW098758–58 | TATTTGCTCCAGGATGCTGATGG | GGCTTCATCGTCAGTCACGC | 65 | 64 | 319 |
| AW086711–314 | TGAATCCGGCTGTCAAATCG | GCGAGCTGCTGGTTCTGAGG | 64 | 65 | 300 |
| AW086711–405 | TAGATGAGGTCACTCAAACGCTGC | AAATCGGTATGGATGATGCG | 64 | 60 | 438 |
| AW086711–484 | TTGCGCATCATCCATACCG | AGTTGCAAAACTCTTGCGTGAGG | 63 | 64 | 305 |
| AW098776–59 | GTCAGGCTCTGGCTGACG | GCCAGCCTTTGCTTGATGTCC | 60 | 66 | 314 |
| AW098776–152 | GCCAAGACGAACATGAATGTGG | GCAGGAACCTGATGCTGGC | 64 | 64 | 359 |
| AW098585–138 | CCTCGCCTCATGTCCTGC | AGCACGTGCGTAGTTCCCG | 63 | 64 | 335 |
| AW098746–319 | GGTTTCATCTCTGGCGCTTCG | CCCATGAGGTCAAAGATGAGGG | 66 | 65 | 249 |
| AW098746–414 | CCACCATGGTCCACTTCATAGC | CTGAAGCGATCCCCCACC | 63 | 63 | 421 |
| AW098409–116 | CAGAGATGGTCGTGGGTTGG | CCCAACATCATCGTCTGAGGG | 63 | 64 | 447 |
| Primer <25 nucleotides from intron | |||||
| AW086551–330 | TTCTGTTTCCAACAGGCCG | GTGCGCAAATTCACAGAGCG | 62 | 65 | 307 |
| AW086551–425 | TCTCGCTCTGTGAATTTGCG | CGATGTCAGAAGGCAGGTGG | 62 | 64 | 231 |
| AW086636–421 | TCAGCTGACTTCGCGTTTGC | GTTGGCTCAAGGAAAGGAGC | 65 | 61 | 314 |
| AW086700–323 | AGTCAAGCGGGCCCTTCC | ATGTGAAGTGCCAGGGCTGC | 64 | 65 | 301 |
| AW086752–361 | AACCAGAGCCCCAACCCG | ACAATCAGCGTGACCTCAAACG | 65 | 64 | 395 |
| AW086783–208 | CGTCTCATCATGCGCAACG | CTTCGCGCTCAGACTCAAGG | 65 | 63 | 665 |
| AW086798–387 | GCATGGAACTCCCGGAACC | CATCAAACTCCCACAACTCATTTCC | 65 | 64 | 393 |
| AW086855–57 | GTGATCATGGCGGGGAGG | TCCTCCGTCCTACAGTCGCC | 64 | 64 | 263 |
| AW086855–188 | CCTGTTGTCGCAGCCAGC | CGTCGTCAACGATTGTAGGG | 64 | 61 | 321 |
| AW086856–213 | TCACGGATCTTTGGCCCG | TTGGGGCCCCTCTTCTTACC | 65 | 64 | 276 |
| AW087036–57 | AGAAGGAGTCTACTCTGCATCTGG | CCTCAAGCTGCTTTCCAGCG | 60 | 65 | 425 |
| AW087051–351 | TTCACAACAGGCAGACCCG | GACAGTGGCTCTCTGGAAGGC | 63 | 63 | 328 |
| AW087074–125 | CAGGCTGGTCCGCTTTGG | CCGTTAACCTGAATATCCTTCACACC | 65 | 64 | 395 |
| AW087074–171 | TGTGAAGGATATTCAGGTTAACGGC | TGATGGTTGAGCTCCGACG | 64 | 63 | 237 |
| AW097915–117 | GGGAGCAACCTGACCCTTCC | CGCTCGAGATCGTAGCCTTGC | 65 | 66 | 252 |
| AW097968–35 | AGTGCGGTGCCAACTGCG | CCGCACTTGCCGTTCTCC | 67 | 65 | 418 |
| AW097979–42 | GATGGCTCCAGCCCTTCG | AGCTCTCCCCTGATGTTTTCAGC | 64 | 65 | 318 |
| AW097979–92 | AACTGGCTGAAAACATCAGGGG | AGTGACGCTGGCAACTCCC | 64 | 63 | 361 |
| AW098011–47 | TGTTGCAGCTTGTGTACTTTCTGTACC | ATGGGTGCGCCTGAAATCG | 64 | 66 | 303 |
| AW098051–84 | GATTCCGTCACTGTCGGTGG | GTGCCGAAACTGGTGCCC | 63 | 65 | 519 |
| AW098200–145 | CAATGACACCAGGCTCCAACC | TGCACCTTGTGGGGCAGC | 65 | 66 | 275 |
| AW098203–61 | CAAGTTCTTGCAGGAAACATTTGG | CCATGCTTGCTGACCGGG | 64 | 66 | 464 |
| AW098216–110 | AGGGTGTCAATACGTCCAAGG | TGGCACCGAGAGACGAAGG | 61 | 64 | 460 |
| AW098262–128 | ACTACGTTGCCCACGACGC | TTCGTAAACATCCTTGCCAGC | 64 | 62 | 233 |
| AW098262–171 | GGATGCAACTGGTGCTGGC | GATCAGATCCGAAGTCGAAACTCC | 65 | 64 | 224 |
| AW098263–244 | TAGCATGGAGCACTGATCGGG | CAATCCAACGTCCAAAGTAAACTGC | 65 | 64 | 281 |
| AW098272–223 | CATTTGTGTGTTCCTTCTGCCG | TGAGTCTCTTGAAGTCTCTTCATTTCC | 64 | 62 | 190 |
| AW098291–131 | TGTAGGCCGAATAGCACTTGG | TGGACCAGTAGTCATGTTGAATGC | 62 | 63 | 245 |
| AW098300–108 | TGCAACGGTGTCGTTGTGC | ACCCGTGAATCTTGATGAGGTCC | 65 | 65 | 453 |
| AW098306–52 | GTTGCGCAGGGTTGAGCC | TGGCCTCTCTGTTGCCAGC | 65 | 65 | 450 |
| AW098419–60 | CGCCAGATTCAGGAGCAACC | CTGGGGTACATGAACACCCTTACG | 65 | 65 | 223 |
| AW098448–78 | ATGTCAAGTGCCAAGGATGC | CACAGCACCGTGGAGCAGC | 61 | 66 | 276 |
| AW098461–262 | CACACTCCTTTCCAATGGGGC | ACAATCGGGAGTCATTGTGACC | 66 | 63 | 269 |
| AW098461–303 | GGGGTCACAATGACTCCCG | CAAGCCCTGAGGCGCAAAGC | 63 | 69 | 330 |
| AW098472–354 | GTAGGATACGGTTTGAGGGCTGC | GCTCTTGAAGAAGAAGTTCGGG | 65 | 61 | 493 |
| AW098535–258 | TCGAACAAGCTGAAGCCC | GAAGTTCGCGTCTGTGCCC | 60 | 64 | 431 |
| AW098580–204 | CAAGACCCCACCATCTACAGGC | CCCTTCACCTTCTCCACAGAGC | 65 | 64 | 315 |
| AW098597–63 | GGAGCTGGTGACAGTGTGAAGG | AGCAGCCATCAGACCCCC | 64 | 63 | 273 |
| AW098717–46 | TCACGGCTCAAACTCTGATTAAATAGC | AAATCCAAGGCCAGAACCCC | 64 | 64 | 334 |
Melting temperature of the forward primer.
Melting temperature of the reverse primer.
Prod. size indicates the predicted PCR product size on the Physcomitrella patens genome sequence.
To evaluate the full set of 245 loci, we sequenced each of these gene regions in the female laboratory strain GG1 (collected from Gross Gerunds, Austria, by D. J. Cove), the male laboratory strains WT4 (collected in Wispertal, Austria, by E. Hartmann) and R40 (collected by S.F.M. in Rensselaer County, New York, USA), and an isolate from Otavalo, Ecuador (collected by S.F.M.). Live cultures of all of these individuals are available from the authors. DNA was extracted from 7-d-old protonemal grown under standard conditions (Cove et al., 2009) using the Nucleon PhytoPure Genomic DNA Extraction Kit (Amersham Biosciences, Piscataway, New Jersey, USA) following the manufacturer’s instructions. PCR was accomplished using GoTaq Green Master Mix (Promega Corporation, Madison, Wisconsin, USA) in 16-μL reactions. The cycling conditions were 94°C for 120 s, then 10 cycles of 94°C for 15 s, an annealing temperature of 65°C that decreased one degree each cycle, and 72°C for 60 s, followed by 20 cycles of 94°C for 15 s, 56°C for 30 s, and 72°C for 60 s. The PCR products were cleaned using the QIAquick PCR Purification Kit (QIAGEN Sciences, Germantown, Maryland, USA). Sequencing used BigDye Terminator version 3.1 chemistry and was accomplished on an ABI 3100 capillary sequencer (Applied Biosystems, Carlsbad, California, USA). Forward and reverse sequence fragments were edited and assembled using Sequencher 4.0 (Gene Codes Corporation, Ann Arbor, Michigan, USA), and all polymorphisms were checked from the chromatograms.
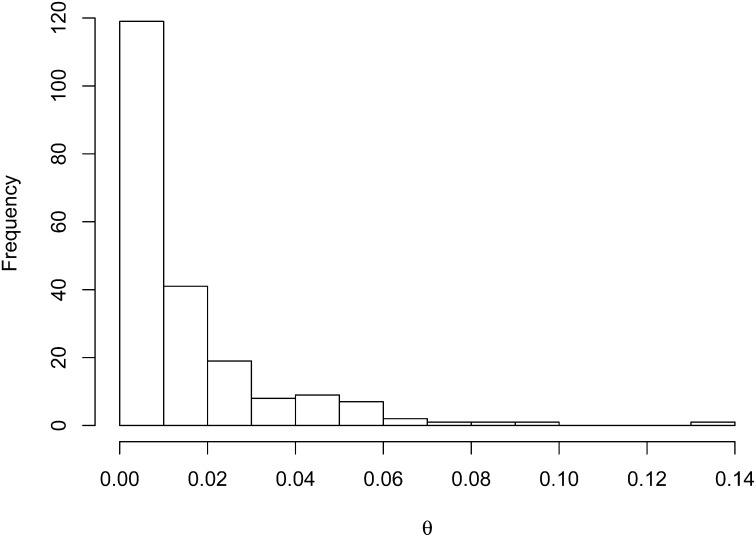
We generated high-quality sequence data for 218 of the 245 loci. We used the software DnaSP (Librado and Rozas, 2009) to estimate the distribution of the per-site genome-wide nucleotide variation (θ, an estimate of Neµ [where Ne is the effective population size and µ is the per-site nucleotide mutation rate]) in C. purpureus (mean: 0.014, median: 0.008, range: 0.0–0.14; Fig. 1, Table 2). Although these data were generated from a modest sample, this stands as the most complete estimate of this fundamental parameter in any bryophyte, and forms a benchmark for further comparisons. It is possible that this estimate of θ is biased upward, by cryptic population structure in our sample, or downward by our small sample size. However, many loci showed no variation among intercontinentally disjunct samples, consistent with previous work (McDaniel and Shaw, 2005), suggesting that the loci that are more diverged reflect locus-specific rather than genome-wide evolutionary processes. For example, loci at the low end of the distribution may be linked to loci that have experienced a selective sweep (McDaniel and Shaw, 2005), while loci on the high end of the distribution may be linked to the sex chromosomes or loci linked to local adaptation (McDaniel et al., 2007, 2008). This degree of variation illustrates the among-locus heterogeneity in evolutionary history within this species. While sampling more individuals would quantitatively improve this estimate, the concordance between this and previous estimates suggests that the median value is unlikely to be qualitatively improved without a much larger sample.
Fig. 1.
A frequency histogram of θ (an estimate of 4Neµ) from the 218 loci in Ceratodon purpureus.
Table 2.
Nucleotide diversity at sequenced loci in four geographically widespread accessions of Ceratodon purpureus.a
| EST accession | Gene name | Lengthb | Thetac | GenBank accession no. |
| AF233229 | auxin binding protein 1-like protein (abp1) | 1138 | 0.000843 | JY262836, JY262996, JY263192, JY262676 |
| AF309562 | hemoglobin mRNA | 820 | 0.007737 | JY262947, JY263101, JY263271, JY262786 |
| AJ250735 | delta 6-fatty acid desaturase | 1264 | 0.043478 | JY262851, JY263008, JY263206, JY262691 |
| AW086517 | similar to SW:IM30_PEA Q03943 CHLOROPLAST MEMBRANE-ASSOCIATED 30 KD PROTEIN PRECURSOR | 1164 | 0 | JY262832, JY262991, JY263187, JY262671 |
| AW086519 | similar to gb:gb|U77939.1|PVU77939 Phaseolus vulgaris ubiquitin-like (PLANT) | 357 | 0.019055 | JY262883, JY263033, JY263228, JY262722 |
| AW086525 | similar to TR:O76968 O76968 RIBOSOMAL PROTEIN L18A | 1171 | 0.042882 | JY262920, JY263071, JY263259, JY262758 |
| AW086531 | similar to SW:SYY_BACST P00952 TYROSYL-TRNA SYNTHETASE | 436 | 0.00489 | JY262928, JY263079, JY262765 |
| AW086539 | Moss EST library CPU Ceratodon purpureus cDNA clone PEP_SOURCE_ID:CPU010318 3-, mRNA sequence | 364 | 0.057971 | JY263102, JY263272, JY262787 |
| AW086546 | similar to SW:RL13_ARATH P41127 60S RIBOSOMAL PROTEIN L13 | 628 | 0.015856 | JY262894, JY263042, JY263239, JY262731 |
| AW086548 | similar to SW:RK24_TOBAC Q02764 50S RIBOSOMAL PROTEIN L24, CHLOROPLAST PRECURSOR | 468 | 0.009183 | JY262834, JY262993, JY263189, JY262673 |
| AW086549 | similar to SW:SYRP_LACBI P87068 SYMBIOSIS-RELATED PROTEIN | 286* | 0.008734 | JY262855, JY262696 |
| AW086551 | similar to TR:O48891 O48891 ATP-DEPENDENT CLP PROTEASE PROTEOLYTIC SUBUNIT. | 787 | 0.001285 | JY263156, JY263316 |
| AW086555 | similar to TR:O04619 O04619 SIMILAR TO MITOCHONDRIAL CARRIER FAMILY | 509 | 0.006367 | JY262870, JY263022, JY263219, JY262710 |
| AW086556 | similar to TR:O65731 O65731 40S RIBOSOMAL PROTEIN S5 | 392 | 0.020997 | JY262973, JY263135, JY262814 |
| AW086566 | similar to TR:O65059 O65059 PROBABLE 40S RIBOSOMAL PROTEIN S15 | 762 | 0.035361 | JY262939, JY263090 |
| AW086575 | similar to TR:P93133 P93133 NADP-ISOCITRATE DEHYDROGENASE | 830 | 0.003851 | JY263138, JY263302 |
| AW086579 | similar to SW:LDLC_HUMAN Q14746 LDLC PROTEIN | 595 | 0.001778 | JY262854, JY263010, JY262695 |
| AW086590 | similar to SW:RL2B_FRIAG O22644 60S RIBOSOMAL PROTEIN L23A | 483 | 0 | JY262886, JY263231, JY262725 |
| AW086594 | similar to TR:O80626 O80626 PUTATIVE RIBOSOMAL PROTEIN L35 | 594 | 0 | JY262830, JY262989, JY263185, JY262669 |
| AW086594 | similar to TR:O80626 O80626 PUTATIVE RIBOSOMAL PROTEIN L35 | 429 | 0 | JY262950, JY263105, JY262790 |
| AW086618 | similar to SW:SYV_BACSU Q05873 VALYL-TRNA SYNTHETASE | 604 | 0.002098 | JY262937, JY263088, JY263268, JY262774 |
| AW086619 | similar to TR:O82413 O82413 HISTIDYL-TRNA SYNTHETASE | 524 | 0.046074 | JY262906, JY263056, JY263251, JY262745 |
| AW086636 | Moss EST library CPU Ceratodon purpureus cDNA clone PEP_SOURCE_ID:CPU010708 3-, mRNA sequence | 283 | 0.021739 | JY263160, JY263320 |
| AW086641 | 2similar to TR:Q9ZRS8 Q9ZRS8 RIBOSOMAL PROTEIN L37A | 981 | 0.012953 | JY262923, JY263074, JY263262, JY262761 |
| AW086647 | similar to SW:SPEE_COFAR O82147 SPERMIDINE SYNTHASE | 327 | 0.008439 | JY262916, JY263067, JY262754 |
| AW086659 | similar to SW:RS13_ARATH P49203 40S RIBOSOMAL PROTEIN S13 | 502 | 0.011655 | JY262882, JY263032, JY263227, JY262721 |
| AW086668 | similar to SW:RS3A_BRARA P49396 40S RIBOSOMAL PROTEIN S3A | 277 | 0.013468 | JY262995, JY263191, JY262675 |
| AW086669 | similar to SW:COXG_YEAST Q01519 CYTOCHROME C OXIDASE POLYPEPTIDE VIB | 535 | 0 | JY263106, JY262791 |
| AW086674 | similar to SW:RL27_PYRST Q02984 60S RIBOSOMAL PROTEIN L27 | 439 | 0.018667 | JY262926, JY263077, JY263263 |
| AW086675 | similar to TR:O48691 O48691 F3I6.17 PROTEIN | 286 | 0.003115 | JY262867, JY263217, JY262707 |
| AW086686 | similar to TR:P93359 P93359 PUTATIVE PRE-PRO-CYSTEINE PROTEINASE | 306 | 0.007755 | JY262889, JY263037, JY263234, JY262727 |
| AW086692 | similar to TR:O16619 O16619 F36H9.3 PROTEIN | 356 | 0.002924 | JY263146, JY263308 |
| AW086694 | similar to TR:O65088 O65088 TAT-BINDING PROTEIN HOMOLOG | 339 | 0.004295 | JY262864, JY263017, JY263215, JY262703 |
| AW086700 | similar to TR:Q9ZNS3 Q9ZNS3 RIBOSOMAL PROTEIN S27 | 322 | 0.090909 | JY263164, JY263324 |
| AW086710 | similar to SW:BAS1_HORVU Q96468 2-CYS PEROXIREDOXIN BAS1 PRECURSOR | 388 | 0.011459 | JY262982, JY263149, JY263311, JY262824 |
| AW086736 | similar to TR:O81925 O81925 40S RIBOSOMAL PROTEIN S6 | 608 | 0 | JY262868, JY263020, JY262708 |
| AW086737 | similar to TR:O23719 O23719 MAP3K DELTA-1 PROTEIN KINASE | 421 | 0.004364 | JY262971, JY263133, JY263298, JY262812 |
| AW086738 | similar to SW:RS21_ORYSA P35687 40S RIBOSOMAL PROTEIN S21 | 806 | 0.055219 | JY263096, JY263270, JY262781 |
| AW086752 | similar to TR:Q9ZU52 Q9ZU52 PUTATIVE ALDOLASE | 285* | 0 | JY263168, JY263328 |
| AW086753 | similar to TR:O04202 O04202 26S PROTEASOME REGULATORY SUBUNIT S12 ISOLOG | 319 | 0.009603 | JY262967, JY263128, JY263295, JY262809 |
| AW086758 | similar to SW:SYK_CRILO P37879 LYSYL-TRNA SYNTHETASE | 316 | 0.004399 | JY262968, JY263129, JY263296, JY262810 |
| AW086759 | similar to SW:YGLA_SYNP2 P28606 HYPOTHETICAL 34.1 KD PROTEIN IN GLNA 3-REGION | 729 | 0.017016 | JY262856, JY263011, JY263210, JY262697 |
| AW086765 | similar to TR:O82229 O82229 PUTATIVE SERINE CARBOXYPEPTIDASE | 420 | 0.025048 | JY262941, JY263092, JY262777 |
| AW086768 | similar to TR:O04820 O04820 HYPOTHETICAL 9.1 KD PROTEIN | – | JY262985 | |
| AW086770 | similar to TR:Q55649 Q55649 ABC TRANSPORTER | 619 | 0.005089 | JY262876, JY263028, JY262715 |
| AW086781 | similar to TR:Q9ZU75 Q9ZU75 UBIQUITIN-CONJUGATING ENZYME E2 | 751 | 0.015385 | JY263151, JY263312 |
| AW086783 | similar to SW:RS28_MAIZE P46302 40S RIBOSOMAL PROTEIN S28 | 663 | 0.017572 | JY263172, JY263331 |
| AW086786 | similar to TR:O65583 O65583 PUTATIVE URACIL PHOSPHORIBOSYL TRANSFERASE | 271 | 0.005742 | JY262874, JY263026, JY263222, JY262713 |
| AW086789 | similar to SW:RS24_HUMAN P16632 40S RIBOSOMAL PROTEIN S24 | 552 | 0.020856 | JY262895, JY263043, JY263240, JY262732 |
| AW086790 | similar to SW:RS3A_CATRO P33444 40S RIBOSOMAL PROTEIN S3A | 506 | 0.026438 | JY262960, JY263119, JY263286, JY262802 |
| AW086798 | similar to TR:Q55953 Q55953 HYPOTHETICAL 18.6 KD PROTEIN | 595 | 0.003401 | JY263176, JY263335 |
| AW086824 | Moss EST library CPU Ceratodon purpureus cDNA clone PEP_SOURCE_ID:CPU020404 5-, mRNA sequence | 230 | 0 | JY262983, JY263152, JY263313, JY262825 |
| AW086827 | similar to SW:RL31_PICMA O65071 60S RIBOSOMAL PROTEIN L31 | 704 | 0.005418 | JY262917, JY263068, JY263257, JY262755 |
| AW086830 | similar to SW:RL4_ARATH P49691 60S RIBOSOMAL PROTEIN L4 | 580 | 0.003736 | JY262927, JY263078, JY263264, JY262764 |
| AW086833 | similar to gb:gb|AF016284.1|AF016284 Arabidopsis thaliana (PLANT) | 323 | 0.002403 | JY262915, JY263066, JY263256, JY262753 |
| AW086837 | similar to SW:SYL_BACSU P36430 LEUCYL-TRNA SYNTHETASE | 540 | 0.001699 | JY262853, JY263009, JY263209, JY262694 |
| AW086841 | similar to TR:O04002 O04002 CDSP32 PROTEIN | 264 | 0.004587 | JY262953, JY262795 |
| AW086845 | similar to SW:RS11_SOYBN P17093 40S RIBOSOMAL PROTEIN S11 | 700 | 0.062613 | JY262871, JY263023, JY263220, JY262711 |
| AW086855 | similar to TR:Q9ZRT5 Q9ZRT5 GLUTATHIONE TRANSFERASE ATGST 11 | 564 | 0.002141 | JY263157, JY263317 |
| AW086858 | similar to gb:emb|Z23165.1|ATRBPS18A A.thaliana ribosomal protein gene (PLANT) | 719 | 0.012759 | JY262881, JY263031, JY263226, JY262720 |
| AW086868 | similar to SW:RS8_ORYSA P49199 40S RIBOSOMAL PROTEIN S7 | 417 | 0.019656 | JY262878, JY263029, JY263224, JY262717 |
| AW086870 | similar to TR:Q43548 Q43548 GOLDEN DELICIOUS APPLE FRUIT EXPRESSED | 304 | 0.007576 | JY262964, JY263125, JY263292, JY262806 |
| AW086877 | similar to SW:CG1C_ORYSA P93411 G1/S-SPECIFIC CYCLIN C-TYPE | 397 | 0 | JY262838, JY262998, JY263194, JY262678 |
| AW086877 | similar to SW:CG1C_ORYSA P93411 G1/S-SPECIFIC CYCLIN C-TYPE | 336 | 0.003704 | JY262847, JY263005, JY262687 |
| AW086878 | similar to SW:RL6_MESCR P34091 60S RIBOSOMAL PROTEIN L6 | 527 | 0.010703 | JY263048, JY263245, JY262737 |
| AW086917 | similar to SW:ARF_ORYSA P51823 ADP-RIBOSYLATION FACTOR | 189* | 0.011364 | JY262931, JY263081, JY263266, JY262767 |
| AW086924 | Moss EST library CPU Ceratodon purpureus cDNA clone PEP_SOURCE_ID:CPU030116 3-, mRNA sequence | 425 | 0.031648 | JY262948, JY263103, JY263273, JY262788 |
| AW086929 | similar to WP:F10G7.1 CE02624 | 416 | 0.045455 | JY262962, JY263123, JY263290, JY262804 |
| AW086934 | similar to SW:YKQ0_YEAST P36053 HYPOTHETICAL 16.2 KD PROTEIN IN PIR3-APE2 INTERGENIC REGION | 220 | 0.00813 | JY263084, JY262770 |
| AW086944 | similar to TR:O48773 O48773 HYPOTHETICAL 47.8 KD PROTEIN | 342 | 0.005782 | JY262965, JY263126, JY263293, JY262807 |
| AW086962 | similar to TR:O81846 O81846 PHOSPHATIDYLINOSITOL SYNTHASE | 699 | 0.011111 | JY262908, JY263059, JY262747 |
| AW086969 | similar to TR:Q43275 Q43275 PLASMA MEMBRANE H+-ATPASE | 185 | 0 | JY263137, JY263301 |
| AW086973 | similar to TR:O22972 O22972 HSP90 ISOLOG | 142* | 0 | JY263154, JY263315 |
| AW086975 | similar to WP:F17C11.8 CE05655 | 708 | 0 | JY262840, JY263000, JY263196, JY262680 |
| AW086979 | similar to TR:Q9ZQN8 Q9ZQN8 PUTATIVE GLUCOSYLTRANSFERASE | 316 | 0.012397 | JY262949, JY263104, JY263274, JY262789 |
| AW086989 | similar to TR:Q14692 Q14692 KIAA0187 PROTEIN | 570 | 0.007792 | JY262885, JY263035, JY263230, JY262724 |
| AW086990 | Moss EST library CPU Ceratodon purpureus cDNA clone PEP_SOURCE_ID:CPU030623 5-, mRNA sequence | 572 | 0.017001 | JY262918, JY263069, JY262756 |
| AW086999 | similar to SW:SMD2_HUMAN P43330 SMALL NUCLEAR RIBONUCLEOPROTEIN SM D2 | 352 | 0.012636 | JY262890, JY263038, JY263235, JY262728 |
| AW087002 | similar to gb:gb|AF068690.1|AF068690 Citrullus lanatus peroxisomal (PLANT) | 458 | 0 | JY263094, JY262779 |
| AW087005 | similar to TR:O65606 O65606 HYPOTHETICAL 23.9 KD PROTEIN | 253 | 0.035573 | JY262835, JY262994, JY263190, JY262674 |
| AW087013 | similar to gb:dbj|D00571.1|PYPLHABBP Pyrus pyrifolia mRNA for light harvesting (PLANT) | 192 | 0.047619 | JY262946, JY263100, JY262785 |
| AW087017 | similar to TR:O82341 O82341 PUTATIVE TGF-BETA RECEPTOR INTERACTING PROTEIN | 306 | – | JY263058 |
| AW087018 | similar to SW:PPCE_HUMAN P48147 PROLYL ENDOPEPTIDASE | 606 | 0.052876 | JY262837, JY262997, JY263193, JY262677 |
| AW087021 | Moss EST library CPU Ceratodon purpureus cDNA clone PEP_SOURCE_ID:CPU031003 5-, mRNA sequence | 578 | 0.016376 | JY262952, JY263109, JY263276, JY262794 |
| AW087023 | Moss EST library CPU Ceratodon purpureus cDNA clone PEP_SOURCE_ID:CPU031009 5-, mRNA sequence | 760 | 0.004662 | JY262850, JY263007, JY263205, JY262690 |
| AW087027 | Moss EST library CPU Ceratodon purpureus cDNA clone PEP_SOURCE_ID:CPU031019 5-, mRNA sequence | 817 | 0.006417 | JY262956, JY263112, JY263279, JY262798 |
| AW087029 | similar to SW:RS8_ORYSA P49199 40S RIBOSOMAL PROTEIN S8 | 482 | 0.005482 | JY262914, JY263065, JY263255, JY262752 |
| AW087030 | similar to WP:C41C4.4 CE01519 PUTATIVE SERINE/THREONINE-PROTEIN KINASE C41C4.4 IN CHROMOSOME II PRECURSOR | 498 | 0.010417 | JY262943, JY263097, JY262782 |
| AW087034 | similar to SW:YML4_ARATH O22815 HYPOTHETICAL MLO-LIKE PROTEIN | 226 | – | JY263139 |
| AW087043 | similar to SW:RS8_ORYSA P49199 40S RIBOSOMAL PROTEIN S8 | 349 | 0.086154 | JY263093, JY262778 |
| AW087048 | similar to TR:O80383 O80383 98B | 511 | 0.007026 | JY263114, JY263281 |
| AW087051 | similar to TR:O80644 O80644 F12L6.23 PROTEIN | 74* | 0 | JY263165, JY263325 |
| AW087053 | similar to SW:IFE1_WHEAT P29557 EUKARYOTIC TRANSLATION INITIATION FACTOR 4E | 227 | 0.017408 | JY262975, JY263140, JY263303, JY262816 |
| AW087065 | similar to SW:SYS_HELAN O81983 SERYL-TRNA SYNTHETASE | 524 | 0.004272 | JY262972, JY263134, JY263299, JY262813 |
| AW087074 | similar to TR:O65686 O65686 PUTATIVE RIBOSOMAL PROTEIN S16 | 700 | 0 | JY263169, JY263329 |
| AW087075 | Moss EST library CPU Ceratodon purpureus cDNA clone PEP_SOURCE_ID:CPU031418 5-, mRNA sequence | 646 | 0.014107 | JY262905, JY263054, JY263250, JY262743 |
| AW097915 | Moss EST library CPU Ceratodon purpureus cDNA clone PEP_SOURCE_ID:CPU011109 5- | 379 | 0.028902 | JY263177, JY263336 |
| AW097924 | Moss EST library CPU Ceratodon purpureus cDNA clone PEP_SOURCE_ID:CPU011114 5-, mRNA sequence | 631 | 0.009688 | JY262875, JY263027, JY263223, JY262714 |
| AW097946 | similar to TR:O81362 O81362 SULFITE REDUCTASE | 176* | 0.004545 | JY262912, JY263063, JY263254, JY262751 |
| AW097948 | similar to SW:RL5_ORYSA P49625 60S RIBOSOMAL PROTEIN L5 | 641 | 0.009762 | JY262936, JY263087, JY263267, JY262773 |
| AW097953 | similar to SW:R13A_PICMA O65055 60S RIBOSOMAL PROTEIN L13A | 183* | 0 | JY262877, JY262716 |
| AW097956 | similar to SW:RL1X_ARATH P51418 60S RIBOSOMAL PROTEIN L18A | 901 | 0.004234 | JY262919, JY263070, JY263258, JY262757 |
| AW097975 | similar to TR:O65068 O65068 60S RIBOSOMAL PROTEIN L17 | 700 | 0.008681 | JY262865, JY262704 |
| AW097976 | similar to TR:O24447 O24447 CARBAMOYL PHOSPHATE SYNTHETASE SMALL SUBUNIT | 512 | 0.007593 | JY262860, JY263015, JY263213 |
| AW097979 | similar to TR:O82808 O82808 F17H15.2 PROTEIN | 609 | 0 | JY263158, JY263318 |
| AW097983 | similar to SW:GYP7_YARLI P09379 PROBABLE GYP7 PROTEIN | 733 | 0.004071 | JY262930, JY263080, JY263265, JY262766 |
| AW097984 | similar to TR:O65059 O65059 PROBABLE 40S RIBOSOMAL PROTEIN S15 | 554 | 0.002037 | JY262873, JY263025, JY263221, JY262712 |
| AW097987 | similar to TR:Q96337 Q96337 AMP-BINDING PROTEIN | 1031 | 0.034648 | JY262934, JY263085, JY262771 |
| AW097994 | similar to TR:O65462 O65462 RECEPTOR LIKE PROTEIN | 359 | 0 | JY262880, JY262719 |
| AW098004 | similar to TR:O66573 O66573 ALDEHYDE DEHYDROGENASE | 673 | 0.008141 | JY262942, JY263095, JY263269, JY262780 |
| AW098011 | similar to TR:Q14997 Q14997 KIAA0077 PROTEIN | 491 | 0.010417 | JY263161, JY263321 |
| AW098019 | similar to SW:VATA_MAIZE P49087 VACUOLAR ATP SYNTHASE CATALYTIC SUBUNIT A | 1081 | 0.008511 | JY262954, JY263110, JY263277, JY262796 |
| AW098020 | similar to TR:O82204 O82204 F6F22.24 PROTEIN | 601 | 0.02139 | JY262869, JY263021, JY263218, JY262709 |
| AW098024 | similar to TR:Q9ZWB8 Q9ZWB8 F21M11.8 PROTEIN | 687 | 0.010737 | JY262957, JY263116, JY263283, JY262799 |
| AW098026 | similar to SW:RS3_MOUSE P17073 40S RIBOSOMAL PROTEIN S3 | 397 | 0.036855 | JY262958, JY263117, JY263284, JY262800 |
| AW098048 | similar to TR:O65023 O65023 HYPOTHETICAL 41.8 KD PROTEIN | 534 | 0.003115 | JY263055, JY262744 |
| AW098048 | similar to TR:O65023 O65023 HYPOTHETICAL 41.8 KD PROTEIN | 57* | 0 | JY262913, JY263064 |
| AW098051 | Moss EST library CPU Ceratodon purpureus cDNA clonePEP_SOURCE_ID:CPU011520 5- | 424 | 0.131034 | JY263166, JY263326 |
| AW098056 | similar to TR:O23984 O23984 EXPRESSED SEQUENCE TAG | 388 | 0.053872 | JY262909, JY263060, JY262748 |
| AW098058 | similar to SW:RM24_YEAST P36525 60S RIBOSOMAL PROTEIN L24, MITOCHONDRIAL PRECURSOR | 439 | 0.033613 | JY262845, JY263004, JY263201, JY262685 |
| AW098074 | similar to gb:gb|L28831.1|SOYRIPR Glycine max ribosomal protein S11 gene, (PLANT) | 706 | 0.037273 | JY262974, JY263136, JY263300, JY262815 |
| AW098078 | similar to SW:RS20_ORYSA P35686 40S RIBOSOMAL PROTEIN S20 | 470 | 0.01982 | JY262935, JY263086, JY262772 |
| AW098097 | similar to TR:O65583 O65583 PUTATIVE URACIL PHOSPHORIBOSYL TRANSFERASE | 210* | 0.005076 | JY262827, JY262665 |
| AW098140 | similar to SW:RS21_ORYSA P35687 40S RIBOSOMAL PROTEIN S21 | 106* | 0.014815 | JY262925, JY263076, JY262763 |
| AW098153 | similar to TR:O82505 O82505 F2P3.12 PROTEIN | 529 | 0 | JY263113, JY263280 |
| AW098157 | similar to SW:RL37_ARATH Q43292 60S RIBOSOMAL PROTEIN L37 | 801 | 0.026701 | JY262842, JY263002, JY263198, JY262682 |
| AW098158 | similar to SW:RL44_GOSHI Q96499 60S RIBOSOMAL PROTEIN L44 | 688 | 0.017544 | JY262843, JY263003, JY263199, JY262683 |
| AW098158 | similar to SW:RL44_GOSHI Q96499 60S RIBOSOMAL PROTEIN L44 | 446 | 0.054795 | JY262858, JY263013, JY262699 |
| AW098185 | Moss EST library CPU Ceratodon purpureus cDNA clone PEP_SOURCE_ID:CPU030823 3-, mRNA sequence | 151* | 0 | JY262901, JY263050, JY262739 |
| AW098187 | similar to SW:GLYM_PEA P34899 SERINE HYDROXYMETHYLTRANSFERASE, MITOCHONDRIAL PRECURSOR | * | – | JY263131 |
| AW098191 | similar to TR:O49336 O49336 T11J7.10 PROTEIN | 330 | 0 | JY262970, JY263132 |
| AW098191 | similar to TR:O49336 O49336 T11J7.10 PROTEIN | 392 | 0 | JY262976, JY263141, JY262817 |
| AW098197 | similar to TR:O49337 O49337 T11J7.11 PROTEIN | 747 | 0.002204 | JY262969, JY263130, JY263297, JY262811 |
| AW098200 | Moss EST library CPU Ceratodon purpureus cDNA clone PEP_SOURCE_ID:CPU021119 5- | – | JY263170 | |
| AW098203 | similar to TR:Q9ZTW0 Q9ZTW0 ABA-RESPONSIVE PROTEIN | 479 | 0.004264 | JY263173, JY263332 |
| AW098216 | similar to SW:R33B_YEAST P41056 60S RIBOSOMAL PROTEIN L33-B | 194* | 0 | JY263178, JY263337 |
| AW098225 | similar to SW:ERD1_ARATH P42762 ERD1 PROTEIN PRECURSOR | 454 | 0.008905 | JY262841, JY263001, JY263197, JY262681 |
| AW098233 | similar to TR:O80526 O80526 F19J9.9 PROTEIN | 708 | 0.006442 | JY262959, JY263118, JY263285, JY262801 |
| AW098234 | similar to SW:GLYM_PEA P34899 SERINE HYDROXYMETHYLTRANSFERASE, MITOCHONDRIAL PRECURSOR | 278 | 0.053292 | JY262857, JY263012, JY263211, JY262698 |
| AW098247 | similar to SW:ALFD_PEA Q01517 FRUCTOSE-BISPHOSPHATE ALDOLASE 2, CHLOROPLAST | 569 | 0.005981 | JY262961, JY263122, JY263289, JY262803 |
| AW098252 | similar to SW:RS2_ARATH P49688 40S RIBOSOMAL PROTEIN S2 | 86* | 0 | JY262872, JY263024 |
| AW098256 | similar to TR:O80799 O80799 T8F5.5 PROTEIN | 517 | 0.002727 | JY262911, JY263062, JY263253, JY262750 |
| AW098258 | similar to TR:O22215 O22215 PUTATIVE ESTERASE D | 619 | 0.043062 | JY262981, JY263148, JY263310, JY262823 |
| AW098262 | similar to TR:Q9ZV56 Q9ZV56 PUTATIVE PHOSPHOCHOLINE CYTIDYLYLTRANSFERASE | 540 | 0.001946 | JY263159, JY263319 |
| AW098272 | similar to TR:O49379 O49379 HYPOTHETICAL 16.4 KD PROTEIN | 211 | 0.023952 | JY263162, JY263322 |
| AW098279 | Moss EST library CPU Ceratodon purpureus cDNA clone PEP_SOURCE_ID:CPU021506 5-, mRNA sequence | 141* | 0 | JY262900, JY263049, JY263246, JY262738 |
| AW098281 | similar to TR:O65068 O65068 60S RIBOSOMAL PROTEIN L17 | 348 | 0.044199 | JY263107, JY263275, JY262792 |
| AW098283 | similar to TR:O64720 O64720 PUTATIVE TBP-BINDING PROTEIN | 431 | 0.004885 | JY262977, JY263142, JY263304, JY262818 |
| AW098284 | similar to TR:Q40922 Q40922 PSEUDOTZAIN | 154* | 0 | JY262966, JY263127, JY263294, JY262808 |
| AW098291 | similar to SW:YAUB_SCHPO Q10166 HYPOTHETICAL 35.7 KD PROTEIN C26A3.11 IN CHROMOSOME I | 286 | 0.02952 | JY263167, JY263327 |
| AW098300 | similar to SW:SUI1_MAIZE P56330 PROTEIN TRANSLATION FACTOR SUI1 HOMOLOG | 537 | 0.072549 | JY263171, JY263330 |
| AW098304 | similar to SW:PRL1_ARATH Q42384 PP1/PP2A PHOSPHATASES PLEIOTROPIC REGULATOR PRL1 | 385 | 0.019139 | JY262893, JY263041, JY263238, JY262730 |
| AW098306 | similar to SW:RL7_ARATH Q42208 60S RIBOSOMAL PROTEIN L7 | 203* | 0 | JY263174, JY263333 |
| AW098317 | Moss EST library CPU Ceratodon purpureus cDNA clone PEP_SOURCE_ID:CPU030319 5-, mRNA sequence | 508 | 0 | JY262980, JY262822 |
| AW098318 | Moss EST library CPU Ceratodon purpureus cDNA clone PEP_SOURCE_ID:CPU030321 5-, mRNA sequence | 128* | 0 | JY262852, JY263208, JY262693 |
| AW098322 | similar to TR:O23601 O23601 HYPOTHETICAL 40.2 KD PROTEIN | 735 | 0.023511 | JY262896, JY263044, JY263241, JY262733 |
| AW098328 | similar to TR:Q23920 Q23920 PEPA | 429 | 0.024938 | JY262938, JY263089, JY262775 |
| AW098337 | similar to SW:P2A_HELAN P48579 SERINE/THREONINE PROTEIN PHOSPHATASE PP2A CATALYTIC SUBUNIT | 293 | 0.004065 | JY262933, JY263083, JY262769 |
| AW098340 | Moss EST library CPU Ceratodon purpureus cDNA clone PEP_SOURCE_ID:CPU030420 5-, mRNA sequence | 434 | 0.011223 | JY262839, JY262999, JY263195, JY262679 |
| AW098349 | Moss EST library CPU Ceratodon purpureus cDNA clone PEP_SOURCE_ID:CPU030717 5-, mRNA sequence | 329 | 0 | JY263207, JY262692 |
| AW098359 | similar to SW:RL29_RAT P25886 60S RIBOSOMAL PROTEIN L29 | 334 | 0.023392 | JY262986, JY263182, JY262666 |
| AW098361 | similar to TR:O80800 O80800 T8F5.6 PROTEIN | 849 | 0.012285 | JY262903, JY263052, JY263248, JY262741 |
| AW098364 | similar to TR:O65636 O65636 HYPOTHETICAL 15.1 KD PROTEIN | 442 | 0.004902 | JY262940, JY263091, JY262776 |
| AW098367 | similar to TR:Q42809 Q42809 GMCK1P | 453 | 0.010195 | JY262910, JY263061, JY263252, JY262749 |
| AW098372 | Moss EST library CPU Ceratodon purpureus cDNA clone PEP_SOURCE_ID:CPU030814 5-, mRNA sequence | 147* | 0.008658 | JY262866, JY263018, JY262705 |
| AW098391 | similar to TR:O30618 O30618 ACYL-COA OXIDASE | 530 | 0.010414 | JY262921, JY263072, JY263260, JY262759 |
| AW098393 | similar to SW:RL7A_ORYSA P35685 60S RIBOSOMAL PROTEIN L7A | 211 | 0.020408 | JY262887, JY263232 |
| AW098407 | similar to TR:O81046 O81046 AXI 1-LIKE PROTEIN | 727 | 0.022222 | JY262863, JY263214, JY262702 |
| AW098418 | similar to SW:RS25_LYCES P46301 40S RIBOSOMAL PROTEIN S25 | 312 | 0.011988 | JY262888, JY263036, JY263233, JY262726 |
| AW098419 | similar to SW:R35A_MOUSE O55142 60S RIBOSOMAL PROTEIN L35A | 550 | 0.01165 | JY263179, JY263338 |
| AW098432 | Moss EST library CPU Ceratodon purpureus cDNA clone PEP_SOURCE_ID:CPU031514 5-, mRNA sequence | 598 | 0 | JY262891, JY263039, JY263236, JY262729 |
| AW098442 | similar to TR:P93321 P93321 CDC2 KINASE HOMOLOGUE, CDC2MSD | 290 | 0.011252 | JY262844, JY263200, JY262684 |
| AW098442 | similar to TR:P93321 P93321 CDC2 KINASE HOMOLOGUE, CDC2MSD | 473 | 0.029591 | JY262892, JY263040, JY263237 |
| AW098448 | similar to TR:Q9ZQX9 Q9ZQX9 40S RIBOSOMAL PROTEIN S27 HOMOLOG | 274 | 0 | JY263181, JY263340 |
| AW098462 | similar to SW:RS1A_ARATH P42798 40S RIBOSOMAL PROTEIN S15A | 215 | 0.044709 | JY262879, JY263030, JY263225, JY262718 |
| AW098472 | Moss EST library CPU Ceratodon purpureus cDNA clone PEP_SOURCE_ID:CPU011207 3- | 446 | 0 | JY263163, JY263323 |
| AW098479 | similar to TR:O48649 O48649 ADP-RIBOSYLATION FACTOR | 251 | 0.029268 | JY262907, JY263057, JY262746 |
| AW098512 | similar to SW:ILV5_SPIOL Q01292 KETOL-ACID REDUCTOISOMERASE PRECURSOR | 645 | 0.019108 | JY262884, JY263034, JY263229, JY262723 |
| AW098515 | similar to TR:O04556 O04556 T7N9.9 | 442 | 0.00391 | JY262846, JY263202, JY262686 |
| AW098521 | similar to TR:O82203 O82203 PUTATIVE RIBOSOMAL PROTEIN | 286 | 0 | JY263145, JY263307, JY262821 |
| AW098560 | similar to TR:O13870 O13870 PROBABLE TRANSCRIPTIONAL REGULATOR C1B3.05 | 458 | 0.008354 | JY262944, JY263098, JY262783 |
| AW098568 | similar to SW:DHE3_RHISN Q53199 PROBABLE GLUTAMATE DEHYDROGENASE | 330 | 0.007305 | JY262831, JY262990, JY263186, JY262670 |
| AW098572 | similar to TR:Q9ZUL5 Q9ZUL5 PUTATIVE DNA-BINDING PROTEIN | 619 | 0.002573 | JY262859, JY263014, JY263212, JY262700 |
| AW098585 | similar to SW:FKB7_WHEAT Q43207 70 KD PEPTIDYLPROLYL ISOMERASE | – | JY263155 | |
| AW098587 | similar to SW:OAT_EMENI Q92413 ORNITHINE AMINOTRANSFERASE | 547 | 0.017842 | JY262902, JY263051, JY263247, JY262740 |
| AW098597 | similar to TR:Q56987 Q56987 HYPOTHETICAL 23.2 KD PROTEIN | 254 | 0 | JY263175, JY263334 |
| AW098620 | similar to TR:Q9ZRI8 Q9ZRI8 FORMATE DEHYDROGENASE | 606 | 0 | JY262929, JY263341 |
| AW098621 | similar to SW:RS1A_ARATH P42798 40S RIBOSOMAL PROTEIN S15A | 732 | 0.044058 | JY263019, JY263216, JY262706 |
| AW098624 | similar to SW:RL44_GOSHI Q96499 60S RIBOSOMAL PROTEIN L44 | – | JY262862 | |
| AW098631 | similar to SW:SPEE_COFAR O82147 SPERMIDINE SYNTHASE | 161* | 0 | JY262861, JY263016, JY262701 |
| AW098643 | similar to TR:Q9ZQP2 Q9ZQP2 PUTATIVE ACYL COENZYME A OXIDASE, PEROXISOMAL COMPONENT | 456 | 0.019697 | JY262955, JY263111, JY263278, JY262797 |
| AW098651 | similar to TR:Q45073 Q45073 HYPOTHETICAL 23.3 KD PROTEIN | 337 | 0.008584 | JY262951, JY263108, JY262793 |
| AW098679 | similar to TR:O82342 O82342 PUTATIVE TGF-BETA RECEPTOR INTERACTING PROTEIN | 807 | 0.054728 | JY262828, JY262987, JY263183, JY262667 |
| AW098700 | similar to TR:Q9ZQR3 Q9ZQR3 PUTATIVE RECEPTOR PROTEIN KINASE | 272 | 0.023622 | JY262848, JY263203, JY262688 |
| AW098717 | similar to TR:O82204 O82204 F6F22.24 PROTEIN | 173* | 0.006211 | JY263180, JY263339 |
| AW098746 | similar to TR:O22662 O22662 PROTEIN PHOSPHATASE U | 532 | 0 | JY263115, JY263282 |
| AW098758 | similar to SW:TRP2_ARATH P25269 TRYPTOPHAN SYNTHASE BETA CHAIN 2 PRECURSOR | 369 | 0 | JY263121, JY263288 |
| AW098761 | similar to TR:Q9ZDU0 Q9ZDU0 30S RIBOSOMAL PROTEIN S9 | 134* | 0.031169 | JY262898, JY263046, JY263243, JY262735 |
| AW098770 | similar to WP:D1054.3 CE05522 | 603 | 0.003924 | JY262978, JY263143, JY263305, JY262819 |
| AW098776 | similar to TR:Q41024 Q41024 SMALL GTP-BINDING PROTEIN | 599 | 0.003643 | JY263147, JY263309 |
| AW098780 | similar to SW:IMB3_HUMAN O00410 IMPORTIN BETA-3 SUBUNIT | 656 | 0 | JY262963, JY263124, JY263291, JY262805 |
| AW098782 | similar to TR:Q9ZVS6 Q9ZVS6 F15K9.15 PROTEIN | 310 | 0.012212 | JY262899, JY263047, JY263244, JY262736 |
| AW098786 | similar to SW:LE22_ARCFU O28084 PUTATIVE 3-ISOPROPYLMALATE DEHYDRATASE LARGE SUBUNIT | 569 | 0.006545 | JY262897, JY263045, JY263242, JY262734 |
| AW098797 | similar to SW:CAHC_HORVU P40880 CARBONIC ANHYDRASE, CHLOROPLAST PRECURSOR | 410 | 0.001855 | JY262984, JY263153, JY263314, JY262826 |
| AW098799 | similar to WP:F41C3.4 CE02732 | 347 | 0.004489 | JY262945, JY263099, JY262784 |
| AW098804 | similar to TR:O81808 O81808 HYPOTHETICAL 62.6 KD PROTEIN | 512 | 0.017094 | JY262922, JY263073, JY263261, JY262760 |
| AW098812 | similar to TR:O81468 O81468 T15F16.12 PROTEIN | 747 | 0.007407 | JY263120, JY263287 |
| AW098823 | similar to SW:RLA1_MAIZE P52855 60S ACIDIC RIBOSOMAL PROTEIN P1 | 852 | 0.005929 | JY262904, JY263053, JY263249, JY262742 |
| AW098831 | similar to TR:O75865 O75865 R32611_2 | 485 | 0.005742 | JY262979, JY263144, JY263306, JY262820 |
| AW098839 | similar to SW:RS19_ORYSA P40978 40S RIBOSOMAL PROTEIN S18 | 857 | 0 | JY262849, JY263006, JY263204, JY262689 |
| AW098949 | similar to TR:O04033 O04033 F7G19.16 | 389 | 0.002045 | JY262924, JY263075, JY262762 |
| AW098949 | similar to TR:O04033 O04033 F7G19.15 | 411 | 0.003956 | JY262932, JY263082, JY262768 |
| AW098974 | similar to SW:RL34_TOBAC P41098 60S RIBOSOMAL PROTEIN L34 | 514 | 0.063425 | JY262829, JY262988, JY263184, JY262668 |
| BI894286 | similar to SW:ATP2_ACTCH P43395 ATP SYNTHASE BETA CHAIN, MITOCHONDRIAL | 272 | – | JY263150 |
| BI894288 | similar to SW:RS23_FRAAN P46297 40S RIBOSOMAL PROTEIN S23 | 565 | 0 | JY262833, JY262992, JY263188, JY262672 |
| CZ893596 | CEPU109 | – | ||
| CZ894207 | CEPU105 | 86 | 0.026316 | |
| CZ894357 | CEPU108 | 243 | 0.0125 |
Accessions studied were: C. purpureus: WT4 (Wispertal, Austria), GG1 (Grossgerunds, Austria), R40 (Petersburg Pass, Renssalaer, New York), and Otavalo, Ecuador (McDaniel E112, DUKE). Additional isolates were sequenced for the loci in bold in C. purpureus (Ros29.10.2009-1 [UF], Ros29.10.2009-2 [UF], Antarctica, Robinson&Wasley 99/00 [DUKE]); T. cylindricus (DUKE11365, DUKE65082, DUKE65084); and C. chloropus (Werner&Ros 14024 [DUKE], Guerra15.4.2001 [DUKE], Ros29.10.2009-1 [UF], Ros29.10.2009-2 [UF]).
Length of the amplified PCR product in C. purpureus; an “*” indicates that C. purpureus lacks the P. patens intron.
A “–” indicates that a single PCR product was generated; a “0” indicates an absence of variation among isolates.
We have identified more than 50 loci with θ = 0.02, a value more than twice the species median. This value is also equivalent to the most variable nuclear loci used for phylogeographic inference in any bryophyte species to date. Using the PCR and sequencing strategy outlined above, we chose 12 loci to sequence in isolates of C. purpureus from the Sierra Nevadas, Spain; Casey Station, Antarctica; and Wollongong, Australia, and 1–2 isolates of the sister groups to C. purpureus, Trichodon cylindricus (Hedw.) Schimp., and Cheilothela chloropus (Brid.) Broth. (Table 2). The PCR products were nearly the same length in all three species, and produced sequences with unambiguous chromatograms. In all cases, the introns were alignable among the three species, but the species differed at ∼10–20% of the intron sites, suggesting that these loci may be useful for phylogeographic and species-level phylogenetic studies. In the complete panel of loci, we also found 23 introns that were present in the P. patens genome that were absent in the C. purpureus genome (Table 2). Using a PCR length variation test, we determined that the intron absence was shared by many species in the Dicranidae (McDaniel and Neubig, unpublished data). These presence/absence polymorphisms may be useful phylogenetic markers (Goffinet et al., 2007). We expect that this panel of primers will be valuable for the bryophyte evolutionary genetics community as a whole.
CONCLUSIONS
In this study, we have generated primers for more than 200 loci, based on comparisons from ESTs from C. purpureus and the genome of P. patens. We have used these loci to estimate the genome-wide distribution of nucleotide diversity within C. purpureus. Because these primers were designed to be homologous to exonic regions that are conserved between species that diverged long ago, these primers may amplify the target region in a wide variety of mosses. We anticipate that these loci will form a valuable addition to the bryophyte molecular ecology toolkit, enabling more detailed phylogeographic and population genetic studies of a variety of focal species.
LITERATURE CITED
- Cove D., Perroud P.-F., Charron A., McDaniel S., Khandelwal A., Quatrano R. 2009. The moss Physcomitrella patens: A novel model system for plant development and genomic studies, Emerging Model Organisms. Cold Spring Harbor Laboratory Press, New York, New York, USA: [DOI] [PubMed] [Google Scholar]
- Cox C. J., Goffinet B., Wickett N. J., Boles S. B., Shaw A. J. 2010. Moss diversity: A molecular phylogenetic analysis of genera. Phytotaxa 9: 175–195 [Google Scholar]
- Goffinet B., Wickett N. J., Werner O., Ros R. M., Shaw A. J., Cox C. J. 2007. Distribution and phylogenetic significance of the 71-kb inversion in the plastid genome in Funariidae (Bryophyta). Annals of Botany 99: 747–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent W. J. 2002. BLAT—The BLAST-Like Alignment Tool. Genome Research 12: 656–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Librado P., Rozas J. 2009. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25: 1451–1452 [DOI] [PubMed] [Google Scholar]
- McDaniel S. F., Shaw A. J. 2005. Selective sweeps and intercontinental migration in the cosmopolitan moss Ceratodon purpureus (Hedw.) Brid. Molecular Ecology 14: 1121–1132 [DOI] [PubMed] [Google Scholar]
- McDaniel S. F., Willis J. H., Shaw A. J. 2007. A linkage map reveals a complex basis for segregation distortion in an interpopulation cross in the moss Ceratodon purpureus. Genetics 176: 2489–2500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDaniel S. F., Willis J. H., Shaw A. J. 2008. The genetic basis of developmental abnormalities in interpopulation hybrids of the moss Ceratodon purpureus. Genetics 179: 1425–1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDaniel S. F., von Stackelberg M., Richardt S., Quatrano R. S., Reski R., Rensing S. A. 2010. The speciation history of the Physcomitrium–Physcomitrella species complex. Evolution 64: 217–231 [DOI] [PubMed] [Google Scholar]
- Rozen S., Skaletsky H. J. 2000. Primer3 on the WWW for general users and for biologist programmers. In S. Misener and S. A. Krawetz [eds.], Methods in molecular biology, vol. 132: Bioinformatics methods and protocols, 365–386. Humana Press, Totowa, New Jersey, USA. [DOI] [PubMed] [Google Scholar]
- Stech M., Quandt D. 2010. 20,000 species and five key markers: The status of molecular bryophyte phylogenetics. Phytotaxa 9: 196–228 [Google Scholar]
- Vanderpoorten A., Shaw A. J. 2010. The application of molecular data to the phylogenetic delimitation of species in bryophytes: A note of caution. Phytotaxa 9: 229–237 [Google Scholar]