Abstract
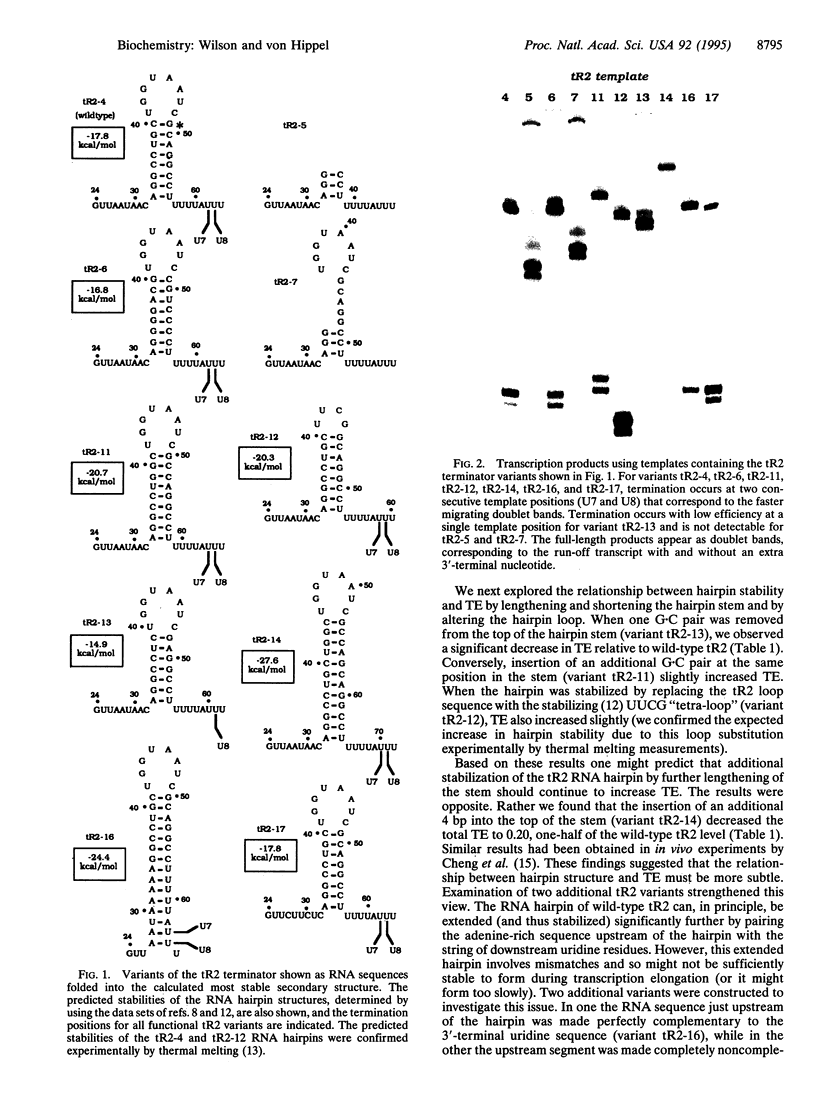
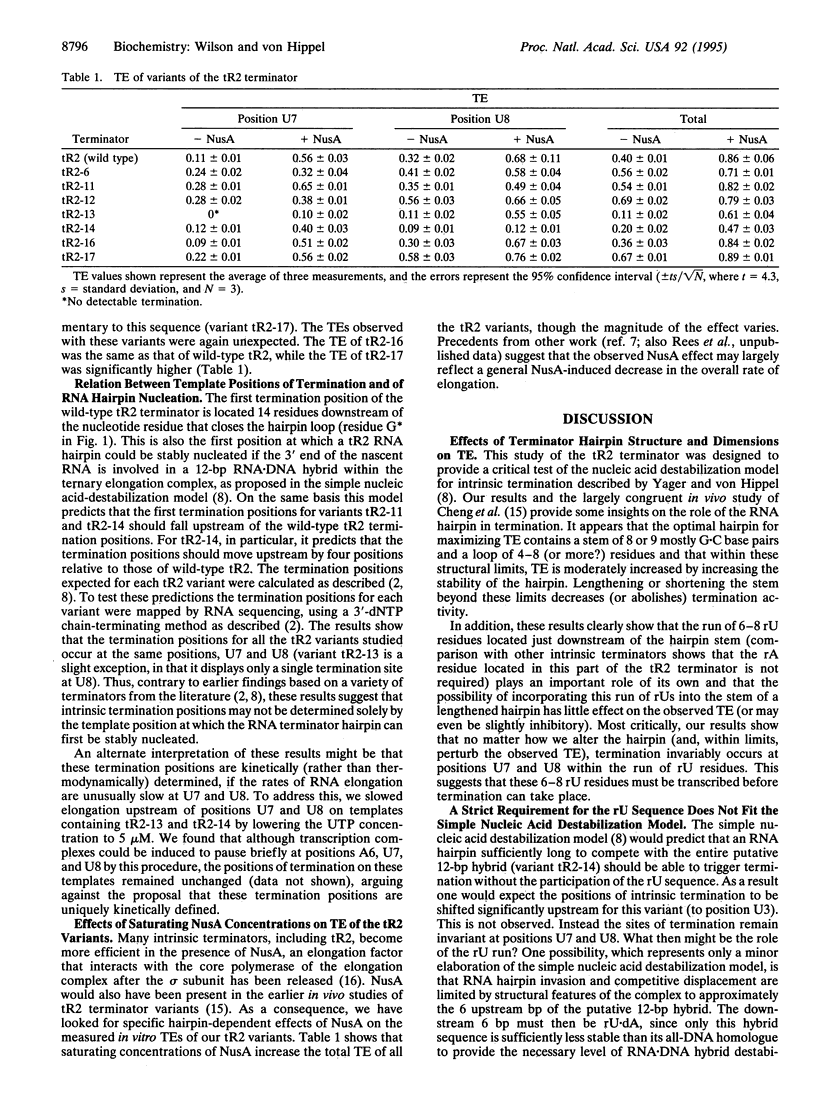
Intrinsic termination of transcription in Escherichia coli involves the formation of an RNA hairpin in the nascent RNA. This hairpin plays a central role in the release of the transcript and polymerase at intrinsic termination sites on the DNA template. We have created variants of the lambda tR2 terminator hairpin and examined the relationship between the structure and stability of this hairpin and the template positions and efficiencies of termination. The results were used to test the simple nucleic acid destabilization model of Yager and von Hippel and showed that this model must be modified to provide a distinct role for the rU-rich sequence in the nascent RNA, since a perfect palindromic sequence that is sufficiently long to form an RNA hairpin that could destabilize the entire putative 12-bp RNA-DNA hybrid does not trigger termination at the expected positions. Rather, our results show that both a stable terminator hairpin and the run of 6-8 rU residues that immediately follows are required for effective intrinsic termination and that termination occurs at specific and invariant template positions relative to these two components. Possible structural or kinetic modifications of the simple model are proposed in the light of these findings and of recent results implicating "inchworming" and possible conformational heterogeneity of transcription complexes in intrinsic termination. Thus, these findings argue that the structure and dimensions of the hairpin are important determinants of the termination-elongation decision and suggest that a complete mechanism is likely to involve specific interactions of the polymerase, the RNA terminator hairpin, and, perhaps, the dT-rich template sequence that codes for the run of rU residues at the 3' end of the nascent transcript.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Antao V. P., Lai S. Y., Tinoco I., Jr A thermodynamic study of unusually stable RNA and DNA hairpins. Nucleic Acids Res. 1991 Nov 11;19(21):5901–5905. doi: 10.1093/nar/19.21.5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlin M. J. New models for the mechanism of transcription elongation and its regulation. Harvey Lect. 1992 1993;88:1–21. [PubMed] [Google Scholar]
- Cheng S. W., Lynch E. C., Leason K. R., Court D. L., Shapiro B. A., Friedman D. I. Functional importance of sequence in the stem-loop of a transcription terminator. Science. 1991 Nov 22;254(5035):1205–1207. doi: 10.1126/science.1835546. [DOI] [PubMed] [Google Scholar]
- Farnham P. J., Platt T. Rho-independent termination: dyad symmetry in DNA causes RNA polymerase to pause during transcription in vitro. Nucleic Acids Res. 1981 Feb 11;9(3):563–577. doi: 10.1093/nar/9.3.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill S. C., Weitzel S. E., von Hippel P. H. Escherichia coli sigma 70 and NusA proteins. I. Binding interactions with core RNA polymerase in solution and within the transcription complex. J Mol Biol. 1991 Jul 20;220(2):307–324. doi: 10.1016/0022-2836(91)90015-x. [DOI] [PubMed] [Google Scholar]
- Hager D. A., Jin D. J., Burgess R. R. Use of Mono Q high-resolution ion-exchange chromatography to obtain highly pure and active Escherichia coli RNA polymerase. Biochemistry. 1990 Aug 28;29(34):7890–7894. doi: 10.1021/bi00486a016. [DOI] [PubMed] [Google Scholar]
- Jin D. J., Burgess R. R., Richardson J. P., Gross C. A. Termination efficiency at rho-dependent terminators depends on kinetic coupling between RNA polymerase and rho. Proc Natl Acad Sci U S A. 1992 Feb 15;89(4):1453–1457. doi: 10.1073/pnas.89.4.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell J. C., Roberts J. W., Jin D. J., Gross C. Determination of intrinsic transcription termination efficiency by RNA polymerase elongation rate. Science. 1994 Nov 4;266(5186):822–825. doi: 10.1126/science.7526463. [DOI] [PubMed] [Google Scholar]
- Nudler E., Goldfarb A., Kashlev M. Discontinuous mechanism of transcription elongation. Science. 1994 Aug 5;265(5173):793–796. doi: 10.1126/science.8047884. [DOI] [PubMed] [Google Scholar]
- Platt T. Transcription termination and the regulation of gene expression. Annu Rev Biochem. 1986;55:339–372. doi: 10.1146/annurev.bi.55.070186.002011. [DOI] [PubMed] [Google Scholar]
- Reynolds R., Bermúdez-Cruz R. M., Chamberlin M. J. Parameters affecting transcription termination by Escherichia coli RNA polymerase. I. Analysis of 13 rho-independent terminators. J Mol Biol. 1992 Mar 5;224(1):31–51. doi: 10.1016/0022-2836(92)90574-4. [DOI] [PubMed] [Google Scholar]
- Rhodes G., Chamberlin M. J. Ribonucleic acid chain elongation by Escherichia coli ribonucleic acid polymerase. I. Isolation of ternary complexes and the kinetics of elongation. J Biol Chem. 1974 Oct 25;249(20):6675–6683. [PubMed] [Google Scholar]
- Wilson K. S., von Hippel P. H. Stability of Escherichia coli transcription complexes near an intrinsic terminator. J Mol Biol. 1994 Nov 18;244(1):36–51. doi: 10.1006/jmbi.1994.1702. [DOI] [PubMed] [Google Scholar]
- Yager T. D., von Hippel P. H. A thermodynamic analysis of RNA transcript elongation and termination in Escherichia coli. Biochemistry. 1991 Jan 29;30(4):1097–1118. doi: 10.1021/bi00218a032. [DOI] [PubMed] [Google Scholar]
- d'Aubenton Carafa Y., Brody E., Thermes C. Prediction of rho-independent Escherichia coli transcription terminators. A statistical analysis of their RNA stem-loop structures. J Mol Biol. 1990 Dec 20;216(4):835–858. doi: 10.1016/s0022-2836(99)80005-9. [DOI] [PubMed] [Google Scholar]
- von Hippel P. H., Yager T. D. The elongation-termination decision in transcription. Science. 1992 Feb 14;255(5046):809–812. doi: 10.1126/science.1536005. [DOI] [PubMed] [Google Scholar]
- von Hippel P. H., Yager T. D. Transcript elongation and termination are competitive kinetic processes. Proc Natl Acad Sci U S A. 1991 Mar 15;88(6):2307–2311. doi: 10.1073/pnas.88.6.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]




