Abstract
Objectives
Uridine abrogates mitochondrial toxicities of nucleoside reverse transcriptase inhibitor in adipocyte cell culture. We aim to study the effect of uridine supplementation on human adipocyte mitochondrial DNA (mtDNA) levels in subjects with human immunodeficiency (HIV) lipoatrophy.
Methods
Sixteen patients with lipoatrophy on stavudine-containing antiretroviral therapy were enrolled, and received NucleomaxX, a dietary supplement with a high bioavailability of uridine (36 g TID every other day for 16 weeks). Patients were then followed off-uridine for another 16 weeks. Highly active antiretroviral therapy remained unchanged during the trial.
Results
Fourteen patients completed the study. Two subjects dropped out before week 4 for study-unrelated reasons. No adverse events were noted throughout the study. HIV-1 RNA, CD4 counts, liver enzymes and hemoglobin remained unchanged. Body mass index, lactate, lipids, insulin and homeostasis model assessment of insulin resistance were unaltered. Fat and peripheral blood and mononuclear cell mtDNA levels did not correlate with each other and exhibited no changes throughout the study. Lipoatrophy scores by patients and physician improved significantly at weeks 16 and 32 compared to study entry.
Conclusion
In this pilot study, NucleomaxX was safe, well tolerated without apparent deleterious effect on HIV indices. In contrast to in vitro data, NucleomaxX did not lead to changes in fat or blood mtDNA levels.
Keywords: mitochondria, uridine, lipoatrophy, lipodystrophy, mitochondrial DNA
Introduction
Lipoatrophy of the face, arms, buttocks and/or legs is a distressing and stigmatizing long-term effect of antiretroviral therapy. Lipoatrophy has been described in human immunodeficiency (HIV)-infected persons with or without simultaneous fat accumulation in central areas of the body. The pathogenesis of lipoatrophy is not fully understood but several lines of evidence point toward an important role of nucleoside reverse transcriptase inhibitors (NRTIs) in general, and the mitochondrial toxicity of stavudine in particular (Mallal et al., 2000; Joly et al., 2002). In vitro data suggest that clinically relevant concentrations of some NRTIs impair DNA polymerase-gamma and therefore interfere with the replication of mitochondrial DNA (mtDNA) (Martin et al., 1994). Consistent with such mitochondrial toxicity, reduced copy numbers of mtDNA (mtDNA-depletion) were found in HIV-infected individuals treated with NRTIs (mostly stavudine) and linked with increased adipocyte apoptosis and ultrastructural abnormalities of adipocyte mitochondria (Walker et al., 2002; Nolan et al., 2003).
Lipoatrophy can be partially reversed by substituting stavudine with abacavir or tenofovir or by switching to NRTI-sparing regimens (McComsey et al., 2004; Tebas et al., 2005; Moyle et al., 2006). Unfortunately, the reversal of lipoatrophy in these switch studies has been slow and only partial. Thus, current medical treatment options for lipoatrophy are limited and incomplete.
The supplementation of uridine is a new promising strategy in the treatment of lipoatrophy (Walker et al., 2003). In cultured adipocytes for example, uridine supplementation prevented and treated stavudine-related mtDNA-depletion and mitochondrial dysfunction (Walker et al., 2006). Furthermore, uridine abrogated consequent adipocyte apoptosis and most importantly normalized the intracellular lipid content (Walker et al., 2006). One recently reported placebo-controlled trial showed significant improvement of peripheral fat by magnetic resonance imaging but with decrease in high-density lipoprotein (HDL) cholesterol (Sutinen et al., 2005). However, no measurements of mtDNA levels in fat or blood were reported, and only nine subjects in that study received active NucleomaxX.
The exact mechanism of action of uridine has not yet been delineated. It is possible, that uridine or its metabolites may compete at various steps ofmitochondrial pyrimidine import, degradation or phosphorylation. On the other hand, a second mechanismmay also be operated: mtDNA encodes respiratory chain subunits. NRTI-induced mtDNA-depletion thus impairs the electron transport through the respiratory chain. The latter in turn is crucial for the function of dihydroorotate-dehydrogenase, a key enzyme in the de novo synthesis of all intracellular pyrimidines. Thus, stavudine-related mitochondrial toxicity may lead to depletion in building blocks for mtDNA synthesis and thus increase the relative amounts of pyrimidine NRTIs. Through this mechanism, pyrimidine NRTIs may then compete more efficiently at DNA polymerase-gamma. A vicious circle is closed and may contribute to a further loss of mtDNA and oxidative capacity. By providing exogenous uridine from which all intracellular pyrimidines can be salvaged, this vicious circle may be disrupted and mtDNA and functions be restored. It has also been shown that the depletion of intracellular pyrimidine pools can lead to cell-cycle arrest and apoptosis (Bodnar et al., 1995; Fox et al., 1999). By replenishing intracellular pyrimidine pools and interfering with adipocyte apoptosis, uridine may therefore also restore adipose tissue in the absence of mtDNA recovery.
High doses of oral uridine or of uridine prodrugs are well tolerated in humans with mild osmotic diarrhea being dose-limiting (Leyva et al., 1984; van Groeningen et al., 1986). Phenotypic HIV-resistance assays and animal data do not indicate that uridine or its metabolites interfere with NRTI at HIV reverse transcriptase and thus with the antiretroviral efficacy of nucleoside analogues (Sommadossi et al., 1988; Calabresi et al., 1990; Koch et al., 2003). Only a case report (Walker et al., 2004) and a small 3-month trial (n=9 on NucleomaxX) (Sutinen et al., 2005) have assessed the safety of uridine supplementation in NRTI-treated HIV-infected subjects.
In this study, we hypothesized that dietary supplementation with uridine will increase mtDNA content in subcutaneous adipose tissue without significant adverse events. As a source of uridine, we used NucleomaxX (Pharma Nord, Vojens, Denmark), a dietary supplement which has been shown to increase the serum concentration of uridine in humans from an average of about 5 μM to more than 150 μM due to its high content (17%) of nucleosides (Venhoff et al., 2005) (www.nucleomaxX.com).
Methods
Study design and population
This single-site, prospective and open-labeled single arm study was approved by the Institutional Review Board at the University Hospitals of Cleveland, Cleveland, Ohio. An investigational new drug application for the use of NucleomaxX in HIV lipoatrophy was obtained. Subjects were enrolled at the John T Carey Special Immunology Unit, University Hospitals of Cleveland, Cleveland, Ohio provided that they gave written informed consent. Enrollment occurred between August and December 2004. Eligibility criteria included HIV-infected men and women of at least 18 years of age, established lipoatrophy and plasma HIV-1 RNA levels below 400 copies/ml at screening. Patients had to be on a stable stavudine-containing antiretroviral regimen for a minimum of 12 consecutive weeks before study entry, with no plan to alter antiretrovirals for the duration of the study. In addition, the study required a past cumulative duration of stavudine treatment of at least 9 consecutive months before study entry. For this study, lipoatrophy was defined as patient self-reported and investigator-confirmed fat wasting of at least moderate severity in at least two of the following areas: face, arms, legs or buttocks. Exclusion criteria were pregnancy, breastfeeding, the diagnosis of liver cirrhosis or decompensated liver disease, active gastrointestinal symptoms at screening, lactose intolerance, diabetes requiring hypoglycemic agents, as well as the use of didanosine, valproic acid, hormonal or immunomodulating therapies. Exclusionary labs were serum creatinine >2.0 mg/dl, aspartate aminotransferase and/or alanine aminotransferase (ALT)>2.5 × upper limit of normal (ULN), platelets ≤100 000, lipase >2 ULN or hemoglobin ≤9.0 g/dl.
The total study duration was 32 weeks. For the first 16 weeks of the study, all subjects received open-label NucleomaxX at a dose of 36 g TID every other day. For the last 16 weeks, patients were followed on-study but uridine was discontinued. Study participants were instructed to drink the entire contents of one sachet of NucleomaxX (36 g) after thoroughly mixing it with milk, juice or water. Since the dosing was on an every other day schedule, instructions were given to study participants that if all three sachets are not taken in one day (first 24-h period), the unused sachet(s) can be taken the next day (second 24-h period), so that subjects would end up receiving a total of three sachets (36 g each) in a 48-h period. There were no food restrictions.
Clinical assessments were obtained at screening, at entry, at weeks 4, 8, 12, 16 and 32. During each of these study visits, subjects were specifically asked about adverse events, with special emphasis on the presence or absence of nausea, vomiting, anorexia, changes in stool pattern, diarrhea (with frequency and consistency recorded) and abdominal pain. Also, at study entry, and weeks 16 and 32, each subject privately filled out a questionnaire with emphasis on changes in body habitus and had a physical examination with special emphasis on signs of fat loss. Metabolic parameters also were assessed at study entry, week 16 and 32. At these visits, subjects had to be fasting for at least 8 h before the study visit. Serum electrolytes, liver enzymes, CD4 cell count, HIV-1 RNA, glucose, insulin, lipid panel, blood lactate were measured per AIDS Clinical Trials Group (ACTG) guidelines (http://aactg.s-3.com/members/download/other/Metabolic/VenousLactateSOP.doc). The homeostasis model assessment of insulin resistance (HOMA-IR) index was calculated from fasting plasma glucose and insulin values (Matthews et al., 1985). Peripheral blood and mononuclear cell (PBMC) were isolated and a 6-mm skin punch biopsy in the upper outer quadrant of the buttocks was performed. The fat samples were separated from the overlying skin and placed in a −70°C freezer until batch processing at the end of study. Additional visits at weeks 4, 8 and 12 were for the purpose of clinical and laboratory monitoring for toxicities or intolerance. NucleomaxX was provided by Pharma Trade Healthcare AB, Spanga, Sweden and was given free of charge to study participants.
Clinical lipoatrophy and lipohypertrophy scores
The questionnaire had to be filled separately and independently by patients and physicians and asked for a rating of fat loss or fat accumulation in predefined body areas. The lipoatrophy score was based on questions about fat loss in the arms, legs, buttocks and the face. Assessments within each of these sites were rated as ‘0’ for ‘absent’; ‘1’ for ‘mild’; ‘2’ for ‘moderate’ and ‘3’ for ‘severe’. Thus, the lipoatrophy score could vary between 0 and 12.
For the lipohypertrophy score, a similar scoring was derived by the addition of the points for answers about fat gain in the following areas (abdomen, neck, breasts and body lipomas). The total lipohypertrophy score could vary between 0 and 12.
DNA isolation and mtDNA assays
The mtDNA assays were performed on batched specimens at the end of the study as described previously (Setzer et al., 2005). The laboratory personnel were blinded to all sample characteristics. Briefly, total DNA was extracted with the QIAamp DNA isolation kit (Quiagen, Hilden, Germany). MtDNA and nuclear DNA (nDNA) copy numbers were determined by quantitative PCR using the ABI 7700 sequence detection system (Applied Biosystems, Foster City, CA, USA). The mtDNA ATP-6 gene was amplified and quantified with a FAM-fluorophore-labeled probe. For the quantification of nDNA, exon 8 of the glyceraldehyde- 3-phosphate dehydrogenase gene was amplified and quantified using a VIC-fluorophore-labeled probe. Amplifications of mitochondrial and nuclear products were performed separately in optical 96-well plates (Applied Biosystems). All samples were run in triplicate. Absolute mtDNA and nDNA copy numbers were calculated using serial dilutions of plasmids with known copy numbers of mtDNA and nDNA (Setzer et al., 2005). The standard deviation of triplicates was between 2 and 10% maximum. The standard deviation of measurements of triplicates repeated from the same template after 3 months was between 10 and 20%.
Statistical considerations and methods
Continuous measures are summarized with medians and ranges; nominal variables are expressed as frequencies and percents. Differences from baseline at both 16 and 32 weeks were calculated for each subject. The significance of change was tested using the Wilcoxon signed rank test, a non-parametric paired test. Spearman rank correlation coefficients were used to describe the relationship between two continuous variables. The level of significance was set at 0.05 for all analyses. All analyses were carried out using SAS, v.8.2 (The SAS Institute, Carey, NC, USA).
Results
Patient characteristics
A total of 16 patients were screened and entered the study. The demographics, clinical and laboratory parameters of the study participants are shown in Table 1. Most were male (69%), 50% were white. The median (range) age was 49 years (range 39–63). Median (range) baseline body mass index was 25 (19–30) kg/m2 and CD4+ cell count was 556 cells/mm3 (range 222–1636). HIV-1 RNA was <50 copies/ml in 13 patients; 88 copies/ml in one and 2100 copies/ml in another subject with very resistant virus. Another patient had 14 000 copies/ml; during the study, he admitted to a breach in adherence to antiretrovirals between screening and study entry.
Table 1.
Baseline demographic and laboratory characteristics of all study subjects (n=16)
| n (%) or median (range) | |
|---|---|
| Age (years) | 49 (39–63) |
| Sex (male) | 11 (69%) |
| Race | |
| White | 8 (50%) |
| African-Americans | 7 (44%) |
| Asian | 1 (6%) |
| Duration of HIV (months) | 105 (33–235) |
| Duration of NRTI therapy (months) | 82 (33–216) |
| Duration of stavudine therapy (months) | 48 (9–120) |
| ALT (U/l) | 47 (30–108) |
| Creatinine kinase (U/l) | 126 (33–369) |
| Hemoglobin (g/dl) | (12.9–18.6) |
| CD4+ cell count (cells/mm3) | 556 (222–1636) |
| HIV-1 RNA <50 copies/ml | 13 (81%) |
| Antiretrovirals used at study entry | |
| Stavudine | 16 (100%) |
| Lamivudine | 10 (62.5%) |
| Abacavir | 2 (12.5%) |
| Tenofovir | 2 (12.5%) |
| PI | 8 (50%) |
| NNRTI | 9 (56%) |
| Enfuvirtide | 1 (6%) |
Abbreviations: ALT, alanine aminotransferase; HIV-1, human immunodeficiency- 1; NNRTI, non-nucleoside reverse transcriptase inhibitor, NRTI, nucleoside reverse transcriptase inhibitor.
Six subjects had chronic hepatitis C, one of them with concurrent hepatitis B. In addition, one individual had a prior episode of cholestatic hepatitis of unclear etiology. Three subjects had systemic arterial hypertension and were on antihypertensives. Two subjects were on lipid-lowering agents including a statin (with a fibrate in one and fish oil in the other).
Fourteen subjects completed the study. Two subjects dropped out before week 4 due to schedule conflicts. No subject modified his antiretrovirals during the trial. Subjects who were receiving antihypertensives and hypolipemic agents continued them unchanged throughout the study. Adherence to study medication, as assessed by sachet count was >95% in all but one subject. The latter is the same subject who enrolled with HIV-1 RNA of 14 000 copies/ml and who also reported intermittent suboptimal adherence to antiretrovirals. His adherence to NucleomaxX was calculated by sachet counts at 50%.
Clinical and laboratory results
At study entry, fat mtDNA was reduced; the values for fat mtDNA from HIV-negative controls is reported in the literature as 851–921 copies/cell (Nolan et al., 2003;McComsey et al., 2005) and the values in subjects with HIV lipoatrophy as 194 copies/cell (Nolan et al., 2003; Setzer et al., 2005). Fat mtDNA correlated negatively with HIV duration (rs = −0.56; P=0.02) and with NRTI duration (rs = −0.50; P=0.04). There was no correlation between PBMC and fat mtDNA levels (rs = −0.19; P=0.47).
There was a significant positive correlation between the patient-generated lipoatrophy score and the physician lipoatrophy score (rs= −0.80; P=0.0002). There were strong negative correlations between fat mtDNA levels and both lipoatrophy scores generated by the patients or the physician (rs= −0.82; P<0.0001 and rs= −0.64; P=0.008, respectively, Figure 1). No correlation was found between PBMC mtDNA and either of the lipoatrophy scores.
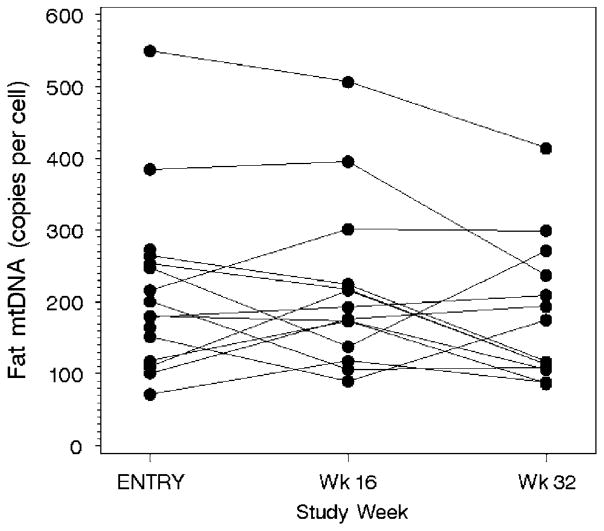
Figure 1.
Fat mtDNA content at baseline, at week 16 (on NucleomaxX) and at week 32 (off NucleomaxX). MtDNA, mitochondrial DNA.
No adverse events were noted throughout the study. In particular, no diarrhea was observed, even in two subjects with previous irritable colon and intermittent chronic diarrhea. These subjects entered the study without diarrhea and remained diarrhea-free throughout the study. Systolic and diastolic blood pressure was not affected. The values of the main metabolic indices are summarized in Table 2. Creatinine kinase, ALT and hemoglobin levels did not change. CD4+ cell counts (mean±s.d.) were 556 (222–1636) cells/mm3 at baseline, 582 (171–1156) cells/mm3 at week 16 and 562 (206–1112) cells/mm3 at week 32 (P>0.1). Overall, HIV-1 RNA did not change significantly; all subjects with HIV-1 RNA below 50 copies/ml remained at the same virologic level throughout the study. The patients with HIV-1 RNA of 88 and 2100 copies/ml at study entry finished the study at 119 and 2930 copies/ml, respectively. The patient with HIV-1 RNA of 14 000 copies/ml finished the study at 123 copies/ml. All subjects remained on the same antiretroviral throughout the study.
Table 2.
Metabolic variables at baseline, at week 16 (on NucleomaxX) and at week 32 (off NucleomaxX)
| Parameter median (range) | Baseline (n=16) | Week 16 (n=14) | Week 32 (n=14) |
|---|---|---|---|
| BMI (kg/m2) | 25.0 (19–30) | 25.0 (21.5–29.3) | 24.6 (20.5–29.3) |
| Lipoatrophy score (by patient) | 7.5 (4–12) | 4 (0–12)* | 4 (0–12)* |
| Lipoatrophy score (by physician) | 9.5 (4–12) | 6.5 (0–10)* | 5.5 (0–12)* |
| Lipohypertrophy score (by patient) | 2.0 (0–12) | 1 (0–10)* | 1.5 (0–9)* |
| Lipohypertrophy score (by physician) | 1.0 (0–9) | 0 (0–7) | 1.0 (0–6)* |
| Lactate (mmol/l) | 0.9 (0.7–2.0) | 1.1 (0.6–2.0) | 1.0 (0.5–2.2) |
| Insulin (mIU/l) | 7.8 (3.0–24.0) | 8.0 (2.4–28.0) | 7.2 (4–23) |
| HOMA-IR | 0.95 (0.40–1.80) | 1.00 (0.40–3.60) | 0.90 (0.50–3.10) |
| Glucose (mg/dl) | 83 (66–99) | 81 (64–100) | 80 (63–119) |
| Cholesterol (mg/dl) | 196 (123–333) | 189 (134–358) | 178 (133–350) |
| Non-HDL cholesterol(mg/dl) | 146 (101–292) 143 | (95–315) | 136 (97–302) |
| Triglycerides (mg/dl) | 142 (53–297) | 130 (48–634) | 113 (52–265) |
| Fat mtDNA (copies/cell) | 190 (72–549) | 185 (90–506) | 146 (86–414) |
| PBMC mtDNA (copies/cell) | 95 (44–253) | 117 (40–256) | 120 (50–229) |
Abbreviations: BMI, body mass index; HDL, high-density lipoprotein; HOMA-IR, homeostasis model assessment of insulin resistance; mtDNA, mitochondrial DNA; PBMC, peripheral blood mononuclear cell.
P<0.05 for change from baseline.
Lipoatrophy scores improved noticeably. Median (range) changes in lipoatrophy scores from baseline to week 16 and from baseline to week 32 were both significant (−3 (−8 to +3); P=0.02 and −2.5 (−8 to +3); P=0.02, respectively for patient-generated scores and −3 (−7 to +1); P<0.001 and −4 (−6 to 0); P=0.001, respectively for physician-generated scores). There were no significant changes in lipoatrophy scores between week 16 and 32. At these time points, the strong initial correlation between fat mtDNA and lipoatrophy scores was no longer detectable.
Lipohypertrophy scores also improved during the study, regardless of whether subjects were on concomitant protease inhibitor therapy. Median (range) reductions in lipohypertrophy scores from baseline to week 16 and to week 32 were both significant (−1 (−5 to +1); P=0.008 and −1(−4 to 0); P=0.04, respectively for patient-generated scores and −1 (−4 to 0); P=0.004 and −1(−3 to +1); P=0.03, respectively for physician-generated scores). Between weeks 16 and 32, the lipohypertrophy scores did not change significantly.
Figure 1 shows a subject-by-subject plot of fat mtDNA levels during the study. Fat mtDNA was unchanged throughout the study from median (range) of 190 (72–549) at study entry to 185 (90–506) and 146 (86–414) at week 16 and week 32, respectively (P=0.86 for changes from week 0 to 16; P=0.76 for changes from baseline to week 32; P=0.24 for changes from week 16 to 32). Similarly, no changes were seen in PBMC mtDNA copy numbers; from median (range) of 95 (44–253) at study entry to 117 (40–256) at week 16 and 120 (50–229) at week 32 (P=0.98 for changes from week 0 to 16; P=0.95 for changes from baseline to week 32; P=0.08 for changes from week 16 to 32).
Fasting triglycerides, total and HDL cholesterol levels and the use of lipid-lowering agents remained unchanged, as were glucose, insulin levels and HOMA-IR (Table 2). One subject had fasting glucose of 119 mg/dl at week 32, but repeat glucose performed within a week was 98 mg/dl.
Discussion
This is a pilot study to explore the safety and effect on mitochondrial indices of pyrimidine precursors in HIV-infected subjects with lipoatrophy. We found that uridine supplementation was well tolerated, safe and did not affect HIV or metabolic indices. Contrary to in vitro observations, uridine supplementation did not lead to changes in mtDNA levels in blood or subcutaneous adipose tissue.
So far, only switching away from thymidine analogues (namely stavudine) has been shown to be effective in reversing lipoatrophy (McComsey et al., 2004; Moyle et al., 2006). The effects in these studies were small and only detected by sensitive objective fat measurements (Martin et al., 2004; McComsey et al., 2004). Thus, interventions such as uridine supplementation are needed and a large study (ACTG 5229) investigating its use for HIV lipoatrophy setting has started.
We recognize the limitations of the pilot study due to its open-labeled nature, its small sample size, the lack of a control group and objective body fat measurements. However, the study was powered to look at changes in fat mtDNA levels. Other investigators have analyzed the diagnostic accuracy of lipoatrophy questionnaires and found strong correlations with objective measurements of fat content (Bacchetti, 2005). In a separate study conducted by one of the authors (GM), there was a strong negative correlation between dual-energy X-ray absorptiometry-measured limb fat and lipoatrophy scores generated either by the patients or the physician (r= −0.58; P=0.0075 and r= −0.59; P=0.0058, respectively) (McComsey et al., 2005). The fact that fat mtDNA in our study strongly correlated with the initial lipoatrophy score provides additional validity to the clinical assessment. Additionally, the patient- and physician-derived lipoatrophy scores correlated strongly despite their independent assessment. Thus, we believe that although no objective measurements were performed in this pilot study, the finding that patients and physicians reported clinical improvement is interesting and deserves further investigations of NucleomaxX in lipoatrophy.
The absence of change in fat mtDNA during the study is at first glance surprising. However, the results do not exclude the possibility that pyrimidine depletion (and not other consequences of the respiratory chain impairment such as decreased ATP-synthesis and altered redox-potential) may be the limiting factor for the survival and/or fat storage of adipocytes. This view may open new avenues in the understanding of the pathogenesis of lipoatrophy and is also supported by the observation that stavudine-exposed adipocytes exhibited fat recovery at lower uridine concentrations (10–50 μM), compared to those needed for improvement of mtDNA content (100 μM) (Walker et al., 2006). A similar temporal behavior of mitochondrial toxicity by uridine treatment was also observed in stavudine-exposed hepatocytes and adipocytes (Walker et al., 2003, 2006). In vitro, uridine improved the toxic phenotype rapidly (within 5 days), whereas the onset of the toxic phenotype was much more prolonged (15 days). The experimental results are therefore compatible with the view that uridine resets the ‘mitochondrial clock’ and together with the long intracellular half-life of uridine (13–18 h) (Walker and Venhoff, 2005) provided the rationale for the intermittent dosing of NucleomaxX. Given the known safety profile of uridine (van Groeningen et al., 1986; Fox et al., 1999) and the uridine pharmacokinetics of NucleomaxX, dosing was three times per day in order to maximize uridine serum concentrations during the ‘on’ cycle (Venhoff et al., 2005).
As noted above, mechanisms independent of mtDNA changes need to be explored. This work is currently ongoing and includes obtaining larger samples of fat tissue in order to assess fat apoptosis, tissue morphology, oxidative biomarkers and mitochondrial function (NIH AI60484; G McComsey, PI).
Although the safety profile of oral uridine or its prodrugs was reportedly excellent in HIV-negative subjects (Webster et al., 1979; Manis et al., 1997), it is important to rule out a negative effect of uridine or its metabolites on the ability of antiretroviral nucleoside analogues to inhibit HIV reverse transcriptase. Although such effect was previously excluded for a number of NRTI and their combinations in phenotypic HIV-resistance assays and for zidovudine in an animal model (Sommadossi et al., 1988; Calabresi et al., 1990; Koch et al., 2003), studies in HIV-infected subjects are needed. Consistent with the prior preclinical data, our pilot study failed to detect a significant interaction between uridine and the antiretroviral efficacy. The long-term safety of NucleomaxX on the antiviral activity of NRTIs will need to be confirmed in larger and longer studies.
Although lipoatrophy is frequently associated with insulin resistance and dyslipidemia, the sugarcane-derived dietary supplement did not have effects on glucose metabolism or serum lipid levels. The good safety profile of NucleomaxX in our study is consistent with the findings of another recently reported small study of NucleomaxX in HIV lipoatrophy (Sutinen et al., 2005); in which in contrast to what we are reporting HDL cholesterol decreased slightly.
In summary, in this pilot study, NucleomaxX was given for 16 weeks to a group of HIV-infected subjects with lipoatrophy and found to be well tolerated, safe, without deleterious effect on lipids or glucose metabolism. Subjective improvement of lipoatrophy was noted by study subjects and physicians. In contrast, no changes in fat or PBMC mtDNA levels were detected. A large randomized, doubleblind and placebo-controlled study using objective measurements of body composition is ongoing.
Acknowledgments
This work was supported by NIAID AI-060484 (GM), and was partially funded by Bristol Myers Squibb Co. This study was presented at the 7th International Workshop on Adverse Drug Reactions and Lipodystrophy in HIV, 13–17 November 2005, Dublin, Ireland.
Footnotes
Role of the funding source: The funding source had no role in the collection, analysis or interpretation of the data or in the decision to submit the paper for publication. Dr McComsey serves as consultant for Bristol Myers Squibb Co. Dr UA Walker has applied for patents regarding the use of uridine or its precursors in lipodystrophy. He also serves as a consultant for the company that produces NucleomaxX.
References
- Bacchetti P, Gripshover B, Grunfeld C, Heymsfield S, McCreath H, Osmond D, et al. Study of fat redistribution and metabolic change in HIV infection (FRAM): Fat distribution in men with HIV infection. J Acquir Immune Defic Syndr. 2005;40:119–120. doi: 10.1097/01.qai.0000182230.47819.aa. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodnar AG, Cooper JM, Leonard JV, Schapira AH. Respiratory-deficient human fibroblasts exhibiting defective mitochondrial DNA replication. Biochem J. 1995;305:817–822. doi: 10.1042/bj3050817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabresi P, Falcone A, St Clair MH, Wiemann MC, Chu SH, Darnowski JW. For the trial to assess the regression of hyperlactatemia to evaluate the regression of established lipodystrophy in HIV-1-positive subjects (TARHEEL; ESS 4001 0) Study Team (1990). Benzylacyclouridine reverses azidothymidine-induced marrow suppression without impairment of anti-human immunodeficiency virus activity. Blood. 76:2210–2215. [PubMed] [Google Scholar]
- Fox RI, Herrmann ML, Frangou CG, Wahl GM, Morris RE, Strand V, et al. Mechanism of action of leflunamide in rheumatoid arthritis. Clin Immunol. 1999;93:198–208. doi: 10.1006/clim.1999.4777. [DOI] [PubMed] [Google Scholar]
- http://aactg.s-3.com/members/download/other/Metabolic/Venous-LactateSOP.doc.
- Joly V, Flandre P, Meiffredy V, Leturque N, Harel M, Aboulker JP, et al. Increased risk of lipoatrophy under stavudine in HIV-1- infected patients: results of a substudy from a comparative trial. AIDS. 2002;16:2447–2454. doi: 10.1097/00002030-200212060-00010. [DOI] [PubMed] [Google Scholar]
- Koch EC, Schneider J, Weiss R, Penning B, Walker UA. Uridine excess does not interfere with the antiretroviral efficacy of nucleoside analogue reverse transcriptase inhibitors. Antivir Ther. 2003;8:485–487. [PubMed] [Google Scholar]
- Leyva A, van Groeningen CJ, Kraal I, Gall H, Peters GJ, Lankelma J, et al. the Rave Study Group. Phase I and pharmacokinetic studies of high-dose uridine intended for rescue from 5-fluorouracil toxicity. Cancer Res. 1984;44:5928–5933. [PubMed] [Google Scholar]
- Mallal SA, John M, Moore CB, James IR, McKinnon EJ the Adult AIDS Clinical Trials Group. Contribution of nucleoside analogue reverse transcriptase inhibitors to subcutaneous fat wasting in patients with HIV infection. AIDS. 2000;14(10):1309–1316. doi: 10.1097/00002030-200007070-00002. [DOI] [PubMed] [Google Scholar]
- Manis FR, Cohn LB, McBride-Chang C, Wolff JA, Kaufman FR. A longitudinal study of cognitive functioning in patients with classical galactosemia, including a cohort treated with oral uridine. J Inherit Metab Dis. 1997;20:549–555. doi: 10.1023/a:1005357622551. [DOI] [PubMed] [Google Scholar]
- Martin JL, Brown CE, Matthews-Davis N, Reardon JE. Effects of antiviral nucleoside analogues on human DNA polymerases and mitochondrial DNA synthesis. Antimicrob Agents Chemother. 1994;38:2743–2749. doi: 10.1128/aac.38.12.2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A, Smith D, Carr A, Ringland C, Amin J, Emery S, et al. Reversibility of lipoatrophy in HIV-infected patients 2 years after switching from a thymidine analogue to abacavir: the MITOX Extension Study. AIDS. 2004;18:1029–1036. doi: 10.1097/00002030-200404300-00011. [DOI] [PubMed] [Google Scholar]
- Matthews DR, Hosker JP, Rudenski AS, Naylor BA, Treacher DF, Turner RC. Homeostasis model assessment: insulin resistance and beta cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia. 1985;28:412–419. doi: 10.1007/BF00280883. [DOI] [PubMed] [Google Scholar]
- McComsey GA, Ward DJ, Hessenthaler SM, Sension MG, Shalit P, Lonergan TJ, et al. Improvement in HAART-associated lipoatrophy in HIV-infected patients switched from stavudine to abacavir or zidovudine: The results of TARHEEL. Clin Infect Dis. 2004;38:263–270. doi: 10.1086/380790. [DOI] [PubMed] [Google Scholar]
- McComsey GA, O’Riordan M, Storer N, Goldman S, Ganz J, Libutti DE, et al. Clinical lipoatrophy assessment strongly correlates with DEXA-measured limb fat and subcutaneous fat mitochondrial DNA levels. Abstract 142. Program and Abstracts of the 13th Conference on Retroviruses and Opportunistic Infections; 5–9 February 2005; Denver, Colorado. 2005. [Google Scholar]
- McComsey GA, Paulsen DM, Lonergan TJ, Hessenthaler SM, Hoppel CL, Williams VC, et al. Improvements in lipoatrophy, mitochondrial DNA content and adipose tissue apoptosis levels after replacement of stavudine with either abacavir or zidovudine. AIDS. 2005;19:15–23. doi: 10.1097/00002030-200501030-00002. [DOI] [PubMed] [Google Scholar]
- Moyle GJ, Sabin CA, Cartledge J, Johnson M, Wilkins E, Churchill D, et al. RAVE (Randomized Abacavir versus Viread Evaluation) Group UK: a randomized comparative trial of tenofovir DF or abacavir as replacement for a thymidine analogue in persons with lipoatrophy. AIDS. 2006;20:2043–2050. doi: 10.1097/01.aids.0000247574.33998.03. [DOI] [PubMed] [Google Scholar]
- Nolan D, Hammond E, Martin A, Taylor L, Hermann S, McKinnon E, et al. Mitochondrial DNA depletion and morphologic changes in adipocytes associated with nucleoside reverse transcriptase inhibitor therapy. AIDS. 2003;17:1329–1338. doi: 10.1097/00002030-200306130-00007. [DOI] [PubMed] [Google Scholar]
- Setzer B, Schlesier M, Walker UA. Effects of didanosine-related depletion on mtDNA in human T lymphocytes. J Infect Dis. 2005;191:848–855. doi: 10.1086/427655. [DOI] [PubMed] [Google Scholar]
- Sommadossi JP, Carlisle R, Schinazi RF, Zhou Z. Uridine reverses the toxicity of 3′-azido-3′ deoxythymidine in normal human granulocyte-macrophage progenitor cells in vitro without impairment of antiretroviral activity. Antimicrob Agents Chemother. 1988;32:997–1001. doi: 10.1128/aac.32.7.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutinen J, Walker UA, Sevastianova K, Hakkinen AM, Ristola M, Yki-Jarvinen H. Uridine supplementation increases subcutaneous fat in patients with HAART-associated lipodystrophy: a randomized placebo controlled trial. Abstract 7. The 7th Workshop on Adverse Drug Reactions and Lipodystrophy in HIV; 13–16 November 2005; Dublin, Ireland. 2005. [Google Scholar]
- Tebas P, Zhang J, Yarasheski K, Evans S, Fischl M, Shevitz A, et al. Switch to a protease inhibitor-containing/nucleoside reverse transcriptase inhibitor-sparing regimen increases appendicular fat and serum lipid levels without affecting glucose metabolism or bone mineral density. The results of a prospective randomized trial, ACTG 5125s. Program and abstracts of the 12th Conference on Retroviruses and Opportunistic Infections; 22–25 February 2005; Boston, Massachusetts. 2005. Abstract 40. [Google Scholar]
- van Groeningen CJ, Leyva A, Kraal I, Peters GJ, Pinedo HM. Clinical and pharmacokinetic studies of prolonged administration of high-dose uridine intended for rescue from 5-FU toxicity. Cancer Treat Rep. 1986;70:745–750. [PubMed] [Google Scholar]
- Venhoff N, Zilly M, Lebrecht D, Schirmer D, Klinker H, Thoden J, et al. Uridine pharmacokinetics of mitocnol a sugar cane extract. AIDS. 2005;19:739–740. doi: 10.1097/01.aids.0000166101.44262.52. [DOI] [PubMed] [Google Scholar]
- Walker UA, Auclair M, Lebrecht D, Kornprobst M, Capeau J, Caron M. Uridine abrogates the adverse effects of antiretroviral pyrimidine analogues on adipose cell functions. Antivir Ther. 2006;11:25–34. [PubMed] [Google Scholar]
- Walker UA, Bickel M, Lütke Volksbeck SI, Ketelsen UP, Schöfer H, Setzer B, et al. Evidence of nucleoside analogue reverse transcriptase inhibitor-associated genetic and structural defects of mitochondria in adipose tissue of HIV-infected patients. J Acquir Immune Defic Syndr. 2002;29:117–121. doi: 10.1097/00042560-200202010-00002. [DOI] [PubMed] [Google Scholar]
- Walker UA, Langmann P, Miehle N, Zilly M, Klinker H, Petschner F. Beneficial effects of oral uridine in mitochondrial toxicity. AIDS. 2004;18:1085–1086. doi: 10.1097/00002030-200404300-00025. [DOI] [PubMed] [Google Scholar]
- Walker UA, Venhoff N. Uridine in the prevention and treatment of NRT1-induced mitochondrial toxicity. Antivir Ther. 2005;10:M117–M123. [PubMed] [Google Scholar]
- Walker UA, Venhoff N, Koch E, Olschweski M, Schneider J, Setzer B. Uridine abrogates mitochondrial toxicity related to nucleoside analogue reverse transcriptase inhibitors in HepG2 cells. Antivir Ther. 2003;8:463–470. [PubMed] [Google Scholar]
- Webster DR, Simmonds HA, Potter CF, Becroft DM. Purine and pyrimidine metabolism in hereditary orotic aciduria during a 15 year follow up study. Adv Exp Med Biol. 1979;122B:203–208. doi: 10.1007/978-1-4684-8559-2_34. [DOI] [PubMed] [Google Scholar]