Abstract
This study was conducted to isolate Salmonella Enteritidis from poultry samples and compare their virulence and antibiotic resistance profiles to S. Enteritidis isolated from outbreaks in northern Jordan. Two hundred presumptive isolates were obtained from 302 raw poultry samples and were subjected to further analysis and confirmation. A phylogenic tree based on 16S rRNA sequencing was constructed and selected isolates representing each cluster were further studied for their virulence in normal adult Swiss white mice. The most virulent strains were isolated from poultry samples and had an LD50 of 1.55 × 105 CFU, while some of the outbreak isolates were avirulent in mice. Antibiotic resistance profiling revealed that the isolates were resistant to seven of eight antibiotics screened with each isolate resistant to multiple antibiotics (from two to six). Of the poultry isolates, 100%, 88.9%, 77.8%, 66.7%, and 50% showed resistance to nalidixic acid, ciprofloxacin, ampicillin, cephalothin, and cefoperazone, respectively. Two outbreak isolates were sensitive to all tested antibiotics, while 71.4% were resistant to cefoperazone and only 28.6% showed resistance to nalidixic acid. Salmonella outbreak isolates were genetically related to poultry isolates as inferred from the 16S rRNA sequencing, yet were phenotypically different. Although outbreak strains were similar to poultry isolates, when tested in the mouse model, some of the outbreak isolates were highly virulent while others were avirulent. This might be due to a variation in susceptibility of the mouse to different S. Enteritidis isolates.
Keywords: Salmonella Enteritidis, poultry, antimicrobial susceptibility, virulence, LD50, PCR detection
Introduction
Foodborne salmonellosis is a persistent concern, causing public health problems leading to major economic losses in almost all countries despite vast improvements in hygienic processing of poultry and its products.1,2 Salmonella enterica and Campylobacter are considered the most important zoonotic agents implicated in foodborne illness as inferred from the high number of outbreaks caused by these pathogens.3,4 There are 6 subspecies of S. enterica with the vast majority of human infections caused by strains belonging to subspecies I, which exhibit high variability in virulence, host adaptation and host specificity among its members.5 Therefore, depending on serovars, cases of salmonellosis can differ in severity.6
The worldwide incidence of non-typhoidal salmonellosis is estimated at 1.3 billion cases with 3 million deaths annually.7 In 2009, about 36 000 salmonellosis cases were reported to the Centers for Disease Control and Prevention (CDC).8 In Europe, in 2006 and 2007 Salmonella was the second most commonly reported pathogen with 160 649 and 151 995 cases of salmonellosis respectively.9,10 Within the genus Salmonella, S. Enteritidis and S. Typhimurium serovars are considered the major etiologic agents of foodborne illnesses.11 Poultry products still represent one of the major sources of salmonellosis worldwide12 with horizontal transmission as the major contributing factor of Salmonella contamination of the poultry products and their processing plants.13
In Jordan, several Salmonella outbreaks associated with consumption of contaminated poultry and mayonnaise are reported yearly (Jordan Food and Drug Administration [JFDA], personal communication). However, no exact figures can be obtained from Jordanian health authorities.
Although salmonellosis is a self-limiting disease among healthy people, it causes serious illness and fatalities among the immunocompromised and elderly, especially if the pathogen is multidrug-resistant.14 If transmission of Salmonella occurs between different hosts, genetic changes affecting the virulence of these pathogens could arise.15 Isolating and characterizing pathogenic bacteria is essential for understanding and managing bacterial diseases. When outbreaks of human salmonellosis occur, it is important for clinicians to be able to verify the virulence of associated isolates, particularly isolates from the suspected vehicle of infection.16 LD50 assays are often used as a standard measure of virulence.16,17 This test becomes important particularly when multiple, potentially pathogenic microorganisms are isolated from the implicated samples. To pinpoint an implicated pathogen, the mouse pathogenicity testing is used as a reliable means for differentiating virulent from avirulent strains. In this test, the pathogen dose causing 50% mortality of test animals (often mice) within a week is calculated.
Resistance to antibiotics can occur in any bacterial species where the pathogen might alter its permeability to the antibiotic, degrade the antibiotic, cause its efflux or lead to its inactivation by enzymatic means.18 Antibiotic resistance rates are reported to be rising for Salmonella worldwide with differing rates for various countries.19 The widespread use of antibiotics in poultry feed at suboptimal doses as growth promoting agents and their prophylactic use have likely contributed to the emergence of multidrug-resistant zoonotic pathogens, including Salmonella, which can be transmitted to humans via the food chain.20 To better understand this issue, it is important to study the patterns and prevalence of bacterial antimicrobial resistance toward the more commonly used antibiotics.
The objectives of this study were to: (1) compare the poultry isolates with S. Enteritidis isolates from recent salmonellosis outbreaks in Jordan for their virulence potential using a mouse pathogenicity assay, and (2) study and compare the antibiotic resistance patterns of confirmed S. Enteritidis isolates from poultry to those of outbreak isolates from humans.
Results
Isolation and biochemical identification of Salmonella spp.
Isolation of Salmonella starts by using the selective and differential media which is then followed by biochemical identification. In this study, two hundred (66%) of the 302 fresh poultry meat samples gave typical Salmonella colony morphologies on XLD, BGA, and S-S agar plates. Confirmation of bacterial identities using the Remel RapID™ ONE system indicated that 145 (72.5%) isolates were likely (≥60% probability) Salmonella serovars while the remaining 48 isolates (24%) were identified as Citrobacter freundii and seven isolates (3.5%) were Proteus spp. All Salmonella serovars were S. enterica subspecies enterica (i.e., subspecies I) and utilized glucose, mannitol, and dulcitol; were unable to use lactose, sucrose, salicin, or urea and were H2S as well as ONPG positive.21 Of 43 isolates from this group (Salmonella other than subspecies I), 28 were tentatively identified as S. Choleraesuis, seven as S. Paratyphi, and there were four isolates each of S. Typhi and S. Pullorum. Similarly, all the seven outbreak isolates were subjected to the same procedure and were confirmed as S. Enteritidis.
Identification of Salmonella and S. Enteritidis by PCR
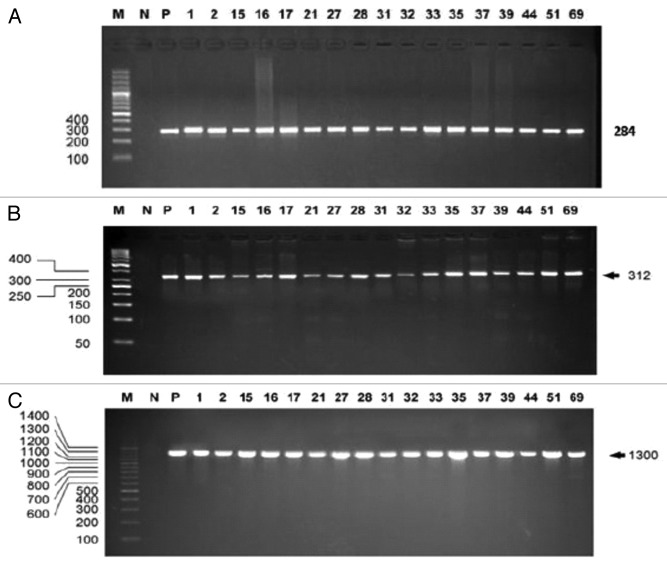
Strain identification is essential for effective investigation of outbreak sources as well as the types of microorganisms contaminating a food product. To confirm the identity of presumptive Salmonella isolates, PCR amplification was performed using universal primers targeting the InvA gene present in all Salmonella species as described by Rahn et al.22 Of the 200 presumptive Salmonella food isolates and the seven outbreak isolates, 180 tested positive using Salmonella-specific PCR assay targeting the InvA gene and amplifying a 284-bp fragment (Fig. 1A). In addition, primers targeting part of the sefA gene23 of S. Enteritidis (Fig. 1B) confirmed 45 isolates as S. Enteritidis while the rest tested negative (Table 1).
Figure 1. A 2% agarose gel showing the PCR amplification of a 284 bp, 312 bp and 1300 bp fragment from the InvA gene (A), SefA gene (B), and the 16S rRNA gene (C) respectively. M, ladder marker (Biobasic); N, non-template negative control; P, positive control (DNA from S. Enteritidis).
Table 1. A summary of the RemelRapID™ ONE results, PCR and 16S rRNA sequencing of Salmonella isolates from poultry and outbreak.
| RemelRapID™ ONE | Salmonella universal PCR | S. Enteritidis specific PCR | 16S rRNA identification | |
|---|---|---|---|---|
| No. of positive samples (%) | 152 (73.4%) | 180 (87%) | 45 (21.7%) | 25 (55.6%) |
| No. of negative samples (%) | 55 (26.6%) | 27 (13%) | 162 (78.3%) | 20 (44.4%) |
DNA sequencing of the 16S rRNA gene
To further confirm the identity of the 45 S. Enteritidis, the 16S rRNA gene was amplified using the universal primers Lwp58 and Lwp57.24 A fragment of approximately 1300 bp of the 16S rRNA gene was amplified from all isolates (Fig. 1C). The 16S rRNA gene PCR products of all S. Enteritidis isolates were sequenced using the same primers and their sequences were aligned and compared with Salmonella and non-Salmonella reference sequences. Based on a sequence match of 95% or more with any of the reference sequences, 18 of 38 poultry isolates were identified as S. Enteritidis in addition to all seven outbreak isolates.
Phylogenetic analysis of the confirmed S. Enteritidis isolates
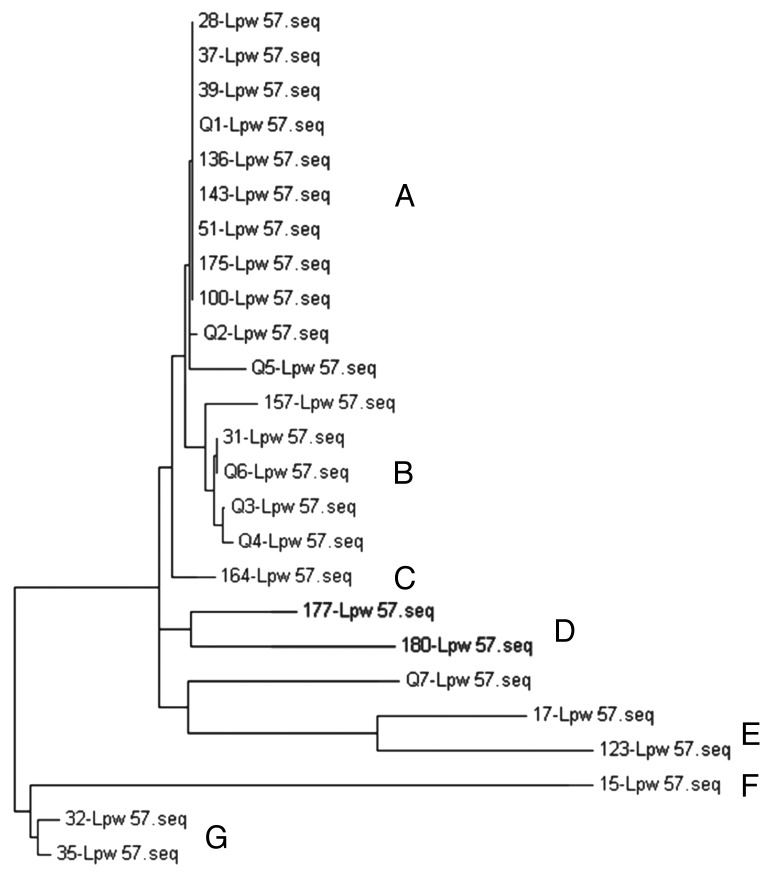
To understand the diversity of the S. Enteritidis isolates, phylogenetic analysis based on the 16S rRNA sequences was conducted. The 25 S. Enteritidis isolates fall into 7 distinct 16S rRNA clusters designated A–G (Fig. 2). Clusters A, B, D, E, and G contained more than one isolate, while clusters C and F each contained only one isolate. Cluster A was a group of 11 S. Enteritidis strains that contained 44% of the isolates. The remaining clusters contained 20% (cluster B), 12% (cluster E), 16% (clusters D and G), and 8% (cluster F and C) of the isolates. Both the outbreak and poultry isolates co-clustered in groups A, B, and E indicating similar phylogeny, while the other groups contained only the food isolates.
Figure 2. 16S rRNA gene phylogenetic tree of S. Enteritidis isolates. A neighbor-joining analysis was used with Felsenstein correction (500 bootstrap replicates). Letters indicate the phylogenetic group while Lpw denotes the name of the primer used for the 16S rRNA sequencing.
Virulence testing in mice
To understand variation in virulence of the S. Enteritidis isolates, representatives from both poultry and outbreak S. Enteritidis isolates were tested in a normal mouse model for their virulence by LD50 determination. The experiment was conducted at three separate times (Table 2) and results were the average of the three experiments. Four of the seven poultry isolates (15, 17, 28, and 32) were avirulent while three isolates (157, 164, and 180) exhibited strong virulence with percentages of killed mice ranging from 86.7 to 100 in each group. In contrast, only one isolate (Q1) was virulent among the chosen three outbreak isolates, which killed an average of 86.7% mice in each group. The other two outbreak strains were avirulent since isolate Q3 failed to kill more than two mice (average 46.7%) in any group while Q5 failed to kill more than one mouse (20%) in any group. When the LD50 was calculated for these strains, the values for the poultry isolates ranged from 1.07 × 106 CFU for isolate 15 to a non-lethal dose (failed to kill more than one mouse) for isolate 35, while the LD50 for the virulent poultry strains ranged from 1.55 × 105 (isolate 157) to 3.24 × 105 CFU for isolate 164. When the LD50 values were determined for outbreak isolates, the values ranged from 2.65 × 106 CFU for isolate Q5 to a non-lethal dose (failed to kill mice) for isolate Q3. Additionally, the LD50 for the virulent outbreak strain Q1 was 4.5 × 105 CFU. In general, the death rate of mice injected with isolate 157 and 164 was significantly (P < 0.05) higher than that of mice injected with other isolates. Moreover, the effect of isolates 35 and Q3 on the viability of injected mice was insignificant (P < 0.05).
Table 2. Results for mouse virulence test and LD50 for selected outbreak and poultry strains of S. Enteritidis in normal mice.
| Isolate ID | Origin of isolate | *Average % of deaths of mice given 107 CFU/mouse and observed for 1 wk£ | Virulence§ | LD50 |
|---|---|---|---|---|
| 15 | Poultry | 53.3abc ± 50.3 | None virulent | 1.07 × 106 |
| 17 | Poultry | 46.7abc ± 50.3 | None virulent | 4.73 × 106 |
| 28 | Poultry | 33.3bc ± 57.7 | None virulent | 8 × 106 |
| 35 | Poultry | 0c ± 0 | None virulent | No LD50€ |
| 157 | Poultry | 93.3a ± 11.5 | virulent | 1.55 × 105 |
| 164 | Poultry | 100a ± 0 | virulent | 3.24 × 105 |
| 180 | Poultry | 86.7ab ± 11.5 | virulent | 2.28 × 105 |
| Q1 | Outbreak¥ | 86.7ab ± 11.5 | virulent | 4.5 × 105 |
| Q3 | Outbreak | 20c ± 34.6 | None virulent | No LD50 |
| Q5 | Outbreak | 46.7abc ± 46.2 | None virulent | 2.65 × 106 |
£Data are the average of three separate experiments (n = 3). §Strains were considered virulent if at least 60% of the five mice injected i.p. with 107 CFU/mouse died within 1 wk. ¥Salmonella isolates were isolated from food which was implicated in foodborne Salmonella outbreaks in Jordan. €No lethal dose was detected. *Percentages with different letters means are statistically significant at P < 0.05.
Antimicrobial testing
When the 25 confirmed S. Enteritidis isolates were tested for their susceptibility to eight of the commonly used antimicrobial agents in agriculture/human medicine, resistance patterns varied considerably (Table 3). Twenty of the 25 tested S. Enteritidis isolates were resistant to two or more antibiotics while 16 isolates (65%) were resistant to at least three antibiotics. No resistance was observed for CAZ, a member of the cephem family, while only four poultry isolates showed resistance to SXT, a folate pathway inhibitor. Resistance to other cephem member (CEF) and phenicols (C) was detected in nine and eight poultry isolates, respectively, while five of seven outbreak isolates were resistant to CEF, and no outbreak isolate was resistant to C. The highest resistance frequency was observed to NA (quinolones), CIP (fluoroquinolones), AM (Penicillin), and CEP (Cephem) with 18, 16, 14, and 12 of the poultry isolates exhibiting resistance to these antibiotics, respectively. In contrast, the outbreak isolates did not exhibit high resistance profiles, with 2, 1, 1, and 1 isolates exhibiting resistance NA, CIP, AM, and CEP, respectively. Table 4 shows a summary of differences in antibiotic resistance between poultry and outbreak strains. Furthermore, two of the outbreak isolates did not show any resistance to any of the tested antibiotics, three isolates showed only intermediate resistance to one antibiotic, and only two isolates exhibited multidrug resistance. In contrast, all 18 poultry isolates exhibited multidrug resistance.
Table 3. Antibiograms of all confirmed S. Enteritidis isolates tested against commonly used antibiotics in Jordan.
| No. | Isolate ID | Antibiotic | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| AMa | CEF | CEP | CAZ | C | CIP | NA | SXT | Totald | ||
| 1 | 15b | I | S | R | S | S | S | I | R | 4 |
| 2 | 17b | R | R | R | S | S | R | R | S | 4 |
| 3 | 28b | S | S | S | S | S | R | R | S | 2 |
| 4 | 31b | S | S | S | S | S | R | R | S | 2 |
| 5 | 32b | S | S | S | S | S | R | R | S | 2 |
| 6 | 35b | S | S | S | S | S | R | R | S | 2 |
| 7 | 37b | R | S | R | S | S | I | R | S | 4 |
| 8 | 39b | R | I | I | S | R | R | R | S | 5 |
| 9 | 51b | I | S | R | S | S | S | R | S | 3 |
| 10 | 100b | R | S | R | S | S | I | R | S | 4 |
| 11 | 123b | R | R | I | S | R | R | R | I | 6 |
| 12 | 136b | R | I | I | S | R | R | R | S | 5 |
| 13 | 143b | R | I | I | S | R | R | R | S | 5 |
| 14 | 157b | R | I | I | S | R | R | R | S | 5 |
| 15 | 164b | R | S | S | S | S | R | R | R | 4 |
| 16 | 175b | R | R | R | S | R | R | R | I | 6 |
| 17 | 177b | R | I | S | S | R | R | R | S | 5 |
| 18 | 180b | R | R | I | S | R | R | R | S | 5 |
| 19 | Q1c | S | S | S | S | S | S | S | S | 0 |
| 20 | Q2c | S | I | S | S | S | S | S | S | 1 |
| 21 | Q3c | S | I | S | S | S | S | S | S | 1 |
| 22 | Q4c | R | R | R | S | S | S | R | S | 3 |
| 23 | Q5c | S | I | S | S | S | R | R | S | 3 |
| 24 | Q6c | S | S | S | S | S | S | S | S | 0 |
| 25 | Q7c | S | I | S | S | S | S | S | S | 1 |
a AM, ampicillin (penicillin family); CEF, cefoperazone (cephem family); CEP, cephalothin (cephem family); CAZ, ceftazidime (cephem family); C, chloramphenicol (phenicols family); CIP, ciprofloxacin (fluoroquinolones family); NA, nalidixic acid (quinolones family); SXT, sulphamethoxazone–trimethoprim (folate pathway inhibitors family); bpoultry isolates; chuman outbreak isolates; dtotal number of antibiotic families to which each isolate is exhibiting intermediate or full resistant.
Table 4. Summary of antimicrobial resistance profiles among the outbreak and poultry isolates of S. Enteritidis.
| Antimicrobial agents | No. of resistant isolates (%) | |
|---|---|---|
| Outbreaka | Poultryb | |
| Ampicillin (AMP) | 1 (14.3) | 14 (77.8) |
| Cefoperazone (CEF) | 5 (71.4) | 9 (50) |
| Cephalothin (CEP) | 1 (14.3) | 9 (66.7) |
| Ceftazidime (CAZ) | 0 (0) | 0 (0) |
| Chloramphenicol (C) | 0 (0) | 8 (44.4) |
| Ciprofloxacin (CIP) | 1 (14.3) | 16 (88.9) |
| Nalidixic acid (NA) | 2 (28.6) | 18 (100) |
| Sulphamethoxazone–Trimethoprime (SXT) | 0 (0) | 4 (22.2) |
an = 7; bn = 18.
Discussion
To study the virulence and LD50 and antibiotic resistance of the confirmed S. Enteritidis isolates, the isolates were subjected to a final confirmation step based on 16S rRNA sequencing and a phylogenetic tree was constructed with seven clusters (Fig. 2). The 16S rRNA gene is considered the most useful housekeeping genetic marker for studying the phylogenetic and evolutionary relationships among closely related bacteria, and thus is fundamental for analysis of phylogenetic relationship among bacterial taxa.25-27 However, 25 of the 45 (55.6%) PCR-confirmed isolates were confirmed as S. Enteritidis, while the other 20 isolates were found to be other Salmonella spp. Nevertheless, none of the 16S rRNA-confirmed isolates that scored low in RapID™ ONE was found positive with 16S rRNA in the present work. Interestingly, among the isolates there were 11 typhoid Salmonella strains (seven S. Paratyphi and four S. Typhi), indicating that poultry might possess multiple Salmonella strains. It is noteworthy mentioning that among the isolated strains was S. Typhi. Since S. Typhi enters the food chain through the fecal-oral route, these isolates could have been transmitted from infected workers to the chicken carcass or due to human wastes contaminating processing water.28
Representatives from each cluster of the phylogenetic tree were tested for their LD50 in normal white Swiss mice. The mouse was chosen as a model for the virulence test according to accepted practices, recognizing that there are similarities between mice and human immune systems although there were instances where results using this model can be different.16,29 Nearly half of the poultry isolates (3/7) were more virulent than the single outbreak strain found pathogenic here. As expected, some of the poultry isolates were avirulent to mice which could be explained by the fact that some isolates exhibit different host specificity; while causing a systemic infection in one host they might cause asymptomatic colonization in other host (such as the mouse in this study).30,31 Alternatively, these differences in virulence might be attributed to deferential regulation of virulence genes. However, it was interesting to see that only one outbreak isolate was virulent while two failed to show any virulence or ability to kill 50% of tested mice. This could be due to physiological differences between humans and animals, which might have led to the selection of bacterial strains that are better adapted to one host vs. other.5 It has also been reported that some S. Enteritidis mutants can cause salmonellosis in humans, but fail to cause any form of infection in mice. Such mutants might lose the ability to survive in the macrophages of the host and thus lose its virulence.32
The development of multiple antimicrobial resistances among foodborne pathogens has emerged as a major public health concern worldwide. This has led to rethinking the use of antibiotics in animal feed as growth promoters, since it is possible that this use of antibiotics may contribute to the emergence of multidrug-resistant organisms and facilitate the transfer of resistance genes to human pathogens.33 In general, all 18 S. Enteritidis isolates from poultry were multidrug-resistant, showing resistance toward two to six antibiotics while only two isolates of the seven outbreak strains showed multidrug resistance. The high percentage of multidrug-resistant Salmonella might be consistent with the fact that stable resistance elements do exist and might be genetically linked to other resistance determinants and therefore, drug use might not only result in resistance against the drug used but also yield multidrug-resistant phenotypes that confer selective advantage to such strains.19
Results obtained in the present study differ from those reported by Dias de Oliveira et al.34 and Fernandez et al.35 who found a smaller percentage of tested isolates which showed multiple resistances, while the rest exhibited complete sensitivity. However, they reported susceptibility of all their isolates to ceftazidime (cephem family) which is identical to the present results, and may reflect the lower use of this antibiotic in both countries. The other two tested members of the cephem family showed around 30% resistance. In the present study, the highest number of resistant isolates was obtained for poultry isolates against nalidixic acid. Similarly, Hur et al.36 reported 89% resistance of S. Enteritidis to nalidixic acid. Others reported somewhat lower resistance profiles to S. Enteritidis. For instance, Antunes et al.20 and Ribeiro et al.37 reported resistance to nalidixic acid in 50% and 60% of tested isolates, respectively. In contrast, Han et al.,38 Dias de Oliveira et al.,34 and Fernandez et al.35 reported a strikingly low level (7.4%, 7.7%, and 10%, respectively) of isolates resistant to nalidixic acid in USA, Brazil, and Vietnam, respectively. The very high frequency of resistance to nalidixic acid observed in Jordan and in the other studies conducted in Brazil, Portugal, and Korea,20,37,39 could be due to its use in agriculture, possibly in poultry feed. This is of particular importance as quinolones family are used for treatment of invasive Salmonella infections.39
It is worth mentioning here that a relatively high level of resistance (78% of poultry isolates) was observed toward ampicillin, which is important because it was the drug of choice for treatment of systemic salmonellosis in humans until late 1980s.40 Nonetheless, higher resistance to ampicillin was reported in a study conducted in Turkey where it was reported that 85.2% of Salmonella isolates were resistant to this antibiotic.41 In contrast, Zou et al.42 reported that only 2.35% of the tested isolates were resistant to ampicillin while Fernandez et al.35 reported a 13.6% resistance to the same antibiotic.
Resistance to ciprofloxacin was high, with 89% of the isolates resistant to this antibiotic. In contrast, Hanson et al.43 and Fernandez et al.35 did not observe Salmonella isolates resistant to ciprofloxacin. Chloramphenicol resistance was also found in 45% of the isolates, a number far higher than that reported by Dias de Oliveira et al.,34 where only 1.1% of isolates were resistant to this antibiotic, and in results reported by Van et al.44 and Yildirim et al.41 who found chloramphenicol resistance in 8% and 10.2% of isolates, respectively. The very low percentage of isolates resistant to this antibiotic in Brazil or Vietnam is probably due to the ban on using this antibiotic in animal feed put in place in the 1970s. The relatively high proportion of resistance to this antibiotic in Jordan suggests that such a ban on the use of this antibiotic in animal feed is not enforced or does not exist. Resistance of the poultry isolates to cephalothin and cefoperozone, both members of the cephem family, was 67% and 50%, respectively. These rates appeared far higher than results reported in Brazil35 where complete sensitivity to cefoperozone and only low resistance to cephalothin was reported. The differences in antibiotic resistance between countries likely reflect the frequency of using these antimicrobials and the nature of rules and regulations that govern their use.
When the outbreak isolates were examined for resistance to the same antibiotics tested for the poultry isolates, outbreak isolates were sensitive to most of the antibiotics except for cefoperozone, to which 71% were resistant. These results were consistent with results reported by the US Food and Drug Administration report45 where percentages of resistance were greater in isolates of veterinary origin than those from human origin. Similarly, Denny et al.9 found that all S. Enteritidis isolates from humans in Europe were sensitive to all tested antimicrobials including nalidixic acid, sulphonamides, and ampicillin. In contrast, Fernandez et al.35 reported that S. Enteritidis isolates from hospitalized patients in Brazil exhibited a broader spectrum of antibiotic resistance profiles than the non-human isolates.
In conclusion, not all S. Enteritidis isolates were virulent in mice, regardless of the host. However, there was variation in pathogenicity among the outbreak as well as poultry isolates. The antibiotic resistances of Salmonella isolates in Jordan were both similar and different from those in other countries, but resistance was regarded as being more frequent than desirable. It is apparent that more strict rules regarding the use of antibiotics in animal agriculture and the hygienic operation of abattoirs and poultry meat processing plants should be observed. Indeed, these results can be used to establish epidemiological baseline data set concerning pathogenicity and antibiotic resistance profiles, which will be pivotal for the establishment of Salmonella surveillance program in Jordan.
Materials and Methods
Salmonella outbreak isolates
Seven Salmonella outbreak strains isolated by the Jordan Food and Drug Administration (JFDA) from foodborne salmonellosis cases which occurred in Northern Jordan between 2006 and 2008 were used. Initially, the isolates were identified and characterized as S. Enteritidis in JFDA laboratories and were further confirmed in our laboratory at the Jordan University of Science and Technology.
Collection of poultry samples
A total of 302 fresh poultry meat samples were collected between October 2008 and June 2009 from many local retail markets in northern Jordan. The samples were procured from fresh poultry carcasses, either whole chicken or chicken parts and packaged individually in sterile plastic bags. The individual samples were transported in insulated boxes with frozen ice packs, stored at 4 °C and were examined within 24 h for the presence of Salmonella and other closely related species.
Culture identification of presumptive Salmonella isolates from poultry samples
The Iso46 method was used to identify presumptive Salmonella from poultry samples. Briefly, approximately 30 g of each poultry meat sample were aseptically cut into small pieces using sterile disposable blades and placed into sterile stomacher bags containing 100 mL of lactose broth (Oxoid) for pre-enrichment. Samples were pummeled 2 min in a stomacher (Model 400, A.J. Seward), and incubated for 18 h at 37 °C. Then 2 mL were transferred into sterile conical tubes containing 8 mL of Selenite Cystine broth (Merck) and incubated at 37 °C for 48 h. Upon completion of the pre-enrichment, one loopful of the Selenite Cystine broth was streaked in quadrants onto Xylose Lysine Deoxycholate agar (XLD, Oxoid), Salmonella–Shigella agar (S-S, Oxoid), and Brilliant Green Agar (BGA, Oxoid) for isolation of Salmonella. All plates were incubated at 37 °C for 24 h. Typical colonies of Salmonella on XLD, S-S, and BGA were identified by their morphological characteristics and were then streaked onto Nutrient Agar (Oxoid) plates and incubated at 37 °C for 24 h.
Biochemical identification and molecular confirmation of Salmonella spp.
Presumptive Salmonella isolates were subjected to RapIDTM Biochemical testing (Remel) as per manufacturer’s instructions followed by molecular confirmation of the isolates by universal PCR using primers for the InvA gene (Table 5) that detects Salmonella in general. Positively identified isolates were subjected to S. Enteritis specific PCR primers for sefA gene (Table 5). PCR-confirmed isolates were further subjected to final confirmation method using the 16S rRNA sequencing following the method described by Woo et al.24 The universal primer pair Lpw57/58 (Table 5) and the BigDye Termination Kit (Life Technologies) were used in this method. These primers amplify the 7 16S rRNA genes in Salmonella. These genes are SEN_r020, SEN_r001, SEN_r006, SEN_r014, SEN_r017, SEN_r011, and SEN_r010. Confirmed Salmonella enterica Enteritidis isolate (ATCC 13076) was used as a positive control, while an E. coli isolate was used as a negative control.
Table 5. Primer sequences of amplified genes, PCR conditions used and product size of each amplicon.
| Gene | Primers 5′…….3′ | Amplification conditions | Product size (bp) | References | ||
|---|---|---|---|---|---|---|
| Temp | Time | No. of cycles | ||||
| InvA |
139: GTGAAATTAT CGCCACGTTC GGGCAA 141: TCATCGCACC GTCAAAGGAA CC |
94 °C | 5 min | 1 | 284 | 22 |
| 94 °C | 30 s | 35 | ||||
| 55 °C | 45 s | |||||
| 72 °C | 45 s | |||||
| 72 °C | 10 min | 1 | ||||
| SefA |
Sef167: AGGTTCAGGC AGCGGTTACT Sef478: GGGACATTTA GCGTTTCTT |
94 °C | 45 s | 1 | 312 | 23 |
| 94 °C | 20 s | 35 | ||||
| 57 °C | 15 s | |||||
| 72 ° | 15 s | |||||
| 72 °C | 2 min | 1 | ||||
| 16S rRNA |
Lwp57: AGTTTGATCC TGGCTCAG Lwp58: AGGCCCGGGA ACGTATTCAC |
94 °C | 2 min | 1 | 1381 | 24 |
| 94 °C | 2 min | 40 | ||||
| 55 °C | 2 min | |||||
| 72 ° | 1 min | |||||
| 72 °C | 7 min | 1 | ||||
Phylogenetic analysis of the isolates based on 16S rRNA sequencing
Aligned 16S rRNA sequences were used for constructing the phylogenetic tree by the neighbor-joining method47 using the MEGA Align package. Maximum likelihood phylogenetic analysis with bootstrap values for n = 500 replicates48 was performed to estimate the confidence of tree topologies. Representatives from these clusters were tested in a mouse model for their pathogenicity by LD50 determination.
Mouse LD50 determination
Mouse pathogenicity was determined as described by Stelma et al.16 with modifications. Briefly, Salmonella isolates representing food and outbreak strains as inferred from the phylogenetic tree were grown overnight at 37 °C in 5 mL nutrient broth (HiMedia). The original CFU/mL of overnight cultures was determined using McFarland standards and diluted to 108 CFU/mL. This was followed by serial 10-fold dilutions (102–107) in sterile PBS. Then 0.1 mL of each dilution was injected intraperitonially into five Swiss white mice (approx. 20 g each). Two groups of control mice were used in each experiment; one group was given 0.1 mL sterile PBS while a second group was not injected. The mice were observed for one week, and deaths were recorded. Strains that killed three (60%) or more mice (received a dose of 107 CFU/ mouse) within the first week were considered virulent. The virulence of selected isolates of S. Enteritidis was estimated by determining the 50% lethal dose (LD50) as described by Reed and Muench.49 The results were the average of three separate experiments with 310 mice used for each experiment. Experiments in mice were conducted upon the approval of the Jordan University of Science and Technology (JUST) Animal Care and Welfare committee. Mice were cared for as per the protocols of JUST Animal Care and Welfare Committee. Statistical analysis was performed using the GLM procedure. Least Significant Differences (LSD) was calculated and P < 0.05 was considered statistically significant.
Antimicrobial susceptibility testing
The antibiotic susceptibility of the 25 confirmed S. Enteritidis isolates (16S RNA sequencing) was determined for 8 antibiotics (ceftazidime [CAZ], cephalothin [CEP], cefoperazone [CEF], sulphamethoxazone-trimethoprim [STX], nalidixic acid [NA], ciprofloxacin [CIP], ampicillin [AM], and chloramphenicol [C]) using the disk diffusion method on Muller Hinton Agar plates (Oxoid) as described in the Clinical and Laboratory Standards Institute guidelines.50
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors would like to acknowledge the Deanship of Research at the Jordan University of Science and Technology for funding the project (25/2009).
Glossary
Abbreviations:
- LD
lethal dose
- CFU
colony forming unit
- PCR
polymerase chain reaction
- CDC
Centers for Disease Control and Prevention
- JFDA
Jordan Food and Drug Administration
- S-S
Salmonella Shegella agar
- BGA
brilliant green agar
- ATCC
American Type Culture Collection
- JUST
Jordan University of Science and Technology
- CAZ
ceftazidime
- CEP
cephalothin
- CEF
cefoperazone
- STX
sulphamethoxazone–trimethoprim
- NA
nalidixic acid
- CIP
ciprofloxacin
- AM
ampicillin
- C
chloramphenicol
- FDA
US Food and Drug Administration
References
- 1.Domínguez C, Gómez I, Zumalacárregui J. Prevalence of Salmonella and Campylobacter in retail chicken meat in Spain. Int J Food Microbiol. 2002;72:165–8. doi: 10.1016/S0168-1605(01)00638-9. [DOI] [PubMed] [Google Scholar]
- 2.Suresh T, Hatha AAM, Harsha HT, Lakshmanaperumalsamy P. Prevalence and distribution of Salmonella serotypes in marketed broiler chickens and processing environment in Coimbatore city of southern India. Food Res Int. 2011;44:823–5. doi: 10.1016/j.foodres.2011.01.035. [DOI] [Google Scholar]
- 3.Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson M-A, Roy SL, Jones JL, Griffin PM. Foodborne illness acquired in the United States--major pathogens. Emerg Infect Dis. 2011;17:7–15. doi: 10.3201/eid1701.P11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barton Behravesh C, Jones TF, Vugia DJ, Long C, Marcus R, Smith K, Thomas S, Zansky S, Fullerton KE, Henao OL, et al. FoodNet Working Group Deaths associated with bacterial pathogens transmitted commonly through food: foodborne diseases active surveillance network (FoodNet), 1996-2005. J Infect Dis. 2011;204:263–7. doi: 10.1093/infdis/jir263. [DOI] [PubMed] [Google Scholar]
- 5.Heithoff DM, Shimp WR, Lau PW, Badie G, Enioutina EY, Daynes RA, Byrne BA, House JK, Mahan MJ. Human Salmonella clinical isolates distinct from those of animal origin. Appl Environ Microbiol. 2008;74:1757–66. doi: 10.1128/AEM.02740-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huehn S, Bunge C, Junker E, Helmuth R, Malorny B. Poultry-associated Salmonella enterica subsp. enterica serovar 4,12:d:- reveals high clonality and a distinct pathogenicity gene repertoire. Appl Environ Microbiol. 2009;75:1011–20. doi: 10.1128/AEM.02187-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thong K-L, Ngeow Y-F, Altwegg M, Navaratnam P, Pang T. Molecular analysis of Salmonella Enteritidis by pulsed-field gel electrophoresis and ribotyping. J Clin Microbiol. 1995;33:1070–4. doi: 10.1128/jcm.33.5.1070-1074.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jackson BR, Griffin PM, Cole D, Walsh KA, Chai SJ. Outbreak-associated Salmonella enterica serotypes and food Commodities, United States, 1998-2008. Emerg Infect Dis. 2013;19:1239–44. doi: 10.3201/eid1908.121511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Denny J, Boelaert F, Borck B, Heuer DE, Ammon A, Mäkelä P. Zoonotic infections in Europe: trends and figures - a summary of the EFSA-ECDC annual report. Euro Surveill. 2007;12:3336. doi: 10.2807/esw.12.51.03336-en. [DOI] [PubMed] [Google Scholar]
- 10.Westrell T, Ciampa N, Boelaert F, Helwigh B, Korsgaard H, Chríel M, Ammon A, Mäkelä P. Zoonotic infections in Europe in 2007: a summary of the EFSAECDC annual report. Euro Surveillance 2009; 14(3):pii=19100. Available online: http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19100 [PubMed]
- 11.Schrank IS, Mores MAZ, Costa JL, Frazzon APG, Soncini R, Schrank A, Vainstein MH, Silva SC. Influence of enrichment media and application of a PCR based method to detect Salmonella in poultry industry products and clinical samples. Vet Microbiol. 2001;82:45–53. doi: 10.1016/S0378-1135(01)00350-9. [DOI] [PubMed] [Google Scholar]
- 12.Nesbitt A, Ravel A, Murray R, McCormick R, Savelli C, Finley R, Parmley J, Agunos A, Majowicz SE, Gilmour M, Canadian Integrated Program for Antimicrobial Resistance Surveillance Public Health Partnership. Canadian Public Health Laboratory Network Integrated surveillance and potential sources of Salmonella Enteritidis in human cases in Canada from 2003 to 2009. Epidemiol Infect. 2012;140:1757–72. doi: 10.1017/S0950268811002548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bouzidi N, Aoun L, Zeghdoudi M, Bensouilah M, Elgroud R, Oucief I, Granier SA, Brisabois A, Desquilbet L, Millemann Y. Salmonella contamination of laying-hen flocks in two regions of Algeria. Food Res Int. 2012;45:897–904. doi: 10.1016/j.foodres.2011.05.027. [DOI] [Google Scholar]
- 14.Gordon MA. Salmonella infections in immunocompromised adults. J Infect. 2008;56:413–22. doi: 10.1016/j.jinf.2008.03.012. [DOI] [PubMed] [Google Scholar]
- 15.Saeed AM, Walk ST, Arshad M, Whittam TS. Clonal structure and variation in virulence of Salmonella Enteritidis isolated from mice, chickens, and humans. J AOAC Int. 2006;89:504–11. [PubMed] [Google Scholar]
- 16.Stelma GN, Jr., Reyes AL, Peeler JT, Francis DW, Hunt JM, Spaulding PL, Johnson CH, Lovett J. Pathogenicity test for Listeria monocytogenes using immunocompromised mice. J Clin Microbiol. 1987;25:2085–9. doi: 10.1128/jcm.25.11.2085-2089.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carter PB. Pathogenecity of Yersinia enterocolitica for mice. Infect Immun. 1975;11:164–70. doi: 10.1128/iai.11.1.164-170.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Livermore DM. Bacterial resistance: origins, epidemiology, and impact. Clin Infect Dis. 2003;36(Suppl 1):S11–23. doi: 10.1086/344654. [DOI] [PubMed] [Google Scholar]
- 19.Mayrhofer S, Paulsen P, Smulders FJM, Hilbert F. Antimicrobial resistance profile of five major food-borne pathogens isolated from beef, pork and poultry. Int J Food Microbiol. 2004;97:23–9. doi: 10.1016/j.ijfoodmicro.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 20.Antunes P, Réu C, Sousa JC, Peixe L, Pestana N. Incidence of Salmonella from poultry products and their susceptibility to antimicrobial agents. Int J Food Microbiol. 2003;82:97–103. doi: 10.1016/S0168-1605(02)00251-9. [DOI] [PubMed] [Google Scholar]
- 21.Jones YE, McLaren IM, Wray C. Laboratory aspects of Salmonella In C. Wray, and A. Wray (EDs.) Salmonella in Domestic Animals. New York, NY: CABI publishing Inc. 2000; pp. 393-405. [Google Scholar]
- 22.Rahn K, De Grandis SA, Clarke RC, McEwen SA, Galán JE, Ginocchio C, Curtiss R, 3rd, Gyles CL. Amplification of an invA gene sequence of Salmonella Typhimurium by polymerase chain reaction as a specific method of detection of Salmonella. Mol Cell Probes. 1992;6:271–9. doi: 10.1016/0890-8508(92)90002-F. [DOI] [PubMed] [Google Scholar]
- 23.Soumet C, Ermel G, Rose V, Rose N, Drouin P, Salvat G, Colin P. Identification by a multiplex PCR-based assay of Salmonella Typhimurium and Salmonella Enteritidis strains from environmental swabs of poultry houses. Lett Appl Microbiol. 1999;29:1–6. doi: 10.1046/j.1365-2672.1999.00559.x. [DOI] [PubMed] [Google Scholar]
- 24.Woo PCY, Fung AMY, Wong SSY, Tsoi HW, Yuen KY. Isolation and characterization of a Salmonella enterica serotype Typhi variant and its clinical and public health implications. J Clin Microbiol. 2001;39:1190–4. doi: 10.1128/JCM.39.3.1190-1194.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Christensen H, Nordentoft S, Olsen JE. Phylogenetic relationships of Salmonella based on rRNA sequences. Int J Syst Bacteriol. 1998;48:605–10. doi: 10.1099/00207713-48-2-605. [DOI] [PubMed] [Google Scholar]
- 27.Janda JM, Abbott SL. 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: pluses, perils, and pitfalls. J Clin Microbiol. 2007;45:2761–4. doi: 10.1128/JCM.01228-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kumar S, Balakrishna K, Singh GP, Batra HV. Rapid detection of Salmonella Typhi in foods by combination of immunomagnetic separation and polymerase chain reaction. World J Microbiol Biotechnol. 2005;21:625–8. doi: 10.1007/s11274-004-3553-x. [DOI] [Google Scholar]
- 29.Guard-Petter J, Henzler DJ, Rahman MM, Carlson RW. On-farm monitoring of mouse-invasive Salmonella enterica serovar Enteritidis and a model for its association with the production of contaminated eggs. Appl Environ Microbiol. 1997;63:1588–93. doi: 10.1128/aem.63.4.1588-1593.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Badie G, Heithoff DM, Sinsheimer RL, Mahan MJ. Altered levels of Salmonella DNA adenine methylase are associated with defects in gene expression, motility, flagellar synthesis, and bile resistance in the pathogenic strain 14028 but not in the laboratory strain LT2. J Bacteriol. 2007;189:1556–64. doi: 10.1128/JB.01580-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chan K, Baker S, Kim CC, Detweiler CS, Dougan G, Falkow S. Genomic comparison of Salmonella enterica serovars and Salmonella bongori by use of an S. enterica serovar Typhimurium DNA microarray. J Bacteriol. 2003;185:553–63. doi: 10.1128/JB.185.2.553-563.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suzuki S. Pathogenicity of Salmonella Enteritidis in poultry. Int J Food Microbiol. 1994;21:89–105. doi: 10.1016/0168-1605(94)90203-8. [DOI] [PubMed] [Google Scholar]
- 33.Simmons M, Donovan DM, Siragusa GR, Seal BS. Recombinant expression of two bacteriophage proteins that lyse Clostridium perfringens and share identical sequences in the C-terminal cell wall binding domain of the molecules but are dissimilar in their N-terminal active domains. J Agric Food Chem. 2010;58:10330–7. doi: 10.1021/jf101387v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dias de Oliveira S, Siqueira Flores F, dos Santos LR, Brandelli A. Antimicrobial resistance in Salmonella Enteritidis strains isolated from broiler carcasses, food, human and poultry-related samples. Int J Food Microbiol. 2005;97:297–305. doi: 10.1016/j.ijfoodmicro.2004.04.022. [DOI] [PubMed] [Google Scholar]
- 35.Fernandes SA, Ghilardi ACR, Tavechio AT, Machado AMO, Pignatari ACC. Phenotypic and molecular characterization of Salmonella Enteritidis strains isolated in São Paulo, Brazil. Rev Inst Med Trop Sao Paulo. 2003;45:59–63. doi: 10.1590/S0036-46652003000200001. [DOI] [PubMed] [Google Scholar]
- 36.Hur J, Kim JH, Park JH, Lee Y-J, Lee JH. Molecular and virulence characteristics of multi-drug resistant Salmonella Enteritidis strains isolated from poultry. Vet J. 2011;189:306–11. doi: 10.1016/j.tvjl.2010.07.017. a. [DOI] [PubMed] [Google Scholar]
- 37.Ribeiro A, Kellermann A, Santos L, Bessa M, Nascimento V. Salmonella spp. in raw broiler parts; occurrence, antimicrobial resistance profile and phage typing of the Salmonella Enteritidis isolates. Braz J Microbiol. 2007;38:296–9. doi: 10.1590/S1517-83822007000200021. [DOI] [Google Scholar]
- 38.Han J, Gokulan K, Barnette D, Khare S, Rooney AW, Deck J, Nayak R, Stefanova R, Hart ME, Foley SL. Evaluation of virulence and antimicrobial resistance in Salmonella enterica serovar Enteritidis isolates from humans and chicken- and egg-associated sources. Foodborne Pathog Dis. 2013;10:1008–15. doi: 10.1089/fpd.2013.1518. [DOI] [PubMed] [Google Scholar]
- 39.Hur J, Choi YY, Park JH, Jeon BW, Lee HS, Kim AR, Lee JH. Antimicrobial resistance, virulence-associated genes, and pulsed-field gel electrophoresis profiles of Salmonella enterica subsp. enterica serovar Typhimurium isolated from piglets with diarrhea in Korea. Can J Vet Res. 2011;75:49–56. b. [PMC free article] [PubMed] [Google Scholar]
- 40.Karki S, Shakya P, Cheng AC, Dumre SP, Leder K. Trends of etiology and drug resistance in enteric fever in the last two decades in Nepal: a systematic review and meta-analysis. Clin Infect Dis. 2013;57:e167–76. doi: 10.1093/cid/cit563. [DOI] [PubMed] [Google Scholar]
- 41.Yildirim Y, Gonulalan Z, Pamuk S, Ertas N. Incidence and antibiotic resistance of Salmonella spp. on raw chicken carcasses. Food Res Int. 2011;44:725–8. doi: 10.1016/j.foodres.2010.12.040. [DOI] [Google Scholar]
- 42.Zou M, Keelara S, Thakur S. Molecular characterization of Salmonella enterica serotype Enteritidis isolates from humans by antimicrobial resistance, virulence genes, and pulsed-field gel electrophoresis. Foodborne Pathog Dis. 2012;9:232–8. doi: 10.1089/fpd.2011.1012. [DOI] [PubMed] [Google Scholar]
- 43.Hanson R, Kaneene JB, Padungtod P, Hirokawa K, Zeno C. Prevalence of Salmonella and E. coli, and their resistance to antimicrobial agents, in farming communities in northern Thailand. Southeast Asian J Trop Med Public Health. 2002;33(Suppl 3):120–6. [PubMed] [Google Scholar]
- 44.Van TTH, Moutafis G, Istivan T, Tran LT, Coloe PJ. Detection of Salmonella spp. in retail raw food samples from Vietnam and characterization of their antibiotic resistance. Appl Environ Microbiol. 2007;73:6885–90. doi: 10.1128/AEM.00972-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.FDA. National antimicrobial resistance monitoring system-enteric bacteria (NARMS): 2003 Executive report. US Department of Health and Human Services, US Food and Drug Administration, Rockville, MD. [Google Scholar]
- 46.ISO. (2002): Microbiology of food and animal feeding stuffs - Horizontal method for the detection of Salmonella spp. International Standard 6573 Fourth edition. 2002-07-15. International Organization for Standardization. Geneva, Switzerland. [Google Scholar]
- 47.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–25. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 48.Felsentein J. Confidence limits on phylogenesis; an approach using the bootstrap. Evolution. 1985;39:783–91. doi: 10.2307/2408678. [DOI] [PubMed] [Google Scholar]
- 49.Reed LJ, Muench HA. Simple method of estimating fifty percent end points. Am J Hyg. 1938;27:493–7. [Google Scholar]
- 50.CLSI. Performance standards for antimicrobial susceptibility testing; 15th informational supplement. CLSI document M100-S15. Clinical and Laboratory Standards Institute 2005. [Google Scholar]