Abstract
• Premise of the study: We have optimized a version of a microsatellite loci isolation protocol for first-generation sequencing (FGS) technologies. The protocol is optimized to reduce the cost and number of steps, and it combines some procedures from previous simple sequence repeat (SSR) protocols with several key improvements that significantly affect the final yield of the SSR library. This protocol may be accessible for laboratories with a moderate budget or for which next-generation sequencing (NGS) is not readily available.
• Methods and Results: We drew from classic protocols for library enrichment by digestion, ligation, amplification, hybridization, cloning, and sequencing. Three different systems were chosen: two with very different genome sizes (Galdieria sulphuraria, 10 Mbp; Pancratium maritimum, 30 000 Mbp), and a third with an undetermined genome size (Kochia saxicola). Moreover, we also report the optimization of the sequencing reagents. A good frequency of the obtained microsatellite loci was achieved.
• Conclusions: The method presented here is very detailed; comparative tests with other SSR protocols are also reported. This optimized protocol is a promising tool for low-cost genetic studies and the rapid, simple construction of homemade SSR libraries for small and large genomes.
Keywords: double PCR enrichment, first-generation sequencing, microsatellites, optimized protocol, patchwork, SSR library
To date, many different strategies for obtaining microsatellite DNA loci or simple sequence repeat (SSR) libraries have been described. The basic approach typically involves digestion, hybridization, cloning, and sequencing, with several variations (Zane et al., 2002; Kalia et al., 2011). The most recent trend in SSR library development is the use of next-generation sequencing (NGS) or pyrosequencing (454 Life Sciences, Roche Diagnostics, Indianapolis, Indiana, USA) technologies (Zalapa et al., 2012). Although pyrosequencing offers the potential to rapidly sequence whole genomes, it is costly (e.g., price >$5000 in Zalapa et al. [2012]). A real reduction in cost is possible with the new Illumina technology (Synthesis Bridge PCR; e.g., Illumina, San Diego, California, USA) (Zalapa et al., 2012), but the best improvements in cost will be possible with the advent of new pH-change sequencing, such as Ion Proton technology (Life Technologies, Paisley, Renfrewshire, United Kingdom). A recent review (Glenn, 2011) summarizes the major characteristics of each commercially available platform to enable direct comparisons. However, even if these new technologies present a greater cost-savings in the long run because they yield many more SSR loci, SSR libraries with a moderate number of loci, which can be produced using standard procedures, can also be useful. However, the majority of these protocols are not cost competitive, and their low efficiency or lack of optimization can restrict their efficacy (Squirrell et al., 2003).
In this study, we optimized an inexpensive protocol that greatly reduces the cost and number of steps while using first-generation sequencing (FGS; Sanger sequencing) technology. To establish this optimized protocol, we drew from classic enrichment protocols (e.g., Kandpal et al., 1994; Edwards et al., 1996; Hamilton et al., 1999; Glenn and Schable, 2005; Techen et al., 2010) as well as the new generation of hybrid enrichment protocols, such as Fast Isolation by AFLP of Sequences COntaining repeats (FIASCO) (Zane et al., 2002). We have called this protocol “SSR-patchwork” because it is a mixture of the best parts of previous SSR protocols and several improvements that are fundamental to the final yield of the SSR library (see Methods and Results and Appendix S1 (158.5KB, pdf) ). Unlike other published SSR protocols, this protocol does not require further optimization by the reader, saving considerable time and money.
METHODS AND RESULTS
Three different systems were chosen: two with very different genome sizes and a third with an undetermined genome size. Of the three systems chosen, two are angiosperms, and the third is a red algae. Kochia saxicola Guss. (Amaranthaceae) has an undetermined genome size, Pancratium maritimum L. (Amaryllidaceae) has a genome of approximately 30 000 Mbp (Zonneveld et al., 2005), and Galdieria sulphuraria (Galdieri) Merola (Cyanidiaceae) has a genome of approximately 10 Mbp (Muravenko et al., 2001) (Appendix 1).
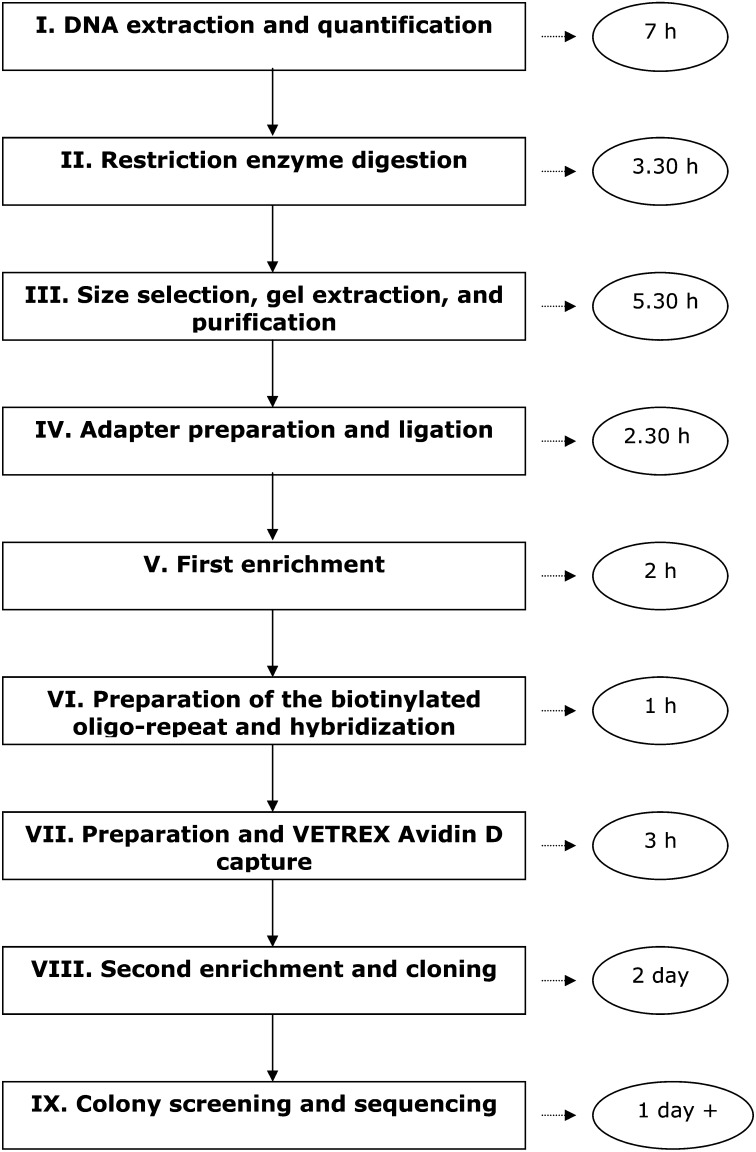
Details of the different steps of the protocol are accessible in Appendix S1 (158.5KB, pdf) . We briefly explain the most important steps here; a relative timeline of the procedure is shown in Fig. 1.
Fig. 1.
Schematic representation of the timeline of the SSR-patchwork protocol. Further details are presented in Appendix S1 (158.5KB, pdf) .
Step 1: DNA extraction and quantification
A total of 2 μg nondegraded DNA from a fresh sample was used for each species (Kochia saxicola, Galdieria sulphuraria, and Pancratium maritimum). The Doyle and Doyle (1990) method was used to produce high-quality genomic DNA, and an RNase step is recommended to improve the cleanliness of the sample. It is imperative to check the concentration and especially the quality of the obtained DNA before proceeding to the other steps. An agarose gel evaluation is sufficient to check the DNA quality (i.e., nondegraded) and concentration using a suitable marker (e.g., Marker II; AppliChem GmbH, Darmstadt, Germany).
Step 2: Restriction enzyme digestion
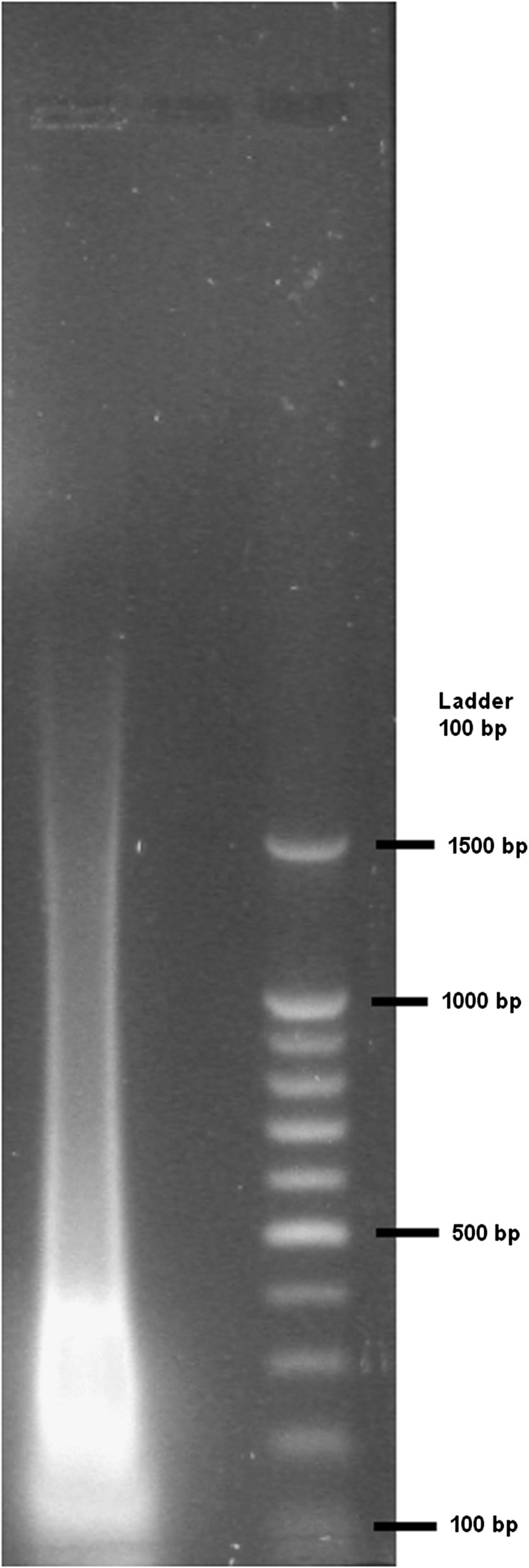
The EcoRI and MseI enzymes were used, as in the amplified fragment length polymorphism (AFLP) procedure (Vos et al., 1995) (Invitrogen, Life Technologies, Paisley, Renfrewshire, United Kingdom). A successful reaction should yield a smear of fragments ranging from 200 to 1000 bp (Fig. 2).
Fig. 2.
Image of a gel representing a successful restriction enzyme digestion using both EcoRI and MseI (Invitrogen, Life Technologies, Paisley, Renfrewshire, United Kingdom) in a Pancratium maritimum template. The first lane is genomic DNA (2 μg), and the second lane is a 100-bp DNA ladder (BenchTop 100-bp DNA Ladder, Promega Corporation, Madison, Wisconsin, USA).
Step 3: Size selection, gel extraction, and purification
After precipitation of the digested samples, the DNA was loaded onto a 1% agarose gel to separate the DNA fragments. The DNA from 250 to 500 bp was then isolated. Although several kits and protocols for gel purification are available, we propose a simple and economical method that does not require a low melting agarose gel to perform this step (see Appendix S1 (158.5KB, pdf) ).
Step 4: Adapter preparation and ligation
The EcoRI-adapter and MseI-adapter (Macrogen, Seoul, Korea) were prepared using a touchdown/hold PCR. A ligation was then performed according to the standard protocol recommended by the manufacturer (Invitrogen, Life Technologies).
Step 5: First enrichment
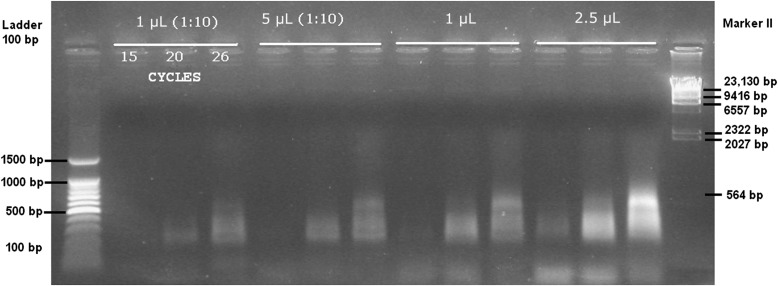
The ligation reaction product was amplified using modified AFLP adapter-specific primers (Macrogen) (without selective terminal base, Pre_EcoI-0 and Pre_MseI-0) (Vos et al., 1995). It is important to note that T4 DNA ligase (Invitrogen, Life Technologies) only ligates one of the strands of the adapter to the digested DNA fragment, while the other is held to the first adapter strand by base pairing. Thus, the first PCR reaction step is a hold at 72°C, which allows the Taq DNA polymerase to ligate the other strand. Several tests can be performed to optimize the ligation pattern (Fig. 3). The best amplification in our study was achieved at 26 cycles and using 2.5 or 5 μL of the ligation template.
Fig. 3.
Image of several patterns amplified after the first enrichment (step 5) using different concentrations of template (ligation reaction) and the primers EcoI-0 and MseI-0 (preselective modified primers; Vos et al., 1995). For each amplification template, 4 μL was loaded. The first lane is a 100-bp DNA ladder (BenchTop 100-bp DNA Ladder, Promega Corporation, Madison, Wisconsin, USA), and the last lane is Marker II (AppliChem GmbH, Darmstadt, Germany). The numbers above the white line indicate the quantity of ligation template used for the PCR enrichment, and numbers below the white line indicate the number of PCR cycles.
Step 6: Preparation of the biotinylated oligo-repeat and hybridization
Several microsatellite motifs can be used (e.g., TG12, GA12, GAG10, CAA10, or AAGT8). Here, for example, the GA12 motif repeat was employed. For the hybridization reaction, 500 ng of oligo-repeat biotinylated probe was used (Macrogen) for 250 ng of enrichment. The reaction was performed entirely by PCR. It is possible to perform this step with a mixture of microsatellite motifs that have the same melting temperature to increase the variety of the hybridization products.
Based on the biotin binding results, it is preferable for the biotin to be in the 3′ position of the oligo-repeat. The 3′ position is preferable to the 5′ position because the biotinylating reaction occurs more efficiently at this position (i.e., more DNA molecules incorporate the biotin when 3′ biotin is used). The labeled oligos can be purified using either the standard desalting method or an additional purification step (e.g., via high-performance liquid chromatography [HPLC]). If the standard desalting method is used, the solution will contain an excess of free biotin molecules that must be taken into account in the next step (Avidin calculation).
It is very important to perform the hybridization reaction with a PCR designed to have an initial denaturing step and a gradual touchdown near the probe’s melting temperature (Tm) (–2°C). It is ideal to calculate the real Tm, taking into consideration the salt (Na+) concentration present in the hybridization buffer. Several Tm calculators are freely available online (e.g., http://www.basic.northwestern.edu/biotools/OligoCalc.html).
Step 7: Preparation and VETREX Avidin D capture
Vectrex Avidin D (Vector Laboratories, Burlingame, California, USA) was employed to capture the hybridization mixture. For a biotinylated oligo-repeat purified by desalting, ∼40 μL of Vectrex Avidin D is required, i.e., about twice the quantity recommended by the manufacturer (binding capacity = 25 ng biotin/μL Vectrex Avidin D).
Step 8: Second enrichment and cloning
Triplicate PCR reactions were performed to amplify the selected DNA fragments to a final concentration of roughly >10 ng/μL. The amplified template was cloned after DNA purification/concentration, according to the manufacturer’s protocol. Several cloning kits are available. We have tested both the Clone Jet PCR cloning kit (Fermentas, Thermo Fisher Scientific, Waltham, Massachusetts, USA) and the PMosBlue blunt-ended cloning kit (GE Healthcare Europe GmbH, Vienna, Austria), which provided comparable results. No differences were observed, except for the inclusion of competent cells in the second kit.
Step 9: Colony screening and sequencing
The amplified colonies were sequenced directly by the modified Sanger method using BigDye Terminator Cycle Sequencing Kit version 3.1 (Applied Biosystems, Life Technologies, Paisley, Renfrewshire, United Kingdom) and a 3130 Genetic Analyzer (Applied Biosystems, Life Technologies). A modified protocol for the optimization of sequencing reagents is reported in Appendix S1 (158.5KB, pdf) .
Microsatellites were defined considering the minimum repeat unit as six for di- and five for tri-, tetra-, penta-, and hexanucleotides. The frequency of microsatellites in the sequenced colonies ranged from approximately 20–32% in P. maritimum to 58–71% in G. sulphuraria and 42–55% in K. saxicola. Approximately 80–84% of the microsatellites obtained are usable for primer design. The types and frequencies of the repeats obtained using the GA12 motif repeat in the hybridization reaction are reported in Table 1. According to these results, the efficiency of this method differed among the different species studied.
Table 1.
Information regarding the microsatellites obtained from the GA-enriched library through the sequencing of 130 colonies in Pancratium maritimum and 60 colonies in Kochia saxicola and Galdieria sulphuraria.
| Pancratium maritimuma | Kochia saxicolab | Galdieria suphurariac | ||||
| Type of repeat | Size range | Frequency (%) | Size range | Frequency (%) | Size range | Frequency (%) |
| AG | 7–33 | 6 | 8–27 | 8.4 | 10 | 1.7 |
| AT | 11 | 0.8 | — | — | 6 | 1.7 |
| CA | 7 | 0.8 | — | — | 6–19 | 16.7 |
| CT | — | — | 10–16 | 5 | 8–13 | 3.4 |
| GA | 6–27 | 10 | 6–25 | 15 | 7–25 | 13.3 |
| GT | 6–24 | 4.6 | 8–24 | 11.7 | 6–17 | 8.4 |
| TC | 12–20 | 2.3 | 12–22 | 8.4 | 16–34 | 16.7 |
| TG | 12–23 | 3.1 | 6–18 | 5 | 13–15 | 5 |
| Total | 27.6 | 53.3 | 66.9 | |||
Genome = ∼30 000 Mbp (Zonneveld et al., 2005).
Undetermined genome size.
Genome = ∼10 Mbp (Muravenko et al., 2001).
DISCUSSION
An essential prerequisite for developing an SSR library protocol is a knowledge of the type of genome being studied; there are many differences between animal and plant genomes as well as among plant species. Plants have a lower proportion of SSR sequences than vertebrates and a higher proportion of SSR sequences than both fungi and invertebrates (Toth et al., 2000; Morgante et al., 2002). The variety of plant SSR frequencies is correlated with the variation of the amount of single/low copy DNA and nonrepetitive DNA (e.g., retrotransposons), which is widely represented. Unlike animals, plants show an extreme variation in genome size, and genomes are generally larger, especially because of the large amounts of repetitive DNA (San Miguel et al., 1998; Morgante et al., 2002).
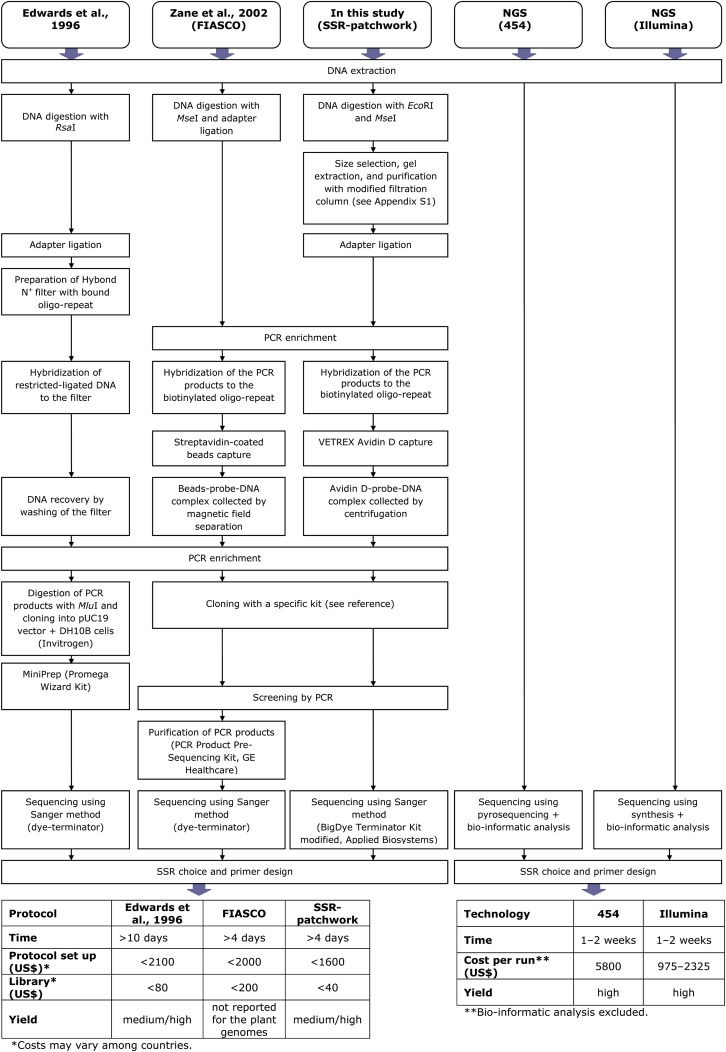
In this study, we describe a simple and optimized protocol to expedite the production of highly enriched SSR libraries using small fragments of plant genomic DNA with different genome sizes. Data from the current literature enabled the improvement of this procedure (see Introduction). We improved various steps, such as restriction enzyme digestion, hybridization reaction, cloning efficiency, and other smaller modifications with the aim of obtaining a better efficiency : cost ratio. A comparison with two previously published FGS-enrichment protocols is shown in Fig. 4. The two SSR protocols selected for comparison are those of Edwards et al. (1996), which was developed for plants (barley, maize, rhododendron, sunflower, sugar beet, wheat, and willow), and FIASCO (Zane et al., 2002), which has been used for animals (rock sparrow, gilt head bream, American anglerfish, horned krill, and red coral). Both procedures reported high yields (>50%), although we were unable to reproduce them, especially for the Edwards et al. (1996) (<2%) protocol, without colony hybridization screening. In addition, the selective hybridization step of this protocol was very time consuming (this was also confirmed by Zane et al., 2002).
Fig. 4.
Comparative schemes of costs and times in the SSR-patchwork protocol and other microsatellite isolation protocols, including both traditional approaches (enrichment and selective hybridization) and NGS (454 and Illumina sequencing technologies). The estimates were provided in the references Edwards et al. (1996); Fiasco: Zane et al. (2002); 454 and Illumina: Zalapa et al. (2012), except for the costs of the protocol by Edwards et al. (1996), which have been deduced by the authors.
The most crucial improvements of our protocol compared with Edwards et al. (1996) and Zane et al. (2002) are discussed below. First, the choice of restriction enzymes was the most crucial step influencing the final yield. Very frequently, the enzymes indicated in the protocols can be changed in the event of inefficient digestion. We tested the enzyme used in the Edwards protocol (RsaI, a four-base cutter) using our three genome templates without good results (i.e., inefficient or partial digestion), as also reported by Fischer and Bachmann (1998) and King et al. (2008). To reduce the time required for protocol setup, a good strategy is to use enzymes known to be good cutters on a variety of templates, such as those employed in AFLP, i.e., EcoRI (a six-base cutter) and MseI (a four-base cutter) (Vos et al., 1995). The FIASCO procedure uses only MseI. In our SSR-patchwork protocol, we preferred to use the classic and well-tested pair EcoRI + MseI. Furthermore, the obtained genome fragments (MseI-MseI, EcoRI-MseI, and EcoRI-EcoRI) were predominantly in the appropriate size range for the next steps (i.e., amplification and cloning).
Another key step of the SSR-patchwork protocol was the selection of small digested DNA fragments (250–500 bp) through size selection, gel extraction, and purification (Fig. 2), which is ideal for successful cloning using an inexpensive kit. In contrast, the FIASCO protocol recommends a very expensive cloning kit because it produces larger DNA fragments (200–1000 bp).
Neither Edwards et al. (1996) or FIASCO report details about the importance of annealing temperature in the hybridization reaction. In fact, as discussed above, optimization of the selective hybridization step was very time-consuming, according to Edwards et al. (1996). In this protocol, the hybridization temperature was very low (37°C for 24 h), producing a high level of nonspecific signal, while in FIASCO, the DNA is hybridized according to the protocol first published online by Travis Glenn. Unfortunately, this protocol is no longer available online, but the official SSR protocol published by the author (Glenn and Schable, 2005) used a moderate temperature (50°C for 10 min), emphasizing the importance of optimizing the annealing temperature for the probe.
In addition, several new “tricks” are implemented in the SSR-patchwork protocol compared with previous SSR protocols, e.g.: (1) an initial extension step in the first enrichment amplification (step 5) to fill the nicks present in the ligase reaction products (step 4); (2) the use of Vectrex Avidin D (Vector Laboratories) vs. streptavidin-coated beads in FIASCO, allowing the use of a normal centrifuge rather than magnetic field for the capture of the hybridization mixture (step 7); (3) the effective cost of the Sanger sequencing is very low (step 9) because the optimization is performed using a BigDye Terminator Cycle Sequencing Kit (Applied Biosystems, Life Technologies); and (4) the very detailed/optimized protocol provided (Appendix S1 (158.5KB, pdf) ) greatly reduces both the time required for procedure setup and costs.
Finally, the advantage of the SSR-patchwork protocol over existing techniques in time and cost is illustrated in Fig. 4.
CONCLUSIONS
The SSR-patchwork protocol presented here is simple, fast, inexpensive, does not require complicated experimental steps (Fig. 4), and is effective for both small and large genomes (Table 1). The general nature of this protocol makes it suitable for microsatellite library construction in a large variety of plant or animal taxa with a variety of genome sizes.
Supplementary Material
Appendix 1.
Voucher information for taxa in this study. Voucher information and algal strain codes were deposited at the Department of Biology, University of Naples Federico II (Italy). Information presented: taxon, geographical locality with GPS coordinates, voucher information–herbarium, and/or strain accession code–Algal Strains Collection.
| Kochia saxicola, Capri (southern Italy) (40°33′30.40″N 14°14′05.40″E), Vallariello-KS11-NAP; |
| Pancratium maritimum, Botanical Garden of Naples (southern Italy) (40°51′39.74″N 14°15′43.31″E), Vallariello-PMO-NAP (SPB); |
| Galdieria sulphuraria, Landmannalaugar (Iceland) (63°59′23.34″N 19°03′32.58″W), Ciniglia-ACUF 420. |
LITERATURE CITED
- Doyle J. J., Doyle J. L. 1990. Isolation of plant DNA from fresh tissue. Focus (San Francisco, Calif.) 12: 13–15 [Google Scholar]
- Edwards K. J., Barker A., Daly A., Jones C., Karp A. 1996. Microsatellite libraries enriched for several microsatellite sequences in plants. BioTechniques 20: 758–760 [DOI] [PubMed] [Google Scholar]
- Fischer D., Bachmann K. 1998. Microsatellite enrichment in organisms with large genomes (Allium cepa L.). BioTechniques 24: 796–802 [DOI] [PubMed] [Google Scholar]
- Glenn T. C. 2011. Field guide to next-generation DNA sequencers. Molecular Ecology Resources 11: 759–769 [DOI] [PubMed] [Google Scholar]
- Glenn T. C., Schable N. A. 2005. Isolating microsatellite DNA loci. Methods in Enzymology 395: 202–222 [DOI] [PubMed] [Google Scholar]
- Hamilton M. B., Pincus E. L., Di Fiore A., Fleischer R. C. 1999. Universal linker and ligation procedures for construction of genomic DNA libraries enriched for microsatellites. BioTechniques 27: 500–507 [DOI] [PubMed] [Google Scholar]
- Kalia R. K., Rai M. K., Kalia S., Singh R., Dhawan A. K. 2011. Microsatellite markers: An overview of the recent progress in plants. Euphytica 177: 309–334 [Google Scholar]
- Kandpal R. P., Kandpal G., Weissman S. M. 1994. Construction of libraries enriched for sequence repeats and jumping clones, and hybridization selection for region-specific markers. Proceedings of the National Academy of Sciences, USA 91: 88–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King J., Thorogood D., Edwards K. J., Armstead I. P., Roberts L., Skøt K., Hanley Z., King I. P. 2008. Development of a genomic microsatellite library in perennial ryegrass (Lolium perenne) and its use in trait mapping. Annals of Botany 101: 845–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgante M., Hanafey M., Powell W. 2002. Microsatellites are preferentially associated with nonrepetitive DNA in plant genome. Nature Genetics 30: 194–200 [DOI] [PubMed] [Google Scholar]
- Muravenko O., Selyakh A. I., Kononenko B. N., Stadnichuk C. I. 2001. Chromosome numbers and nuclear DNA contents in the red microalgae Cyanidium caldarium and three Galdieria species. European Journal of Phycology 36: 227–232 [Google Scholar]
- San Miguel P., Gaut B. S., Tikhonov A., Nakajima Y., Bennetzen J. L. 1998. The paleontology of intergene retrotransposons of maize. Nature Genetics 20: 43–45 [DOI] [PubMed] [Google Scholar]
- Squirrell J., Hollingsworth P. M., Woodehead M., Russel J., Lowe J., Gibby M., Powell W. 2003. How much effort is required to isolate nuclear microsatellites from plants? Molecular Ecology 12: 1339–1348 [DOI] [PubMed] [Google Scholar]
- Techen N., Arias R. E. S., Glynn N. C., Pan Z., Khan I. A., Scheffler B. E. 2010. Optimized construction of microsatellite-enriched libraries. Molecular Ecology Resources 10: 508–515 [DOI] [PubMed] [Google Scholar]
- Toth G., Gáspari Z., Jurka J. 2000. Microsatellites in different eukaryotic genomes: Survey and analysis. Genome Research 10: 967–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos P., Hogers R., Bleeker M., Reijans M., van de Lee T., Hornes M., Friters A., et al. 1995. AFLP: A new technique for DNA fingerprinting. Nucleic Acids Research 23: 4407–4414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalapa J. E., Cuevas H., Zhu H., Steffan S., Senalik D., Zeldin E., McCown B., et al. 2012. Using next-generation sequencing approaches to isolate simple sequence repeat (SSR) loci in the plant sciences. American Journal of Botany 99: 193–208 [DOI] [PubMed] [Google Scholar]
- Zane L., Bargelloni L., Patarnello T. 2002. Strategies for microsatellite isolation: A review. Molecular Ecology 11: 1–16 [DOI] [PubMed] [Google Scholar]
- Zonneveld B. J. M., Leitch I. J., Bennett M. D. 2005. First nuclear DNA amounts in more than 300 angiosperms. Annals of Botany 96: 229–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.