Abstract
• Premise of the study: We present a protocol for the annotation of transcriptome sequence data and the identification of candidate genes therein using the example of the nonmodel conifer Abies alba.
• Methods and Results: A normalized cDNA library was built from an A. alba seedling. The sequencing on a 454 platform yielded more than 1.5 million reads that were de novo assembled into 25149 contigs. Two complementary approaches were applied to annotate gene fragments that code for (1) well-known proteins and (2) proteins that are potentially adaptively relevant. Primer development and testing yielded 88 amplicons that could successfully be resequenced from genomic DNA.
• Conclusions: The annotation workflow offers an efficient way to identify potential adaptively relevant genes from the large quantity of transcriptome sequence data. The primer set presented should be prioritized for single-nucleotide polymorphism detection in adaptively relevant genes in A. alba.
Keywords: Abies alba, adaptation, annotation, candidate genes, de novo sequencing, Pinaceae
To gain insights into the molecular level of adaptation, attention has turned to the investigation of adaptively relevant genes (candidate genes). For nonmodel organisms, access to candidate genes is limited and the transfer of primers, e.g., from expressed sequence tag (EST) libraries, if available, requires high labor costs. For instance, the resequencing of 800 genes selected from more than 7000 ESTs from Pinus taeda L. yielded only 70 candidate genes for Abies alba Mill. (Mosca et al., 2012). Because sequencing costs are decreasing rapidly, de novo sequencing in nonmodel organisms is now achievable. For the identification of candidate genes in de novo–sequenced organisms, the use of differential expression profiling (e.g., Street et al., 2006; Huang et al., 2012) can be performed, but it requires the sequencing of several samples. The sequencing of a single transcriptome, in contrast, is very cost-effective. However, the reduction of the data remains challenging. Blasting against available databases is the standard method, which results in outputs of large quantities and is therefore mainly used for annotation only (e.g., Parchman et al., 2010). Here, we present a protocol for the efficient reduction of transcriptomic data down to 283 candidate gene sequences that were used for immediate primer development. The protocol is applicable for species that lack genomic resources. It combines a standard and a specific annotation approach and led to the resequencing of 88 gene fragments in A. alba.
METHODS AND RESULTS
A normalized transcriptome of a 1-yr-old A. alba seedling from the Black Forest (Forest District Calw, Germany; voucher MB-P-001007, Herbarium Marburgense, University of Marburg) was sequenced on a 454 GS FLX Titanium platform (cDNA library preparation: Vertis Biotechnology AG, Freising, Germany; sequencing: Genoscreen, Lille, France). The 454 run yielded 1521698 reads with an average length of 359 nucleotides (nt). Trimming and de novo assembly of the raw reads into contigs using Newbler software version 2.3 (454 Life Sciences, Branford, Connecticut, USA) resulted in 25149 contigs consisting of 381808 complete and 619615 partially assembled reads. The contig length was between 100 nt and 2394 nt, with an average length of 498 nt. A total of 484576 reads remained as singletons (Table 1). Contigs were submitted to the Transcriptome Shotgun Assembly database (TSA) at the National Center for Biotechnology Information (NCBI) (accession no.: JV134525–JV157085).
Table 1.
Statistics of the 454 transcriptome sequencing run and metrics of the Newbler assembly software.
| Size (nt) in quantiles | |||||||||
| Sequence type | Number | % | Nucleotides | Average (nt) | 0% | 25% | 50% | 75% | 100% |
| Reads trimmed | 1521698 | 100 | 546346058 | 359.0 | <21 | <303 | <395 | <444 | <1088 |
| Reads assembled | 381808 | 25.1 | |||||||
| Reads partial | 619615 | 40.7 | |||||||
| Reads singleton | 484576 | 31.8 | 175198711 | 361.6 | <50 | <307 | < 397 | < 443 | <876 |
| Reads repeat | 1617 | 0.1 | |||||||
| Reads outlier | 20389 | 1.3 | |||||||
| Reads too short | 13693 | 0.9 | |||||||
| Contigs | 25149 | 12511848 | 498 | <100 | <365 | <468 | <601 | <2394 | |
| N50 Contiga | 704 | ||||||||
Half of all bases are assembled in contigs of this size or longer.
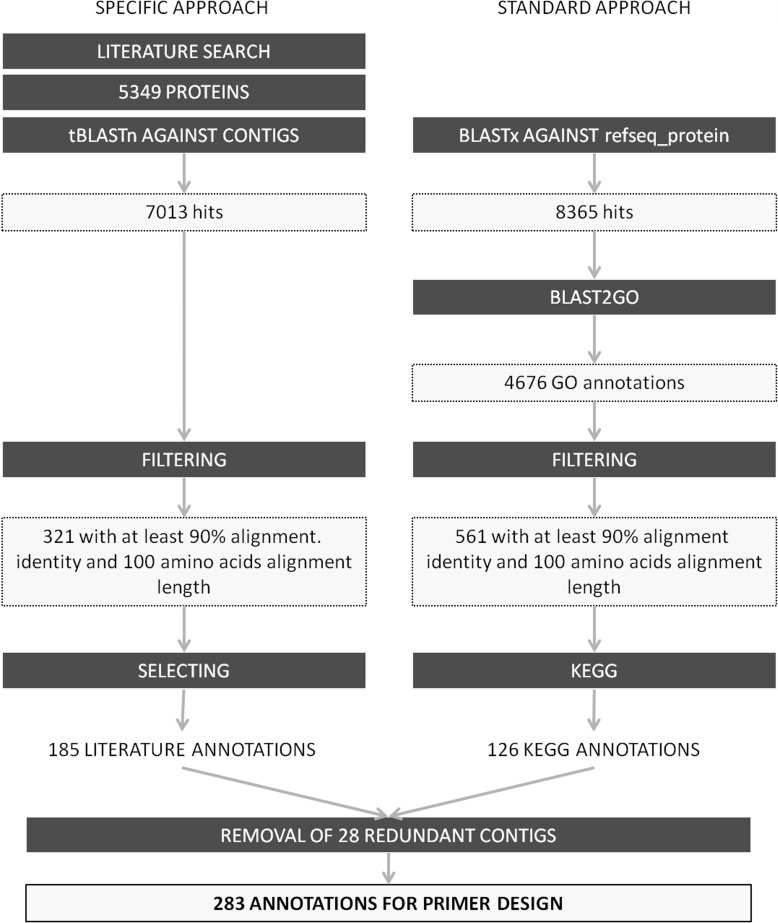
In the specific approach (Fig. 1), we tested a novel annotation protocol: After a literature survey with key words “adaptation,” “candidates,” “drought,” “evolution,” “RT-PCR,” and “selection” in various combinations using the Web of Science database, we selected 5349 unique proteins and downloaded them from UniProt or NCBI (downloaded in November 2011). The proteins were subsequently searched against the contigs coming from the de novo transcriptome sequencing that were formatted as the reference database using the BLAST+ 2.2.24 toolkit (tBLASTn parameters: softmasking = threshold 15 max_target_seqs 10000). To increase reliability of alignments and to avoid too-short amplicons, only alignments with a length of at least 100 amino acids and an identity of at least 90% were considered further. From the contigs that passed the filter, 157 were selected for primer design. In the standard approach (Fig. 1), contigs were searched against the refseq_protein database (downloaded from NCBI 14 June 2011) with strict BLAST-settings (BLASTx parameters: threshold 999, window-size 4, gapopen 32767, gapextend 32767, E-value 1e−20) (Altschul et al., 1990). Gene ontologies (Ashburner et al., 2000) were assigned to contig-protein hits using Blast2GO 2.5.0 (Conesa et al., 2005) and subsequently filtered as described above. To select for well-described proteins, contig sequences were used for primer design if they could be assigned to enzyme IDs with the Kyoto Encyclopedia of Genes and Genomes (KEGG) (Ogata et al., 1999) in the final annotation step. Primers were developed specifying the amplified range according to the contig-protein alignment boundaries using default standard PCR settings of PerlPrimer (version 1.1.12; Marshall, 2004). Primers were tested in a 30 μL PCR reaction with 17.28 μL double-distilled water, 3 μL 10× PCR buffer with MgCl2 (20 mM), 1.2 μL MgCl2 (25 mM), 3 μL Primermix (forward and reverse each 2 μM), 1.44 μL dNTPs (each 5 mM), 0.24 μL bovine serum albumin (BSA) (20 mg mL−1), 0.24 μL Dream Taq polymerase (5 U μL−1, Fermentas, St. Leon-Rot, Germany), and 3.6 μL DNA (10 ng μL−1). The PCR was performed with 5 min initial denaturation at 94°C followed by 35 repetitions of 45 s denaturation at 94°C, 45 s annealing at 52–59°C, 45 s elongation at 72°C, and a 10 min final elongation at 72°C. For the amplification test, four samples were randomly chosen for each gene from a set of 80 different silver fir trees that were sampled in May 2011 in Mont Ventoux (44°10′44.35″N, 5°14′32.29″E, France). Amplification was evaluated by electrophoresis in 1% agarose gels. When amplification was too weak, the volume of MgCl2 was increased to 1.8 μL. When faint ancillary bands appeared, no additional magnesium was added to the mastermix. If PCR products occurred as a single band, one sample was chosen for sequence analysis in each case to ensure that the region of interest was amplified (LGC Genomics GmbH, Berlin, Germany). Gene sequences were aligned to the corresponding contigs using the CodonCode Aligner software (default large gap settings) to reveal the location of the introns. The gene sequences were searched against the nr nucleotide database of NCBI (default discontiguous megaBLAST settings, web application).
Fig. 1.
Workflow of the annotation protocol. Numbers of the output after each step are given. The standard approach starts with 25149 contigs. The specific approach uses them as the reference database for the tBLASTn step.
In the specific approach, tBLASTn and subsequent sorting led to 321 contigs. For primer development, 185 contigs were picked. In the standard approach, the initial number of contigs was decreased to one third after the BLASTx step. Approximately half of the hits could be further annotated with Gene Ontologies. After filtering, 126 contigs were successfully assigned to enzyme-IDs and used for primer design (Fig. 1). In combination, 283 different contigs were annotated and only 28 were annotated with both approaches. Primer testing and sequencing resulted in 88 gene sequences (Table 2). Fifty-seven genes were annotated using the specific approach, and 42 using the standard approach. Eleven were annotated by both approaches. The assembly of the gene sequences and the corresponding cDNA contigs revealed 43 introns in 26 genes. The length of the gene sequences ranged from 262 to 1486 nt. All gene sequences aligned to sequences from the nr nucleotide database (NCBI) where the highest E-value was 5.00e−32. Twelve gene sequences hit organelle DNA (10 chloroplast, one mitochondrial, and one ribosomal). The remaining 76 are involved in the biosynthesis of different compounds (21), regulation (20), primary metabolism (14), growth (11), stress response (8), and water transport (2). In the biosynthesis group, enzymes from the auxin pathways, the phenylpropanoid pathways, and the tetrapyrrol pathways were dominant. With the exception of the primary metabolism group, all groups included candidates for the analysis of adaptation at gene level that had been investigated in previous studies of conifers (e.g., González-Martínez et al., 2006).
Table 2.
Primers for resequencing of annotated gene fragments in Abies alba.a
| Gene Locus ID | Primer sequences (5′–3′) | Ta (°C) | No. of introns | Intron length (nt) | Total length (nt) | Annotation approach | BLASTn of gene sequences against nr nucleotide database (E-value) | GO-ID biological process |
| 95 | F: ACAGAAACTAAAGCTAGTGTCG | 57 | 0 | — | 696 | 1 | Keteleeria davidiana chloroplast DNA, complete sequence (0) | reductive pentose-phosphate cycle, photorespiration, oxidation reduction |
| R: CCTTAATTTCACCCGTCTCAG | ||||||||
| 215 | F: CCAAGGACTCTGATCGAATCC | 56 | 2 | 411 | 1486 | 2 | Abies firma clone 1 4-coumarate:CoA ligase (4CL) gene, partial cds (0) | response to UV, response to wounding, phenylpropanoid metabolic process, response to fungus |
| R: GAAGCCAGCATTCAAAGACTC | ||||||||
| 241 | F: AACGTCCGTTAATACTTCGG | 56 | 3 | 256 | 1370 | 2 | Arabidopsis thaliana fructose-bisphosphate aldolase, class I (FBA1) mRNA, complete cds (1E-125) | glycolysis |
| R: AGTAAGTGTAGCCCTTCACG | ||||||||
| 323 | F: AAGCAAGCTTCTGAAATTCC | 53 | 2 | 278 | 804 | 1 | A. thaliana plasma membrane H+-ATPase gene, complete cds (1E-90) | auxin biosynthetic process, ATP biosynthetic process, proton transport |
| R: TGGTAGAGTCTACCAAATGAG | ||||||||
| 1362 | F: GAAGAGGTAGCTGCATTGGT | 59 | 0 | — | 871 | 1 | Ricinus communis processing-splicing factor, putative, mRNA (0.0) | response to hypoxia, sucrose biosynthetic process, nuclear mRNA splicing, via spliceosome |
| R: GGGCTTATACCGTAAATATACCCA | ||||||||
| 1704 | F: CAACTACTTCAGAGACAGAC | 52 | 2 | 327 | 858 | 2 | Pinus taeda mitogen-activated protein kinase 13 (MAPK13) mRNA, complete cds (2E-84) | |
| R: AAAGATTCCCTCCAAATCAG | ||||||||
| 2387 | F: TAAATGGCTCAATTCCTCCTACTG | 61 | 1 | 128 | 624 | 1 | Medicago truncatula Alpha-1,4-galacturonosyltransferase (MTR_7g075840) mRNA, complete cds (8E-99) | |
| R: GTTCCAAGCTTCCACAATACTC | ||||||||
| 2565 | F: GTGTCTGGAAGGGAATACAAGG | 58 | 0 | — | 432 | 1 | PREDICTED: Vitis vinifera adenosylhomocysteinase-like, transcript variant 1 (LOC100253872), mRNA (1E-109) | embryonic development ending in seed dormancy, one-carbon metabolic process, posttranscriptional gene silencing, methylation-dependent chromatin silencing |
| R: CCTTGACTCCTTCATGGATCAG | ||||||||
| 2774 | F: GTTACAGGAAGCCTTTCTGG | 55 | 0 | — | 502 | 2 | Citrus sinensis pectinesterase mRNA, complete cds (5E-32) | cell wall modification |
| R: GCGGGATGAATTATCTTGTC | ||||||||
| 2937 | F: TGAGCTGATTGCTAATGCGG | 58 | 0 | — | 622 | 2 | Solanum tuberosum clone 154D06 fructose-bisphosphate aldolase-like mRNA, complete cds (5E-120) | glycolysis |
| R: GGACATGGTGGTCATTGAGG | ||||||||
| 2986 | F: CTGTCTGTGACGGATCTAGC | 57 | 0 | — | 355 | 1 | Populus trichocarpa arogenate/prephenate dehydratase (PDT1), mRNA (1E-52) | l-phenylalanine biosynthetic process |
| R: TGAGGATGGCTTACAACACG | ||||||||
| 3421 | F: CTCATCTCTGCCAGAAAGAC | 55 | 0 | — | 324 | 2 | Picea sitchensis isolate CR201 phenylalanine ammonia lyase-like protein mRNA, partial cds (0.0) | phenylpropanoid metabolic process, biosynthetic process, l-phenylalanine catabolic process |
| R: GTAGAGCTTCATCTACGAGG | ||||||||
| 3593 | F: AGGACCTGAAATACCTTGCT | 56 | 0 | — | 337 | 2 | Abies firma chloroplast, partial genome (6E-170) | transport, respiratory electron transport chain, photosynthesis |
| R: TCCGTGTTTATCTCACAGGT | ||||||||
| 3689 | F: CGATTGCATCTCTGTACGCC | 58 | 0 | — | 619 | 2 | Pseudotsuga menziesii var. menziesii haplotype Pm-TBE_412m2 thiazole biosynthetic enzyme (TBE) gene, complete cds (0.0) | thiamin biosynthetic process |
| R: GCTCTTGAGCCTCTTGACAC | ||||||||
| 3918 | F: TTCCAAGGTCTTCTCAAGGT | 55 | 0 | — | 400 | 2 | Pinus taeda cellulose synthase catalytic subunit (CesA1) mRNA, complete cds (0.0) | cellulose biosynthetic process, cellular cell wall organization, secondary cell wall biogenesis, rhamnogalacturonan I side chain metabolic process |
| R: TGAAGAGTAGGAGTTTCGGT | ||||||||
| 3942 | F: GTATGATACCGATGTGACGA | 55 | 0 | — | 273 | 2 | Ricinus communis proteasome subunit alpha type, putative, mRNA (8E-48) | ubiquitin-dependent protein catabolic process |
| R: TTTGTAATGGATGCACTCGG | ||||||||
| 3981 | F: GGAGAAGTCTACAGTTCCAG | 54 | 0 | — | 918 | 1 | Pinus radiata UDP-glucose dehydrogenase gene, partial sequence (0.0) | oxidation reduction |
| R: ATAGTCCAGTGTCTTGAACTC | ||||||||
| 4103 | F: ATGGCCACCTTACTAAGAAGC | 57 | 0 | — | 841 | 1 | Pinus pinaster mRNA for S-adenosylmethionine synthase 1 (sams1 gene) (0.0) | auxin biosynthetic process, one-carbon metabolic process |
| R: CCACTTAAGGACCTTTACAGTCTC | ||||||||
| 4492 | F: TGGGTGCAACTGAAGATAGAG | 57 | 0 | — | 698 | 1 | Medicago truncatula magnesium-chelatase subunit chlI (MTR_2g015390) mRNA, complete cds (4E-160) | auxin biosynthetic process, chlorophyll biosynthetic process, photosynthesis |
| R: TTTCTACAACTAGCAAGCCTGAG | ||||||||
| 4921 | F: GAAGGTCGGCTATATCAGGT | 56 | 0 | — | 664 | 2 | PREDICTED: Glycine max proteasome subunit alpha type-4-like, transcript variant 1 (LOC100786457), mRNA (2E-147) | response to cadmium ion, ubiquitin-dependent protein catabolic process |
| R: AGCTTAGACAGAGACTCAGG | ||||||||
| 5004 | F: CAGATGTGAGCCATTACTTTGAC | 57 | 0 | — | 461 | 1 | Picea sitchensis isolate VD401 magnesium chelatase H-like protein mRNA, partial cds (0) | chlorophyll biosynthetic process |
| R: CAACCTCTGAATATAGCTGCCT | ||||||||
| 5823 | F: TGCTTGATATACGTCCTGGG | 57 | 0 | — | 293 | 2 | Picea sitchensis isolate VD401 phytochrome A-like protein mRNA, partial cds (0) | regulation of transcription, photomorphogenesis, tryptophan biosynthetic process |
| R: CTAGACAGTGTTGCTCCACG | ||||||||
| 5945 | F: CTGTCACTCAGATCTTCAGC | 55 | 0 | — | 339 | 2 | P. abies (L.) Karst. Lhcb1*2-2 mRNA for light-harvesting chlorophyll a/b-binding protein (0.0) | photosynthesis, light harvesting, protein-chromophore linkage |
| R: AGATGATCAGCGAGATTCTC | ||||||||
| 6119 | F: AGAGGATGTTGGGCATTATGG | 57 | 0 | — | 567 | 1 | Picea mariana pyruvate dehydrogenase E1 beta subunit (Sb68) mRNA, partial cds (0.0) | pollen tube development, oxidation reduction |
| R: CATCACATGGTATCTCATCCGA | ||||||||
| 6594 | F: TGGCTTTATCTTGGAGACTTCAC | 58 | 1 | 348 | 712 | 1 | Ricinus communis phosphatidylinositol 4-kinase, putative, mRNA (5E-51) | phosphoinositide biosynthetic process, phosphoinositide phosphorylation, signal transduction, phosphoinositide-mediated signaling |
| R: GAATAAGGTCATAGCCTGCCG | ||||||||
| 6757 | F: TATCATGCCCTGAAAGCGTC | 58 | 5 | 177 | 939 | 1 | Arabidopsis thaliana ribonucleoside-diphosphate reductase subunit M1 (RNR1) mRNA, complete cds (1E-39) | |
| R: ACTTCCACAAGCAAGACACTC | ||||||||
| 7098 | F: CTTTACTGTTGGAGGTAGATCAG | 55 | 0 | — | 782 | 1 | Arabidopsis thaliana UDP-glucuronic acid decarboxylase (AUD1) mRNA, complete cds (1E-153) | dTDP-rhamnose biosynthetic process, d-xylose metabolic process |
| R: GTTTGTTTGTCTTTGTACTCCC | ||||||||
| 7208 | F: GTTACATTCGTAAGTAGCTTGG | 54 | 0 | — | 326 | 1 | Pinus thunbergii NADH dehydrogenase subunit 5 (nad5) gene, partial cds; mitochondrial (0) | transport, ATP synthesis coupled electron transport |
| R: AAATGGTCGAGAAGTCTACTG | ||||||||
| 7324 | F: ATTGGAGATGGAGCCATGAC | 57 | 0 | — | 471 | 1 | Picea abies 1-deoxy-d-xylulose 5-phosphate synthase type I (DXS1) mRNA, complete cds (0) | terpenoid biosynthetic process, thiamin biosynthetic process |
| R: TCTCTGCATATGGGTAACCC | ||||||||
| 8248 | F: CAAGTATTCCGAAAGGCAGC | 57 | 1 | 601 | 1128 | 2 | P. abies mRNA for porin Mip1 (3E-154) | response to water deprivation, water transport, transmembrane transport, response to salt stress |
| R: ACAAAGGTGCCCACAATCTC | ||||||||
| 8583 | F: TCTCCTACATTGACGATCCC | 56 | 0 | — | 393 | 2 | Picea sitchensis isolate VA301 phenylalanine ammonia lyase-like protein mRNA, partial cds (5E-162) | phenylpropanoid metabolic process, biosynthetic process, l-phenylalanine catabolic process |
| R: CCATCCAAGCACTTGAAGAG | ||||||||
| 8855 | F: TATTTGCTGGTCGGGATTCG | 58 | 2 | 275 | 926 | 2 | P. sylvestris Lhca4*1-2 mRNA encoding Lhca4 protein (type 4 protein of light-harvesting complex of photosystem I) (partial) (7E-179) | photosynthesis, light harvesting |
| R: CTGCACTAGGTTCTCGAACG | ||||||||
| 9366 | F: AGTGAAAGCAACAACTTAGG | 53 | 0 | — | 598 | 1 | Tamarix hispida peroxiredoxin 2 (Prx2) mRNA, complete cds (5E-139) | cell redox homeostasis, oxidation reduction |
| R: TCTGGCTTCATTGATTTGTC | ||||||||
| 9512 | F: GTACTGGAGTAGCTGCACGA | 59 | 1 | 99 | 415 | 2 | Cycas revoluta class III HD-Zip protein HDZ32 gene, partial cds (4E-52) | regulation of transcription, DNA-dependent |
| R: TACAAAGTGCTGCACAGCAG | ||||||||
| 9652 | F: TGCAAAGAAAGTCAAGGCGA | 58 | 2 | 418 | 913 | 2 | Pinus pinaster COBRA-like protein gene, partial cds (0) | |
| R: CCCATACGGTGTTAATGGCT | ||||||||
| 11301 | F: GATGTTGTTCGTGCAAAGAC | 54 | 0 | — | 490 | 2 | Pinus pinaster mRNA for malate dehydrogenase (MDH gene) (0.0) | malate metabolic process, oxidation reduction, tricarboxylic acid cycle, glycolysis |
| R: GCGAACTTAATTCCCTTCTC | ||||||||
| 13329 | F: GATATGTGCCCAAGAACATTCTG | 57 | 0 | — | 350 | 1 | PREDICTED: Glycine max probable rhamnose biosynthetic enzyme 1-like (LOC100789909), mRNA (7E-87) | |
| R: CCTTGCATGCTTCAAGAAGG | ||||||||
| 13536 | F: CTGCTGATTCTGATCAGTCC | 56 | 0 | — | 368 | 2 | Pinus thunbergii PtANTL1 mRNA for AINTEGUMENTA-like protein, complete cds (0.0) | regulation of transcription, DNA-dependent |
| R: TCCACAATGCAAACATAGGC | ||||||||
| 14455 | F: GAACAAGATCGACTACTGCC | 56 | 0 | — | 834 | 2 | Pinus taeda mRNA for alpha-1,6-xylosyltransferase (x34.1 gene) (0.0) | root hair elongation, xyloglucan biosynthetic process |
| R: TTTGATGGCCTTGAAAGCAG | ||||||||
| 14479 | F: CCACTCCCAAGTACTCAAAGG | 57 | 0 | — | 588 | 1 | Picea abies mRNA for translation elongation factor-1 alpha, partial (0.0) | translational elongation |
| R: CAAGTGTGGCAATCCAACAC | ||||||||
| 14514 | F: GGGTTCTGATTCTCCAAAGG | 56 | 0 | — | 322 | 2 | Metasequoia glyptostroboides fructose-1,6-diphosphate aldolase mRNA, complete cds (2E-74) | pentose-phosphate shunt, response to salt stress, glycolysis, response to cadmium ion |
| R: CTGCATACTTGGCCAAAGTG | ||||||||
| 14585 | F: TCTTGAATTCTTCCTATGTCCCAG | 57 | 1 | 193 | 915 | 1 | PREDICTED: Vitis vinifera galacturonosyltransferase 8-like (LOC100258818), mRNA (6E-119) | homogalacturonan biosynthetic process |
| R: AATTGCACATCTGCACAAACTC | ||||||||
| 14887 | F: GGTTAGACCAGTTCATAACC | 53 | 0 | — | 1156 | 2 | PREDICTED: Glycine max elongation factor 2-like (LOC100788357), mRNA (0.0) | |
| R: GTCTTCAAACTCTGACAAGG | ||||||||
| 15135 | F: TTGCAGGACTTCTTTAATGG | 53 | 0 | — | 657 | 2 | Ricinus communis heat shock protein, putative, mRNA (0.0) | oxidation reduction, response to stress, auxin biosynthetic process |
| R: TCTTCTTGTCAGATGGATCC | ||||||||
| 15337 | F: TTTATTGTATTCCTCCTAGGCCAG | 57 | 1 | 232 | 1086 | 1 | Picea glauca isolate D8411049-162 cellulose synthase family protein gene, partial sequence (0.0) | cellulose biosynthetic process, cellular cell wall organization |
| R: CACAATCTAAGCCACATTCTTCC | ||||||||
| 15484 | F: TTCGACGCCAACGTTATCTG | 58 | 0 | — | 663 | 2 | Pinus pinaster phenylalanine ammonia-lyase (pal2) mRNA, complete cds (0.0) | phenylpropanoid metabolic process, biosynthetic process, l-phenylalanine catabolic process |
| R: GGCCCAGAGAATTGACATCC | ||||||||
| 15727 | F: CACTGAAGGTTGTGGACGAG | 58 | 0 | — | 325 | 2 | Pinus pinaster mRNA for cytosolic serine hydroxymethyltransferase (cshmt gene) (2E-138) | l-serine metabolic process, one-carbon metabolic process, glycine metabolic process |
| R: GTTCAGAAGGGCTGTGTAGG | ||||||||
| 15811 | F: TTCGAGATCATCTGGACTGC | 57 | 0 | — | 438 | 2 | Abies alba genotype Lamacce 1 chalcone synthase (CHS) gene, CHS-A8 allele, complete cds (0) | biosynthetic process |
| R: CGACTGTTTCGACAGTGAGG | ||||||||
| 15969 | F: GGAAACCTTCTTGTTCACATCTG | 57 | 0 | — | 990 | 1 | Pinus contorta S-adenosylmethionine synthetase (sams2) mRNA, complete cds (0.0) | auxin biosynthetic process, one-carbon metabolic process |
| R: CTTGTCTGGAATCCTCCCTG | ||||||||
| 16727 | F: GGTGACTGTGAAGGCAATGG | 58 | 0 | — | 331 | 2 | Populus EST from severe drought-stressed opposite wood (0.000000003) | lipid transport |
| R: TCCCACATTTCTTTCCAGCT | ||||||||
| 16816 | F: CATCTTGGCTTCGTGATTGTC | 57 | 3 | 132 | 562 | 2 | Pseudotsuga menziesii class III homeodomain-leucine zipper (C3HDZ1) gene, complete cds (0) | |
| R: TGCAATTTGGCGTAATCGAC | ||||||||
| 16883 | F: CTCACAGAGGTCAGAAAGAATGG | 58 | 0 | — | 710 | 1 | Pinus contorta S-adenosylmethionine synthetase (sams2) mRNA, complete cds (0.0) | auxin biosynthetic process, one-carbon metabolic process |
| R: CTGCTTCAAAGGTTTGACAATCTC | ||||||||
| 16979 | F: CCTGGATAGTGAAATTGGAGG | 55 | 0 | — | 535 | 1 | Keteleeria davidiana chloroplast DNA, complete sequence (0) | auxin biosynthetic process |
| R: ATCCTTCTCTGAATGAGTTTCG | ||||||||
| 17340 | F: CTTGGTTAATTTCCGTCCTG | 54 | 0 | — | 281 | 2 | Abies firma chloroplast, partial genome (0) | transport, photosynthesis, electron transport chain |
| R: CAGCTCCTACATTTAAACCC | ||||||||
| 17637 | F: TGCTGAGAAAGTTGATTCTTCC | 56 | 0 | — | 424 | 1 | Ricinus communis transferase, transferring glycosyl groups, putative, mRNA (2E-69) | |
| R: GTATTCGAGGTGTAGATTGCTG | ||||||||
| 17975 | F: CAAACATTGCTGCAAAGCTC | 56 | 2 | 94 | 547 | 1 | Ricinus communis cysteine synthase, putative, mRNA (1E-65) | cysteine biosynthetic process from serine |
| R: CCTATTCCAGCAACCAATATGTC | ||||||||
| 18135 | F: GAGACTTTGGATTCGATCCC | 55 | 1 | 132 | 683 | 2 | Picea abies mRNA for putative chlorophyll A-B binding protein, (pPA0001 gene) (0) | photosynthesis, light harvesting in photosystem I |
| R: AGAAGGCCGCAAATATAGTG | ||||||||
| 18444 | F: ATTAATCTTTGCAGGGAAGC | 54 | 0 | — | 313 | 2 | P. sylvestris mRNA for polyubiquitin (3E-116) | |
| R: AGACGAGATGAAGTGTAGAC | ||||||||
| 18599 | F: GGAATGCATGATCCATTTCTG | 55 | 0 | — | 678 | 1 | S. tuberosum mRNA for NADH dehydrogenase, NADH-binding subunit (complex I) (0.0) | oxidation reduction |
| R: TACCTGAATTGTTCTTGCGA | ||||||||
| 18680 | F: CTGCGATGGATAAACTACCT | 55 | 1 | 214 | 465 | 2 | Picea glauca isolate D761009-28 myb family protein gene, partial sequence (1E-140) | regulation of transcription |
| R: GCTAGTGTTGCTATTGTGGG | ||||||||
| 19005 | F: GGAGATTGAGCAACGAAGAG | 56 | 0 | — | 368 | 1 | Abies firma chloroplast, partial genome (0) | auxin biosynthetic process |
| R: TTTGAATCCCTGAAATCCTGG | ||||||||
| 19173 | F: AGAACCAATCCCTGTTACAC | 55 | 0 | — | 343 | 2 | Ricinus communis proteasome subunit alpha type, putative, mRNA (2E-84) | defense response to bacterium, ubiquitin-dependent protein catabolic process, response to zinc ion |
| R: GATCAGTTCCAATCACACCT | ||||||||
| 19540 | F: ACCAATTCTCTTGTTCTCGG | 55 | 0 | — | 634 | 1 | Cedrus deodara chloroplast DNA, complete sequence (0) | plasma membrane ATP synthesis coupled proton transport, auxin biosynthetic process |
| R: CGAACCATGTAAAGATCATTCC | ||||||||
| 20156 | F: ATGGATCCCTGGAATTTATGC | 55 | 0 | — | 386 | 1 | Picea sitchensis isolate VD401 magnesium chelatase H-like protein mRNA, partial cds (3E-110) | RNA processing, chlorophyll biosynthetic process |
| R: ATACTCTACCTACTACAGAATCCC | ||||||||
| 20318 | F: ACAGCTCCCATTAATCTGAC | 55 | 0 | — | 356 | 1 | PREDICTED: Glycine max cellulose synthase-like protein D3-like (LOC100785985), mRNA (6E-69) | root hair elongation, cellulose biosynthetic process, response to cold, cellular cell wall organization, plant-type cell wall biogenesis |
| R: CCAGAATTGTTCATTTCTCCAC | ||||||||
| 20694 | F: GTCGAACAATGAAGACGAGG | 56 | 0 | — | 346 | 1 | PREDICTED: Vitis vinifera zinc finger CCCH domain-containing protein 49-like (LOC100259323), mRNA (6E-43) | cell wall modification, regulation of transcription |
| R: TGTGAGCGAAGAAACAAACC | ||||||||
| 21136 | F: AGACTGGTGTTACATTTGCGT | 57 | 1 | 229 | 535 | 1 | P. taeda gene for protochlorophyllide reductase (3E-168) | oxidation reduction, chlorophyll biosynthetic process, photosynthesis |
| R: CCAACAAGCTTCTCACTAATTTCC | ||||||||
| 21165 | F: ATGCACGATGTTCTTGATGC | 57 | 2 | 204 | 644 | 1 | PREDICTED: Glycine max premRNA-processing-splicing factor 8-like (LOC100804026), mRNA (4E-46) | response to hypoxia, sucrose biosynthetic process |
| R: GGTGTCATGTTTATATGACAGTGG | ||||||||
| 21173 | F: ACATTGTTGCTAACGATCCG | 56 | 0 | — | 333 | 2 | Picea sitchensis isolate VA100 basic endochitinase-like protein mRNA, partial cds (1E-138) | cell wall macromolecule catabolic process, chitin catabolic process |
| R: AGACGAGGTAGAGATTGAGC | ||||||||
| 21890 | F: GAAAGCTTACAGGGAAGCAG | 55 | 1 | 358 | 607 | 2 | Picea sitchensis isolate VD401 SWAP domain-containing protein-like protein mRNA, partial cds (2E-116) | RNA processing |
| R: ACGATATCCAAGCATCATCC | ||||||||
| 21957 | F: AACAACTTCACAGTTTCTCC | 54 | 0 | — | 292 | 2 | Abies firma chloroplast, partial genome (2E-157) | auxin biosynthetic process, chlorophyll biosynthetic process, oxidation reduction, photosynthesis, dark reaction |
| R: GGAATCGGTAAATCAACGAC | ||||||||
| 22174 | F: GATGATCCGGTTCGAATACC | 55 | 0 | — | 334 | 1 | Abies firma chloroplast, partial genome (6E-157) | regulation of apoptosis, transcription, DNA-dependent |
| R: AAACGTAAGATACAAGTGGGTG | ||||||||
| 23660 | F: AGGAAGATGTTAGGCTCGGG | 58 | 1 | 781 | 1232 | 2 | P. abies mRNA for porin Mip1 (6E-157) | response to water deprivation, water transport, transmembrane transport |
| R: GAAGCCCTTCACAACTCCAG | ||||||||
| 23809 | F: ATGCGCTCTATGTTAGAACG | 55 | 0 | — | 1058 | 2 | Abies firma chloroplast, partial genome (9E-168) | oxidation reduction, chlorophyll biosynthetic process, photosynthesis, dark reaction |
| R: AATCTCAAGACGTTTACCGA | ||||||||
| 23850 | F: GAAGATTTATTCGGCAACTG | 52 | 1 | 449 | 695 | 2 | Pinus taeda mitogen-activated protein kinase 6 (MAPK6) mRNA, complete cds (4E-65) | auxin biosynthetic process, protein amino acid phosphorylation, conjugation, mitosis, cell division |
| R: ATCTGATCCTCTGTTAAGGT | ||||||||
| 23982 | F: TGAGACTTGCTTGGGAAGAG | 57 | 2 | 586 | 921 | 1 | Pisum sativum nonphosphorylating glyceraldehyde-3-phosphate dehydrogenase (gapN) gene, complete cds (4E-33) | metabolic process |
| R: AGCCCATTGTAAACGAAGGA | ||||||||
| 24523 | F: TTCAGACTCGAACGTTTGCA | 58 | 0 | — | 449 | 2 | Ginkgo biloba catalase mRNA, complete cds (3E-98) | hydrogen peroxide catabolic process, oxidation reduction |
| R: AAGCTTTCATTCCCAGACGG | ||||||||
| 24699 | F: AAGATAAGCAGTTTGCTGCA | 56 | 0 | — | 262 | 2 | Ageratina adenophora heat shock protein 70.58 mRNA, complete cds (2E-81) | auxin biosynthetic process, response to stress |
| R: AACATTCTTCTCGCCAACAG | ||||||||
| 24902 | F: CCCTCTCAATCTTGAGGATGC | 58 | 1 | 240 | 662 | 1 | Arabidopsis thaliana ferredoxin–NADP+ reductase (RFNR2) mRNA, complete cds (3E-54) | electron transport chain |
| R: CAGATGGACCTGTAATTTGAACCT | ||||||||
| 25060 | F: CTGCAAGATACTTCAAAGATGCAC | 58 | 2 | 163 | 624 | 1 | PREDICTED: Glycine max ATP-citrate synthase beta chain protein 1-like (LOC100800904), mRNA (2E-74) | acetyl-CoA biosynthetic process, cellular carbohydrate metabolic process |
| R: ATTTGGTGTAGAGAACATCTTCCC | ||||||||
| 26089 | F: GATTATTGATTCTACCACCGGA | 55 | 1 | 281 | 1233 | 1 | Rosa multiflora elongation factor 1-alpha mRNA, complete cds (0.0) | translational elongation |
| R: TTTCTCAACAGCCTTGATGAC | ||||||||
| 26764 | F: GGGAATTGGCTCGTATCTGG | 58 | 0 | — | 359 | 1 | Cucumis sativus 6-phosphogluconate dehydrogenase (6PGDH) mRNA, complete cds (7E-55) | response to glucose stimulus, response to sucrose stimulus, response to fructose stimulus, response to salt stress, pentose-phosphate shunt, oxidation reduction, response to cadmium ion |
| R: GTTCTGCTTAGCAATCTTTGTCC | ||||||||
| 27033 | F: TTTACTCCACCATTACGAGG | 55 | 0 | — | 948 | 2 | Medicago truncatula heat shock protein (MTR_7g024390) mRNA, complete cds (0) | response to virus, auxin biosynthetic process, protein folding, response to heat, response to bacterium, response to cadmium ion, response to high light intensity, response to hydrogen peroxide, protein amino acid phosphorylation |
| R: TTCGCAATGATAGGATTGCA | ||||||||
| 27963 | F: TAGGCCCATAGCTAACAAACC | 57 | 0 | — | 318 | 1 | Keteleeria davidiana chloroplast DNA, complete sequence (0) | transcription, DNA-dependent |
| R: TCGAATTGTTTCATCCTCCCA | ||||||||
| 28203 | F: TGTGGACGAGGAGATATTCG | 56 | 0 | — | 315 | 2 | Pinus pinaster mRNA for cytosolic serine hydroxymethyltransferase (cshmt gene) (1E-128) | l-serine metabolic process, one-carbon metabolic process, glycine metabolic process |
| R: TTCAGAAAGGGCTGTGTAGG | ||||||||
| 28456 | F: GATTTCGAGAGCTGGTATCCC | 58 | 0 | — | 853 | 1 | Ricinus communis oligosaccharyl transferase, putative, mRNA (7E-169) | protein amino acid glycosylation |
| R: AGCTGTCGGTTGATGTTCTG | ||||||||
| 28639 | F: GTAGAATAAGTGGGAGCCGT | 57 | 0 | — | 438 | 2 | Abies fabri 26S ribosomal RNA gene, partial sequence (0) | |
| R: ATAGGAAGAGCCGACATCGA | ||||||||
| 29437 | F: CTTCAGGTGCTCGATATCGT | 56 | 0 | — | 403 | 2 | Populus trichocarpa argonaute protein group (AGO911), mRNA (2E-99) | |
| R: TCAACTGGAAACGTTAGCTC |
Note: — = not available; T = annealing temperature.
Values are based on the sequence of one sample randomly chosen from a sample set of 80 trees from a population at Mont Ventoux (France).
CONCLUSIONS
The two approaches of the workflow are complementary, each contributing approximately half of the annotations in the final set of sequences. The standard approach can be run rapidly, but targets only well-known genes. The specific approach based on a review of the relevant literature is novel and provided a substantial amount of nonredundant annotations. As an advantage, it can be easily adjusted and extended freely to the researcher’s interest. The quality-tested primers can be used for assessing the degree of gene polymorphism in ecological genetics studies.
LITERATURE CITED
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. 1990. Basic local alignment search tool. Journal of Molecular Biology 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H., Cherry J. M., Davis A. P., et al. 2000. Gene Ontology: Tool for the unification of biology. Nature Genetics 25: 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conesa A., Götz S., García-Gómez J. M., Terol J., Talón M., Robles M. 2005. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics (Oxford, England) 21: 3674–3676 [DOI] [PubMed] [Google Scholar]
- González-Martínez S. C., Ersoz E., Brown G. R., Wheeler N. C., Neale D. B. 2006. DNA sequence variation and selection of tag single-nucleotide polymorphisms at candidate genes for drought-stress response in Pinus taeda L. Genetics 172: 1915–1926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H.-R., Yan P.-C., Lascoux M., Ge X.-J. 2012. Flowering time and transcriptome variation in Capsella bursa‐pastoris (Brassicaceae). New Phytologist 194: 676–689 [DOI] [PubMed] [Google Scholar]
- Marshall O. 2004. PerlPrimer: Cross-platform, graphical primer design for standard, bisulphite, and real-time PCR. Bioinformatics 20: 2471–2472 [DOI] [PubMed] [Google Scholar]
- Mosca E., Eckert A. J., Liechty J. D., Wegrzyn J. L., La Porta N., Vendramin G. G., Neale D. B. 2012. Contrasting patterns of nucleotide diversity for four conifers of Alpine European forests. Evolutionary Applications 5: 762–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata H., Goto S., Sato K., Fujibuchi W., Bono H., Kanehisa M. 1999. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Research 27: 29–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parchman T. L., Geist K. S., Grahnen J. A., Bankman C. W., Buerkle C. A. 2010. Transcriptome sequencing of an ecologically important tree species: Assembly, annotation, and marker discovery. BMC Genomics 11: 180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Street N. R., Skogström O., Sjödin A., Tucker J., Rodríguez-Acosta M., Nilsson P., Jansson S., Taylor G. 2006. The genetics and genomics of the drought response in Populus. Plant Journal 48: 321–341 [DOI] [PubMed] [Google Scholar]