Abstract
Hand, Foot and Mouth Disease (HFMD) is a self-limiting viral disease that mainly affects infants and children. In contrast with other HFMD causing enteroviruses, Enterovirus71 (EV71) has commonly been associated with severe clinical manifestation leading to death. Currently, due to a lack in understanding of EV71 pathogenesis, there is no antiviral therapeutics for the treatment of HFMD patients. Therefore the need to better understand the mechanism of EV71 pathogenesis is warranted. We have previously reported a human colorectal adenocarcinoma cell line (HT29) based model to study the pathogenesis of EV71. Using this system, we showed that knockdown of DGCR8, an essential cofactor for microRNAs biogenesis resulted in a reduction of EV71 replication. We also demonstrated that there are miRNAs changes during EV71 pathogenesis and EV71 utilise host miRNAs to attenuate antiviral pathways during infection. Together, data from this study provide critical information on the role of miRNAs during EV71 infection.
Introduction
Hand, Foot and Mouth Disease (HFMD) is a contagious viral disease that commonly affects infants and children, with blisters and flu like symptoms [1]–[6]. HFMD is caused by a group of enteroviruses such as enterovirus 71 (EV71) and coxsackievirus A16 (CA16) [7]–[9]. However, unlike other HFMD causing enteroviruses, EV71 is also commonly associated with severe clinical diseases, including neurological diseases such as aseptic meningitis, brainstem and cerebellar encephalitis leading to cardiopulmonary failure and death [3], [10]–[13]. Due in part to a lack of understanding of viral pathogenesis of EV71 causing HFMD, there are no antiviral therapies or vaccines approved by the United States Food and Drug Administration (FDA) to prevent HFMD infections. There is therefore a need to gain a better understanding of the mechanism of EV71 pathogenesis. During infection, viruses attenuate host gene expression to enhance their survival [14]–[18]. Such activities include altering the cellular microenvironment to allow successful virus replication and evasion of the host immune system [19].
The host innate system is the first line of defence with the interferon (IFNs) being a key component against viral infection [17], [18], [20]–[25]. The release of IFNs begins with the recognition of pathogen-associated molecular patterns (PAMPs) which include Toll like receptors (TLRs), NOD like receptors (NLRs), retinoic acid-inducible gene I (RIG-I) like receptors [24], [26]. This family of cytokines act in an autocrine and paracrine manner to induce expression of transcription factors, gamma activated sequence and IFN stimulated response element and the consequent induction of IFN-stimulated genes (ISGs) which function to slow down viral infection [20]–[22]. Antiviral pathways such as the programmed cell death or apoptosis could also be activated by the host cells in attempt to control the infection by limiting viral replication [19].
Many viruses have evolved to counteract with strategies by host cells to establish infection. One of the mediators for such regulation includes genes that encode for microRNA (miRNAs). microRNAs are small single-stranded RNA species of approximately 20–24 bases in length that play a pivotal role in the post transcriptional regulation of gene expression [27], [28]. These small RNA are first discovered in Caenorhabditis elegans but was later discovered in a wide range of organisms from uni to multicellular eukaryotes [29]–[31]. miRNAs are found to regulate many cellular functions including cell proliferation, differentiation, homeostasis, immune activation and apoptosis [28], [29], [32]–[34]. As such, miRNAs can contribute to the repertoire of host-pathogen interactions during infection by modulating and directing host and viral gene expression thereby playing a pivotal role during infection. Elucidating the mechanism that the virus uses to bypass host defence systems and establish infections will provide us with a better understanding of the pathogenesis mechanism of the EV71 virus, allowing development of potent antiviral therapeutics for HFMD patients.
In this study, we explore the role of miRNAs in the pathogenesis of EV71 infected HT29 cells and identified key miRNA changes due to EV71 infection which might play a key role during EV71 pathogenesis.
Results
We have previously reported a human colorectal adenocarcinoma cell line (HT29) with epithelioid morphology as a useful model to study the pathogenesis of EV71 [35]. Utilising the HT29 as a model, this study further elucidate the role of miRNAs during EV71 infection.
Knockdown of DGCR8 resulted in the reduction of EV71 replication
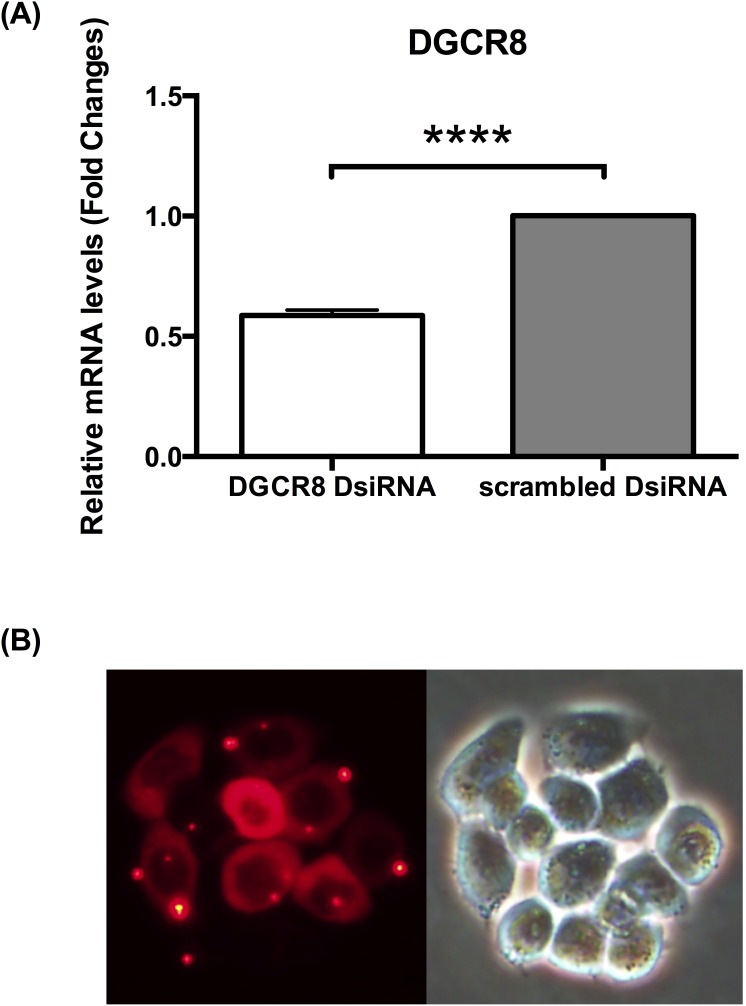
In order to characterise the role of miRNAs during EV71 pathogenesis, DGCR8 was knocked down prior to infection. HT29 cells were transfected with DsiRNA specific to DGCR8, an essential cofactor for miRNAs biogenesis. As control, HT29 cells were transfected with NC1, negative control duplex-scrambled DsiRNA. Knockdown of DGCR8 was verified using qPCR (Figure 1a). Transfection was shown to be successful using TYE 563 DS Transfection Control, fluorescent-labelled transfection control duplex (Figure 1b).
Figure 1. Knockdown of DGCR8 using DsiRNA.
(A) Relative expression of DGCR8 gene measured by qPCR verifying knockdown. (n = 3, **** = p values of <0.0001) (B) Transfection Control using fluorescent-labeled transfection control duplex TYE 563 DS to monitor transfection efficiency.
miRNAs is essential for EV71 replication
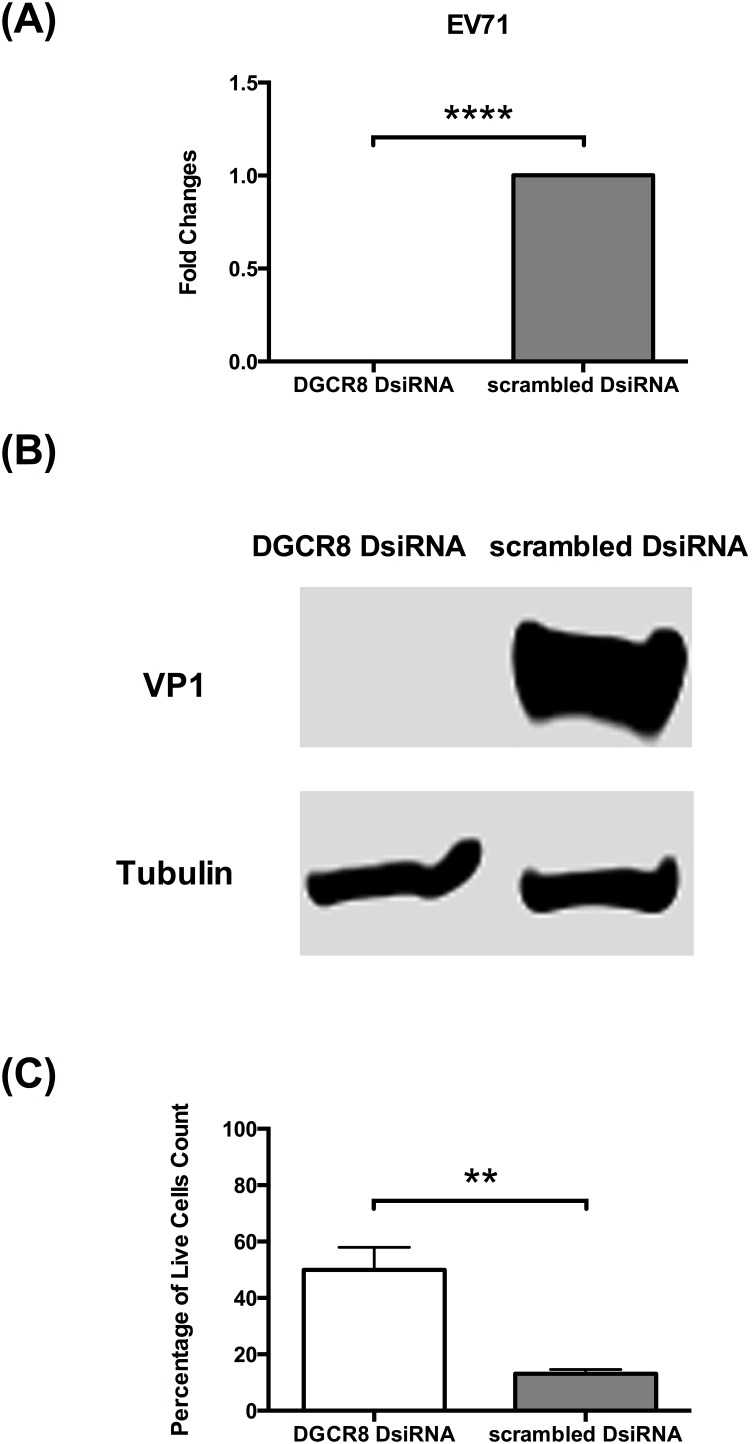
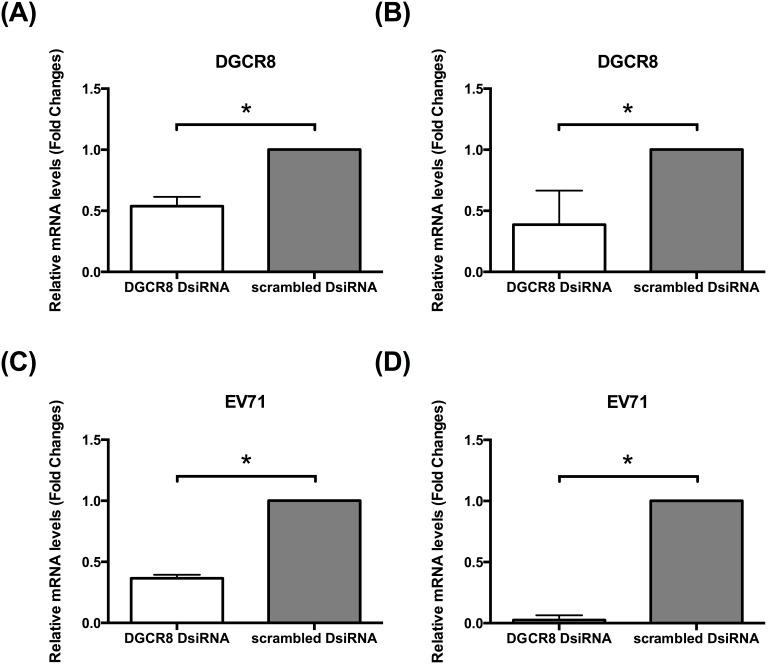
To investigate the role of miRNAs in EV71 infection, we compare EV71 replication in miRNAs depleted HT29 cells against control cells (scrambled DsiRNA). Using established protocols, the kinetics of EV71 RNA synthesis in both infected DGCR8 knockdown cells and control cells were examined quantitatively using real time quantitative polymerase chain reaction (qPCR) at 36h post infection (hpi). Interestingly, DGCR8 knockdown (miRNAs depleted) HT29 cells were unable to support EV71 proliferation compared to control cells (Figure 2). There is a significant decrease in viral RNA between DGCR8 knockdown HT29 cell in contrast with control cells (scrambled DsiRNA) (Figure 2a). Consistent with our qPCR analysis, protein analysis of viral VP1 protein representation of EV71 replication, were shown to be highly downregulated in EV71 infected DGCR8 knockdown cells (Figure 2b). To further elucidate the effect of miRNAs during EV71 infection, we asked if the host cells survivability was enhanced in EV71 infected miRNAs depleted cells. Indeed, it was observed that at 72 hpi, there was a statistically significant percentage of living cells in EV71 infected DGCR8 knockdown cells compared to control cells infected with EV71 (Figure 2c and Figure S1). In order to validate our findings in HT29 cells, we further test this on another colorectal cell line (RKO) and skeleton muscle cell line (RD) where we knock down DGCR8 and examine the effect on EV71 replication (Figure 3). Consistent with our findings in HT29 model, EV71 is unable to establish infection in miRNAs depleted cells (HT29, RKO and RD cells) (Figure 2 and 3).
Figure 2. miRNA depleted HT29 cells is not able to support EV71 replication.
DGCR8 knockdown HT29 cells and control HT29 cells (scrambled DsiRNA) were infected with EV71 (MOI of 1). Total intracellular RNA were harvested at 36hpi, converted to cDNA and measured by qPCR with primers specific to viral VP1. Total intracellular protein were harvested at at 36hpi and measured by western blot (A) Relative expression of EV71 viral RNA measured by qPCR. (n = 3, **** = p values of <0.0001) (B) Western blot analysis for EV71 VP1 viral protein. (C) Cell viability assessed at 72hpi using vital dye trypan blue. (n = 3, ** = p values of <0.01).
Figure 3. EV71 is unable to establish infection in both RKO and RD miRNAs depleted cells.
DGCR8 knockdown cells and control cells (scrambled DsiRNA) in both RKO and RD cell line were infected with EV71 (MOI of 1). Total intracellular RNA were harvested at 12hpi, converted to cDNA and measured by qPCR with primers specific to viral VP1. (A) Relative expression of DGCR8 gene in miRNAs depleted RKO cells measured by qPCR verifying knockdown. (n = 3, * = p values of <0.05) (B) Relative expression of DGCR8 gene in miRNAs depleted RD cells measured by qPCR verifying knockdown. (n = 3, * = p values of <0.05) (C) Relative expression of EV71 viral RNA in miRNAs depleted RKO cells measured by qPCR. (n = 3, * = p values of <0.05) (D) Relative expression of EV71 viral RNA in miRNAs depleted RKO cells measured by qPCR. (n = 3, * = p values of <0.05).
Taken together, DGCR8 or rather miRNAs affects EV71 replication and recused its detrimental phenotype.
Identification of deregulated miRNAs during EV71 infection
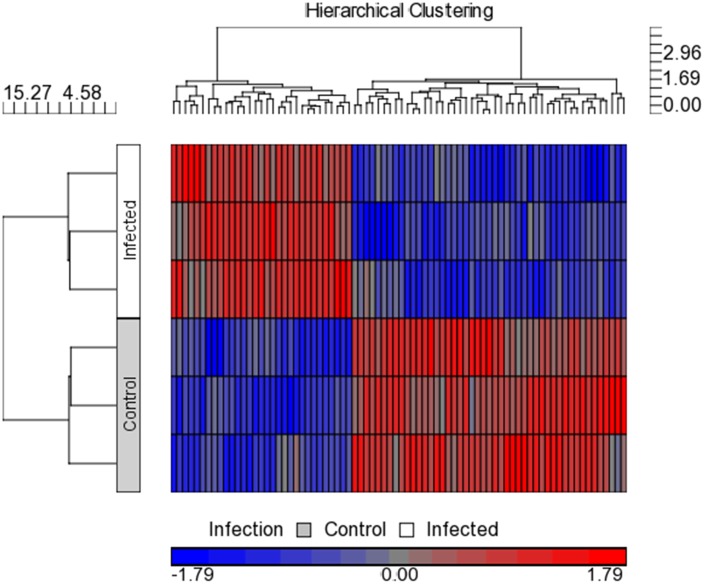
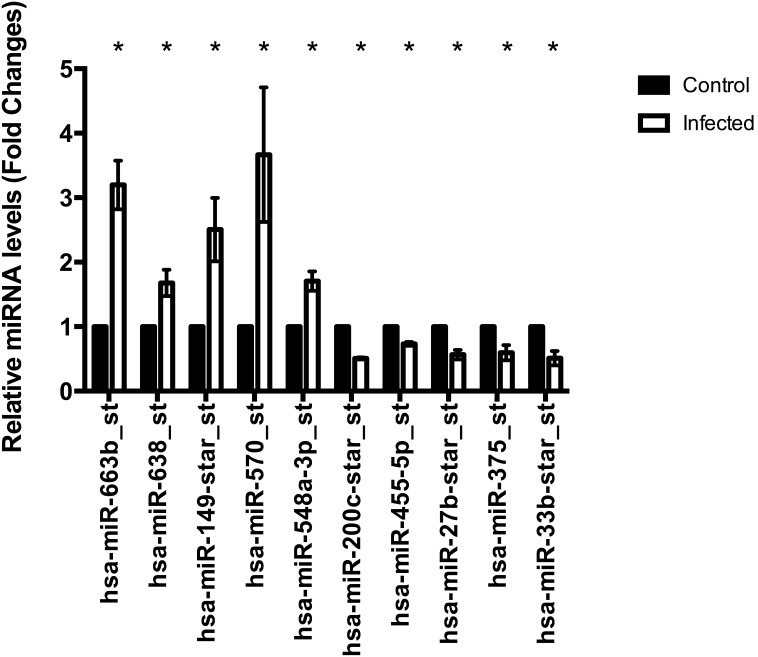
To pinpoint on the differentially transcribed miRNAs used by EV71 during infection, we perform miRNAs profiling using Affymetrix GeneChip miRNA array. Specifically, comparative miRNAs expression between EV71 infected cells and control non-infected cells were performed. Microarray hybridisation identified 78 miRNAs being differentially expressed during EV71 infection (p<0.05) (Figure 4 and Table S1). To confirm the microarray results, relative abundance of the selected miRNAs were assayed using qPCR. The expression pattern from 10 selected miRNAs qPCR data showed similar direction of response in microarray analysis data with both methodologies showing similar trends (Figure 5 and 6). This indicates high quality and reliability of the microarray data analysis.
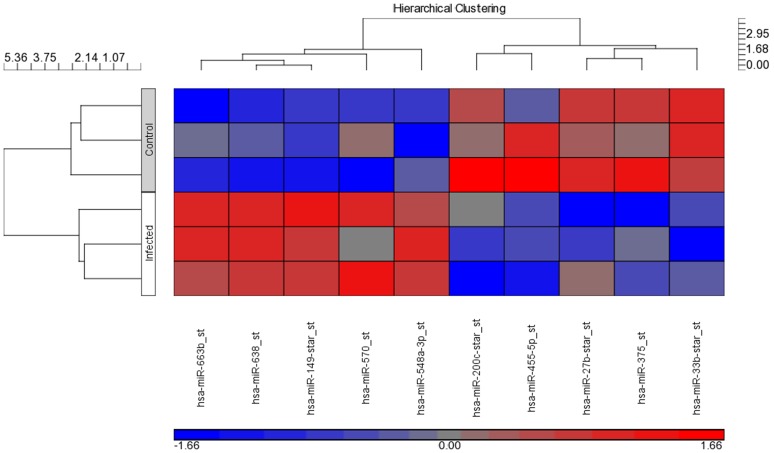
Figure 4. Heat map of the microarray miRNA expression profile of EV71 infected and non-infected control colorectal cells, HT29.
The two-way hierarchical cluster heat map showed differential expressed miRNAs of two groups of samples. The miRNAs were chosen according to the cut off p<0.05 where blue represents miRNAs with decreased expression and red represent miRNAs with increased expression. (n = 3, * = p values of <0.05).
Figure 5. Validation of differential expression of selected miRNAs by qPCR.
Ten expression levels of selected miRNAs from microarray assay were validated using qPCR. U6snRNA was used as reference RNA for the normalization of miRNAs and relative gene expression was quantified based on 2−ΔΔCT. (n = 3, * = p values of <0.05).
Figure 6. Heat map of differential expression of selected miRNAs by microarray.
The two-way hierarchical cluster heat map showed differential expressed selected miRNAs of two groups of samples. The miRNAs from both methodology qPCR data showed similar direction of response in microarray analysis data with both methodologies showing similar trends where blue represents miRNAs with decreased expression and red represent miRNAs with increased expression.
EV71 down regulate host immune system using host miRNAs during infection
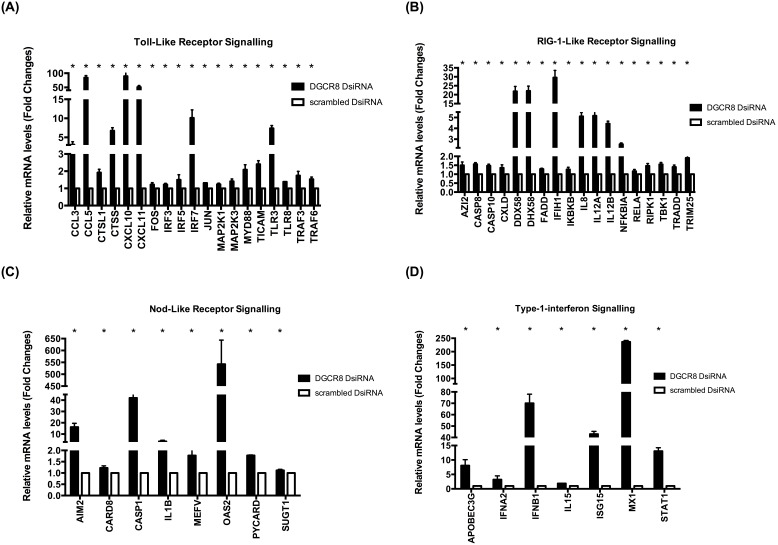
To understand why miRNAs depleted cells resulted in the inability of EV71 to established infection; we examine the expression of 84 key genes involved in the innate antiviral immune response between EV71 infected miRNAs depleted HT29 cells in comparison with control cells (scrambled DsiRNA). In particular, we examine the host antiviral expression profile between infected DGCR8 knockdown cells against control cells. Using qPCR, quantitative expression of 52 antiviral genes from various antiviral signalling pathways such as Toll-like receptor signalling, Nod-like receptor signalling, RIG-1-like receptor signalling and type 1 interferon signalling were statistically up regulated in DGCR8 knockdown cells (Figure 7a, 7b, 7c, 7d). Noteworthy, CASP1, OAS2, DDX58, DHX58, CCL5, CXCL10, CXCL11, IRF7, IFBB1, ISG15, MX1 and STAT1 have at least 10–550 fold increase (Figure 7a, 7b, 7c, 7d).
Figure 7. EV71 is unable to down regulate host antiviral response in miRNA depleted cells during infection.
DGCR8 knockdown cells and control cells (scrambled DsiRNA) were infected with EV71 (MOI of 1). Total intracellular RNA were harvested at 36hpi, converted to cDNA and measured by quantitative real time polymerase chain reaction (qPCR) (n = 3, * = p values of <0.05). A) Relative expression of Toll-like receptor signalling measured by qPCR. (B) Relative expression of RIG-1 like receptor signalling measured by qPCR. (C) Relative expression of Nod-like receptor signalling measured by qPCR. (D) Relative expression of Type 1 interferon signalling measured by qPCR.
Discussion
Viruses have developed highly sophisticated gene-silencing mechanisms to evade host-immune response in order to establish infection. One of such mediators used by viruses is miRNAs. miRNAs is a new paradigm of RNA-directed gene expression regulation which contribute to the repertoire of host-pathogen interactions during viral infection. Cellular miRNAs could be involved during viral infection with either positive or negative effects on viral replication kinetics [36], [37]. Indeed, computational prediction reveal that one miRNAs may regulate hundreds of genes while one gene may be in turn regulated by hundreds of miRNAs [38]. However, the role of cellular miRNAs in the defence against viral infection has thus remained elusive. Given the scope of miRNA-mediated gene regulation in the mammalian system, cellular miRNAs may directly or indirectly affect virus pathogenesis. Cellular miRNA expression could be remodelled by viral infection, which can alter the host cellular microenvironment and attributing to both host antiviral defence and viral factors [25], [32], [36], [39], [40].
The biogenesis of miRNAs begins with the transcription of a long primary transcript known as pri-miRNA by RNA polymerase II [27]–[29], [32], [39], [41]. Pri-miRNAs are cleaved/processed by RNase III enzyme (Drosha), double-stranded-RNA-binding protein, (DiGeorge Syndrome Critical Region; DGCR8) and RNAse III-like Dicer to become functional miRNAs. In this study, we have demonstrated that abolishing functional miRNAs in human epithelial colorectal cell line (HT29) impair EV71 replication while this was not observed in control cells treated with scrambled DsiRNA. On the contrary, global knockdown of miRNAs have been previously shown to have enhance HIV-1 infection [42], [43]. Knock down of DGCR8, a precursor to miRNAs biogensis were shown to enhance human immunodeficiency virus (HIV) replication [42], [43]. This implies that EV71 utilise miRNAs during infection and in the absent of miRNAs, EV71 was unable to successfully mount the infection.
Another study reported by Otsuka and colleagues showed a similar finding in Vesicular stomatitis virus (VSV) infection [44]. Otsuka and colleagues observed that impaired miRNA production in mice with dicer deficient mice resulted in the hypersensitivity toward infection by VSV [44]. It was observed there is no alternation of interferon-mediated antiviral response by dicer deficiency instead the change in virus replication is due to miR-24 and miR-93 [44]. Otsuka and colleagues shown that miR-24 and miR-93 could target VSV viral protein and thus in miRNA depleted cells, VSV replication was enhanced in the absence of miR-24 and miR-93 [44].
In recent years, there are reported evidences of specific host-cellular miRNAs modulating host-pathogen interaction [44]–[51]. Cytomegalovirus (CMV) for example was observed to down regulate miR-27a [45]. Overexpression of miR-27a using miRNAs mimics was shown to inhibit CMV proliferation and virus titres suggesting that miR-27a or miR-27a-regulated genes plays a role during CMV infection [45], [52]. Hepatitis C virus (HCV), an enveloped RNA virus of the Flavivirus family, responsible for both acute and chronic hepatitis in humans induced a liver-specific cellular miRNA, miR-122 for the facilitation of HCV replication [17], [48], [51], [53]–[55]. It was suggested that miR-122 is a valuable target for antiviral intervention and therapeutics. In vivo treatment of HCV-infected chimpanzees with antisense miR-122 successfully reduced HCV virus titres demonstrating promising antiviral treatment against virus induced diseases [48], [49], [53], [54].
Indeed, various miRNAs have been identified to play a role during EV71 pathogenesis. Recent publications of investigations into the role of miRNA in enterovirus infection have reported that enterovirus-induced miRNAs such as miR-23b, miR-296-5p, miR-141 and miR-146a play a role in EV71 replication [8], [56]–[58]. The use of miRNA inhibitors or mimics have demonstrated that miRNAs plays an important role in EV71 pathogenesis. miRNA inhibitors or mimics on these miRNAs either inhibited or enhanced EV71 replication during infection.
In particular, our results revealed miR-548 to be significantly up regulated in EV71 infected cells at 36hpi in comparison with control non infected cells. Further bioinformatics analysis revealed that miR-548 was involved in antiviral response during infection [17], [23], [25], [38], [59]. Consistent with our findings, Li and colleagues have recently identified the involvement of miR-548 in IFN signalling pathways during viral infection [59]. Li and colleagues demonstrated that miR-548 down-regulates host antiviral response via direct targeting of IFN-λ1 [59]. In contrast, in EV71 infected rhabdomyosarcoma (RD) cells, miR-548 was observed to decrease in a time dependent manner [59]. In view that the host innate system is the first line of defence with IFNs being a key player during viral infection, it is atypical that miR-548 was up regulated to down regulate the host immune system [17], [18], [20]–[25]. The use of miR-548 mimics and inhibitors were shown to enhance or inhibit EV71 replication in RD cells respectively [59].
The innate immune system has evolved an arsenal of mechanism to sense and destroy invading pathogen. An interesting question that arises is whether EV71 is able to regulate host cellular miRNA transcription and processing during infection thereby leading to the down-regulation of deleterious cellular miRNA and/or the up-regulation of advantageous cellular miRNAs to enhance its replication in the host cells. To add to this intriguing interplay between cellular miRNAs and viral pathogens, Huang and colleagues has demonstrated that HIV, an enveloped RNA virus of the Retreoviridae family utilise host cellular miR-28, miR-125b, miR-150, miR-223 and miR-382 to control viral protein synthesis in order to evade host immune system [47]. Epstein-Barr virus (EBV), a DNA virus of the Herpes family, for example induce the expression of miR-29b, miR-155 and miR-146a to mediate the down regulation of innate immune response and IFN signalling pathways to enhances its survival [46], [60], [61]. Lecellier and colleagues have shown that in primate foamy virus type 1 (PFV-1), overexpression of cellular miRNA miR-32 could induce an antiviral response to inhibit virus replication [37].
To test this reasoning that EV71 down regulate host miRNAs during infection to enhance its survival, we next ask if the innate response and signalling pathways were altered in EV71 infected miRNA-depleted cells. Therefore, we analysed the expression of 84 key genes involved in the innate antiviral immune response between EV71 infected miRNAs depleted HT29 cells in comparison with control cells (scrambled DsiRNA). Quantitative analysis\revealed 52 antiviral genes from various antiviral signalling pathways such as Toll-like receptor signalling, Nod-like receptor signalling, RIG-1-like receptor signalling and type 1 interferon signalling were significantly up regulated in EV71 infected miRNAs depleted HT29 cells. In particular, CASP1, OAS2, DDX58, DHX58, CCL5, CXCL10, CXCL11, IRF7, IFBB1, ISG15, MX1 and STAT1 have at least 10–550 fold increase. The enhanced host antiviral response in miRNA-depleted cells during EV71 infection may result in the inability for EV71 to replicate during infection (Figure 8).
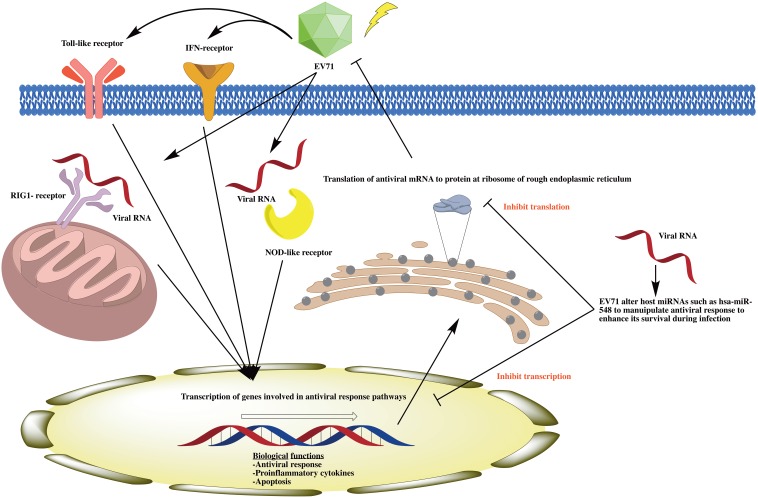
Figure 8. Schematic illustration of the interplay between EV71 and host miRNAs.
EV71 altered host miRNAs such as has-miR-548 to manipulate host antiviral responses such as Toll-like signalling, NOD-like signalling, RIG-1 signalling and Type I interferon pathways to enhance its survival during pathogenesis.
The host immune response is responsible for the eradication of invading pathogens, as such viruses has developed means to counteract with the induction, signalling and antiviral pathways [16], [20], [21], [25], [40]. RNA viruses generally block and inhibit IFN and antiviral molecules detrimental to virus replication [20], [24]. Despite all the sophisticated mechanism used by viruses to inhibit host immune system, host has been under evolutionary pressure against viral antagonism of IFN system [20], [21], [24], [40]. Our result suggests that EV71 attenuate host miRNAs to mediate host immune system during infection (Figure 8). As a result of DGCR8 knockdown, EV71 is unable to regulate host miRNAs to enhance cellular microenvironment leading to the inability to establish infection (Figure 8). Notwithstanding, IFN signalling pathways such as type I interferon was previously demonstrated to play an important role during EV71 pathogenesis [62], [63]. These results warrant further studies for the identification of specific miRNAs which play a pivotal role during EV71 pathogenesis specifically targeting the host antiviral signalling pathway.
Conclusion
In conclusion, we provided evidence for an intricate physiological interplay between host miRNAs machinery and invading pathogen, EV71. We reported three salient findings. First, DGCR8, an essential cofactor for microRNAs biogenesis contribute to a central role during EV71 pathogenesis. We have shown that EV71 is not able to replicate efficiently in miRNAs depleted cells. Secondly, our results suggest that EV71 may remodel the cellular miRNAs composition thereby utilising the host cellular miRNAs to enhance cellular microenvironment for viral replication. Specifically, our result suggests that EV71 may be at least partially responsible for the up regulation of host miR-548 to enhance cellular microenvironment for viral replication. Thirdly, remodelling of miRNAs during EV71 infection plays a significant role specifically in antiviral response. We have demonstrated that in EV71 infected miRNAs depleted cells, host antiviral responses from various antiviral signalling pathways such as Toll-like receptor signalling, Nod-like receptor signalling, RIG-1-like receptor signalling and type 1 interferon signalling were significantly higher in comparison with EV71 infected control cells. This may explain our result on the inability for EV71 to infect miRNAs depleted cells given the enhanced host antiviral response. Taken together, our study has provide valuable knowledge toward the study of host-pathogen interaction during EV71 pathogenesis and this could lead to future investigation for the development of antiviral therapeutics using miRNAs inhibitors against EV71. This will open the door toward novel host-defence mechanism that exists in mammalian cells as well as to the antiviral mechanisms employed during EV71 pathogenesis.
Materials and Methods
Cell culture and virus propagation
The EV71 strain used in this study was isolated from a fatal case of HFMD during October 2000 outbreak in Singapore, Enterovirus 5865/sin/000009 strain from subgenogroup B4 (accession number 316321; hereby designated as Strain 41) (a gift from Mrs Phoon Meng Chee, Department of Microbiology, National University of Singapore). Human colorectal cell line (HT29) (ATCC catalog no. HTB-38) was maintained in Roswell Park Memorial Institute medium (RPMI) (PAA Laboratories, Austria) supplemented with 10% (v/v) Fetal Bovine Serum (FBS) (PAA Laboratories, Austria) and 2% penicillin–streptomycin (PAA Laboratories, Austria) at 37°C with 5% CO2. Human skeleton muscle cell line (RD) (ATCC catalog no. CCL-136) and Human colorectal cell line (RKO) (ATCC catalog no. CRL-2577) were maintained in Eagle’s minimum essential medium (EMEM) (PAA Laboratories, Austria) supplemented with 10% (v/v) Fetal Bovine Serum (FBS) (PAA Laboratories, Austria) and 2% penicillin–streptomycin (PAA Laboratories, Austria) at 37°C with 5% CO2. The virus stock was prepared by propagation of viruses using 90% confluent HT29 cells monolayer in RPMI with 10% FBS at 37°C with 5% CO2. The virus titres were determined using 50% tissue culture infective dose (TCID50) per millilitre (mL) according to Reed and Muench method [64].
DsiRNA transfection and EV71 infection
HT29, RD and RKO cells were seeded at a concentration of 2×105 cells/ml in 6-well plates and incubated for 8 h at 37°C with 5% CO2. Cells were washed twice with phosphate buffered saline (PBS) and transfected using Lipofectamine RNAiMAX (Invitrogen Corporation, CA, USA) with TriFECTa Dicer-Substrate RNAi KIT (Integrated DNA Technology, Coralville, IA, USA). Briefly, 10nM of DGCR8 DsiRNA (NM_001190326 duplex 2), NC1, negative control duplex-scrambled DsiRNA (NM_001190326) and TYE 563 DS Transfection Control, fluorescent-labeled transfection control duplex (NM_001190326) were used to transfect the cells respectively (Integrated DNA Technology, Coralville, IA, USA). Following 24 h after transfection, cells were washed twice with PBS and infected with EV71 at multiplicity of infection (MOI) of 1. The culture media were removed and replaced with 2mL of fresh RPMI medium after 1 h of incubation at 37°C with 5% CO2.
EV71 infection for miRNA profiling
HT29 cells were seeded at a concentration of 5×105 cells/ml in 6-well plates and incubated for 24 h at 37°C with 5% CO2. Cells were washed twice with phosphate buffered saline (PBS) and infected with EV71 at multiplicity of infection (MOI) of 1 or nil respectively. The culture media were removed and replaced with 2 mL of fresh RPMI medium after 1 h of incubation at 37°C with 5% CO2.
Total cellular RNA extraction
The respective infected cells were harvest at 12 or 36 hours posts infection (hpi) respectively. The total cellular RNA of HT29, RD and RKO cells were extracted using the miRNeasy mini kit (Qiagen, Hilden, Germany) in accordance to the manufacturer’s instructions. Briefly, the cells were lysed and homogenise using lyses solution provided (Qiagen, Hilden, Germany). Total RNA were harvested using the RNeasy spin column and wash twice before elution (Qiagen, Hilden, Germany). Harvested total RNA was quantitated using Nanodrop 100 spectrophotometer (ThermoScientific, Waltham, USA).
miRNA microarray analysis
The extracted total RNA was labelled with FlashTag Biotin HSR RNA Labeling Kits for the Affymetrix GeneChip miRNA array according to manufacturer’s instruction. Briefly, one microgram of total RNA will be incubated with ATP and Poly A polymerase at 37°C for 15m to add a 3′ polyA tail. A ligation reaction will then be performed to covalently attach to the miRNA population a multiple-biotin molecule containing a 3DNA dendrimer. Labelled samples will subsequently be processed according to manufacturer’s instructions for the Affymetrix miRNA Array 2.0 (Affymetrix, Santa Clara, CA). After hybridisation for 16 h at 48°C, the arrays will be washed and stained in an Affymetrix Fluidics station 450, then scanned in an Affymetrix 3000 7G scanner. Raw data for the microarray (GSE57372) was analysed with Partek Genomics Suite (Partek Incorporated, Saint Louis, USA) where p<0.05 is considered significant.
cDNA synthesis
For the conversion of mRNA to cDNA, 1 µg of the total RNA was reverse transcripted using RT2 First Strand kit (Qiagen, Hilden, Germany) in accordance to the manufacturer’s instructions. Briefly, 1 µg of the extracted RNA was first mixed with Buffer GE to a total volume of 10 µl for genomic DNA elimination and incubated at 42°C for 5 m and on ice for 1 m. Reverse-transcription mix which consist of enzyme reverse transcriptase and buffer was then added to a volume of 20 µl and subjected to thermal profile of 42°C for 15 m followed by 95°C for 5 m. 91 µl of RNase free water was then added to each reaction in accordance to the manufacturer’s instructions.
For conversion of microRNA to cDNA, 10 ng of the total RNA was reverse transcripted using the Universal cDNA Synthesis kit (Exiqon, Denmark) the manufacturer’s instructions. Briefly, 10ng of the extracted RNA will be mixed with enzyme reverse transcriptase and buffer to a volume of 10 µl. The reaction mix will then subject to thermal profile of 25°C for 5 m, 42°C for 30 m followed by 85°C for 5 m according to the manufacturer’s instructions.
Quantitative real time polymerase chain reaction
The EV71 specific primers targeting the conserve VP1 regions were 5′-GCTCTATAGGAGATAGTGTGAGTAGGG-3′ and the reverse primer 5′-ATGACTGCTCACCTGCGTGTT-3′ [65]. Primers for DGCR8 were 5′-ACTTGTGCATGTTAGCTGTGTAGA-3′ and the reverse primer 5′-GCTTAAGACTAGTTTACAAGACCAGAGT-3′ were designed to span exon-exon boundaries. The primers for the house keeping gene actin (β-ACT) used were 5′-ACCAACTGGGACGACATGGAGAAA-3′ and the reverse primer 5′-TAGCACAGCCTGGATAGCAACGTA-3′. The qPCR was performed using the iTaq Universal SYBR Green Supermix (Bio-Rad Laboratories, CA, USA) on the BioRad CFX96 Real-Time PCR system (Bio-Rad Laboratories, CA, USA). Briefly, 1 µl of cDNA and 1 µl of the forward and the reverse primers were added iTaq Universal SYBR Green Supermix. The reaction mix was then subjected to thermal profile of denaturation at 95°C for 30s, followed by amplification and quantification in 40 cycles at 95°C for 5s followed by 60°C for 30 s. At the end of amplification cycles, melting temperature analysis was performed by the BioRad CFX96 Real-Time PCR system (Bio-Rad Laboratories, CA, USA). Relative gene expression was quantified based on 2−ΔΔCT method [66].
For RT2 profiler PCR array, PCR arrays were performed with customized PCR containing pre-dispensed primers to monitor Human antiviral response PAHS-122ZD-2 (Qiagen, Hilden, Germany). qPCR was performed using RT2 SYBR Green qPCR mastermix (Qiagen, Hilden, Germany). Briefly, a total volume of 25 µl of cDNA and RT2 SYBR Green qPCR mastermix were added into each well of the Human antiviral response array 96 wells plate PAHS-122ZD-2 (Qiagen, Hilden, Germany) in accordance to the manufacturer’s instructions. The reaction mix was then subjected to thermal profile of denaturation at 95°C for 10m, followed by amplification and quantification in 40 cycles at 95°C for 15 s followed by 60°C for 1m. At the end of amplification cycles, melting temperature analysis was performed by the BioRad CFX96 Real-Time PCR system (Bio-Rad Laboratories, CA, USA).
For the detection of microRNA, qPCR was performed using the ExiLENT SYBR Green Master mix (Exiqon, Denmark) on the BioRad CFX96 Real-Time PCR system (Bio-Rad Laboratories, CA, USA). Briefly, 2.5 µl of cDNA and 1 µl of the forward and the reverse primers was added to ExiLENT SYBR Green Master mix (Exiqon, Denmark) according to the manufacturer’s instructions. The reaction mix will then subjected to thermal profile of denaturation at 95°C for 10 m, followed by amplification and quantification in 40 cycles at 95°C for 10 s, 60°C for 30 s followed by 50°C for 30 s. At the end of amplification cycles, melting temperature analysis will be performed by the BioRad CFX96 Real-Time PCR system (Bio-Rad Laboratories, CA, USA). Primers for microRNA qPCR were designed and purchased from Exiqon, Denmark. U6snRNA was used as reference RNA for the normalization of microRNA (miRNA) qPCR data. Relative gene expression was quantified based on 2−ΔΔCT method [66].
Western blot
Total cellular protein for HT29 cells and control cells were collected using a lysis mix in mammalian cell lysis solution–CelLytic M (Sigma-Aldrich Pte Ltd, USA) in accordance with manufacturer’s instructions. Equal protein concentration (20 µg) from each samples were added onto 10% Mini-PROTEAN TGX (Bio-Rad Laboratories, CA, USA) and separated by electrophoresis. Separated proteins were transferred onto nitrocellulose membrane (Bio-Rad Laboratories, CA, USA). The membranes were incubated with mouse anti EV71 antibody (AbD serotech, Oxford, UK) or anti tubulin antibody (Santa Cruz Biotechnology inc, California, USA) respectively in shaker 4°C overnight. The membranes were washed three times in Tris-buffered saline with 0.05% tween-20 before and incubating with secondary antibodies conjugated to horseradish peroxidase (HRP) for 30m. (ThermoScientific, Waltham, USA). After three wash with Tris-buffered saline and 0.05% tween-20, membrane was subjected to Clarity Western ECL Substrate (Bio-Rad Laboratories, CA, USA) in accordance with manufacturer’s instructions. The membranes were then scanned using the LI-COR C-DiGit Blot Scanner (LI-COR Biosciences, Lincoln, Nebraska, USA).
Cell viability and counts
Cell count and viability was performed at 0 h, 48 h and 72 h on the Luna Automated Cell Counter system (Logos Biosystem, USA) in accordance to the manufacturer’s instructions. Briefly, the cells were trypsinised and topped up with fresh media to a total volume of 1000 µl of media and 10 µl of this cell suspension were mixed with 10 µl of tryphan blue. 10 µl of this diluted cell suspension were then loaded onto the Luna counting slide for analysis.
Statistical analysis
All statistical analysis was performed on GraphPad Prism Version 6.0c (GraphPad Software, USA). Student t test was used to compare two groups. p values of <0.05 were considered statistically significant.
Supporting Information
Cell viability assessed 24 h after transfection and throughout 72 h post EV71 infection using vital dye trypan blue. (n = 3, * = p values of <0.05).
(TIFF)
Altered miRNA expression profile of EV71 infected and non-infected control colorectal cells, HT29.
(DOCX)
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. Data are within the paper and its Supporting Information files.
Funding Statement
This research was supported by Singapore Polytechnic. Yan Long Edmund Lui was supported by Queensland University of Technology Higher Degree Research Award Scholarship. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Lui YLE, Lin Z, Lee JJ, Chow VTK, Poh CL, et al. (2013) Beta-actin variant is necessary for Enterovirus 71 replication. Biochemical and Biophysical Research Communications 433: 607–610. [DOI] [PubMed] [Google Scholar]
- 2. McMinn PC (2002) An overview of the evolution of enterovirus 71 and its clinical and public health significance. FEMS Microbiol Rev 26: 91–107. [DOI] [PubMed] [Google Scholar]
- 3. McMinn PC (2012) Recent advances in the molecular epidemiology and control of human enterovirus 71 infection. Curr Opin Virol 2: 199–205. [DOI] [PubMed] [Google Scholar]
- 4. Ooi MH, Wong SC, Lewthwaite P, Cardosa MJ, Solomon T (2010) Clinical features, diagnosis, and management of enterovirus 71. Lancet Neurol 9: 1097–1105. [DOI] [PubMed] [Google Scholar]
- 5. Patel KP, Bergelson JM (2009) Receptors identified for hand, foot and mouth virus. Nat Med 15: 728–729. [DOI] [PubMed] [Google Scholar]
- 6. Solomon T, Lewthwaite P, Perera D, Cardosa MJ, McMinn P, et al. (2010) Virology, epidemiology, pathogenesis, and control of enterovirus 71. Lancet Infect Dis 10: 778–790. [DOI] [PubMed] [Google Scholar]
- 7. Wong SS, Yip CC, Lau SK, Yuen KY (2010) Human enterovirus 71 and hand, foot and mouth disease. Epidemiol Infect 138: 1071–1089. [DOI] [PubMed] [Google Scholar]
- 8. Ho BC, Yu SL, Chen JJ, Chang SY, Yan BS, et al. (2011) Enterovirus-induced miR-141 contributes to shutoff of host protein translation by targeting the translation initiation factor eIF4E. Cell Host Microbe 9: 58–69. [DOI] [PubMed] [Google Scholar]
- 9. Huang HI, Weng KF, Shih SR (2012) Viral and host factors that contribute to pathogenicity of enterovirus 71. Future Microbiol 7: 467–479. [DOI] [PubMed] [Google Scholar]
- 10. Chu PY, Lin KH, Hwang KP, Chou LC, Wang CF, et al. (2001) Molecular epidemiology of enterovirus 71 in Taiwan. Arch Virol 146: 589–600. [DOI] [PubMed] [Google Scholar]
- 11. Lee JJ, Seah JB, Chow VT, Poh CL, Tan EL (2011) Comparative proteome analyses of host protein expression in response to Enterovirus 71 and Coxsackievirus A16 infections. J Proteomics 74: 2018–2024. [DOI] [PubMed] [Google Scholar]
- 12. Lee MS, Chang LY (2010) Development of enterovirus 71 vaccines. Expert Rev Vaccines 9: 149–156. [DOI] [PubMed] [Google Scholar]
- 13. Lee MS, Tseng FC, Wang JR, Chi CY, Chong P, et al. (2012) Challenges to licensure of enterovirus 71 vaccines. PLoS Negl Trop Dis 6: e1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Viswanathan K, Fruh K, DeFilippis V (2010) Viral hijacking of the host ubiquitin system to evade interferon responses. Curr Opin Microbiol 13: 517–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. tenOever BR (2013) RNA viruses and the host microRNA machinery. Nat Rev Microbiol 11: 169–180. [DOI] [PubMed] [Google Scholar]
- 16. Taylor KE, Mossman KL (2013) Recent advances in understanding viral evasion of type I interferon. Immunology 138: 190–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Randall RE, Goodbourn S (2008) Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J Gen Virol 89: 1–47. [DOI] [PubMed] [Google Scholar]
- 18. Rajsbaum R, Garcia-Sastre A (2013) Viral evasion mechanisms of early antiviral responses involving regulation of ubiquitin pathways. Trends Microbiol 21: 421–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Munday DC, Surtees R, Emmott E, Dove BK, Digard P, et al. (2012) Using SILAC and quantitative proteomics to investigate the interactions between viral and host proteomes. Proteomics 12: 666–672. [DOI] [PubMed] [Google Scholar]
- 20. Versteeg GA, Garcia-Sastre A (2010) Viral tricks to grid-lock the type I interferon system. Curr Opin Microbiol 13: 508–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Thompson MR, Kaminski JJ, Kurt-Jones EA, Fitzgerald KA (2011) Pattern recognition receptors and the innate immune response to viral infection. Viruses 3: 920–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Oudshoorn D, Versteeg GA, Kikkert M (2012) Regulation of the innate immune system by ubiquitin and ubiquitin-like modifiers. Cytokine Growth Factor Rev 23: 273–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. de Weerd NA, Nguyen T (2012) The interferons and their receptors–distribution and regulation. Immunol Cell Biol 90: 483–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Brennan K, Bowie AG (2010) Activation of host pattern recognition receptors by viruses. Curr Opin Microbiol 13: 503–507. [DOI] [PubMed] [Google Scholar]
- 25. Boss IW, Renne R (2011) Viral miRNAs and immune evasion. Biochim Biophys Acta 1809: 708–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kanarek N, Ben-Neriah Y (2012) Regulation of NF-kappaB by ubiquitination and degradation of the IkappaBs. Immunol Rev 246: 77–94. [DOI] [PubMed] [Google Scholar]
- 27. Sullivan CS, Ganem D (2005) MicroRNAs and viral infection. Mol Cell 20: 3–7. [DOI] [PubMed] [Google Scholar]
- 28. Scaria V, Hariharan M, Maiti S, Pillai B, Brahmachari SK (2006) Host-virus interaction: a new role for microRNAs. Retrovirology 3: 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ghosh Z, Mallick B, Chakrabarti J (2009) Cellular versus viral microRNAs in host-virus interaction. Nucleic Acids Res 37: 1035–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lau NC, Lim LP, Weinstein EG, Bartel DP (2001) An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294: 858–862. [DOI] [PubMed] [Google Scholar]
- 31. Lee RC, Feinbaum RL, Ambros V (1993) The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75: 843–854. [DOI] [PubMed] [Google Scholar]
- 32. Skalsky RL, Cullen BR (2010) Viruses, microRNAs, and host interactions. Annu Rev Microbiol 64: 123–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nathans R, Chu CY, Serquina AK, Lu CC, Cao H, et al. (2009) Cellular microRNA and P bodies modulate host-HIV-1 interactions. Mol Cell 34: 696–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mack GS (2007) MicroRNA gets down to business. Nat Biotechnol 25: 631–638. [DOI] [PubMed] [Google Scholar]
- 35. Lui Y, Timms P, Hafner L, Tan T, Tan K, et al. (2013) Characterisation of enterovirus 71 replication kinetics in human colorectal cell line, HT29. SpringerPlus 2: 267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Berkhout B, Jeang KT (2007) RISCy business: MicroRNAs, pathogenesis, and viruses. J Biol Chem 282: 26641–26645. [DOI] [PubMed] [Google Scholar]
- 37. Lecellier CH, Dunoyer P, Arar K, Lehmann-Che J, Eyquem S, et al. (2005) A cellular microRNA mediates antiviral defense in human cells. Science 308: 557–560. [DOI] [PubMed] [Google Scholar]
- 38. Gottwein E, Cullen BR (2008) Viral and cellular microRNAs as determinants of viral pathogenesis and immunity. Cell Host Microbe 3: 375–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sullivan CS (2008) New roles for large and small viral RNAs in evading host defences. Nat Rev Genet 9: 503–507. [DOI] [PubMed] [Google Scholar]
- 40. Boss IW, Renne R (2010) Viral miRNAs: tools for immune evasion. Curr Opin Microbiol 13: 540–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Roberts AP, Jopling CL (2010) Targeting viral infection by microRNA inhibition. Genome Biol 11: 201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chable-Bessia C, Meziane O, Latreille D, Triboulet R, Zamborlini A, et al. (2009) Suppression of HIV-1 replication by microRNA effectors. Retrovirology 6: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Triboulet R, Mari B, Lin Y, Chable-Bessia C, Bennasser Y, et al. (2007) Suppression of microRNA-silencing pathway by HIV-1 during virus replication. Science 315: 1579–1582. [DOI] [PubMed] [Google Scholar]
- 44. Otsuka M, Jing Q, Georgel P, New L, Chen J, et al. (2007) Hypersusceptibility to vesicular stomatitis virus infection in Dicer1-deficient mice is due to impaired miR24 and miR93 expression. Immunity 27: 123–134. [DOI] [PubMed] [Google Scholar]
- 45. Buck AH, Perot J, Chisholm MA, Kumar DS, Tuddenham L, et al. (2010) Post-transcriptional regulation of miR-27 in murine cytomegalovirus infection. RNA 16: 307–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cameron JE, Yin Q, Fewell C, Lacey M, McBride J, et al. (2008) Epstein-Barr virus latent membrane protein 1 induces cellular MicroRNA miR-146a, a modulator of lymphocyte signaling pathways. J Virol 82: 1946–1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Huang J, Wang F, Argyris E, Chen K, Liang Z, et al. (2007) Cellular microRNAs contribute to HIV-1 latency in resting primary CD4+ T lymphocytes. Nat Med 13: 1241–1247. [DOI] [PubMed] [Google Scholar]
- 48. Jopling CL, Schutz S, Sarnow P (2008) Position-dependent function for a tandem microRNA miR-122-binding site located in the hepatitis C virus RNA genome. Cell Host Microbe 4: 77–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lanford RE, Hildebrandt-Eriksen ES, Petri A, Persson R, Lindow M, et al. (2010) Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science 327: 198–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Martinez I, Gardiner AS, Board KF, Monzon FA, Edwards RP, et al. (2008) Human papillomavirus type 16 reduces the expression of microRNA-218 in cervical carcinoma cells. Oncogene 27: 2575–2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yoshikawa T, Takata A, Otsuka M, Kishikawa T, Kojima K, et al. (2012) Silencing of microRNA-122 enhances interferon-alpha signaling in the liver through regulating SOCS3 promoter methylation. Sci Rep 2: 637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wiertz E, Hill A, Tortorella D, Ploegh H (1997) Cytomegaloviruses use multiple mechanisms to elude the host immune response. Immunol Lett 57: 213–216. [DOI] [PubMed] [Google Scholar]
- 53. Jopling CL, Norman KL, Sarnow P (2006) Positive and negative modulation of viral and cellular mRNAs by liver-specific microRNA miR-122. Cold Spring Harb Symp Quant Biol 71: 369–376. [DOI] [PubMed] [Google Scholar]
- 54. Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P (2005) Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science 309: 1577–1581. [DOI] [PubMed] [Google Scholar]
- 55. Randall G, Panis M, Cooper JD, Tellinghuisen TL, Sukhodolets KE, et al. (2007) Cellular cofactors affecting hepatitis C virus infection and replication. Proc Natl Acad Sci U S A 104: 12884–12889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wen BP, Dai HJ, Yang YH, Zhuang Y, Sheng R (2013) MicroRNA-23b Inhibits Enterovirus 71 Replication through Downregulation of EV71 VPl Protein. Intervirology 56: 195–200. [DOI] [PubMed] [Google Scholar]
- 57.Zheng Z, Ke X, Wang M, He S, Li Q, et al. (2013) Human microRNA hsa-miR-296-5p suppresses Enterovirus 71 replication by targeting the viral genome. J Virol. [DOI] [PMC free article] [PubMed]
- 58. Ho BC, Yu IS, Lu LF, Rudensky A, Chen HY, et al. (2014) Inhibition of miR-146a prevents enterovirus-induced death by restoring the production of type I interferon. Nat Commun 5: 3344. [DOI] [PubMed] [Google Scholar]
- 59. Li Y, Xie J, Xu X, Wang J, Ao F, et al. (2013) MicroRNA-548 down-regulates host antiviral response via direct targeting of IFN-lambda1. Protein Cell 4: 130–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lo AK, To KF, Lo KW, Lung RW, Hui JW, et al. (2007) Modulation of LMP1 protein expression by EBV-encoded microRNAs. Proc Natl Acad Sci U S A 104: 16164–16169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Xiao C, Rajewsky K (2009) MicroRNA control in the immune system: basic principles. Cell 136: 26–36. [DOI] [PubMed] [Google Scholar]
- 62. Liu ML, Lee YP, Wang YF, Lei HY, Liu CC, et al. (2005) Type I interferons protect mice against enterovirus 71 infection. J Gen Virol 86: 3263–3269. [DOI] [PubMed] [Google Scholar]
- 63. Lui YLE, Tan TL, Timms P, Hafner LM, Tan KH, et al. (2014) Elucidating the host–pathogen interaction between human colorectal cells and invading Enterovirus 71 using transcriptomics profiling. FEBS Open Bio 4: 426–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Reed LJ, Muench H (1938) A simple method of estimating fifty percent endpoints. The American Journal of Hygiene 27: 493–497. [Google Scholar]
- 65. Tan EL, Chow VT, Quak SH, Yeo WC, Poh CL (2008) Development of multiplex real-time hybridization probe reverse transcriptase polymerase chain reaction for specific detection and differentiation of Enterovirus 71 and Coxsackievirus A16. Diagn Microbiol Infect Dis 61: 294–301. [DOI] [PubMed] [Google Scholar]
- 66. Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Cell viability assessed 24 h after transfection and throughout 72 h post EV71 infection using vital dye trypan blue. (n = 3, * = p values of <0.05).
(TIFF)
Altered miRNA expression profile of EV71 infected and non-infected control colorectal cells, HT29.
(DOCX)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. Data are within the paper and its Supporting Information files.