Abstract
Genetic variation in the genome of a given species is the basis for natural selection and genetic improvement through selective breeding. We applied 29 microsatellites located on 11 linkage groups to study genetic variation in 276 accessions of J. curcas collected from nine locations in five countries in South America, Asia and Africa to initiate a breeding program. To our surprise, we did not detect any genetic diversity at all 29 microsatellites loci. All the 276 accessions were homozygous at all loci and shared the same genotype at each locus, suggesting no microsatellite variation in the genome of Jatropha curcas. This result is quite unusual, and may have a profound influence on the breeding strategies and genome study of this species.
Keywords: Jatropha, DNA markers, genetic vitiation, epigenetics, breeding.
Introduction
Jatropha curcas (Euphorbiaceae) is oil-bearing tree species tree and grows naturally in countries of the equatorial Americas 1. It has been introduced to other tropical countries in Africa and Southeast Asia (Indian, Indonesia, Malaysia and China) by Portuguese sailors 400 years ago 1. J. curcas is considered a potential bioenergy tree. It does not compete with food crops for fertile land resources 2. Therefore, it was introduced rapidly in various rural programmes with the expectation of producing biodiesel sustainably and profitably 3,4. Genetic variation is the basis for natural selection and genetic improvement through selective breeding. Genetic variation can be measured at DNA level 5. Microsatellites are one of the most preferred co-dominant DNA markers for studying genetic diversity 6 because of their abundance in a given genome, high polymorphism and the ease of genotyping with automatic DNA sequencers. In J. curcas, microsatellites have been recently developed 7,8 and used in constructing a linkage map 8, and studying genetic diversity within and among populations 7,9,10. Results of studies on genetic diversity of J. curcas are controversial 11. Some reported no genetic variation at microsatellite loci 8,10,12, while some reported limited variation 13,14.
To initiate a breeding program for genetic improvement of J. curcas, we collected seeds from nine locations in five countries in South America, Asia and Africa since 2006 and 2007 (Table 1). The collected seeds were germinated in Singapore, and 276 trees were planted and managed under the same condition in our experimental farm in 2007. We observed substantial variations of phenotypic traits, such branch number, tree height and seed yield 10. We isolated DNA from fresh leaves of each tree using a plant DNA isolation kit (Qiagen, Hilden, Germany). DNA was examined in 1% agarose gels to ensure sufficient quality and quantity for PCR. We selected 29 microsatellites from over 1000 markers developed in our lab 8 because they were able to amplify DNA from J. curcas and the related species J. integerrima. Among the 29 microsatellites, 21 named with Jatr- were developed from a J. curcas individual, while the remaining 8 named with Jcuint- were cloned from a hybrid (J. curcas X J. integerrima) individual. Sixteen markers were already mapped onto 11 linkage groups of jatropha 8, and the remaining 13 had not been mapped yet (Table 2). We applied the reference families, which were used to construct the first generation linkage map of jatropha 8, to examine the inheritance patterns of alleles at the 29 loci. All microsatellites were genotyped using the DNA sequencer ABI3730xl (Applied Biosystems, CA, USA) as described previously 8.
Table 1.
Sampling locations of Jatropha curcas, Jatropha integerrima and their hybrid (J. curcas x J. integerrima) on three continents
| Population | Species | Location | Country/Continent | Number |
|---|---|---|---|---|
| Jcu01 | J. curcas | Java | Indonesia/Asia | 32 |
| Jcu02 | J. curcas | Sumatera | Indonesia/Asia | 48 |
| Jcu03 | J. curcas | Yunnan | China/Asia | 48 |
| Jcu04 | J. curcas | Sichuan | China/Asia | 32 |
| Jcu05 | J. curcas | Tamil Nadu | India/Asia | 32 |
| Jcu06 | J. curcas | Sulawesi | Indonesia/Asia | 8 |
| Jcu07 | J. curcas | Suriname | Suriname/South America | 32 |
| Jcu08 | J. curcas | Tanzania | Tanzania/Africa | 32 |
| Jcu09 | J. curcas | Mixed | Not known | 12 |
| Jint01 | J. integerrima | Singapore | Singapore/Asia | 1 |
| Hybrid | J. cu. x J. int. | Singapore | Singapore/Asia | 1 |
Table 2.
Genotypes of 276 J. curcas individuals, one J. integerrima individual and one hybrid (J. curcas x J. integerrima) individual in 29 microsatellite loci*
| LG | Locus | Genbank Accession no. | J. curcas | J. integerrima | J. curcas x J. integerrima |
|---|---|---|---|---|---|
| 1 | Jcuint091 | JF502118 | 215/215 | 189/189 | 189/215 |
| 2 | Jatr691 | EU664836 | 142/142 | 173/177 | 142/173 |
| 3 | Jatr901 | EU664920 | 159/159 | 163/169 | 159/163 |
| 3 | Jcuint221 | JF502150 | 394/394 | 373/375 | 373/394 |
| 4 | Jcuint034 | JF502095 | 338/338 | 334/338 | 334/338 |
| 4 | Jcuint298 | JF502166 | 229/229 | 225/227 | 225/229 |
| 5 | Jcuint061 | JF502107 | 358/358 | 340/340 | 340/358 |
| 5 | Jcuint192 | JF502143 | 214/214 | 196/208 | 196/214 |
| 6 | Jatr152 | EU664742 | 192/192 | 188/204 | 188/192 |
| 6 | Jatr190 | EU664749 | 95/95 | 75/75 | 75/95 |
| 7 | Jatr220 | EU664757 | 223/223 | 223/235 | 223/235 |
| 8 | Jatr679 | EU664757 | 171/171 | 186/186 | 171/186 |
| 9 | Jcuint079 | JF502114 | 162/162 | 145/171 | 145/162 |
| 10 | Jatr153 | EU664743 | 260/260 | 242/242 | 242/260 |
| 11 | Jatr154 | JF502114 | 462/462 | 454/454 | 454/462 |
| 11 | Jatr229 | EU664743 | 151/151 | 161/163 | 151/163 |
| UN | Jatr161 | EU664646 | 250/250 | 234/246 | 234/250 |
| UN | Jatr211 | EU664755 | 283/283 | 259/283 | 259/283 |
| UN | Jatr219 | KC934904 | 356/356 | 362/362 | 356/362 |
| UN | Jcuint240 | KC934905 | 393/393 | 435/435 | 393/435 |
| UN | Jatr194 | KC934906 | 126/126 | 114/114 | 114/126 |
| UN | Jatr221 | EU664758 | 133/133 | 146/146 | 133/146 |
| UN | Jatr030 | EU664686 | 178/178 | 170/170 | 170/178 |
| UN | Jatr079 | KC934907 | 195/195 | 204/206 | 195/206 |
| UN | Jatr093 | KC934908 | 87/87 | 89/89 | 87/89 |
| UN | Jatr095 | KC934909 | 185/185 | 185/185 | 185/185 |
| UN | Jatr102 | EU664717 | 390/390 | 382/394 | 382/390 |
| UN | Jatr103 | KC934910 | 139/139 | 135/137 | 135/139 |
| UN | Jatr105 | KC934911 | 315/315 | 319/325 | 315/319 |
*: All the 276 J. curcas trees showed the same genotypes at each of the 29 microsatellite loci. LG: Linkage group where the markers were mapped. UN: Unmapped. Primer sequences of mapped microsatellites can be found in the paper of Wang et al (2011) 8, while the primer sequences of unmapped markers were submitted to GenBank. The numbers (e.g. 215/215) under each species are the sizes of alleles in base pairs (bp) at each locus.
To further examine microsatellite variation, we blasted the sequences of the 29 selected microsatellites against the draft genome sequence of a J. curcas individual from Japan 15. We also examined the reproduction patterns of J. curcas using artificial pollination. We tested whether the female flowers developed into fruits by covering the female flowers with paper bags to prevent pollinations and by pollinating the female flowers with pollens from individuals of J. curcas or closely related species J. integerrima. If seeds were obtained, we germinated the seeds and genotyped them with the microsatellites to see whether the offspring obtained alleles from the father side.
All the 29 microsatellites amplified specific PCR products in J. curcas and J. integerrima (Table 2). Surprisingly, all the 276 J. curcas individuals were identical and homozygous in all 29 loci, whereas in the individuals from the species J. integerrima and the hybrid, most microsatellites were heterozygous and polymorphic (Table 2 and Figure 1). These results suggest that (A) there is no genetic variation in the 29 microsatellite loci in J. curcas, (B) technical limitations prevent detection of genetic variation; or (C) the J. curcas accessions are cloned from a single tree recently, or (D) J. curcas is haploid or double haploid or (E) J. curcas is capable of apomixes. However, we can rule out the possibilities B-E. As mentioned above, we used one (i.e. ABI3730xl sequencer) of the most advanced genotyping platforms for genotyping microsatellites. The markers showed Mendelian inheritance in the two reference families used for constructing the linkage map of jatropha 8, thus excluding the possibility of errors in genotyping. The 276 trees used in this study were collected from five countries on three continents, and some seeds were directly collected from the wild. Therefore, it is impossible that they were cloned from single individuals recently. Even if all individuals evolved from a single ancestor, after the first introduction 400 years ago (ca. 400 generations), microsatellites should have shown some degree of variation. This is because microsatellites usually show a higher degree of mutation (10-4/generation) than other regions (10-7/generation) of the genome 6. Karyotype analysis showed that J. curcas was diploid, containing 11 chromosome pairs 16, thus excluding the possibility of it being haploid. When the female flowers were covered with paper bags to prevent pollination, the female flowers aborted, whereas using artificial pollination, the female flowers developed into fruits. Microsatellite genotyping showed that the offspring inherited an allele at each locus from the father side (Table 2 and Figure 1). These data indicate that J curcas can reproduce sexually. Thus, our results suggest that there is no microsatellite variation among the 276 individuals.
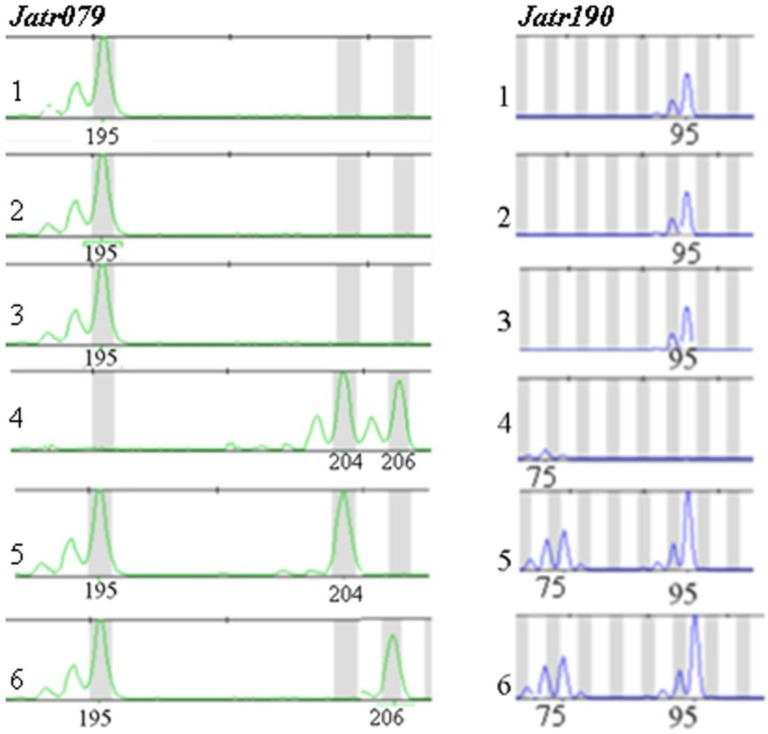
Fig 1.
No microsatellite variation in Jatropha curcas, but some variation in one Jatropha integerrima and two hybrid (J. curcas x J. integerrima) individuals at microsatellite loci Jatr079 and Jatr190. 1-3: thee Jatropha curcas individuals, 4: one J. integerrima individual and 5-6: two hybrid (J. curcas x J. integerrima) individuals. The numbers of each peak are the sizes of allele in base pairs (bp).
Furthermore, in the linkage map of jatropha 8, all 216 mapped microsatellites and 290 SNPs were homozygous in the J. curcas parent, while they were heterozygous in the two hybrid parents. By blasting the sequences of the 22 microsatellites isolated from J. curcas against the draft genome sequence of an individual from Japan 17, we did not find any difference in the number of repeat unit (Figure 2), indicating that there is no microsatellite variation between our J. curcas individual and the individual from Japan. By blasting the eight microsatellites isolated from the hybrid individual against the J. curcas genome sequence, we detected variation of both sequence and repeat length (Figure 2) in 7 of the eight microsatellites. Only in the locus Jcuint221, there was no repeat length difference, probably the sequence of Jcuint221 originated from J. curcas. The sequence similarity of the seven microsatellites ranged from 89% for Jcuint091 to 97% for Jcuint079, while the number of repeat unit changed by 1-10 (Figure 2), indicating that there is microsatellite variation between J. curcas and J. integerrima and within J. integerrima. Yi et al. 10 used 12 SSR primer pairs developed by Sudheer et al 7 to genotype 162 J. curcas samples from five countries. They did not detect any polymorphism and found that all the individuals were homozygous, sharing the same genotype in each locus. In a recent study 12 using 28 microsatellites, no microsatellite variation was detected in J. curcas accessions collected from many places outside Mexico, strongly supporting that there are no microsatellite variation in the genome of J. curcas.
Fig 2.
Alignment of DNA sequences of two microsatellite markers (Jatr161 and Jcuint221) against the genome sequence of a J. curcas individual from Japan. At the locus Jatr161 isolated from J. curcas, there is not repeat and sequence variance, whereas at the locus Jcuint221 isolated from the hybrid (J. curcas x J. integerrima), both DNA sequences and the number of repeats are different.
The phenomenon of no microsatellite variation, but high phonotypic variation in J. curcas is quite unusual and worthy further studying. Epigenetic variations may play an important role in the phenotypic difference 10. Examining epigenetic patterns across the whole genome and in many individuals of J. curcas using whole genome bisulfite sequencing 18 may shield new insights into how the epigenetic variations contribute to the phenotypic variation. In fact, understanding more about the genetic and epigenetic variations in J. curcas and the mechanism underlying phenotypic variations are surely helpful in designing breeding programs to improve the oil yield of J. curcas for sustainable and profitable production of biodiesel.
References
- 1.Heller J. Physic nut, Jatropha curcas L. Rome, Italy: Bioversity International; 1996. [Google Scholar]
- 2.Fairless D. Biofuel: The little shrub that could - maybe. Nature. 2007;449:652–655. doi: 10.1038/449652a. [DOI] [PubMed] [Google Scholar]
- 3.Pan BZ, Xu ZF. Benzyladenine treatment significantly increases the seed yield of the biofuel plant Jatropha curcas. J Plant Grow Reg. 2011;30:166–174. [Google Scholar]
- 4.Ariza-Montobbio P, Lele S. Jatropha plantations for biodiesel in Tamil Nadu, India Viability, livelihood trade-offs, and latent conflict. Ecol Econom. 2010;70:189–195. [Google Scholar]
- 5.Weiner MP, Gabriel SB, Stephens JC. Genetic variation: a laboratory manual. USA: Cold Spring Harbor Laboratory Press; 2007. [Google Scholar]
- 6.Goldstein DB, Schlotterer C. Microsatellites: Evolution and Applications. Oxford: Oxford University Press; 1999. [Google Scholar]
- 7.Sudheer P, Rahman H, Mastan SG, Reddy MP. Isolation of novel microsatellites using FIASCO by dual probe enrichment from Jatropha curcas L. and study on genetic equilibrium and diversity of Indian population revealed by isolated microsatellites. Mol Bio Report. 2010;37:3785–3793. doi: 10.1007/s11033-010-0033-2. [DOI] [PubMed] [Google Scholar]
- 8.Wang CM, Liu P, Yi CX, Gu KY, Sun F. et al. A first generation microsatellite-and SNP-based linkage map of Jatropha. PloS one. 2011;6:e23632. doi: 10.1371/journal.pone.0023632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun QB, Li LF, Wu Y, Ge GJ. SSR and AFLP markers reveal low genetic diversity in the biofuel plant in China. Crop Sci. 2008;48:1865–1871. [Google Scholar]
- 10.Yi CX, Zhang SL, Liu XK, Bui HTN, Hong Y. Does epigenetic polymorphism contribute to phenotypic variances in Jatropha curcas L.? BMC Plant Bio. 2010;10:295. doi: 10.1186/1471-2229-10-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Surwenshi A, Kumar V, Shanwad V, Jalageri B. Critical review of diversity in Jatropha curcas for crop improvement: A candidate biodiesel crop. Res J Agric Sci. 2011;2:193–198. [Google Scholar]
- 12.Vischi M, Raranciuc S, Baldini M. Evaluation of genetic diversity between toxic and non toxic Jatropha curcas L. accessions using a set of simple sequence repeat (SSR) markers. Afri J Biotechnol. 2013;12:265–274. [Google Scholar]
- 13.Rosado TB, Laviola BG, Faria DA, Pappas MR, Bhering LL. et al. Molecular markers reveal limited genetic diversity in a large germplasm collection of the biofuel crop Jatropha curcas L. in Brazil. Crop Sci. 2010;50:2372–2382. [Google Scholar]
- 14.Zhang ZY, Guo XL, Liu BY, Tang L, Chen F. Genetic diversity and genetic relationship of Jatropha curcas between China and Southeast Asian revealed by amplified fragment length polymorphisms. Afri J Biotechnol. 2011;10:2825–2832. [Google Scholar]
- 15.Sato T, Pal S. A two-step site and mRNA-level model for predicting microRNA targets. BMC Bioinformatics. 2010;11:612. doi: 10.1186/1471-2105-11-612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carvalho CR, Clarindo WR, Praca MM, Araujo FS, Carels N. Genome size, base composition and karyotype of Jatropha curcas L, an important biofuel plant. Plant Sci. 2008;174:613–617. [Google Scholar]
- 17.Sato S, Hirakawa H, Isobe S, Fukai E, Watanabe A. et al. Sequence analysis of the genome of an oil-bearing tree, Jatropha curcas L. DNA Res. 2011;18:65–76. doi: 10.1093/dnares/dsq030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li N, Ye M, Li Y, Yan Z, Butcher LM. et al. Whole genome DNA methylation analysis based on high throughput sequencing technology. Methods. 2010;52:203–212. doi: 10.1016/j.ymeth.2010.04.009. [DOI] [PubMed] [Google Scholar]