Abstract
Diverse mechanisms contribute to the evolution of reproductive barriers, a process that is critical in speciation. Amongst these are alterations in gene products and in gene dosage that affect development and reproductive success in hybrid offspring. Because of its strict parent-of-origin dependence, genomic imprinting is thought to contribute to the aberrant phenotypes observed in interspecies hybrids in mammals and flowering plants, when the abnormalities depend on the directionality of the cross. In different groups of mammals, hybrid incompatibility has indeed been linked to loss of imprinting. Aberrant expression levels have been reported as well, including imprinted genes involved in development and growth. Recent studies in humans emphasize that genetic diversity within a species can readily perturb imprinted gene expression and phenotype as well. Despite novel insights into the underlying mechanisms, the full extent of imprinted gene perturbation still remains to be determined in the different hybrid systems. Here we review imprinted gene expression in intra- and interspecies hybrids and examine the evolutionary scenarios under which imprinting could contribute to hybrid incompatibilities. We discuss effects on development and reproduction and possible evolutionary implications.
In many plants and animals, interspecific hybridization events yield offspring that are phenotypically different from either of the parent species. Such hybrids typically display developmental abnormalities and, in animals, often have reduced fertility or complete sterility, particularly in males. Hybrid incompatibilities arise because, although the parental species may be genetically similar, the genomes are still too divergent to sustain normal development, physiology and reproduction when mixed in the hybrid offspring (Wu and Ting, 2004). Extensive research has been performed on genetic incompatibilities in plant and animal hybrids (Ishikawa and Kinoshita, 2009; Johnson, 2010). Key loci have been mapped and characterized in experimental model species, providing important insights into the aberrant phenotypes such as male hybrid sterility (Maheshwari and Barbash, 2011).
Phenotypic abnormalities in interspecies hybrids often differ greatly between the reciprocal crosses. The classic example of such an asymmetry is seen in reciprocal crosses between donkeys and horses, where both directions of the cross produce sterile offspring, but the gross phenotype of the progeny (that is, ‘mule' versus ‘hinny') depends on the direction of the cross. Horses and donkeys have a different chromosome number, but this cannot explain the differential hybrid phenotypes that depend on the direction of the cross (as the reciprocal crosses have the same autosomal karyotype). More than 50 years ago serum concentrations of a placental hormone were reported to be markedly higher in mule than in hinny conceptuses, suggestive of parental genome-specific gene expression (Allen, 1969).
The North-American genus Peromyscus (‘deer mice') has been studied extensively to explore hybrid incompatibilities in mammals (see Table 1 for a list of species and terms). Pronounced cross-direction-dependent phenotypes were reported in hybrids between the closely-related allopatric species P. maniculatis and P. polionotus (Vrana et al., 1998). Also in interspecies hybrids in Mus (house mouse), between the sympatric species M. musculus and M. spretus, morphological differences are apparent between reciprocal hybrids (Zechner et al., 2004). These hybrid effects were observed in crosses between a mixed M. musculus domesticus strain and lab stocks of M. spretus. To be definitive about where the incompatibilities lie between M. musculus and M. spretus (or M. m. castaneus, see below), reciprocal crosses between several different wild-derived stocks (or wild caught animals) of M. musculus and M. spretus populations would be needed.
Table 1. Terminology and abbreviations.
| Mules | Progeny of a male donkey and a female horse |
| Hinnies | Progeny of female donkeys and male horses |
| Peromyscus | North-American genus of mice (‘deer mice') |
| P. maniculatis (‘M') | A species with polygamous mating behaviour |
| P. polionotus (‘P') | Species with apparent monogamous mating behaviour |
| P × M | Hybrid produced by a female P. maniculatis paired with male P. polionotus |
| M × P | Hybrid produced by a male P. maniculatis paired with female P. polionotus |
| Mus musculus (‘MU') | Widely studied mouse species |
| M. spretus (‘S') | Species related to M. musculus, in the Mediterranean, that diverged over one million years ago |
| (MU × S) F1 | Hybrid produced by a male M. musculus paired with a female M. spretus |
| (S × MU) F1 | Hybrid produced by a female M. musculus paired with a female M. spretus |
| C57Bl/6J (‘B') | A mixed M. M. domesticus laboratory mouse inbred strain |
| CAST/EiJ (‘C') | M. M. castaneus laboratory mouse strain |
| Arabidopsis | Genus of small flowering plants of the mustard family (Brassicaceae) |
| A. thaliana, A. arenosa | Related Arabidopsis species used in imprinting studies |
| DMR | ‘Differentially methylated region': here, a sequence element with allele-specific CpG methylation |
| ICRs | ‘Imprinting control regions': essential regulatory DMRs, which have germ line-derived, mono-allelic DNA methylation and mediate imprinted gene expression in cis. |
| D–M model | Dobzhansky–Muller model |
| AmAp | Alleles derived from the mother and father, respectively |
Besides other candidate mechanisms—such as the maternal inheritance of mitochondrial DNA and its interactions with the nuclear genome, or possible maternal effects (Turelli and Moyle, 2007; Johnson, 2010)—the epigenetic phenomenon of genomic imprinting is thought to be one of the contributors to the phenotypic differences between reciprocal hybrids. Genomic imprinting evolved convergently in flowering plants and mammals (Feil and Berger, 2007) and mediates mono-allelic expression at selected genes, in a parent-of-origin-dependent manner. Imprinted genes contribute to diverse processes in development and growth, including that of nourishing the extra-embryonic tissues (placenta in mammals/endosperm in plants). In mammals, imprinted genes also have important roles in brain development and function (Wilkinson et al., 2007).
In interspecies hybrids, differences between the parental species in the genetic control and patterns of imprinting may have different effects dependent on the orientation of the cross, including epigenetic perturbation of imprinting control leading to ‘loss of imprinting' (biallelic expression). Studies in mammals have provided clear evidence for perturbed imprinting in inter- and intraspecies hybrids (reviewed below). However, as many imprinted genes have been discovered in these same interspecies hybrids, and polymorphisms are necessary to identify allele-specific expression differences, it is possible that hybridization itself could induce imprinting depending on the location of the polymorphism(s) between strains, for instance in cis-acting elements.
Crosses between different Arabidopsis species have provided evidence that perturbed imprinted gene expression occurs also in plant hybrids (Josefsson et al., 2006; Jullien and Berger, 2010). Particularly, the imprinted expression of MEDEA (MEA) and PHERES (PHE) in endosperm is perturbed in hybrids between A. thaliana and A. arenosa and this could contribute to the endosperm overgrowth seen in these hybrids (Josefsson et al., 2006). As ploidy was often altered in these existing studies, the results have been somewhat difficult to interpret considering the mechanisms involved (Walia et al., 2009; Jullien and Berger, 2010).
Here we focus on the animal systems, which have provided most insights into imprinting in hybrids. We also discuss the extent to which intraspecies polymorphisms may perturb imprinted gene expression and hence phenotype.
Imprinted gene expression in mammalian interspecies hybrids
The best-studied mammalian hybrids are in rodents, particularly those generated by crossing P. maniculatis and P. polionotus. These deer mouse species are similar in size and evolutionarily separated by ∼100 000 years. Whereas P. maniculatis (‘M') is polygamous, P. polionotus (‘P') is thought to be largely monogamous (Foltz, 1981). This difference in mating system could potentially be relevant with regards to the evolution of genomic imprinting because it should lead to different patterns of intrafamilial conflict. The most widely accepted theory for the evolution of imprinting—the ‘conflict hypothesis' (Moore and Haig, 1991)— assumes that competition over maternally provided resources between offspring fathered by different males can favour imprinted expression (Haig, 2013; see also Burt and Trivers, 2008). Hence, difference in mating system could alter patterns of relatedness and change patterns of conflict within families.
Although both deer mouse species show allelic expression at the imprinted genes analysed, the reciprocal crosses yield phenotypically different offspring (Rogers and Dawson, 1970). A female P. maniculatis crossed with a P. polionotus male produces offspring that are smaller than either of the parents. Remarkably, the reciprocal cross produces conceptuses that are oversized, with placentomegaly, and often die before birth (Loschiavo et al., 2007; Duselis and Vrana, 2010). In the latter, (P × M)F1 hybrids, the overgrowth phenotype is linked to loss of imprinting at Peg3, Mest (Peg1), Snrpn, H19 and Grb10 (Vrana et al., 1998, 2000). Genetic studies showed that placental overgrowth correlates with loss of imprinting at Peg3 (Loschiavo et al., 2007; Vrana et al., 2000), specifically loss of (maternal) DNA methylation at the imprinting control region (ICR) of this locus (Wiley et al., 2008). Loss of methylation was also observed at the imprinted H19 locus (Vrana et al., 1998) and at the ICR controlling the growth-related Kcnq1 domain. The latter correlated with aberrant, biallelic expression of the Kcnq1ot1 long non-coding RNA that mediates allelic repression along this imprinted domain, particularly in the placenta. The X chromosome is also genetically linked to the overgrowth phenotype (Vrana et al., 2000; Duselis et al., 2005). In female (P × M)F1 embryos, X chromosome inactivation was not random but was heavily skewed towards the paternal X (Vrana et al., 2000). This pronounced deviation from random X inactivation likely contributes to the phenotype in females, and could be due to polymorphisms at the Xce locus, which influences the choice of the X chromosome to be inactivated (Cattanach and Williams, 1972). In the reciprocal, phenotypically almost normal (M × P)F1 hybrids, only some imprinted genes displayed a varying degree of loss of imprinting (that is, Igf2r, Ascl2 and Grb10), in some tissues only (Vrana et al., 1998).
To explore the genus Mus, hybrids between M. spretus (S) and M. musculus (MU) lines have been studied in most detail. Although some imprinted genes are affected in an asymmetric manner, the data so far suggest that loss of imprinting occurs similarly in the hybrids derived from the reciprocal crosses (Shi et al., 2004, 2005). It is unknown whether this is responsible for the less pronounced phenotypic difference between the reciprocal hybrids than in the Peromyscus crosses. (MU × S)F1 adults are smaller than the reciprocal, (S × MU)F1, hybrids. However, both are larger than the parental species and show placental overgrowth (Zechner et al., 2004). The increased body weight and large internal organs are associated with loss of imprinting at Peg1(Mest) in the (S × MU)F1 hybrids, and less so in the (MU × S)F1 hybrids, and the extent differs between individual animals (Shi et al., 2004). The Mit1(Lb9)- and Copg2-imprinted genes located close to Peg1 on chromosome 6, and the Nnat, Slc22a18 and Peg3 genes also showed evidence for loss of imprinting, again mostly in the (MU × S)F1 hybrids (Shi et al., 2005). A tissue-specific effect occurred at the imprinted gene Sgce, with complete loss of imprinting in kidney and skeletal muscle in the (S × MU)F1 hybrids. The brain-specific imprinted gene Rasgrf1, in contrast, showed tissue-specific loss of imprinting in the (MU × S)F1 hybrids.
It is unclear to what extent the loss of imprinting in interspecies Mus hybrids correlates with epigenetic changes at the corresponding ICRs. Similarly as in the Peromyscus hybrids, however, there is loss of DNA methylation at the Peg3 differentially methylated region in (MU × S)F1 but not in (S × MU)F1 hybrids (Shi et al., 2005). Earlier studies analysed allelic DNA methylation at different ICRs in (MU × S)F1 embryos, but did not provide evidence for extensive methylation losses (Paulsen et al., 1998). Global methylation levels are normal in the hybrid placentae, with no evidence for loss of methylation at repeat elements (Schutt et al., 2003). Similar data were obtained in hybrids between antelope species (Robinson et al., 2000). This contrasts with marsupials, in which placentae in interspecies hybrids were reported to have a global loss of DNA methylation and aberrant activation of retro-elements (O'Neill et al., 1998).
A recent study on M. m. domesticus × M. m. castaneus inter sub-species hybrids reported strain-dependent changes in imprinted expression at the Kcnq1 locus on chromosome 7 (Korostowski et al., 2012). Detailed analysis in reciprocal inter sub-species hybrid hearts revealed an altered relative abundance of parental-specific transcripts depending on the direction of the cross. Essentially, expression from the CAST/EiJ (C) allele was higher than from the C57BL/6J (B) allele, which continued throughout heart development, but was not contingent on differential DNA methylation. The timing of a developmental switch from mono- to biallelic expression was delayed in (C × B)F1 hybrids compared with (B × C)F1 hybrids. The strain-specific genetic factor involved (such as a cis-regulatory polymorphism, for example, affecting transcription factor binding) must be located on chromosome 7 because these studies used a strain of mice with a single CAST/Ei chromosome 7 on an otherwise C57Bl/6J background.
Although imprinting is perturbed in Mus and Peromyscus hybrids, a recent study provides little evidence for such perturbation in the genus Equus. Of the genes imprinted in trophoblast, all are expressed from the same parental allele predominantly in both the hinny and mule. In contrast to the Mus and Peromyscus hybrids, the H19- and PEG3-imprinted loci showed no evidence for loss of DNA methylation (Wang et al., 2013). However, individuals varied in expression levels at imprinted genes. A newly discovered imprinted gene controlling chromatin, HAT1 (histone acetyltransferase-1), was more highly expressed in mule than in hinny trophoblast. It would be interesting to explore imprinted gene expression in mule and hinny embryos to complement these extensive studies on trophoblast.
Genetic polymorphisms and the control of mono-allelic gene expression in humans
Human populations show considerable genetic diversity, including variation in whether or not a specific gene is imprinted. For example, at the IGF2-receptor (IGF2R) gene, mono-allelic expression from the maternal allele occurs in a minority of individuals only (Monk et al., 2006; Yotova et al., 2008). Unexpectedly, the maternal DNA methylation imprint at this locus was normally present in all individuals (Vu et al., 2006). The regulatory long non-coding RNA produced from the unmethylated allele of the imprinting control region (ICR), however, was expressed in some individuals only. Lack of expression of this long non-coding RNA correlates with biallelic expression (‘loss of imprinting') of IGF2R and of close-by placenta-specific genes (Monk et al., 2006). The growth-related PEG1/MEST gene on human chromosome 7q32 is polymorphically imprinted (Huntriss et al., 2013); its isoform-2 in placenta and early embryos is highly variable between conceptuses and this correlates with differential DNA methylation levels at an exonic differentially methylated region (McMinn et al., 2006).
Recent studies in humans have uncovered that SNPs may induce pronounced changes in locus-specific DNA methylation levels (Hellman and Chess, 2010). Whether polymorphic loss of imprinting also results from genetic diversity remains unknown. However, single-nucleotide mutations were shown to prevent binding of pluripotency factors (OCT4 and SOX2) to the H19 ICR. Interestingly, these mutations induced aberrant acquisition of DNA methylation at this ICR and biallelic expression of the locus' IGF2 gene, resulting in an overgrowth upon maternal, but not upon paternal, transmission (Demars et al., 2010; Poole et al., 2012). Mouse studies have shown that Igf2 overexpression induces fetal and postnatal overgrowth, and Peg1 overexpression leads to reduced growth and fat mass expansion. Biallelic Igf2r expression leads to reduced fetal growth as well. In experimental rat studies, levels of IGF2R expression were linked to an increased risk of liver tumours (Mills et al., 1998), presenting another level at which selection pressure may arise. It should be interesting to explore more broadly whether specific human genes have a tendency to become imprinted, or, conversely, to lose their imprinted gene expression status in human populations.
Differences between the parental genomes affect imprinted gene expression in hybrids
The developmental establishment and maintenance of imprinting involve complex interactions between cis-regulatory elements and trans-acting factors, some of which are themselves subject to imprinting (Kelsey and Feil, 2013; Wolf, 2013). The transcription factor gene ZAC1 is imprinted in both mice and humans. This maternally repressed growth-related gene controls the expression of multiple other imprinted genes, including Igf2, H19, Cdkn1c and Dlk1. ZAC1 binds to regulatory sequences at these other loci, and its expression was proposed to be important in the control of an imprinted gene network (Varrault et al., 2006). Further evidence for interdependence of imprinted gene expression comes from targeting studies on the H19 gene (Gabory et al., 2009) and from data indicating that the imprinted IGF2-H19 locus physically interacts with several other imprinted loci in the nucleus (Murrell et al., 2004; Zhao et al., 2006). Genetic studies on Peg3 using congenic lines formally showed that its frequently observed loss of imprinting in (MU × S)F1 hybrids is due to a yet unknown spretus-specific trans-acting factor (Shi et al., 2005).
The Polycomb group gene Sfmbt2 is expressed only from the paternal allele in early embryos and extra-embryonic tissues (Lehnert et al., 2011; Wang et al., 2011). The precise function of this polycomb group protein in gene regulation and whether it affects other imprinted genes remains to be explored.
Several of the trans-acting factors that control imprinting maintenance are organized in multi-protein complexes (Kacem and Feil, 2009; Kelsey and Feil, 2013). The composition and/or efficiency of such complexes may be compromised in hybrids due to polymorphisms, which could, for instance, interfere with stabilizing protein–protein interactions. Imprinted regulatory ncRNAs may also contribute to trans-regulation between the parental chromosomes. For example, the maternally inherited copy of the imprinted Dlk1-Dio3 domain on mouse chromosome 12 expresses two imprinted microRNAs (miRNA) (mir127 and mir136) that are antisense to a paternally expressed developmental gene called Rtl1. The imprinted miRNAs induce RISC-mediated cleavage of Rtl1 mRNA, and thereby affect gene expression on the opposite parental chromosome (Davis et al., 2005). During recent years it has become apparent that hundreds of small regulatory RNAs are imprinted. As small RNAs and their targets are often specific to individual species (Ha et al., 2008; Ng et al., 2012), for imprinted clusters of miRNAs this could induce aberrant effects on target gene expression in interspecies hybrids. Significantly, several of the clusters of imprinted miRNAs discovered in mammals are present in specific species only (Noguer-Dance et al., 2010; Girardot et al., 2012), indicating that their imprinting status and regulatory function(s) have arisen recently during evolution.
The somatic maintenance of methylation imprints in the early embryo involves non-histone proteins that are in part maternally provided through the oocyte (Kelsey and Feil, 2013). Amongst these is ZFP57, a KRAB-domain zinc-finger protein, that binds preferentially to methylated DNA at a hexanucleotide sequence motif. Many ICRs comprise this sequence motif. Loss of ZFP57 expression leads to loss of the DNA methylation imprints at multiple ICRs (Quenneville et al., 2011). Concordantly, genetic mutations at ZFP57 in humans are causally involved in imprinting disorders and are linked to reduced DNA methylation at specific ICRs (Mackay et al., 2008). Other maternal factors, including DPPA3 (also called PGC7, Stella), also contribute to the methylation maintenance at ICRs in the zygote and early embryo, on both the maternally and paternally inherited copies of the genome (Nakamura et al., 2007). In interspecies hybrids, incompatibilities between maternal factors and target sequences on the paternal genome might affect genomic imprinting during the early stages of development. Intriguingly, in the Peromyscus hybrids, perturbation of imprinting was most pronounced in the extra-embryonic lineages. Possibly, the maintenance of differential DNA methylation is less stable in these lineages and could therefore be more susceptible to incompatibilities between the parental genomes.
Genetic polymorphisms in promoters and enhancers could have pronounced effects on the measured allele specificity of expression at imprinted genes in hybrids. Different alleles at a given imprinted gene may show different expression levels from the repressed allele, which will influence the perceived allele specificity of expression in the reciprocal hybrids. Indeed, many imprinted genes show markedly different expression levels in reciprocal hybrids. In female rodents, the trophectoderm shows imprinted X inactivation, with gene expression occurring on the maternally inherited X chromosome, but not the paternally inherited X chromosome (Takagi and Sasaki, 1975). In female rodent hybrids, consequently, placental development could be affected by polymorphisms at X-linked genes. One candidate is the imprinted Esx1 gene, at which loss of expression leads to placental overgrowth (Salas et al., 2004). The placental dysplasia in M. musculus × M. spretus hybrids is indeed genetically linked to the X chromosome, to a region comprising Esx1 (Hemberger and Zechner, 2004; Zechner et al., 2004). Also in Peromyscus, placental overgrowth in hybrids is linked to the X chromosome (Vrana et al., 2000) and is associated with abnormal levels of Esx1 expression (Duselis and Vrana, 2010). Whether the differential Esx1 expression between reciprocal hybrids is caused by genetic polymorphisms at the locus itself is not known. Data from the placenta-specific PHLDA2 gene in humans, however, indicate that genetic polymorphisms may affect phenotype (Ishida et al., 2012). In European birth cohorts, a 15-base-pair repeat sequence variant at the promoter is linked to reduced PHLDA2 expression on maternal transmission, resulting in increased birth weight. Given the importance of dosage in imprinted gene expression, particularly in mammals, intra- and interspecific hybrids are expected to display phenotypic effects. As several imprinted genes appear to be part of an expression network, perturbed expression levels at one gene may also induce altered expression at other imprinted loci.
The imprinting status of a gene is not a fixed property, and hence, as the imprinting status of genes evolves there are genes that will be imprinted in some species, but not in other closely related species. In plants, systematic studies show little overlap of imprinted gene expression between different species (Berger et al., 2012). In mammals, there is evidence for recent acquisition of imprinting in the primate and rodent lineages. One example is Sfbmt2, a gene that became imprinted in the genus Mus (but not in Peromyscus) owing to an evolutionarily recent insertion of a miRNA cluster (Lehnert et al., 2011). Several clusters of imprinted small regulatory RNA have been detected in specific groups of mammals only, pinpointing their recent appearance during evolution (Girardot et al., 2012). Among the group of imprinted genes derived by retrotransposition from the X chromosome, Mcts2 retrotransposed and became imprinted around the supra-primate clade (Wood et al., 2008) and more recently still, U2af1-rs1 (or Zrsr1) was formed from the most recent retrotransposition event among the known imprinted retrogenes. It is present and imprinted in mouse and rat but absent in kangaroo rat (Dipodomys ordii), guinea pig (Cavia porcellus) and humans (McCole et al., 2011). One might expect the recently evolved imprinted genes to be particularly prone to perturbation in reciprocal hybrids.
Genomic imprinting, an interspecies barrier?
Although the overall importance of perturbed imprinting as a barrier to hybridization remains unclear, there are several intriguing results. Several of the genes at which loss of imprinting was detected (Slc22a18, Igf2r, Peg1/Mest, Peg3, Cd81, H19, Kcnq1ot1) in Mus and Peromyscus interspecies hybrids are important in placental and embryonic development, and in these mouse hybrids, placental development is particularly affected (Vrana et al., 2000). Other perturbed imprinted genes are important in brain development and function (Ndn, Snrpn, Nnat, Rasgrf1, Copg2), and this could be relevant to the hybrid phenotype as well. As mammalian imprinted genes are organized in clusters, each controlled by a differentially methylated region, it is interesting that both in Mus and Peromyscus hybrids, abnormal DNA methylation was detected at some germline differentially methylated regions, in some of the analysed tissues. However, in other genera, perturbation of imprinting in interspecies hybrids could be much less pronounced than in rodents, as suggested by the recent research on mule and hinny conceptuses (Wang et al., 2013).
In hybrids between M. musculus and M. spretus, the biallelic expression of the Rasgrf1 and Peg1 imprinted genes is associated with higher expression levels (Shi et al., 2005). Also in Peromyscus hybrids, expression levels are abnormally high at the growth-related Igf2, Grb10, Cdkn1c and Phlda2 genes (Duselis and Vrana, 2007). In humans, perturbed expression of IGF2 is involved in the fetal growth syndromes such as Beckwith–Wiedemann syndrome and Silver–Russell syndrome (Hirasawa and Feil, 2010), and reduced expression of PHLDA2 is associated with increased birth weight (Ishida et al., 2012). This interesting parallel suggests that the loci that are susceptible to loss of imprinting are also those that are most readily perturbed because of genetic differences between the parental genomes in interspecific hybrids (Shi et al., 2005). Another parallel with imprinting-related disorders in humans is the apparent embryonic occurrence of the loss of imprinting in the interspecies animal hybrids.
Loss of imprinting in hybrids might imply that cis-regulatory regions are not conserved between species and undergo rapid evolution, but recent studies in mammals indicate that this is generally not the case. CpG islands at imprinted loci do not generally show a lower level of conservation compared with autosomal CpG islands. At the maternally methylated ICRs, there is even a tendency to strongly maintain CpG density. At paternal ICRs, in contrast, CpG density is lost during evolution, which indicates that these ICR regions evolve more quickly than the maternal ICRs (Schulz et al., 2010).
The above reviewed studies evoke different hypothetical scenarios through which imprinted gene expression may become perturbed in hybrid offspring. Even without alteration of the imprinting status, there may in some cases be marked differences in gene expression levels between the reciprocal hybrids (Figure 1). In cases where trans-acting factors are perturbed, some of which may be subject to imprinting themselves, different scenarios predict that this affects imprinted gene expression in hybrid offspring (Figure 2). Although empirical studies presented in this review demonstrated or strongly suggested the involvement of imprinted genes in hybrid incompatibilities, their role(s) in population differentiation and speciation has not received much attention from a theoretical perspective. Below, we present several evolutionary considerations that may be relevant in the context of this question.
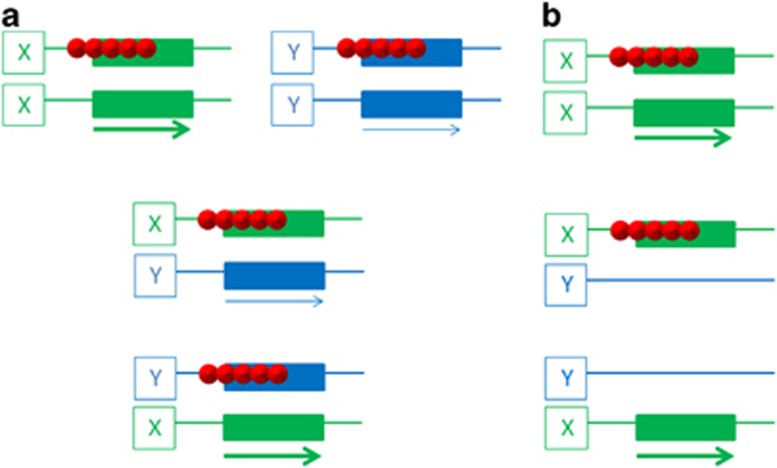
Figure 1.
Scenarios of imprinted gene perturbation in hybrids. (a) A hypothetical imprinted gene (rectangles) has a different level of expression in one species (‘genotype X') than in another (genotype Y). In the reciprocal X × Y and Y × X hybrids, this leads to different expression levels, with possible phenotypic consequences. A similar differential expression arises in hybrid offspring in case a gene is imprinted in one species, but not in the other. (b) Some imprinted genes are present in certain analysed species only. Shown is a recently evolved imprinted gene present in species X (top of the figure) but not in species Y (not shown). In the X × Y and Y × X reciprocal hybrids, this leads to either normal expression or a complete lack of expression. Small red spheres indicate DNA methylation. Arrows depict RNA expression; their thickness reflects levels of transcription.
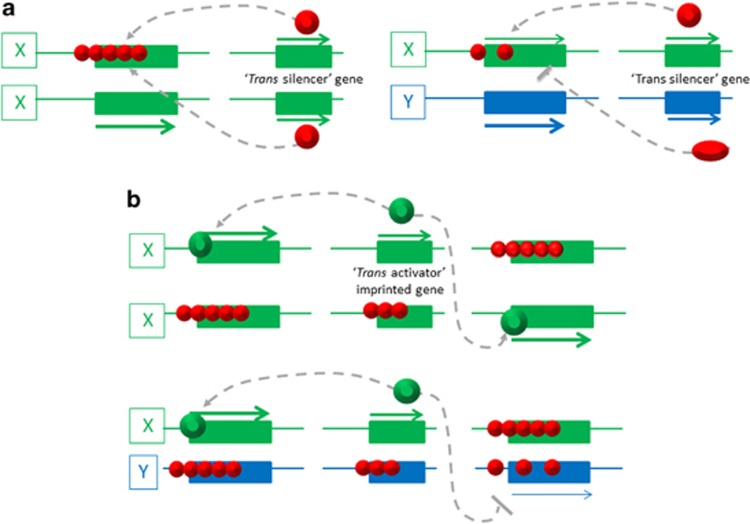
Figure 2.
Scenarios through which trans-acting factors may affect imprinted gene expression in hybrids. (a) Trans-acting factors involved in the somatic maintenance of allelic repression at imprinted genes. To the left, the situation in a wild-type embryo, in which the two paternal genomes are of the same genotype (‘X' genotype). A trans-acting repressor (large red sphere) maintains allelic repression at an imprinted target gene elsewhere in the genome. In embryos of the hybrid X × Y genotype, the Y genome-encoded repressor protein does not repress efficiently the imprinted gene of the X genotype, leading to loss of imprinting. (b) A trans-activator-imprinted gene encodes an activating factor (green spheres), which maintains allelic gene transcription at multiple imprinted target genes, on both the parental genomes. In embryos of hybrid genotype—shown here is the example of hybrid X × Y genotype—the factor activates imprinted genes on its own parental (X) genome, but not on the opposite parental (Y) genome. This leads to an aberrant acquisition of repressive modifications and loss of expression at these imprinted genes. Imprinted trans-acting factors can not only be proteins but also non-coding RNAs.
Evolutionary considerations
Models of hybrid incompatibility have mostly been built on the framework of the Dobzhansky–Muller (D–M) model (Dobzhansky 1937; Muller 1942), which explains how lineages could evolve incompatibilities without any lineage having to pass through the low-fitness state observed in hybrids (Turelli and Orr, 2000). The D–M model posits that evolution must have proceeded through a series of changes that were either beneficial in one or more lineages (adaptive divergence) or were at least not deleterious (neutral divergence). The D–M model typically assumes that differentiation requires more than one locus because a one-locus system cannot move between high-fitness states (fixation for different alleles) that are separated by a low-fitness intermediate without evolving through that low-fitness state. For example, under-dominance can produce an incompatibility, but divergence from fixation of one allele (for example, A1) to an alternative allele (for example, A2) requires a population to pass through the low-fitness heterozygote state (A1A2). The D–M model's solution to this problem is that incompatibilities evolve in a multi-locus system with epistasis (Johnson, 2000). The logic of the model should extend to imprinted genes, but the presence of imprinting offers other scenarios for the evolution of incompatibilities that are not captured in the classic D–M model. We explore a few simple scenarios to understand how imprinting could potentially lead to the evolution of hybrid incompatibilities.
A diversity of studies generally support the tenet of the D–M model that epistasis is involved in hybrid incompatibilities (Johnson, 2000). However, although the simple D–M model appears to explain the presence and types of incompatibilities, it does not directly explain asymmetries that are commonly observed in reciprocal crosses, where the outcome depends on the direction of the cross (that is, on the sex of the parent from each species) (Turelli and Moyle, 2007). Direction-dependent patterns require either asymmetrical inheritance (sex chromosomes or cytoplasmic factors) or the presence of an epigenetic asymmetry such as genomic imprinting or a maternal effect. Turelli and Moyle (2007) provide a very general analysis of the evolution of asymmetrical D–M isolation in the presence of asymmetrical inheritance and, although they do not examine imprinting per se, they suggest that the parent-of-origin effects associated with imprinting could provide the necessary inheritance asymmetry to produce asymmetrical incompatibilities. Here we build on their conclusions by explicitly examining some scenarios to understand how imprinted genes can evolve incompatibilities through a D–M-like process (that is, with few genetic steps needed to cause incompatibilities without passing through a fitness valley).
Cis-regulated imprinting
Consider a locus that evolves imprinting through a cis process (that is, that a locus determines its own imprinting status). Assume that two populations start with a non-imprinted allele (A1) and one population evolves to fix an allele, A2, that is imprinted and expressed from the paternal copy only. This scenario would lead to an asymmetry in the reciprocal hybrids, as the alleles would be epigenetically different depending on the direction of the cross. Hybrids that inherit the A2 allele from their mothers (A1A2) would inherit a silenced copy of the allele, and therefore would show mono-allelic expression of the A1 allele, whereas the hybrids inheriting the A2 allele from their fathers (A2A1) would show biallelic expression (as the A2 allele would not have been silenced by their father, and the A1 allele is not imprinted regardless of its parent-of-origin). Although we can imagine that the two reciprocal hybrid genotypes could have different phenotypes (as neither expression pattern in hybrids appears in the parentals), neither of the hybrid genotypes can presumably be deleterious, as both configurations would have to have appeared in the ancestral population along the evolutionary path from fixation of A1 to A2. Therefore, a cis-regulated imprinted locus is unlikely to contribute to population incompatibility through such a one-step process, implying that there would need to be a second evolutionary step to produce an incompatibility through this mechanism.
For imprinted genes, there is a general expectation that additional evolutionary steps will occur after the establishment of imprinted expression, potentially leading to D–M incompatibilities. These additional changes are expected because, when a locus evolves to become imprinted, the change in expression sets the stage for selection to favour alleles that are adapted to being imprinted (either because of the presence of mono-allelic expression or because selection now operates on only one parent-of-origin configuration) (Haig, 2002). Thus, the outcome is that the alleles present at imprinted loci have co-evolved with their imprinting status.
To understand how the evolutionary origin of imprinting at a locus drives further evolution at the locus, we can consider how the presence of imprinting may favour alleles with different expression levels. Such a case may occur when selection favours overexpression of the gene to compensate for the lack of biallelic expression or when the presence of imprinting removes counteracting selection associated with parental conflict (that is, evolution proceeds by the ‘loudest voice prevails' principle predicted by the parental conflict hypothesis; see Haig, 1997). If a population fixes the imprinted allele (A2), selection may then favour some new allele (A3) that shows higher expression. Under this scenario, there would not only be an asymmetry in hybrids but also a dosage problem in the reciprocal hybrids. The A1A3 would show mono-allelic expression of the A1 allele, which is not deleterious in this scenario, but the A3A1 would show biallelic expression, and hence would have very high expression levels because the A3 allele is highly expressed. In a more complex scenario (where the A1 allele was not present when imprinting originated), it is possible that mono-allelic expression of the A1 allele could be deleterious, and the two reciprocal heterozygotes could both have low fitness and show asymmetries. Thus, this process is similar to that envisioned by the D–M model in that a population could evolve incompatibility through a two-step process that did not require a low-fitness intermediate, but the two changes would have to occur at the same locus, rather than at two different loci.
A more extreme scenario could appear if a locus diverges to show opposing patterns of imprinting in two evolutionary lineages. For example, if one population fixes for a paternally expressed allele Ap and the other population fixes for a maternally expressed allele Am, then one configuration of the cross (AmAp) would lead to null expression, whereas the other configuration (ApAm) would lead to diploid expression of a pair of alleles that evolved to be mono-allelically expressed. Such a scenario could lead to extreme asymmetries in the outcome of reciprocal crosses, with completely different phenotypic consequences associated with null expression versus biallelic expression. Apart from some imprinted genes that show marked tissue-specific differences between species (Prickett and Oakey, 2012), however, there is limited evidence that the same gene shows different patterns of imprinting in different species (other than the simple pattern of being imprinted in one species and not another).
Trans-effects in imprinting evolution
Although the single-locus scenarios (above) can result in reduced hybrid fitness and asymmetries, there is no simple single-step evolutionary path from the fixation of one allele to another that would also lead to an incompatibility. However, we can imagine a scenario akin to the basic D–M in which changes happen at a pair of loci, but in this case we consider a target gene and a locus that regulates its imprinting status in trans. With trans-regulation, species may diverge for recognition sites or can diverge in the properties of trans-acting factors such that trans-acting factors are unable to imprint a gene in one lineage or maintain an imprint in the hybrid once established in the parental genotypes. The two sides may also diverge in tandem, such that species differ in recognition sites and regulatory machinery (for example, sequences of miRNAs or other trans-acting factors).
With trans-regulation there are two-step scenarios similar to the D–M model that could potentially lead to asymmetries in a cross as in the single-locus model. For example, consider a case where populations start out fixed for the two-locus genotype A1A1B1B1, where the A locus affects some fitness-related trait and the B locus is a trans-acting regulator of imprinting. We can imagine a diversity of scenarios in such a system depending on how the establishment and maintenance of imprinting works. For example, a new allele (A2) may arise and fix in one population and subsequently, a new allele (B2) may be favoured and fixed at the B locus because it leads to imprinting at the A locus (leading to one population fixed for A1A1B1B1 and one fixed for A2A2B2B2). In one direction of the cross (which direction depends on the pattern of imprinting), individuals would express both the A1 and A2 allele, which presumably is not deleterious because that genotype was present when the A2 allele arose and fixed. However the other direction of the cross individuals would express only the A1 allele. If mono-allelic expression of that allele is deleterious (perhaps because it shows a much lower expression level than A2), then that cross direction could lead to an incompatibility. The pattern in the hybrids would follow that of the single-locus model where the imprinting status of an allele depends on its parent-of-origin in the cross, which is the ultimate origin of the asymmetry. However, the two-locus scenario allows for one direction to show reduced fitness because that configuration never appeared in the evolutionary history of the populations.
Finally, it is possible that imprinting contributes to hybrid incompatibilities through a trans-acting maternal effect on imprinting. Maternal effects, in which the influence of genes expressed in the mother (that is, the maternal effect) depends on the genotype of the offspring, can potentially be a major contributor to hybrid incompatibilities when the two evolve to be complementary (Wolf and Brodie, 1998). Under such conditions, a mismatch between the maternal effect and offspring genotype can produce hybrid incompatibilities (Wolf, 2000). Such a scenario can extend to imprinted genes (Wolf and Hager, 2006). Some trans-acting factors like ZFP57 and DPPA3 appear to have their influence, at least to some degree, via a maternal effect, where the maternally loaded factors contribute to the maintenance or establishment of imprints in the early embryo (Nakamura et al., 2007; Li et al., 2008). Because ZFP57 binds to methylated DNA at a hexanucleotide sequence motif, changes in that motif across lineages would lead to a mismatch between the maternally derived factor like ZFP57 and the target sequence motif, resulting in a scenario where imprinting cannot be established or maintained (though ZFP57 itself may be a poor candidate for such a process given that alleles appear to be functionally interchangeable between humans and mice, at least in cultured cells; Takikawa et al., 2013). Consider a case as above where the B locus controls establishment of an imprint with maternal expression at the A locus, but in this case the B locus trans-effect is via a maternal effect (so it is the B locus genotype of the mother that matters in the establishment/maintenance of the imprint). If lineages are fixed for the A1A1B1B1 and A2A2B2B2 genotypes because of a co-evolutionary process wherein the motif present at the A locus has evolved in concert with the recognition sequence at the B locus, then the two could show an incompatibility where the conflicting pairs of alleles (A1 with B2 and A2 with B1) would fail to maintain the imprint. In this case, the A1A2B1B2 hybrid would have a mother who is homozygous for the B2 allele, and so the paternally inherited A1 allele, which should be silenced, would not have its imprint maintained in the zygote and the hybrid would show biallelic expression. The reciprocal hybrid would follow this same scenario and lead to a loss of imprinting. In both cases, imprinting is lost because the paternally inherited allele is incompatible with the trans-acting maternal factor that is required to establish or maintain the imprint. This scenario does not necessarily lead to an asymmetry, but is a clear mechanism through which trans-regulation of imprinting could contribute to hybrid incompatibility. More simply, the reciprocal heterozygotes at the A locus (A1A2 and A2A1) will necessarily experience different maternal effects (B2B2 versus B1B1, respectively, in this case) and so any genetically based epistatic interaction in which the influence of the parent-of-origin of alleles at the A locus (presumably caused by imprinting) depends on the maternal genotype at the B locus could potentially create an asymmetry in the reciprocal cross.
In conclusion, although it remains unclear whether hybrid incompatibilities match the expectations under any of these simple scenarios, it is likely that the evolution of incompatibilities involving imprinted genes requires a multi-step scenario. These multi-step scenarios share the feature that the system is ‘fine tuned' through additional evolutionary steps. Such fine tuning may, for instance, occur through changes in gene expression levels (dosage) at the imprinted genes in question or at other loci that evolve changes in response to the appearance of imprinting at other loci (that is, compensatory changes that evolve to accommodate the changes in gene expression levels in the system). Fine tuning may also occur when trans-acting factors involve some matching mechanisms, such as when the trans-factor binds to a particular target sequence. The target sequences and trans-acting factors can co-evolve and diverge across lineages, leading to a scenario in which the trans-factors are unable to bind the target sequence from the other lineage. Empirical data from hybrids remain limited and to begin to fully dissect these complex multi-step scenarios, we need comparative data across variably incompatible lineages in a phylogeny that can be crossed and compared genomically to identify the order of changes and their contributions to the patterns of hybrid phenotypes.
Data archiving
There were no data to deposit.
Acknowledgments
This material is based on work supported by the National Science Foundation through the National Evolutionary Synthesis Center (NESCent) under grant number NSF #EF-0905606. RF is funded by the Agence Nationale de la Recherche and the EPIGENMED ‘Laboratory of Excellence' (Montpellier), and is affiliated to the European network of excellence EPIGENESYS and the Marie Curie training network INGENIUM. JBW acknowledges grant support from the Natural Environment Research Council and the Biotechnology and Biological Sciences Research Council. RJO is funded by the Medical Research Council (grant number G1001689) and the Wellcome Trust (grant number 084358/Z/07/Z).
The authors declare no conflict of interest.
References
- Allen WR. Factors influencing pregnant mare serum gonadotrophin production. Nature. 1969;223:64–65. doi: 10.1038/223064a0. [DOI] [PubMed] [Google Scholar]
- Berger F, Vu TM, Li J, Chen B. Hypothesis: selection of imprinted genes is driven by silencing deleterious gene activity in somatic tissues. Cold Spring Harb Symp Quant Biol. 2012;77:23–29. doi: 10.1101/sqb.2012.77.014514. [DOI] [PubMed] [Google Scholar]
- Burt A, Trivers R. The Biology of Selfish Genetic Elements. Harvard University Press: Cambridge, MA, USA; 2008. Genes in conflict. [Google Scholar]
- Cattanach BM, Williams CE. Evidence of non-random X chromosome activity in the mouse. Genet Res. 1972;19:229–240. doi: 10.1017/s001667230001449x. [DOI] [PubMed] [Google Scholar]
- Davis E, Caiment F, Tordoir X, Cavaille J, Ferguson-Smith A, Cockett N, et al. RNAi-mediated allelic trans-interaction at the imprinted Rtl1/Peg11 locus. Curr Biol. 2005;15:743–749. doi: 10.1016/j.cub.2005.02.060. [DOI] [PubMed] [Google Scholar]
- Demars J, Shmela ME, Rossignol S, Okabe J, Netchine I, Azzi S, et al. Analysis of the IGF2/H19 imprinting control region uncovers new genetic defects, including mutations of OCT-binding sequences, in patients with 11p15 fetal growth disorders. Hum Mol Genet. 2010;19:803–814. doi: 10.1093/hmg/ddp549. [DOI] [PubMed] [Google Scholar]
- Dobzhansky T. Genetics and the Origin of Species. Columbia University Press: New York, NY, USA; 1937. [Google Scholar]
- Duselis AR, Wiley CD, O'Neill MJ, Vrana PB. Genetic evidence for a maternal effect locus controlling genomic imprinting and growth. Genesis. 2005;43:155–165. doi: 10.1002/gene.20166. [DOI] [PubMed] [Google Scholar]
- Duselis AR, Vrana PB. Assessment and disease comparisons of hybrid developmental defects. Hum Mol Genet. 2007;16:808–819. doi: 10.1093/hmg/ddm025. [DOI] [PubMed] [Google Scholar]
- Duselis AR, Vrana PB. Aberrant growth and pattern formation in Peromyscus hybrid placental development. Biol Reprod. 2010;83:988–996. doi: 10.1095/biolreprod.110.085654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil R, Berger F. Convergent evolution of genomic imprinting in plants and mammals. Trends Genet. 2007;23:192–199. doi: 10.1016/j.tig.2007.02.004. [DOI] [PubMed] [Google Scholar]
- Foltz DW. Genetic evidence for long-term monogamy in a small rodent, Peromyscus polionotus. Am Nat. 1981;117:665–675. [Google Scholar]
- Gabory A, Ripoche MA, Le Digarcher A, Watrin F, Ziyyat A, Forne T, et al. H19 acts as a trans regulator of the imprinted gene network controlling growth in mice. Development. 2009;136:3413–3421. doi: 10.1242/dev.036061. [DOI] [PubMed] [Google Scholar]
- Girardot M, Cavaille J, Feil R. Small regulatory RNAs controlled by genomic imprinting and their contribution to human disease. Epigenetics. 2012;7:1341–1348. doi: 10.4161/epi.22884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha M, Pang M, Agarwal V, Chen ZJ. Interspecies regulation of microRNAs and their targets. Biochim Biophys Acta. 2008;1779:735–742. doi: 10.1016/j.bbagrm.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haig D. Parental antagonism, relatedness asymmetries, and genomic imprinting. Proc Biol Sci. 1997;264:1657–1662. doi: 10.1098/rspb.1997.0230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haig D. Genomic Imprinting and Kinship. Rutgers University Press: New Brunswick, NJ, USA; 2002. [Google Scholar]
- Haig D.2013Coadaptation and conflict, misconception and muddle, in the evolution of genomic imprinting Heredity (Edinb)(advance online publication 16 October 2013 doi: 10.1038/hdy.2013.97 [DOI] [PMC free article] [PubMed]
- Hellman A, Chess A. Extensive sequence-influenced DNA methylation polymorphism in the human genome. Epigenetics Chromatin. 2010;3:11. doi: 10.1186/1756-8935-3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemberger M, Zechner U. Genetic and genomic approaches to study placental development. Cytogenet Genome Res. 2004;105:257–269. doi: 10.1159/000078197. [DOI] [PubMed] [Google Scholar]
- Hirasawa R, Feil R. Genomic imprinting and human disease. Essays Biochem. 2010;48:187–200. doi: 10.1042/bse0480187. [DOI] [PubMed] [Google Scholar]
- Huntriss JD, Hemmings KE, Hinkins M, Rutherford AJ, Sturmey RG, Elder K, et al. Variable imprinting of the MEST gene in human preimplantation embryos. Eur J Hum Genet. 2013;21:40–47. doi: 10.1038/ejhg.2012.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida M, Monk D, Duncan AJ, Abu-Amero S, Chong J, Ring SM, et al. Maternal inheritance of a promoter variant in the imprinted PHLDA2 gene significantly increases birth weight. Am J Hum Genet. 2012;90:715–719. doi: 10.1016/j.ajhg.2012.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa R, Kinoshita T. Epigenetic programming: the challenge to species hybridization. Mol Plant. 2009;2:589–599. doi: 10.1093/mp/ssp028. [DOI] [PubMed] [Google Scholar]
- Johnson NA.2000Gene interaction and the origin of speciesIn: Wolf JB, Brodie ED III, Wade MJ (eds)Epistasis and the Evolutionary Process Oxford University Press: New York, NY, USA; 97–212. [Google Scholar]
- Johnson NA. Hybrid incompatibility genes: remnants of a genomic battlefield. Trends Genet. 2010;26:317–325. doi: 10.1016/j.tig.2010.04.005. [DOI] [PubMed] [Google Scholar]
- Josefsson C, Dilkes B, Comai L. Parent-dependent loss of gene silencing during interspecies hybridization. Curr Biol. 2006;16:1322–1328. doi: 10.1016/j.cub.2006.05.045. [DOI] [PubMed] [Google Scholar]
- Jullien PE, Berger F. Parental genome dosage imbalance deregulates imprinting in Arabidopsis. PLoS Genet. 2010;6:e1000885. doi: 10.1371/journal.pgen.1000885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kacem S, Feil R. Chromatin mechanisms in genomic imprinting. Mamm Genome. 2009;20:544–556. doi: 10.1007/s00335-009-9223-4. [DOI] [PubMed] [Google Scholar]
- Kelsey G, Feil R. New insights into establishment and maintenance of DNA methylation imprints in mammals. Philos Trans R Soc Lond B Biol Sci. 2013;368:20110336. doi: 10.1098/rstb.2011.0336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korostowski L, Sedlak N, Engel N. The Kcnq1ot1 long non-coding RNA affects chromatin conformation and expression of Kcnq1, but does not regulate its imprinting in the developing heart. PLoS Genet. 2012;8:e1002956. doi: 10.1371/journal.pgen.1002956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laschiavo M, Nguyen QK, Duelis AR, Vrana PB. Maping and identification of candidate loci responsible for Peromyscus hybrid overgrowth. Mamm Genome. 2007;18:75–85. doi: 10.1007/s00335-006-0083-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehnert S, Kapitonov V, Thilakarathne PJ, Schuit FC. Modeling the asymmetric evolution of a mouse and rat-specific microRNA gene cluster intron 10 of the Sfmbt2 gene. BMC Genomics. 2011;12:257. doi: 10.1186/1471-2164-12-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Ito M, Zhou F, Youngson N, Zuo X, Leder P, et al. A maternal-zygotic effect gene, Zfp57, maintains both maternal and paternal imprints. Dev Cell. 2008;15:547–557. doi: 10.1016/j.devcel.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay DJ, Callaway JL, Marks SM, White HE, Acerini CL, Boonen SE, et al. Hypomethylation of multiple imprinted loci in individuals with transient neonatal diabetes is associated with mutations in ZFP57. Nat Genet. 2008;40:949–951. doi: 10.1038/ng.187. [DOI] [PubMed] [Google Scholar]
- Maheshwari S, Barbash DA. The genetics of hybrid incompatibilities. Annu Rev Genet. 2011;45:331–355. doi: 10.1146/annurev-genet-110410-132514. [DOI] [PubMed] [Google Scholar]
- McCole RB, Loughran NB, Chahal M, Fernandes LP, Roberts RG, Fraternali F, et al. A case-by-case evolutionary analysis of four imprinted retrogenes. Evolution. 2011;65:1413–1427. doi: 10.1111/j.1558-5646.2010.01213.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMinn J, Wei M, Sadovsky Y, Thaker HM, Tycko B. Imprinting of PEG1/MEST isoform 2 in human placenta. Placenta. 2006;27:119–126. doi: 10.1016/j.placenta.2004.12.003. [DOI] [PubMed] [Google Scholar]
- Mills JJ, Falls JG, De Souza AT, Jirtle RL. Imprinted M6p/Igf2 receptor is mutated in rat liver tumors. Oncogene. 1998;16:2797–2802. doi: 10.1038/sj.onc.1201801. [DOI] [PubMed] [Google Scholar]
- Monk D, Arnaud P, Apostolidou S, Hills FA, Kelsey G, Stanier P, et al. Limited evolutionary conservation of imprinting in the human placenta. Proc Natl Acad Sci USA. 2006;103:6623–6628. doi: 10.1073/pnas.0511031103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore T, Haig D. Genomic imprinting in mammalian development: a parental tug-of-war. Trends Genet. 1991;7:45–49. doi: 10.1016/0168-9525(91)90230-N. [DOI] [PubMed] [Google Scholar]
- Muller HJ. Isolating mechanisms, evolution, and temperature. Biol Symp. 1942;6:71–125. [Google Scholar]
- Murrell A, Heeson S, Reik W. Interaction between differentially methylated regions partitions the imprinted genes Igf2 and H19 into parent-specific chromatin loops. Nat Genet. 2004;36:889–893. doi: 10.1038/ng1402. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Arai Y, Umehara H, Masuhara M, Kimura T, Taniguchi H, et al. PGC7/Stella protects against DNA demethylation in early embryogenesis. Nat Cell Biol. 2007;9:64–71. doi: 10.1038/ncb1519. [DOI] [PubMed] [Google Scholar]
- Ng DW, Lu J, Chen ZJ. Big roles for small RNAs in polyploidy, hybrid vigor, and hybrid incompatibility. Curr Opin Plant Biol. 2012;15:154–161. doi: 10.1016/j.pbi.2012.01.007. [DOI] [PubMed] [Google Scholar]
- Noguer-Dance M, Abu-Amero S, Al-Khtib M, Lefevre A, Coullin P, Moore GE, et al. The primate-specific microRNA gene cluster (C19MC) is imprinted in the placenta. Hum Mol Genet. 2010;19:3566–3582. doi: 10.1093/hmg/ddq272. [DOI] [PubMed] [Google Scholar]
- O'Neill RJ, O'Neill MJ, Graves JA. Undermethylation associated with retroelement activation and chromosome remodelling in an interspecific mammalian hybrid. Nature. 1998;393:68–72. doi: 10.1038/29985. [DOI] [PubMed] [Google Scholar]
- Paulsen M, Davies KR, Bowden LM, Villar AJ, Franck O, Fuermann M, et al. Syntenic organization of the mouse distal chromosome 7 imprinting cluster and the Beckwith-Wiedemann syndrome region in chromosome 11p15.5. Hum Mol Genet. 1998;7:1149–1159. doi: 10.1093/hmg/7.7.1149. [DOI] [PubMed] [Google Scholar]
- Poole RL, Leith DJ, Docherty LE, Shmela ME, Gicquel C, Splitt M, et al. Beckwith-Wiedemann syndrome caused by maternally inherited mutation of an OCT-binding motif in the IGF2/H19-imprinting control region, ICR1. Eur J Hum Genet. 2012;20:240–243. doi: 10.1038/ejhg.2011.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prickett AR, Oakey RJ. A survey of tissue-specific genomic imprinting in mammals. Mol Genet Genomics. 2012;287:621–630. doi: 10.1007/s00438-012-0708-6. [DOI] [PubMed] [Google Scholar]
- Quenneville S, Verde G, Corsinotti A, Kapopoulou A, Jakobsson J, Offner S, et al. In embryonic stem cells, ZFP57/KAP1 recognize a methylated hexanucleotide to affect chromatin and DNA methylation of imprinting control regions. Mol Cell. 2011;44:361–372. doi: 10.1016/j.molcel.2011.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson TJ, Wittekindt O, Pasantes JJ, Modi WS, Schempp W, Morris-Rosendahl DJ. Stable methylation patterns in interspecific antelope hybrids and the characterization and localization of a satellite fraction in the Alcelaphini and Hippotragini. Chromosome Res. 2000;8:635–643. doi: 10.1023/a:1009294226213. [DOI] [PubMed] [Google Scholar]
- Rogers JF, Dawson WD. Foetal and placental size in a Peromyscus species cross. J Reprod Fertil. 1970;21:255–262. doi: 10.1530/jrf.0.0210255. [DOI] [PubMed] [Google Scholar]
- Salas M, John R, Saxena A, Barton S, Frank D, Fitzpatrick G, et al. Placental growth retardation due to loss of imprinting of Phlda2. Mech Dev. 2004;121:1199–1210. doi: 10.1016/j.mod.2004.05.017. [DOI] [PubMed] [Google Scholar]
- Schulz R, Proudhon C, Bestor TH, Woodfine K, Lin CS, Lin SP, et al. The parental non-equivalence of imprinting control regions during mammalian development and evolution. PLoS Genet. 2010;6:e1001214. doi: 10.1371/journal.pgen.1001214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schutt S, Florl AR, Shi W, Hemberger M, Orth A, Otto S, et al. DNA methylation in placentas of interspecies mouse hybrids. Genetics. 2003;165:223–228. doi: 10.1093/genetics/165.1.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi W, Krella A, Orth A, Yu Y, Fundele R. Widespread disruption of genomic imprinting in adult interspecies mouse (Mus) hybrids. Genesis. 2005;43:100–108. doi: 10.1002/gene.20161. [DOI] [PubMed] [Google Scholar]
- Shi W, Lefebvre L, Yu Y, Otto S, Krella A, Orth A, et al. Loss-of-imprinting of Peg1 in mouse interspecies hybrids is correlated with altered growth. Genesis. 2004;39:65–72. doi: 10.1002/gene.20027. [DOI] [PubMed] [Google Scholar]
- Takagi N, Sasaki M. Preferential inactivation of the paternally derived X chromosome in the extraembryonic membranes of the mouse. Nature. 1975;256:640–642. doi: 10.1038/256640a0. [DOI] [PubMed] [Google Scholar]
- Takikawa S, Wang X, Ray C, Vakulenko M, Bell FT, Li X. Human and mouse ZFP57 proteins are functionally interchangeable in maintaining genomic imprinting at multiple imprinted regions in mouse ES cells. Epigenetics. 2013;8:1268–1279. doi: 10.4161/epi.26544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turelli M, Orr HA. Dominance, epistasis and the genetics of postzygotic isolation. Genetics. 2000;154:1663–1679. doi: 10.1093/genetics/154.4.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turelli M, Moyle LC. Asymmetric postmating isolation: Darwin's corollary to Haldane's rule. Genetics. 2007;176:1059–1088. doi: 10.1534/genetics.106.065979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varrault A, Gueydan C, Delalbre A, Bellmann A, Houssami S, Aknin C, et al. Zac1 regulates an imprinted gene network critically involved in the control of embryonic growth. Dev Cell. 2006;11:711–722. doi: 10.1016/j.devcel.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Vrana PB, Guan XJ, Ingram RS, Tilghman SM. Genomic imprinting is disrupted in interspecific Peromyscus hybrids. Nat Genet. 1998;20:362–365. doi: 10.1038/3833. [DOI] [PubMed] [Google Scholar]
- Vrana PB, Fossella JA, Matteson P, del Rio T, O'Neill MJ, Tilghman SM. Genetic and epigenetic incompatibilities underlie hybrid dysgenesis in Peromyscus. Nat Genet. 2000;25:120–124. doi: 10.1038/75518. [DOI] [PubMed] [Google Scholar]
- Vrana PB. Genomic imprinting as a mechanism of reproductive isolation in mammals. J Mammal. 2007;88:5–23. [Google Scholar]
- Vu TH, Jirtle RL, Hoffman AR. Cross-species clues of an epigenetic imprinting regulatory code for the IGF2R gene. Cytogenet Genome Res. 2006;113:202–208. doi: 10.1159/000090833. [DOI] [PubMed] [Google Scholar]
- Walia H, Josefsson C, Dilkes B, Kirkbride R, Harada J, Comai L. Dosage-dependent deregulation of an AGAMOUS-LIKE gene cluster contributes to interspecific incompatibility. Curr Biol. 2009;19:1128–1132. doi: 10.1016/j.cub.2009.05.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Chow J, Hong J, Smith AF, Moreno C, Seaby P, et al. Recent acquisition of imprinting at the rodent Sfmbt2 locus correlates with insertion of a large block of miRNAs. BMC Genomics. 2011;12:204. doi: 10.1186/1471-2164-12-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Miller DC, Harman R, Antczak DF, Clark AG. Paternally expressed genes predominate in the placenta. Proc Natl Acad Sci USA. 2013;110:10705–10710. doi: 10.1073/pnas.1308998110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiley CD, Matundan HH, Duselis AR, Isaacs AT, Vrana PB. Patterns of hybrid loss of imprinting reveal tissue- and cluster-specific regulation. PLoS One. 2008;3:e3572. doi: 10.1371/journal.pone.0003572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson LS, Davies W, Isles AR. Genomic imprinting effects on brain development and function. Nat Rev Neurosci. 2007;8:832–843. doi: 10.1038/nrn2235. [DOI] [PubMed] [Google Scholar]
- Wolf JB. Gene interactions from maternal effects. Evolution. 2000;54:1882–1898. doi: 10.1111/j.0014-3820.2000.tb01235.x. [DOI] [PubMed] [Google Scholar]
- Wolf JB. Evolution of genomic imprinting as a coordinator of coadapted gene expression. Proc Natl Acad Sci USA. 2013;110:5085–5090. doi: 10.1073/pnas.1205686110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf JB, Brodie ED., III The coadaptation of parental and offspring characters. Evolution. 1998;52:535–544. doi: 10.1111/j.1558-5646.1998.tb01632.x. [DOI] [PubMed] [Google Scholar]
- Wolf JB, Hager R. A maternal-offspring coadaptation theory for the evolution of genomic imprinting. PLoS Biol. 2006;4:e380. doi: 10.1371/journal.pbio.0040380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood AJ, Schulz R, Woodfine K, Koltowska K, Beechey CV, Peters J, et al. Regulation of alternative polyadenylation by genomic imprinting. Genes Dev. 2008;22:1141–1146. doi: 10.1101/gad.473408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CI, Ting CT. Genes and speciation. Nat Rev Genet. 2004;5:114–122. doi: 10.1038/nrg1269. [DOI] [PubMed] [Google Scholar]
- Yotova IY, Vlatkovic IM, Pauler FM, Warczok KE, Ambros PF, Oshimura M, et al. Identification of the human homolog of the imprinted mouse Air non-coding RNA. Genomics. 2008;92:464–473. doi: 10.1016/j.ygeno.2008.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zechner U, Shi W, Hemberger M, Himmelbauer H, Otto S, Orth A, et al. Divergent genetic and epigenetic post-zygotic isolation mechanisms in Mus and Peromyscus. J Evol Biol. 2004;17:453–460. doi: 10.1046/j.1420-9101.2003.00656.x. [DOI] [PubMed] [Google Scholar]
- Zhao Z, Tavoosidana G, Sjolinder M, Gondor A, Mariano P, Wang S, et al. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat Genet. 2006;38:1341–1347. doi: 10.1038/ng1891. [DOI] [PubMed] [Google Scholar]