Abstract
The epigenetic phenomenon of genomic imprinting has motivated the development of numerous theories for its evolutionary origins and genomic distribution. In this review, we examine the three theories that have best withstood theoretical and empirical scrutiny. These are: Haig and colleagues' kinship theory; Day and Bonduriansky's sexual antagonism theory; and Wolf and Hager's maternal–offspring coadaptation theory. These theories have fundamentally different perspectives on the adaptive significance of imprinting. The kinship theory views imprinting as a mechanism to change gene dosage, with imprinting evolving because of the differential effect that gene dosage has on the fitness of matrilineal and patrilineal relatives. The sexual antagonism and maternal–offspring coadaptation theories view genomic imprinting as a mechanism to modify the resemblance of an individual to its two parents, with imprinting evolving to increase the probability of expressing the fitter of the two alleles at a locus. In an effort to stimulate further empirical work on the topic, we carefully detail the logic and assumptions of all three theories, clarify the specific predictions of each and suggest tests to discriminate between these alternative theories for why particular genes are imprinted.
The discovery of genomic imprinting, where the expression of an allele depends on its parental origin, motivated a diversity of theories attempting to explain its existence (Spencer and Clark, 2014). Three main theories have withstood scrutiny and are the focus of this review: Haig and colleagues' kinship theory (Haig and Westoby, 1989; Haig, 2000a, 2004); Day and Bonduriansky's (2004) sexual antagonism theory (see also Bonduriansky, 2007); and Wolf and Hager's (2006) maternal–offspring coadaptation theory (see also Wolf and Hager, 2009; Wolf, 2013). Although these theories rest on different logic and fundamental assumptions, they share a critical common feature: some process creates a selective asymmetry between the maternally and paternally inherited allelic copies at a locus that causes selection to favor differential expression of the alleles (typically silencing of one of the copies) (Figures 1, 2, 3).
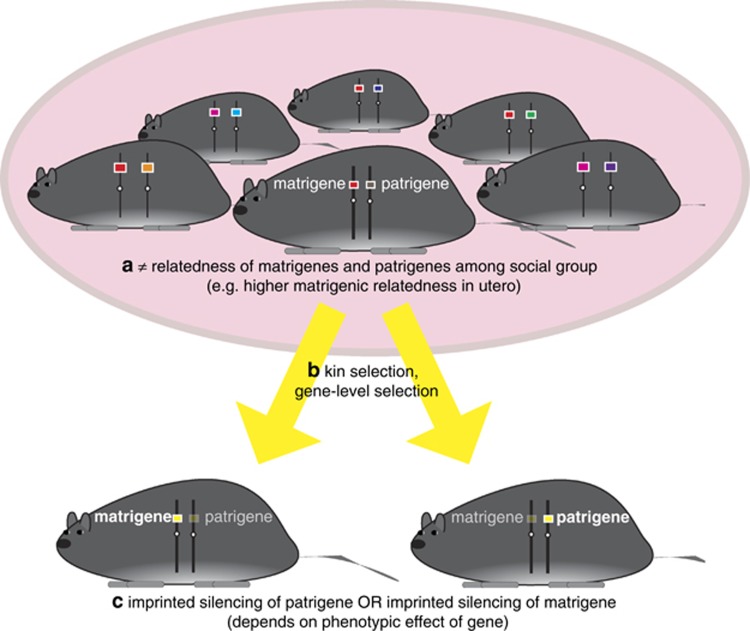
Figure 1.
The kinship theory of genomic imprinting has two prerequisites: first, epigenetic marks that differentiate matrigenes from patrigenes; second, a difference in the relatedness of matrigenes and patrigenes to the social group. (a) The social group in the example depicted is a single litter of offspring, and multiple mating produces a relatedness asymmetry between half-siblings. The relatedness for matrigenes is ½ and the relatedness for patrigenes is 0. (Other sources of relatedness asymmetry are possible—e.g., sex-biased dispersal or high fitness variance in one sex—and social interactions are not limited to the juvenile period only). (b) The kinship theory envisions kin selection acting independently on genes of maternal and paternal origin and solves for the evolutionarily stable gene expression strategy for matrigenes and patrigenes. (c) For genes where the matrigenic allele's optimum expression level is higher than that of the patrigene's (e.g., a fetal growth inhibitor), the kinship theory predicts silencing of the patrigenic allele; for genes with the opposite effect (e.g., a fetal growth enhancer), the prediction is for patrigenic expression.
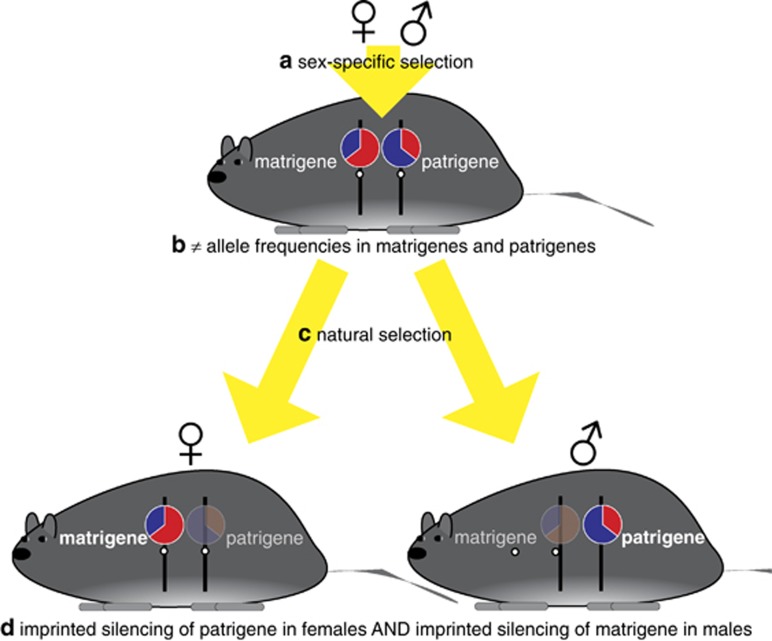
Figure 2.
(a, b) The sexual antagonism theory of genomic imprinting starts with sexually antagonistic selection, which produces different allele frequencies, shown as pie charts, for genes of maternal and paternal origin. (c, d) Natural selection favors individuals that are able to express the fitter of the two alleles at a locus, which for males will be the patrigenic allele and for females will be the matrigenic allele. (In addition, the sexual antagonism theory may predict matrigenic or patrigenic expression in both sexes, such that the expressed allele derives from the parental sex that experiences stronger selection pressure. This scenario is not depicted).
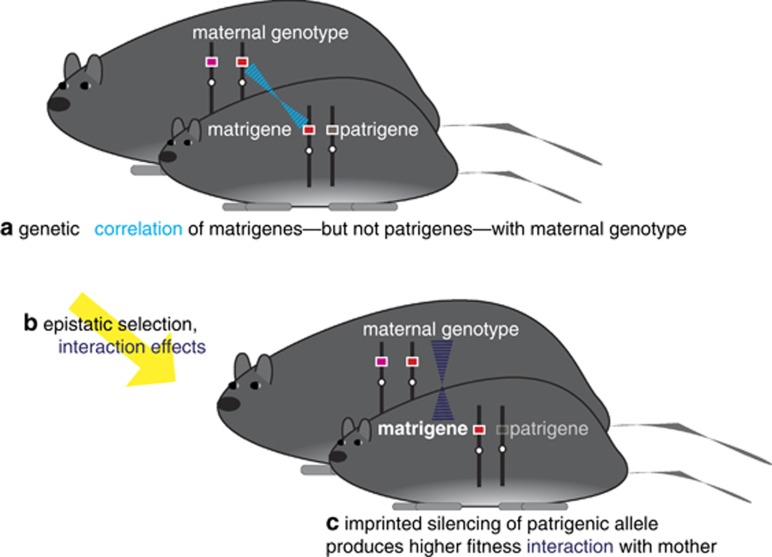
Figure 3.
(a) The maternal–offspring coadaptation theory of genomic imprinting relies on the correlation of genes in the mother and genes of maternal origin in the offspring (shown in light blue). (b) Fitness of offspring is determined by the interaction (shown in dark purple) between the phenotypes of mothers and offspring. (c) Imprinted silencing of the patrigenic allele can be favored for either of two reasons, depending on the genetic architecture of the interacting phenotypes. First, when a single gene governs the interaction and phenotypic matching between mothers and their offspring produces high fitness, then silencing of the patrigenic allele is beneficial to offspring because it raises the probability of producing a match. Second, if different loci are involved in the phenotypic interaction, past correlational selection will have produced a covariance between them, generating haplotypes with combinations of alleles that interact well together. (N.B. This multi-locus interaction is not depicted in the figure.) The offspring is more likely to inherit from its mother an allele that interacts well with the alleles in the mother's genotype. This also favors the imprinted silencing of the patrigenic allele because it raises the probability that the offspring expresses an allele that makes for a good interaction with the maternal phenotype.
Here we provide an overview of the fundamental logic and critical assumptions of these models. We then derive predictions that can be used to distinguish between theories. In doing so, we also highlight ambiguities in and overlap between the predictions they make, with a goal of motivating further research. In addition, we suggest some areas for future work that will test some of these predictions.
Theories for the evolution of genomic imprinting
A brief note on terminology: throughout this section we refer to ‘matrigenic' and ‘patrigenic' allelic copies to describe the alleles of a haploid genotype that derive from a diploid mother or father, respectively (Queller, 2003). This additional vocabulary helps to clarify where the selective forces are acting. One should take note of the important distinction between genes deriving from parents and genes belonging to parents. Because of their ploidies and because of their expected relatedness to other individuals, ‘mother's genes' and ‘matrigenes' do not refer to the same things, nor do ‘paternal' and ‘patrigenic' (Úbeda and Haig, 2003; Wolf and Wade, 2009). ‘Matrigenic' and ‘patrigenic' are equivalent to Haig's (1996) less euphonious ‘madumnal' and ‘padumnal'.
The kinship theory
The kinship theory is a kin selection model, where matrigenic and patrigenic alleles experience different patterns of relatedness in the social environment (for example, individuals tend to encounter more matrilineal than patrilineal relatives), and as a result, their expression has different consequences for their respective inclusive fitnesses (Haig, 2002, pp 5–6; Figure 1). The kinship theory focuses on genes whose expression level governs the extent of some physiological or behavioral interaction between individuals. As an example, fetal expression of a growth factor not only influences the fetus' development but may also indirectly affect the growth, and potentially the fitness, of siblings through its demand for shared maternal resources. Variation in the fetal expression level of this growth factor gene can have different, and opposing, inclusive fitness effects for the two alleles if these have unequal relatedness to individuals with whom the fetus interacts (for example, because the fetuses are maternal half-sibs, which share matrigenic but not patrigenic alleles). Because of this relatedness asymmetry, increased expression might increase the inclusive fitness of the patrigenic allele but decrease the inclusive fitness of the matrigenic allele. Haig (1997) termed this selection pressure parental antagonism, drawing the analogy with sexual antagonism, in which a trait is favored in males but disfavored in females (and vice versa).
In the face of parental antagonism, an allele that adjusts its expression level in cis to bring about that parental origin's optimum is selectively favored over an alternative allele that does not adjust its expression (Haig and Westoby, 1989). Assuming parental origin-specific expression is possible, the kinship theory then seeks an evolutionarily stable strategy (ESS) for gene expression from the two parental origins (Haig, 1997). This ESS corresponds to an expression level where only the copy from the parental origin that originally favored a higher level of expression is expressed, with the copy from the parental origin that originally favored reduced expression being completely silenced—that is, the canonical pattern of genomic imprinting. Further, the expressing parental origin expresses at that parental origin's optimal level. At the ESS in this example, the growth factor gene will be expressed exclusively from the patrigenic allele at the patrigene's optimal level. This sketch of the logic of the kinship theory covers both the strong and the weak versions of the theory (sensu Wilkins and Haig, 2003). These versions correspond to the familiar distinction in evolutionary biology between origin and maintenance: the strong version of the kinship theory claims that the origin of imprinted expression is a consequence of parental antagonism; the weak version submits that once imprinted expression has already been established at a locus, an imprinted gene evolves subject only to the inclusive fitness consequences for the matrigene or patrigene, whichever copy remains expressed.
Although Haig and Westoby (1989) originally focused on multiple mating as the source of relatedness asymmetry for matrigenes and patrigenes, later work has extended the kinship theory to cover other sources, including haplodiploidy (Haig, 1992; Queller, 2003) and sex-biased dispersal or sex-biased fitness variance (Haig, 2000b; Úbeda and Gardner, 2010, 2011, 2012; Van Cleve et al., 2010; Brandvain et al., 2011). Further, the kinship theory can be applied to any behavior that influences the fitness of kin and is not limited to the solicitation behaviors of offspring. For example, the provisioning of shared or potentially shared resources by adults is another social behavior that has parentally antagonistic inclusive fitness effects when the two alleles of the provider are asymmetrically related to the recipients of the resource (Úbeda and Gardner, 2011, 2012).
The sexual antagonism theory
The sexual antagonism theory for the evolution of genomic imprinting relies on sex-specific selection pressure (Day and Bonduriansky, 2004; Bonduriansky, 2007; Figure 2). When selection differs between males and females—be it in magnitude or in direction—the allele frequencies in eggs and sperm (and, therefore, in matrigenes and patrigenes of the next generation) become unequal. Two scenarios can generate this non-equivalence. First, if selection is sexually antagonistic for a gene, patrigenes will be enriched for alleles of male benefit and matrigenes will be enriched for alleles of female benefit. There is thus a selective advantage to a novel modifier that adjusts expression level in an imprinted and sex-specific way—that is, patrigenic expression of the gene when in males and matrigenic expression when in females. Second, there is a net selective advantage to modifying expression in the standard imprinted fashion—that is, independently of the sex of bearer—provided that the allele deriving from the parent that experienced stronger selection in the previous generation is expressed.
Analogous logic underlies Spencer and Clark's (2006) ‘Chip Off the Old Block' theory for the origin of imprinting, where selective asymmetry arises because the haploid genome inherited from the parent of the less dispersive sex will tend to be enriched for locally adapted alleles (see also Spencer and Clark, 2014). Spencer and Clark's (2006) model therefore predicts the imprinted silencing of alleles inherited from the more dispersive parental sex for loci with key functional roles in the exploitation of the local ecological niche.
In the sexual antagonism theory, selection favors imprinted expression because the two alleles at a diploid locus carry, on average, different information or instructions, and one of these copies provides more adaptive information than the other. Population variation for such alleles, and the absence of other epigenetic mechanisms that temper their sex-specific effects on fitness, results in intralocus sexual conflict, whereby a sex is prevented from reaching its phenotypic optimum as a result of a different pattern of selection on the same locus in the other sex (Bonduriansky and Chenoweth, 2009). Whenever there is differential selection between the sexes, the parent of the same sex as the offspring will, on average, provide fitter alleles to that offspring compared with the other parent. Imprinted silencing, therefore, provides a simple mechanism through which individuals can express the more adaptive set of instructions for their sex. For the sexual antagonism theory, imprinting mitigates intralocus sexual conflict by allowing each sex to approach more closely its sex-specific phenotypic optimum.
The requirement for the evolution of imprinted expression under this theory is simple: sex-specific selection pressure on a gene. Any species with two sexes is subject to imprinting of genes under sex-specific selection. The requirements of the sexual antagonism theory are less restrictive than any of the other theories' requirements, and, as such, the sexual antagonism theory is expected to apply more broadly.
The maternal–offspring coadaptation theory
The fitness of a genotype depends not only on its direct phenotypic effect on its bearer but also on the interactions between the phenotypes of different individuals (Moore et al., 1997; Wolf et al., 1999a; McGlothlin et al., 2010). Certain of these interactions are more (or less) than the sum of their parts, producing a form of epistasis (Wolf, 1999). Whenever interactions between traits affect fitness, genetic correlations between the interacting traits are expected to evolve (Sinervo and Svensson, 2002), even when the phenotypes belong to different individuals, as in the case of mothers and offspring (Wolf and Brodie, 1998).
Wolf and Hager's (2006, 2009) maternal–offspring coadaptation theory supposes that the two alleles in an offspring code for non-equivalent fitness interactions with the mother, favoring imprinted expression (Figure 3). They examine two scenarios, one where the same locus pleiotropically influences both maternal and offspring traits (the single-locus model) and a second where different loci affect the maternal and offspring traits (the two-locus model). If the interaction confers higher fitness when there is a greater genetic covariance (which, for the single-locus case, is equivalent to phenotypic ‘matching') between the two actors, then selection favors expression of the matrigenic allele and silencing of the patrigenic allele in the offspring. In the single-locus model, the two alleles in the offspring at a diploid locus are non-equivalent for fitness because an offspring's maternally derived allele is necessarily more likely to be present in the mother than its paternally derived allele, and expression of the maternally derived allele is more likely to produce a phenotypic ‘match' between mother and offspring. In the two-locus model, past correlational selection on the maternal–offspring interaction results in a scenario where the matrigenic portion of the offspring's genotype is positively genetically correlated with any maternal effects with which the offspring's phenotype must interact; that is, past selection will have built linkage disequilibrium between the two loci. The interaction is therefore more likely to be a high fitness one when expression is limited to the matrigenic allele, which will have evolved some degree of coadaptation with the alleles present in the mother. If instead the highest fitness interaction is achieved by a more negative genetic covariance (or heightened mismatch) between mother and offspring, then the prediction is for the matrigenic allele to be silenced and the patrigenic allele to be expressed.
Predictions of the theories
Direction of expression and silencing
Each theory makes a strong prediction about the direction of imprinted expression and silencing—that is, which of the two alleles at a diploid locus is expressed and which is silenced (Table 1). Recent advances in next generation sequencing technology mean that genome-wide estimates of both the numbers of imprinted loci as well as the direction of expression in these will soon be available across a wide taxonomic range (for example, Gehring et al., 2011; Wang et al., 2011; Zhang et al., 2011; Ikeda, 2012; Wang and Clark, 2014).
Table 1. Predictions on imprinting of the kinship, coadaptation and sexual antagonism theories.
| Taxon | Context/tissue | Kinship | Coadaptation | Sexual antagonism |
|---|---|---|---|---|
| Mammals, plants, angiosperms, other placental viviparous animals (e.g., some reptiles and fish) | Resource acquisition from mother (placenta, endosperm) | +Patrigene −matrigene | ±Matrigene | a |
| Birds, fish, monotremes, gymnosperms | Embryo, resource acquisition in egg | None | ±Matrigene | None |
| Birds, fish | Resource acquisition from parent, post hatching | +Patrigene −matrigene | ±Matrigene | a |
| Sex-role-reversed birds or fish | Resource acquisition from father | −Patrigene +matrigene | ±Patrigene | a |
| Social insect workers | Queen–worker interactions; nervous tissue, sensory, communication | Variableb | Variableb | None |
| Moss | Sporophyte resource acquisition from gametophyte | +Patrigene −matrigene | ±Matrigene | None |
| Insects | Sibling competition—mixed paternity broods | +Patrigene −matrigene | None in most species | a |
| Many birds | Post-dispersal competition, females disperse | −Patrigene +matrigene | None | None |
| Many mammals | Post-dispersal competition, males disperse | +Patrigene −matrigene | None | +Patrigene −matrigene |
| Any sexually reproducing species | Male-function traits (e.g., secondary sexual traits, including high growth ratea) | None | None | +Patrigene −matrigene |
| Any sexually reproducing species | Female-function traits (e.g., metabolic pathways and behaviors involved in offspring production, crypsis adaptations) | None | None | −Patrigene +matrigene |
| Hermaphrodite plants, fish | Male traits | None | None | +Patrigene −matrigene |
| Hermaphrodite plants, fish | Female traits | None | None | −Patrigene +matrigene |
| Species with female multiple mating | Cooperative breeding and resource sharing between maternal siblings | −Patrigene +matrigene | None | None |
| Species with female defense or harem polygyny | Cooperative breeding and resource sharing between paternal siblings | +Patrigene −matrigene | None | None |
| Species with male-biased juvenile dispersal | Cooperative breeding and resource sharing between neighbors | −Patrigene +matrigene | None | None |
| Species with male-biased reproductive skew | Cooperative breeding and resource sharing between neighbors | +Patrigene −matrigene | None | None |
The + and − indicate whether a gene is expected to increase or decrease the trait listed in the context/tissue column. ‘None' indicates that no imprinting is generally expected in the absence of pleiotropy (side effects of imprinting for another context). (Brandvain et al. (2011) present a table with similar predictions).
Note that if sexual selection favors large body size in males (as is the case in many mammals, and some other taxa), then male embryos and neonates may be selected to extract maternal resources at a greater rate than their sisters. In this case, the sexual antagonism model has somewhat overlapping predictions with the kinship theory. For example, if sexually dimorphic imprinting is not possible, then the sexual antagonism theory would predict paternal expression and maternal silencing of genes that regulate growth rate.
Many different predictions for the kinship theory (Queller, 2003). For the maternal–offspring coadaptation theory, the prediction is ‘± matrigene', but an analogous parallel worker–worker coadaptation theory would be ‘±patrigene' for singly mated haplodiploid species where workers are more related through patrigenes and ‘±matrigene' for multiply mated.
Generally speaking, the kinship theory predicts that genes that promote growth will tend to be paternally expressed and those that inhibit growth will be maternally expressed (Moore and Haig, 1991; Brandvain et al., 2011). More precisely, the theory predicts that the allelic copy (or the parental origin context) that gains greater inclusive fitness from higher relative expression of the gene is the copy that should be expressed. In the case of growth-promoting genes, if the gain by one offspring comes at the expense of siblings, the prediction is for the allele with lower relatedness, which is typically the patrigenic allele, to be expressed. As a corollary, the kinship theory predicts that two different imprinted genes whose phenotypic effects oppose one another should show reciprocal expression patterns (Wilkins and Haig, 2001). Igf2 and Igf2r are the classic examples that meet both of these strong predictions of the kinship theory (Haig and Graham, 1991); other imprinted genes have also upheld the predictions of the kinship theory (Haig, 2004).
The sexual antagonism theory makes a strong prediction about the direction of imprinted expression. For all imprinted genes that influence a single phenotype (that is, a single target of selection), regardless of how these genes influence the trait, the parental origin of expression should be the same within a sex. Genes with segregating polymorphism have alleles with varying effects on male and female fitness, and, in the sexual antagonism theory, imprinting exists to increase the probability that the fitter allele is expressed in each sex (or in the sex under stronger selection). From an offspring's perspective, the fittest alleles derive from the same-sex parent. Thus, in contrast to the kinship theory, reciprocally imprinted genes with opposing phenotypic effects within a sex, such as Igf2 and Igf2r, are not predicted under the sexual antagonism theory. When selection is stronger on one sex than the other, the sexual antagonism theory predicts that more imprinted genes will be expressed from the parental sex that experiences stronger selection.
The coadaptation theory similarly predicts that all genes involved in a single social interaction are expressed from the same parental origin. For maternal–offspring interactions, the choice to express the matrigenic or patrigenic allele is determined by whether a positive or a negative genetic covariance between the traits in mothers and their offspring produces a higher fitness interaction for the offspring. Initially, the coadaptation theory purported to explain the enrichment of maternally expressed imprinted genes in placenta and early seeds, the prime sites of maternal–offspring interaction (Wolf and Hager, 2006). But this may have been too coarse a prediction. Neither the placenta nor the seed is a single tissue, nor does either represent a single interaction in the sense that the model uses. The observation of both kinds of imprinted genes in placenta (Proudhon and Bourc'his, 2010; Wang et al., 2011) and a more complicated picture of parental origin-specific expression in seeds (Autran et al., 2011; McKeown et al., 2011) do not argue against the coadaptation theory.
Specificity of imprinting
We assume below that the requisite machinery for establishing and maintaining imprints is available to each tissue and in each taxon (for the progress on which, see: Yi and Goodisman, 2009; Glastad et al., 2011; Drewell et al., 2012). In other words, our predictions stem from the evolutionary pressures—not mechanistic constraints—faced by organisms.
Tissue specificity
The kinship and coadaptation theories both derive fitness variation from social interactions (Figures 1 and 3). Not surprisingly, both theories predict imprinted expression in the organs of social interaction (for example, fetal brain, endosperm, placenta), all of which mediate the transfer of resources between generations (Table 1). However, there is no specific prediction of imprinted expression in mothers under the coadaptation theory, for this does not alter the genetic covariance between them and their offspring. Any attempt to explain imprinted gene expression in mothers by an appeal to the coadaptation theory (for example Stringer et al., 2012) therefore misses the mark. For the coadaptation theory, imprinted expression in adults might be a simple matter of inertia: if imprinted expression is adaptive in early life and erasure of the imprint is costly, then adults will bear the same pattern of imprinted expression as young. In contrast, the kinship theory is compatible with imprinted expression in adult tissues involved in social interactions (for example, brains and mammary glands; Brandvain et al., 2011). However, it should be noted that asymmetries of interests between matrigenes and patrigenes are dependent on several species-specific demographic factors, and that the regulation of social behaviors is complex, making clear-cut predictions on the direction of imprinting in adult brains especially challenging (Curley, 2011). Where a gene's expression cannot influence the fitness of kin, the kinship theory does not predict the evolution of imprinted expression.
Under the sexual antagonism theory, genomic imprinting is most likely to evolve in phenotypes under the strongest sexually antagonistic or sex-specific selection, such as primary or secondary sexual traits for which the optimal degree of sexual dimorphism has not evolved (Table 1). Because the rate of evolutionary turnover of secondary sexual traits in particular is expected to be high (Holland and Rice, 1998), some loci affecting their expression are likely to experience sex-specific selection. Empirical studies suggest that sex-specific selection and intralocus sexual conflict are indeed widespread (Bonduriansky and Chenoweth, 2009; Connallon et al., 2010), and identifying the phenotypic and genomic targets of the strongest sexually antagonistic selection will make it possible to derive strong and specific predictions for the phenotypes and loci where imprinting should occur, as well as the pattern of imprinting. A strong prediction of the sexual antagonism theory is that imprinted genes with expression that is confined to sex-limited tissues would not express the opposite sex-of-parent allele. Matrigenic expression of an imprinted testis-specific gene or patrigenic expression of a mammary gland-specific imprinted gene, for example, cannot be explained by this theory, except by invoking pleiotropic effects of the gene in other tissues.
Taxon specificity
Two factors affect the predicted taxonomic distribution of imprinted genes under the kinship theory: (1) the relatedness asymmetry for the two alleles at a diploid locus in interacting kin; and (2) the potential of such kin interactions to affect fitness. Sex-biased dispersal, which also favors imprinting under Spencer and Clark's (2006) ‘Chip Off the Old Block' theory, and multiple mating are the two most obvious ways by which such relatedness asymmetries are generated (Brandvain et al., 2011). Asymmetrically related kin are likely to influence one another's fitness during the period of parental care or communal care of young. A weak prediction is therefore that genomic imprinting will be found in organisms like social insects, seed plants, birds and mammals (Table 1), which exhibit such social interactions and relatedness asymmetries, thus predisposing them to parental antagonism. A strong prediction of the kinship theory is that imprinted expression is unlikely to originate in taxa that lack such social interactions and relatedness asymmetries, for example, plants that are obligate selfers and oviparous animals with limited social interactions.
The sexual antagonism theory applies to a wide range of taxa—indeed, any species subject to sex-specific selection. In gonochoristic animals, species showing pronounced sexual dimorphism suggest a history of sexual antagonism, and so might be more likely to harbor genomic imprinting under the sexual antagonism theory. Taxa subject to strong sexual selection (such as species characterized by a strongly male-biased operational sex ratio, or the opportunity for a male to monopolize access to multiple females) should in general experience the strongest sex-specific selection, making them a likely place to find imprinting (Table 1). Female choice for good genes in males is predicted to lead to the evolution of imprinting (with patrigenic expression) because of the different strength of selection in the two sexes.
The coadaptation theory requires fitness interactions between mothers and offspring. These interactions are predicted to be strongest in species with prolonged maternal care, or at least where maternal traits have important fitness consequences for offspring and have the opportunity to interact with offspring traits. Taxa such as seed plants, birds and mammals, which have an extended period of physical interaction between diploid generations, would be the most likely organisms in which to find imprinting under the coadaptation theory (Table 1). In this way, the coadaptation and kinship theories overlap in their predictions. They differ, however, in that the coadaptation theory can also explain imprinting in organisms that lack an extended period of interaction. For example, in oviparous species without parental care the egg may serve as a reservoir of maternal effects, which, by definition, are maternal phenotypes—not matrigenic phenotypes (Wolf and Wade, 2009). Coadaptation is possible between offspring phenotypes and these egg-derived maternal effects. Likewise, in many systems, oviposition decisions by mothers can interact with offspring traits (for example, adaptation to the environment where they are deposited), generating correlational selection on maternal–offspring combinations (that is, coadaptation between oviposition choice and offspring performance).
There is at present much interest in whether birds and social insects have imprinted genes. Setting aside the issue of mechanistic limitations, both the coadaptation theory and the kinship theory could explain the existence of imprinted genes in these taxa as a consequence of the special social interactions among relatives. The sexual antagonism theory could also make sense of imprinting in these taxa but only in those species with strong sex-specific or sexual selection (Table 1).
Dosage sensitivity
Under both the sexual antagonism and coadaptation theories, selection for imprinted expression relies on there being segregating variation at the imprinted locus. In contrast, in the kinship theory, imprinted expression functions to allow dosage modification rather than an ontogenetic choice between two alleles with alternative phenotypic effects. The kinship theory would predict the evolution of imprinted expression only in cases where the gene's expression level affected inclusive fitness in the ancestor; that is, an imprinted gene would have to be dosage sensitive at the origin of imprinted expression and might therefore exhibit dosage sensitivity in the present. Following this logic, we would also predict that closely related taxa that have not evolved imprinted expression should show dosage sensitivity for genes that have evolved imprinted expression in their sister taxon. Unfortunately, both of these follow-on predictions come with the caveat that dosage sensitivity of a locus in the present does not necessarily mean dosage sensitivity in its past; we do not know how dosage sensitivity evolves at a locus after it acquires imprinted expression.
Loss of imprinting
Each theory posits a unique selection pressure to drive the evolution of imprinted expression. Removing that selection pressure might, in principle, spur the loss of imprinted expression at a locus, although this possibility hasn't been explored in great theoretical detail (but see Wilkins and Haig, 2002). One problem surrounding any discussion of the loss of imprinting is common to all theories: we don't know how the acquisition of imprinted expression affects the subsequent evolution of imprinted genes. This subsequent evolution might influence the likelihood of reversion to biallelic expression. For example, if a locus has evolved some form of dosage compensation, such that the current monoallelic expression is close in its level to the ancestral biallelic expression, then reversion to biallelic expression could be constrained. Keeping this major caveat always in mind, we can still attempt some predictions about the patterns of loss of imprinting under each theory.
According to the kinship theory, the total expression level of an imprinted locus is higher than that of the ancestral, biallelically expressed locus (Haig, 1997). At the ESS in the kinship theory, all of a gene's expression comes from the single active allele, which evolves to express at its parental origin's optimum. This level is higher than the other parental origin's optimum and higher than the ancestral, biallelic level, which presumably would have fallen somewhere between the two parental origin optima. In addition, for reciprocally imprinted genes with opposing phenotypic effects, the kinship theory predicts that expression levels will escalate (Wilkins and Haig, 2001; Frank and Crespi, 2011; Wilkins, 2011). Should parental antagonism cease to exist at a locus, for instance owing to a change in mating system, then the selection pressure in favor of imprinted expression evaporates. However, the sudden loss of imprinted silencing might actually decrease fitness, as biallelic expression would raise the expression level above that of its former imprinted state, which is favorable to neither the matrigene nor the patrigene. Removing imprinting would disrupt normal development, which requires a balance in the dosage of interacting genes (Frank and Crespi, 2011; Wilkins, 2011). Under the kinship theory, the loss of imprinting is therefore unlikely. If anything, selection maintains imprinted expression even after a switch in demography or mating system that removes relatedness asymmetries. An alternative adaptive outcome is for an imprinted gene to become monoallelically expressed in a random fashion (Haig, 2006). This restores some measure of diploidy to the organism at a gross level and avoids the problem of raising the total expression level, but it requires that there is a mechanistic route from imprinted expression to random monoallelic expression. The mechanistic and evolutionary connections between imprinted expression and monoallelic expression have not been explored.
For the other two theories, the loss of imprinting does not entail the same cost, for neither sees imprinting as a way to modify the total expression level from a locus. Under the sexual antagonism theory, an individual is essentially choosing one allele over another because the chosen allele is expected to increase the fitness of its bearer. If sexual antagonism is resolved through the evolution of optimal sexual dimorphism, or if the strength of selection in the two sexes is balanced, there is no longer an expectation that one allele at a locus will produce higher fitness outcomes than the other. Under the sexual antagonism theory, the imprinting status of a gene is predicted to be labile over evolutionary time and, given a clade of organisms that has experienced changes to its mating system, one should predict gains and losses of imprinting.
Loss of imprinting under the coadaptation theory has not been formally explored. The selection pressure that promotes the origin of imprinted expression stems from allelic variation at the locus that produces fitness variance in the interactions between mothers and offspring. Imprinted expression produces more of the high-fitness interactions. Like the sexual antagonism theory, the coadaptation theory employs a standard assumption of quantitative genetic models: that variation is available and persistent. But under selection, this variation can in principle be depleted. Once variation is removed, the selection to retain imprinting is lost. However, a locus at which fitness variance was ever present—owing perhaps to repeated deleterious mutation—could experience selection for imprinted expression for prolonged periods through these repeated episodes. Similar to the other theories, we cannot know how imprinted expression influences the likelihood of reversion to biallelic expression. But, all other things equal, under coadaptation the selection pressure to retain imprinted expression dissipates as genetic variation is consumed by selection.
Tests of the theories
Requisite genetic variation
The kinship theory assumes that gene expression levels have opposing effects on the inclusive fitness of patrigenic and matrigenic alleles. Unlike the premises of the two other theories, this form of selection exists only in hypothesis and has never been demonstrated empirically because of the difficulty of measuring inclusive fitness. If a way to measure inclusive fitness (especially as a function of a change in the level of gene expression) could be devised for an experimental system, an approach that mimics what has been done to detect sexually antagonistic fitness variance could be successful in finding parentally antagonistic fitness variance. Two such approaches are suggested. First, allelic variation in the expression level of parentally antagonistic genes is predicted to be associated with variation in inclusive fitness for those alleles (for the analog in the study of sexual antagonism, see Innocenti and Morrow, 2010). Second, imprinted effects of genes can be detected in a quantitative genetic framework by adding a gametic effect to the model (Tier and Solkner, 1993; de Koning et al., 2002). One would seek a parental origin-specific component of inclusive fitness variation in a similar fashion to Foerster et al.'s (2007) tactic for finding sexually antagonistic fitness variance. Rather than an attempt to find natural variation that influences inclusive fitness, one might be able to generate such variation experimentally. Further, tests of the theory do not always demand the measurement of inclusive fitness. If there were a way to subtly manipulate the expression level of imprinted genes in offspring, there are discriminating predictions for their direct effects on kin. (N.B. The complete knockout of imprinted genes does not provide the subtlety needed for examining fitness effects of expression level.) For instance, upregulation of paternally expressed genes in offspring should impose costs on mothers that will be reflected in maternal total fitness, whereas the downregulation of paternally expressed genes should increase a mother's fitness.
The sexual antagonism theory requires either of two things: sexually antagonistic selection, which is widespread in nature (Bonduriansky and Chenoweth, 2009); or sex biases in selection strength, which are similarly well documented (Connallon et al., 2010). One challenge for the sexual antagonism theory is therefore to address why we don't observe genomic imprinting more widely. An additional prediction of the sexual antagonism theory is that under sexual antagonism, sex-specific imprinting can evolve. That is, males and females can show different patterns of matrigenic and patrigenic bias in expression. This possibility depends on there being the regulatory potential to achieve sex-specific imprinted expression. Single-gene sex-specific imprinted effects (which excludes, for example, chromosome- or genome-wide modifications like paternal X-inactivation in mammals and whole-genome elimination in scale insects) on phenotype have been detected in a quantitative genetic framework (Hager et al., 2008), but more sequencing studies (for example, Wang et al., 2011) will be needed to test for the molecular basis of such phenotypic effects.
The coadaptation theory requires that interactions between individuals affect fitness, for which there is ample evidence (Wolf et al., 1999b). Fitness variance owing to maternal effects is also now widely recognized (Mousseau and Fox, 1998) and fitness variance stemming from the interaction between maternal and offspring phenotypes is also known (Moore et al., 1997, 1998; Wolf and Brodie, 1998; Wolf et al., 1999a; Wolf, 2000). Experimental procedures for documenting maternal–offspring interaction effects typically involve cross-fostering of young, which requires waiting until the offspring is at least to a certain stage of development (although future studies could employ other strategies such as embryo transfer), followed by measurement of the phenotype of interest. In this case the phenotypes of interest are related to fitness (for example, survival, birth and weaning weights, growth rates, so on). This poses a challenge for the study of imprinted genes' potential role in mother–offspring interactions because imprinted genes are expressed in the earliest tissues of both seed plants and mammals, before experimental cross-fostering is possible. Statistical approaches can be used to overcome this constraint, albeit to a limited degree (for example, Wolf and Cheverud, 2012).
Experimental evolution
The natural selection pressures that the theories invoke—parental antagonism, sexual antagonism, coadaptation—can be modulated in a lab setting, allowing for tests of their predictions experimentally. If one supposes that the requisite genetic variation resides in a population at the onset of an artificial selection experiment, then one might be able to evolve novel imprinted genes by boosting the relevant selection pressure. Obtaining de novo imprinted expression of a gene that was biallelically expressed at the onset of an experiment is perhaps too hopeful, although at least over the timescale of such an experiment. Fortunately, the theories provide other predictions about the response to selection that are more easily tested.
The kinship theory derives imprinted expression from parentally antagonistic selection pressure, the strength of which can be most easily manipulated by adjustments to the mating system (for example, Holland and Rice, 1999). Experimental reductions of patrilineal relatedness should cause paternally expressed imprinted genes to increase their expression levels. The original difference in the optimal level of expression for matrigenic and patrigenic alleles can only be exacerbated by further reductions to patrilineal relatedness. At the same time, maternally expressed imprinted genes are not expected to modify their expression level in response to lessened patrilineal relatedness, as the optimal level of expression for these genes is unaffected by this change. To test this, one could select a subset of the suite of imprinted genes to examine. The appeal of this experiment is that the kinship theory provides a clear, strong prediction.
Sexual antagonism and sex biases in the strength of selection can also be manipulated in a lab setting by changing the mating system of an organism. On the phenotypic level, one predicts that the sex-specific parent–offspring covariance for traits under selection evolves after many generations of either selection pressure. Under sexual antagonism, parent–offspring covariance should increase for same-sex parents and offspring and decrease for opposite-sex pairs. Under stronger selection on one sex, males for instance, the father–offspring covariance should increase and the mother–offspring covariance should decrease for both sexes of offspring. These phenotypic responses can also come about by other means (for example, sex-specific recombination rates and sexually dimorphic gene regulation), so care must be taken in interpreting phenotypic data. Alternatively, the response can be studied directly by using genomic techniques to compare differences in the parental origin-dependent expression of genes on a genome-wide scale before and after the period of experimental evolution.
Using an experimental evolution approach to test the coadaptation theory might be particularly challenging. One could impose random cross-fostering experimentally for a number of generations. This would disrupt the interaction between mothers and her offspring and therefore remove the selection pressure on imprinted expression. However, apart from experimental constraints such as the difficulty of cross-fostering early in development, the expected responses to a loss of selection (as opposed to a reversal as described for the other theories) are likely to be very slow and may not be expected over the brief duration of an experiment.
Although experimental evolution approaches might be possible for the kinship and the sexual antagonism theories, the challenge is to find an organism that has two sexes, imprinted genes, high fecundity and a suitably short generation time. Unfortunately, mammals, where much is known about imprinting, may not be a suitable choice. In addition, because the strength of parental antagonism will tend to covary with the strength of sexual antagonism (for example, increased polyandry will lead to an intensification of both types of antagonism), it will be necessary to derive fine-grained predictions that distinguish the patterns of imprinting predicted under the kinship and sexual antagonism theories.
Molecular evolution
Data on DNA sequence divergence and expression level divergence are rapidly becoming available from various taxa. The theories offer predictions about the rate of these two forms of divergence, albeit weak predictions. In general, rapid molecular evolution is thought to be a response to the various genetic conflicts that imprinted genes and imprinted expression generate. The theories differ in the extent to which they predict such conflicts and where they find them.
In the kinship theory, the genome is rife with conflict over genomic imprinting. First, genes with antagonistic effects and reciprocal expression patterns are in conflict over the eventual phenotype (Wilkins and Haig, 2001). If the proteins encoded by the antagonistic genes physically interact, then sequence divergence in the coding regions is a possible outcome, assuming that the amino acid sequences are not prohibitively constrained. Another possible outcome is for cis-regulatory evolution to produce expression level divergence as each gene evolves increased expression. Second, because imprinted genes and the genes underlying the imprinting machinery, which are typically in trans, reap different inclusive fitness returns from an imprinted gene's expression—for example, silencing of matrigenic alleles at an imprinted gene may be advantageous to those matrigenic alleles but disadvantageous on average to an unimprinted gene responsible for maintaining the imprinted gene's methylation state—they may be in conflict over the total expression level from an imprinted locus (Burt and Trivers, 1998). This form of intragenomic conflict is expected to lead to antagonistic coevolution between loci, with the genes involved in establishing and maintaining imprints perhaps evolving resistance to imprinted genes' regulatory cues. Both forms of intragenomic conflict may lead to an elevated rate of divergence for imprinted genes as well as genes involved in applying and interpreting the imprints.
The kinship theory also predicts elevated expression levels for certain imprinted genes. It should be possible to compare the expression level of imprinted genes with the average gene in the genome (or some other appropriate set for comparison). If imprinted genes should prove to be highly expressed, then they may share certain structural properties with other highly expressed genes, such as amino acid bias, codon bias and few, small introns (Akashi, 2001; Castillo-Davis et al., 2002; Urrutia and Hurst, 2003).
Under the sexual antagonism theory, if selection is sexually antagonistic but stronger in one sex, then imprinted expression of the traditional variety (that is, imprinting that is not sex specific) may evolve. In this outcome, the sex experiencing stronger selection gains a benefit from this expression pattern, but the sex that faces weaker selection is pushed farther from its selective optimum, for it expresses an allele that experienced selection most recently in the opposite sex. The genes involved in applying the imprints and interpreting them are therefore expected to be under sexually antagonistic selection pressure. This is a different form of selection pressure on the imprinting machinery from that of the kinship theory. In the kinship theory, the imprinted genes and the imprinting genes have conflicting interests. In the sexual antagonism theory, the two types of genes have concordant interests: imprinting is favored in one sex but disfavored in the other, and the two genes reap the benefits and costs of imprinting equally. In this case, imprinted expression, when it evolves as a response to intralocus sexual antagonism, is itself sexually antagonistic. Whereas the intragenomic conflict found in the kinship theory promotes perpetual coevolution and divergence, intralocus sexual antagonism need not do so. Despite this, the sexual antagonism theory predicts some evolution of the imprinting machinery in light of the possibility for sexually antagonistic selection pressure on it.
Progress has already been made on testing these predictions for the molecular evolution of imprinted genes (for example, McVean and Hurst, 1997; Smith and Hurst, 1998; Parker-Katiraee et al., 2007; Spillane et al., 2007; Miyake et al., 2009; Hutter et al., 2010; O'Connell et al., 2010; Wolff et al., 2011), but no consistent pattern has yet emerged.
Comparative data
At present, we have a depth of knowledge on imprinting in only a few mammalian and angiosperm taxa (for example, mouse, human, Arabidopsis and maize), but even then only for a few tissues (for example, brain, placenta, endosperm, early-stage embryos). In addition, we have knowledge of only a select few imprinted genes from a wider taxonomic span (for example, Killian et al., 2001; Renfree et al., 2009). Suppose we had deep knowledge of imprinting in a breadth of taxa for which we had a reliable phylogeny, which due to the development of novel genomic techniques may not be far off, what would this mean for our understanding of the evolution of imprinting?
First, it would allow us to gauge whether there was a core set of imprinted genes common to a particular taxonomic group. If so, this would argue that for these genes, imprinted expression arose early and has been retained through evolutionary time. If instead the pattern were one of little overlap between species, then it would argue that imprinting status is a more labile trait. Such evolutionary lability would allow us to examine the rate and patterns of gain and loss of imprinted expression. The different theories make weakly discriminating predictions about these rates (see above section on loss of imprinting). Third, if any gains or losses prove to be repeated across the phylogeny, we would have phylogenetically independent events to compare, and correlations with other life history or ecological characters could be sought. There is overlap in the ecological and life history correlates that each theory predicts—for example, strength of interaction among kin, breeding system, extent of parental care—but we have at least some ability to discriminate between theories based on such data.
Data Archiving
There were no data to deposit.
Acknowledgments
This manuscript benefited from the efforts of three anonymous reviewers. This material is based on work supported by the National Science Foundation through the National Evolutionary Synthesis Center (NESCent) under grant number NSF #EF-0905606. LR was funded by the Royal Society and the Natural Environment Research Council (NERC). DCQ was funded by the National Science Foundation. RB was funded by the Department of Industry, Innovation, Science, Research and Tertiary Education, Australian Government and Australian Research Council (ARC) through a Future Fellowship and Discovery Grant. JBW is funded by grant support from NERC and the Biotechnology and Biological Sciences Research Council (BBSRC).
The authors declare no conflict of interest.
References
- Akashi H. Gene expression and molecular evolution. Curr Opin Genet Dev. 2001;11:660–666. doi: 10.1016/s0959-437x(00)00250-1. [DOI] [PubMed] [Google Scholar]
- Autran D, Baroux C, Raissig MT, Lenormand T, Wittig M, Grob S, et al. Maternal epigenetic pathways control parental contributions to Arabidopsis early embryogenesis. Cell. 2011;145:707–719. doi: 10.1016/j.cell.2011.04.014. [DOI] [PubMed] [Google Scholar]
- Bonduriansky R.2007The genetic architecture of sexual dimorphism: the potential roles of genomic imprinting and condition dependenceIn: Fairbairn DJ, Blanckenhorn WU, Székely T (eds). Sex, Size, and Gender Roles: Evolutionary Studies of Sexual Dimorphism Oxford University Press: Oxford, UK; 176–184. [Google Scholar]
- Bonduriansky R, Chenoweth SF. Intralocus sexual conflict. Trends Ecol Evol. 2009;24:280–288. doi: 10.1016/j.tree.2008.12.005. [DOI] [PubMed] [Google Scholar]
- Brandvain Y, Van Cleve J, Úbeda F, Wilkins JF. Demography, kinship, and the evolving theory of genomic imprinting. Trends Genet. 2011;27:251–257. doi: 10.1016/j.tig.2011.04.005. [DOI] [PubMed] [Google Scholar]
- Burt A, Trivers R. Genetic conflicts in genomic imprinting. Proc Biol Sci. 1998;265:2393–2397. doi: 10.1098/rspb.1998.0589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo-Davis CI, Mekhedov SL, Hartl DL, Koonin EV, Kondrashov FA. Selection for shorter introns in highly expressed genes. Nat Genet. 2002;31:415–418. doi: 10.1038/ng940. [DOI] [PubMed] [Google Scholar]
- Connallon T, Cox RM, Calsbeek R. Fitness consequences of sex-specific selection. Evolution. 2010;64:1671–1682. doi: 10.1111/j.1558-5646.2009.00934.x. [DOI] [PubMed] [Google Scholar]
- Curley JP. Is there a genomically imprinted social brain. Bioessays. 2011;33:662–668. doi: 10.1002/bies.201100060. [DOI] [PubMed] [Google Scholar]
- Day T, Bonduriansky R. Intralocus sexual conflict can drive the evolution of genomic imprinting. Genetics. 2004;167:1537–1546. doi: 10.1534/genetics.103.026211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Koning DJ, Bovenhuis H, van Arendonk JAM. On the detection of imprinted quantitative trait loci in experimental crosses of outbred species. Genetics. 2002;161:931–938. doi: 10.1093/genetics/161.2.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drewell RA, Lo N, Oxley PR, Oldroyd BP. Kin conflict in insect societies: a new epigenetic perspective. Trends Ecol Evol. 2012;27:367–373. doi: 10.1016/j.tree.2012.02.005. [DOI] [PubMed] [Google Scholar]
- Foerster K, Coulson T, Sheldon BC, Pemberton JC, Clutton-Brock TH, Kruuk LEB. Sexually antagonistic genetic variation for fitness in red deer. Nature. 2007;447:1107–U1109. doi: 10.1038/nature05912. [DOI] [PubMed] [Google Scholar]
- Frank SA, Crespi BJ. Pathology from evolutionary conflict, with a theory of X chromosome versus autosome conflict over sexually antagonistic traits. Proc Natl Acad Sci USA. 2011;108:10886–10893. doi: 10.1073/pnas.1100921108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring M, Missirian V, Henikoff S. Genomic analysis of parent-of-origin allelic expression in Arabidopsis thaliana seeds. PLoS One. 2011;6:e23687. doi: 10.1371/journal.pone.0023687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glastad KM, Hunt BG, Yi SV, Goodisman MAD. DNA methylation in insects: on the brink of the epigenomic era. Insect Mol Biol. 2011;20:553–565. doi: 10.1111/j.1365-2583.2011.01092.x. [DOI] [PubMed] [Google Scholar]
- Hager R, Cheverud JM, Leamy LJ, Wolf JB. Sex dependent imprinting effects on complex traits in mice. BMC Evol Biol. 2008;8:303. doi: 10.1186/1471-2148-8-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haig D. Intragenomic conflict and the evolution of eusociality. J Theor Biol. 1992;156:401–403. doi: 10.1016/s0022-5193(05)80683-6. [DOI] [PubMed] [Google Scholar]
- Haig D. Placental hormones, genomic imprinting, and maternal-fetal communication. J Evol Biol. 1996;9:357–380. [Google Scholar]
- Haig D. Parental antagonism, relatedness asymmetries, and genomic imprinting. Proc Biol Sci. 1997;264:1657–1662. doi: 10.1098/rspb.1997.0230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haig D. The kinship theory of genomic imprinting. Annu Rev Ecol Syst. 2000;31:9–32. [Google Scholar]
- Haig D. Genomic imprinting, sex-biased dispersal, and social behavior. Ann NY Acad Sci. 2000;907:149–163. doi: 10.1111/j.1749-6632.2000.tb06621.x. [DOI] [PubMed] [Google Scholar]
- Haig D. Genomic Imprinting and Kinship. Rutgers University Press: New Brunswick, NJ, USA; 2002. [Google Scholar]
- Haig D. Genomic imprinting and kinship: how good is the evidence. Ann Rev Genet. 2004;38:553–585. doi: 10.1146/annurev.genet.37.110801.142741. [DOI] [PubMed] [Google Scholar]
- Haig D. Self-imposed silence: parental antagonism and the evolution of X-chromosome inactivation. Evolution. 2006;60:440–447. [PubMed] [Google Scholar]
- Haig D, Graham C. Genomic imprinting and the strange case of the insulin-like growth factor-II receptor. Cell. 1991;64:1045–1046. doi: 10.1016/0092-8674(91)90256-x. [DOI] [PubMed] [Google Scholar]
- Haig D, Westoby M. Parent-specific gene-expression and the triploid endosperm. Am Nat. 1989;134:147–155. [Google Scholar]
- Holland B, Rice WR. Chase-away sexual selection: antagonistic seduction versus resistance. Evolution. 1998;52:1–7. doi: 10.1111/j.1558-5646.1998.tb05132.x. [DOI] [PubMed] [Google Scholar]
- Holland B, Rice WR. Experimental removal of sexual selection reverses intersexual antagonistic coevolution and removes a reproductive load. Proc Natl Acad Sci USA. 1999;96:5083–5088. doi: 10.1073/pnas.96.9.5083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutter B, Bieg M, Helms V, Paulsen M. Divergence of imprinted genes during mammalian evolution. BMC Evol Biol. 2010;10:116. doi: 10.1186/1471-2148-10-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda Y. Plant imprinted genes identified by genome-wide approaches and their regulatory mechanisms. Plant Cell Physiol. 2012;53:809–816. doi: 10.1093/pcp/pcs049. [DOI] [PubMed] [Google Scholar]
- Innocenti P, Morrow EH. The sexually antagonistic genes of Drosophila melanogaster. PLoS Biol. 2010;8:e1000335. doi: 10.1371/journal.pbio.1000335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killian JK, Nolan CM, Wylie AA, Li T, Vu TH, Hoffman AR, et al. Divergent evolution in M6P/IGF2R imprinting from the Jurassic to the Quaternary. Hum Mol Genet. 2001;10:1721–1728. doi: 10.1093/hmg/10.17.1721. [DOI] [PubMed] [Google Scholar]
- McGlothlin JW, Moore AJ, Wolf JB, Brodie ED. Interacting phenotypes and the evolutionary process. III. Social evolution. Evolution. 2010;64:2558–2574. doi: 10.1111/j.1558-5646.2010.01012.x. [DOI] [PubMed] [Google Scholar]
- McKeown PC, Laouielle-Duprat S, Prins P, Wolff P, Schmid MW, Donoghue MTA, et al. Identification of imprinted genes subject to parent-of-origin specific expression in Arabidopsis thaliana seeds. BMC Plant Biol. 2011;11:113. doi: 10.1186/1471-2229-11-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVean GT, Hurst LD. Molecular evolution of imprinted genes: no evidence for antagonistic coevolution. Proc R Soc Lond B. 1997;264:739–746. doi: 10.1098/rspb.1997.0105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake T, Takebayashi N, Wolf DE. Possible diversifying selection in the imprinted gene, MEDEA, in Arabidopsis. Mol Biol Evol. 2009;26:843–857. doi: 10.1093/molbev/msp001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore AJ, Brodie ED, Wolf JB. Interacting phenotypes and the evolutionary process.1. Direct and indirect genetic effects of social interactions. Evolution. 1997;51:1352–1362. doi: 10.1111/j.1558-5646.1997.tb01458.x. [DOI] [PubMed] [Google Scholar]
- Moore AJ, Wolf JB, Brodie ED.1998The influence of direct and indirect genetic effects on the evolution of behavior: social and sexual selection meet maternal effectsIn: Mousseau TA, Fox CW (eds)Maternal Effects as Adptations Oxford University Press: New York, NY, USA; 22–41. [Google Scholar]
- Moore T, Haig D. Genomic imprinting in mammalian development - a parental tug-of-war. Trends Genet. 1991;7:45–49. doi: 10.1016/0168-9525(91)90230-N. [DOI] [PubMed] [Google Scholar]
- Mousseau TA, Fox CW. Maternal Effects as Adaptations. Oxford University Press: New York, NY, USA; 1998. [Google Scholar]
- O'Connell MJ, Loughran NB, Walsh TA, Donoghue MTA, Schmid KJ, Spillane C. A phylogenetic approach to test for evidence of parental conflict or gene duplications associated with protein-encoding imprinted orthologous genes in placental mammals. Mamm Genome. 2010;21:486–498. doi: 10.1007/s00335-010-9283-5. [DOI] [PubMed] [Google Scholar]
- Parker-Katiraee L, Carson AR, Yamada T, Arnaud P, Feil R, Abu-Amero SN, et al. Identification of the imprinted KLF14 transcription factor undergoing human-specific accelerated evolution. PLoS Genet. 2007;3:665–678. doi: 10.1371/journal.pgen.0030065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudhon C, Bourc'his D. Identification and resolution of artifacts in the interpretation of imprinted gene expression. Brief Funct Genomics. 2010;9:374–384. doi: 10.1093/bfgp/elq020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queller DC. Theory of genomic imprinting conflict in social insects. BMC Evol Biol. 2003;3:15. doi: 10.1186/1471-2148-3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renfree MB, Hore TA, Shaw G, Graves JAM, Pask AJ. Evolution of genomic imprinting: insights from marsupials and monotremes. Annu Rev Genomics Hum Genet. 2009;10:241–262. doi: 10.1146/annurev-genom-082908-150026. [DOI] [PubMed] [Google Scholar]
- Sinervo B, Svensson E. Correlational selection and the evolution of genomic architecture. Heredity. 2002;89:329–338. doi: 10.1038/sj.hdy.6800148. [DOI] [PubMed] [Google Scholar]
- Smith NGC, Hurst LD. Molecular evolution of an imprinted gene: repeatability of patterns of evolution within the mammalian insulin-like growth factor type II receptor. Genetics. 1998;150:823–833. doi: 10.1093/genetics/150.2.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer HG, Clark AG. A chip off the old block: a model for the evolution of genomic imprinting via selection for parental similarity. Genetics. 2006;174:931–935. doi: 10.1534/genetics.106.060475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer HG, Clark AG.2014Non-conflict theories for the evolution of genomic imprinting Hereditye-pub ahead of print 8 January 2014; doi: 10.1038/hdy.2013.129 [DOI] [PMC free article] [PubMed]
- Spillane C, Schmid KJ, Laoueille-Duprat S, Pien S, Escobar-Restrepo JM, Baroux C, et al. Positive Darwinian selection at the imprinted MEDEA locus in plants. Nature. 2007;448:349–352. doi: 10.1038/nature05984. [DOI] [PubMed] [Google Scholar]
- Stringer JM, Suzuki S, Pask AJ, Shaw G, Renfree MB. Selected imprinting of INS in the marsupial. Epigenetics Chromatin. 2012;5:14. doi: 10.1186/1756-8935-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tier B, Solkner J. Analyzing gametic variation with an animal model. Theor Appl Genet. 1993;85:868–872. doi: 10.1007/BF00225031. [DOI] [PubMed] [Google Scholar]
- Úbeda F, Gardner A. A model for genomic imprinting in the social brain: juveniles. Evolution. 2010;64:2587–2600. doi: 10.1111/j.1558-5646.2010.01015.x. [DOI] [PubMed] [Google Scholar]
- Úbeda F, Gardner A. A model for genomic imprinting in the social brain: adults. Evolution. 2011;65:462–475. doi: 10.1111/j.1558-5646.2010.01115.x. [DOI] [PubMed] [Google Scholar]
- Úbeda F, Gardner A. A model for genomic imprinting in the social brain: elders. Evolution. 2012;66:1567–1581. doi: 10.1111/j.1558-5646.2011.01517.x. [DOI] [PubMed] [Google Scholar]
- Úbeda F, Haig D. Dividing the child. Genetica. 2003;117:103–110. doi: 10.1023/a:1022320801661. [DOI] [PubMed] [Google Scholar]
- Urrutia AO, Hurst LD. The signature of selection mediated by expression on human genes. Genome Res. 2003;13:2260–2264. doi: 10.1101/gr.641103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Cleve J, Feldman MW, Lehmann L. How demography, life history, and kinship shape the evolution of genomic imprinting. Am Nat. 2010;176:440–455. doi: 10.1086/656277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Clark AG.2014Using next-generation RNA sequencing to identify imprinted genes Hereditye-pub ahead of print 12 March 2014; doi: 10.1038/hdy.2014.18 [DOI] [PMC free article] [PubMed]
- Wang X, Soloway PD, Clark AG. A survey for novel imprinted genes in the mouse placenta by mRNA-seq. Genetics. 2011;189:109–122. doi: 10.1534/genetics.111.130088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins JF. Genomic imprinting and conflict-induced decanalization. Evolution. 2011;65:537–553. doi: 10.1111/j.1558-5646.2010.01147.x. [DOI] [PubMed] [Google Scholar]
- Wilkins JF, Haig D. Genomic imprinting of two antagonistic loci. Proc Biol Sci. 2001;268:1861–1867. doi: 10.1098/rspb.2001.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins JF, Haig D. Parental modifiers, antisense transcripts and loss of imprinting. Proc Biol Sci. 2002;269:1841–1846. doi: 10.1098/rspb.2002.2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins JF, Haig D. What good is genomic imprinting: the function of parent-specific gene expression. Nat Rev Genet. 2003;4:359–368. doi: 10.1038/nrg1062. [DOI] [PubMed] [Google Scholar]
- Wolf JB.1999Indirect genetic effects and gene interactionsIn: Wolf JB, Brodie ED, Wade MJ (eds). Epistasis and the Evolutionary Process Oxford University Press: New York, NY, USA; 158–176. [Google Scholar]
- Wolf JB. Gene interactions from maternal effects. Evolution. 2000;54:1882–1898. doi: 10.1111/j.0014-3820.2000.tb01235.x. [DOI] [PubMed] [Google Scholar]
- Wolf JB. Evolution of genomic imprinting as a coordinator of coadapted gene expression. Proc Natl Acad Sci USA. 2013;110:5085–5090. doi: 10.1073/pnas.1205686110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf JB, Brodie ED. The coadaptation of parental and offspring characters. Evolution. 1998;52:299–308. doi: 10.1111/j.1558-5646.1998.tb01632.x. [DOI] [PubMed] [Google Scholar]
- Wolf JB, Brodie ED, Moore AJ. Interacting phenotypes and the evolutionary process. II. Selection resulting from social interactions. Am Nat. 1999;153:254–266. doi: 10.1086/303168. [DOI] [PubMed] [Google Scholar]
- Wolf JB, Brodie ED, Wade MJ. Epistasis and the Evolutionary Process. Oxford University Press: New York, NY, USA; 1999. [Google Scholar]
- Wolf JB, Cheverud JM. Detecting maternal effects by statistical cross fostering. Genetics. 2012;191:261–277. doi: 10.1534/genetics.111.136440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf JB, Hager R. A maternal-offspring coadaptation theory for the evolution of genomic imprinting. PLoS Biol. 2006;4:2238–2243. doi: 10.1371/journal.pbio.0040380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf JB, Hager R. Selective abortion and the evolution of genomic imprinting. J Evol Biol. 2009;22:2519–2523. doi: 10.1111/j.1420-9101.2009.01874.x. [DOI] [PubMed] [Google Scholar]
- Wolf JB, Wade MJ. What are maternal effects (and what are they not) Philos Trans R Soc Lond B Biol Sci. 2009;364:1107–1115. doi: 10.1098/rstb.2008.0238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff P, Weinhofer I, Seguin J, Roszak P, Beisel C, Donoghue MTA, et al. High-resolution analysis of parent-of-origin allelic expression in the Arabidopsis Endosperm. PLoS Genet. 2011;7:e1002126. doi: 10.1371/journal.pgen.1002126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi SV, Goodisman MAD. Computational approaches for understanding the evolution of DNA methylation in animals. Epigenetics. 2009;4:551–556. doi: 10.4161/epi.4.8.10345. [DOI] [PubMed] [Google Scholar]
- Zhang M, Zhao HN, Xie SJ, Chen J, Xu YY, Wang KK, et al. Extensive, clustered parental imprinting of protein-coding and noncoding RNAs in developing maize endosperm. Proc Natl Acad Sci USA. 2011;108:20042–20047. doi: 10.1073/pnas.1112186108. [DOI] [PMC free article] [PubMed] [Google Scholar]