Abstract
At the heart of genomic imprinting in mammals are imprinting control regions (ICRs), which are the discrete genetic elements that confer imprinted monoallelic expression to several genes in imprinted gene clusters. A characteristic of the known ICRs is that they acquire different epigenetic states, exemplified by differences in DNA methylation, in the sperm and egg, and these imprint marks remain on the sperm- and oocyte-derived alleles into the next generation as a lifelong memory of parental origin. Although there has been much focus on gametic marking of ICRs as the point of imprint specification, recent mechanistic studies and genome-wide DNA methylation profiling do not support the existence of a specific imprinting machinery in germ cells. Rather, ICRs are part of more widespread methylation events that occur during gametogenesis. Instead, a decisive component in the specification of imprints is the choice of which sites of gamete-derived methylation to maintain in the zygote and preimplantation embryo at a time when much of the remainder of the genome is being demethylated. Among the factors involved in this selection, the zinc-finger protein Zfp57 can be regarded as an imprint-specific, sequence-specific DNA binding factor responsible for maintaining methylation at most ICRs. The recent insights into the balance of gametic and zygotic contributions to imprint specification should help understand mechanistic opportunities and constraints on the evolution of imprinting in mammals.
Introduction
Genomic imprinting in mammals provides the classic example of lifelong epigenetic memory of parental origin (Ferguson-Smith, 2011). One hundred and fifty imprinted genes have been identified in mammals so far (Williamson et al., 2013). Although these genes have diverse functions in mammalian development, growth, physiology and behaviour, an underlying assumption is that their imprinting depends upon a common mechanism that marks them differentially in the male and female germline and perpetuates this epigenetic difference in the next generation (Kelsey and Feil, 2013). Many imprinted genes are found in clusters (Wan and Bartolomei, 2008; Ferguson-Smith, 2011) and the correct imprinted expression of all the genes in a cluster depends on the existence and appropriate epigenetic marking of a discrete genetic element, the imprinting control region (ICR). These are the critical elements, so much of the following discussion will focus on the properties of ICRs, what leads them to become epigenetically distinguished in the sperm and egg and how such epigenetic asymmetry is maintained after fertilisation (Proudhon et al., 2012). We also consider how imprint marks figure in the wider epigenetic landscape of the gametes and the recent evidence that specification of imprints depends in a major way on events after fertilisation. We hope this provides a backdrop against which to consider the possible mechanistic constraints on and opportunities for the evolution of imprinted loci. We should point out that most of what follows relates to what we know about in the mouse. Certainly, the general properties of imprinted gene clusters and their regulation is conserved between mouse and human (Ferguson-Smith, 2011) and in the few other eutherian mammals in which imprinted genes have started to be characterised; however, the current knowledge of DNA methylation dynamics in the germline and the mechanisms of imprint establishment and maintenance stems largely from work in the mouse. It will be an important future goal to assess the degree to which the mouse as a model organism is representative of other mammalian species in these regards.
Epigenetic properties of imprinted genes
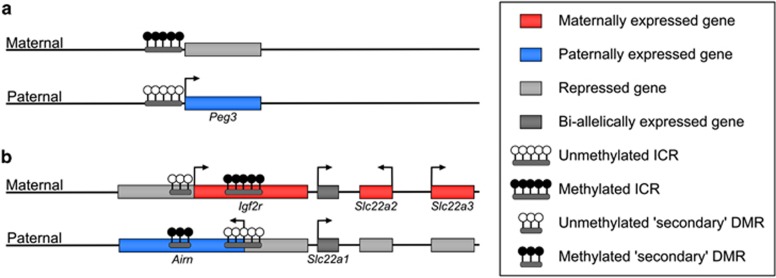
Imprinted, parent-allele-specific expression in eutherian mammals is dependent on the existence of epigenetic differences between the two parental alleles that allow them to be transcribed differently in the same nucleus. In many cases, this equates to complete silencing of one allele of an imprinted gene. These epigenetic differences are conferred in the gametes, or very early in embryogenesis, and, in general, will remain with and distinguish the parentally-derived alleles for the lifetime of the individual (Kelsey and Feil, 2013). However, while gamete-specific epigenetic marks are fundamental to imprinting, it is important to recognise that not all imprinted genes are regulated in the same way, and it may be useful to think of genes that are controlled ‘directly' and ‘indirectly' by the primary imprint marks. This distinction is most pertinent in imprinted gene clusters (Wan and Bartolomei, 2008; Ferguson-Smith, 2011), in which only one gene or genetic element carries the primary imprint mark and is thus directly imprinted—this element corresponds to the ICR—whereas the imprinted expression of the remaining genes in a cluster can be considered to be indirect, as it depends on the existence of the primary epigenetic mark at the ICR (Figure 1).
Figure 1.
Examples of directly and indirectly regulated imprinted regions. Schematic representation of the (a) Peg3 imprinted gene on chromosome 7 and (b) the Igf2r imprinted cluster on chromosome 17. The expression status of the genes on the maternal and paternal alleles is illustrated; active promoters are represented by horizontal arrows. (a) The differentially methylated ICR established during germ cell development is located at the promoter of the Peg3 gene and directly regulates the monoallelic transcription of this gene. (b) The maternally methylated ICR indirectly regulates the monoallelic expression of the adjacent genes at this locus, partially mediated by the monoallelic methylation acquired at the nearby secondary DMR at the Igf2r promoter.
One characteristic of ICRs is the profound difference in DNA methylation on the maternally- and paternally-derived copies; one copy being highly methylated and the other unmethylated (Smallwood and Kelsey, 2012; Tomizawa et al., 2011). This methylation difference can be traced back to the sperm and egg, hence the term gametic or germline differentially methylated region (gDMR). (The term ICR is more properly reserved for those gDMRs that have been functionally demonstrated, either through targeted mutagenesis in the mouse or the occurrence of mutations in imprinted gene syndromes, to control imprinted expression in cis of several genes in a cluster.) Allele-specific methylation at the ICR controls the properties of the element: for instance, the methylated copy may promote, whereas the unmethylated copy represses, expression of the genes in the cluster. There are a variety of mechanisms by which ICRs set up and maintain the allelic regulation of the clusters they control: the precise details do not really concern us and we do not intend to catalogue all possible mechanisms, which have been reviewed elsewhere (Wan and Bartolomei, 2008; Ferguson-Smith, 2011). One illustrative example is provided by the Igf2r cluster (Figure 1b), in which the ICR is methylated on the maternal allele and coincides with a promoter for a non-coding transcript, Airn. It is the Airn non-coding transcript, or the process of monoallelic transcription initiating from the ICR, that gives rise to monoallelic expression of the remaining genes in the cluster by silencing genes in cis. The process initiating from the ICR sets up additional parental allele epigenetic differences in the cluster, such as ‘secondary' DMRs or allele-specific histone modifications, that determine monoallelic expression at the individual loci, and these secondary differences can themselves become stably propagated and may even be maintained if differential methylation at the ICR is lost later in development (Lewis et al., 2004). Outside of the extensive imprinted clusters, there are also smaller, much simpler clusters as well as ‘singleton' imprinted genes. Monoallelic expression of these genes also depends upon allelic epigenetic differences, but the mechanisms may be more direct, for example, the presence of a gDMR directly overlying the imprinted gene promoter (Figure 1a). Although gDMRs are found in almost all imprinted gene clusters and singleton imprinted genes, it is conceivable that there are imprinted loci whose monoallelic expression is truly independent of DNA methylation, reflecting other possible allelic epigenetic asymmetries inherited from the gametes or conferred in the early embryo.
The developmental dynamics of DNA methylation
At the present time, some 20 gDMRs at imprinted loci have been identified in the mouse whose methylation originates in the egg and 3 whose methylation originates in the sperm (Tomizawa et al., 2011; Proudhon et al., 2012). This disparity might suggest fundamentally different mechanisms by which DNA methylation marks are established in the male and female germlines and/or differences in how oocyte- and sperm-derived methylation is treated after fertilisation in the zygote. At a molecular level too, there appear to be differences between maternal gDMRs and paternal gDMRs: maternal gDMRs can be considered to be differentially methylated CpG islands overlapping a promoter for a coding or non-coding transcript (the majority of genes in the genome possess a CpG island at their transcription start site, which is usually unmethylated), while paternal gDMRs are CG-rich elements, which may not fit the definition of CpG islands, and are located intergenically (Kobayashi et al., 2006; Schulz et al., 2010).
It is important to consider the marking of gDMRs in relation to the global changes in DNA methylation that occur across the genome during germ cell specification, gametogenesis and within the embryo after fertilisation. In mammals, germ cells arise in the gastrulating embryo from a small number of somatic cells (in the mouse, this occurs from embryonic day 6, e6). Concomitant with germ cell specification and proliferation are extensive changes in the epigenetic state of these cells. There is almost wholesale removal of pre-existing DNA methylation such that, soon after primordial germ cells (PGCs) have migrated to the genital ridge (from e10.5), DNA methylation is reduced to a very low level throughout the genome (Seisenberger et al., 2012). This includes the erasure of imprints, so that the differences in methylation between the parental alleles are eliminated. Loss of DNA methylation is likely to occur by a combination of ‘passive' (dilution with repeated rounds of DNA replication, because factors required to maintain methylation are absent or repressed) and ‘active' processes (including oxidation of 5-methylcytosine to 5-hydroxymethycytosine and further derivatives by the action of enzymes of the Tet family) (Hackett et al., 2013; Yamaguchi et al., 2013).
The re-establishment of DNA methylation in germ cells occurs at different times in development and in different cellular contexts in the male and female gonads (Sasaki and Matsui, 2008; Smallwood and Kelsey, 2012). In the male gonad, de novo DNA methylation, including the three known paternally methylated gDMRs, initiates around e13.5 in germ cells arrested in mitosis (known as prospermatogonia) and is largely complete by e17.5 (Davis et al., 2000; Seissenberger et al., 2012). De novo methylation occurs before male germ cells undergo meiosis and, moreover, multiple cell divisions occur between the onset of methylation and formation of mature sperm. Therefore, faithful maintenance of methylation marks is required during spermatogonial stem cell division and renewal and the further rounds of DNA replication before and after meiosis. In the female gonad, PGCs enter meiosis from e13.5 and arrest in prophase 1, in which state they remain until maturation and ovulation. De novo DNA methylation occurs in meiotically arrested cells and no further DNA replication takes place between the onset of methylation and formation of the mature gamete. De novo methylation initiates after activation of follicles and during the later stages of oocyte growth in the postnatal ovary—in the mouse, around the transition from the primary to secondary follicle—and is completed by the time oocytes are fully grown and enter the transcriptionally quiescent, germinal vesicle (GV) stage (Smallwood et al., 2011; Shirane et al., 2013). There is evidence for oocyte-size-dependent initiation and progression of methylation of maternal gDMRs, suggesting that different factors influence the timing of establishment of imprints at different loci (Lucifero et al., 2004; Hiura et al., 2006).
The de novo DNA methylation machinery
There are two de novo DNA methylation enzymes in mammals—Dnmt3a and Dnmt3b—that are able to methylate cytosines, predominantly but not exclusively at CpG dinucleotides, in unmethylated DNA, and these are the activities responsible for setting up imprints. A third enzyme, Dnmt1, is the ‘maintenance' enzyme responsible for copying DNA methylation onto the nascent DNA strand at ‘hemimethylated' sites that have arisen from symmetrically methylated CpG dinucleotides during DNA replication. Together with other factors, Dnmt1 is required for faithful maintenance of methylation at DMRs during the later stages of spermatogenesis as well as in the embryo after fertilisation. Dnmt1 also has a subsidiary role in completing de novo methylation in oocytes by filling in gaps (hemimethylated CpG sites) left by Dnmt3a (Shirane et al., 2013). There is a third member of the Dnmt3 family, Dnmt3L (Bourc'his et al., 2001; Hata et al., 2002). Dnmt3L lacks DNA methyltransferase activity, but forms complexes with Dnmt3a and Dnmt3b, modulating their activity and attenuating their intrinsic sequence preferences (Wienholz et al., 2010). In oocytes in the mouse, Dnmt3a is the only de novo activity (Kaneda et al., 2004 and 2010), but it has an obligate requirement for Dnmt3L (Smallwood et al., 2011; Kobayashi et al., 2012; Shirane et al., 2013). In mouse male germ cells, Dnmt3a and Dnmt3b both contribute to de novo methylation in combination with Dnmt3L (Kato et al., 2007). Although often referred to as being ‘essential for imprints in oocytes', which has given rise to the impression that Dnmt3L is specific for imprinted sequences and some repetitive elements, and particularly as it has no orthologues outside of the mammalian lineage, it is now apparent that Dnmt3L is a generic factor and required for all de novo methylation in oocytes (Smallwood et al., 2011; Kobayashi et al., 2012; Shirane et al., 2013).
Although Dnmt3a and Dnmt3L are essential for de novo methylation in oocytes, they are dispensable for oocyte development and viability. The majority of DNA methylation is put in place late in oocyte growth, such that entry into meiosis, chromosome pairing and synapsis occur on a largely hypomethylated genome. There is also no obvious impact of DNA methylation on gene transcription (Kobayashi et al., 2012). However, while Dnmt3a- and Dnmt3L-deficient oocytes appear to be unimpaired and can be fertilised normally, they cannot support embryonic development beyond e10.5 (Kaneda et al., 2004). To a large extent, this is because of the absence of maternally-derived imprints, although abnormal expression of non-imprinted genes could also contribute to the failure of such conceptuses. Therefore, it appears that the functional relevance of de novo methylation in oocytes is primarily to ensure that maternal gDMRs acquire methylation, although much more of the genome becomes methylated. In male germ cells, in contrast, failure of de novo methylation has a devastating effect. Dnmt3L, which is expressed only in prospermatogonia, is required to assist Dnmt3a and Dnmt3b in the methylation and silencing of retrotransposons (long interspersed nuclear elements, LINEs, and intra-cisternal A particles, IAPs) in addition to imprints (Bourc'his and Bestor, 2004; Kato et al., 2007). Deficiency of Dnmt3L gives rise to subsequent meiotic failure in spermatocytes, characterised by multiple non-homologous synapses, suggesting a role of Dnmt3L in pre-meiotic genome scanning that results in methylation of interspersed repeats. (In the female germline, retrotransposon silencing is accomplished by multiple other mechanisms operating at different stages during oocyte development and growth, Lim et al., 2013). As such, imprinted gDMRs rely on the same machinery as that required for genome defence, but it need not follow that imprinting arose from a genome defence mechanism in mammals; it rather depends on how imprinted gDMRs fit within the wider DNA methylation landscape of the gametes.
Imprints in gametes: a targeted or a default process?
Until recently, there was a general assumption that imprints (ICRs, gDMRs) were specialised sequence elements that would be recognised by specific trans-acting factors that would target the de novo methylation machinery during gametogenesis. To some extent, this concept arose with the proposition that imprinting evolved from a genome defence mechanism (Sleutels and Barlow, 2002), which was supported by early observations of similarities between imprinted loci, transgenes displaying imprinted behaviour and repetitive elements, owing to the common presence of reiterated sequence motifs (Neumann et al., 1995). Recent work has led to a revision of this hypothesis in favour of the conclusion that DMRs become methylated in germ cells as part of a more universal DNA methylation mechanism. This is important, because it means that we can probably dispense with the notion that a specific imprinting machinery operates in the germline.
As noted before, maternal gDMRs are CpG islands and associated with promoters; the three known paternal gDMRs are CG-rich elements enriched in reiterated sequences and intergenic in location (Schulz et al., 2010). This apparent distinction may well reflect the general distribution of DNA methylation in the gametes rather than the existence of distinct mechanisms acting specifically on gDMRs in the male and female germlines. The intragenic location of many maternal gDMRs led to the proposal that transcription would be required for de novo methylation in oocytes; for example, the first gDMR/ICR identified, in the Igf2r locus, is within an intron of the gene, a relatively unusual location for a CpG island (Stöger et al., 1993). This possibility was also suggested by the discovery of ‘imprinted retrogenes'. Several imprinted retrogenes, such as U2af1-rs1 and Nap1l5, have integrated into the introns of larger transcription units and have their own maternal gDMRs, their intragenic location seeming to be a characteristic (Smith et al., 2003; Wood et al., 2007). A role for transcription in gDMR methylation in oocytes has now been demonstrated genetically for the Gnas and SNRPN imprinted loci (Chotalia et al., 2009; Smith et al., 2011); in many cases, the transcription events required for gDMR methylation appear to initiate from upstream, oocyte-specific promoters.
These findings raised the possibility that gDMR marking was actually part of a more widespread mechanism and that other CpG islands should become methylated if they were located within active transcription units in oocytes. This prediction was confirmed by profiling DNA methylation of a large number of CpG islands in non-growing and GV oocytes, which identified >1000 de novo methylated CpG islands, the majority of which map within genes (Smallwood et al., 2011). This subset of methylated CpG islands in the maternal germline contrasts with the broader observation that promoter-associated CpG islands were largely unmethylated, which is thought to be at least partially mediated by active chromatin marks associated with promoter activity, such as trimethylation of lysine 4 on histone 3 (H3K4me3). Moreover, there were no apparent discriminating sequence features of the methylated CpG islands in comparison with unmethylated CpG islands. Most of these CpG islands lost methylation after fertilisation suggesting that there is a generic mechanism of de novo methylation in oocytes, of which gDMRs are a part, and that the specification of imprinting occurs after fertilisation with the decision at which CpG islands to maintain gamete-derived methylation.
It is likely that a greater diversity of mechanisms of de novo methylation operate in male germ cells, because both Dnmt3a and Dnmt3b are active and have partially redundant functions as well as preferences for different sets of sequence elements. Dnmt3L appears to be an essential co-factor, as lack of Dnmt3L in male germ cells has a greater effect than ablation of either Dnmt3a or Dnmt3b (Bourc'his and Bestor 2004; Kato et al., 2007). It is possible that there is a transcription-based mechanism by which the Dnmt3a/Dnmt3L complex is recruited, analogous to that in oocytes, because transcripts traversing some of the paternal gDMRs have been detected in prospermatogonia (Henckel et al., 2011; Singh et al., 2013). It will be important to consider the context of gDMRs in the methylation landscape of mature sperm and whether they are methylated as part of a generic mechanism(s), as appears to be the case in the female germline. For one of the paternal gDMRs, Rasgrf1, both Dnmt3a and Dnmt3b participate in methylation in a mechanism that also involves piRNAs, a specific class of small non-coding RNA molecules involved in gene repression, particularly of retrotransposon-derived sequences. The methylation of this gDMR appears to occur because it contains a copy of a solo long terminal repeat-type retrotransposon for which a closely related element is located in a piRNA cluster on another chromosome (Watanabe et al., 2011). piRNAs have been implicated as guides in de novo methylation of transposable elements in male germ cells, although the underlying mechanism is unclear, demonstrating that some incidences of imprinting utilise components of a genome defence mechanism (Aravin and Bourc'his, 2008)
The methylation landscapes of the gametes
Recent whole-genome methylation profiling has revealed in exquisite detail the methylation landscapes of the gametes and the extent of the differences between oocyte and sperm that exist up to the point of fertilisation (Kobayashi et al., 2012; Erkek et al., 2013; Shirane et al., 2013). This knowledge has also generated new insights into the mechanisms of de novo methylation with implications for imprinting. With the exception of some CG-rich components of interspersed repeats that escape reprogramming in PGCs, the entire methylation landscape in oocytes is established de novo in the later stages of oocyte growth (Kobayashi et al., 2012; Shirane et al., 2013). In the male germline, de novo methylation may progress from its onset in pre-meiotic prospermatogonia across multiple cell divisions until spermiogenesis; the details of the progression in methylation across the genome remain to be elucidated. Ultimately, the sperm genome is more highly methylated than the oocyte genome; more accurately put, the distribution of methylation differs. In mature sperm, the genome is almost universally hypermethylated, with the notable exceptions of CpG islands, most of which remain unmethylated (Kobayashi et al., 2012; Erkek et al., 2013). In oocytes, in contrast, DNA methylation appears largely to be confined to active gene bodies, and this has clear resonance with the finding that the gDMRs of imprinted genes require transcription events for de novo methylation in oocytes (Chotalia et al., 2009). By this analysis, about half of the genome is differentially methylated between sperm and oocyte, so the potential for imprinting extends over a substantial fraction of the genome. Put another way, maternal gDMRs are among a class of CpG islands that are located within transcription-associated methylated domains in oocytes (about 6% of all CpG islands; Smallwood et al., 2011), but are unmethylated in sperm owing to the widespread hypomethylation of CpG islands. Paternal gDMRs are methylated in sperm because they are within the almost universal hypermethylated compartment of the sperm genome, but the corresponding regions in oocytes are within hypomethylated domains. These observations focus attention on how gamete-derived methylation is processed after fertilisation as being the basis for imprint specification.
Selection of imprints after fertilisation
The genome experiences wholesale epigenetic upheaval after fertilisation and during preimplantation development, a phenomenon known as reprogramming. The mechanisms responsible for reprogramming of DNA methylation are likely to be complex and involve a combination of ‘active' and ‘passive' processes, but are not yet fully understood. Moreover, oocyte- and sperm-derived genomes are subject to different demethylation regimes. Passive demethylation refers to the dilution of methylation at symmetric CpG sites because of failure to reinstate methylation on the nascent DNA strand at DNA replication owing to absence of the maintenance DNA methyltransferase Dnmt1. It is thought that demethylation of the oocyte-derived genome from the two-cell stage onwards can be accounted for largely by this passive route. The sperm-derived genome undergoes a more precipitate decline in methylation that is evident in the zygote even before pronuclear fusion (Santos et al., 2002). This early loss of methylation is an active process; subsequent reduction in methylation can be achieved passively. The mechanisms responsible for active demethylation of the sperm-derived genome are still hotly debated. It is clear that oxidation of 5-methylcytosine to 5-hydroxy-methylcytosine and further oxidation derivatives by the Tet3 enzyme is involved (Gu et al., 2011); complete removal of modified cytosine then requires activities of the base-excision repair pathway (Santos et al., 2013). However, there is evidence that the very first phase of active demethylation in the zygote is accomplished by another mechanism(s), the details of which remain to be elucidated (Santos et al., 2013). The lowest point in DNA methylation is attained by the time of blastocyst formation. Thereafter, de novo methylation takes place in a cell lineage-specific fashion (for example, the inner cell mass of the mouse blastocyst gaining methylation in advance of the trophectoderm) with both parental genomes being treated equally (Smith et al., 2012).
Imprinted genes are among the exceptions to these genome-wide demethylation events. Both maternal and paternal gDMRs require the presence of Dnmt1 for the maintenance of methylation (Howell et al., 2001; Hirasawa et al., 2008), suggesting the existence of factors that can selectively target Dnmt1 to DMRs. In addition, factors are necessary to prevent erasure of methylation of gDMRs, and different factors may be involved in protecting sperm-derived gDMRs in the zygote, and in protecting sperm- and oocyte-derived gDMRs from passive demethylation. One critical protein is PGC7 (also known as Stella and Dppa3). PGC7-null zygotes lose methylation at a selection of oocyte- and sperm-derived gDMRs (Nakamura et al., 2007). PGC7 is required to protect the maternal genome in the zygote against Tet3-dependent oxidation of 5-methylcytosine (Wossidlo et al., 2011); its targeting to specific sites in the maternal genome appears to rely on binding to histone 3 dimethylated at lysine 9 (H3K9me2) (Nakamura et al., 2012).
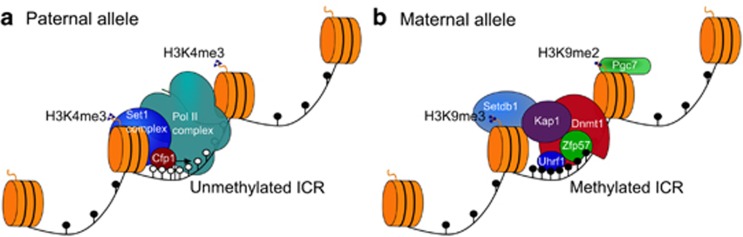
The selection to maintain methylation in the embryo is especially evident at the CpG islands that were methylated in gametes. Given that ∼2000 CpG islands are differentially methylated between the oocyte and sperm (Smallwood et al., 2011; Kobayashi et al., 2012), but a much smaller number (20–30) of gDMRs maintain allele-specific methylation after fertilisation and throughout life, there must be an effective, regulated and highly specific process by which imprints are selected in the early embryo. It has recently been shown that the process of selection and protection does involve a sequence-specific DNA binding factor, Zfp57, so that Zfp57 could be claimed to be a truly ‘imprint-specific' factor (Figure 2). Zfp57 is a Krüppel-associated box-containing zinc-finger protein whose involvement in imprinting was identified through genetic experiments in mice (Li et al., 2008) and from cases of the human imprinting disorder 6q24 transient neonatal diabetes mellitus that presented with loss of methylation at multiple imprinted loci (Mackay et al., 2008). Subsequently, it has been shown in embryonic stem cells that Zfp57 binds a hexanucleotide sequence found within all gDMRs, but only binds when the hexamer is methylated at the central CpG, and that ablation of Zfp57 causes loss of methylation of gDMRs (Quenneville et al., 2011). Zfp57 interacts with the multifunctional Kap1 repressor protein (also known as Trim28, Tif1β), which appears to act as a scaffold for other epigenetic modifying factors at the gDMR, including Dnmt1, its auxiliary factor Uhrf1/Np95, and the H3K9 methyltransferase Setdb1. In fact, Zfp57 binds hemimethylated DNA (Liu et al., 2012), so it seems to act as a lynchpin for the recruitment of the Kap1 complex to ensure post-replicative maintenance of DNA methylation and the reinstatement of the repressive histone modification H3K9me3 at gDMRs. Oocyte-derived Kap1 is essential for the maintenance of imprinted DMR methylation in the single-cell embryo before it is provided at zygotic genome activation (Messerschmidt et al., 2012). Therefore, the Zfp57:Kap1 complex may be one of the factors needed to target Dnmt1 to gDMRs at a time when it is largely excluded from the nucleus in the zygote and early preimplantation stages. Human and mouse Zfp57 are functionally interchangeable (Takikawa et al., 2013), but the possibility of partial redundancy is suggested by the fact that not all gDMRs are equally affected in human patients with ZFP57 mutations (Boonen et al., 2013). Zfp57 is part of the large superfamily of Krüppel-associated box-containing-zinc-finger proteins in mammals, so it is conceivable that other members of this family are involved in the maintenance of epigenetic states of gDMRs at later developmental stages or in tissues in which Zfp57 is no longer expressed. Additional factors, aside from the Zfp57:Kap1 complex, may contribute to gDMR methylation maintenance during the critical stages of preimplantation methylation reprogramming, but the extent to which they are specific for imprinted gDMRs is unclear.
Figure 2.
Factors involved in the maintenance of DNA methylation at imprinting control regions (ICRs) during early embryonic development. In the preimplantation embryo, the parental alleles of ICRs are bound by specific complexes to maintain their differential methylation status during the genome-wide epigenetic reprogramming. (a) The unmethylated ICR has the properties typical of a CpG island. It coincides with an expressed promoter and is, therefore, associated with the RNA polymerase II (pol II) complex. Unmethylated CpG sites are bound by the Cfp1 protein, which recruits the Set1 complex to trimethylate H3K4 which, in turn, reinforces the DNA hypomethylated state, as methylated H3K4 is antagonistic to binding of Dnmt3a/Dnmt3L. Protection against de novo DNA methylation of the unmethylated ICR allele could, therefore, be accomplished by generic factors acting at CpG islands, rather than imprint-specific factors. (b) The methylated ICR is bound by the Zfp57:Kap1 complex and Uhrf1 to direct the action of Dnmt1; the complex also interacts with the Setdb1 methyltransferase, which deposits the repressive histone modification H3K9me3. In addition, recognition of H3K9me2 by the PGC7/Stella protein may protect against demethylation in the zygote. Maintenance of DNA methylation at an ICR, therefore, involves the interplay between the imprint-specific factor Zfp57 and generic factors, such as Dnmt1.
An intriguing class of proteins required for imprinted methylation are members of the NLRP (nucleotide-binding oligomerisation domain, leucine-rich repeat and pyrin domain containing protein) family. Although this protein family is known for its role in innate immunity, several NLRP proteins are highly expressed in oocytes, are required for progression beyond the two-cell stage and define a sub-family of reproduction-related NLPRs that appear to have evolved rapidly in mammals (Tian et al., 2009). In humans, maternal-effect mutations in NLRP7 have been identified as a cause of biparental hydatidiform mole (Murdoch et al., 2006). Biparental hydatidiform moles are conceptuses that adopt an androgenetic-like pathology of excessive proliferation of extraembryonic trophoblast; although biparental, they are deficient in methylation at many or all maternal DMRs, with sperm-derived gDMRs apparently unaffected. Mutations have also been reported in NLRP2 as a cause of loss of methylation of the KvDMR in a Beckwith—Wiedemann syndrome kindred (Meyer et al., 2009), whereas in additional cases of biparental hydatidiform mole mutations have been found in C6orf221 (Parry et al., 2011). Although uncharacterised, the C6orf221 protein may interact in the same complex as NLRP7 or NLRP2. How a NLRP7/NLRP2/C6orf221-containing complex is involved in DNA methylation events is rather obscure at this time, and it has not been possible to conclude whether they have a role in establishment of methylation states in oocytes or faithful maintenance in preimplantation embryos. It is intriguing that these proteins could be involved in the formation of structural components of the oocyte cytoplasm (Li et al., 2008). It is possible that this complex ensures the correct localisation or nuclear translocation of the factors required for controlling DNA methylation events in the zygote or early cleavage embryo. Alternatively, NLRP7 and associated proteins may directly interact with DNA binding and DNA methylation modifying factors, such as YY1 (Mahadevan et al., 2013). Although the focus of the effects of these factors has been very much on imprinted gDMRs, they might have a wider role, as methylation anomalies at non-imprinted loci have been identified in a patient from a mother carrying an NLRP7 mutation (Beygo et al., 2013).
As well as ensuring the maintenance of gamete-derived methylation of one parental allele, selection for imprinting necessarily also involves the protection from gain of methylation of the opposing allele, particularly during developmental phases in which de novo methy-lation is most active. As the default state of a CpG island is hypomethylation, particularly for CpG islands associated with transcription start sites, and hypomethylated CpG islands are thought to be bound by multiple factors that provide a chromatin environment hostile to DNA methylation (such as methylated H3K4) (Blackledge and Klose, 2011; Deaton and Bird, 2011), it might be considered that protection against de novo methylation of the unmethylated copy could be accomplished by generic rather than imprint-specific protection factors (Figure 2). However, the situation might be more complex, with the identification of ‘transient' gDMRs. These are CpG islands that were methylated specifically in oocytes and protected during early embryogenesis by Zfp57 binding, but whose paternal allele gains methylation after implantation (Proudhon et al., 2012). This propensity to gain methylation may suggest that these elements are not conventional CpG islands, for example, in relation to their location within genes. Such transient DMRs may be associated with parental-allele-specific or skewed allelic expression early in embryogenesis, but there is also evidence that they can give rise to stable imprinting of linked genes, although the DMRs themselves lose monoallelic methylation during embryogenesis (Duffié et al., 2014).
Conservation of imprinting
As mentioned above, most of what we know about methylation landscapes and dynamics in germ cells and preimplantation embryos comes from studies done in the mouse. The assumption has been that the principles established in the mouse will be conserved across the eutherian lineage. Although the DNA methylation landscape of the oocyte is known so far only in the mouse, aside from an early report on the human SNRPN DMR (El-Maarri et al., 2001), the few DMRs tested in human oocytes or those from other mammals (for example, bovine) have shown the expected gametic methylation pattern (Geuns et al., 2003, 2007a, 2007b; El Hajj et al., 2011; Heinzmann et al., 2011; O'Doherty et al., 2012). It is also the case that the methylation maintenance factor Zfp57 is required for DMR methylation in both mouse and human (Li et al., 2008; Mackay et al., 2008). On the other hand, there are substantial differences among mammals in epigenetic and gene regulation mechanisms, particularly in the early preimplantation development period. For example, the mouse is relatively unusual in having zygotic genome activation at the two-cell stage, in comparison with many other mammals (eight-cell stage); mouse, but not human, has imprinted X-inactivation in extraembryonic tissues, and this imprinted phase is present prior to blastocyst formation (Patrat et al., 2009); the degree of methylation reprogramming and the contributions of active and passive processes may differ among mammalian species (Beaujean et al., 2004). Therefore, it should not be discounted that there could be differences in mechanisms and perhaps timing of imprint establishment among mammals.
Mechanistic opportunities for the evolution of new imprinted genes
With the new knowledge of the DNA methylation landscapes of the gametes, insights into the mechanisms responsible for de novo DNA methylation, and recognition of the importance of post-zygotic events for selecting and maintaining gDMR methylation, we can envisage a number of scenarios by which new imprinted genes could arise and which could contribute to species-specific imprinting. The guiding principle is that the opportunity to imprint is provided by differential methylation in the gametes, from which the selection for imprinting is made in the embryo. New opportunities to imprint could arise from alterations in the DNA methylation landscape in the oocyte or sperm, within a species or between species. As we have seen, DNA methylation in the oocyte is driven primarily by transcription events; consequently, activation of new transcription units or recruitment of new promoters would be predicted to create new hypermethylated domains, which could include new intragenic, methylated CpG islands. If such a CpG island contained a motif recognised by Zfp57, the new methylated state would provide the opportunity to maintain methylation allele-specifically in the embryo as a potential new gDMR. (The assumption here is that the default state of the CpG island in sperm is hypomethylation and that, as a CpG island, generic factors would tend to maintain the hypomethylated state of the paternally-derived allele in the embryo.) Comparative methylation and transcriptome analysis of oocytes would be useful in identifying whether this has taken place in the mammalian lineage. Imprinted retrogenes may have arisen by a related mechanism: retrotransposition events, that include CG-rich elements, into active transcription units may constitute the origin of new intragenic methylated CpG islands (Wood et al., 2007). Again, presence of Zfp57-motifs would be required to ensure retention of gamete-derived methylation in the embryo. Comparative analysis in multiple species of retrotransposition events in relation to transcription units active in oocytes and distribution of Zfp57 binding motifs would help test this prediction.
Returning to the notion of directly and indirectly imprinted genes, it is interesting to consider implications for the ‘speed' of evolution of imprinting at individual loci. It is possible that directly imprinted genes (Figure 1a) acquire imprinted monoallelic expression in a single step. For example, if a new methylated CpG island emerged as a result of an altered transcription unit in oocytes or from a retrotransposition event, the expectation is that there would be an immediate, absolute difference in methylation of the two parental alleles in resulting offspring (providing Zfp57 binding sites are present). In fact, for such events, it is more difficult to conceive how differential gametic methylation could arise gradually over generations. A consequence would be that any promoter associated with the newly methylated CpG island would be monoallelically expressed, and the yield of transcript from that promoter would be half of the non-imprinted state. For a dosage-sensitive protein-coding gene, this is more likely to be sustainable if the new gDMR did not overlie the sole promoter (many genes have multiple promoters)—further evolution of the contributions of the imprinted and non-imprinted promoters could occur depending on selective pressures, with imprinted expression becoming the predominant mode as one possible outcome. Alternatively, the newly emerged primary imprint could provide the source of a non-coding transcript that could indirectly imprint the gene in which it resides and/or influence the allelic balance in expression of flanking genes (Figure 1b). This indirect imprinting could provide the opportunity for gradual evolution of monoallelic expression at a locus and accrual of imprinting across a cluster of genes until an evolutionary stable state is attained. Indeed, there have been observations of polymorphic imprinting in humans that could support this process. Monoallelic maternal expression of the IGF2R gene has been observed in a subset of human placental samples (Xu et al., 1993; Monk et al., 2006); however, the differential methylation at an intronic CpG island (corresponding to the ICR in mouse) is present irrespective of monoallelic or biallelic expression of IGF2R (Monk et al., 2006), suggesting that secondary epigenetic modifications may be acquired in some individuals. The possibilities of immediate and progressive acquisition of imprinted monoallelic expression might be useful to consider in relation to theories seeking to explain the evolution of imprinting.
Data Archiving
There were no data to deposit.
Acknowledgments
We thank members of NESCent and the working group on ‘Testing theories for the evolution of genomic imprinting' for a stimulating atmosphere for discussion, and Anne Ferguson-Smith in particular for encouragement. Research in GK's group is funded by the Biotechnology and Biological Sciences Research Council and the Medical Research Council of the UK, and by the European Union through the INGENIUM Initial Training Network.
The authors declare no conflicts of interest.
References
- Aravin AA, Bourc'his D. Small RNA guides for de novo DNA methylation in mammalian germ cells. Genes Dev. 2008;22:970–975. doi: 10.1101/gad.1669408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaujean N, Hartshorne G, Cavilla J, Taylor J, Gardner J, Wilmut I, et al. Non-conservation of mammalian preimplantation methylation dynamics. Curr Biol. 2004;14:R266–R267. doi: 10.1016/j.cub.2004.03.019. [DOI] [PubMed] [Google Scholar]
- Beygo J, Ammerpohl O, Gritzan D, Heitmann M, Rademacher K, Richter J, et al. Deep bisulfite sequencing of aberrantly methylated Loci in a patient with multiple methylation defects. PLoS One. 2013;8:e76953. doi: 10.1371/journal.pone.0076953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackledge NP, Klose R. CpG island chromatin: a platform for gene regulation. Epigenetics. 2011;6:147–152. doi: 10.4161/epi.6.2.13640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boonen SE, Mackay DJ, Hahnemann JM, Docherty L, Grønskov K, Lehmann A, et al. Transient neonatal diabetes, ZFP57, and hypomethylation of multiple imprinted loci: a detailed follow-up. Diabetes Care. 2013;36:505–512. doi: 10.2337/dc12-0700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourc'his D, Bestor TH. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature. 2004;431:96–99. doi: 10.1038/nature02886. [DOI] [PubMed] [Google Scholar]
- Bourc'his D, Xu GL, Lin CS, Bollman B, Bestor TH. Dnmt3L and the establishment of maternal genomic imprints. Science. 2001;294:2536–2539. doi: 10.1126/science.1065848. [DOI] [PubMed] [Google Scholar]
- Chotalia M, Smallwood SA, Ruf N, Dawson C, Lucifero D, Frontera M, et al. Transcription is required for establishment of germline methylation marks at imprinted genes. Genes Dev. 2009;23:105–117. doi: 10.1101/gad.495809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis TL, Yang GJ, McCarrey JR, Bartolomei MS. The H19 methylation imprint is erased and re-established differentially on the parental alleles during male germ cell development. Hum Mol Genet. 2000;9:2885–2894. doi: 10.1093/hmg/9.19.2885. [DOI] [PubMed] [Google Scholar]
- Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25:1010–1022. doi: 10.1101/gad.2037511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffié R, Ajjan S, Greenberg MV, Zamudio N, Escamilla Del Arenal M, Iranzo J, et al. The Gpr1/Zdbf2 locus provides new paradigms for transient and dynamic genomic imprinting in mammals. Genes Dev. 2014;28:463–478. doi: 10.1101/gad.232058.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Hajj N, Trapphoff T, Linke M, May A, Hansmann T, Kuhtz J, et al. Limiting dilution bisulfite (pyro)sequencing reveals parent-specific methylation patterns in single early mouse embryos and bovine oocytes. Epigenetics. 2011;6:1176–1188. doi: 10.4161/epi.6.10.17202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Maarri O, Buiting K, Peery EG, Kroisel PM, Balaban B, Wagner K, et al. Maternal methylation imprints on human chromosome 15 are established during or after fertilization. Nat Genet. 2001;27:341–344. doi: 10.1038/85927. [DOI] [PubMed] [Google Scholar]
- Erkek S, Hisano M, Liang CY, Gill M, Murr R, Dieker J, et al. Molecular determinants of nucleosome retention at CpG-rich sequences in mouse spermatozoa. Nat Struct Mol Biol. 2013;20:868–875. doi: 10.1038/nsmb.2599. [DOI] [PubMed] [Google Scholar]
- Ferguson-Smith AC. Genomic imprinting: the emergence of an epigenetic paradigm. Nat Rev Genet. 2011;12:565–575. doi: 10.1038/nrg3032. [DOI] [PubMed] [Google Scholar]
- Geuns E, De Rycke M, Van Steirteghem A, Liebaers I. Methylation imprints of the imprint control region of the SNRPN-gene in human gametes and preimplantation embryos. Hum Mol Genet. 2003;12:2873–2879. doi: 10.1093/hmg/ddg315. [DOI] [PubMed] [Google Scholar]
- Geuns E, De Temmerman N, Hilven P, Van Steirteghem A, Liebaers I, De Rycke M. Methylation analysis of the intergenic differentially methylated region of DLK1-GTL2 in human. Eur J Hum Genet. 2007;15:352–361. doi: 10.1038/sj.ejhg.5201759. [DOI] [PubMed] [Google Scholar]
- Geuns E, Hilven P, Van Steirteghem A, Liebaers I, De Rycke M. Methylation analysis of KvDMR1 in human oocytes. J Med Genet. 2007;44:144–147. doi: 10.1136/jmg.2006.044149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu TP, Guo F, Yang H, Wu HP, Xu GF, Liu W, et al. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature. 2011;477:606–610. doi: 10.1038/nature10443. [DOI] [PubMed] [Google Scholar]
- Hackett JA, Sengupta R, Zylicz JJ, Murakami K, Lee C, Down TA, et al. Germline DNA demethylation dynamics and imprint erasure through 5-hydroxymethylcytosine. Science. 2013;339:448–452. doi: 10.1126/science.1229277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata K, Okano M, Lei H, Li E. Dnmt3L cooperates with the Dnmt3 family of de novo DNAmethyltransferases to establish maternal imprints in mice. Development. 2002;129:1983–1893. doi: 10.1242/dev.129.8.1983. [DOI] [PubMed] [Google Scholar]
- Heinzmann J, Hansmann T, Herrmann D, Wrenzycki C, Zechner U, Haaf T, et al. Epigenetic profile of developmentally important genes in bovine oocytes. Mol Reprod Dev. 2011;78:188–201. doi: 10.1002/mrd.21281. [DOI] [PubMed] [Google Scholar]
- Henckel A, Chebli K, Kota SK, Arnaud P, Feil R. Transcription and histone methylation changes correlate with imprint acquisition in male germ cells. EMBO J. 2011;31:606–615. doi: 10.1038/emboj.2011.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirasawa R, Chiba H, Kaneda M, Tajima S, Li E, Jaenisch R, et al. Maternal and zygotic Dnmt1 are necessary and sufficient for the maintenance of DNA methylation imprints during preimplantation development. Genes Dev. 2008;22:1607–1616. doi: 10.1101/gad.1667008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiura H, Obata Y, Komiyama J, Shirai M, Kono T. Oocyte growth-dependent progression of maternal imprinting in mice. Genes Cells. 2006;11:353–361. doi: 10.1111/j.1365-2443.2006.00943.x. [DOI] [PubMed] [Google Scholar]
- Howell CY, Bestor TH, Ding F, Latham KE, Mertineit C, Trasler JM, et al. Genomic imprinting disrupted by a maternal effect mutation in the Dnmt1 gene. Cell. 2001;104:829–838. doi: 10.1016/s0092-8674(01)00280-x. [DOI] [PubMed] [Google Scholar]
- Kaneda M, Hirasawa R, Chiba H, Okano M, Li E, Sasaki H. Genetic evidence for Dnmt3a-dependent imprinting during oocyte growth obtained by conditional knockout with Zp3-Cre and complete exclusion of Dnmt3b by chimera formation. Genes Cells. 2010;15:169–179. doi: 10.1111/j.1365-2443.2009.01374.x. [DOI] [PubMed] [Google Scholar]
- Kaneda M, Okano M, Hata K, Sado T, Tsujimoto N, Li E, et al. Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature. 2004;429:900–903. doi: 10.1038/nature02633. [DOI] [PubMed] [Google Scholar]
- Kato Y, Kaneda M, Hata K, Kumaki K, Hisano M, Kohara Y, et al. Role of the Dnmt3 family in de novo methylation of imprinted and repetitive sequences during male germ cell development in the mouse. Hum Mol Genet. 2007;16:2272–2280. doi: 10.1093/hmg/ddm179. [DOI] [PubMed] [Google Scholar]
- Kelsey G, Feil R. New insights into establishment and maintenance of DNA methylation imprints in mammals. Philos Trans R Soc Lond B Biol Sci. 2013;368:20110336. doi: 10.1098/rstb.2011.0336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi H, Sakurai T, Imai M, Takahashi N, Fukuda A, Yayoi O, et al. Contribution of intragenic DNA methylation in mouse gametic DNA methylomes to establish oocyte-specific heritable marks. PLoS Genet. 2012;8:e1002440. doi: 10.1371/journal.pgen.1002440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi H, Suda C, Abe T, Kohara Y, Ikemura T, Sasaki H. Bisulfite sequencing and dinucleotide content analysis of 15 imprinted mouse differentially methylated regions (DMRs): paternally methylated DMRs contain less CpGs than maternally methylated DMRs. Cytogenet Genome Res. 2006;113:130–137. doi: 10.1159/000090824. [DOI] [PubMed] [Google Scholar]
- Lewis A, Mitsuya K, Umlauf D, Smith P, Dean W, Walter J, et al. Imprinting on distal chromosome 7 in the placenta involves repressive histone methylation independent of DNA methylation. Nat Genet. 2004;36:1291–1295. doi: 10.1038/ng1468. [DOI] [PubMed] [Google Scholar]
- Li L, Baibakov B, Dean J. A subcortical maternal complex essential for preimplantation mouse embryogenesis. Dev Cell. 2008;15:416–425. doi: 10.1016/j.devcel.2008.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim AK, Lorthongpanich C, Chew TG, Tan CW, Shue YT, Balu S, et al. The nuage mediates retrotransposon silencing in mouse primordial ovarian follicles. Development. 2013;140:3819–3825. doi: 10.1242/dev.099184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Ito M, Zhou F, Youngson N, Zuo X, Leder P, et al. A maternal-zygotic effect gene, Zfp57, maintains both maternal and paternal imprints. Dev Cell. 2008;15:547–557. doi: 10.1016/j.devcel.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucifero D, Mann MR, Bartolomei MS, Trasler JM. Gene-specific timing and epigenetic memory in oocyte imprinting. Hum Mol Genet. 2004;13:839–849. doi: 10.1093/hmg/ddh104. [DOI] [PubMed] [Google Scholar]
- Liu Y, Toh H, Sasaki H, Zhang X, Cheng X. An atomic model of Zfp57 recognition of CpG methylation within a specific DNA sequence. Genes Dev. 2012;26:2374–2379. doi: 10.1101/gad.202200.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay DJ, Callaway JL, Marks SM, White HE, Acerini CL, Boonen SE, et al. Hypomethylation of multiple imprinted loci in individuals with transient neonatal diabetes is associated with mutations in ZFP57. Nat Genet. 2008;40:949–951. doi: 10.1038/ng.187. [DOI] [PubMed] [Google Scholar]
- Mahadevan S, Wen S, Wan YW, Peng HH, Otta S, Liu Z, et al. NLRP7 affects trophoblast lineage differentiation, binds to overexpressed YY1 and alters CpG methylation. Hum Mol Genet. 2013. [DOI] [PMC free article] [PubMed]
- Meyer E, Lim D, Pasha S, Tee LJ, Rahman F, Yates JR, et al. Germline mutation in NLRP2 (NALP2) in a familial imprinting disorder (Beckwith-Wiedemann Syndrome) PLoS Genet. 2009;5:e1000423. doi: 10.1371/journal.pgen.1000423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messerschmidt DM, de Vries W, Ito M, Solter D, Ferguson-Smith A, Knowles BB. Trim28 is required for epigenetic stability during mouse oocyte to embryo transition. Science. 2012;335:1499–1502. doi: 10.1126/science.1216154. [DOI] [PubMed] [Google Scholar]
- Monk D, Arnaud P, Apostolidou S, Hills FA, Kelsey G, Stanier P, et al. Limited evolutionary conservation of imprinting in the human placenta. Proc Natl Acad Sci USA. 2006;103:6623–6628. doi: 10.1073/pnas.0511031103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murdoch S, Djuric U, Mazhar B, Seoud M, Khan R, Kuick R, et al. Mutations in NALP7 cause recurrent hydatidiform moles and reproductive wastage in humans. Nat Genet. 2006;38:300–302. doi: 10.1038/ng1740. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Arai Y, Umehara H, Masuhara M, Kimura T, Taniguchi H, et al. PGC7/Stella protects against DNA demethylation in early embryogenesis. Nat Cell Biol. 2007;9:64–71. doi: 10.1038/ncb1519. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Liu YJ, Nakashima H, Umehara H, Inoue K, Matoba S, et al. PGC7 binds histone H3K9me2 to protect against conversion of 5mC to 5hmC in early embryos. Nature. 2012;486:415–419. doi: 10.1038/nature11093. [DOI] [PubMed] [Google Scholar]
- Neumann B, Kubicka P, Barlow DP. Characteristics of imprinted genes. Nat Genet. 1995;9:12–13. doi: 10.1038/ng0195-12. [DOI] [PubMed] [Google Scholar]
- O'Doherty AM, O'Shea LC, Fair T. Bovine DNA methylation imprints are established in an oocyte size-specific manner, which are coordinated with the expression of the DNMT3 family proteins. Biol Reprod. 2012;86:67. doi: 10.1095/biolreprod.111.094946. [DOI] [PubMed] [Google Scholar]
- Parry DA, Logan CV, Hayward BE, Shires M, Landolsi H, Diggle C, et al. Mutations causing familial biparental hydatidiform mole implicate c6orf221 as a possible regulator of genomic imprinting in the human oocyte. Am J Hum Genet. 2011;89:451–458. doi: 10.1016/j.ajhg.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrat C, Okamoto I, Diabangouaya P, Vialon V, Le Baccon P, Chow J, et al. Dynamic changes in paternal X-chromosome activity during imprinted X-chromosome inactivation in mice. Proc Natl Acad Sci USA. 2009;106:5198–5203. doi: 10.1073/pnas.0810683106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudhon C, Duffié R, Ajjan S, Cowley M, Iranzo J, Carbajosa G, et al. Protection against de novo methylation is instrumental in maintaining parent-of-origin methylation inherited from the gametes. Mol Cell. 2012;47:909–920. doi: 10.1016/j.molcel.2012.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quenneville S, Verde G, Corsinotti A, Kapopoulou A, Jakobsson J, Offner S, et al. In embryonic stem cells, ZFP57/KAP1 recognize a methylated hexanucleotide to affect chromatin and DNA methylation of imprinting control regions. Mol Cell. 2011;44:361–372. doi: 10.1016/j.molcel.2011.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos F, Peat J, Burgess H, Rada C, Reik W, Dean W. Active demethylation in mouse zygotes involves cytosine deamination and base excision repair. Epigenetics Chromatin. 2013;6:39. doi: 10.1186/1756-8935-6-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos F, Hendrich B, Reik W, Dean W. Dynamic reprogramming of DNA methylation in the early mouse embryo. Dev Biol. 2002;241:172–182. doi: 10.1006/dbio.2001.0501. [DOI] [PubMed] [Google Scholar]
- Sasaki H, Matsui Y. Epigenetic events in mammalian germ-cell development: reprogramming and beyond. Nat Rev Genet. 2008;9:129–140. doi: 10.1038/nrg2295. [DOI] [PubMed] [Google Scholar]
- Schulz R, Proudhon C, Bestor TH, Woodfine K, Lin CS, Lin SP, et al. The parental non-equivalence of imprinting control regions during mammalian development and evolution. PLoS Genet. 2010;6:e1001214. doi: 10.1371/journal.pgen.1001214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seisenberger S, Andrews S, Krueger F, Arand J, Walter J, Santos F, et al. The dynamics of genome-wide DNA methylation reprogramming in mouse primordial germ cells. Mol Cell. 2012;48:849–862. doi: 10.1016/j.molcel.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirane K, Toh H, Kobayashi H, Miura F, Chiba H, Ito T, et al. Mouse oocyte methylomes at base resolution reveal genome-wide accumulation of non-CpG methylation and role of DNA methyltransferases. PLoS Genet. 2013;9:e1003439. doi: 10.1371/journal.pgen.1003439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P, Li AX, Tran DA, Oates N, Kang ER, Wu X, et al. De novo DNA methylation in the male germ line occurs by default but is excluded at sites of H3K4 methylation. Cell Rep. 2013;4:205–219. doi: 10.1016/j.celrep.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleutels F, Barlow DP. The origins of genomic imprinting in mammals. Adv Genet. 2002;46:119–163. doi: 10.1016/s0065-2660(02)46006-3. [DOI] [PubMed] [Google Scholar]
- Smallwood SA, Kelsey G. De novo DNA methylation: a germ cell perspective. Trends Genet. 2012;28:33–42. doi: 10.1016/j.tig.2011.09.004. [DOI] [PubMed] [Google Scholar]
- Smallwood SA, Tomizawa S, Krueger F, Ruf N, Carli N, Segonds-Pichon A, et al. Dynamic CpG island methylation landscape in oocytes and preimplantation embryos. Nat Genet. 2011;43:811–814. doi: 10.1038/ng.864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith EY, Futtner CR, Chamberlain SJ, Johnstone KA, Resnick JL. Transcription is required to establish maternal imprinting at the Prader-Willi syndrome and Angelman syndrome locus. PLoS Genet. 2011;7:e1002422. doi: 10.1371/journal.pgen.1002422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RJ, Dean W, Konfortova G, Kelsey G. Identification of novel imprinted genes in a genome-wide screen for maternal methylation. Genome Res. 2003;13:558–569. doi: 10.1101/gr.781503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ZD, Chan MM, Mikkelsen TS, Gu H, Gnirke A, Regev A, et al. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature. 2012;484:339–344. doi: 10.1038/nature10960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stöger R, Kubicka P, Liu CG, Kafri T, Razin A, Cedar H, et al. Maternal-specific methylation of the imprinted mouse Igf2r locus identifies the expressed locus as carrying the imprinting signal. Cell. 1993;73:61–71. doi: 10.1016/0092-8674(93)90160-r. [DOI] [PubMed] [Google Scholar]
- Takikawa S, Wang X, Ray C, Vakulenko M, Bell FT, Li X. Human and mouse ZFP57 proteins are functionally interchangeable in maintaining genomic imprinting at multiple imprinted regions in mouse ES cells. Epigenetics. 2013;8:1268–1279. doi: 10.4161/epi.26544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X, Pascal G, Monget P. Evolution and functional divergence of NLRP genes in mammalian reproductive systems. BMC Evol Biol. 2009;9:202. doi: 10.1186/1471-2148-9-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomizawa S, Kobayashi H, Watanabe T, Andrews S, Hata K, Kelsey G, et al. Dynamic stage-specific changes in imprinted differentially methylated regions during early mammalian development and prevalence of non-CpG methylation in oocytes. Development. 2011;138:811–820. doi: 10.1242/dev.061416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan LB, Bartolomei MS. Regulation of imprinting in clusters: noncoding RNAs versus insulators. Adv Genet. 2008;61:207–223. doi: 10.1016/S0065-2660(07)00007-7. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Tomizawa S, Mitsuya K, Totoki Y, Yamamoto Y, Kuramochi-Miyagawa S, et al. Role of piRNA and non-coding RNA in de novo DNA methylation of the imprinted mouse Rasgrf1 locus. Science. 2011;332:848–852. doi: 10.1126/science.1203919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienholz BL, Kareta MS, Moarefi AH, Gordon CA, Ginno PA, Chédin F. DNMT3L modulates significant and distinct flanking sequence preference for DNA methylation by DNMT3A and DNMT3B in vivo. PLoS Genet. 2010;6:e1001106. doi: 10.1371/journal.pgen.1001106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson CM, Blake A, Thomas S, Beechey CV, Hancock J, Cattanach BM, et al. 2013Mouse Imprinting Data and References. MRC Harwell: Oxfordshire, UK http://www.har.mrc.ac.uk/research/genomic_imprinting/ .
- Wood AJ, Schulz R, Woodfine K, Koltowska K, Beechey CV, Peters J, et al. A screen for retrotransposed imprinted genes reveals an association between X chromosome homology and maternal germ-line methylation. PLoS Genet. 2007;3:e20. doi: 10.1371/journal.pgen.0030020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wossidlo M, Nakamura T, Lepikhov K, Marques CJ, Zakhartchenko V, Boiani M, et al. 5-Hydroxymethylcytosine in the mammalian zygote is linked with epigenetic reprogramming. Nat Commun. 2011;2:241. doi: 10.1038/ncomms1240. [DOI] [PubMed] [Google Scholar]
- Xu Y, Goodyer CG, Deal C, Polychronakos C. Functional polymorphism in the parental imprinting of the human IGF2R gene. Biochem Biophys Res Commun. 1993;197:747–754. doi: 10.1006/bbrc.1993.2542. [DOI] [PubMed] [Google Scholar]
- Yamaguchi S, Shen L, Liu Y, Sendler D, Zhang Y. Role of Tet1 in erasure of genomic imprinting. Nature. 2013;504:460–464. doi: 10.1038/nature12805. [DOI] [PMC free article] [PubMed] [Google Scholar]