Aaron Jex and colleagues report whole-genome sequencing of adult male and female whipworm, Trichuris suis, from experimentally infected pigs. They also report stage-, sex- and tissue-specific transcriptomes and provide insights into host-parasite interactions.
Supplementary information
The online version of this article (doi:10.1038/ng.3012) contains supplementary material, which is available to authorized users.
Subject terms: Genomics, Transcriptomics, Parasite biology
Abstract
Trichuris (whipworm) infects 1 billion people worldwide and causes a disease (trichuriasis) that results in major socioeconomic losses in both humans and pigs. Trichuriasis relates to an inflammation of the large intestine manifested in bloody diarrhea, and chronic disease can cause malnourishment and stunting in children. Paradoxically, Trichuris of pigs has shown substantial promise as a treatment for human autoimmune disorders, including inflammatory bowel disease (IBD) and multiple sclerosis. Here we report whole-genome sequencing at ∼140-fold coverage of adult male and female T. suis and ∼80-Mb draft assemblies. We explore stage-, sex- and tissue-specific transcription of mRNAs and small noncoding RNAs.
Supplementary information
The online version of this article (doi:10.1038/ng.3012) contains supplementary material, which is available to authorized users.
Main
Soil-transmitted helminths, including whipworm (Trichuris), hookworms (Necator and Ancylostoma) and the large roundworm (Ascaris), are among the most prevalent and devastating parasites of humans globally and predominate in impoverished nations1. Trichuris infects 1 billion people, and chronic infection of high intensity can lead to typhlitis, colitis, chronic dysentery and malnutrition through malabsorption as well as reduced physical and cognitive development2. Consequently, trichuriasis, which disproportionately affects children, has an estimated global burden of 1 million–6.5 million disability-adjusted life years, exceeding that of schistosomiasis, trachomiasis, trypanosomiasis or leishmaniasis1. Despite this, Trichuris species are classified by the World Health Organization as neglected parasites in urgent need of improved control3.
Contrasting with the substantial burden of trichuriasis and other neglected helminths is the observation that human populations in endemic countries tend to suffer from substantially fewer immunopathological diseases4, which are common and increasingly prevalent5 in countries in which exposure to pathogens is limited. These observations have inspired the 'hygiene hypothesis'6, which proposes that a lack of exposure of humans to common pathogens impairs immune function and leads to increased autoimmune disease. This hypothesis is supported by clinical data, with routine deworming positively7 and early-childhood helminth infection negatively8 correlating with autoimmune disorders. Recent studies have shown that porcine Trichuris (T. suis) administered to humans suffering from IBD (including Crohn's disease and ulcerative colitis) can reduce clinical symptoms9,10. Similar observations have been made in patients with multiple sclerosis11. Although helminths can alter immune responses in their hosts via a variety of excretory-secretory (ES) molecules12, the specific interactions between T. suis and its host remain unclear. By sequencing the T. suis genome and transcriptomes (mRNAs and small RNAs), we provide deep insights into the molecular biology of this parasite and its modulation of host immune responses. These data provide a solid basis for exploring human trichuriasis, developing new anti-parasitic drugs and elucidating how helminths suppress autoimmune disorders.
Results
Sequencing, assembly and synteny
We sequenced the genomes of single adult female and male T. suis at ∼140-fold coverage, producing draft assemblies of 76 and 81 Mb, respectively (Table 1, Supplementary Figs. 1 and 2 and Supplementary Tables 1 and 2). Matches to conserved eukaryotic genes indicated that each assembly is 96% complete, with minimal redundancy (Supplementary Table 3). Alignment of these assemblies showed high similarity, with 68 Mb aligning as direct one-to-one matches in blocks of a mean length of 2.5 kb. Overall, sequence identity was 99.2% (38,854 SNPs). Despite the reported XX and XY karyotypes for female and male Trichuris, respectively13, we found no evidence for a Y chromosome among the male-specific scaffolds, suggesting that this chromosome contains largely repetitive sequences common to both sexes; this finding is consistent with observations made of the T. suis karyotype, suggesting that the sex chromosomes were the smallest chromosomal pair and were morphologically very similar in both sexes13. Repetitive sequences comprise ∼32% of the genome, including 8% DNA transposable elements, 2.9% long tandem repeats and 3.3% retrotransposons (Supplementary Table 4). Each genome encoded ∼1,000 transfer RNA (tRNA) genes, with copy numbers reflecting codon usage in protein-encoding regions (Supplementary Fig. 3 and Supplementary Tables 5 and 6).
Table 1.
Features of the scaffolded assembly of the adult male and female T. suis genomes
| Male genome | Female genome | |
|---|---|---|
| k-mer (17 nucleotides) estimated genome size (in Mb) | 83.6 | 87.2 |
| Total read data; estimated coverage | 11.73 Gb; 140× | 12.36 Gb; 142× |
| Total scaffolded assembly size (Mb); total scaffolds | 81.3; 60,856 | 76.0; 42,663 |
| Total scaffolds of >200 bp: length (Mb); no. of scaffolds | 74.2; 4,293 | 71.0; 3,288 |
| Largest scaffold (Mb) | 1.59 | 1.44 |
| N50 in kb (scaffolds >200 bp)a | 500 | 440 |
| N90 in kb (scaffolds >200 bp); total number for >N90a | 81.0; 185 | 104; 168 |
| % GC content, whole genome; scaffolds >200 bp) | 43.9; 43.6 | 43.6; 43.5 |
| % repetitive sequence (scaffolds >200 bp) | 31.7 | 32.3 |
| Proportion of the genome that is coding, exonic; including introns | 26.2; 66.2 | 29.2; 73.4 |
| Number of protein-encoding genes | 14,781 | 14,470 |
| Mean gene length (kb) | 3.7 | 3.9 |
| Mean number of exons per gene | 5.4 | 5.7 |
| Mean exon length (bp) | 271 | 270 |
| Mean number of introns per gene | 4.4 | 4.7 |
| Mean intron length (bp) | 511 | 509 |
| Number of transfer RNAs | 991 | 1,021 |
aN50 and N90 denote that 50% and 90% of assembly, respectively, is represented by scaffolds of at least this size.
Protein-encoding gene set
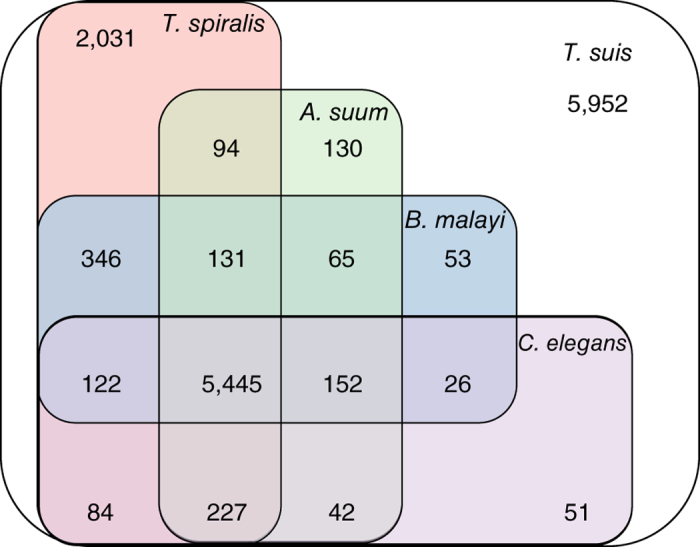
The female and male T. suis genomes encode at least 14,470 and 14,781 protein-encoding genes, respectively, representing ∼70% of each genome, including introns and exons. We identified 14,356 and 14,315 female and male genes with an ortholog or homolog in the opposite sex, with 10,403 genes being defined as unambiguous one-to-one orthologs. Evidence for sex-specific genes was limited, with just one and 41 supported as female and male specific, respectively (see Supplementary Note). Of these sex-specific genes, only three male genes have a predicted function, having homology to C. elegans frk-1 (encoding a receptor tyrosine-kinase), gpc-1 (encoding a G protein-coupled receptor (GPCR)) and his-66 (encoding a histone protein), respectively. The sex-specific genes show no clustering among scaffolds, providing little evidence for their association with the sex chromosomes. Most of the remaining differences in the genes of the two genders relate to a higher copy number of some genes in the male. From both assemblies, we defined a unified set of 14,820 genes for T. suis (Table 1 and Supplementary Table 7), with 12,910 (87.1%) supported by high-throughput RNA sequencing (RNA-seq) data. The majority (59.8%) of these genes have homologs (BLASTp cut-off: 1 × 10−5) in other nematodes, including 6,286 (42.4%), 6,340 (42.7%), 6,149 (41.5%) and 8,480 (57.2%) in Ascaris suum14, Brugia malayi15, Caenorhabditis elegans16 and Trichinella spiralis17, respectively (Fig. 1). Functions were assigned to 9,342 (63.0%) protein-encoding genes (Supplementary Tables 8, 9, 10, 11, 12, 13). Focusing on key functional or druggable proteins14, we predicted 653 peptidases and 288 peptidase inhibitors. Peptidase classes S1 (116) and S8 (42) are expanded in T. suis compared with those represented in other nematode genomes14,15,16,17. The T. suis genome also encodes 269 phosphatases and 232 kinases. We identified a large complement of receptors and transporters18; these molecules include 228 GPCRs, as well as 1,962 channel, pore and transporter proteins. Among the last group are 133 peroxisomal protein importers, more than in A. suum (n = 74) (ref. 14), which suggests a greater importance of fatty-acid digestion and metabolism in T. suis.
Figure 1.
Homologs shared between T. suis (class Enoplea, order Trichocephalida) and related nematode species.
We predicted 618 canonical ES proteins in adult T. suis (Supplementary Tables 14 and 15), including 165 proteases, many of which might have a role in disrupting intestinal epithelial cells in the host19,20 and in the formation of the syncytial tunnel around the Trichuris stichosome21. Notable among these molecules are 33 chymotrypsin-like serine proteases, which have key roles in helminths associated with host invasion22, immunosuppression23 and tissue destruction24. In addition to proteases, non–membrane-bound transporters comprise a major component of the secretome. These transporters include 41 pore-forming toxins (porins), 25 of which have homology to the Trichuris trichiura porin TT47, which induces ion-conducting pores in planar lipid bilayers and assists in the formation of the syncytial tunnel in the intestinal epithelium25. Helminth-mediated immunomodulation by ES products is well documented12. Among the predicted T. suis ES proteins, we found a variety of immunomodulators (Supplementary Table 16). On the basis of these findings and available literature for helminths12,26,27, we propose a Trichuris-driven immunomodulation model (Supplementary Fig. 4), in which the parasite suppresses inflammation by secreting (i) serpins to inhibit neutrophil cathepsins and elastases; (ii) apyrases to prevent conversion of regulatory T cells to pro-inflammatory T cells; (iii) cystatins to promote anti-inflammatory (producing interleukin-4 (IL-4) and IL-10) T cells by disrupting antigen presentation by dendritic and B cells; (iv) calreticulins that bind to dendritic cells and stimulate IL-4 production and limit inflammation by binding free calcium ions; and (v) molecular mimics12 of host galectins, mammalian macrophage inhibitory factor and tumor growth factor-β that stimulate apoptosis in activated T cells, promote alternative activation of macrophages and block the stimulation of (proinflammatory) Toll-like receptor pathways. This model is consistent with the pathophysiology described for Trichuris infection26, and probably operates in tandem with immunosuppressive processes linked to glycans27 and lipids (for example, sphingolipids; see Supplementary Note).
Transcriptome and differential transcription
We explored stage-, sex- and tissue-specific transcription (mRNAs), with a focus on parasite-host interactions. We predicted 36,763 transcripts, with 15,174 of them being perfect matches to the intron/exon chain (excluding UTRs), as annotated in the genome, and 21,589 representing novel splice isoforms (Supplementary Fig. 5 and Supplementary Table 17). In total, 6,293 (43.7%) T. suis genes were predicted to encode at least two isoforms. The number of splice-isoforms per gene only moderately correlated with exon number (R2 = 0.44; Supplementary Figs. 6 and 7). Alternative splicing correlated with gene function, reflected in an enrichment (P ≤ 0.05; Pearson's chi-squared analysis) of protein catabolism (i.e., proteases), membrane-bound ion transport and kinase activity among alternatively spliced genes and apoptosis among single splice-isoform genes. Genes with multiple and single splice isoforms also differed in their conservation and predicted essentiality. Of the 6,307 T. suis genes with C. elegans homologs, alternatively spliced genes predominated by a three-to-one margin (4,510 versus 1,875) and represented 204 (75.8%) of the 269 essential genes predicted. The latter observation needs to be considered when mining helminth genomes for novel drug targets. If a novel inhibitor targeting products of such genes interacts with one of the spliced domains, isoform switching may be sufficient to overcome its effect.
Some protein domains were significantly associated (P ≤ 0.05; Pearson's chi-squared analysis) with specific, alternative splice events, with exon skipping and the use of alternative first or last exons appearing to differ in their functional implications for transcription (Fig. 2a). Most notable was an over-representation of substrate-binding motifs (for example, immunoglobin, EGF-like or DnaJ) for genes biased toward transcripts with skipped exons. Remodeling of binding-motif structure through alternative splicing affects binding specificity in other organisms28, and we propose that exon skipping is important in regulating binding specificity of proteins in T. suis. Given the varied functions associated with alternative first- or last-exon splicing events (Fig. 2a), we hypothesize that these specific modifications might play a part in regulating protein localization, another known role for alternative splicing28.
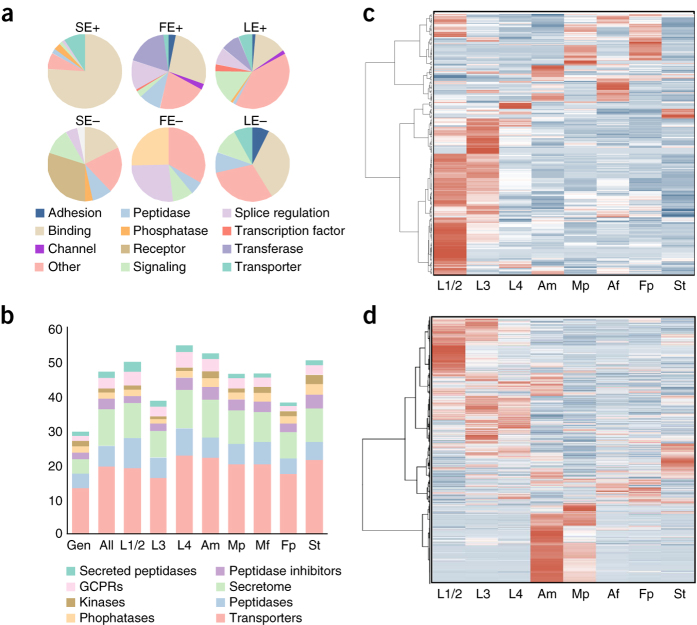
Figure 2. Stage- and tissue-specific small-RNA and mRNA transcriptome of T. suis.
(a) Association between gene function and alternative splice variation. Charts show the inferred function of protein domains encoded by genes showing a statistically significant (P ≤ 0.05; Pearson's chi-squared analysis) positive (+) or negative (−) bias toward skipped exon (SE), alternative first (FE) or last exon (LE) splice events. Only genes encoding ten or more transcripts are included in this analysis. (b) Proportional representation of major protein classes or groups encoded by the genome (Gen), and their proportional abundance in all transcriptomic data (All) and in larval (L1/2, L3 and L4), adult male (Am), female (Af) and tissue-specific libraries, including in the male (Mp) and female posterior body (Fp) and the stichosome (St). (c) Self-organizing heatmap (transcripts per million (TPM) values normalized by gene) clustering miRNAs by their transcription abundance (represented as log2-transformed reads per kilobase per million reads (RPKM) values) in each larval, adult and tissue-specific library. (d) Self-organizing heatmap (TPM values normalized by gene) clustering 22A-RNAs by their transcription abundance (represented as log2-transformed RPKM values) in each larval, adult and tissue-specific library.
T. suis undergoes substantial developmental changes throughout its direct life cycle29. To understand developmental processes in this parasite, we used RNA-seq to characterize transcription in various stages, sexes or body portions (stichosomal versus all non-stichosomal tissues from male and female adult worms): first and second (L1/L2), third (L3) and fourth (L4) larval stages adult male and female; stichosome and adult male posterior body and female posterior body excluding the stichosome (Supplementary Fig. 8 and Supplementary Table 18). Overall, a number of major functional classes of proteins showed higher representation in the transcriptome of T. suis than in the genome (Fig. 2b). Secretory proteins were notable in this regard, making up ∼4% of the T. suis gene set but representing ∼10% of the transcriptional abundance in all libraries. Peptidases, particularly secreted peptidases, were also over-represented in the transcriptome and, notably, were upregulated during larval development and in the stichosome.
The stichosome is the thin, elongate anterior end of Trichuris embedded tightly within a syncytial tunnel21 in the superficial layer of the large intestinal mucosa. Within this tunnel, the parasite secretes proteins and other molecules and absorbs nutrients from cell cytoplasm and surrounding tissue fluids, probably through thousands of bacillary cells30. Given its central importance in feeding and interaction with the host, we focused on transcription in the stichosome relative to the rest of the worm body (Supplementary Table 18; see Supplementary Note for detailed comparisons among other stages or tissues). Transcription was enriched for 2,210 genes (encoding 3,721 transcripts) in the stichosome relative to both the male and female posterior bodies (Supplementary Table 18). Among these genes are 160 peptidases (encoding 256 transcripts) and 41 porins (85 transcripts), supporting their role in host-tissue degradation and syncytial tunnel formation19,25 (Supplementary Fig. 9). Also notable is the enrichment of a large number of secreted and membrane-bound transporters (222 genes encoding 371 transcripts) of various ions (for example, sodium, phosphate and calcium) and small molecules (for example, glucose and nucleosides). Sugar metabolism is enriched in the stichosome, suggesting that absorbed glucose is rapidly metabolized in the stichocytes. Also upregulated in the stichosome are transcripts associated with endocytosis and vesicle formation, lysozyme and peroxisome pathways as well as fatty acid and amino acid (cysteine and methionine; lysine) degradation. At least one isoform of each putative immunomodulatory gene encoded by T. suis is transcribed in the stichosome, with 22 transcripts encoding galactins, serpins, venom allergen–like proteins, apyrase or calreticulin specifically enriched in the stichosome relative to both the male and female posterior bodies.
Chymotrypsin-like (S1) serine proteases (n = 28 of 31 genes, and 51 of 135 transcripts) are also upregulated in the stichosome. Many are homologs of vertebrate plasmin, which is thought to regulate blood clotting in the host31. A poorly understood consequence of trichuriasis is bloody diarrhea32, and some evidence suggests that Trichuris ingests blood33. It may be that some T. suis chymotrypsin-like serine proteases act as anticoagulants or assist in digesting blood, serum and tissue components (for example, fibrinogen). Notably, T. muris infection alters the mucus barrier in the host's gut epithelium, leading to an increased susceptibility to nematode infections34 through the degradation of mucin 2 (Muc2) polymers35. Muc2 depolymerization by T. muris is blocked by chymostatin and antipain35, suggesting a probable role for chymotrypsin-like and other serine proteases. Several of the secreted serine proteases enriched in the stichosome are homologs of Schistosoma mansoni serine protease 1 (SP1) and human kallikrein. The latter molecule regulates the degradation of kininogen to bradykinin, stimulating vasodilation, the cytosolic release of Ca2+, neutrophil recruitment and increased inflammation36. SP1 is a potent vasodilator in mice37, suggesting that it has an ability to convert vertebrate kininogen to bradykinin. Given the anti-inflammatory capacity of T. suis, we propose that some of these chymotrypsin-like serine proteases might degrade host kininogen but do not enable bradykinin production, thereby preventing bradykinin receptor stimulation and, thus, inhibiting inflammation. Notably, bradykinin receptors have key roles in various autoimmune disorders, including IBD38 and multiple sclerosis36.
Genetic regulatory networks
The T. suis gene set has complete RNA-interference machinery, suggesting potential for functional genomic studies and indicating a role for small noncoding RNAs in gene regulation. We explored these small RNAs in T. suis (Supplementary Figs. 10 and 11 and Supplementary Tables 19, 20, 21) and produced ∼435 million sequence reads. Approximately 92% of these reads mapped to the T. suis genome, with 16%, 23% and 9% classified as microRNAs (miRNAs), small interfering RNAs (siRNAs) and tiny noncoding RNAs (tncRNAs), respectively. Approximately 4% of the small-RNA reads mapped with an antisense (>80% of reads) bias to transposable elements, consistent with Piwi-interacting RNAs (piRNAs)39. However, similarly to small RNAs in Ascaris suum40, < 0.01% had characteristics consistent with 21U-RNAs, which function as piRNAs in C. elegans41.
We identified 319 miRNAs, with 132 having close homologs in other nematodes (Supplementary Table 22). These miRNAs accounted for 16% of all small-RNA reads sequenced, with tsu-let-7 (50% of all miRNA reads), tsu-miR-1 (17%), tsu-novel-51 (8%; a homolog of tsp-novel-51 miRNA from T. spiralis) and tsu-miR-228 (4%) the most highly transcribed. Approximately two-thirds of the miRNAs were most abundant in larval stages, suggesting a central role in development, with a diminishing number of miRNAs enriched in adults (Fig. 2c). This trend was reversed in the transition from L4 to adult female. To explore the functional implications of differential transcription of these miRNAs, we predicted miRNA-binding sites linked to 3′ UTRs among 22,954 of the 23,824 mRNA isoforms (representing 7,180 genes) for which at least part of the 3′ UTR could be identified on the basis of RNA-seq data. We focused on miRNA-mRNA interactions recognized or proposed for C. elegans. Of the 785,143 predicted binding sites with homology to C. elegans (both miRNA and mRNA), 300,042 were supported by information in public databases and 3,238 by experimental findings42. Owing to differences in gene copy number, these 300,042 binding sites represented 45 and 62 miRNAs as well as 3,205 and 3,877 coding genes in C. elegans and T. suis, respectively.
For T. suis, the shift from L3 to L4 coincides with a universal downregulation of 24 of these conserved miRNAs, including tsu-miR-1, tsu-miR-252 and tsu-miR-236—the second, seventh and eighth most abundant miRNAs, respectively, in T. suis overall. We identified 69 transcripts enriched in L4 (relating to 62 T. suis genes and 61 C. elegans homologs) with binding sites for each of these miRNAs. Many of the C. elegans genes with inferred homology to these transcripts are involved in larval or embryonic development (for example, rol-3, slt-1 and sox-3), growth (for example, egl-4, unc-44 and lin-39) or early sexual determination (for example, sex-1; WormBase), suggesting similar functional roles in T. suis. The maturation of T. suis to adulthood coincides with a variety of sex-specific changes in miRNA levels (relative to L4s). In both sexes, tsu-miR-228 (the fourth most abundantly transcribed miRNA in T. suis) and several isoforms of tsu-miR-61 were downregulated, and tsu-miR-34 and two 'minor' miRNAs—tsu-miR-256 and tsu-miR-50—were upregulated. Many of the coding genes (n = 447) upregulated in male and female adults are predicted to be co-regulated by tsu-miR-61 and tsu-miR-228. Homologs of these coding genes in C. elegans are enriched in GO terms (biological process) for embryonic and genital development, reproduction, morphogenesis and growth and metabolism, and include tbx-2, vps-16, xnp-1 and dyci-1 (WormBase). Notable among predicted tsu-miR-34–regulated genes were homologs of srp-2 (encoding serpin-2, an anti-inflammatory protein in helminths)12, which is downregulated in the adult worm (with the exception of the stichosome) compared with larval stages.
When we compared male and female T. suis adults, we found that major differences in miRNA transcription also related to tsu-miR-61, tsu-miR-228, tsu-miR-236 and tsu-miR-252, highlighting their importance in this nematode. Enriched in males are tsu-miR-228 and four copies of tsu-miR-61, and in females, tsu-miR-236, tsu-miR-252 and one copy of tsu-miR-61 (with closest homology to cel-miR-61-5p). Considering the ambiguity associated with the enrichment of different tsu-miR-61 isoforms in both males and females, we focused on tsu-miR-228, tsu-miR-236 and tsu-miR-252. In males, the enrichment of tsu-miR-228 coincides with a downregulation of 412 transcripts (representing 320 T. suis genes) with a predicted tsu-miR-228 binding site. On the basis of their function in C. elegans homologs, we infer many of these transcripts to be linked to vulva development (for example, exc-4, mys-1, nekl-2 and sem-4), egg production (for example, cbd-1, nsy-1, ppt-1, unc-29 and unc-58) and embryogenesis and germline development (for example, bcat-1, lars-1, rnp-4, rpt-5, slt-1 and tbp-1). In females, enriched transcription of tsu-miR-236 and tsu-miR-252 coincides with a downregulation of 262 transcripts (representing 205 genes) predicted to be co-regulated by these miRNAs. Homologs of these 'female-suppressed' coding genes in C. elegans include genes involved in spermatogenesis (for example, cogc-5 and cpb-1), male mating or fertility (for example, goa-1 and odc-1), the regulation of germline specification or apoptosis (for example, glp-1, him-1, rpt-5, let-60, vps-16 and vps-41) and chemosensation (for example, crh-1, grk-2 and lys-2). Collectively, these data suggest that sexual dimorphism in T. suis might relate, at least partially, to post-transcriptional sex suppression by miRNAs rather than exclusive transcriptional promotion by mRNAs.
In addition to miRNAs, we identified 1,028,808 putative small RNAs mapping to coding regions of the genome. Of these RNAs, 673,355 mapped antisense (≥80% of reads at each location) to exons, suggesting a potential role as siRNAs41. Most abundant among the siRNAs predicted for T. suis (except those derived from males) were sequences of 24–25 nt with a 5′ guanine (i.e., 24G and 25G), compared with 22G and 26G sequences predominating among siRNAs predicted for other nematodes to date40,41. Putative siRNAs were predicted for 3,497 protein-encoding genes. Many siRNAs have key roles in germline tissues40,41. In T. suis, we identified transcripts for 508 coding genes, for which putative siRNAs were uniquely transcribed in the adult female and the female posterior body relative to the stichosome, the adult male and the male posterior body (Supplementary Tables 7 and 18; see URLs). These coding genes were enriched for transposable elements/transposases, histones or histone methytransferases, DNA- or RNA-binding, chromatin folding and homeodomain-related proteins. Similarly, but in lower abundance, these functions were also enriched in relation to the 69 coding genes associated with siRNAs uniquely transcribed in the adult male and the male posterior body. We hypothesize that these highly transcribed siRNAs protect chromatin in the T. suis germline, and this hypothesis is supported by the observation that 162 of the female-enriched siRNAs are absent from the larval stages studied here (Supplementary Tables 7 and 18).
Novel class of tncRNAs
Conspicuous among the T. suis small RNAs is an abundance of 22-nt sequences with a 5′ adenine cap. Although representing just 2.9% (n = 58,307) of consensus small RNAs, these sequences represent 9.2% of all small-RNA transcription. By location, 22-nt 5′-adenylated sequences are evenly distributed between coding and noncoding regions, and within noncoding regions, between annotated (such as transposable elements, tRNAs or other noncoding RNAs) and un-annotated spaces. However, 89% of transcription attributed to these sequences relates to un-annotated, noncoding space in the genome. On the basis of their size and abundance, these sequences are consistent with tncRNAs43; however, they have characteristics not previously attributed to this class. For instance, in T. suis, they have a clear strand bias, with 83% of their transcription occurring on the Watson (i.e., antisense) strand. These 'antisense'-biased tncRNAs (henceforth called 22A-RNAs) form 1,208 clusters (ranging from 22 to 11,831 nt) among 238 assembly scaffolds, with a median of three 22A-RNA sequences per cluster at a median spacing of 97 bp. Although 40% of multicopy 22A-RNA sequences are found in the same cluster, clusters comprising one repeated 22A-RNA sequence are rare, and sequences are often shared among clusters and genomic scaffolds, indicating that tandem duplication is not the only mechanism associated with cluster formation. Few transposable elements are found within 25 kb of these clusters, suggesting that their insertion or translocation within the genome is not recent.
At this stage, we can only speculate about the function(s) and mechanism(s) of action of these sequences. Their 100-nt genomic neighborhoods vary: some regions resemble (but do not overlap with) known protein-encoding sequences, others resemble known noncoding RNAs such as tRNAs, and still others resemble neither. Eleven of these neighborhoods show partial similarities to cryptic tRNAs, ∼100 nt in size, discovered in C. elegans by the modENCODE Consortium44. The sequences of the 22A-RNAs themselves are also heterogeneous, with only one over-represented 8-nt sequence motif (5′-A[CA]GATAT[GT]-3′) occurring in 4.5% (245 of 5,457) of 22A-RNA sequences (Supplementary Fig. 12). Given these findings, we propose that 22A-RNAs may be processed from larger noncoding RNAs of diverse types, some of which are highly conserved and familiar, others of which are both hypothetical and unfamiliar. Despite having no obvious promoter motif (such as that proposed for 21U-RNAs)41, these sequences seem to be transcriptionally regulated, and their abundance varies substantially among stages, sexes and tissues in T. suis; including, notably, an enrichment in the adult male body and male posterior body relative to all other stages and tissues (Fig. 2d), which may suggest a role in the male germline. As a proportion of overall small RNA transcription, 22A-RNAs are most abundant in the stichosome, wherein they comprise ∼22% of all small-RNA reads determined. Indeed, the stichosome is notably restricted in its classes of small RNAs, with miRNAs (39% of all small-RNA reads from the stichosome) and 22A-RNAs dominating the small-RNA population in this organ. Whether this finding points to an involvement of these novel T. suis noncoding RNAs in host interactions deserves detailed investigation.
Discussion
Globally, helminthiases are seriously neglected causes of morbidity and mortality. Genomic and transcriptomic explorations of T. suis should enable the design of urgently needed therapeutics against human trichuriasis, one of the world's most important and neglected helminthiases. An intriguing feature of T. suis is its possible use as a therapy for human autoimmune disorders9,10,11. A detailed characterization of how this parasite modulates the host immune response is thus a key priority. Secreted proteins (including cystatins and serpins, thioredoxin peroxidase and various putative mimics of host proteins) seem to have a central role in this process, primarily through inhibiting inflammation. Our findings indicate a role for parasite-derived lipids, including the inferred synthesis of β-glucosylceramide, a known anti-inflammatory and putative therapy for IBD45, during the early developmental phase of T. suis (see Supplementary Note). It is likely that both proteins and lipids work in concert with N-linked glycans, which are known immunomodulators produced by T. suis27, particularly L4 and adult stages, in which pathways associated with their synthesis are transcriptionally enriched (see Supplementary Note). The detailed characterization of these molecules in vitro and in vivo, using existing models of IBD and other autoimmune disorders, might pave the way for parasite-derived therapies5. Indeed, a better understanding of the T. suis–host interactions might shed new light on why helminth exposure seems crucial for the development of a healthy immune system in humans. This is the first study to characterize the genomes of male and female individuals of a dioecious nematode. We found little evidence for sex-specific genes or assembly contigs, despite the reported XY karyotype of this species. However, intriguingly, miRNAs seem to have a major role in regulating sexual development in this species, with tsu-miR-228 in male, and tsu-miR-236 and tsu-miR-252 in female worms predicted to regulate and suppress key feminizing and masculinizing developmental genes, respectively. This is the first time that this has been observed for a metazoan.
Methods
Sample preparation and storage.
Trichuris suis were isolated from experimentally infected pigs inoculated orally with a single dose of 5,000–50,000 embryonated eggs (Animal Ethics Permission No. 2010/561-1914; University of Copenhagen). Individuals of T. suis were isolated at 10 (L1/L2 larvae), 18 (L3s), 28 (L4s) and 49 (adulthood) d after inoculation (p.i.)29,46 and washed in physiological saline (37 °C) and RPMI 1640 (GIBCO) with antibiotic-antimycotic (GIBCO). Adult male and female T. suis were separated. Stichosomes were excised from whole adult worms (n = 10, irrespective of sex), pooled and frozen, as were the posterior portions of the worms (n = 10 of each sex). All stages or tissues were snap frozen in liquid nitrogen and stored at −80 °C.
Genomic sequencing and assembly.
Total genomic DNA was isolated each from a single adult male or female T. suis47,48. Paired-end (insert sizes, 170 bp and 500 bp) and mate-paired (800 bp, 2 kb, 5 kb and 10 kb) libraries were constructed from total and whole-genomic amplified (WGA) genomic DNA, respectively14,49 and sequenced using a HiSeq 2000 machine (Illumina). Low-quality sequences, base-calling duplicates and adapters were removed using standard approaches. Sequence quality and heterozygosity were assessed by 17-mer frequency distribution50 and genome sizes estimated51. Corrected and filtered data were assembled into contigs using SOAPdenovo v2.0 (ref. 50) and assessed for accuracy using SOAP2aligner52. Assembly completeness and redundancy were assessed using CEGMA53 and RNA-seq data using Bowtie2 (ref. 54).
RNA isolation and RNA-seq.
Total RNAs from L1/L2 (n = 50,000 from five pigs), L3 (n = 15,000 from four pigs) and L4 (n = 3,000 from two pigs), adult male (n = 10), adult female (n = 10), and stichosomal (mixed sex; n = 10) and nonstichosomal portions of adult females (n = 10) and males (n = 10) were individually purified using TriPure reagent (Roche). Polyadenylated (polyA+) RNA was purified from 10 μg of total RNA for each library using Sera-mag oligo(dT) beads and fragmented, purified and sequenced using HiSeq 2000 (refs. 14,49). Small noncoding RNAs (∼18–30 nt) were isolated from 10 μg of total RNA for each library by size fractionation on polyacrylamide gels, purified, adaptor-ligated, reverse transcribed, amplified by PCR and sequenced using HiSeq 2000. All RNA-seq data were adaptor trimmed and length and quality filtered using standard approaches.
Synteny and polymorphism analysis, and annotation of repeat content.
For comparative analysis, the assemblies for adult male or female T. suis were aligned using MUMmer3 (ref. 55). Repetitive sequences in each assembly were identified14 using Tandem Repeats Finder (TRF)56, RepeatMasker57, LTR_FINDER58, PILER59 and RepeatScout60, with a consensus population of predicted repetitive elements constructed in RepeatScout using fit-preferred alignment scores. Transfer RNAs were predicted using tRNA-SCAN61. The male assembly was explored for scaffolds likely to represent the male-specific Y chromosome13,62. Reads from all genomic sequence libraries each for male and female T. suis were aligned to their own and the opposite sex (both repeat unmasked and hard-masked) assembly (i.e., male-to-male, male-to-female, female-to-male and female-to-female) using Bowtie2 (ref. 54). Contigs with >80% coverage in same-sex but <20% coverage in opposite-sex read alignments were deemed 'sex-specific'.
Prediction and functional annotation of the protein-encoding gene set.
The male and female protein-encoding gene set of T. suis was inferred in MAKER2 (ref. 63). Briefly, (i) the nonredundant T. suis transcriptome was aligned each assembly using BLAT64 and filtered for full-length ORFs, which were used (ii) to train hidden Markov models (HMM) for de novo gene prediction using SNAP65 and AUGUSTUS66, with these models supplemented using (iii) homologous genes from T. spiralis17 and C. elegans16; and (iv) all T. suis RNA-seq data from all libraries used to infer each transcript using Tophat2 (ref. 67) and Cufflinks2 (refs. 68,69); (v) all HMM-predicted, homology and evidence-based information was then combined into a single consensus gene set, and (vi) genes overlapping with predicted repetitive regions of the genome and/or having significant E < 1 × 10-5, BLASTn homology to known repetitive sequences (i.e., transposable elements) in RepBase57 and no close homology to C. elegans or T. spiralis protein-encoding genes were removed. The male and female T. suis gene sets were unified by orthology prediction using InParanoid70, with T. spiralis17 as an out-group.
Conserved protein domains encoded by each gene were identified using InterProScan71, with these data used to infer Gene Ontology72. Using Reciprocal BLASTp and OrthoMCL73, the T. suis inferred proteome was clustered with predicted homologs or orthologs for other nematodes, including Ascaris suum14, Brugia malayi15, C. elegans16 and T. spiralis17. Each contig was assessed for a known functional ortholog in the Kyoto Encyclopedia of Genes and Genomes (KEGG) using the KEGG orthology bases annotation system (kobas)74. In addition, T. suis inferred proteins were compared by BLASTx/BLASTp with protein sequences available for A. suum, B. malayi, C. elegans and T. spiralis, and in the databases UniProt75, SwissProt and TREMBL76, as well as specialist databases for key protein groups represented in MEROPS77, WormBase78, KS-SARfari and GPCR-SARfari, and the Transporter Classification database (TCDB)79. ES proteins were predicted using Phobius80 and by BLASTp comparison with the validated signal peptide database (SPD)81 and proteomic data for the nematodes B. malayi82,83 and Meloidogyne incognita84 and the trematode Schistosoma mansoni85.
Differential transcription analysis of mRNA.
Reconstruction and quantification (in fragments per kilobase per million reads (FPKMs)) of the T. suis transcriptome was conducted using TopHat2 (ref. 67) and Cufflinks2 (refs. 68,69). Predicted alternative splice events were classified86. Comparisons of splice events and gene function (based on encoded Pfam domains) were conducted by pairwise Pearson chi-squared analysis (P value ≤ 0.05). We also compared the relationship between gene essentiality and being a single or multi-isoform gene, with essentiality predicted14. Differential transcription was assessed using NOISeq87, with 20% of the evaluated reads for each library used in five iterations to simulate technical replicates.
Annotation and differential transcription analysis of small noncoding RNAs.
Canonical miRNAs were identified and quantified in miRDeep2 (refs. 88, 89, 90), and supported using miRNAs published for A. suum40, C. elegans91, Haemonchus contortus92, Brugia pahangi92 and T. spiralis93. The 3′ UTR for each Cufflinks-predicted T. suis transcript was identified by comparison with the T. suis genome annotation. Each 3′ UTR was screened for miRNA binding sites using PITA94. These binding sites were filtered on the basis of homologous miRNA-transcript binding interactions predicted for C. elegans in curated databases (microRNA.org, RNA22 and TargetScan) or demonstrated empirically42. All non-miRNA reads from each small-RNA library for T. suis were then aligned to the male T. suis genome using Bowtie2 (ref. 54) and clustered using ShortStack95, with a minimum cluster depth cutoff of 10. Small-RNA reads having perfect alignment overlap (i.e., the same start and stop position) were defined as homologous and condensed into a consensus sequence by majority rule. Each consensus small RNA was classified41 and nucleotide diversity within homologous small-RNA reads was assessed using custom Perl scripts. Specific small-RNA sequences (for example, 21U-RNAs)41 and their 5′ and 3′ flanking regions were explored for sequence motifs using MEME96. Differential transcription among stage- or tissue-specific libraries was assessed (in reads per million mapped reads, RPKM) for miRNAs, siRNAs and 22A-RNAs using NOISeq87, and clustered by stage- or tissue-specific transcription pattern using R.
Analysis of the genomic neighborhoods and primary sequences of 22A-RNAs.
We extracted 22A-RNAs with 100-nt flanks lacking scaffolding (N) residues, merged those with spatial overlaps along the genome, and further condensed them to 80% sequence identity with CD-HIT-EST97. We probed resultant nonredundant 22A-RNA regions for protein-encoding exons with BlastX79, and known noncoding RNAs (ncRNAs) with INFERNAL 1.1/cmscan98. BlastX was run against predicted proteomes from all published nematode genomes in WormBase WS240 (ref. 87), as well as the T. suis male proteome from this study; cmscan was run against the ncRNA database RFAM 11.0 (ref. 99). Regions passing these filters were tested for similarity to (i) genomic DNA from other nematode species, (ii) other 22A-RNA neighborhoods and (iii) novel ncRNAs from C. elegans. The first was assayed by BlastN against published nematode genomic sequences in WormBase WS240 (ref. 87). The second was assayed by BlastN against 22A-RNA regions spatially merged for genomic overlaps but not condensed with CD-HIT-EST. The third was assayed by BlastN against a set of 8,126 C. elegans ncRNAs taken from the WS240 release of WormBase87. All searches used E-value thresholds of ≤10−3.
A set of 25,259 C. elegans ncRNAs was obtained from WormBase WS240. We filtered out those named 'asRNA', 'rRNA', 'scRNA', 'snoRNA', 'snRNA' or 'tRNA', leaving 8,126 ncRNAs with no official similarity to well-known structures. Most of these ncRNAs had been discovered by modENCODE45; 176 others were long noncoding RNAs100. We then checked for previously undescribed motifs via RFAM 11.0 and cmscan.
To discover whether 22A-RNA sequences contained novel motifs, we extracted 22A-RNA sequences without flanking protein-encoding or ncRNA similarities, merged them spatially and for 80% identity, and scanned with MEME96, using a first-order Markov model from the adult male T. suis genome (via MEME's fasta-get-markov). We ran MEME with arguments: '-dna -revcomp -nsites 100 -bfile TS_M_200bpormore.1markov.txt -nmotifs 10 -evt 0.05 -minw 6 -maxw 22 -mod anr'. For one resulting 8-nt motif, we used FIMO101 to determine where it occurred in original, unmerged 22A-RNA sequences, with arguments: '–bgfile TS_M_200bpormore.1markov.txt–output-pthresh 0.001'. The motif was displayed as a logarithmic WebLogo102.
URLs.
WormBase, http://www.wormbase.org/; microRNA.org, http://www.microrna.org/microrna/home.do; RNA22, https://cm.jefferson.edu/rna22v1.0/; TargetScanWorm, http://www.targetscan.org/worm_52/; salient data files are accessible via http://gasser-research.vet.unimelb.edu.au/Trichuris_suis/ and ftp://ftp.wormbase.org/pub/wormbase/species/t_suis/; browsable male and female genomes are accessible via http://gasser-research.vet.unimelb.edu.au/jbrowse/JBrowse-1.11.2/index.html?data=TsuisMale/ and http://gasser-research.vet.unimelb.edu.au/jbrowse/JBrowse-1.11.2/index.html?data=TsuisFemale/, respectively, or through WormBase via ftp://ftp.wormbase.org/pub/wormbase/species/t_suis/.
Accession numbers.
All short-read data are available via Sequence Read Archive: SRR1041639, SRR1041640, SRR1041641, SRR1041642, SRR1041643, SRR1041644 (genomic DNA male); SRR1041645, SRR1041646, SRR1041647, SRR1041648, SRR1041649, SRR1041650 (genomic DNA female); SRR1041651, SRR1041652, SRR1041653, SRR1041654, SRR1041655, SRR1041656, SRR1041657, SRR1041658 (mRNA); SRR1041659, SRR1041660, SRR1041661, SRR1041662, SRR1041663, SRR1041664, SRR1041669, SRR1041670 (small RNA). Annotated assemblies of each genome are accessible via BioProject PRJNA208415 (male) and PRJNA208416 (female).
Supplementary Information
Integrated supplementary information
Calculated as GC% per 10-kb nonoverlapping sliding window.
Data generated using 500-bp sliding windows using a 250-bp overlap. The x axis is GC content (%), and the y axis is the proportion of the windows (bins) corresponding to each GC measurement.
(a) tRNA copy number; (b) frequency of amino acids encode by the inferred T. suis proteome.
Based on enriched transcripts found in larval stages of T. suis embedded in the host gut epithelium and up-regulated in the stichosome relative to non-stichosomal tissues in adult males and females. Mechanisms of action (adapted from 1–4) include secretion of (1) cysteine protease inhibitors, N-glycosylated proteins and a putative TGF-β mimic to suppress antigen presentation by host dendritic and B-cells and limit prolifation/stimulation of proinflammatory T-cells; (2) calreticulins believed to bind to scavenger receptors on dendritic cells and stimulate production of 'anti-inflammatory' IL-4 and IL-10, leading to proliferation of Fox3+ regulatory T-cells; (3) calreticulins and apyrases that bind free Ca+ and ATP respectively and prevent conversion of Fox3+ to pro-inflammatory T-cells; (4) thioredoxin peroxidase (TPX) and a putative macrophage initiation factor (MIF) mimic to stimulate alternative activation of host macrophages, inhibiting inflammatory nitrous oxide (NO) production and toll-like receptor (TLR) pathways and stimulating the production of IL-4 and IL-10; and (5) serine protease inhibitors that block neutrophilsecreted cathepsin G and elastases, thereby limiting tissue destruction and reducing inflammation. Red X represents the blocking of a pathway or mechanism owing to Trichuris-generated immunomodulation.
Splice events predicted for each transcript are available in Supplementary Table 17.
(a) Exons per gene; (b) transcripts per gene.
Blue-banding highlight major clusters based on transcriptional profile for stichosome, larval, made and female enriched transcripts. Each row represents one transcript. Highest transcription represented by white coloration. Lowest transcription represented by red coloration. Color scale used allows comparison between/among libraries for each transcript, but not between/among distinct transcripts. FPKM values for each transcript available in Supplementary Table 18. Dendrogram clustering cropped for presentation.
All parasite-derived molecules are enriched in the stichosome tissues relative to nonstichosomal tissues in both male and female adult T. suis. In this schematic, we propose that secreted proteases, including S1 (chymotrypsin), S9, M12 (astacin) and C1 (cathepsin) families, play an early role in formation of the syncitial tunnel in the host gut epithelium. Among the secreted S1 proteases are several that degrade Muc2 polymers in the host intestinal mucus barrier, exposing the gut epithelial cell membranes. These membranes are then breached by T. suis secreted porins, allowing small molecule transporters to enter the host cell cytoplasm and harvest simple sugars, and nucleic and amino acids, which are then taken back into the bacilliary cells through endocytosis or another unknown mechanism. In addition, S1 proteases degrade long protein polymers found in the host blood/plasma including fibrinogen (thus inhibiting co-agulation and possibly contributing to the gut hemorrhaging) and kallikrien (possibly providing a means to inhibit inflammation by blocking bradykinin formation). While this is occurring a potent mix of immunomodulatory proteins (Supplementary Fig. 4 and Supplementary Table 16) are secreted and specifically regulate or suppress an inflammatory response. We hypothesize also that larger peptide are taken up into the stichocyte by endocytosis and vesicle transport through the bacillary cells and further degraded in the peroxisomes in the stichocytes themselves. Increased lysosome activity in the stichocytes may also suggest degradation of lipids acquired from the host cells, but we cannot propose a source for these molecules based on our current data. CU, cuticle; MU, muscle; STC, stichocyte; OL, esophageal lumen; BC, bacillary cell.
Mp, male nonstichosomal (i.e., posterior body) tissue; MALE, adult male; F.POST, female nonstichosomal (i.e., posterior body) tissue; FEMALE, adult female; L10, larvae at 10 d after infection (d.p.i.); L18, larvae at 18 d.p.i.; L28, larvae at 28 d.p.i.; STICH, adult (mixed sex) stichosomal tissue.
This represents the one significant motif, among possible motifs ranging in size from 6 to 22 nt, that was shared between 1,174 nonredundant 22A-RNA sequences. The logarithmic probability distribution of individual nucleotides in the motif is shown as a WebLogo (see Online Methods).
Supplementary information
Supplementary Figures 1–12, Supplementary Tables 1–4, 7, 19 and 20, and Supplementary Note (PDF 3403 kb)
Male-encoded transfer RNAs. The annotated location and identity of each tRNA predicted for the male Trichuris suis genome. (XLSX 62 kb)
Female-encoded transfer RNAs. The annotated location and identity of each tRNA predicted for the female Trichuris suis genome. (XLSX 64 kb)
G protein–coupled receptors (GPCRs). Inferred GPCRs encoded by Trichuris suis based on BLASTp comparison with homologous protein sequences in the GPCR databases, IUPHAR and gpcr-sarfari (see Online Methods). (XLSX 21 kb)
Kinases. Inferred kinases encoded by Trichuris suis based on BLASTp comparison with homologous protein sequences in the ks-sarfari and Uniprot databases (see Online Methods). (XLSX 29 kb)
Phosphatases. Inferred phosphatases encoded by Trichuris suis based on BLASTp comparison with homologous protein sequences in Uniprot (see Online Methods). (XLSX 25 kb)
Peptidases. Inferred peptidases encoded by Trichuris suis based on BLASTp comparison with homologous protein sequences in the MEROPs protease domain database (see Online Methods). (XLSX 48 kb)
Peptidase inhibitors. Inferred peptidase inhibitors encoded by Trichuris suis based on BLASTp comparison with homologous protein sequences in the MEROPs protease inhibitor domain database (see Online Methods). (XLSX 24 kb)
Transporters and channels. Inferred peptidase inhibitors encoded by Trichuris suis based on BLASTp comparison with homologous protein sequences in the Transporter Classification Database (TC-DB; see Online Methods). (XLSX 125 kb)
Secretome. Inferred secreted proteins encoded by Trichuris suis based on encoding a signal peptide and lacking a recognizable transmembrane domain and having BLASTp homology to sequences in the Secreted Proteins Database (SPD) or to proteomic data available for the secretome of at least one helminth species (see Online Methods). (XLSX 52 kb)
Secreted peptidases. Inferred secreted peptidases based on their inclusion in the predicted Trichuris suis secretome and inferred (BLASTp) homology to the MEROPs protease domain database. (XLSX 19 kb)
Immunomodulatory proteins. Trichuris suis–encoded proteins with sequence homology to known immunomodulatory proteins secreted by this or any other species of helminth and their stage-, sex- and tissue-specific transcription (in FPKMs). (XLSX 17 kb)
Alternative splicing. A summary of predicted alternative splicing events of the Trichuris suis transcriptome and the functional (Pfam) domains encoded by each transcript splice isoform. (XLSX 1559 kb)
Stage- and tissue-specific transcription FPKM values. Normalized transcriptional levels (expressed as fragments per kilobase per million reads (FPKM)) of each Trichuris suis messenger RNA in each stage-, sex- and tissue-specific library. (XLSX 3472 kb)
Small RNA read clusters. A summary of small RNA read clusters mapping to the Trichuris suis male assembly (used for all small RNA mappings owing to its larger size), showing, for each cluster, its location in the male assembly, whether the cluster is predicted to form a hair-pin fold (HP), the dominant DICER-derived read length among the reads mapping to each cluster, whether the proportion of reads mapping to the anti-sense strand, whether the cluster is found with 500 bp of an annotated transcription start site (i.e., transcription start site associated (TSSa)), the number of reads mapping to each cluster, and the number and ratio of these reads mapping to the 'forward' or 'reverse' stand, whether the cluster has a substantial forward or reverse strand bias (≥ 80% of reads mapping to one strand), the number of reads, categorized by their 5' and 3' nucleotides, mapping to each cluster, the transcription associated with each cluster in each stage-, sex- or tissue-specific library (normalized as FPKM values), and the number of reads mapping to each cluster by read length. (XLSX 7089 kb)
Micro-RNAs. The micro-RNAs (miRNAs) predicted for Trichuris suis, their scoring parameters, the number of reads in the total dataset mapping to their predicted mature, star and hairpin regions, nearest homolog in another nematode, and predicted mature, star and precursor sequence and their stage and tissue-specific transcription (in total reads and FPKMs). (XLSX 92 kb)
Acknowledgements
We are indebted to the staff of the BGI-Shenzhen who assisted this study but whose names are not included in the authorship. We also acknowledge the continued contributions of staff at WormBase. This project was funded by the Australian Research Council (ARC), the National Health and Medical Research Council (NHMRC) of Australia and BGI-Shenzhen. This research was supported by a Victorian Life Sciences Computation Initiative (VLSCI) grant number VR0007 on its Peak Computing Facility at the University of Melbourne, an initiative of the Victorian Government (A.R.J. and R.B.G.). Other support to R.B.G. from the Alexander von Humboldt Foundation, Australian Academy of Science, the Australian-American Fulbright Commission, Melbourne Water Corporation and the IBM Research Collaboratory for Life Sciences – Melbourne is gratefully acknowledged. P.N. was supported by Danish Agency for Science, Technology and Innovation. N.D.Y. holds an NHMRC Early Career Research Fellowship. E.M.S. was supported by startup funds from Cornell University. P.W.S. is an investigator with the Howard Hughes Medical Institute (HHMI) and acknowledges support from the US National Institutes of Health (NIH).
Author Contributions
P.N. and S.T. provided T. suis, and N.D.Y. purified nucleic acids for sequencing. L.H. and S.W. coordinated sequencing. A.R.J., S.L., P.K.K., R.S.H., J.X. and P.X. undertook the assembly, annotation and analyses of genomic and transcriptomic data. A.R.J., R.S.H., A.H., P.K.K. and E.M.S. planned and performed additional, detailed bioinformatic analyses. A.R.J., P.W.S., J.-P.Y.S. and R.B.G. drafted and edited the manuscript, tables, figures and supplementary information. A.R.J., N.D.Y., J.W. and R.B.G. conceived and planned the project. A.R.J., N.D.Y. and R.B.G. supervised and coordinated the research.
Accession codes
Primary accessions
BioProject
Sequence Read Archive
Competing interests
The authors declare no competing financial interests.
Contributor Information
Aaron R Jex, Email: ajex@unimelb.edu.au.
Jun Wang, Email: wangjun@genomics.org.cn.
Robin B Gasser, Email: robinbg@unimelb.edu.au.
References
- 1.Hotez PJ, Fenwick A, Savioli L, Molyneux DH. Rescuing the bottom billion through control of neglected tropical diseases. Lancet. 2009;373:1570–1575. doi: 10.1016/S0140-6736(09)60233-6. [DOI] [PubMed] [Google Scholar]
- 2.Stephenson LS, Holland CV, Cooper ES. The public health significance of Trichuris trichiura. Parasitology. 2000;121(suppl.):S73–S95. doi: 10.1017/s0031182000006867. [DOI] [PubMed] [Google Scholar]
- 3.Anonymous. Schistosomiasis and soil-transmitted helminth infections (World Health Assembly Resolution WHA54.19) http://apps.who.int/gb/archive/pdf_files/WHA54/ea54r19.pdf (2001).
- 4.Okada H, Kuhn C, Feillet H, Bach JF. The 'hygiene hypothesis' for autoimmune and allergic diseases: an update. Clin. Exp. Immunol. 2010;160:1–9. doi: 10.1111/j.1365-2249.2010.04139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bach JF. The effect of infections on susceptibility to autoimmune and allergic diseases. N. Engl. J. Med. 2002;347:911–920. doi: 10.1056/NEJMra020100. [DOI] [PubMed] [Google Scholar]
- 6.Elliott DE, Summers RW, Weinstock JV. Helminths as governors of immune-mediated inflammation. Int. J. Parasitol. 2007;37:457–464. doi: 10.1016/j.ijpara.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 7.Endara P, et al. Long-term periodic anthelmintic treatments are associated with increased allergen skin reactivity. Clin. Exp. Allergy. 2010;40:1669–1677. doi: 10.1111/j.1365-2222.2010.03559.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rodrigues LC, et al. Early infection with Trichuris trichiura and allergen skin test reactivity in later childhood. Clin. Exp. Allergy. 2008;38:1769–1777. doi: 10.1111/j.1365-2222.2008.03027.x. [DOI] [PubMed] [Google Scholar]
- 9.Summers RW, Elliott DE, Urban JF, Jr., Thompson RA, Weinstock JV. Trichuris suis therapy for active ulcerative colitis: a randomized controlled trial. Gastroenterology. 2005;128:825–832. doi: 10.1053/j.gastro.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 10.Summers RW, et al. Trichuris suis seems to be safe and possibly effective in the treatment of inflammatory bowel disease. Am. J. Gastroenterol. 2003;98:2034–2041. doi: 10.1111/j.1572-0241.2003.07660.x. [DOI] [PubMed] [Google Scholar]
- 11.Fleming JO, et al. Probiotic helminth administration in relapsing-remitting multiple sclerosis: a phase 1 study. Mult. Scler. 2011;17:743–754. doi: 10.1177/1352458511398054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hewitson JP, Grainger JR, Maizels RM. Helminth immunoregulation: the role of parasite secreted proteins in modulating host immunity. Mol. Biochem. Parasitol. 2009;167:1–11. doi: 10.1016/j.molbiopara.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spakulová M, Kralova I, Cutillas C. Studies on the karyotype and gametogenesis in Trichuris muris. J. Helminthol. 1994;68:67–72. doi: 10.1017/s0022149x0001350x. [DOI] [PubMed] [Google Scholar]
- 14.Jex AR, et al. Ascaris suum draft genome. Nature. 2011;479:529–533. doi: 10.1038/nature10553. [DOI] [PubMed] [Google Scholar]
- 15.Ghedin E, et al. Draft genome of the filarial nematode parasite Brugia malayi. Science. 2007;317:1756–1760. doi: 10.1126/science.1145406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.C. elegans Sequencing Consortium. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science282, 2012–2018 (1998). [DOI] [PubMed]
- 17.Mitreva M, et al. The draft genome of the parasitic nematode Trichinella spiralis. Nat. Genet. 2011;43:228–235. doi: 10.1038/ng.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaminsky R, et al. A new class of anthelmintics effective against drug-resistant nematodes. Nature. 2008;452:176–180. doi: 10.1038/nature06722. [DOI] [PubMed] [Google Scholar]
- 19.Cooper ES, Whyte-Alleng CA, Finzi-Smith JS, MacDonald TT. Intestinal nematode infections in children: the pathophysiological price paid. Parasitology. 1992;104(suppl.):S91–S103. doi: 10.1017/s0031182000075272. [DOI] [PubMed] [Google Scholar]
- 20.Drake LJ, Bianco AE, Bundy DA, Ashall F. Characterization of peptidases of adult Trichuris muris. Parasitology. 1994;109:623–630. doi: 10.1017/s0031182000076502. [DOI] [PubMed] [Google Scholar]
- 21.Tilney LG, Connelly PS, Guild GM, Vranich KA, Artis D. Adaptation of a nematode parasite to living within the mammalian epithelium. J. Exp. Zool. A Comp. Exp. Biol. 2005;303:927–945. doi: 10.1002/jez.a.214. [DOI] [PubMed] [Google Scholar]
- 22.Toubarro D, et al. Serine protease-mediated host invasion by the parasitic nematode Steinernema carpocapsae. J. Biol. Chem. 2010;285:30666–30675. doi: 10.1074/jbc.M110.129346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Balasubramanian N, Toubarro D, Simoes N. Biochemical study and in vitro insect immune suppression by a trypsin-like secreted protease from the nematode Steinernema carpocapsae. Parasite Immunol. 2010;32:165–175. doi: 10.1111/j.1365-3024.2009.01172.x. [DOI] [PubMed] [Google Scholar]
- 24.Toubarro D, et al. An apoptosis-inducing serine protease secreted by the entomopathogenic nematode Steinernema carpocapsae. Int. J. Parasitol. 2009;39:1319–1330. doi: 10.1016/j.ijpara.2009.04.013. [DOI] [PubMed] [Google Scholar]
- 25.Drake L, et al. The major secreted product of the whipworm, Trichuris, is a pore-forming protein. Proc. Biol. Sci. 1994;257:255–261. doi: 10.1098/rspb.1994.0123. [DOI] [PubMed] [Google Scholar]
- 26.Kringel H, Iburg T, Dawson H, Aasted B, Roepstorff A. A time course study of immunological responses in Trichuris suis–infected pigs demonstrates induction of a local type 2 response associated with worm burden. Int. J. Parasitol. 2006;36:915–924. doi: 10.1016/j.ijpara.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 27.Klaver EJ, et al. Trichuris suis–induced modulation of human dendritic cell function is glycan-mediated. Int. J. Parasitol. 2013;43:191–200. doi: 10.1016/j.ijpara.2012.10.021. [DOI] [PubMed] [Google Scholar]
- 28.Stamm S, et al. Function of alternative splicing. Gene. 2005;344:1–20. doi: 10.1016/j.gene.2004.10.022. [DOI] [PubMed] [Google Scholar]
- 29.Beer RJ. Studies on the biology of the life-cycle of Trichuris suis Schrank, 1788. Parasitology. 1973;67:253–262. doi: 10.1017/s0031182000046497. [DOI] [PubMed] [Google Scholar]
- 30.Sheffield HG. Electron microscopy of the bacillary band and stichosome of Trichuris muris and T. vulpis. J. Parasitol. 1963;49:998–1009. [PubMed] [Google Scholar]
- 31.Hoover-Plow J. Does plasmin have anticoagulant activity? Vasc. Health Risk Manag. 2010;6:199–205. doi: 10.2147/vhrm.s9358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.MacDonald TT, et al. Histopathology and immunohistochemistry of the caecum in children with the Trichuris dysentery syndrome. J. Clin. Pathol. 1991;44:194–199. doi: 10.1136/jcp.44.3.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burrows RB, Lillis WG. The whipworm as a blood sucker. J. Parasitol. 1964;50:675–680. [PubMed] [Google Scholar]
- 34.Hasnain SZ, et al. Mucin gene deficiency in mice impairs host resistance to an enteric parasitic infection. Gastroenterology. 2010;138:1763–1771. doi: 10.1053/j.gastro.2010.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hasnain SZ, McGuckin MA, Grencis RK, Thornton DJ. Serine protease(s) secreted by the nematode Trichuris muris degrade the mucus barrier. PLoS Negl. Trop. Dis. 2012;6:e1856. doi: 10.1371/journal.pntd.0001856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Golias C, Charalabopoulos A, Stagikas D, Charalabopoulos K, Batistatou A. The kinin system–bradykinin: biological effects and clinical implications. Hippokratia. 2007;11:124–128. [PMC free article] [PubMed] [Google Scholar]
- 37.Da'dara A, Skelly PJ. Manipulation of vascular function by blood flukes? Blood Rev. 2011;25:175–179. doi: 10.1016/j.blre.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marceau F, Regoli D. Therapeutic options in inflammatory bowel disease: experimental evidence of a beneficial effect of kinin B1 receptor blockade. Br. J. Pharmacol. 2008;154:1163–1165. doi: 10.1038/bjp.2008.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat. Rev. Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang J, et al. Deep small RNA sequencing from the nematode Ascaris reveals conservation, functional diversification, and novel developmental profiles. Genome Res. 2011;21:1462–1477. doi: 10.1101/gr.121426.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ruby JG, et al. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell. 2006;127:1193–1207. doi: 10.1016/j.cell.2006.10.040. [DOI] [PubMed] [Google Scholar]
- 42.Zisoulis DG, et al. Comprehensive discovery of endogenous Argonaute binding sites in Caenorhabditis elegans. Nat. Struct. Mol. Biol. 2010;17:173–179. doi: 10.1038/nsmb.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ambros V, Lee RC, Lavanway A, Williams PT, Jewell D. MicroRNAs and other tiny endogenous RNAs in C. elegans. Curr. Biol. 2003;13:807–818. doi: 10.1016/s0960-9822(03)00287-2. [DOI] [PubMed] [Google Scholar]
- 44.Lu ZJ, et al. Prediction and characterization of noncoding RNAs in C. elegans by integrating conservation, secondary structure, and high-throughput sequencing and array data. Genome Res. 2011;21:276–285. doi: 10.1101/gr.110189.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zigmond E, et al. β-glucosylceramide: a novel method for enhancement of natural killer T lymphoycte plasticity in murine models of immune-mediated disorders. Gut. 2007;56:82–89. doi: 10.1136/gut.2006.095497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kringel H, Roepstorff A, Murrell KD. A method for the recovery of immature Trichuris suis from pig intestine. Acta Vet. Scand. 2002;43:185–189. [PubMed] [Google Scholar]
- 47.Gasser RB. Molecular tools—advances, opportunities and prospects. Vet. Parasitol. 2006;136:69–89. doi: 10.1016/j.vetpar.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 48.Sambrook, J. & Russell, D.W. Molecular Cloning: A Laboratory Manual 3rd edn. (Cold Spring Harbor Laboratory Press, 2001).
- 49.Young ND, et al. Whole-genome sequence of Schistosoma haematobium. Nat. Genet. 2012;44:221–225. doi: 10.1038/ng.1065. [DOI] [PubMed] [Google Scholar]
- 50.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lander ES, Waterman MS. Genomic mapping by fingerprinting random clones: a mathematical analysis. Genomics. 1988;2:231–239. doi: 10.1016/0888-7543(88)90007-9. [DOI] [PubMed] [Google Scholar]
- 52.Li R, et al. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics. 2009;25:1966–1967. doi: 10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- 53.Parra G, Bradnam K, Ning Z, Keane T, Korf I. Assessing the gene space in draft genomes. Nucleic Acids Res. 2009;37:289–297. doi: 10.1093/nar/gkn916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Delcher AL, Salzberg SL, Phillippy AM. Using MUMmer to identify similar regions in large sequence sets. Curr. Protoc. Bioinformatics. 2003;10:10.3. doi: 10.1002/0471250953.bi1003s00. [DOI] [PubMed] [Google Scholar]
- 56.Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tarailo-Graovac M, Chen N. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr. Protoc. Bioinformatics. 2009;25:4.10. doi: 10.1002/0471250953.bi0410s25. [DOI] [PubMed] [Google Scholar]
- 58.Xu Z, Wang H. LTR_FINDER: an efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res. 2007;35:W265–W268. doi: 10.1093/nar/gkm286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Edgar RC, Myers EW. PILER: identification and classification of genomic repeats. Bioinformatics. 2005;21(suppl. 1):i152–i158. doi: 10.1093/bioinformatics/bti1003. [DOI] [PubMed] [Google Scholar]
- 60.Price AL, Jones NC, Pevzner PA. De novo identification of repeat families in large genomes. Bioinformatics. 2005;21(suppl. 1):i351–i358. doi: 10.1093/bioinformatics/bti1018. [DOI] [PubMed] [Google Scholar]
- 61.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Carvalho AB, Clark AG. Efficient identification of Y chromosome sequences in the human and Drosophila genomes. Genome Res. 2013;23:1894–1907. doi: 10.1101/gr.156034.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Holt C, Yandell M. MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics. 2011;12:491. doi: 10.1186/1471-2105-12-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kent WJ. BLAT—the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Korf I. Gene finding in novel genomes. BMC Bioinformatics. 2004;5:59. doi: 10.1186/1471-2105-5-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stanke M, Morgenstern B. AUGUSTUS: a web server for gene prediction in eukaryotes that allows user-defined constraints. Nucleic Acids Res. 2005;33:W465–W467. doi: 10.1093/nar/gki458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim D, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Trapnell C, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.O'Brien KP, Remm M, Sonnhammer EL. Inparanoid: a comprehensive database of eukaryotic orthologs. Nucleic Acids Res. 2005;33:D476–D480. doi: 10.1093/nar/gki107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Quevillon E, et al. InterProScan: protein domains identifier. Nucleic Acids Res. 2005;33:W116–W120. doi: 10.1093/nar/gki442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Harris MA, et al. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32:D258–D261. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fischer S, et al. Using OrthoMCL to assign proteins to OrthoMCL-DB groups or to cluster proteomes into new ortholog groups. Curr. Protoc. Bioinformatics. 2011;35:6.12. doi: 10.1002/0471250953.bi0612s35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wu J, Mao X, Cai T, Luo J, Wei L. KOBAS server: a web-based platform for automated annotation and pathway identification. Nucleic Acids Res. 2006;34:W720–W724. doi: 10.1093/nar/gkl167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wu CH, et al. The Universal Protein Resource (UniProt): an expanding universe of protein information. Nucleic Acids Res. 2006;34:D187–D191. doi: 10.1093/nar/gkj161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Boeckmann B, et al. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003;31:365–370. doi: 10.1093/nar/gkg095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rawlings ND, Barrett AJ, Bateman A. MEROPS: the peptidase database. Nucleic Acids Res. 2010;38:D227–D233. doi: 10.1093/nar/gkp971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Harris TW, et al. WormBase 2014: new views of curated biology. Nucleic Acids Res. 2014;42:D789–D793. doi: 10.1093/nar/gkt1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Saier MH, Jr., Yen MR, Noto K, Tamang DG, Elkan C. The Transporter Classification Database: recent advances. Nucleic Acids Res. 2009;37:D274–D278. doi: 10.1093/nar/gkn862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Käll L, Krogh A, Sonnhammer EL. Advantages of combined transmembrane topology and signal peptide prediction—the Phobius web server. Nucleic Acids Res. 2007;35:W429–W432. doi: 10.1093/nar/gkm256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chen Y, et al. SPD—a web-based secreted protein database. Nucleic Acids Res. 2005;33:D169–D173. doi: 10.1093/nar/gki093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bennuru S, et al. Brugia malayi excreted/secreted proteins at the host/parasite interface: stage- and gender-specific proteomic profiling. PLoS Negl. Trop. Dis. 2009;3:e410. doi: 10.1371/journal.pntd.0000410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hewitson JP, et al. The secretome of the filarial parasite, Brugia malayi: proteomic profile of adult excretory-secretory products. Mol. Biochem. Parasitol. 2008;160:8–21. doi: 10.1016/j.molbiopara.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 84.Bellafiore S, et al. Direct identification of the Meloidogyne incognita secretome reveals proteins with host cell reprogramming potential. PLoS Pathog. 2008;4:e1000192. doi: 10.1371/journal.ppat.1000192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cass CL, et al. Proteomic analysis of Schistosoma mansoni egg secretions. Mol. Biochem. Parasitol. 2007;155:84–93. doi: 10.1016/j.molbiopara.2007.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang ET, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tarazona S, Garcia-Alcalde F, Dopazo J, Ferrer A, Conesa A. Differential expression in RNA-seq: a matter of depth. Genome Res. 2011;21:2213–2223. doi: 10.1101/gr.124321.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Friedländer MR, et al. Discovering microRNAs from deep sequencing data using miRDeep. Nat. Biotechnol. 2008;26:407–415. doi: 10.1038/nbt1394. [DOI] [PubMed] [Google Scholar]
- 89.Friedländer MR, Mackowiak SD, Li N, Chen W, Rajewsky N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012;40:37–52. doi: 10.1093/nar/gkr688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mackowiak SD. Identification of novel and known miRNAs in deep-sequencing data with miRDeep2. Curr. Protoc. Bioinformatics. 2011;36:12.10. doi: 10.1002/0471250953.bi1210s36. [DOI] [PubMed] [Google Scholar]
- 91.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Winter AD, et al. Diversity in parasitic nematode genomes: the microRNAs of Brugia pahangi and Haemonchus contortus are largely novel. BMC Genomics. 2012;13:4. doi: 10.1186/1471-2164-13-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chen MX, et al. Identification and characterization of microRNAs in Trichinella spiralis by comparison with Brugia malayi and Caenorhabditis elegans. Parasitol. Res. 2011;109:553–558. doi: 10.1007/s00436-011-2283-x. [DOI] [PubMed] [Google Scholar]
- 94.Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nat. Genet. 2007;39:1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- 95.Axtell MJ. ShortStack: comprehensive annotation and quantification of small RNA genes. RNA. 2013;19:740–751. doi: 10.1261/rna.035279.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bailey TL, Elkan C. The value of prior knowledge in discovering motifs with MEME. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1995;3:21–29. [PubMed] [Google Scholar]
- 97.Fu L, Niu B, Zhu Z, Wu S, Li W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics . 2012;28:3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nawrocki EP, Eddy SR. Bioinformatics . 2013;29:2933–2935. doi: 10.1093/bioinformatics/btt509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Burge SW, et al. Rfam 11.0: 10 years of RNA families. Nucleic Acids Res. 2012;41:D226–D232. doi: 10.1093/nar/gks1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Nam JW, Bartel DP. C. elegans. Genome Res. 2012;22:2529–2540. doi: 10.1101/gr.140475.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Grant CE, Bailey TL, Noble WS. FIMO: scanning for occurrences of a given motif. Bioinformatics. 2011;27:1017–1018. doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Calculated as GC% per 10-kb nonoverlapping sliding window.
Data generated using 500-bp sliding windows using a 250-bp overlap. The x axis is GC content (%), and the y axis is the proportion of the windows (bins) corresponding to each GC measurement.
(a) tRNA copy number; (b) frequency of amino acids encode by the inferred T. suis proteome.
Based on enriched transcripts found in larval stages of T. suis embedded in the host gut epithelium and up-regulated in the stichosome relative to non-stichosomal tissues in adult males and females. Mechanisms of action (adapted from 1–4) include secretion of (1) cysteine protease inhibitors, N-glycosylated proteins and a putative TGF-β mimic to suppress antigen presentation by host dendritic and B-cells and limit prolifation/stimulation of proinflammatory T-cells; (2) calreticulins believed to bind to scavenger receptors on dendritic cells and stimulate production of 'anti-inflammatory' IL-4 and IL-10, leading to proliferation of Fox3+ regulatory T-cells; (3) calreticulins and apyrases that bind free Ca+ and ATP respectively and prevent conversion of Fox3+ to pro-inflammatory T-cells; (4) thioredoxin peroxidase (TPX) and a putative macrophage initiation factor (MIF) mimic to stimulate alternative activation of host macrophages, inhibiting inflammatory nitrous oxide (NO) production and toll-like receptor (TLR) pathways and stimulating the production of IL-4 and IL-10; and (5) serine protease inhibitors that block neutrophilsecreted cathepsin G and elastases, thereby limiting tissue destruction and reducing inflammation. Red X represents the blocking of a pathway or mechanism owing to Trichuris-generated immunomodulation.
Splice events predicted for each transcript are available in Supplementary Table 17.
(a) Exons per gene; (b) transcripts per gene.
Blue-banding highlight major clusters based on transcriptional profile for stichosome, larval, made and female enriched transcripts. Each row represents one transcript. Highest transcription represented by white coloration. Lowest transcription represented by red coloration. Color scale used allows comparison between/among libraries for each transcript, but not between/among distinct transcripts. FPKM values for each transcript available in Supplementary Table 18. Dendrogram clustering cropped for presentation.
All parasite-derived molecules are enriched in the stichosome tissues relative to nonstichosomal tissues in both male and female adult T. suis. In this schematic, we propose that secreted proteases, including S1 (chymotrypsin), S9, M12 (astacin) and C1 (cathepsin) families, play an early role in formation of the syncitial tunnel in the host gut epithelium. Among the secreted S1 proteases are several that degrade Muc2 polymers in the host intestinal mucus barrier, exposing the gut epithelial cell membranes. These membranes are then breached by T. suis secreted porins, allowing small molecule transporters to enter the host cell cytoplasm and harvest simple sugars, and nucleic and amino acids, which are then taken back into the bacilliary cells through endocytosis or another unknown mechanism. In addition, S1 proteases degrade long protein polymers found in the host blood/plasma including fibrinogen (thus inhibiting co-agulation and possibly contributing to the gut hemorrhaging) and kallikrien (possibly providing a means to inhibit inflammation by blocking bradykinin formation). While this is occurring a potent mix of immunomodulatory proteins (Supplementary Fig. 4 and Supplementary Table 16) are secreted and specifically regulate or suppress an inflammatory response. We hypothesize also that larger peptide are taken up into the stichocyte by endocytosis and vesicle transport through the bacillary cells and further degraded in the peroxisomes in the stichocytes themselves. Increased lysosome activity in the stichocytes may also suggest degradation of lipids acquired from the host cells, but we cannot propose a source for these molecules based on our current data. CU, cuticle; MU, muscle; STC, stichocyte; OL, esophageal lumen; BC, bacillary cell.
Mp, male nonstichosomal (i.e., posterior body) tissue; MALE, adult male; F.POST, female nonstichosomal (i.e., posterior body) tissue; FEMALE, adult female; L10, larvae at 10 d after infection (d.p.i.); L18, larvae at 18 d.p.i.; L28, larvae at 28 d.p.i.; STICH, adult (mixed sex) stichosomal tissue.
This represents the one significant motif, among possible motifs ranging in size from 6 to 22 nt, that was shared between 1,174 nonredundant 22A-RNA sequences. The logarithmic probability distribution of individual nucleotides in the motif is shown as a WebLogo (see Online Methods).
Supplementary Figures 1–12, Supplementary Tables 1–4, 7, 19 and 20, and Supplementary Note (PDF 3403 kb)
Male-encoded transfer RNAs. The annotated location and identity of each tRNA predicted for the male Trichuris suis genome. (XLSX 62 kb)
Female-encoded transfer RNAs. The annotated location and identity of each tRNA predicted for the female Trichuris suis genome. (XLSX 64 kb)
G protein–coupled receptors (GPCRs). Inferred GPCRs encoded by Trichuris suis based on BLASTp comparison with homologous protein sequences in the GPCR databases, IUPHAR and gpcr-sarfari (see Online Methods). (XLSX 21 kb)
Kinases. Inferred kinases encoded by Trichuris suis based on BLASTp comparison with homologous protein sequences in the ks-sarfari and Uniprot databases (see Online Methods). (XLSX 29 kb)
Phosphatases. Inferred phosphatases encoded by Trichuris suis based on BLASTp comparison with homologous protein sequences in Uniprot (see Online Methods). (XLSX 25 kb)
Peptidases. Inferred peptidases encoded by Trichuris suis based on BLASTp comparison with homologous protein sequences in the MEROPs protease domain database (see Online Methods). (XLSX 48 kb)
Peptidase inhibitors. Inferred peptidase inhibitors encoded by Trichuris suis based on BLASTp comparison with homologous protein sequences in the MEROPs protease inhibitor domain database (see Online Methods). (XLSX 24 kb)
Transporters and channels. Inferred peptidase inhibitors encoded by Trichuris suis based on BLASTp comparison with homologous protein sequences in the Transporter Classification Database (TC-DB; see Online Methods). (XLSX 125 kb)
Secretome. Inferred secreted proteins encoded by Trichuris suis based on encoding a signal peptide and lacking a recognizable transmembrane domain and having BLASTp homology to sequences in the Secreted Proteins Database (SPD) or to proteomic data available for the secretome of at least one helminth species (see Online Methods). (XLSX 52 kb)
Secreted peptidases. Inferred secreted peptidases based on their inclusion in the predicted Trichuris suis secretome and inferred (BLASTp) homology to the MEROPs protease domain database. (XLSX 19 kb)
Immunomodulatory proteins. Trichuris suis–encoded proteins with sequence homology to known immunomodulatory proteins secreted by this or any other species of helminth and their stage-, sex- and tissue-specific transcription (in FPKMs). (XLSX 17 kb)
Alternative splicing. A summary of predicted alternative splicing events of the Trichuris suis transcriptome and the functional (Pfam) domains encoded by each transcript splice isoform. (XLSX 1559 kb)
Stage- and tissue-specific transcription FPKM values. Normalized transcriptional levels (expressed as fragments per kilobase per million reads (FPKM)) of each Trichuris suis messenger RNA in each stage-, sex- and tissue-specific library. (XLSX 3472 kb)
Small RNA read clusters. A summary of small RNA read clusters mapping to the Trichuris suis male assembly (used for all small RNA mappings owing to its larger size), showing, for each cluster, its location in the male assembly, whether the cluster is predicted to form a hair-pin fold (HP), the dominant DICER-derived read length among the reads mapping to each cluster, whether the proportion of reads mapping to the anti-sense strand, whether the cluster is found with 500 bp of an annotated transcription start site (i.e., transcription start site associated (TSSa)), the number of reads mapping to each cluster, and the number and ratio of these reads mapping to the 'forward' or 'reverse' stand, whether the cluster has a substantial forward or reverse strand bias (≥ 80% of reads mapping to one strand), the number of reads, categorized by their 5' and 3' nucleotides, mapping to each cluster, the transcription associated with each cluster in each stage-, sex- or tissue-specific library (normalized as FPKM values), and the number of reads mapping to each cluster by read length. (XLSX 7089 kb)
Micro-RNAs. The micro-RNAs (miRNAs) predicted for Trichuris suis, their scoring parameters, the number of reads in the total dataset mapping to their predicted mature, star and hairpin regions, nearest homolog in another nematode, and predicted mature, star and precursor sequence and their stage and tissue-specific transcription (in total reads and FPKMs). (XLSX 92 kb)