Abstract
Memory B cells (MBCs) are long-lived sources of rapid, isotype-switched secondary antibody-forming cell (AFC) responses. Whether MBCs homogeneously retain the ability to self-renew and terminally differentiate or if these functions are compartmentalized into MBC subsets has been unclear. It was previously suggested that antibody isotype controls MBC differentiation upon restimulation. Here we demonstrate that subdividing MBCs based on expression of CD80 and PD-L2, independent of isotype, identified MBC subsets with distinct functional behaviors upon rechallenge. CD80+PD-L2+ MBCs differentiated rapidly into AFCs but did not generate germinal centers (GCs); conversely CD80−PD-L2− MBCs generated few early AFCs but robustly seeded GCs. Gene expression patterns of subsets support both the identity and function of these distinct MBC types. Hence, MBC differentiation and regeneration are compartmentalized.
Introduction
Memory B cells (MBCs), which provide protection against antigen re-exposure1–3, can differentiate into antibody-forming cells (AFCs) and make new antibodies, or enter germinal centers (GCs) and provide a renewed source of lasting B cell immunity. Despite the importance of MBCs for vaccine- and infection-induced protection4–6, we have a limited understanding of the nature of these cells and how they participate in secondary responses.
Based on expression microarray comparisons between MBCs and naïve B cells, we previously identified several surface proteins—including CD80, PD-L2 and CD73—that are expressed exclusively on MBCs and serve to divide MBCs into multiple phenotypic subsets7. We have focused on subpopulations of MBCs defined by expression of the two B7 family members, CD80 and PD-L2. These subsets differ in a number of properties: CD80−PD-L2−, double-negative (DN) MBCs, have relatively very few mutations7,8. CD80+PD-L2+, double-positive (DP) MBCs have the most mutations, and CD80−PD-L2+ single-positive (SP) MBCs have an intermediate mutational content7,8. Although all subsets contain cells expressing surface B cell receptors of the immunoglobulin M (IgM) or switched IgG isotypes, the DN subset is predominantly IgM+, and the SP and DP populations contain progressively more IgG+ cells. These two features—mutation and isotype switch—which are both irreversible DNA alterations that occur during the primary response, indicate that the memory populations are stable and that cells do not move from one population to another (otherwise mutational content and switching would equalize between the populations).
Classically, B cell secondary responses generate rapid effector function, most likely by quickly converting MBCs to AFCs9. This raises the question of how the memory compartment undergoes self-renewal in the face of terminal differentiation of MBCs into AFCs. Though it is unclear how MBCs are homeostatically maintained, stem cell gene expression signatures have been identified in MBCs10–12. It has been proposed that self-renewing MBCs represent a discrete population that can differentiate into both plasma cells and GC B cells after antigen re-exposure10,11. If this were the case, it is possible that either all MBCs retain self-renewal as well as terminal differentiation potential, with the fate of the cell being determined by environmental cues13. Alternatively, these functions may be segregated into different dedicated subsets of MBCs, which may be pre-programmed to respond differently even upon identical stimuli.
Recently two groups have suggested that the MBC pool is functionally divided by antibody isotype expression, either IgM or switched IgG14,15. They found that isotype-switched MBCs differentiated into AFCs while IgM+ MBCs generated new GCs. From these results they proposed that surface isotype reflects fundamental differences in MBC potential, and suggested that signaling differences between IgG+ and IgM+ cells could govern different functional responses16,17. On a parallel track, we proposed that the subsets defined by CD80 and PD-L2 expression represent a spectrum of MBC commitment, with the DN cells being more “naïve-like” and the DP cells more “memory-like”9. Expression of these subset markers on murine MBCs has been confirmed by others in different systems17–20. We hypothesized that upon reactivation the more memory-like DP MBCs will differentiate quickly into effector cells that function by providing new AFCs and not GCs, and that more naïve-like DN MBCs will make new GCs thus renewing the memory pool by providing a new source of cellular immunity.
Here we have tested these hypotheses by examining the function after reactivation in vivo of MBC populations distinguished by CD80 and PD-L2 expression, while controlling for isotype expression. We generated, purified and transferred these MBC subsets with and without T cells and assessed their ability to make AFCs and GCs upon reexposure to antigen. We found substantial functional heterogeneity that was independent of isotype, but dependent on subset markers. Hence, MBC functional heterogeneity is not determined by BCR isotype, as thought, but rather by cell intrinsic features that are captured by the expression of key surface markers. This view of the composition of the MBC compartment has implications for monitoring immune states and hence for vaccine development.
Results
Generating, purifying and testing MBC subsets
Wild-type mice generate exceedingly small populations of MBCs (2–4 × 104 per spleen)21,22. Though such mice develop the MBC subpopulations we are studying19, there are too few MBCs in wild-type mice to permit purification and subsequent retransfer. Thus, to generate more robust numbers of MBCs, we used a transfer system similar to that previously used7, based on the Igh-targeted B1.8 knockin (KI) mouse developed by Rajewsky and colleagues23. The Igλ-expressing B cells in such mice recognize the haptens 4-hydroxy-3-nitrophenyl acetyl (NP) and 4-hydroxy-5-iodo-3-nitrophenyl acetyl (NIP). Specifically, to generate MBCs we transferred 1 × 106 NIP-binding (NIP+) B cells from B1.8+/− Jκ+/− BALB/c mice into gene-targeted BCR transgenic recipient BALB/c mice that bear an irrelevant BCR (AM14 Tg x Vκ8R KI BALB/c; Supplementary Fig. 1). These recipients have normal lymphoid architecture and composition, but only transferred B cells can respond to NP, since recipient mice have no NP-specific B cells. We immunized recipients with the T-dependent antigen NP-chicken gamma globulin (CGG) precipitated in alum. Assuming that <5% of transferred cells stably home to the spleen, this gives a precursor frequency pre-immunization of about 5 × 104 cells, a number that we have validated to be in a similar range to that seen in wild-type mice7.
At 8 weeks post-immunization, ~1–3% of the cells in the recipient spleen were NP-binding (Fig. 1a and unpublished), but not of GC phenotype (Supplementary Fig. 2); these cells were presumptive MBCs. Approximately 20–30% of NIP+ MBCs generated were IgG1+ (Fig. 1a and unpublished). These subsets are stably observed at 26 weeks post-immunization (Supplementary Fig. 3).
Figure 1.
Generation and purification of MBC subsets. (a) Flow cytometry analysis of splenic cells from AM14 Tg x Vκ8R KI recipient mice that received NP-specific B cell and were at 8 weeks post-immunization with NP-CGG in alum. Left shows gating on antigen-specific B cells (CD19+NIP+) after gating on live cells and right shows staining for IgG1. Numbers indicate percentage of the parent-gated cells in the indicated population. Data are from one mouse from one experiment, representative of nine independent experiments with twenty to thirty mice per experiment. (b) Flow cytometry analysis of splenic B cells from AM14 Tg x Vκ8R KI recipient mice that did not receive NP-specific B cells but were immunized with NP-CGG in alum 8 weeks prior. Numbers indicate percentage of the parent-gated cells in the indicated population. Data are from one mouse from one experiment, representative of three independent experiments with three mice per experiment. (c) MBC subset distribution and frequency after gating as in (a), after staining for either IgG1 or IgM: (left) CD19+ NIP+ IgG1− MBCs (right) CD19+ NIP+ IgM− MBCs. Subsets were identified according to expression of CD80 and PD-L2 that separates up to three populations: DP, SP and DN. Numbers indicate percentage of the parent gated cells in the indicated population. Data are from one mouse from one experiment, representative of nine independent experiments with twenty to thirty mice per experiment (left) or two independent experiments with forty-three to forty-five mice per experiment (right).
Identity of MBC subsets
To functionally define MBC subsets, we sorted cells by flow cytometry from each subset, along with naïve precursor cells for subsequent re-transfer or in vitro analysis. For retransfers, we purified MBC subsets by flow cytometry by gating either on IgG1− MBCs for DP, SP and DN expression patterns, or in separate sorts by gating on IgM− MBCs for DP and SP expression patterns (Fig. 1c). We used these strategies to avoid engaging the B cell receptor of the sorted cells. Separately, we found nearly all of IgG1− MBCs were IgM+, and conversely that the IgM− MBCs were mostly IgG1+ (Supplementary Fig. 4a). Critically for interpretation of the subsequent experiments, the only types of MBCs that could generate IgG1+ AFCs, assayed in our study, were those that express IgM, IgG1, or IgG3; however, the latter only represents ~2% of the MBCs (Supplementary Fig. 4b).
Initially, to better define subset identities, we performed microarray-based transcriptome analysis on cells sorted as above but without regard to BCR isotype. All three MBC subsets were transcriptionally more similar to each other than they were to naïve cells (Supplementary Fig. 5a). However, MBC subsets as defined by CD80 and PD-L2 have clearly distinct gene expression patterns (Supplementary Fig. 5b). Notably, genes encoding a number of transcription factors that are candidates for specification of subset identity—including Mef2b, Uhrf1, Zbtb32, Bcl6, Satb1 and Klf2—were differentially expressed. While the functional significance of specific expressed genes remains to be determined, these data provide further support for exploring the overall functional capacities of these cell types.
Functional differences among subsets upon immunization
To test MBC subset function upon secondary immunization, we transferred 5 × 104 sort-purified MBCs into AM14 KI x Vκ8R KI BALB/c recipient mice, allowing comparison of the MBC subset function between animals without confounding host effects. Given the ~5% recovery of transferred B cells, the precursor frequency of the responding cells was at or below what would expected in an intact wild-type mouse. Of note, the numbers of Igλ variable region (V) mutations per B cell was low in all of these populations, consistent with limited affinity maturation in most MBCs (compared to long-lived plasma cells that contain many more V region mutations)24 (F.J.W. and M.J.S., manuscript submitted). In particular, all populations contain few mutations, including MBCs that are unmutated7,24, and notably more than half of DP B cells have no replacement (R) mutations throughout their light chain V regions, and that 70% lack any R mutations specifically in CDRs where such changes might affect affinity (Supplementary Fig. 6). This finding suggests that starting affinities of MBC populations will be relatively similar.
We immunized recipients of MBCs one day post-transfer with NP-ovalbumin (OVA) precipitated in alum (Supplementary Fig. 7a). In some groups we also included in vitro generated rested-effector DO11.10 T cell receptor (TCR) transgenic T cells, which have memory cell properties and thus ought to be the most physiologic partner for MBCs4. These T cells are OVA-specific and were obtained from DO11.10 Tcra−/− BALB/c mice They had a surface phenotype of CD25−, PD-1−, and CD44+ (Supplementary Fig. 7b), similar in phenotype to memory T cells as reported4. We refer below to these T cells simply as “memory T cells”. Such cells represent a source of homogeneous and synchronous memory T cells that could not have been generated via in vivo priming, as this generates inadequate numbers of recoverable cells (our unpublished observations).
Early burst of AFCs generated by IgG1− DP and IgG1+ MBCs
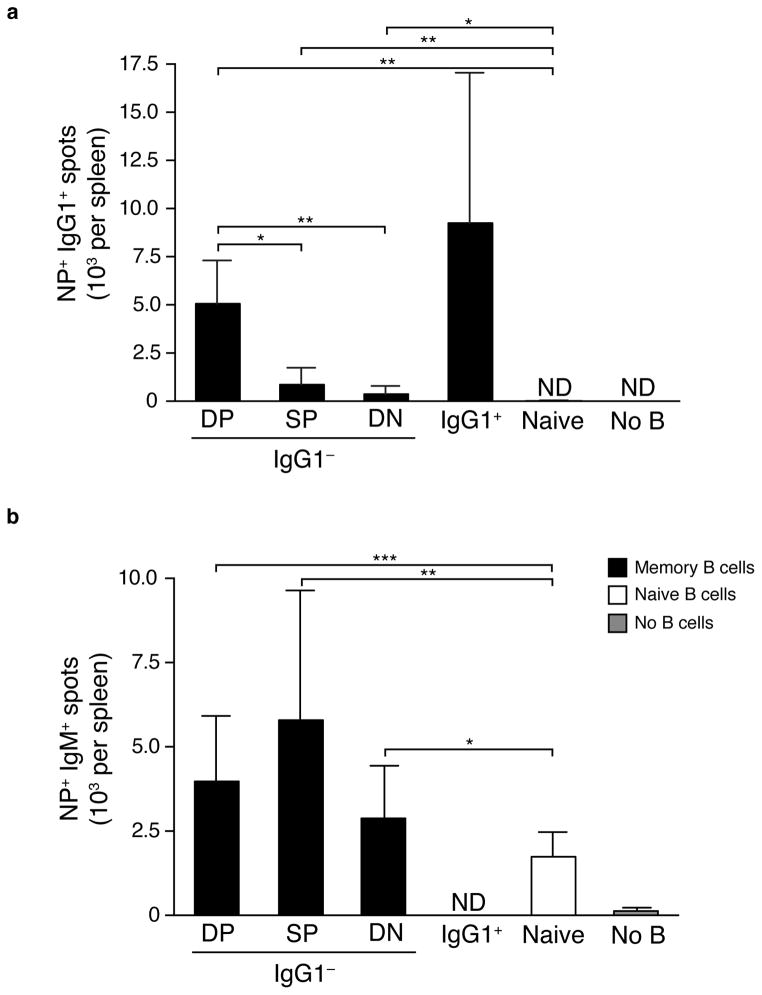
We assessed the capacity of IgG1− or IgG1+ MBC subsets in the spleen to differentiate into AFCs 3.5 days post-immunization. Of note, DP IgG1− MBCs produced numbers of IgG1+ AFCs similar to those produced by the total population of IgG1+ MBCs (Fig. 2a). Hence, at least one MBC subset comprised almost completely of IgM+ MBCs is just as efficient a source of isotype-switched AFCs as previously switched MBCs. Among IgG1− MBCs, the DP subset generated 5- and 13-fold more NP+ IgG1+ AFCs than SP or DN B cells, respectively (Fig. 2a), indicating that these memory-like MBCs have a greater capacity to rapidly undergo isotype switch and differentiate into AFCs. DP MBCs also generated AFCs with the highest relative affinity of any subset of MBCs (Supplementary Fig. 8).
Figure 2.
DP IgG1− and IgG1+ MBCs are the major producers of early IgG1+ AFCs. Numbers of AFCs per spleen were determined by ELISPOT after transfer of DP, SP or DN IgG1− MBCs; IgG1pos MBCs or naïve B cells in recipient mice 3.5 days post immunization with NP-OVA in alum. (a) Numbers of NP+ IgG1+ AFCs generated in mice transferred with the indicated populations of cells. ND, not detected. * p<0.001, ** p<0.0001. (Mann Whitney nonparametric, two-tailed test). Data are combined from two independent experiments (error bars represent standard deviation) with five to fourteen mice per group. (b) Numbers of IgM+ AFCs. ND, not detected. * p<0.05, ** p<0.001, *** p<0.0001. (Mann Whitney nonparametric, two-tailed test). Data are combined from two independent experiments (error bars represent standard deviation) with three to sixteen mice per group.
At day 3.5, IgG1− DN MBCs still produced more IgG1+ AFCs than did naïve B cells. IgM+ NP-specific AFCs were generated to the same extent among IgG1− MBC subsets, with the exception of IgG1− SP MBCs that generated slightly more AFCs (Fig. 2b). As with IgG1+ AFCs, all three MBC subsets also generated more IgM+ AFCs than did naïve B cells (Fig. 2b). Hence, with respect to IgG1 AFC generation, even DN MBC, which are inferior to other MBCs, are still substantially more capable than naïve B cells.
IgM MBCs do not depend on memory T cells to make AFCs
In the above experiments, MBCs were co-transferred with antigen-specific memory T cells. However, to establish the degree to which MBC responses depend on such T cells, we repeated the above experiments without adding them. We found that IgG1− DP MBCs generated more IgG1+ AFCs than DN MBCs, even in the absence of a preexisting memory T cell population (Fig. 3a). Nonetheless, IgG1− DN MBCs generated fewer IgG1+ AFCs (one-sixth the frequency) and IgM+ AFCs (roughly half) when only recipient naive T cell help was available (Fig. 3), compared to experiments that cotransferred memory T cells (Fig. 2). These data indicate that IgG1− MBCs need specific T cells for optimal expansion and/or differentiation into IgG1+ AFCs but that such T cells do not alter the underlying nature of the MBC subset responses.
Figure 3.
MBC requirement of T cells to generate early AFC responses. Numbers of AFCs per spleen were determined by ELISPOT analysis after transfer of DP, SP or DN IgG1− MBC; IgG1pos MBC or naïve B cells in recipient mice 3.5 days post immunization with NP-OVA in alum. Recipient mice were treated with anti-CD4 or PBS as a control before transfer of B cells. (a) Numbers of NP+ IgG1+ spleen AFCs. (b) Numbers of NP+ IgM+ spleen AFCs. Six to nineteen mice per group for PBS treated mice, and five to thirteen mice per group for anti-CD4 treated mice. ND, not detected. * p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001. (Mann Whitney nonparametric, two-tailed test). Data are combined from two (for anti-CD4 treated mice) or three (for PBS treated mice) independent experiments (error bars represent standard deviation).
T cell requirement for MBCs to generate AFCs
To test whether MBC responses required any type of T cell, recipient mice were pre-treated with an anti-CD4 monoclonal antibody to deplete recipient T cells before transferring MBC subsets (Supplementary Fig. 9). Virtually no IgG1+ AFCs were generated from any IgG1− MBCs in T cell-depleted recipients (Fig. 3a), indicating that some form of T cell help is required for IgM+ MBCs to rapidly make switched antibodies. IgG1+ MBCs generated 18-fold more NP+ IgG1+ AFCs in recipients with naïve T cells compared to when T cells were depleted (Fig. 3a). These findings indicate that even IgG MBCs greatly, though not absolutely, depend on T cells to generate IgG1+ AFCs.
Furthermore, IgG1− DP MBCs generated 3-fold more IgM+ AFCs and IgG1− DN MBCs generated 3.7-fold more IgM+ AFCs (Fig. 3b) in intact recipients compared to T cell-depleted recipients. Due to the limited numbers of IgG1− SP MBCs sorted, we were only able to study this subset in recipients that were T-depleted or that had received memory T cells: IgG1− SP MBCs generated 1.7-fold more IgM+ AFCs with memory T cells (Fig. 2b) than without T cells (Fig. 3b). IgG1− SP MBCs were able to make the most IgM+ AFCs in either recipient. These observations indicate that IgG1− MBCs do not require T cells to differentiate into IgM+ AFCs. However, T cells contribute to numerical expansion of IgM AFCs and generation of IgG1 AFCs, though such T cell help could derive from either polyclonal naïve T cells or memory antigen-specific T cells.
Memory T cells expand more with IgG1− DP MBCs
Both CD80 and PD-L2, used to define MBC subsets, directly interact with molecules expressed on helper T cells25–27. Therefore, we sought to determine if MBC subsets had a differential effect on antigen-specific memory-like T cells. Indeed, antigen-specific T cells proliferated to a significantly greater extent in recipients that received IgG1− DP MBCs relative to recipients that received other MBC types or naïve B cells (Fig. 4). That DP MBCs were superior in this regard might indicate that expression of both B7 family member molecules could contribute to such an outcome, though this remains to be tested directly.
Figure 4.
DP IgG1− MBCs promote more cognate T cell expansion than do other B cell types. Numbers of KJ1-26+ CD4+ OVA-specific T cells in mice that received memory T cells with DP, SP or DN IgG1− MBCs; IgG1+ MBCs; naïve B cell or no B cells, 3.5 days after immunization with NP-OVA in alum. * p<0.01, ** p<0.001, *** p<0.0001. (two-tailed t-test). Data are combined from two independent experiments. Error bars represent standard deviation, with six to sixteen mice per group.
DN and SP, but not DP, MBCs generate new GCs
Previous studies suggested that only IgM+ MBCs generate new GCs14,15. However, these studies did not separate MBCs according to subset as defined by CD80 and PD-L2. Given the heterogeneity we observed among IgM+ MBCs we hypothesized that they could also differ in their capacity to make GCs upon reimmunization. To test this, mice were sacrificed 10.5 days after immunizing secondary recipients that had received different subsets of IgG1− MBCs with memory T cells. IgG1− DP MBCs did not generate GC B cells, while IgG1− DN MBCs produced almost as many GC B cells as did naïve B cells (Fig. 5a). Conversely, we assessed the capacity of IgM− MBCs, which had previously been thought to be unable to generate secondary GCs. In this case, DN IgM− MBCs were too rare to study, but we were able to test SP and DP MBCs. IgM− SP MBCs were able to generate GC B cells, and such cells were similar in number to those derived from IgG1− SP MBCs (Fig. 5b). These results indicate that the capacity for MBCs to make GCs is not dependent specifically on their isotype, since SP MBCs of either isotype and IgM+ DN MBCs can make GCs with similar efficiency. Rather, subset identity is a better predictor of GC-forming capacity of MBCs.
Figure 5.
GC B cells are generated from SP or DN IgG1− MBCs or naïve B cells, but not DP MBCs. Numbers of CD95+ CD38− NIP+ GC B cells 10.5 days after immunization generated after transfer of: (a) DP, SP or DN IgG1− MBCs; IgG1+ MBCs; naïve B cells or no B cells, along with memory T cells, or (b) DP or SP IgM− MBCs, naive B cells or no B cells, along with memory T cells. ND, not detected. * p<0.05, ** p<0.01, *** p<0.0001. (two-tailed t-test). Data are combined from three independent experiments with eight to twenty-two mice per group for NP-OVA in alum immunized mice and one to fifteen mice per group for alum immunized mice (a), or two independent experiments with six to thirteen mice per group for NP-OVA in alum immunized mice or two to six mice per group for alum immunized mice (b). Error bars represent standard deviation.
DN MBCs make the most late AFCs
At day 10.5 post-immunization, in contrast to day 3.5, IgG1− DN MBCs contributed most of the IgG1+ NP-specific AFCs in the spleen (Fig. 6a). IgG1− DN MBCs generated 7-fold more IgG1+ AFCs than IgG1− DP MBCs and 2.5-fold more AFCs even than IgG1+ MBCs at day 10.5 (Fig. 6a). Since GCs were readily generated by IgG1− DN MBCs (Fig. 5a), this finding suggests that AFCs derived from IgG1− DN MBC could emanate from secondary GCs. In keeping with this observation, IgG1− DN MBCs and naïve B cells, which both generated the largest numbers of NP+ GC B cells, also generated the most IgG1+ AFCs 10.5 days post immunization (Fig. 6a). At day 10.5, both DP and SP subsets of IgM− MBCs generated fewer IgG1+ AFCs than did naïve B cells or IgG1− DN MBCs (Fig. 6a). There were few IgM+ AFCs in the spleen at day 10.5, with little difference in the numbers of AFCs generated by any of the IgG1− MBC subsets (Fig. 6b). Overall, MBC subsets that generated an early large burst of AFCs did not proliferate further and were overtaken by progeny of MBC subsets that were more capable of proliferation and secondary GC generation.
Figure 6.
Late IgG1+, but not IgM+, AFC formation is dominated by DN IgG1− MBCs and naïve B cells. Numbers of AFC per spleen were determined by ELISPOT analysis after transfer of DP, SP or DN IgG1− MBC; DP or SP IgM− MBC; naïve B cell or no B cell responses in spleens of mice that received memory T cells 10.5 days post immunization with NP-OVA in alum or alum alone (not shown in a as there were no detectable responses). (a) Numbers of NP+ IgG1+ spleen AFCs. (b) Numbers of NP+ IgM+ spleen AFCs.* p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001. (Mann Whitney nonparametric, two-tailed test). Data are combined from four independent experiments with seven to twenty-four mice per group (a) or two independent experiments with six to sixteen mice per group for alum immunized mice or one to eleven mice per group for alum immunized mice (b). ND, not detected. Error bars represent standard deviation.
Gene expression patterns of MBC subsets
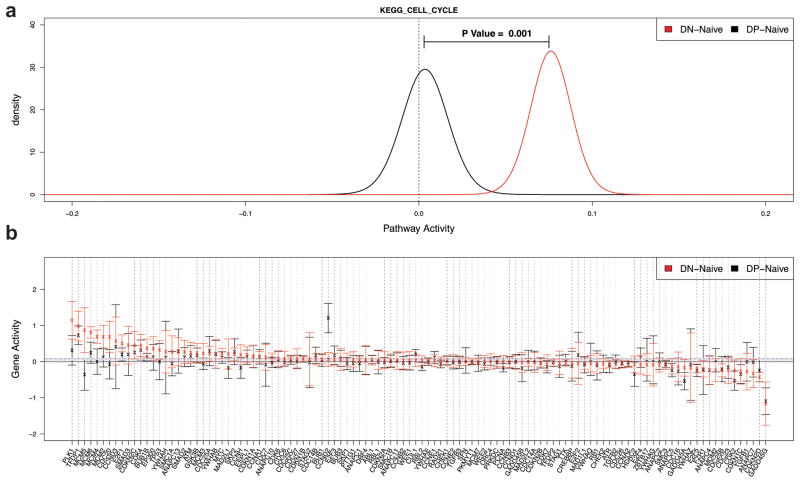
The above data demonstrate that DP MBCs differentiate rapidly into AFCs whereas DN MBCs undergo a substantial early proliferative burst, then differentiate into GC B cells. We sought clues to these behaviors in the microarray data as described above. Notably, the transcriptome of DN MBCs demonstrated strong signatures of cell cycle-promoting genes, when compared to naïve B cells, whereas this was not the case for DP MBCs. This is based on KEGG cell cycle annotation (Fig. 7), as well as several related REACTOME pathways including E2F-regulated genes28 (not shown). To further bolster the conclusion that cell cycle-related genes were more highly expressed in DN than DP cells, we used quantitative PCR on sorted MBC subsets obtained 45 weeks post-immunization. Three of five tested genes from the KEGG cell cycle list, Cdc20, Mcm5, and Plk1—but not E2f and Ccn2—were confirmed to be more abundantly expressed in this assay (Supplementary Fig. 10 and data not shown). The transcriptional repressor Zbtb32, which is associated with plasma cell differentiation29, was expressed by over 10-fold higher in DP MBCs as compared to DN MBCs as determined by q-PCR (Supplementary Fig. 9). Together these data provide initial clues as to why DN MBCs may be more prone to proliferate rather than differentiate, whereas DP MBCs are destined for AFC differentiation upon reimmunization.
Figure 7.
The cell cycle pathway is significantly activated in the DN subset. QuSAGE was used to quantify the activity of the KEGG CELL CYCLE gene set. Data are derived from three biological replicates per cell type. (a) Activity Probability Density Functions (PDFs) of the DN subset relative to naïve B cells (red) and DP subset relative to naïve B cells (black). (b) Activity of individual genes in the KEGG CELL CYCLE pathway in the DN subset (red) and DP subset (black). Activity is quantified by the log2 fold-change relative to naïve B cells. Points indicate the mean and bars show the 95% confidence intervals.
DISCUSSION
In this study we elucidate the distinct functions of defined, stable MBC subsets that were previously described by us7,8 and others17–20,30. Studying these three types of MBCs, we found that there was a spectrum of behavior ranging from more “naïve-like” to more “memory-like”. DN MBCs, phenotypically resembling naïve B cells, were the MBCs capable of forming substantial secondary GC responses with similar efficiency to naïve B cells. Importantly, they also differed from naïve B cells, since DN IgM+ MBCs but not naïve B cells were able to generate switched AFCs at critical early time points. In contrast, DP IgM+ MBCs produced no GC B cells but generated remarkably quick and large isotype-switched secondary AFC responses. SP MBCs had intermediate properties, with some capability to make secondary GCs and a robust ability to spawn AFCs, albeit to a lesser degree than DP MBCs.
This work should revise current views on heterogeneity within the MBC compartment. It has previously been posited that expression of IgM and IgG per se define functional MBC subsets14,15. However, our data indicate that these are surrogate markers, although they correlate to some degree with function. This conclusion is substantiated by our demonstration of functional heterogeneity within both IgM+ and IgG1+ MBC compartments, in which the markers CD80 and PD-L2 delineate unique functional capacities. Conversely, across IgM+ and IgG1+ phenotypes, the MBCs expressing similar B7-family member profiles have similar functions, as most clearly highlighted by the equal GC-forming capacity of SP MBC regardless of isotype.
Nonetheless, the earlier findings can be reconciled with our current observations. Previously IgM+ MBCs were reported to generate secondary GCs. In agreement with this finding, IgM+ MBCs do include a substantial fraction of DN MBCs, which are capable of re-entering GCs. However, among IgM+ MBCs are both SP and DP MBC, which generate a rapid isotype-switched AFC response, with DP MBCs generating essentially no secondary GC response. Hence responses of IgM+ MBCs are truly heterogeneous and expression of IgM by itself is not predictive of MBC behavior upon restimulation. Similarly, IgG+ MBCs were reported to only make AFCs. However, IgG1+ MBCs were also heterogeneous with respect to CD80 and PD-L2 expression. SP and DP IgG1+ MBC subsets parallel the behavior of the similar IgM MBC subsets, most notably with respect to the ability of SP to make secondary GC responses. These findings elucidate unsuspected functional heterogeneity, even among IgG1+ MBCs.
We also previously found that subsets of MBCs express CD73 (refs. 7,12). From these data we inferred that there could be as many as 5 different MBC subsets just defined by CD80, PD-L2 and CD73, notwithstanding diversity of isotype expression. Thus, there could be yet more functional heterogeneity to be discovered.
While this work focused on intrinsic function of MBCs, we also studied effects of T cells on MBCs and of MBCs on T cells. A strength of our study involved the use of memory T cells, which normally would be the collaborating partner for MBCs, thus we believe our results are likely to reflect the physiological condition. The addition of T cells, and in particular rested effectors that are qualitatively similar to memory T cells, greatly augmented responses of all subsets, allowing for the clearest demonstration of the subset differences. Notably, the nature of the differences between the subsets was consistent regardless of the T cells being used. In contrast, when we depleted all T cells, including polyclonal host T cells, there were minimal MBC responses, except by IgG1+ MBCs, which generated some (though substantially fewer) AFCs. This result was in contrast to responses to virus-like particles, which do not require T cells for MBC responses22.
Another advantage of using defined T cell populations to collaborate with MBCs was that we could track T cell responses to immunization in the context of different types of MBCs. One important finding was that antigen-specific T cells expanded to a greater degree when collaborating with DP MBCs. This result occurred despite the fact that DP MBCs themselves actually do not form robust GCs. It is intriguing to speculate that expression of CD80 and PD-L2 modulated T cell responses, since T cells should express the receptors for both ligands (i.e. CD28, CTLA-4 and PD-1)31,32. This finding would be reminiscent of effects upon T follicular helper cells by the expression of these B7 family members on B cells in a primary GC reaction27,33. Our results point to an added layer of complexity and tuning occurring in T–B interactions during secondary responses.
An important implication of our findings is that effector function and self-renewal are segregated into two distinct cell types within the MBC compartment. This scenario is unlike the situation for both naïve B and T cells, in which a single cell has the capacity to form all of the dedicated progeny, including effector and memory type cells. In this regard, the MBC compartment adopts a configuration more like a stem cell and committed progenitor model34. In this view, DN MBCs resemble secondary stem cells while DPs would correspond to committed progenitors. One might predict that there will be parallels to well-studied stem cell systems35,36. Indeed, others have noted that both bulk memory T and memory B cells share some gene expression patterns with hematopoietic stem cells11. Others and we have observed in unseparated MBCs the expression of Bmpr1a10–12, which encodes a receptor that commonly regulates stem cell maintenance versus differentiation37. Segregation of function into different MBC subsets may ensure self-renewal while still enabling robust responses to external signals. This suggests that a layered or multi-subset approach to secondary responses has evolved, which may be particularly suited for pathogens that mutate, as the DN subset contains MBCs that can respond to such adaptations, even if the antibody secreted from progeny of DP MBCs possesses little cross-reactivity with a mutant pathogen6,38,39.
Additionally, our findings have implications for vaccine design and monitoring. Different pathogens may require different types of MBC responses for effective clearance. Rational vaccine design would involve optimizing adjuvants, routes, timing and composition to elicit the desired MBC subsets that would in turn match the need for effective clearance and immunity to each specific pathogen. Monitoring of vaccine responses ideally would include measurement of antigen-specific B cells in all relevant MBC subsets; currently only serologic responses at sometimes arbitrary time points are the accepted measures of success2,40. Incorporation of this new knowledge and approach could shed light on why some vaccines yield protection of limited duration or efficacy. In this regard, exactly how pathogens and other immunologic contexts (including adjuvants, routes, molecular structure of the antigen) may influence subset development and even inherent characteristics, is not yet known, but would be important to determine.
Finally, how the subsets of MBCs we have defined achieve their different functions needs to be resolved. The detailed molecular basis for such differentiation at the level of transcription factor expression, epigenetic programming and signaling remains to be defined.–Nonetheless, as an initial step, our microarray analysis highlights differential expression of key transcription factors, including Zbtb-family members that are associated broadly with lineage specification. We have also uncovered transcriptional signatures related to cell cycle progression that may underlie DN MBC propensity for initial expansion rather than differentiation. The current results are an important advance in demonstrating differential functionality of stable, authentic subsets; intriguingly, expression of CD80 on some human MBCs is already reported41. Our results help elucidate a rapidly evolving literature on heterogeneity within MBCs. In particular they explain how the MBC compartment overall both provides immediate effector function and replenishes itself, which has been a central question in the field.
Online METHODS
Mice and immunizations
B1.8 KI BALB/c mice were generated as described23 and maintained on the Jκ KO strain42 to enrich the frequency of λ+ NP-specific B cells. B1-8 KI +/+ Jκ KO −/− mice were crossed to BALB/c mice from The Jackson Laboratory to generate B1.8+/−Jκ+/− BALB/c mice, which were used for naïve controls and for transfers of NP+ B cells used to generate MBCs. AM14 Tg x Vκ8R KI BALB/c mice were generated as described43–45, which were used as recipient mice for primary immunization, sorting of MBCs, and week 4 secondary responses.
AM14 KI BALB/c x Vκ8R KI BALB/c mice were generated as described45,46, and were used as recipient mice for secondary responses. DO11.10 Tcra−/− BALB/c mice were generated as described47, which were used as a source for T cells to make memory T cells. Their TCR recognizes the amino acids 323–339 (ISQAVHAAHAEINEAGR) of OVA.BALB/c mice from The Jackson Laboratory were used as a source of APCs to make memory T cells. All mice were maintained under specific pathogen-free conditions. The Yale Institutional Animal Care and Use Committee approved all animal experiments.
For generating MBCs in a primary response, mice were immunized intra-peritoneally with 50 μg of NP-CGG precipitated in alum. The ratio of NP to CGG ranged between 26 and 33. For secondary responses mice were immunized intra-peritoneally with 50 μg of NPOVA precipitated in Alum. The ratio of NP to OVA ranged between 8 to 10. Precipitated alum alone was also used as a control in day 10.5 experiments. All mice were immunized at 6–12 wk of age.
Antibodies and detection reagents
The following staining reagents were prepared in our laboratory: NIP-binding reagents (NIP-allophycocyanin), monoclonal anti-CD4 (GK1.5), monoclonal anti-IgM (B7-6), anti-CD19 (1D3.2), anti-CD44 (1M7) as described48. Anti-PD-L2 (TY-25), anti-CD80 (16-10A1), anti-CD38 (90), anti-TCR DO11.10 (KJ126), anti-CD62L (Mel-14) were from BioLegend. Anti-IgG1 (A85-1), anti-CD4 (RM4-4) were from BD Biosciences. Anti-CD19 (1D3) and anti-CD95 (Jo2) were from BD Pharmingen.
Flow cytometry, cell sorting and ELISpot assay
Flow cytometry and the ELISPOT assay for AFC formation were performed as described27. Dead cell exclusion in all flow cytometry experiments used propidium iodide or ethidium monoazide (Molecular Probes). Doublets were excluded in gating strategy.
For analysis of the production of AFCs by ELISPOT assay or antibody by ELISA, plates were coated overnight at 4 °C with 5 μg NP-BSA conjugated at the appropriate ratio (NP4-BSA for IgM, NP16-BSA for IgG1, and NP1.9–2.0-BSA for IgG1 affinity studies). For all ELISPOTs the mean of triplicates for each mouse is reported.
For flow cytometry sorting, spleens were pooled from AM14 x Vκ8R BALB/c mice, kept on ice in buffers without sodium azide and treated with unlabeled anti-FcγRII/II (24.G2) and stained with the relevant antibodies. Propidium iodide was used for live/dead discrimination. Cells were sorted on a FACSAria (BD Biosciences). Data were analyzed with FlowJo software (TreeStar).
Adoptive transfer for memory generation
Splenic B cells from B1-8 mice were prepared using the EasySep Mouse B Cell Enrichment Kit following the manufacturer’s protocol (StemCell Technologies). Single cell suspensions were transferred intravenously into tail veins of recipient mice. Approximately 1 × 106 NIP+ B cells were transferred per mouse. The purity of B cells was typically 90%. Approximately 12–24 h post transfer mice were immunized as described above.
Adoptive transfer for secondary responses
MBC subsets from AM14 Tg x Vκ8R KI BALB/c mice or naïve B cells from B1-8 mice were sorted, as described below. Single cell suspensions of 5 × 104 B cells and, when noted, 2 × 105 memory T cells” were transferred intravenously into tail veins of recipient AM14 KI x Vκ8R KI BALB/c mice. The purity of sorted cells was upwards of 98%. Approximately 20–24 h post transfer mice were immunized as described above.
Naïve CD4 T cell isolation and preparation of APCs
Naïve CD4+ T cells were purified from DO11.10 Tcra −/− BALB/c mice. Cell suspensions from spleen after RBC lysis were negatively selected using the EasySep CD4+ Mouse T Cell Enrichment Kit following the manufacturer’s protocol (StemCell Technologies). Resulting cells were routinely >95% CD4+ KJ-126+.
Splenic APCs from BALB/c mice were prepared by complement depletion of BALB/c using anti-Thy1.2 supernatant (30H12) in PBS for 30 min on ice and rabbit complement for 30 min at 37°C, and then irradiated (2000 cGy).
Generation of memory T cells
CD4+ T cell effectors were generated in vitro, as previously described4,49. In brief, 2 × 105/ml naïve cells were cultured for four days with 2 × 105/ml T-depleted irradiated BALB/c splenocytes as APCs in the presence of 5.6 μM OVA 323–339 peptide (Genescript RP10610) and 50 U/ml interleukin 2 for 4 d in Clicks medium (EHAA) supplemented with 5% FCS (HyClone), 2 mM L-glutamine, 10 mM HEPES buffer (pH 7), Pen/Strep, 1% NEAA, and 2-mercaptoethanol. After 4 d, effectors were washed thoroughly and re-cultured in fresh Clicks medium for 4 d in the absence of antigen and cytokine. Live cells were isolated by Percoll gradient separation. The resulting population was routinely >95% CD4+ KJ-126+.
Depletion of CD4+ T cells
For CD4+ T cell depletion of AM14 KI x Vκ8R KI BALB/c knock-in recipients, GK1.5 was produced and purified as described43. For most experiments, mice were injected intraperitoneally with 300 μg of GK1.5 or PBS once 4 d before transfer of B cells. In one experiment mice were injected twice, 2–4 d before transfer of B cells. T cell depletion was monitored by checking one mouse on the day of transfer and at the time of spleen harvest.
Microarray generation and data analysis
mRNA from memory B cell subset samples defined by CD80 and PD-L2 surface expression were isolated using the Qiagen RNeasy Micro kit per manufacturer’s instructions and hybridized to Illumina MouseWG-6 v2.0 Expression BeadChip arrays at the Yale Keck Microarray Facility. The data analyses were carried out using packages in R. Raw expression data were normalized using the quantile method provided by the lumi package in R/Bioconductor. Differentially expressed genes between DN subset and DP subsets were defined by two criteria: (1) an absolute log2 fold-change ≥1, and (2) a statistically significant change in expression as determined by LIMMA using a Benjamani-Hochberg false discovery rate cutoff of q < 0.05.
Principal Component Analysis (PCA) was performed on gene expression profiles of the 350 most variable probes with Coefficient of Variation (CV) greater than 0.05. Gene Set Enrichment Tests were performed using QuSAGE version 1.3.150 and n.points was set to 214 for PDF convolution. The KEGG_CELL_CYCLE gene set was downloaded from MSigDB database v4.0 (http://www.broadinstitute.org/gsea/msigdb/collections.jsp#C2).
Quantitative PCR
Total RNA was isolated from sort-purified populations using the RNeasy Plus Micro kit (Qiagen, 74034) and first-strand synthesis was performed with the SuperScript III First-Strand Synthesis System for RT-PCR (Invitrogen, 18080-051). Quantitative PCR was performed with the SYBR FAST qPCR Kit (KAPA Biosystems, KK4600) on a LightCycler 96 system (Roche). Primer sequences were as follows: Zbtb32 sense, 5′-GGTACAGTTAGCGGCTAGACT-3′, and antisense, 5′-GGAAGGGCTTATGTCTTCAACC-3′ ; Plk1 sense, 5′-CTTCGCCAAATGCTTCGAGAT-′3, and antisense, 5′-TAGGCTGCGGTGAATTGAGAT-3′ ; Cdc20 sense, 5′-CAGCCTGGAGACTACATATCCT-3′, and antisense, 5′-CGGAGTGACTGGTCATGTTTC-3′ ; Mcm5 sense, 5′-CAGAGGCGATTCAAGGAGTTC-3′, and antisense, 5′-CGATCCAGTATTCACCCAGGT-3′ ; Gapdh sense, 5′-TCCCACTCTTCCACCTTCGA-3′, and antisense, 5′-AGTTGGGATAGGGCCTCTCTT-3′ ; Ccnd2 sense, 5′-TGAATTACCTGGACCGTTTCTTG-3′, and antisense, 5′-AGAGTTGTCGGTGTAAATGCAC-3′, and E2f1 sense, 5′-TGCAGAAACGGCGCATCTAT-3′, and antisense, 5′-CCGCTTACCAATCCCCACC-3′.
To quantify the fold-change of key genes in MBC subsets compared to naive B cells, ΔΔCt was calculated for each gene of interest (GOI) according to the following formula: . Results were calculated as relative change (2(−ΔΔCt) for each gene.
Statistics
The Mann-Whitney test or the t-test were used for statistical analyses, as indicated; all comparisons were two-tailed. Results were analyzed with Prism software (GraphPad) and significance was determined at the 95% confidence level. All bar graphs use the mean as a center value. No specific randomization or blinding protocol was used.
Supplementary Material
Acknowledgments
We thank L. Conter, J. Cullen and E. Song for technical assistance; the Yale Cell Sorter Facility for cell sorting; and the Yale Animal Resource Center and A. Durso for expert animal care. Supported by the National Institutes of Health (R01-AI46303 to M.J.S. and K08- AI078533 to M.M.T), DFG Research Fellowship (WE 4752/1-1 to F.J.W.), National Health and Medical Research Council Arthritis Australia fellowships (K.L.G.-J.), and NSF Graduate Research fellowships (G.V.Z.-C. and S.S.).
Footnotes
ACCESSION CODES
The microarray data from memory B cell subsets defined by CD80 and PD-L2 surface expression have been deposited in NCBI’s Gene Expression Omnibus51 and are accessible through GEO Series accession number GSE51604 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE51604).
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
AUTHOR CONTRIBUTIONS
G.V.Z.-C., M.M.T, K.L.G.-J., F.J.W. and M.J.S. designed research; G.V.Z.-C. and F.J.W. did research; S.S. performed the T cell enumeration experiment; H.M. and S.H.K. analyzed microarray data; K.L.G.-J. and F.J.W. gave technical support and conceptual advice; G.V.Z.-C. and M.J.S. analyzed data and wrote the manuscript.
References
- 1.Ahmed R, Gray D. Immunological memory and protective immunity: understanding their relation. Science. 1996;272:54–60. doi: 10.1126/science.272.5258.54. [DOI] [PubMed] [Google Scholar]
- 2.Sallusto F, Lanzavecchia A, Araki K, Ahmed R. From vaccines to memory and back. Immunity. 2010;33:451–463. doi: 10.1016/j.immuni.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Radbruch A, et al. Competence and competition: the challenge of becoming a long-lived plasma cell. Nat Rev Immunol. 2006;6:741–750. doi: 10.1038/nri1886. [DOI] [PubMed] [Google Scholar]
- 4.McKinstry KK, et al. Rapid default transition of CD4 T cell effectors to functional memory cells. J Exp Med. 2007;204:2199–2211. doi: 10.1084/jem.20070041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bevan MJ. Understand memory, design better vaccines. Nat Immunol. 2011;12:463–465. doi: 10.1038/ni.2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wrammert J, et al. Broadly cross-reactive antibodies dominate the human B cell response against 2009 pandemic H1N1 influenza virus infection. J Exp Med. 2011;208:181–193. doi: 10.1084/jem.20101352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tomayko MM, Steinel NC, Anderson SM, Shlomchik MJ. Cutting edge: Hierarchy of maturity of murine memory B cell subsets. J Immunol. 2010;185:7146–7150. doi: 10.4049/jimmunol.1002163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anderson SM, Tomayko MM, Ahuja A, Haberman AM, Shlomchik MJ. New markers for murine memory B cells that define mutated and unmutated subsets. J Exp Med. 2007;204:2103–2114. doi: 10.1084/jem.20062571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Good-Jacobson KL, Shlomchik MJ. Plasticity and Heterogeneity in the Generation of Memory B Cells and Long-Lived Plasma Cells: The Influence of Germinal Center Interactions and Dynamics. J Immunol. 2010;185:3117–3125. doi: 10.4049/jimmunol.1001155. [DOI] [PubMed] [Google Scholar]
- 10.Bhattacharya D, et al. Transcriptional profiling of antigen-dependent murine B cell differentiation and memory formation. J Immunol. 2007;179:6808–6819. doi: 10.4049/jimmunol.179.10.6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luckey CJ, et al. Memory T and memory B cells share a transcriptional program of self-renewal with long-term hematopoietic stem cells. Proc Natl Acad Sci USA. 2006;103:3304–3309. doi: 10.1073/pnas.0511137103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tomayko MM, et al. Systematic comparison of gene expression between murine memory and naive B cells demonstrates that memory B cells have unique signaling capabilities. J Immunol. 2008;181:27–38. doi: 10.4049/jimmunol.181.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tangye SG, Hodgkin PD. Divide and conquer: the importance of cell division in regulating B-cell responses. Immunology. 2004;112:509–520. doi: 10.1111/j.1365-2567.2004.01950.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dogan I, et al. Multiple layers of B cell memory with different effector functions. Nat Immunol. 2009;10:1292–1299. doi: 10.1038/ni.1814. [DOI] [PubMed] [Google Scholar]
- 15.Pape KA, Taylor JJ, Maul RW, Gearhart PJ, Jenkins MK. Different B cell populations mediate early and late memory during an endogenous immune response. Science. 2011;331:1203–1207. doi: 10.1126/science.1201730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reynaud CA, et al. IgM memory B cells: a mouse/human paradox. Cell Mol Life Sci. 2012;69:1625–1634. doi: 10.1007/s00018-012-0971-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kometani K, et al. Repression of the transcription factor Bach2 contributes to predisposition of IgG1 memory B cells toward plasma cell differentiation. Immunity. 2013;39:136–147. doi: 10.1016/j.immuni.2013.06.011. [DOI] [PubMed] [Google Scholar]
- 18.Bemark M, et al. A unique role of the cholera toxin A1-DD adjuvant for long-term plasma and memory B cell development. J Immunol. 2011;186:1399–1410. doi: 10.4049/jimmunol.1002881. [DOI] [PubMed] [Google Scholar]
- 19.Onodera T, et al. Memory B cells in the lung participate in protective humoral immune responses to pulmonary influenza virus reinfection. Proc Natl Acad Sci USA. 2012:1–6. doi: 10.1073/pnas.1115369109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yates JL, Racine R, McBride KM, Winslow GM. T cell-dependent IgM memory B cells generated during bacterial infection are required for IgG responses to antigen challenge. J Immunol. 2013;191:1240–1249. doi: 10.4049/jimmunol.1300062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu AH, Jena PK, Wysocki LJ. Tracing the development of single memory-lineage B cells in a highly defined immune response. J Exp Med. 1996;183:2053–2063. doi: 10.1084/jem.183.5.2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weisel FJ, et al. Unique requirements for reactivation of virus-specific memory B lymphocytes. J Immunol. 2010;185:4011–4021. doi: 10.4049/jimmunol.1001540. [DOI] [PubMed] [Google Scholar]
- 23.Sonoda E, et al. B cell development under the condition of allelic inclusion. Immunity. 1997;6:225–233. doi: 10.1016/s1074-7613(00)80325-8. [DOI] [PubMed] [Google Scholar]
- 24.Takahashi Y, Ohta H, Takemori T. Fas is required for clonal selection in germinal centers and the subsequent establishment of the memory B cell repertoire. Immunity. 2001;14:181–192. doi: 10.1016/s1074-7613(01)00100-5. [DOI] [PubMed] [Google Scholar]
- 25.Ramiscal RR, Vinuesa CG. T-cell subsets in the germinal center. Immunol Rev. 2013;252:146–155. doi: 10.1111/imr.12031. [DOI] [PubMed] [Google Scholar]
- 26.Latchman Y, et al. PD-L2 is a second ligand for PD-1 and inhibits T cell activation. Nat Immunol. 2001;2:261–268. doi: 10.1038/85330. [DOI] [PubMed] [Google Scholar]
- 27.Good-Jacobson KL, et al. PD-1 regulates germinal center B cell survival and the formation and affinity of long-lived plasma cells. Nat Immunol. 2010;11:535–542. doi: 10.1038/ni.1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wong JV, Dong P, Nevins JR, Mathey-Prevot B, You L. Network calisthenics: control of E2F dynamics in cell cycle entry. Cell Cycle. 2011;10:3086–3094. doi: 10.4161/cc.10.18.17350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yoon HS, et al. ZBTB32 is an early repressor of the CIITA and MHC class II gene expression during B cell differentiation to plasma cells. J Immunol. 2012;189:2393–2403. doi: 10.4049/jimmunol.1103371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taylor JJ, Pape KA, Jenkins MK. A germinal center-independent pathway generates unswitched memory B cells early in the primary response. J Exp Med. 2012;209:597–606. doi: 10.1084/jem.20111696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ferguson SE, Han S, Kelsoe G, Thompson CB. CD28 is required for germinal center formation. J Immunol. 1996;156:4576–4581. [PubMed] [Google Scholar]
- 32.Sharpe AH, Freeman GJ. The B7-CD28 superfamily. Nat Rev Immunol. 2002;2:116–126. doi: 10.1038/nri727. [DOI] [PubMed] [Google Scholar]
- 33.Good-Jacobson KL, Song E, Anderson S, Sharpe AH, Shlomchik MJ. CD80 Expression on B Cells Regulates Murine T Follicular Helper Development, Germinal Center B Cell Survival, and Plasma Cell Generation. J Immunol. 2012;188:4217–4225. doi: 10.4049/jimmunol.1102885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fearon DT, Manders P, Wagner SD. Arrested differentiation, the self-renewing memory lymphocyte, and vaccination. Science. 2001;293:248–250. doi: 10.1126/science.1062589. [DOI] [PubMed] [Google Scholar]
- 35.Copley MR, Beer PA, Eaves CJ. Hematopoietic stem cell heterogeneity takes center stage. Cell Stem Cell. 2012;10:690–697. doi: 10.1016/j.stem.2012.05.006. [DOI] [PubMed] [Google Scholar]
- 36.He S, Nakada D, Morrison SJ. Mechanisms of stem cell self-renewal. Annu Rev Cell Dev Biol. 2009;25:377–406. doi: 10.1146/annurev.cellbio.042308.113248. [DOI] [PubMed] [Google Scholar]
- 37.Li Z, Chen YG. Functions of BMP signaling in embryonic stem cell fate determination. Exp Cell Res. 2013;319:113–119. doi: 10.1016/j.yexcr.2012.09.016. [DOI] [PubMed] [Google Scholar]
- 38.Corti D, et al. Heterosubtypic neutralizing antibodies are produced by individuals immunized with a seasonal influenza vaccine. J Clin Invest. 2010;120:1663–1673. doi: 10.1172/JCI41902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ekiert DC, et al. Antibody recognition of a highly conserved influenza virus epitope. Science. 2009;324:246–251. doi: 10.1126/science.1171491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pulendran B, Ahmed R. Immunological mechanisms of vaccination. Nat Immunol. 2011;131:509–517. doi: 10.1038/ni.2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bar-Or A, et al. Immunological memory: contribution of memory B cells expressing costimulatory molecules in the resting state. J Immunol. 2001;167:5669–5677. doi: 10.4049/jimmunol.167.10.5669. [DOI] [PubMed] [Google Scholar]
- 42.Chen J, et al. B cell development in mice that lack one or both immunoglobulin kappa light chain genes. EMBO J. 1993;12:821–830. doi: 10.1002/j.1460-2075.1993.tb05722.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shlomchik MJ, Zharhary D, Saunders T, Camper SA, Weigert MG. A rheumatoid factor transgenic mouse model of autoantibody regulation. Int Immunol. 1993;5:1329–1341. doi: 10.1093/intimm/5.10.1329. [DOI] [PubMed] [Google Scholar]
- 44.Hannum LG, Ni D, Haberman AM, Weigert MG, Shlomchik MJ. A disease-related rheumatoid factor autoantibody is not tolerized in a normal mouse: implications for the origins of autoantibodies in autoimmune disease. J Exp Med. 1996;184:1269–1278. doi: 10.1084/jem.184.4.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Prak EL, Weigert M. Light chain replacement: a new model for antibody gene rearrangement. J Exp Med. 1995;182:541–548. doi: 10.1084/jem.182.2.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sweet RA, et al. A new site-directed transgenic rheumatoid factor mouse model demonstrates extrafollicular class switch and plasmablast formation. Autoimmunity. 2010;43:607–618. doi: 10.3109/08916930903567500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dittrich AM, et al. A new mechanism for inhalational priming: IL-4 bypasses innate immune signals. J Immunol. 2008;181:7307–7315. doi: 10.4049/jimmunol.181.10.7307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hannum LG, Haberman AM, Anderson SM, Shlomchik MJ. Germinal center initiation, variable gene region hypermutation, and mutant B cell selection without detectable immune complexes on follicular dendritic cells. J Exp Med. 2000;192:931–942. doi: 10.1084/jem.192.7.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hu H, et al. CD4(+) T cell effectors can become memory cells with high efficiency and without further division. Nat Immunol. 2001;2:705–710. doi: 10.1038/90643. [DOI] [PubMed] [Google Scholar]
- 50.Yaari G, Bolen CR, Thakar J, Kleinstein SH. Quantitative set analysis for gene expression: a method to quantify gene set differential expression including gene-gene correlations. Nucleic Acids Res. 2013;41:e170–e170. doi: 10.1093/nar/gkt660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.