The U-turn is a classical three-dimensional RNA folding motif first identified in the anticodon and T-loops of tRNAs. It is demonstrated, using NMR spectroscopy, that a protonated cytidine can functionally and structurally replace the uridine at the first position of the canonical U-turn motif in the apical loop of the neomycin riboswitch.
Keywords: RNA structure, U-turn, protonated cytidine, NMR, hydrogen bond
Abstract
The U-turn is a classical three-dimensional RNA folding motif first identified in the anticodon and T-loops of tRNAs. It also occurs frequently as a building block in other functional RNA structures in many different sequence and structural contexts. U-turns induce sharp changes in the direction of the RNA backbone and often conform to the 3-nt consensus sequence 5′-UNR-3′ (N = any nucleotide, R = purine). The canonical U-turn motif is stabilized by a hydrogen bond between the N3 imino group of the U residue and the 3′ phosphate group of the R residue as well as a hydrogen bond between the 2′-hydroxyl group of the uridine and the N7 nitrogen of the R residue. Here, we demonstrate that a protonated cytidine can functionally and structurally replace the uridine at the first position of the canonical U-turn motif in the apical loop of the neomycin riboswitch. Using NMR spectroscopy, we directly show that the N3 imino group of the protonated cytidine forms a hydrogen bond with the backbone phosphate 3′ from the third nucleotide of the U-turn analogously to the imino group of the uridine in the canonical motif. In addition, we compare the stability of the hydrogen bonds in the mutant U-turn motif to the wild type and describe the NMR signature of the C+-phosphate interaction. Our results have implications for the prediction of RNA structural motifs and suggest simple approaches for the experimental identification of hydrogen bonds between protonated C-imino groups and the phosphate backbone.
INTRODUCTION
In order to carry out their biological functions, many RNA molecules, such as tRNAs, ribosomal RNAs, self-splicing introns, RNaseP-RNA, or riboswitches, adopt intricate three-dimensional globular structures rivaling or even surpassing protein structures in their complexity. The steadily rising number of novel X-ray- or NMR-derived structures determined for functional RNAs continuously reveals novel architectures of three-dimensional RNA tertiary structures. While all these structures are very different from each other, many of them share smaller three-dimensional RNA folding motifs or submotifs as common structural building blocks. Such motifs are, for example, stable tetraloops (Woese et al. 1990; Allain and Varani 1995; Jucker and Pardi 1995a; Jucker et al. 1996), tetraloop-receptor interactions (Costa and Michel 1995; Cate et al. 1996), kink-turn motifs (Klein et al. 2001), ribose-zippers (Cate et al. 1996), E-loop motifs (Wimberly et al. 1993; Wimberly 1994), A-minor motifs (Nissen et al. 2001), and different types of turn-motifs. A classic example for an RNA folding submotif is the so called “U-turn” motif, which was initially discovered in the early tRNA structures solved by Rich and coworkers and Klug and coworkers in the 1970s (Klug et al. 1974; Quigley and Rich 1976). The U-turn motif occurs in the anticodon- as well as the TΨC-loop of many tRNAs. Subsequently, the U-turn motif was found to be widespread not only in tRNAs but also in many other functional RNAs, including ribosomal RNAs (e.g., Huang et al. 1996; Nagaswamy et al. 2001, pdb entry 1k5i; Zhang et al. 2001, pdb entry 1hs2), the hammerhead ribozyme (Pley et al. 1994, pdb entry 1hmh; Doudna 1995), and snRNAs (Stallings and Moore 1997, pdb entry 2u2a), as well as viral RNA elements (Puglisi and Puglisi 1998, pdb entry 1bvj; Liu et al. 2007). As already implied by their name, U-turns induce a sharp turn in the RNA backbone and therefore present one possibility to introduce a hairpin loop in an RNA structure. In general, U-turn motifs share the 3-nt consensus sequence 5′-UNR-3′, where N stands for any nucleotide and R for a purine nucleotide. The change in the direction of the RNA backbone occurs at the phosphate group 3′ from the U residue of the consensus sequence. Canonical U-turns are stabilized by two hydrogen bonds. One hydrogen bond is formed between the uridine N3H3 imino group and the phosphate group at the 3′ site of the purine nucleotide. The second hydrogen bond involves the 2′-OH group of the uridine as the hydrogen bond donor and the N7 nitrogen of the purine base as the hydrogen bond acceptor group. Despite their 3-nt consensus sequence, U-turns rarely occur as isolated trinucleotide hairpin loops closed by stable Watson-Crick base pairs. In most cases, they are part of larger loop structures often closed by noncanonical sheared G:A, reversed Hoogsteen A:U, or pyrimidine:pyrimidine base pairs (Gutell et al. 2000). Furthermore, additional nucleotides can be inserted mostly 3′ from the U-turn consensus sequence. Importantly, these additional nucleotides are often solvent-exposed and therefore available for long-range tertiary interactions with other RNA elements. The highly stable GNRA-tetraloop motifs which are also nucleation sites for long-range RNA-RNA interactions share many structural features with the U-turn motif (Jucker and Pardi 1995b; Gutell et al. 2000), such as a base-phosphate hydrogen bonding interaction, a hydrogen bond between a 2′-OH-group and the N7-nitrogen of the conserved purine base, as well as the backbone geometry at the turning phosphate group.
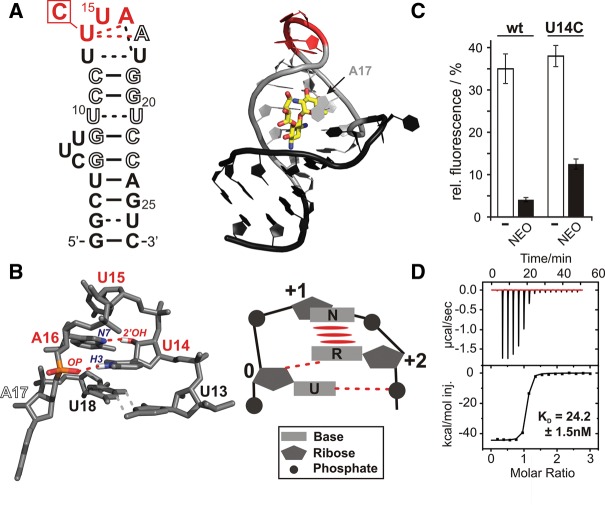
The neomycin sensing riboswitch is a synthetic gene regulatory element developed by a combined in vitro and in vivo selection procedure for the translational control of gene expression in response to the aminoglycoside neomycin in Saccharomyces cerevisiae (Weigand et al. 2008). With a minimal size of only 27 nt, it is the smallest known ligand-dependent RNA-based regulatory element. The riboswitch adopts a hairpin-like secondary structure (Fig. 1A) and is able to bind its ligand neomycin with an affinity in the low nanomolar range (Weigand et al. 2011). Upon insertion into the 5′ UTR of mRNAs in front of the AUG-start codon, it interferes with the scanning small ribosomal subunit in a ligand-dependent manner. When no neomycin is present, the scanning small ribosomal subunit reads through the hairpin structure and reaches the start codon. In the presence of neomycin, the hairpin structure is stabilized and presents a road block for the small ribosomal subunit. The hairpin-like secondary structure of the neomycin riboswitch is formally capped by a strongly conserved hexaloop with the sequence 5′-U13U14U15A16A17U18-3′, which adopts a well-defined and rigid structure (Fig. 1A; Duchardt-Ferner et al. 2010, pdb entry 2kxm). The first and the last nucleotide of the hexaloop U13 and U18 form an asymmetric cis Watson-Crick/Watson-Crick U:U base pair stabilized by two direct hydrogen bonds between the two pyrimidine bases. Nucleotides U14, U15, and A16 fold into a canonical U-turn structure with hydrogen bonds between the N3H3-imino group of U14 and the 3′ phosphate group of A16, as well as between the 2′-OH group of U14 and the N7-nitrogen of A16 (Fig. 1B). A17 is looped out of the structure and reaches down to the ligand binding site. Importantly, it is the only nucleotide in the apical hexaloop involved in direct contact with neomycin, and its conformation is stabilized by stacking and electrostatic interactions between its base and the ligand (Fig. 1A). In principle, the formation of the U13:U18 base pair and the folding of the U-turn motif encompassing nucleotides U14 to A16 can already be observed in the absence of the ligand at low temperatures (Duchardt-Ferner et al. 2010). However, ligand binding strongly stabilizes all elements of the loop structure by interacting with A17 and, through indirect effects, by stabilizing the helical element below the loop.
FIGURE 1.
Structural and functional characteristics of the neomycin riboswitch and its U14C mutant. (A) Secondary (left) and tertiary (right) structure of the WT neomycin riboswitch bound to ribostamycin. Residues which are involved in ligand binding are shown as open letters, and the residues forming the U-turn are indicated in red. The site of the U14C mutation is indicated. In the 3D structure, the ligand ribostamycin is shown in stick representation. The position of the base of A17, the only nucleotide from the apical loop in contact with the ligand, is marked by an arrow. (B) (Left) 3D-structure of the apical loop of the WT RNA-ribostamycin complex comprising the U-turn motif and the closing U13:U18 base pair (Duchardt-Ferner et al. 2010). Individual nucleotides are labeled according to the color code and numbering scheme used in A. Hydrogen bonds between the U14 N3H3 imino group and the phosphate group 3′ from A16 as well as between the U14 2′-OH group and the A16 N7 nitrogen are indicated by dashed red lines, and the donor and acceptor groups are labeled. U14, U15, and A16 adopt a classical U-turn fold showing all the canonical stabilizing interactions shown in the schematic representation of a consensus U-turn motif on the right. Stacking interactions are marked by elongated red spheres. (C) In vivo gene regulatory activity of the U14C mutant compared to the WT riboswitch as monitored in a GFP reporter gene assay in S. cerevisiae in the presence of neomycin (black bars) or in the absence of ligand (white bars). (D) ITC titration of the U14C mutant with the ligand neomycin. The resulting dissociation constant is given.
In a recent screen aimed at identifying functional sequence variants of the apical loop of the neomycin riboswitch, we found that a replacement of U14 by a C residue (Fig. 1A) also results in functional riboswitches (Weigand et al. 2014). Such a mutation should interfere with the proper folding and the stability of the U-turn motif because C lacks an imino proton and is thus not able to form a hydrogen bond analogous to the one between the U imino group and the phosphate backbone in the canonical U-turn motif.
Interestingly, function-retaining replacements of the U by a C in biologically important U-turn motifs have been reported previously. For instance, the stem–loop V in the VS ribozyme folds into a U-turn motif thought to be involved in substrate binding (Campbell and Legault 2005; Campbell et al. 2006, pdb entries 1tbk, 1yn2). The U to C mutation of U696 in the U-turn is the only variant that retains significant ribozyme activity (Rastogi et al. 1996). Another example is the stem–loop IIa of yeast U2 snRNA (SL2), which contains a conserved U-turn motif as part of the consensus sequence 5′-UAAY-3′ (Stallings and Moore 1997, pdb entry 2u2a). The U56C-SL2-mutant with the sequence 5′-CAAC-3′ is the only viable alternative. However, the structural basis for the ability of C to replace the U in these RNAs has not been established, and it is unclear if the mutant RNAs retain canonical U-turn structures.
Assuming that the three-dimensional loop structure in the U to C mutants in these systems must be preserved to retain function, there are a number of ways to rationalize these findings. First, it is possible that the remaining hydrogen bonding and stacking interactions are strong enough to preserve the fold of these loops despite the absence of the hydrogen bond between the imino group and the phosphate. Second, a slight shift of the base position in the C-mutant might bring the C4 amino group of the C into a position to hydrogen bond with the phosphate. Third, the C in the mutant could adopt the rare imino tautomeric form and then form a hydrogen bond equivalent to the U imino group. Fourth, the C might become protonated at the N3 position and form a hydrogen bond as well as an ionic interaction with the negatively charged phosphate group. Recent bioinformatics surveys and quantum chemical studies of base-phosphate interactions found that cytidine amino groups are able to form highly stable hydrogen bonds with the phosphate backbone but did not discuss evidence for interactions between protonated cytosine bases and phosphate groups (Zirbel et al. 2009).
Here, we investigated in detail the structural basis for the conservation of function in the U14C-mutant of the neomycin riboswitch-ligand complex using NMR spectroscopy. We found that C14 is stably protonated and forms a hydrogen bond which is isosteric to the one between the U imino group and the phosphate in the canonical U-turn motif, thereby preserving the overall fold of the apical hexaloop of the riboswitch. Using NMR methods, we characterized the strength of the hydrogen bond between the protonated C and the phosphate backbone and found that it is highly stable over a wide range of temperatures and pH values. By browsing the pdb, we discovered an additional example for the functional and structural replacement of a canonical UNR-type U-turn by a C+NR-type turn. Furthermore, protonated C-residues might play a role in stabilizing long-range RNA-RNA tertiary interactions.
Thus, our findings have consequences for the prediction of three-dimensional RNA structures from sequences since they extend the U-turn consensus sequence and may serve as an example of how protonated cytidine residues and their interactions can be easily identified by modern NMR techniques.
RESULTS AND DISCUSSION
A cytidine is able to functionally replace the uridine in the U-turn of the neomycin riboswitch
During an in vivo screen of a neomycin riboswitch library with a randomized apical hexaloop (nt 13–18), sequences with a U to C mutation at position 14 were identified as functional despite their apparent deviation from the structurally important U-turn consensus sequence (Fig. 1C; Weigand et al. 2014). However, compared to the WT riboswitch, their regulatory power is slightly diminished. The WT riboswitch represses gfp expression 8.6-fold in the presence of 100 μM neomycin in the medium, whereas only a threefold reduction of gfp expression is achieved by the U14C-mutant (Fig. 1C). ITC experiments in conditions as described previously for the WT (Weigand et al. 2011) show that the U14C-mutant binds to neomycin in vitro with a KD of 24.2 nM (Fig. 1D). Thus, the affinity of the mutant is only slightly diminished in comparison to the WT riboswitch, which binds neomycin with a KD of 9.2 nM (Weigand et al. 2011). This already suggests that the structure of the ligand binding pocket of the riboswitch is not strongly altered by the mutation in the U-turn motif of its apical loop.
The C14 is N3-protonated and forms a hydrogen bond with the backbone phosphate group as in the canonical U-turn motif
While in the in vivo functional studies and in the ITC binding experiments, neomycin was used as the ligand, all NMR experiments were performed with ribostamycin as the ligand. As thoroughly demonstrated in our previous NMR studies of the neomycin riboswitch (Duchardt-Ferner et al. 2010), the smaller ligand ribostamycin binds to the riboswitch with the same binding mode as neomycin. However, due to the absence of ring IV in ribostamycin, there is reduced signal overlap in some regions of the NMR spectra. In addition, we could directly transfer NMR signal assignments from our earlier NMR studies of the WT riboswitch-ribostamycin complex to the U14C-mutant.
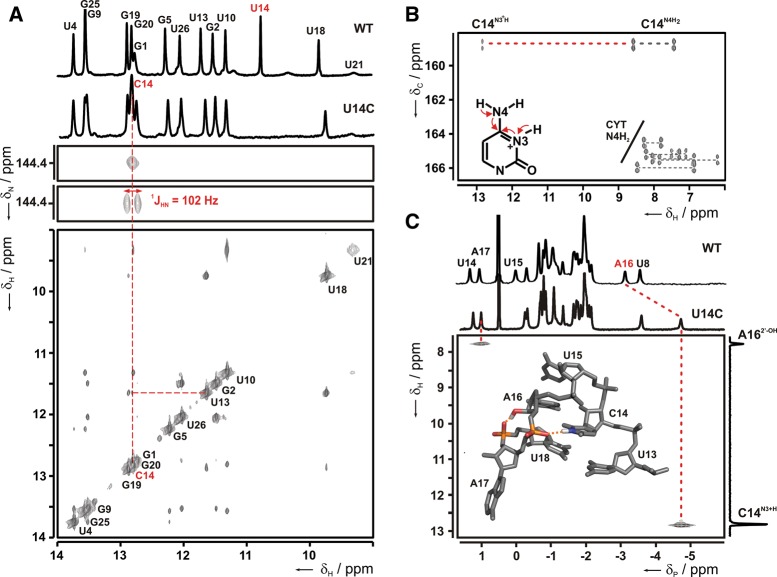
The imino proton 1D-1H-NMR spectra of the WT and the U14C-mutant in complex with ribostamycin are highly similar, suggesting that the three-dimensional structure of the U14C mutant complex closely resembles the WT (Fig. 2A, top). Importantly, for both complexes the resonances of the U13 and U18 imino protons are present and display very similar chemical shifts and line widths. This indicates that the U13:U18 base pair directly adjacent to the U-turn motif and the mutation site is present in both complexes and is of similar stability (Supplemental Fig. 1). The only obvious difference between the two spectra is the absence of the signal corresponding to the U14 imino group at 10.87 ppm in the U14C-mutant, which is to be expected (Fig. 2A, top). However, closer inspection of the U14C-mutant spectrum reveals a novel imino proton signal with a chemical shift of 12.83 ppm at 10°C. Similar chemical shift values are normally observed for guanine imino protons in Watson-Crick base pairs but have also been reported for imino protons of protonated cytidines in base triples when they form hydrogen bonds to guanine carbonyl groups (Nixon et al. 2002a,b; Cornish et al. 2006). In contrast, the imino protons of protonated cytidines in base triples (e.g., Holland and Hoffman 1996; Brodsky et al. 1998; Cornish et al. 2006; Cash et al. 2013) or in the DNA i-motif (Guéron and Leroy 2000; Lieblein et al. 2012) with hydrogen bonds to nitrogen acceptors have chemical shifts larger than 14.5 ppm. 2D-NOESY experiments show that the novel imino proton resonance has NOE cross peaks to the imino protons of U13 and U18 (Fig. 2A, bottom; Supplemental Fig. 2). Similar cross peaks were previously observed for the U14 imino proton resonance in the WT riboswitch ribostamycin complex (Duchardt-Ferner et al. 2010). An 1H,15N-HSQC-experiment with the U14C-mutant RNA selectively labeled with 13C,15N-cytidine shows that the imino proton is connected to a cytidine N3 with a nitrogen chemical shift of 144.4 ppm. The 1JHN scalar coupling constant between the imino proton and the N3 nitrogen is 102 Hz, which is in the same range as the one-bond scalar coupling constants observed in guanosine and uridine imino groups (Fig. 2A, middle). In contrast, the 1JHN scalar coupling measured for the imino group in C+:C base pairs in the i-motif in DNA is considerably smaller (∼45 Hz), since there the proton is delocalized in the N+H…N hydrogen bond (Lieblein et al. 2012). NOE correlations, an 1H,15N-HSQC-experiment for amino groups (Supplemental Fig. 3A), and a 2D-H(N)C-experiment (Fig. 2B) with the selectively cytidine-labeled sample connect the cytidine imino group with the amino group of the same cytidine with proton chemical shifts of 8.58 and 7.43 ppm (Fig. 2B) and a nitrogen chemical shift of 102.1 ppm (Supplemental Fig. 3A). Thus, the N3-protonation of C14 is not due to the presence of the neutral imino tautomeric form, but the C14 residue is in the positively charged, N3-protonated state. The amino group chemical shifts for C14 differ significantly from those reported for other protonated cytidine residues (Holland and Hoffman 1996; Brodsky et al. 1998; Cornish et al. 2006). There, chemical shifts between 11.7 ppm and 8.5 ppm were reported for the amino protons. In addition, amino nitrogen chemical shifts for protonated cytidines reported previously were larger than 111.0 ppm (Nixon et al. 2002b; Cornish et al. 2005, 2006).
FIGURE 2.
Characterization of the protonated cytidine in the U14C mutant riboswitch-ribostamycin complex. (A) (Top) Comparison of the 1D-imino proton spectra of WT and mutant RNA bound to ribostamycin. Resonance assignments are given for the WT. (Middle) Imino region of 1H,15N-HSQC spectra with and without 15N-decoupling in the direct dimension recorded with a selectively 15N-cytidine-labeled U14C-riboswitch-ribostamycin complex. The 1JHN coupling constant is indicated. (Bottom) Imino region of a 2D-NOESY spectrum of the U14C mutant in complex with ribostamycin. Resonance assignments are given for the diagonal peaks. The NOE indicating the spatial proximity between the imino protons of C14 and U13 is highlighted by dashed lines. (B) 2D-H(N)C spectrum recorded with a selectively 13C,15N-cytidine-labeled sample of the U14C mutant bound to ribostamycin. The C4 resonance is correlated to the 1H chemical shift of the amino groups for all cytidines (gray dashed lines). For C14, the C4 resonance is connected simultaneously to the 1H imino and amino frequencies (red dashed line). The magnetization transfer pathway for this experiment is indicated by red arrows in the inset showing the structure of a protonated cytidine. (C) 1D-31P spectra of WT and U14C mutant (top) and 2D-1H,31P-long-range HSQC spectrum of the U14C mutant (bottom). Phosphorous resonances displaying unusual chemical shifts are assigned in the 1D-31P spectrum of the WT. A projection is shown along the hydrogen dimension of the long-range HSQC and resonances giving rise to cross peaks are connected by dashed lines and labeled. A modeled 3D structure of the U-turn motif containing a protonated cytidine at the first position is shown as an inset. Hydrogen bonds observed as cross peaks in the 1H,31P-long-range HSQC are indicated by dashed orange lines in the structure, and the donor and acceptor groups are colored according to atom type.
The unusual RNA backbone structure of the apical hexaloop motif of the WT riboswitch ligand complex with its concatenation of the U13:U18 noncanonical base pair, the U-turn containing U14, U15, and A16, and the bulged-out A17 is reflected in its 31P-NMR spectrum (Fig. 2C, top). In agreement with previous observations for other U-turn structures (Morosyuk et al. 2001; Campbell et al. 2006), the 31P resonances of the phosphate groups 3′ from U14 (the turning phosphate of the U-turn), U15, and A17 are shifted downfield compared to the bulk of the backbone phosphate groups. The phosphate group 3′ from A16 which is hydrogen-bonded to the U14 imino group according to the U-turn consensus is shifted upfield compared to the bulk of the 31P-resonances. Importantly, this 31P chemical shift “fingerprint” is completely conserved in the U14C-mutant RNA (Fig. 2C, top), indicating that the backbone structure of the WT and mutant RNA is very similar. Remarkably, the 31P resonance of the phosphate group 3′ from A16 is shifted even further upfield compared to the WT (Fig. 2C, top), likely due to its spatial proximity and hydrogen bonding to the imino group of the protonated C14. A 2D 1H,31P-long-range HSQC spectrum reveals a direct correlation between the C14 H3 imino proton and this 31P resonance, due to the presence of a small scalar coupling between these two spins (Fig. 2C, bottom). Thereby, this experiment directly demonstrates the existence of a hydrogen bond between the N3H3 imino group of the protonated C14 and the backbone phosphate 3′ from A16. This hydrogen bond conforms exactly to the U-turn consensus structure. The second correlation observed in this spectrum is due to a hydrogen bond between the A16 2′-OH group and the phosphate group 3′ from A17. This correlation was previously observed for the WT RNA-ribostamycin complex, further supporting the close structural similarity between the WT and mutant RNA structure. Notably, despite their small line widths and intense NMR signals, the C14 amino protons show no correlations in the 1H,31P-long-range HSQC, suggesting that they are not involved in additional hydrogen-bonding interactions with the phosphate as is possible, e.g., in a bifurcated hydrogen-bonding arrangement. The absence of hydrogen-bonding interactions involving the C14 amino group might explain the deviation in chemical shifts from those reported previously for C+ residues in other RNAs containing base triples. In these cases, the amino groups are directly hydrogen-bonded either to a C6 carbonyl oxygen (Holland and Hoffman 1996; Brodsky et al. 1998) or an N7 nitrogen of a guanine (Nixon et al. 2002b; Cornish et al. 2006).
Previous studies have established that the chemical shifts of both the nitrogen (Wang et al. 1991) and those of the carbon nuclei (Legault and Pardi 1994, 1997) of the bases are excellent reporters of their protonation state and can even be used for the determination of pKa values of individual bases in complex RNA structures (Legault and Pardi 1994; Cai and Tinoco 1996; Ravindranathan et al. 2000; Lupták et al. 2001; Huppler et al. 2002). In line with these previous observations, we find that the protonation at N3 significantly affects the other chemical shifts associated with C14, in particular those of the hetero-atoms. In agreement with the findings by Legault and Pardi (1997) for protonated 5′-CMP, the most pronounced effects are observed for the chemical shifts of the C4 and C2 carbon nuclei. The C4 carbon resonance frequency is shifted upfield by >4.3 ppm (Fig. 2B), and the C2 resonance frequency is shifted upfield by >4.8 ppm (Supplemental Fig. 3F), respectively, compared to the nonprotonated cytidine moieties in the same molecule. While they still display extreme chemical shift values, the C6, C5, N1, and C1′ chemical shifts of C14 differ less strongly from those of the nonprotonated cytidines (Supplemental Fig. 3B–E). The direction of the chemical shift change caused by cytidine protonation is the same as observed previously for protonated CMP (Legault and Pardi 1997). C6 and C1′ of C14 are shifted downfield, whereas C5 of C14 is shifted upfield in comparison to the chemical shifts found for the nonprotonated cytidine residues (Supplemental Fig. 3). As expected (Wang et al. 1991), the N3 nitrogen chemical shift of C14 (144.4 ppm) is also clearly separated from the bulk of the cytidine N3 resonances (195–200 ppm) and is in a similar chemical shift range as observed for protonated cytidines in base triples (142.7 ppm, 138.4 ppm) stabilizing structurally related pseudoknots (Nixon et al. 2002b; Cornish et al. 2005, 2006).
Since the C4 and C2 chemical shifts show the largest changes upon protonation, we attempted to use these chemical shifts to determine the pKa value for C14 protonation by recording H(N)C spectra at different pH values. However, at pH 8.2, we observe virtually the same C4 and C2 chemical shifts for C14 as at pH 6.2 (Supplemental Fig. 4A,B). Neither did we observe a second set of signals for C14 in a 1H,13C-HSQC spectrum (Supplemental Fig. 4C) as would be expected when protonated and de-prontonated species would coexist in slow chemical exchange (Cai and Tinoco 1996). This indicates that even at pH 8.2, C14 exists almost exclusively in the protonated state. Higher pH values could not be tested since then the functional groups of the ligand become significantly de-protonated (Freire et al. 2007), leading to changes in its RNA-binding properties. However, our data suggest that the pKa value of C14 is larger than 8.2 and thus more than four pH units above the pKa value of isolated CMP (Legault and Pardi 1997; Wilcox et al. 2011) However, such strong pKa shifts due to involvement of the protonated residue in the formation of stable RNA secondary and tertiary structures are not unprecedented (Wilcox and Bevilacqua 2013).
The hydrogen bond between the protonated cytidine and the phosphate backbone is very stable
To qualitatively assess the stability of the hydrogen bond between the protonated C14 and the phosphate backbone in comparison to the other base pairs in the riboswitch-ligand complex, we recorded imino proton 1D-1H NMR- and 1D-1H,15N-HSQC spectra using the selectively cytidine-labeled sample at different temperatures and pH values (Fig. 3A,B; Supplemental Fig. 5). From these experiments, it is immediately obvious that this hydrogen bond is very stable. The C14 imino proton signal is still detectable at 35°C and at pH 6.2 (Fig. 3A). At pH 8.2, the imino proton is detectable (Fig. 3B) at temperatures up to 25°C (Supplemental Fig. 5A). The line width of the C14 imino proton signal under all these conditions is similar to the one of U14 in the WT as well as to many of the other imino proton signals (Fig. 3A; Supplemental Figs. 1, 5). Thus, the stability of the C14+-phosphate interaction is comparable to those of canonical base-pairing interactions.
FIGURE 3.
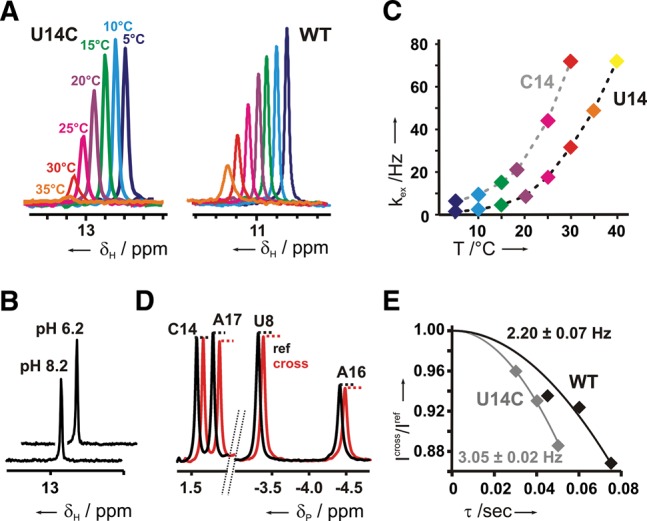
Stability of the cytidine N3H3-phosphate group hydrogen bond. (A) 1D-1H,15N-HSQC spectra of the selectively 15N-cytidine-labeled U14C mutant RNA (left) and of the uniformly 15N-labeled WT RNA (right) in complex with ribostamycin at different temperatures and pH 6.2. (B) 1D-1H,15N- HSQC spectra for the U14C imino proton at pH values of 6.2 and 8.2 and a temperature of 10°C. (C) Imino proton solvent exchange rates for the WT and U14C mutant at different temperatures. Exchange rates in Hz are plotted as a function of temperature. (D) Cross- (red) and reference (black) spectrum of a quantitative 1D 31P{1H} spin-echo experiment for the U14C mutant with an evolution delay of 40 msec at 22°C. The 3′ phosphate groups of U8, A16, and A17 (labeled U8, A16, and A17) are all involved in hydrogen-bonding interactions as indicated by the reduction in signal intensity in the cross experiment whereas the 3′ phosphate group of C14 (labeled C14) is not. (E) Quantification of the scalar 2hJHP coupling constants between the phosphate group 3′ from A16 and C14 H3 or U14 H3 in the mutant or the WT, respectively. Signal intensity ratios between a coupled cross (Icross) and a de-coupled reference experiment (Iref) for the mutant (gray) and the WT (black) are plotted against the duration of the evolution delay τ. Data were fitted using the equation Icross/Iref = cos(π2hJHP τ) to obtain the 2hJHP coupling constant.
The stability of individual hydrogen bonds can be inferred from imino proton solvent exchange experiments (Leroy et al. 1988). In these experiments, magnetization of the bulk water protons is transferred to imino protons by chemical exchange. The rate constant for the transfer of the water magnetization to individual imino protons is correlated to the base-pair opening rate, which is a direct measure of base-pair stability. The water exchange rates for the U14 H3 proton in the WT riboswitch-ribostamycin complex and the C14 H3 proton of the mutant complex were recorded as a function of temperature (Fig. 3C). Our data show that the hydrogen bond involving the imino group of the protonated C14 is only slightly less stable than the canonical U-turn hydrogen bond involving the U14 imino group in the WT.
Another measure of hydrogen-bond strength is the size of the two-bond trans-hydrogen-bond scalar coupling between the hydrogen-bond donor proton and the hydrogen-bond acceptor group. The apparent 2hJHP-scalar coupling constant between the H3 imino proton of C14 and the A16 3′ phosphate group was determined in a quantitative spin-echo 1D-31P{1H}-experiment (Fig. 3D,E; Duchardt-Ferner et al. 2011) to be at least 3.05 Hz. Thus, this coupling constant is similar in size to the 2hJHP-coupling constant involving the U14 imino proton in the WT RNA (lower limit of 2.2 Hz), in agreement with comparable hydrogen-bonding strengths in both RNAs. The apparent 2hJHP-scalar coupling between the 2′-OH proton of A16 and the A17 phosphate group in the WT RNA is slightly smaller (1.8 Hz). In comparison, the 2hJHP-scalar couplings measured between protein amide and hydroxyl protons and the phosphate group of bound FMN range from 0.5 to 1.7 Hz (Löhr et al. 2000), whereas a 1.3-Hz coupling constant was measured between an arginine side chain proton in a small molecule receptor and a noncovalently bound phosphate group (Federwisch et al. 2008).
The replacement of uridine by a protonated cytidine also occurs in other U-turn motifs
To check for the occurrence of other hydrogen bonding interactions between phosphate backbone groups and potentially protonated cytidine residues, we analyzed all X-ray structures of RNAs and RNPs with a nominal resolution better than 2.5 Å present in the pdb for short (<3.0 Å) cytidine N3-phospate group oxygen distances. One such short cytidine N3-phosphate group oxygen distance (2.6 Å) was found in the high-resolution X-ray structure of the large ribosomal subunit from the archaeon Haloarcula marismortui (Klein et al. 2004, pdb entry 1s72). It is observed between nt C1469 and the phosphate group 3′ from nt A1471, which are both located in a 5′-C1469AACU-3′ pentaloop closed by a G-C base pair (Fig. 4A). If C1469 were a U, the first 3 nt of this pentaloop would correspond to the classical U-turn consensus sequence. Both the short N3…O=P distance and the angle between the donor and the acceptor group would agree with the presence of a hydrogen bond if C1469 were protonated at the N3 nitrogen. Importantly, a hydrogen bond between the 2′-OH-group of C1469 and the N7 of A1471 is also present in this loop, which corresponds to the second hydrogen bond of the U-turn consensus structure (Fig. 4A). Measurements of the backbone torsion angles confirm that the turning phosphate in this pentaloop is the phosphate immediately 3′ from C1469, as expected for a U-turn structure. Thus, this loop in the 23S rRNA of H. marismortui is most likely another example for a U-turn structure with a protonated C instead of the U at the first position of its consensus sequence. Remarkably, in the 23S rRNA of the thermophilic archaeon Thermococcus celer, the same loop contains the canonical U-turn sequence 5′-UAACU-3′, while Escherichia coli 23S rRNA contains a stable GNRA tetraloop at this position as expected if the 3D structure is conserved at this position in the rRNA (Fig. 4B).
FIGURE 4.
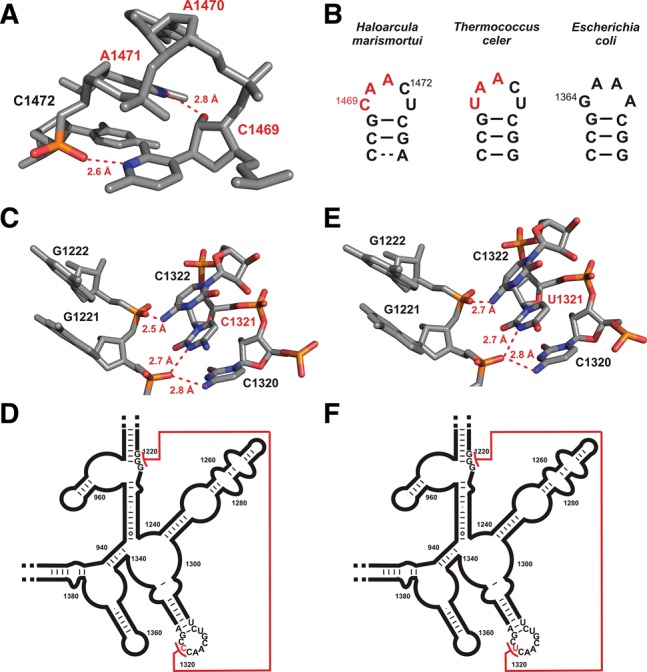
Occurrence of hydrogen bonds between protonated cytidine N3 moieties and backbone phosphate groups in other RNAs. (A) Residues 1469–1472 from the X-ray structure of the 23S large ribosomal subunit of Haloarcula marismortui (pdb entry 1s72), which form a U-turn motif with a cytidine at the first position. Potential hydrogen bonds are indicated by dashed red lines and the distances between the donor nitrogen and the acceptor oxygen are given. Donor and acceptor are colored according to atom type. (B) Primary and secondary structures of the hairpin loop between residues 1466 and 1476 in the 23S RNA in H. marimortui (left) and the respective regions in T. celer (middle) and E. coli (right). (C) Section of the X-ray structure of the small ribosomal subunit from T. thermophilus (pdb entry 2vqe) containing a potential long-range C+-phosphate backbone interaction. Possible hydrogen bonds between the protonated imino group of cytidine C1321 and the amino groups of C1322 and C1320 to the acceptor backbone phosphates are indicated by dashed red lines, and the respective distances are given. (D) Secondary structure of the corresponding region of the 16S RNA from T. thermophilus. The long-range interaction depicted in atomic detail in C is indicated by a red line. (E) Section of the X-ray structure of the small ribosomal subunit from E. coli (pdb entry 2qal). Potential hydrogen bonds of the uridine U1321 imino proton and the amino groups of C1320 and C1322 to the acceptor backbone phosphates are indicated by dashed red lines; distances are given. (F) Secondary structure of the corresponding section of the 16S RNA from E. coli. The long-range interaction shown in detail in E is indicated by a red line.
Hydrogen bonding between protonated cytidines and the phosphate backbone might play a role in mediating long-range tertiary RNA-RNA interactions
The search for short distances between cytidine N3 nitrogens and phosphate group oxygens also revealed a putative hydrogen-bonding interaction between C1321 and the phosphate 3′ from nucleotide G1221 in the structure of the small ribosomal subunit from Thermus thermophilus (Fig. 4C; Kurata et al. 2008, pdb entry 2vqe). In this case, C1321 is part of a larger highly structured loop corresponding to a T-loop motif (Nagaswamy and Fox 2002), while nt G1221 is part of a sequentially and structurally remote A-form helical element (helix 32) (Fig. 4D). Again, the N3…O=P distance and the angle between the donor group atoms and the acceptor are highly suggestive of a hydrogen bond if C1321 were protonated. Interestingly, an equivalent hydrogen-bonding interaction is present at the same position in the 3.0-Å X-ray structure of the E. coli ribosome (Fig. 4E,F), where, remarkably, the C is replaced by a U (Borovinskaya et al. 2007, pdb entry 2qal). Therefore, it appears likely that interactions between protonated cytidines and phosphate backbone groups might play a more general role in stabilizing the tertiary structures of large and structurally complex RNAs. Apparently, protonated cytidines can replace uridines not only in U-turns but also in more complex structure elements.
In summary, we have shown here that a protonated cytidine (C+) can functionally and structurally replace the uridine at the first position of the classical U-turn motif by forming a very stable hydrogen bond with the negatively charged phosphate backbone in the neomycin riboswitch. Another example of the functional and structural equivalence of classical and C+-containing U-turns is found in 23S rRNA. The possibility of replacing the U with a protonated cytidine residue in U-turn structures would also help to explain mutational data for other functional RNAs, as, e.g., for the VS ribozyme (Rastogi et al. 1996) or the SL2 of the U2 snRNA (Stallings and Moore 1997). In addition, protonated cytidine residues may play a more general role in stabilizing RNA tertiary structures by forming strong ionic hydrogen bonds with phosphate backbone groups. The unique heteronuclear chemical shift signature of selected hetero-atoms in the nucleobase moieties of protonated cytidines can aid in their rapid experimental identification even in cases where the H3 imino proton is not directly observable due to fast chemical exchange with the solvent or in cases where cytidine residues are only transiently protonated, as recently postulated in DNA (Nikolova et al. 2011).
MATERIALS AND METHODS
GFP activity assay
The gene regulatory activity of the WT neomycin riboswitch and the U14C mutant in response to neomycin was tested by an in vivo green fluorescent protein reporter gene assay in S. cerevisiae, as described previously (Weigand et al. 2008).
Sample preparation
Unlabeled, selectively 15N-cytidine-, and selectively 13C,15N-cytidine-labeled samples for the U14C mutant of the neomycin riboswitch RNA with the sequence 5′-GGCUGCUUGUCCUCUAAGGUCCAGUC-3′ were synthesized by in vitro transcription with T7 RNA-polymerase and linearized plasmid-DNA as a template. 15N- and 13C,15N-labeled nucleotide triphosphates are commercially available (Silantes GmbH). In order to generate uniform 3′ ends, the primary RNA transcripts contained a hammerhead ribozyme. Processed transcripts of the RNA were purified by preparative denaturing PAGE according to standard protocols. The purified RNA was folded by heating to 95°C for 5 min, followed by injection into five equivalents of ice cold water. Samples were then rebuffered and concentrated using vivaspin concentrators (MW cutoff 3.000 Da). Sample concentrations were 800 µM for the unlabeled, 616 µM for the 15N-cytidine-, and 660 µM for 13C,15N-cytidine-labeled sample. All NMR samples were prepared in NMR buffer containing 25 mM potassium phosphate (pH 6.2 or 8.2) and 50 mM KCl. Samples were titrated with 1.2 equivalents of commercially available ribostamycin (Sigma-Aldrich). Saturation of the RNA with ribostamycin was verified by the disappearance of imino resonances of the free riboswitch in the 1D-1H spectra. Buffer exchange was performed for the 13C,15N-cytidine-labeled sample with NMR buffer at pH 8.2 using vivaspin concentrators (MW cutoff 3.000 Da).
NMR spectroscopy
All NMR spectra were collected with 600 MHz and 800 MHz Bruker Avance spectrometers equipped with 5-mm cryogenic triple resonance HCN and HCP z-gradient probe heads. For the determination of the 2hJHP coupling constants, quantitative spin-echo 31P{1H}-1D spectra were obtained on a Bruker DRX300 spectrometer equipped with a 5-mm room temperature BBO z-gradient probe head as described previously (Duchardt-Ferner et al. 2011). The majority of NMR resonance assignments for the U14C mutant could be transferred from the WT RNA (Duchardt-Ferner et al. 2010; Schmidtke et al. 2010). Additional NMR resonance assignment experiments for the U14C-mutant RNA were recorded at 10°C in 10% (v/v) D2O for the exchangeable proton and in 100% (v/v) D2O at 25°C for the nonexchangeable protons. 2D-1H,1H-NOESY-, 2D-1H, 15N-HSQC- (Bodenhausen and Ruben 1980), and 2D-H(C)N- (Sklenar et al. 1993) experiments were carried out using standard pulse sequences (Fürtig et al. 2003). Assignments for the cytidine C4 resonances were obtained from a 2D-H(N)C experiment with the nitrogen offset centered at 130 ppm. The assignments for the C2 resonances of the nonprotonated cytidines were derived from an 2D-H6(C6N1)C2 experiment (Fürtig et al. 2004). All NMR data were processed with TOPSPIN2.1 (Bruker Biospin).
Solvent exchange measurements for the WT and U14C mutant imino protons were recorded with a modified 2D-1H,15N-HSQC experiment (Rinnenthal et al. 2010) containing a 180°-RE-BURP soft pulse (Geen and Freeman 1991) applied on the water frequency. A variable inversion recovery delay between 4 μsec and 3000 msec and a recycling delay of 6 sec were used. Data were collected within a temperature range between 0°C and 40°C. Curve fitting of signal intensities and determination of the exchange rates was carried out as described (Rinnenthal et al. 2010) using OriginLab 8.1.
Database search
From a list of nonredundant RNA structures in the protein data bank (release 1.16 - 22/06/2013) which was compiled and made publicly available on the RNA 3D Hub web site (http://rna.bgsu.edu/rna3dhub/) by the BGSU RNA Group, all structures with a resolution of 2.5 Å or better were selected. The data set comprised 378 X-ray structures. A macro in the programming language Python was created to visualize the resulting structures in the program PyMol and analyze distances. Hydrogen-bonding patterns between potentially protonated cytidines and phosphate group oxygen atoms of the RNA backbone were analyzed by selecting the N3 atoms of cytidine residues and the OP1 and OP2 phosphate group oxygen atoms of the phosphate backbone. Distances between the nitrogen and oxygen atoms were measured, and all distances smaller than 3.0 Å were stored as putative hydrogen bonds.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
This work was supported by the Aventis Foundation, the Center of Biomolecular Magnetic Resonance (BMRZ), and the Deutsche Forschungsgemeinschaft (SFB 902 A2, SU 402/4-1, WO 901/2-1).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.043083.113.
REFERENCES
- Allain FH, Varani G 1995. Structure of the P1 helix from group I self-splicing introns. J Mol Biol 250: 333–353 [DOI] [PubMed] [Google Scholar]
- Bodenhausen G, Ruben DJ 1980. Natural abundance nitrogen-15 NMR by enhanced heteronuclear spectroscopy. Chem Phys Lett 69: 185–189 [Google Scholar]
- Borovinskaya MA, Pai RD, Zhang W, Schuwirth BS, Holton JM, Hirokawa G, Kaji H, Kaji A, Cate JH 2007. Structural basis for aminoglycoside inhibition of bacterial ribosome recycling. Nat Struct Mol Biol 14: 727–732 [DOI] [PubMed] [Google Scholar]
- Brodsky AS, Erlacher HA, Williamson JR 1998. NMR evidence for a base triple in the HIV-2 TAR C-G.C+ mutant-argininamide complex. Nucleic Acids Res 26: 1991–1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Z, Tinoco I Jr 1996. Solution structure of loop A from the hairpin ribozyme from tobacco ringspot virus satellite. Biochemistry 35: 6026–6036 [DOI] [PubMed] [Google Scholar]
- Campbell DO, Legault P 2005. Nuclear magnetic resonance structure of the Varkud satellite ribozyme stem–loop V RNA and magnesium-ion binding from chemical-shift mapping. Biochemistry 44: 4157–4170 [DOI] [PubMed] [Google Scholar]
- Campbell DO, Bouchard P, Desjardins G, Legault P 2006. NMR structure of varkud satellite ribozyme stem–loop V in the presence of magnesium ions and localization of metal-binding sites. Biochemistry 45: 10591–10605 [DOI] [PubMed] [Google Scholar]
- Cash DD, Cohen-Zontag O, Kim N-K, Shefer K, Brown Y, Ulyanov NB, Tzfati Y, Feigon J 2013. Pyrimidine motif triple helix in the Kluyveromyces lactis telomerase RNA pseudoknot is essential for function in vivo. Proc Natl Acad Sci 110: 10970–10975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cate JH, Gooding AR, Podell E, Zhou K, Golden BL, Kundrot CE, Cech TR, Doudna JA 1996. Crystal structure of a group I ribozyme domain: principles of RNA packing. Science 273: 1678–1685 [DOI] [PubMed] [Google Scholar]
- Cornish PV, Hennig M, Giedroc DP 2005. A loop 2 cytidine-stem 1 minor groove interaction as a positive determinant for pseudoknot-stimulated −1 ribosomal frameshifting. Proc Natl Acad Sci 102: 12694–12699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornish PV, Giedroc DP, Hennig M 2006. Dissecting non-canonical interactions in frameshift-stimulating mRNA pseudoknots. J Biomol NMR 35: 209–223 [DOI] [PubMed] [Google Scholar]
- Costa M, Michel F 1995. Frequent use of the same tertiary motif by self-folding RNAs. EMBO J 14: 1276–1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna JA 1995. Hammerhead ribozyme structure: U-turn for RNA structural biology. Structure 3: 747–750 [DOI] [PubMed] [Google Scholar]
- Duchardt-Ferner E, Weigand JE, Ohlenschläger O, Schmidtke SR, Suess B, Wöhnert J 2010. Highly modular structure and ligand binding by conformational capture in a minimalistic riboswitch. Angew Chem Int Ed Engl 49: 6216–6219 [DOI] [PubMed] [Google Scholar]
- Duchardt-Ferner E, Ferner J, Wöhnert J 2011. Rapid identification of noncanonical RNA structure elements by direct detection of OH…O=P, NH…O=P, and NH2…O=P hydrogen bonds in solution NMR spectroscopy. Angew Chem Int Ed Engl 50: 7927–7930 [DOI] [PubMed] [Google Scholar]
- Federwisch G, Kleinmaier R, Drettwan D, Gschwind RM 2008. The H-bonding network of acylguanidine complexes: Combined intermolecular 2hJH,P and 3hJN,P scalar couplings provide an insight into the geometric arrangement. J Am Chem Soc 130: 16846–16847 [DOI] [PubMed] [Google Scholar]
- Freire F, Cuesta I, Corzana F, Revuelta J, Gonzalez C, Hricovini M, Bastida A, Jimenez-Barbero J, Asensio JL 2007. A simple NMR analysis of the protonation equilibrium that accompanies aminoglycoside recognition: dramatic alterations in the neomycin-B protonation state upon binding to a 23-mer RNA aptamer. Chem Commun (Camb) 2: 174–176 [DOI] [PubMed] [Google Scholar]
- Fürtig B, Richter C, Wöhnert J, Schwalbe H 2003. NMR spectroscopy of RNA. Chembiochem 4: 936–962 [DOI] [PubMed] [Google Scholar]
- Fürtig B, Richter C, Bermel W, Schwalbe H 2004. New NMR experiments for RNA nucleobase resonance assignment and chemical shift analysis of an RNA UUCG tetraloop. J Biomol NMR 28: 69–79 [DOI] [PubMed] [Google Scholar]
- Geen H, Freeman R 1991. Band-selective radiofrequency pulses. J Magn Reson 93: 93–141 [Google Scholar]
- Guéron M, Leroy J-L 2000. The i-motif in nucleic acids. Curr Opin Struct Biol 10: 326–331 [DOI] [PubMed] [Google Scholar]
- Gutell RR, Cannone JJ, Konings D, Gautheret D 2000. Predicting U-turns in ribosomal RNA with comparative sequence analysis. J Mol Biol 300: 791–803 [DOI] [PubMed] [Google Scholar]
- Holland JA, Hoffman DW 1996. Structural features and stability of an RNA triple helix in solution. Nucleic Acids Res 24: 2841–2848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Wang YX, Draper DE 1996. Structure of a hexanucleotide RNA hairpin loop conserved in ribosomal RNAs. J Mol Biol 258: 308–321 [DOI] [PubMed] [Google Scholar]
- Huppler A, Nikstad LJ, Allmann AM, Brow DA, Butcher SE 2002. Metal binding and base ionization in the U6 RNA intramolecular stem–loop structure. Nat Struct Biol 9: 431–435 [DOI] [PubMed] [Google Scholar]
- Jucker FM, Pardi A 1995a. Solution structure of the CUUG hairpin loop: a novel RNA tetraloop motif. Biochemistry 34: 14416–14427 [DOI] [PubMed] [Google Scholar]
- Jucker FM, Pardi A 1995b. GNRA tetraloops make a U-turn. RNA 1: 219–222 [PMC free article] [PubMed] [Google Scholar]
- Jucker FM, Heus HA, Yip PF, Moors EH, Pardi A 1996. A network of heterogeneous hydrogen bonds in GNRA tetraloops. J Mol Biol 264: 968–980 [DOI] [PubMed] [Google Scholar]
- Klein DJ, Schmeing TM, Moore PB, Steitz TA 2001. The kink-turn: a new RNA secondary structure motif. EMBO J 20: 4214–4221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein DJ, Moore PB, Steitz TA 2004. The roles of ribosomal proteins in the structure assembly, and evolution of the large ribosomal subunit. J Mol Biol 340: 141–177 [DOI] [PubMed] [Google Scholar]
- Klug A, Robertus JD, Ladner JE, Brown RS, Finch JT 1974. Conservation of the molecular structure of yeast phenylalanine transfer RNA in two crystal forms. Proc Natl Acad Sci 71: 3711–3715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurata S, Weixlbaumer A, Ohtsuki T, Shimazaki T, Wada T, Kirino Y, Takai K, Watanabe K, Ramakrishnan V, Suzuki T 2008. Modified uridines with C5-methylene substituents at the first position of the tRNA anticodon stabilize U.G wobble pairing during decoding. J Biol Chem 283: 18801–18811 [DOI] [PubMed] [Google Scholar]
- Legault P, Pardi A 1994. In situ probing of adenine protonation in RNA by 13C NMR. J Am Chem Soc 116: 8390–8391 [Google Scholar]
- Legault P, Pardi A 1997. Unusual dynamics and pKa shift at the active site of a lead-dependent ribozyme. J Am Chem Soc 119: 6621–6628 [Google Scholar]
- Leroy JL, Kochoyan M, Huynh-Dinh T, Gueron M 1988. Characterization of base-pair opening in deoxynucleotide duplexes using catalyzed exchange of the imino proton. J Mol Biol 200: 223–238 [DOI] [PubMed] [Google Scholar]
- Lieblein AL, Kramer M, Dreuw A, Fürtig B, Schwalbe H 2012. The nature of hydrogen bonds in cytidine…H+…cytidine DNA base pairs. Angew Chem Int Ed Engl 51: 4067–4070 [DOI] [PubMed] [Google Scholar]
- Liu P, Li L, Millership JJ, Kang H, Leibowitz JL, Giedroc DP 2007. A U-turn motif-containing stem–loop in the coronavirus 5′ untranslated region plays a functional role in replication. RNA 13: 763–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löhr F, Mayhew SG, Rüterjans H 2000. Detection of scalar couplings across NH•••OP and OH•••OP hydrogen bonds in a flavoprotein. J Am Chem Soc 122: 9289–9295 [Google Scholar]
- Lupták A, Ferré-D'Amaré AR, Zhou K, Zilm KW, Doudna JA 2001. Direct pKa measurement of the active-site cytosine in a genomic hepatitis δ virus ribozyme. J Am Chem Soc 123: 8447–8452 [DOI] [PubMed] [Google Scholar]
- Morosyuk SV, SantaLucia J Jr, Cunningham PR 2001. Structure and function of the conserved 690 hairpin in Escherichia coli 16 S ribosomal RNA. III. Functional analysis of the 690 loop. J Mol Biol 307: 213–228 [DOI] [PubMed] [Google Scholar]
- Nagaswamy U, Fox GE 2002. Frequent occurrence of the T-loop RNA folding motif in ribosomal RNAs. RNA 8: 1112–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaswamy U, Gao X, Martinis SA, Fox GE 2001. NMR structure of a ribosomal RNA hairpin containing a conserved CUCAA pentaloop. Nucleic Acids Res 29: 5129–5139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolova EN, Kim E, Wise AA, O'Brien PJ, Andricioaei I, Al-Hashimi HM 2011. Transient Hoogsteen base pairs in canonical duplex DNA. Nature 470: 498–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissen P, Ippolito JA, Ban N, Moore PB, Steitz TA 2001. RNA tertiary interactions in the large ribosomal subunit: the A-minor motif. Proc Natl Acad Sci 98: 4899–4903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon P, Cornish PV, Suram SV, Giedroc DP 2002a. Thermodynamic analysis of conserved loop–stem interactions in P1–P2 frameshifting RNA pseudoknots from plant Luteoviridae. Biochemistry 41: 10665–10674 [DOI] [PubMed] [Google Scholar]
- Nixon P, Rangan A, Kim YG, Rich A, Hoffman DW, Hennig M, Giedroc DP 2002b. Solution structure of a luteoviral P1–P2 frameshifting mRNA pseudoknot. J Mol Biol 322: 621–633 [DOI] [PubMed] [Google Scholar]
- Pley HW, Flaherty KM, McKay DB 1994. Three-dimensional structure of a hammerhead ribozyme. Nature 372: 68–74 [DOI] [PubMed] [Google Scholar]
- Puglisi EV, Puglisi JD 1998. HIV-1 A-rich RNA loop mimics the tRNA anticodon structure. Nat Struct Biol 5: 1033–1036 [DOI] [PubMed] [Google Scholar]
- Quigley GJ, Rich A 1976. Structural domains of transfer RNA molecules. Science 194: 796–806 [DOI] [PubMed] [Google Scholar]
- Rastogi T, Beattie TL, Olive JE, Collins RA 1996. A long-range pseudoknot is required for activity of the Neurospora VS ribozyme. EMBO J 15: 2820–2825 [PMC free article] [PubMed] [Google Scholar]
- Ravindranathan S, Butcher SE, Feigon J 2000. Adenine protonation in domain B of the hairpin ribozyme. Biochemistry 39: 16026–16032 [DOI] [PubMed] [Google Scholar]
- Rinnenthal J, Klinkert B, Narberhaus F, Schwalbe H 2010. Direct observation of the temperature-induced melting process of the Salmonella fourU RNA thermometer at base-pair resolution. Nucleic Acids Res 38: 3834–3847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidtke SR, Duchardt-Ferner E, Weigand JE, Suess B, Wöhnert J 2010. NMR resonance assignments of an engineered neomycin-sensing riboswitch RNA bound to ribostamycin and tobramycin. Biomol NMR Assign 4: 115–118 [DOI] [PubMed] [Google Scholar]
- Sklenar V, Peterson RD, Rejante MR, Feigon J 1993. Two- and three-dimensional HCN experiments for correlating base and sugar resonances in 15N,13C-labeled RNA oligonucleotides. J Biomol NMR 3: 721–727 [DOI] [PubMed] [Google Scholar]
- Stallings SC, Moore PB 1997. The structure of an essential splicing element: stem–loop IIa from yeast U2 snRNA. Structure 5: 1173–1185 [DOI] [PubMed] [Google Scholar]
- Wang C, Gao H, Gaffney BL, Jones RA 1991. Nitrogen-15-labeled oligodeoxynucleotides. 3. Protonation of the adenine N1 in the A•C and A•G mispairs of the duplexes d[CG(15N1)AGAATTCCCG]2 and d[CGGGAATTC(15N1)ACG]2. J Am Chem Soc 113: 5486–5488 [Google Scholar]
- Weigand JE, Sanchez M, Gunnesch EB, Zeiher S, Schröder R, Suess B 2008. Screening for engineered neomycin riboswitches that control translation initiation. RNA 14: 89–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigand JE, Schmidtke SR, Will TJ, Duchardt-Ferner E, Hammann C, Wöhnert J, Suess B 2011. Mechanistic insights into an engineered riboswitch: a switching element which confers riboswitch activity. Nucleic Acids Res 39: 3363–3372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigand JE, Gottstein-Schmidtke SR, Demolli S, Groher F, Duchardt-Ferner E, Wöhnert J, Suess B 2014. Sequence elements distal to the ligand binding pocket modulate the efficiency of a synthetic riboswitch. ChemBioChem 10.1002/cbic.201402067 [DOI] [PubMed] [Google Scholar]
- Wilcox JL, Bevilacqua PC 2013. A simple fluorescence method for pKa determination in RNA and DNA reveals highly shifted pKa’s. J Am Chem Soc 135: 7390–7393 [DOI] [PubMed] [Google Scholar]
- Wilcox JL, Ahluwalia AK, Bevilacqua PC 2011. Charged nucleobases and their potential for RNA catalysis. Acc Chem Res 44: 1270–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimberly B 1994. A common RNA loop motif as a docking module and its function in the hammerhead ribozyme. Nat Struct Biol 1: 820–827 [DOI] [PubMed] [Google Scholar]
- Wimberly B, Varani G, Tinoco I Jr 1993. The conformation of loop E of eukaryotic 5S ribosomal RNA. Biochemistry 32: 1078–1087 [DOI] [PubMed] [Google Scholar]
- Woese CR, Winker S, Gutell RR 1990. Architecture of ribosomal RNA: constraints on the sequence of “tetra-loops”. Proc Natl Acad Sci 87: 8467–8471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Fountain MA, Krugh TR 2001. Structural characterization of a six-nucleotide RNA hairpin loop found in Escherichia coli, r(UUAAGU). Biochemistry 40: 9879–9886 [DOI] [PubMed] [Google Scholar]
- Zirbel CL, Sponer JE, Sponer J, Stombaugh J, Leontis NB 2009. Classification and energetics of the base-phosphate interactions in RNA. Nucleic Acids Res 37: 4898–4918 [DOI] [PMC free article] [PubMed] [Google Scholar]