Aminoacyl-tRNA protein transferases are enzymes that catalyze the tRNA-dependent post-translational addition of amino acids onto the N terminus of a polypeptide. tRNA in this case plays the role of an amino acid donor, independent of a canonical role in protein synthesis. Previous models for the Escherichia coli leucyl/phenylalanyl-tRNA protein transferase argued that recognition of tRNAs by the enzyme was relatively nonspecific, simply docking the D-stem of the tRNA onto a positively charged cluster of the enzyme. Here, the authors define the preferences of this enzyme for sequence features—a G3:C70 base pair and a set of four key nucleotides (C72, A4:U69, and C68)—found in the most abundant leucyl-tRNA (anticodon 5′-CAG-3′) using biochemical approaches and mutational swaps. These provide a clear understanding of how known specificity must be achieved for this important enzyme.
Keywords: N-end rule, quantitative mass spectrometry, L/F transferase, aminoacyl-tRNA protein transferase, tRNA recognition, isoacceptors
Abstract
Escherichia coli leucyl/phenylalanyl-tRNA protein transferase catalyzes the tRNA-dependent post-translational addition of amino acids onto the N-terminus of a protein polypeptide substrate. Based on biochemical and structural studies, the current tRNA recognition model by L/F transferase involves the identity of the 3′ aminoacyl adenosine and the sequence-independent docking of the D-stem of an aminoacyl-tRNA to the positively charged cluster on L/F transferase. However, this model does not explain the isoacceptor preference observed 40 yr ago. Using in vitro-transcribed tRNA and quantitative MALDI-ToF MS enzyme activity assays, we have confirmed that, indeed, there is a strong preference for the most abundant leucyl-tRNA, tRNALeu (anticodon 5′-CAG-3′) isoacceptor for L/F transferase activity. We further investigate the molecular mechanism for this preference using hybrid tRNA constructs. We identified two independent sequence elements in the acceptor stem of tRNALeu (CAG)—a G3:C70 base pair and a set of 4 nt (C72, A4:U69, C68)—that are important for the optimal binding and catalysis by L/F transferase. This maps a more specific, sequence-dependent tRNA recognition model of L/F transferase than previously proposed.
INTRODUCTION
Transfer RNAs (tRNAs), in addition to their prominent role in translation, also participate in alternative functions in many organisms in vivo, including amino acid biosynthesis (Ibba and Soll 2004; Sheppard et al. 2008), antibiotic biosynthesis (Nolan and Walsh 2009), cell envelope remodeling (Villet et al. 2007; Roy and Ibba 2008; Fonvielle et al. 2009; Giannouli et al. 2009), and targeted proteolysis (Abramochkin and Shrader 1996; Mogk et al. 2007; for review and more details, see Banerjee et al. 2010; Francklyn and Minajigi 2010). These alternative functions often utilize aminoacyl-tRNA (aa-tRNA) as a source of activated amino acids, yet deacyl-tRNAs also have regulatory roles in gene expression during amino acid starvation in both prokaryotes and eukaryotes (Wendrich et al. 2002; Zaborske et al. 2009).
Currently, there is a conundrum regarding the precise in vivo mechanism in which the aa-tRNA used for alternative functions evades the protein biosynthesis machinery. Elongation factor Tu (EF-Tu) binds to an aa-tRNA molecule in the cytoplasm where it hydrolyzes a GTP molecule and releases the aa-tRNA to the ribosomal A-site for protein synthesis (Marshall et al. 2008; Agirrezabala and Frank 2009; Schmeing and Ramakrishnan 2009). EF-Tu has an in vivo concentration of ∼100 µM, and it binds to all aa-tRNAs with strong, similar affinities (KD in low nM range) (Andersen and Wiborg 1994; LaRiviere et al. 2001; Schrader et al. 2011). Some possible evasion mechanisms have been described, such as channeling substrates through a protein complex (Bailly et al. 2007), regulating the subcellular localization of tRNAs (Stortchevoi et al. 2003), having competitive binding affinities to aa-tRNAs (Roy and Ibba 2008), utilizing misacylated tRNAs (Stanzel et al. 1994; Becker and Kern 1998), or idiosyncratic features of specific tRNA isoacceptors (Giannouli et al. 2009). However, it remains unclear how canonical aa-tRNA species participate in both translation and alternative functions.
Aminoacyl-tRNA protein transferases catalyze the tRNA-dependent post-translational addition of amino acid from an aa-tRNA to the N-terminus of a protein polypeptide (Soffer 1974). These proteins are ubiquitously expressed in the cytosol of eubacteria (Tobias et al. 1991), yeast (Bachmair and Varshavsky 1989), plants (Potuschak et al. 1998; Graciet and Wellmer 2010), and mammals (Gonda et al. 1989). Aminoacyl-tRNA protein transferases are involved in a wide variety of biological functions (for review, see Dougan et al. 2010; Saha and Kashina 2011). The enzymatic N-terminal addition of an amino acid to a protein has been identified as a molecular marker to target proteins for degradation via the N-end rule pathway, where it describes the relationship between the in vivo half-life of a protein and the identity of its N-terminal amino acid (Bachmair and Varshavsky 1989). Yet it does not always result in protein degradation but may have additional cellular functions (Karakozova et al. 2006; Zhang et al. 2010).
The sole aminoacyl-tRNA protein transferase in Escherichia coli, L/F transferase, has degenerate aa-tRNA specificity in vitro where it utilizes Leu-tRNALeu, Phe-tRNAPhe (Leibowitz and Soffer 1969), and to a lesser extent Met-tRNAMet as substrates (Scarpulla et al. 1976). In vivo studies, however, suggest that leucylation is the dominant modification (Shrader et al. 1993). A model for L/F transferase tRNA recognition has been proposed based on biochemical and structural data (Leibowitz and Soffer 1971; Abramochkin and Shrader 1996; Suto et al. 2006; Watanabe et al. 2007). In vitro studies with misacylated tRNAs (Leibowitz and Soffer 1971; Abramochkin and Shrader 1996) and minimalistic adenosine esters of natural and unnatural amino acids (3′ rA-aa) (Wagner et al. 2011) are sufficient for aminoacyl transfer. This suggests that the major determinant of tRNA recognition by L/F transferase is the 3′ terminal adenosine and the aminoacyl moiety of an aa-tRNA (Leibowitz and Soffer 1971; Abramochkin and Shrader 1996).
Several X-ray crystal structures of L/F transferase with substrate analogs (minimal substrate phenylalanyl adenosine [rA-Phe] and inhibitor puromycin) bound have provided insights into the mechanism of tRNA recognition by L/F transferase (Suto et al. 2006; Dong et al. 2007; Watanabe et al. 2007). However, in closer examination of the crystal structures with the two analogs revealed differences in their mode of binding (Fung et al. 2014). Since there are no crystal structures solved for L/F transferase in complex with an intact aa-tRNA bound, the molecular insights derived from these structures remain within the 3′ rA-aa of an aa-tRNA. Nevertheless, the current L/F transferase tRNA recognition model includes the recognition of the 3′ aminoacyl adenosine, the sequence-independent docking of the tRNA D-stem to the positively charged cluster (R76, R80, K83, R84) of L/F transferase, and the disruption or bending of the 3′ acceptor stem of the tRNA during catalysis (Leibowitz and Soffer 1971; Abramochkin and Shrader 1996; Suto et al. 2006; Watanabe et al. 2007).
It has also been demonstrated that there is a preference for the tRNALeu (anticodon 5′-CAG-3′) isoacceptor for L/F transferase activity (Rao and Kaji 1974). The current tRNA recognition model does not explain this isoacceptor preference. A comparison with the reported apparent Km values for rA-Phe (124 μM) and Phe-tRNAPhe (1.56 μM), suggests that the tRNA body contributes to the L/F transferase recognition significantly (Rao and Kaji 1974; Abramochkin and Shrader 1996; Wagner et al. 2011). In vitro assays with mutant tRNAs suggest that the anticodon and variable loop are not the basis for specificity (Abramochkin and Shrader 1996). Upon meta-analysis of various tRNA recognition studies, Abramochkin et al. observed a correlation between the “strength” of the acceptor stem base pairs and overall L/F transferase activity and hypothesized that tRNAs with weak acceptor stems (i.e., mismatches or more A:U base pairs) are better L/F transferase substrates (Rao and Kaji 1974; Scarpulla et al. 1976; Abramochkin and Shrader 1996).
We hypothesize that the specificity for tRNALeu isoacceptors is a result of previously unidentified sequence elements in the tRNA body that are recognized by L/F transferase. Here, we report on our investigations on L/F transferase aa-tRNA isoacceptor recognition. Specifically, we focus on whether there are discriminator bases for tRNALeu isoacceptors that are specific for L/F transferase recognition. Using in vitro-transcribed tRNAs and a quantitative matrix assisted laser desorption/ ionization time-of-flight mass spectrometry (MALDI-ToF MS) enzyme activity assay developed by our lab (Ebhardt et al. 2009; Fung et al. 2011, 2014), our results demonstrated that there is a preference for the CAG isoacceptor by L/F transferase. Through mutations at the acceptor, D-, and T-stem of tRNALeu isoacceptors, we identified two independent, sequence elements in the acceptor stem of Leu-tRNALeu that are important for optimal L/F transferase binding and catalysis. These include the G3:C70 base pair and a set of 4 nt in the acceptor stem that contribute to optimal tRNA recognition by L/F transferase. This study demonstrates that tRNA recognition by L/F transferase is more specific and sequence dependent than previously hypothesized.
RESULTS
L/F transferase activity assays with tRNALeu, tRNAPhe, and tRNAMet isoacceptors
As the initial investigations with the different tRNALeu isoacceptors was incomplete (Rao and Kaji 1974; Abramochkin and Shrader 1996), we initiated our investigations by examining all five E. coli tRNALeu isoacceptors as L/F transferase substrates using the quantitative MALDI-ToF MS-based activity assay we previously developed (Ebhardt et al. 2009; Fung et al. 2011). Minimal differences using either purified (fully modified) or in vitro-transcribed (unmodified) tRNAs was previously reported (Abramochkin and Shrader 1995), and with our need to generate hybrid tRNAs, we utilized in vitro-transcribed tRNAs as substrates. Sequences reported for E. coli tRNA genes in the genomic tRNA database (Chan and Lowe 2009) were used for template design. tRNALeu isoacceptors (including tRNALeu [anticodon 5′-CAG-3′], tRNALeu [UAG], tRNALeu [CAA], tRNALeu [UAA], and tRNALeu [GAG)]) were in vitro-transcribed and purified. In addition to the tRNALeus, we also transcribed and purified the sole tRNAPhe (GAA) isoacceptor and two representative elongator tRNAMet (CAU) sequences, which were chosen from the ileX and metT genes. We omitted the initiator tRNAMet, as the characteristic 5′ cytosine overhang is not compatible with the T7 promoter site for in vitro transcription. Also, Scarpulla et al. suggested that L/F transferase does not utilize initiator tRNAMets (Scarpulla et al. 1976).
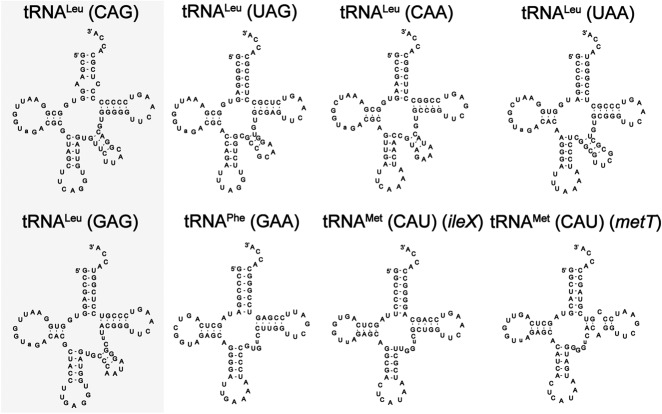
Figure 1 shows the cloverleaf structures of the tRNAs investigated. In addition to differences in the esterified amino acids of the tRNA substrates, cloverleaf structural examination reveals substantial differences. First, all tRNALeu isoacceptors have larger D-loops and diverse, lengthy variable loops compared to tRNAPhe and tRNAMet. Additionally, some individual differences are observed, such as the A:C mismatch at the base of the acceptor stem for tRNALeu (CAG). It had been previously hypothesized that L/F transferase specifically favors a Leu-tRNALeu with weak base pairs in the acceptor stem (mismatches or more A:U pairs) (Abramochkin and Shrader 1996).
FIGURE 1.
Cloverleaf structures of E. coli tRNA isoacceptors for leucine, phenylalanine, and methionine. The two tRNALeu isoacceptor species of importance in this study are highlighted in the gray box. Two representative elongator methionyl-tRNA species are selected for this study with their respective gene names.
Enzymatic analysis of the addition of an amino acid to a peptide substrate in the presence of different tRNA isoacceptors was a modified method from our previously established quantitative MALDI-ToF MS method (Ebhardt et al. 2009; Fung et al. 2011, 2014), where we varied the concentration of tRNA substrate and measured product formation in a continuous aminoacylation system. Figure 2A shows a graph of quantified peptide product formation over time for three representative tRNAs (tRNALeu [CAG], tRNALeu [GAG], and tRNAPhe [GAA]). Initial rates of product formation are calculated from the slope of the linear tangent line to the curve. Initial rates of product formation determined for the eight tRNA isoacceptors (tRNALeu, tRNAPhe, and tRNAMet) are listed in Supplemental Table 1, while the data are summarized graphically in Figure 2B and the kinetic parameters are listed in Table 1.
FIGURE 2.
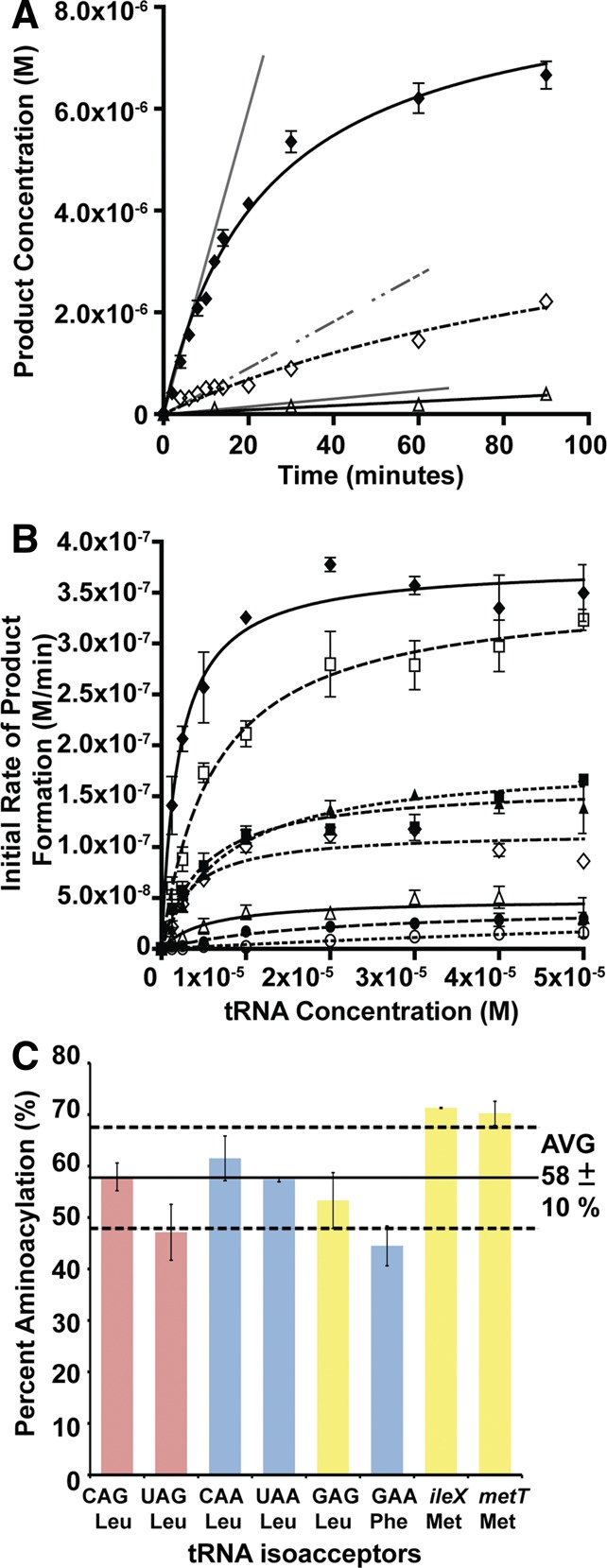
A preference of leucyl-tRNA (CAG) isoacceptor by L/F transferase. (A) Graphical analysis of product formation over time for tRNALeu (CAG) (♦), tRNAPhe (GAA) (◊), and tRNALeu (GAG) (Δ) when using an initial tRNA substrate concentration of 1.25 μM. Errors represented are standard deviation of triplicate measurements of a single independent experiment. Initial rate of product formation is calculated from the slope of the linear tangent line (gray) drawn to the curve. (B) A graphical display of initial rate of product formation vs. tRNA concentration for isoacceptors tRNALeu (CAG) (♦), tRNALeu (UAG) (□), tRNALeu (CAA) (▪), tRNALeu (UAA) (▴), tRNAPhe (GAA) (◊), tRNALeu (GAG) (Δ), tRNAMet (CAU) ileX (•), and tRNAMet (CAU) metT (○). Errors represented are the standard deviation of three independent experiments. (C) A bar graph presenting the maximal percent aminoacylation for natural isoacceptors after 7 min of aminoacylation. Errors represented are the standard deviation of three independent experiments.
TABLE 1.
Kinetic parameters of L/F transferase-catalyzed peptide bond formation on in vitro-transcribed E. coli leucyl-, phenylalanyl-, and methionyl-tRNA isoacceptors
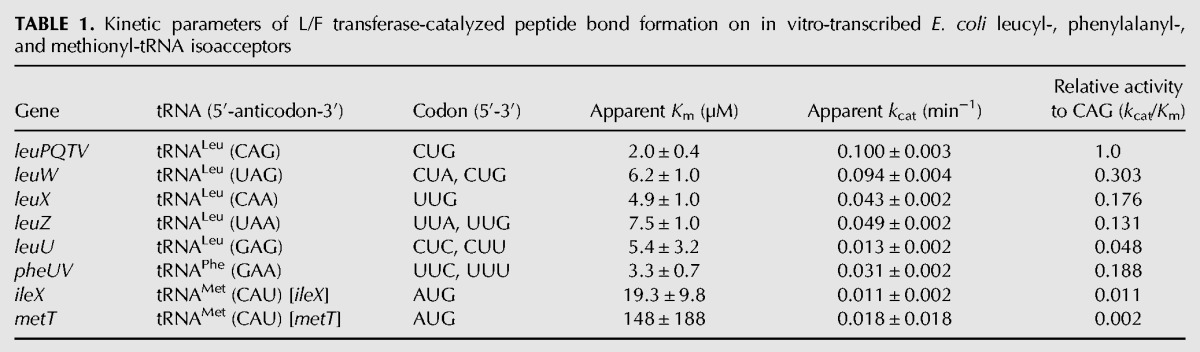
Our data are in agreement with previous investigations reporting that leucylation is the optimal amino acid addition by E. coli L/F transferase in comparison to phenylalanylation and methionylation (Shrader et al. 1993). Our data in Figure 2B and Table 1 demonstrate that L/F transferase is most specific to the Leu-tRNALeu (CAG) isoacceptor with an apparent Km of 2.0 ± 0.4 µM with an apparent kcat of 0.100 ± 0.003 min−1. This is in agreement with the observed isoacceptor preference by Rao and Kaji ( 1974). Although an apparent Km of 0.11 µM has been reported for the CAG isoacceptor under different reaction conditions (Rao and Kaji 1974; Abramochkin and Shrader 1996), the relative Km fold changes among the isoacceptors tested (CAG, UAG, CAA, and UAA) are comparable between the two studies. Subsequent data will be compared to the CAG isoacceptor tRNA as the optimal reference.
Both Met-tRNAMet isoacceptors are poor substrates of L/F transferase with large apparent Km values (9.5- and 74-fold increase) and low relative activity (90- and 500-fold decrease in the kcat/Km ratios). Phe-tRNAPhe is an intermediate substrate with an apparent Km of 3.3 ± 0.7 µM with mid-range relative activity (5.3-fold decrease in the kcat/Km ratio). Interestingly, there is a significant difference between the initial rates among the five leucine isoacceptors (Fig. 2B). Specifically, the Leu-tRNALeu (GAG) isoacceptor, that has not been previously tested, is as poor of a substrate as tRNAMet, with a 21-fold decrease in the kcat/Km ratio when compared to the CAG isoacceptor. Since the leucine isoacceptors have equal amino acid identity contribution, comparison among these tRNAs directly reflects the structural contribution to the recognition and catalytic efficiency by L/F transferase. Among the leucine isoacceptors, the apparent Km values are within a fourfold change, and the apparent kcat values are within a 7.6-fold change when compared to the CAG anticodon-containing isoacceptor. This suggests that L/F transferase recognizes leucine isoacceptors with somewhat similar affinities, but there is optimal recognition of certain isoacceptors over others for their catalytic efficiencies.
The catalytic efficiency differences between the leucine isoacceptors cannot be explained by the current recognition model. The current recognition model of tRNA by L/F transferase suggests that L/F transferase recognizes mainly the 3′-terminal adenosine and the aminoacyl moiety, while the remainder of the tRNA body enhances binding affinity in a sequence-independent manner. We hypothesize that there is a more specific recognition mechanism for tRNA binding. There are two potential explanations for the differences between CAG and GAG isoacceptors. Since our assay uses in vitro-transcribed tRNAs, there is a possibility that some tRNAs are not folded properly and hence may not be aminoacylated properly. Alternatively, the current model regarding the recognition requires modification, and L/F transferase recognizes beyond the 3′-terminal aminoacylated adenosine.
Differences in aminoacylation
To ensure that the differences in L/F transferase product formation rates is due to differences in the RNA sequence and structure but not due to reduced aminoacylation, experiments to test aminoacylation rates for all tRNAs were performed similarly as previously described (Wolfson and Uhlenbeck 2002). Figure 2C shows a bar graph plotting the maximal percent aminoacylation after 7 min for each of the tRNA isoacceptors (for a full time course, see Supplemental Fig. S1). We found that all in vitro-transcribed tRNA isoacceptors were aminoacylated between 45% and 71%. Specifically, tRNAMets were aminoacylated to 71% and tRNAPhe was aminoacylated to 45%. tRNALeu isoacceptors were aminoacylated to between 47% and 62%. Although there are variations in aminoacylation between tRNA isoaccepting species, these differences do not extrapolate to the kinetic differences observed in Figure 2B (see Supplemental Fig. S2). Thus, the specific substrate specificity is not due to differential aminoacylation and is inherent to the sequence and structure of the aa-tRNA.
Our percent aminoacylation values are within the typical range observed for this method, and they reflect the equilibrium state of aminoacylation rate by aaRS and spontaneous deacylation rate (Wolfson and Uhlenbeck 2002). Since the uncharged tRNA fraction remains relatively high, we also examined whether the presence of uncharged tRNA (also a product of the reaction) significantly inhibits L/F transferase. Preliminary competition assays of uncharged tRNAPhe in a leucylation L/F transferase assay and uncharged tRNALeu in a phenylalanylation L/F transferase assay suggest that the uncharged tRNA can compete for binding under high concentrations (Supplemental Fig. S3). Comparing the apparent Ki (uncharged tRNALeu = 31 μM and uncharged tRNAPhe = 25 μM) with the apparent Km (Leu-tRNALeu = 2 μM and Phe-tRNAPhe = 3 μM in this study and rA-Phe = 124 μM [Wagner et al. 2011]) confirms that the recognition of aa-tRNA substrate by L/F transferase is through both the amino acid moiety and the tRNA body. The amino acid moiety contributes to an approximately eightfold difference (comparing Phe-tRNAPhe with uncharged tRNAPhe), while the tRNA body contributes to an ∼40-fold difference (comparing Phe-tRNAPhe with rA-Phe) in affinity, further confirming that the tRNA body does contribute significantly to recognition.
Determining the recognition element by hybrid tRNAs
To identify the specific element that is important for L/F transferase tRNA recognition, we synthesized, purified, and assayed hybrid tRNAs via “step-by-step” mutations to convert the weak substrate Leu-tRNALeu (GAG) into the strong substrate Leu-tRNALeu (CAG). The large differences in the rates of product formation when utilizing these two substrates is apparent in Figure 2B, where the data for these two tRNAs are shown as solid lines. Mutations to the acceptor, D-, and T-stems of the tRNA were investigated. As both tRNAs are aminoacylated by leucyl-tRNA synthetase (LeuRS), it was predicted that the mutations would not inhibit aminoacylation. Previous studies have identified the tRNA nucleotides that are essential for E. coli LeuRS recognition (Asahara et al. 1993a,b, 1998; Larkin et al. 2002), and none of the mutations investigated alter these nucleotides. Our assumptions were validated by performing aminoacylation assays to confirm that these hybrid tRNAs were properly aminoacylated.
tRNALeu (GAG) acceptor stem hybrids
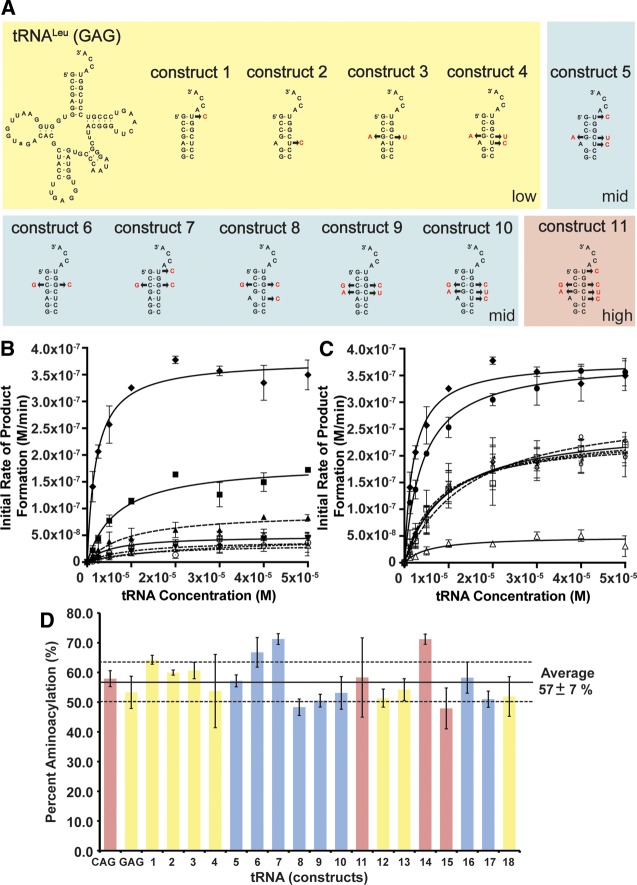
Figure 3A shows the sequences and mutations in the acceptor stem of the tRNA hybrids (constructs 1–11). Mutations in constructs 1–5 focus on the major differences to “step-by-step” convert the weak Leu-tRNALeu (GAG) substrate into the better Leu-tRNALeu (CAG) substrate. Construct 1 with the U72C mutation converts the noncanonical G:U to a canonical G:C pair. Construct 2 with the U68C mutation mimics the A:C mismatch in the isoacceptor of Leu-tRNALeu (CAG). Construct 3 (double mutant) with the G4A and C69U mutations converts a G:C pair to a weaker A:U pair. Construct 4 (triple mutant) combines the mutations in construct 2 and 3, while construct 5 (quadruple mutant) combines the mutations in construct 1 and 4. Initial rates of product formation determined for all hybrid tRNA constructs are listed in Supplemental Table 2, and the kinetic parameters are listed in Table 2. Figure 3B graphically shows the initial rates of product formation vs. tRNA concentration. Subsequent data are compared to the wild-type GAG isoacceptor as a reference. Although constructs 1–4 feature the major acceptor stem sequence differences, they remain poor L/F transferase substrates (1.7- to 4.5-fold increase in apparent Km compared to GAG, and lower catalytic efficiency). Interestingly, construct 5 (quadruple mutant) exhibits an enhanced utilization by L/F transferase activity to a mid-point to that of the optimal substrate CAG when the four nucleotide mutations combined.
FIGURE 3.
Acceptor stem hybrids identify two independent sequence elements for optimal substrate utilization. (A) Cloverleaf structures of tRNA hybrid constructs 1–11. (B) A graphical display of initial rate of product formation vs. tRNA concentration for tRNALeu (CAG) (♦), tRNALeu (GAG) (Δ), constructs 1 (▴), 2 (▾), 3 (∇), 4 (○), and 5 (▪). Constructs 1–4 display low activity similar to GAG, while construct 5 displays a mid-range activity. Errors represented are the standard deviation of three independent experiments. (C) A graphical display of initial rate of product formation vs. tRNA concentration for tRNALeu (CAG) (♦), tRNALeu (GAG) (Δ), constructs 6 (□), 7 (♀), 8 (×), 9 (♂), 10 (☆), and 11 (•). Constructs 6–10 display mid-range activity (overlapping), while construct 11 displays high activity. Errors represented are the standard deviation of three independent experiments. (D) A bar graph presenting the maximal percent aminoacylation of all hybrid constructs after 7 min of aminoacylation. Errors represented are the standard deviation of three independent experiments.
TABLE 2.
Kinetic parameters of L/F transferase-catalyzed peptide bond formation on in vitro-transcribed leucyl-tRNA and phenylalanyl-tRNA hybrids
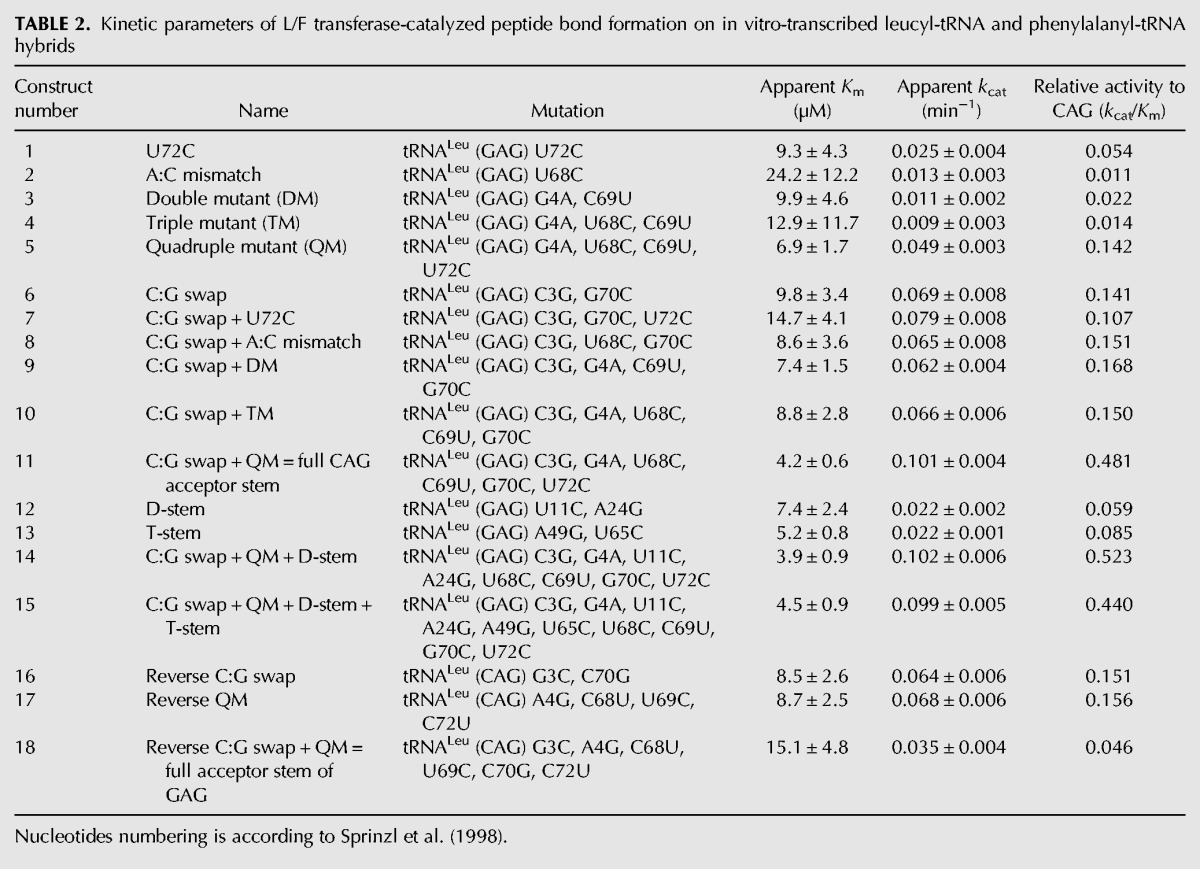
Figure 3C graphically shows the initial rates of product formation vs. tRNA concentration for constructs 6–11 in converting isoacceptor GAG to CAG. Construct 6 (C:G swap) swaps the C:G pair of GAG isoacceptor to a G:C pair of CAG isoacceptor, a relative conserved modification in the acceptor stem. Surprisingly, this conserved C:G swap in the acceptor stem alone enhanced L/F transferase activity to a mid-point level when compared to the optimal substrate CAG. Next, we wanted to determine whether we could improve the activity of GAG with minimal modifications to its acceptor stem. We combined the C:G swap mutation with the mutations in constructs 1–4 and named those constructs 7–10. Kinetic analysis show that the C:G swap in combination with U72C, U68C, double mutant, or the triple mutant does not have any improvement on substrate utilization. Not until construct 11 (C:G swap + quadruple mutant), essentially the full acceptor stem of CAG, does the apparent Km increase slightly and the apparent kcat improve to wild-type CAG levels. A total of a 10-fold improvement in the kcat/Km ratio is achieved with mutations in the acceptor stem of GAG alone.
The maximal percent aminoacylation after 7 min for each tRNA hybrid construct is shown in a bar graph in Figure 3D (for full time course, see Supplemental Fig. S4). Again, there are no significant differences between the aminoacylation of these tRNA hybrids compared to the wild-type isoacceptors. The changes in aminoacylation of the hybrid constructs are not sufficient for changes in the initial reaction rates.
Here, we have demonstrated that mutations in the GAG tRNA acceptor stem alone are able to optimize the utilization of aa-tRNAs by L/F transferase to the maximal CAG isoacceptor levels. We have determined two independent sequence elements—the C:G swap at position 3:70 and the quadruple mutations (G4A, U68C, C69U, U72C)—within the acceptor stem that are important for L/F transferase recognition and catalysis.
tRNALeu (GAG) D- and T-stem hybrids
To further determine whether other parts of the tRNA body contribute to L/F transferase recognition and catalysis, we generated constructs 12–15 with mutations focused on the D- and T-stem of Leu-tRNALeu (GAG) (Fig. 4A). Construct 12 (D-stem), with the mutations U11C and A24G, converts the entire D-stem/D-loop to mimic CAGs. Construct 13 (T-stem), with the mutations A49G and U65C, converts the weaker A:U pair to a stronger G:C pair in the T-stem. Construct 14 (C:G swap + QM + D-stem) and construct 15 (C:G swap + QM + D-stem + T-stem) combine the full acceptor stem mutations with D- and T-stem mutations. Figure 4B clearly shows that D- or T-stem mutations alone do not significantly alter the utilization of the aa-tRNA by L/F transferase. Additionally, full acceptor stem mutations in combination with D-stem and T-stem mutations also do not significantly alter the utilization of construct 11. This suggests that the D- and T-stems do not play a significant role in L/F transferase recognition and catalysis.
FIGURE 4.
No significant recognition contribution by the D-stem and T-stem of the tRNA body. (A) Cloverleaf structures of tRNA hybrid constructs 12–15. (B) A graphical display of initial rate of product formation vs. tRNA concentration for tRNALeu (CAG) (♦), tRNALeu (GAG) (Δ), constructs 12 ( ), 13 (⋄), 14 (⊗), and 15 (⊠). Errors represented are the standard deviation of three independent experiments.
), 13 (⋄), 14 (⊗), and 15 (⊠). Errors represented are the standard deviation of three independent experiments.
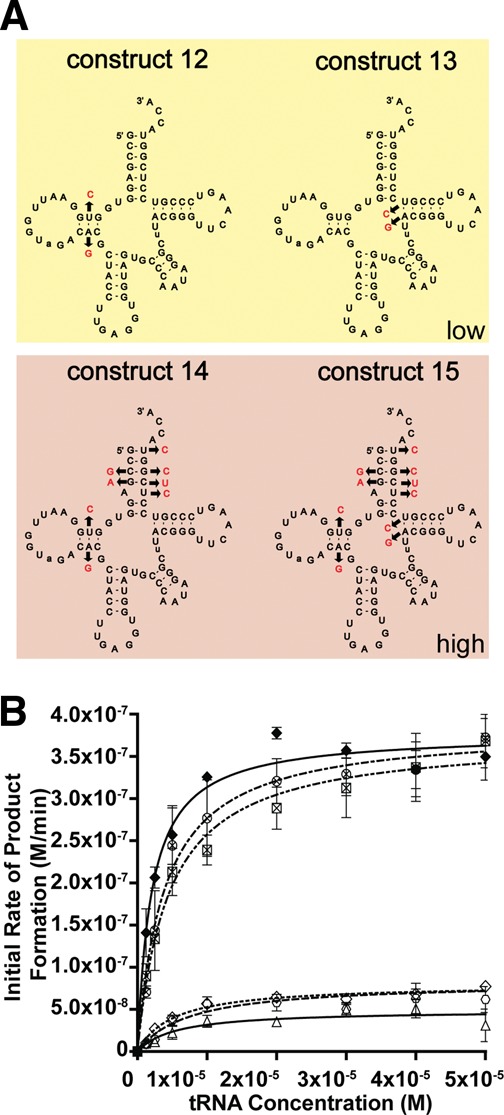
tRNALeu (CAG) reverse hybrids
To validate our findings on the two independent sequence elements recognized by L/F transferase, we generated reverse hybrids to convert the optimal CAG substrate into the poorer GAG substrate. The sequence and mutations are depicted in Figure 5A. Construct 16 (reverse C:G swap) is a reversal of the C:G swap hybrid; meanwhile, construct 17 (reverse quadruple mutant) with the mutation A4G, C68U, U69C, and C72U is a reversal of the quadruple mutant. Construct 18 (reverse C:G + QM) combines the mutations in constructs 16 and 17 to generate the full acceptor stem of the GAG isoacceptor in the context of the CAG isoacceptor. Figure 5B shows the graph of initial rates of product formation vs. tRNA concentration. Subsequent data are compared to the wild-type CAG isoacceptor as a reference. The reverse C:G swap and reverse quadruple mutant, as predicted, independently decrease the affinity (apparent Km increase by 4.3-fold compared to CAG) and decrease the relative catalytic efficiency of the CAG isoacceptor to a midway level (6.4- to 6.6-fold decrease in kcat/Km ratio). The reverse full acceptor stem of GAG (reverse C:G swap + QM) significantly decreases the apparent Km by a 7.5-fold and decreases the relative catalytic efficiency to a lower level similar to that of the GAG isoacceptor (21.7-fold decrease in kcat/Km ratio compared to CAG). These data confirm the importance of the C:G swap and quadruple mutant in L/F transferase recognition and catalysis.
FIGURE 5.
Reverse hybrids validate the identified two independent sequence elements for optimal substrate utilization. (A) Cloverleaf structures of tRNA hybrid construct 16–18. (B) A graphical display of the initial rate of product formation vs. tRNA concentration for tRNALeu (CAG) (♦), tRNALeu (GAG) (Δ), constructs 16 (□), 17 (▪), and 18 (•). Errors represented are the standard deviation of three independent experiments.
DISCUSSION
The acceptor stem of an aa-tRNA is important for L/F transferase recognition
An atypical function for tRNAs is their role in tRNA-dependent post-translational addition of amino acids to the N-terminus of proteins, which leads to protein degradation (Bachmair and Varshavsky 1989). The eubacterial L/F transferase catalyzes the transfer of a leucine or phenylalalanine (or to a lesser extent, methionine) from a cognate aa-tRNA onto the N-terminus of a protein polypeptide (Leibowitz and Soffer 1969; Scarpulla et al. 1976). The protein peptide substrate specificity has been well studied with the aid of X-ray crystal structures and in vitro enzymatic assays (Mogk et al. 2007; Watanabe et al. 2007; Wang et al. 2008a,b; Ninnis et al. 2009; Schuenemann et al. 2009; Kawaguchi et al. 2013). Nonetheless, the aa-tRNA recognition by L/F transferase has remained somewhat elusive. Although there are tRNA substrate analog (3′ rA-Phe and puromycin)-bound X-ray crystal structures, the molecular insights remain within the 3′ aminoacyl adenosine as an intact aa-tRNA: protein structure has not been solved (Suto et al. 2006; Watanabe et al. 2007).
Here, we investigated L/F transferase's preference for a specific tRNALeu isoacceptor and subsequently determined the nucleotides of an aa-tRNA that are optimal for substrate utilization. Our results indicated that the tRNALeu (CAG) isoacceptor is the optimal L/F transferase substrate. Using in vitro-transcribed hybrid tRNAs, we identified two independent sequence elements in this optimal tRNA substrate including the G3:C70 base pair and a set of 4 nt (C72, A4:U69, C68) at the acceptor stem that are shown to be important for binding and catalysis. Our data do not support the weak acceptor stem hypothesis proposed (Abramochkin and Shrader 1996), since individual mutants that generate weak base pairs (i.e., A:U or A:C mismatch in constructs 2 and 3) have no significant effects on activity, while a single conserved C:G or G:C swap at position 3:70 (in constructs 6 and 16) significantly modifies L/F transferase activity. Comparing the 3:70 base pair on wild-type tRNA isoacceptors (Fig. 1), we observed that the two high activity Leu-tRNALeus CAG and UAG contain the G3:C70 base pair, while the remaining tRNAs contain the C3:G70 base pair. Thus, the G3:C70 base pair may serve as a simple predictor for tRNA substrate utilization by L/F transferase. Additionally, the set of 4 nt only enhances binding and catalysis when in combination (construct 5), but individual mutation effects (constructs 1–4) are relatively insignificant. The G1:C72 base pair is common between the CAG, UAG, and CAA isoacceptors, the three better tRNALeu isoacceptors of the five, whereas the A4:U69 and A5:C68 mismatch are unique features of CAG. We hypothesize that the sequence elements may function in combination by contributing to the overall helical shape of the acceptor stem for efficient substrate recognition and catalysis. Future experiments with chemical acylation of various amino acids to various acceptor stem helices may provide additional insights into determining the relative contribution of the amino acid and tRNA to L/F transferase binding affinity and thus the molecular mechanism of aa-tRNA recognition.
Comparison of aa-tRNA recognition to Weissella viridescens FemX transferase
A group of aminoacyl-tRNA protein transferases involved in peptidoglycan biosynthesis have a similar structural fold, but are dissimilar in substrate specificity or function, to L/F transferase (Benson et al. 2002; Biarrotte-Sorin et al. 2004; Rai et al. 2006; Dong et al. 2007). Factors essential for methicillin resistance (Fem) transferase X from Weissella viridescens (FemXWv) transfers L-Ala from L-Ala-tRNAAla to UDP-MurNAc-pentapeptide (Maillard et al. 2005). The C-terminal domain of L/F transferase belongs to the GCN5-related N-acetyltransferase (GNAT) protein superfamily (Rai et al. 2006; Dong et al. 2007) and is similar to the two domains of FemXWv (Biarrotte-Sorin et al. 2004). The Ala-tRNAAla specificity for FemXWv is mainly through steric hindrance of the aminoacyl moiety, where it excludes most amino acids besides glycine (Fonvielle et al. 2009). To distinguish tRNAAla from tRNAGly, FemXWv recognizes the 2:71 base pair in the acceptor stem specifically (tRNAAla with the determinant G2:C71 base pair, and tRNAGly with the anti-determinant C2:G71 base pair) (Villet et al. 2007; Fonvielle et al. 2009).
The aa-tRNA recognition by the structurally similar L/F transferase and FemXWv is more similar than once thought. First, L/F transferase also uses steric hindrance by the C-shaped hydrophobic pocket to select for the aminoacyl moiety (Suto et al. 2006; Watanabe et al. 2007). From this study, we demonstrated that L/F transferase similarly has a major determinant base pair (G3:C70) for efficient utilization of specific tRNALeu isoacceptors. L/F transferase and FemXWv, however, are different in the number of contacts between the protein and the aa-tRNA substrate. FemXWv is suggested to recognize the distal end of an aa-tRNA (up to the first two base pairs) (Villet et al. 2007; Fonvielle et al. 2009); meanwhile, our data suggest that L/F transferase recognizes up to 5 bp in the acceptor stem specifically. The additional tRNA body recognition mechanism by L/F transferase may provide sufficient affinity to compete with EF-Tu.
A proposed model of L/F transferase aa-tRNA recognition
Our investigations have added to the current model of aa-tRNA recognition by L/F transferase that has been developed from previous biochemical and structural studies (Leibowitz and Soffer 1971; Abramochkin and Shrader 1996; Suto et al. 2006; Watanabe et al. 2007). L/F transferase, like other aa-tRNA binding enzymes, recognizes both the esterified amino acid as well as sequence-specific elements within the tRNA body for recognition. The amino acid selectivity is mainly through the C-shaped hydrophobic pocket of L/F transferase, which sterically prevents larger β-branched amino acids (i.e., Ile and Val) and disfavors smaller amino acids (i.e., Ala and Pro), as they are not large enough to make sufficient hydrophobic contacts (Suto et al. 2006; Watanabe et al. 2007). Figure 6 shows a proposed docking model of aa-tRNA recognition by L/F transferase. Based on the close proximity of the positive cluster (R76, R80, K83, and R84) of L/F transferase to the acceptor stem of an aa-tRNA, we suggest that the positive cluster may contribute to the specific recognition of the acceptor stem. Thus, the recognition of the tRNA body involves the 3′ aminoacyl adenosine, the major determinant G3:C70 base pair, the combined set of 4 nt (C72, A4:U69, C68), and the sequence-independent recognition of the D-stem. We hypothesize that the eukaryotic aminoacyl-tRNA protein transferase (ATE1) may similarly depend on both the esterified amino acid and sequence-specific determinants on the tRNA body (i.e., likely the acceptor stem) for efficient aa-tRNA recognition.
FIGURE 6.
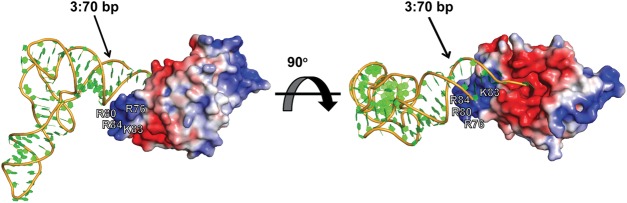
A proposed docking model of aminoacyl-tRNA binding to L/F transferase. Our model suggests that, in addition to the 3′ aminoacyl adenosine recognition and electrostatic interaction, the positive cluster (R76, R80, K83, and R84) of L/F transferase may play a role in the specific recognition of the acceptor stem of an aminoacyl-tRNA. To generate the model, the structure of L/F transferase-rA-Phe complex (shown as electrostatic surface, PDB ID: 2Z3K) (Watanabe et al. 2007) was superimposed on the FemX-peptidyl-RNA complex (PDB ID: 4II9) (Fonvielle et al. 2013) via the conserved core of the GNAT domain. The combined 3′ CCA ends (the cytosines 74 and 75 of the peptidyl-RNA and adenosine of rA-Phe) were then used as references to dock the yeast tRNAPhe (shown as ribbon, PDB ID: 1EHZ) (Shi and Moore 2000). The model was generated using PyMOL (version 1.41), and electrostatic potentials were calculated by APBS (version 1.8).
Codon usage, abundance, and aminoacylation efficiency of tRNALeu isoacceptors
As the tRNALeu (CAG) isoacceptor was determined to be the optimal substrate, we evaluated the literature regarding this isoacceptor and what has been reported with respect to abundance, codon bias, and amino acid-dependent aminoacylation changes. tRNALeu (CAG) is the most abundant leucine isoacceptor in E. coli, representing 50% of all tRNALeu isoacceptors (Dong et al. 1996), and decodes the most frequently used 5′-CUG-3′ codon across various growth rates (Emilsson and Kurland 1990; Dong et al. 1996) and media conditions (Holmes et al. 1977). The CAG isoacceptor is, therefore, widely used during protein synthesis and does not appear to be idiosyncratic for the post-translational addition of amino acids. However, during leucine starvation, the tRNALeu (CAG) aminoacylation level rapidly decreases to 9% of its steady-state level (Elf et al. 2003; Dittmar et al. 2005; Sorensen et al. 2005), yet it also increases rapidly upon restoration of leucine levels. It has been suggested that this differential aminoacylation may serve as a quick response to environmental stress (Elf et al. 2003; Dittmar et al. 2005; Sorensen et al. 2005). These rapid changes in tRNALeu (CAG) aminoacylation levels during environmental stress may regulate L/F transferase activity.
Comparison of aa-tRNA recognition by EF-Tu
Figure 7 summarizes the recognition nucleotides of Leu-tRNALeu (CAG) by E. coli L/F transferase, LeuRS (Asahara et al. 1993a,b, 1998; Larkin et al. 2002), and EF-Tu (Schrader et al. 2009, 2011; Schrader and Uhlenbeck 2011). All three enzymes interact with the single-stranded 3′ CCA end, but their recognition nucleotides are independent and distinct from each other, perhaps adding to the evolutionary selective pressures on these sequences.
FIGURE 7.
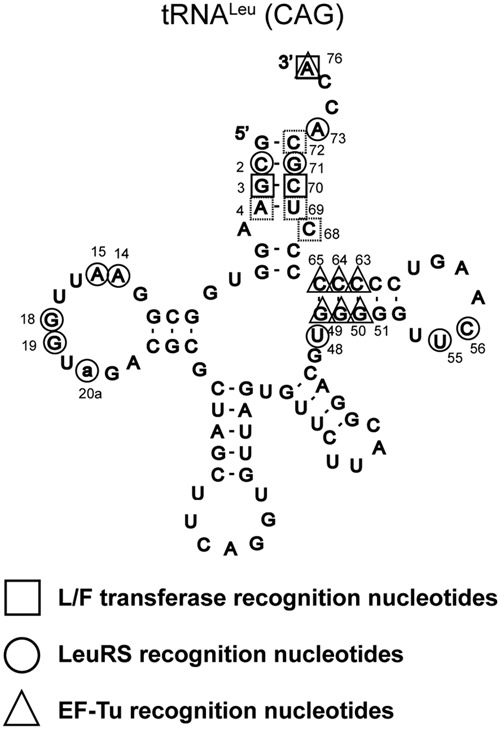
A summary of Leu-tRNALeu (CAG) recognition nucleotides by L/F transferase (□), LeuRS (○), and EF-Tu (Δ). Solid squares represent the major determinant G3:C70 base pair; meanwhile, dashed line squares represent the set of 4 nt (C72, A4:U69, C68) for L/F transferase aa-tRNA recognition. Data for LeuRS recognition were from Asahara et al. (1993a,b, 1998) and Larkin et al. (2002), and data for EF-Tu recognition were from Schrader et al. (2009, 2011) and Schrader and Uhlenbeck (2011). Nucleotides numbering is according to Sprinzl et al. (1998).
All elongator aa-tRNAs bind to EF-Tu with approximately the same affinity (Louie et al. 1984; Louie and Jurnak 1985; Ott et al. 1990; Asahara and Uhlenbeck 2005). It has been shown that there is a thermodynamic compensation mechanism that balances the affinity for the esterified amino acid and the affinity of the tRNA using three adjacent base pairs (49:65, 50:64, and 51:63) in the T-stem of an aa-tRNA (LaRiviere et al. 2001; Asahara and Uhlenbeck 2002; Dale et al. 2004; Sanderson and Uhlenbeck 2007a,b; Schrader et al. 2009, 2011). A model has been proposed and validated, enabling the binding affinity of any tRNA to EF-Tu to be predicted using three T-stem base pairs (Schrader and Uhlenbeck 2011). Based on the model, Schrader et al. identified some aa-tRNAs that bind weakly to EF-Tu and are proposed to be candidates of nontranslational tRNAs (Giannouli et al. 2009; Schrader and Uhlenbeck 2011). However, based on the predictions, all E. coli tRNALeu isoacceptors appears to bind similarly to EF-Tu, suggesting that all E. coli tRNALeu isoacceptors participate in translation with equal efficiency (Schrader and Uhlenbeck 2011). Given that L/F transferase has a low in vivo concentration (L/F ∼0.5 μM vs. EF-Tu ∼100 µM) with a weak affinity for aa-tRNA (L/F KD ∼200 nM vs. EF-Tu KD ∼5 nM) when compared to EF-Tu, this predicts a competition of substrates that does not favor L/F transferase (Leibowitz and Soffer 1969; Scarpulla et al. 1976; Shrader et al. 1993).
Thus, as the tRNALeu CAG isoacceptor seems to participate in both translation and alternative functions, the specific isoacceptor preference and the molecular mechanism in which L/F transferase competes with EF-Tu for aa-tRNA remain elusive. It is possible that there are still unidentified factors that aid in substrate recruitment by L/F transferase in vivo. Or, perhaps L/F transferase needs to be opportunistic and only accesses aa-tRNA substrates when EF-Tu is inactivated through a more general mechanism. For example, during the stringent response, when nutrients are limited, it has been estimated that almost half of the GTP molecules is converted to pentaphosphate guanosine (pppGpp) synthesis (Fiil et al. 1972). Given that EF-Tu binds to an aa-tRNA in its GTP-bound state, the loss of GTP molecules would affect the concentration of free aa-tRNA in the cytosol. pppGpp molecules are hydrolyzed into tetraphosphate guanosine (ppGpp), which is the functional molecule of the stringent response (Wu and Xie 2009). Interestingly, it has been shown that pppGpp can substitute GTP and binds to EF-Tu to form the ternary complex with aa-tRNA, whereas the EF-Tu-ppGpp cannot form the ternary complex (Pingoud and Block 1981). The tetraphosphate ppGpp resembles GDP and has been shown to be able to inhibit EF-Tu directly (Ki = 7 × 10−7 M) and indirectly via trapping the EF-Tu:EF-Ts complex (Ki = 4 × 10−5 M) (Rojas et al. 1984). Therefore, we hypothesize a general mechanism for L/F transferase aa-tRNA utilization where EF-Tu is inactivated during the stringent response and cannot bind an aa-tRNA efficiently, allowing free aa-tRNA to be used for post-translational addition of amino acids.
Concluding remarks
In conclusion, we have demonstrated that the most abundant leucyl-tRNA in E. coli, tRNALeu (CAG), is the most optimal substrate for L/F transferase. We confirmed that the rate differences are not due to differential aminoacylation. Using “step-by-step” hybrid tRNAs, we have identified two independent sequence elements on the acceptor stem of Leu-tRNALeu (CAG) that are important for optimal binding and catalysis by L/F transferase. A G3:C70 base pair and a set of 4 nt in combination (C72, A4:U69, C68) contribute to optimal tRNA recognition by L/F transferase. This maps a more specific, sequence-dependent tRNA recognition model of L/F transferase than previously thought.
MATERIALS AND METHODS
Materials
Unless stated otherwise, all chemicals were purchased from Sigma-Aldrich. α-Cyano-4-hydroxycinnamic acid (CHCA), triethylamine (TEA), and bromoethane were purchased from Acros Organics. The substrate and product peptides (REPGLCTWQSLR, LREPGLCTWQSLR, FREPGLCTWQSLR, and MREPGLCTWQSLR) were purchased from the Institute for Biomolecular Design (University of Alberta, Canada). Peptide stock solutions were made, and their absolute concentrations were determined by amino acid analysis (Institute for Biomolecular Design).
Expression vectors and protein purifications
E. coli L/F transferase wild type with a 6× histidine tag in a pCA24N expression vector was obtained from the ASKA strain collection (Kitagawa et al. 2005) from the National Institute of Genetics (Japan). A clone of a 6× histidine-tagged E. coli leucyl-tRNA synthetase (LeuRS) in a pCA24N expression vector was obtained from the National Institute of Genetics (Japan). A clone of a 6× histidine-tagged E. coli phenylalanyl-tRNA synthetase (PheRS) in a pET28a expression vector was a gift from Jack Szostak (Harvard Medical School). E. coli methionyl-tRNA synthetase (MetRS) was cloned into a pET28a plasmid vector between the NheI and NotI restriction sites which incorporates an N-terminal 6× histidine tag. Cloned 6× histidine-tagged nucleotidyl transferase (CCA adding enzyme) in a pET22b expression plasmid was a generous gift from Allen Weiner (University of Washington). Proteins were purified as described (Fung et al. 2011) with the proteins dialyzed into storage buffer (50 mM Tris–Cl [pH 7.4], 100 mM NaCl, 1 mM DTT, and 10% glycerol), flash frozen, and stored at −80°C.
In vitro transcription of tRNA
Purified, unmodified E. coli tRNAs were prepared by primer extension, in vitro transcription, and preparative denaturing gel electrophoresis as previously described (Fahlman and Uhlenbeck 2004; Fung et al. 2011). Overlapping DNA oligonucleotides (IDT) used for in vitro tRNA and hybrid tRNA transcriptions are listed in Supplemental Tables 3 and 4, respectively.
Stable isotope labeling of peptides
To generate peptides of identical chemical composition that differ in mass by five mass units (m/z), the substrate and product peptides were alkylated using either bromoethane or deuterated (d5)-bromoethane as previously described (Hale et al. 1996, 2004; Ebhardt et al. 2009; Fung et al. 2011).
L/F transferase activity assay
The peptide bond formation reaction was modified from the original procedure described by Ebhardt et al. (2009) and Fung et al. (2011). Briefly, in each strip of eight tubes (Corning Thermowell Gold PCR) was a 90-μL tRNA precharging reaction containing 1× reaction buffer (50 mM Hepes, pH 7.5, 50 mM KCl, 15 mM MgCl2), 2 mM ATP, 0.2 mM CTP, 2 mM β-mercaptoethanol, 2 mM amino acid (leucine, phenylalaine, or methionine), 5.91 μM substrate peptide (REPGLCTWQSLR), 5.91 μM standard product peptide (LREPGLCTWQSLR, FREPGLCTWQSLR, or MREPGLCTWQSLR), 1 μL of 1 mg/mL CCA adding enzyme, and 1 μL of 1 mg/mL aa-tRNA synthetase (LeuRS, PheRS, or MetRS). Additionally, these tubes also contained the refolded (65°C in 5 mM NaOAc and then slow cooled to 37°C) tRNA species (isoacceptors or hybrids) with final concentrations ranging from 1.25 to 50 μM. These reaction mixtures were incubated for 7 min at 37°C to facilitate aminoacylation of the tRNA. The reactions were initiated by the addition of wild-type L/F transferase (final concentration 3.8 µM) to the samples containing the aminoacylated tRNAs and peptide substrate. At increasing time points, 5 μL of the reactions were added to 5 μL quench solution (0.01 µg/µL BSA, 10% acetonitrile, 2% trifluoracetic acid). For analysis, 10 μL matrix solution (saturated R-Cyano-4-hydroxycinnamic acid in 50% acetonitrile and 0.2% TFA) were added to the quenched reaction aliquots, and 1 μL of the mixture was spotted in duplicate on a MALDI-ToF sample plate. The spectra were collected on a Bruker Daltonics Ultraflex MALDI-ToF/ToF at the Institute for Biomolecular Design (University of Alberta, Canada). To quantify product formation, the ratio of the relative intensity of the labeled product peptide to the relative intensity of the labeled internal standard peptide was used as described previously (Fung et al. 2011).
Curve-fit analysis
The initial rates of product formation calculated from the quantitative MALDI-ToF MS enzymatic assay data were plotted vs. tRNA substrate concentration using GraphPad Prism Version 5.02 (GraphPad Software).
Radiolabeling tRNA and aminoacylation assay
To ensure that the differences in L/F transferase product formation rates are due to the RNA structure and not due to reduced aminoacylation, aminoacylation rates were tested. The aminoacylation assay has been modified as previously described (Wolfson and Uhlenbeck 2002). Briefly, 2 µM purified in vitro-transcribed tRNAs were folded by heating for 3 min at 65°C in 10 mM MgCl2. To 32P-label the tRNAs at the 3′-terminal inter-nucleotide linkage, the folded tRNAs were added to a 100-µL reaction containing 50 mM glycine-HCl (pH 9.0), 50 µM NaPPi, 2 µM [α-32P] ATP [3000 Ci/mmol], 0.06 µg/µL CCA adding enzyme, and 80 units of RNase OUT and incubated at 37°C for 5 min. Two microliters of 10 units/mL yeast PPase and 2 µM CTP were added, and the reaction mixture was incubated for an additional 2 min before quenching by phenol/chloroform extraction. After ethanol precipitation, 3′-32P-labeled-tRNAs were purified by a pre-equilibrated desalting column (Thermo Fisher, 7k MWCO). Aminoacylation levels were determined in a 25-µL reaction containing 1× aminoacylation buffer (50 mM HEPES [pH 7.5], 30 mM KCl, 15 mM MgCl2, 2 mM ATP, 0.5 mM DTT), 1 mM amino acid (leucine, phenylalanine, or methionine), 0.8 µM 3′-32P-labeled tRNA (isoaccepting or hybrid species), and 1 µL of 1 mg/mL aa-tRNA synthetase (LeuRS, PheRS, or MetRS) at 37°C. Aliquots were taken at specific time points (0, 0.5, 1, 2, 3, 4, 5, 7, 10, and 20 min) and quenched on ice with 200 mM sodium acetate, pH 5.0 containing 1 unit/µL of nuclease S1 or P1. The quenched aliquots were kept on ice until the aminoacylation time course was completed and then incubated for 10 min at room temperature to digest the tRNA into AMP and aminoacyl-AMP (AMP-AA). One microliter of the digestion reaction was spotted on 9-cm, prewashed polyethylenimine-cellulose plates, and the AMP and AMP-AA were separated by TLC in glacial acetic acid/1 M NH4Cl/H2O (5:10:85). The radioactivity was measured by PhosphorImager (Molecular Dynamics) and quantified by ImageQuant (version 5.2).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
This project was supported by the Natural Sciences and Engineering Research Council (NSERC) of Canada (Discovery Grant 341453-12 to R.P.F.). A.W.S.F. is supported by a NSERC Alexander Graham Bell Canada Graduate Scholarship and Alberta Innovates Technology Futures Graduate Student Scholarship. We would like to thank Jack Moore (Institute for Biomolecular Design) and Fahlman lab members for technical assistance and support.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.044529.114.
REFERENCES
- Abramochkin G, Shrader TE 1995. The leucyl/phenylalanyl-tRNA-protein transferase. Overexpression and characterization of substrate recognition, domain structure, and secondary structure. J Biol Chem 270: 20621–20628 [DOI] [PubMed] [Google Scholar]
- Abramochkin G, Shrader TE 1996. Aminoacyl-tRNA recognition by the leucyl/phenylalanyl-tRNA-protein transferase. J Biol Chem 271: 22901–22907 [DOI] [PubMed] [Google Scholar]
- Agirrezabala X, Frank J 2009. Elongation in translation as a dynamic interaction among the ribosome, tRNA, and elongation factors EF-G and EF-Tu. Q Rev Biophys 42: 159–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen C, Wiborg O 1994. Escherichia coli elongation-factor-Tu mutants with decreased affinity for aminoacyl-tRNA. Eur J Biochem 220: 739–744 [DOI] [PubMed] [Google Scholar]
- Asahara H, Uhlenbeck OC 2002. The tRNA specificity of Thermus thermophilus EF-Tu. Proc Natl Acad Sci 99: 3499–3504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asahara H, Uhlenbeck OC 2005. Predicting the binding affinities of misacylated tRNAs for Thermus thermophilus EF-Tu.GTP. Biochemistry 44: 11254–11261 [DOI] [PubMed] [Google Scholar]
- Asahara H, Himeno H, Tamura K, Hasegawa T, Watanabe K, Shimizu M 1993a. Recognition nucleotides of Escherichia coli tRNALeu and its elements facilitating discrimination from tRNASer and tRNATyr. J Mol Biol 231: 219–229 [DOI] [PubMed] [Google Scholar]
- Asahara H, Himeno H, Tamura K, Nameki N, Hasegawa T, Shimizu M 1993b. Discrimination among E. coli tRNAs with a long variable arm. Nucleic Acids Symp Ser 29: 207–208 [PubMed] [Google Scholar]
- Asahara H, Nameki N, Hasegawa T 1998. In vitro selection of RNAs aminoacylated by Escherichia coli leucyl-tRNA synthetase. J Mol Biol 283: 605–618 [DOI] [PubMed] [Google Scholar]
- Bachmair A, Varshavsky A 1989. The degradation signal in a short-lived protein. Cell 56: 1019–1032 [DOI] [PubMed] [Google Scholar]
- Bailly M, Blaise M, Lorber B, Becker HD, Kern D 2007. The transamidosome: A dynamic ribonucleoprotein particle dedicated to prokaryotic tRNA-dependent asparagine biosynthesis. Mol Cell 28: 228–239 [DOI] [PubMed] [Google Scholar]
- Banerjee R, Chen S, Dare K, Gilreath M, Praetorius-Ibba M, Raina M, Reynolds NM, Rogers T, Roy H, Yadavalli SS, et al. 2010. tRNAs: cellular barcodes for amino acids. FEBS Lett 584: 387–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker HD, Kern D 1998. Thermus thermophilus: a link in evolution of the tRNA-dependent amino acid amidation pathways. Proc Natl Acad Sci 95: 12832–12837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson TE, Prince DB, Mutchler VT, Curry KA, Ho AM, Sarver RW, Hagadorn JC, Choi GH, Garlick RL 2002. X-ray crystal structure of Staphylococcus aureus FemA. Structure 10: 1107–1115 [DOI] [PubMed] [Google Scholar]
- Biarrotte-Sorin S, Maillard AP, Delettre J, Sougakoff W, Arthur M, Mayer C 2004. Crystal structures of Weissella viridescens FemX and its complex with UDP-MurNAc-pentapeptide: insights into FemABX family substrates recognition. Structure 12: 257–267 [DOI] [PubMed] [Google Scholar]
- Chan PP, Lowe TM 2009. GtRNAdb: a database of transfer RNA genes detected in genomic sequence. Nucleic Acids Res 37: D93–D97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale T, Sanderson LE, Uhlenbeck OC 2004. The affinity of elongation factor Tu for an aminoacyl-tRNA is modulated by the esterified amino acid. Biochemistry 43: 6159–6166 [DOI] [PubMed] [Google Scholar]
- Dittmar KA, Sorensen MA, Elf J, Ehrenberg M, Pan T 2005. Selective charging of tRNA isoacceptors induced by amino-acid starvation. EMBO Rep 6: 151–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H, Nilsson L, Kurland CG 1996. Co-variation of tRNA abundance and codon usage in Escherichia coli at different growth rates. J Mol Biol 260: 649–663 [DOI] [PubMed] [Google Scholar]
- Dong X, Kato-Murayama M, Muramatsu T, Mori H, Shirouzu M, Bessho Y, Yokoyama S 2007. The crystal structure of leucyl/phenylalanyl-tRNA-protein transferase from Escherichia coli. Protein Sci 16: 528–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougan DA, Truscott KN, Zeth K 2010. The bacterial N-end rule pathway: expect the unexpected. Mol Microbiol 76: 545–558 [DOI] [PubMed] [Google Scholar]
- Ebhardt HA, Xu Z, Fung AW, Fahlman RP 2009. Quantification of the post-translational addition of amino acids to proteins by MALDI-TOF mass spectrometry. Anal Chem 81: 1937–1943 [DOI] [PubMed] [Google Scholar]
- Elf J, Nilsson D, Tenson T, Ehrenberg M 2003. Selective charging of tRNA isoacceptors explains patterns of codon usage. Science 300: 1718–1722 [DOI] [PubMed] [Google Scholar]
- Emilsson V, Kurland CG 1990. Growth rate dependence of transfer RNA abundance in Escherichia coli. EMBO J 9: 4359–4366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahlman RP, Uhlenbeck OC 2004. Contribution of the esterified amino acid to the binding of aminoacylated tRNAs to the ribosomal P- and A-sites. Biochemistry 43: 7575–7583 [DOI] [PubMed] [Google Scholar]
- Fiil NP, von Meyenburg K, Friesen JD 1972. Accumulation and turnover of guanosine tetraphosphate in Escherichia coli. J Mol Biol 71: 769–783 [DOI] [PubMed] [Google Scholar]
- Fonvielle M, Chemama M, Villet R, Lecerf M, Bouhss A, Valery JM, Etheve-Quelquejeu M, Arthur M 2009. Aminoacyl-tRNA recognition by the FemXWv transferase for bacterial cell wall synthesis. Nucleic Acids Res 37: 1589–1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonvielle M, Li de La Sierra-Gallay I, El-Sagheer AH, Lecerf M, Patin D, Mellal D, Mayer C, Blanot D, Gale N, Brown T, et al. 2013. The structure of FemXWv in complex with a peptidyl-RNA conjugate: mechanism of aminoacyl transfer from Ala-tRNAAla to peptidoglycan precursors. Angew Chem Int Ed Engl 52: 7278–7281 [DOI] [PubMed] [Google Scholar]
- Francklyn CS, Minajigi A 2010. tRNA as an active chemical scaffold for diverse chemical transformations. FEBS Lett 584: 366–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung AW, Ebhardt HA, Abeysundara H, Moore J, Xu Z, Fahlman RP 2011. An alternative mechanism for the catalysis of peptide bond formation by L/F transferase: substrate binding and orientation. J Mol Biol 409: 617–629 [DOI] [PubMed] [Google Scholar]
- Fung AW, Ebhardt HA, Krishnakumar KS, Moore J, Xu Z, Strazewski P, Fahlman RP 2014. Probing the leucyl/phenylalanyl tRNA protein transferase active site with tRNA substrate analogues. Protein Pept Lett 21: 603–614 [DOI] [PubMed] [Google Scholar]
- Giannouli S, Kyritsis A, Malissovas N, Becker HD, Stathopoulos C 2009. On the role of an unusual tRNAGly isoacceptor in Staphylococcus aureus. Biochimie 91: 344–351 [DOI] [PubMed] [Google Scholar]
- Gonda DK, Bachmair A, Wunning I, Tobias JW, Lane WS, Varshavsky A 1989. Universality and structure of the N-end rule. J Biol Chem 264: 16700–16712 [PubMed] [Google Scholar]
- Graciet E, Wellmer F 2010. The plant N-end rule pathway: structure and functions. Trends Plant Sci 15: 447–453 [DOI] [PubMed] [Google Scholar]
- Hale JE, Butler JP, Pourmand RR 1996. Analysis of cysteine residues in peptides and proteins alkylated with volatile reagents. Amino Acids 10: 243–252 [DOI] [PubMed] [Google Scholar]
- Hale JE, Butler JP, Gelfanova V, You JS, Knierman MD 2004. A simplified procedure for the reduction and alkylation of cysteine residues in proteins prior to proteolytic digestion and mass spectral analysis. Anal Biochem 333: 174–181 [DOI] [PubMed] [Google Scholar]
- Holmes WM, Goldman E, Miner TA, Hatfield GW 1977. Differential utilization of leucyl-tRNAs by Escherichia coli. Proc Natl Acad Sci 74: 1393–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibba M, Soll D 2004. Aminoacyl-tRNAs: setting the limits of the genetic code. Genes Dev 18: 731–738 [DOI] [PubMed] [Google Scholar]
- Karakozova M, Kozak M, Wong CC, Bailey AO, Yates JR III, Mogilner A, Zebroski H, Kashina A 2006. Arginylation of β-actin regulates actin cytoskeleton and cell motility. Science 313: 192–196 [DOI] [PubMed] [Google Scholar]
- Kawaguchi J, Maejima K, Kuroiwa H, Taki M 2013. Kinetic analysis of the leucyl/phenylalanyl-tRNA-protein transferase with acceptor peptides possessing different N-terminal penultimate residues. FEBS Open Bio 3: 252–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa M, Ara T, Arifuzzaman M, Ioka-Nakamichi T, Inamoto E, Toyonaga H, Mori H 2005. Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): unique resources for biological research. DNA Res 12: 291–299 [DOI] [PubMed] [Google Scholar]
- LaRiviere FJ, Wolfson AD, Uhlenbeck OC 2001. Uniform binding of aminoacyl-tRNAs to elongation factor Tu by thermodynamic compensation. Science 294: 165–168 [DOI] [PubMed] [Google Scholar]
- Larkin DC, Williams AM, Martinis SA, Fox GE 2002. Identification of essential domains for Escherichia coli tRNAleu aminoacylation and amino acid editing using minimalist RNA molecules. Nucleic Acids Res 30: 2103–2113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibowitz MJ, Soffer RL 1969. A soluble enzyme from Escherichia coli which catalyzes the transfer of leucine and phenylalanine from tRNA to acceptor proteins. Biochem Biophys Res Commun 36: 47–53 [DOI] [PubMed] [Google Scholar]
- Leibowitz MJ, Soffer RL 1971. Enzymatic modification of proteins. VII. Substrate specificity of leucyl,phenylalanyl-transfer ribonucleic acid-protein transferase. J Biol Chem 246: 5207–5212 [PubMed] [Google Scholar]
- Louie A, Jurnak F 1985. Kinetic studies of Escherichia coli elongation factor Tu-guanosine 5′-triphosphate-aminoacyl-tRNA complexes. Biochemistry 24: 6433–6439 [DOI] [PubMed] [Google Scholar]
- Louie A, Ribeiro NS, Reid BR, Jurnak F 1984. Relative affinities of all Escherichia coli aminoacyl-tRNAs for elongation factor Tu-GTP. J Biol Chem 259: 5010–5016 [PubMed] [Google Scholar]
- Maillard AP, Biarrotte-Sorin S, Villet R, Mesnage S, Bouhss A, Sougakoff W, Mayer C, Arthur M 2005. Structure-based site-directed mutagenesis of the UDP-MurNAc-pentapeptide-binding cavity of the FemX alanyl transferase from Weissella viridescens. J Bacteriol 187: 3833–3838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall RA, Aitken CE, Dorywalska M, Puglisi JD 2008. Translation at the single-molecule level. Annu Rev Biochem 77: 177–203 [DOI] [PubMed] [Google Scholar]
- Mogk A, Schmidt R, Bukau B 2007. The N-end rule pathway for regulated proteolysis: prokaryotic and eukaryotic strategies. Trends Cell Biol 17: 165–172 [DOI] [PubMed] [Google Scholar]
- Ninnis RL, Spall SK, Talbo GH, Truscott KN, Dougan DA 2009. Modification of PATase by L/F-transferase generates a ClpS-dependent N-end rule substrate in Escherichia coli. EMBO J 28: 1732–1744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan EM, Walsh CT 2009. How nature morphs peptide scaffolds into antibiotics. Chembiochem 10: 34–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott G, Schiesswohl M, Kiesewetter S, Forster C, Arnold L, Erdmann VA, Sprinzl M 1990. Ternary complexes of Escherichia coli aminoacyl-tRNAs with the elongation factor Tu and GTP: Thermodynamic and structural studies. Biochim Biophys Acta 1050: 222–225 [DOI] [PubMed] [Google Scholar]
- Pingoud A, Block W 1981. The elongation factor Tu ⋅ guanosine tetraphosphate complex. Eur J Biochem 116: 631–634 [DOI] [PubMed] [Google Scholar]
- Potuschak T, Stary S, Schlogelhofer P, Becker F, Nejinskaia V, Bachmair A 1998. PRT1 of Arabidopsis thaliana encodes a component of the plant N-end rule pathway. Proc Natl Acad Sci 95: 7904–7908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai R, Mushegian A, Makarova K, Kashina A 2006. Molecular dissection of arginyltransferases guided by similarity to bacterial peptidoglycan synthases. EMBO Rep 7: 800–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao PM, Kaji H 1974. Utilization of isoaccepting leucyl-tRNA in the soluble incorporation system and protein synthesizing systems from E.coli. FEBS Lett 43: 199–202 [DOI] [PubMed] [Google Scholar]
- Rojas AM, Ehrenberg M, Andersson SG, Kurland CG 1984. ppGpp inhibition of elongation factors Tu, G and Ts during polypeptide synthesis. Mol Gen Genet 197: 36–45 [DOI] [PubMed] [Google Scholar]
- Roy H, Ibba M 2008. RNA-dependent lipid remodeling by bacterial multiple peptide resistance factors. Proc Natl Acad Sci 105: 4667–4672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha S, Kashina A 2011. Posttranslational arginylation as a global biological regulator. Dev Biol 358: 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson LE, Uhlenbeck OC 2007a. The 51–63 base pair of tRNA confers specificity for binding by EF-Tu. RNA 13: 835–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson LE, Uhlenbeck OC 2007b. Directed mutagenesis identifies amino acid residues involved in elongation factor Tu binding to yeast Phe-tRNAPhe. J Mol Biol 368: 119–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpulla RC, Deutch CE, Soffer RL 1976. Transfer of methionyl residues by leucyl, phenylalanyl-tRNA-protein transferase. Biochem Biophys Res Commun 71: 584–589 [DOI] [PubMed] [Google Scholar]
- Schmeing TM, Ramakrishnan V 2009. What recent ribosome structures have revealed about the mechanism of translation. Nature 461: 1234–1242 [DOI] [PubMed] [Google Scholar]
- Schrader JM, Uhlenbeck OC 2011. Is the sequence-specific binding of aminoacyl-tRNAs by EF-Tu universal among bacteria? Nucleic Acids Res 39: 9746–9758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader JM, Chapman SJ, Uhlenbeck OC 2009. Understanding the sequence specificity of tRNA binding to elongation factor Tu using tRNA mutagenesis. J Mol Biol 386: 1255–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader JM, Chapman SJ, Uhlenbeck OC 2011. Tuning the affinity of aminoacyl-tRNA to elongation factor Tu for optimal decoding. Proc Natl Acad Sci 108: 5215–5220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuenemann VJ, Kralik SM, Albrecht R, Spall SK, Truscott KN, Dougan DA, Zeth K 2009. Structural basis of N-end rule substrate recognition in Escherichia coli by the ClpAP adaptor protein ClpS. EMBO Rep 10: 508–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard K, Yuan J, Hohn MJ, Jester B, Devine KM, Soll D 2008. From one amino acid to another: tRNA-dependent amino acid biosynthesis. Nucleic Acids Res 36: 1813–1825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Moore PB 2000. The crystal structure of yeast phenylalanine tRNA at 1.93 Å resolution: a classic structure revisited. RNA 6: 1091–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrader TE, Tobias JW, Varshavsky A 1993. The N-end rule in Escherichia coli: cloning and analysis of the leucyl, phenylalanyl-tRNA-protein transferase gene aat. J Bacteriol 175: 4364–4374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soffer RL 1974. Aminoacyl-tRNA transferases. Adv Enzymol Relat Areas Mol Biol 40: 91–139 [DOI] [PubMed] [Google Scholar]
- Sorensen MA, Elf J, Bouakaz E, Tenson T, Sanyal S, Bjork GR, Ehrenberg M 2005. Over expression of a tRNALeu isoacceptor changes charging pattern of leucine tRNAs and reveals new codon reading. J Mol Biol 354: 16–24 [DOI] [PubMed] [Google Scholar]
- Sprinzl M, Horn C, Brown M, Ioudovitch A, Steinberg S 1998. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res 26: 148–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanzel M, Schon A, Sprinzl M 1994. Discrimination against misacylated tRNA by chloroplast elongation factor Tu. Eur J Biochem 219: 435–439 [DOI] [PubMed] [Google Scholar]
- Stortchevoi A, Varshney U, RajBhandary UL 2003. Common location of determinants in initiator transfer RNAs for initiator-elongator discrimination in bacteria and in eukaryotes. J Biol Chem 278: 17672–17679 [DOI] [PubMed] [Google Scholar]
- Suto K, Shimizu Y, Watanabe K, Ueda T, Fukai S, Nureki O, Tomita K 2006. Crystal structures of leucyl/phenylalanyl-tRNA-protein transferase and its complex with an aminoacyl-tRNA analog. EMBO J 25: 5942–5950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobias JW, Shrader TE, Rocap G, Varshavsky A 1991. The N-end rule in bacteria. Science 254: 1374–1377 [DOI] [PubMed] [Google Scholar]
- Villet R, Fonvielle M, Busca P, Chemama M, Maillard AP, Hugonnet JE, Dubost L, Marie A, Josseaume N, Mesnage S, et al. 2007. Idiosyncratic features in tRNAs participating in bacterial cell wall synthesis. Nucleic Acids Res 35: 6870–6883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner AM, Fegley MW, Warner JB, Grindley CL, Marotta NP, Petersson EJ 2011. N-terminal protein modification using simple aminoacyl transferase substrates. J Am Chem Soc 133: 15139–15147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KH, Oakes ES, Sauer RT, Baker TA 2008a. Tuning the strength of a bacterial N-end rule degradation signal. J Biol Chem 283: 24600–24607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KH, Roman-Hernandez G, Grant RA, Sauer RT, Baker TA 2008b. The molecular basis of N-end rule recognition. Mol Cell 32: 406–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K, Toh Y, Suto K, Shimizu Y, Oka N, Wada T, Tomita K 2007. Protein-based peptide-bond formation by aminoacyl-tRNA protein transferase. Nature 449: 867–871 [DOI] [PubMed] [Google Scholar]
- Wendrich TM, Blaha G, Wilson DN, Marahiel MA, Nierhaus KH 2002. Dissection of the mechanism for the stringent factor RelA. Mol Cell 10: 779–788 [DOI] [PubMed] [Google Scholar]
- Wolfson AD, Uhlenbeck OC 2002. Modulation of tRNAAla identity by inorganic pyrophosphatase. Proc Natl Acad Sci 99: 5965–5970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Xie J 2009. Magic spot: (p) ppGpp. J Cell Physiol 220: 297–302 [DOI] [PubMed] [Google Scholar]
- Zaborske JM, Narasimhan J, Jiang L, Wek SA, Dittmar KA, Freimoser F, Pan T, Wek RC 2009. Genome-wide analysis of tRNA charging and activation of the eIF2 kinase Gcn2p. J Biol Chem 284: 25254–25267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Saha S, Shabalina SA, Kashina A 2010. Differential arginylation of actin isoforms is regulated by coding sequence-dependent degradation. Science 329: 1534–1537 [DOI] [PMC free article] [PubMed] [Google Scholar]