The Piwi-piRNA complex in animal germlines represses transposable elements (TEs) to maintain genome integrity. However, little is known about primate PIWI-piRNA pathways. In this paper, the authors analyze Piwi-piRNA complexes in the adult testes of the common marmoset, a model primate. They find that marmoset piRNAs show characteristics of mouse pachytene piRNAs and mostly derive from conserved piRNA clusters, some of which contain pseudogenes with antisense orientation. In addition, more piRNAs map to TE subfamilies when they have copies in piRNA clusters. Their findings suggest that pachytene piRNA clusters function in both TE traps and regulation of protein-coding gene expression.
Keywords: miRNAs, piRNAs, PIWI, transposable elements, pseudogenes
Abstract
Small RNAs mediate gene silencing by binding Argonaute/Piwi proteins to regulate target RNAs. Here, we describe small RNA profiling of the adult testes of Callithrix jacchus, the common marmoset. The most abundant class of small RNAs in the adult testis was piRNAs, although 353 novel miRNAs but few endo-siRNAs were also identified. MARWI, a marmoset homolog of mouse MIWI and a very abundant PIWI in adult testes, associates with piRNAs that show characteristics of mouse pachytene piRNAs. As in other mammals, most marmoset piRNAs are derived from conserved clustered regions in the genome, which are annotated as intergenic regions. However, unlike in mice, marmoset piRNA clusters are also found on the X chromosome, suggesting escape from meiotic sex chromosome inactivation by the X-linked clusters. Some of the piRNA clusters identified contain antisense-orientated pseudogenes, suggesting the possibility that pseudogene-derived piRNAs may regulate parental functional protein-coding genes. More piRNAs map to transposable element (TE) subfamilies when they have copies in piRNA clusters. In addition, the strand bias observed for piRNAs mapped to each TE subfamily correlates with the polarity of copies inserted in clusters. These findings suggest that pachytene piRNA clusters determine the abundance and strand-bias of TE-derived piRNAs, may regulate protein-coding genes via pseudogene-derived piRNAs, and may even play roles in meiosis in the adult marmoset testis.
INTRODUCTION
Eukaryotic cells express a large number of small noncoding RNAs, 20–35 nucleotides (nt) in length, that are derived from various parts of the genome, including intergenic regions, transposable elements (TEs) and their remnants, and also from viruses (Girard and Hannon 2008; Ghildiyal and Zamore 2009; Siomi and Siomi 2009; Ketting 2011). These small RNAs associate with Argonaute proteins to form regulatory complexes known as RNA-induced silencing complexes (RISCs), which then pair with complementary RNA targets to promote the inactivation of homologous sequences. These pathways are referred to as RNA interference (RNAi), or RNA silencing, and help regulate protein levels and restrain the expression of parasitic and pathogenic invaders such as TEs and viruses (Bartel 2009; Saito and Siomi 2010). The RNAi pathway is present in a diverse range of eukaryotes, including some unicellular organisms, and is thought to have evolved as a form of nucleic acid-based immunity to inactivate TEs and viruses (Buchon and Vaury 2006).
According to their biogenesis and associated Argonaute proteins, small silencing RNAs are classified into three types: microRNAs (miRNAs), endogenous small interfering RNAs (endo-siRNAs), and PIWI-interacting RNAs (piRNAs) (Ghildiyal and Zamore 2009; Kim et al. 2009; Siomi and Siomi 2009). miRNAs are 21–24 nt RNAs that derive from distinctive hairpin precursors and repress target messenger RNAs (mRNAs) either by transcript destabilization, translational inhibition, or both, by base-pairing to the “seed” sequence, which is defined relative to the position of the miRNA 5′ end (Bartel 2009). Endo-siRNAs are 21–22 nt RNAs that mainly derive from TEs and pair to their transcripts to induce cleavage via small RNA-guided endonuclease activity or Slicer activity of associated Argonautes, thereby silencing TEs (Ghildiyal et al. 2008; Kawamura et al. 2008; Okamura et al. 2008; Tam et al. 2008; Watanabe et al. 2008). In addition, some endo-siRNAs are derived from complementary annealed transcripts or long fold-back transcripts (Babiarz et al. 2008; Kawamura et al. 2008; Okamura and Lai 2008; Miyoshi et al. 2010). Most miRNAs and endo-siRNAs are produced by Dicer and loaded onto the AGO subfamily of Argonautes.
In contrast, piRNAs are longer, at 24–35 nt, and their sequences are more diverse than any other known class of cellular RNAs (Siomi et al. 2011; Pillai and Chuma 2012). piRNAs are produced by Dicer-independent mechanisms from long single-stranded precursors mainly derived from active TEs and intergenic unannotated large blocks known as piRNA clusters, after which they are loaded onto the PIWI subfamily of Argonautes (Malone and Hannon 2009; Siomi et al. 2011). Some populations of these primary piRNAs are further amplified by a feed-forward amplification loop, the so-called ping-pong cycle, in which sense and antisense transcripts of TEs are reciprocally cleaved by the Slicer activity of PIWI proteins (Brennecke et al. 2007; Gunawardane et al. 2007; Aravin et al. 2008). The amplified secondary piRNAs have characteristic features with 1U/10A partners and a 10-nt 5′ overlap, which is referred to as the ping-pong signature. Although miRNAs and endo-siRNAs are expressed in both animals and plants, piRNA expression is largely restricted to animal gonads, where they primarily mediate TE silencing for genome integrity and germ cell development (Thomson and Lin 2009; Siomi et al. 2011). piRNAs act as guides for the cleavage of TE transcripts by PIWI protein Slicer activity in the cytoplasm, which often couples with the ping-pong cycle, or for Slicer-independent heterochromatin formation at TE loci in the nucleus (Sienski et al. 2012; Dönertas et al. 2013; Huang et al. 2013; Le Thomas et al. 2013; Muerdter et al. 2013; Ohtani et al. 2013).
In mice, piRNAs are most abundantly expressed in male germ cells, especially during spermatogenesis, and are loaded onto the three PIWI proteins: MIWI (PIWIL1), MILI (PIWIL2), and MIWI2 (PIWIL4) (Thomson and Lin 2009; Siomi et al. 2011; Pillai and Chuma 2012). Depletion of individual Piwi genes causes male-specific sterility because of severe defects in sperm formation, indicating their nonredundant functions of Piwi genes in the testes (Deng and Lin 2002; Kuramochi-Miyagawa et al. 2004; Carmell et al. 2007). Two distinct piRNA populations have been identified: the fetal prepachytene and adult pachytene piRNA pools. Prepachytene piRNAs are enriched in TE- and other repeat-derived sequences and associate with MIWI2 and MILI proteins (Aravin et al. 2008; De Fazio et al. 2011), suggesting that they might play a role in silencing TEs. Moreover, as ping-pong signatures are detected between MIWI2 and MILI, this indicates that they are involved in amplification of prepachytene piRNAs. In contrast, pachytene piRNAs have a higher proportion of intergenic, unannotated sequences, with a diminished contribution from TE-derived sequences (Aravin et al. 2006, 2007; Girard et al. 2006; Beyret et al. 2012). They are loaded onto MILI and MIWI from pachytene spermatocytes to elongating spermatids that are not further amplified. Although the loss of genes required to generate pachytene piRNAs blocks the production of mature sperm and results in TE deregulation (Deng and Lin 2002; Aravin and Hannon 2008; Reuter et al. 2011; Pillai and Chuma 2012; Vourekas et al. 2012), a biological role for pachytene piRNA clusters has yet to be identified. It also remains unknown if the absence of these RNAs causes the severe defects in spermatogenesis observed in mutant mice defective in the pachytene piRNA pathway.
Unlike rodents, primates possess four PIWI genes (PIWIL1–PIWIL4); thus, piRNA-mediated silencing may differ between rodents and primates (Sasaki et al. 2003; Ishizu et al. 2012). However, little is known about primate PIWI-piRNA pathways. Herein, in an effort to understand them, we analyze small RNAs in the adult testes of Callithrix jacchus, the common marmoset. Marmosets are small New World monkeys that have been extensively used in biomedical research and provide an attractive alternative to traditional nonhuman primate species. This is mainly because (1) they reach sexual maturity at 18 months of age, (2) they often bear twins after a relatively short gestation period, and (3) transgenic animals can now be produced (Sasaki et al. 2009; Orsi et al. 2011; Okano et al. 2012). To gain insights into the function of small RNAs in the marmoset testis, we used small RNA sequencing (small RNA-seq) to generate extensive small RNA data, in conjunction with RNA-seq data. We show that approximately 200 of nearly 700 miRNA genes identified are specific to marmosets. The most abundant class of small RNAs in the adult testis is, however, piRNAs. Using a specific monoclonal antibody that recognizes the PIWI protein MARWI (marmoset PIWIL1), a marmoset homolog of mouse MIWI, we generate MARWI-associated piRNA data and find that most piRNAs in adult testes are associated with MARWI, and they show characteristics of mouse pachytene piRNAs. Most MARWI-associated piRNAs derive from conserved intergenic clusters, some of which are located on the X chromosome. This suggests that they escape transcriptional silencing by meiotic sex chromosome inactivation (MSCI), which silences the X and Y chromosomes shortly after the zygotene-to-pacytene transition (Turner 2007; Heard and Turner 2011). Some clusters also contain pseudogenes of an antisense orientation, suggesting regulation of their cognate functional genes. Interestingly, piRNAs tend to map to TEs with copies in piRNA clusters much more than TEs with no copy in piRNA clusters, indicating that pachytene piRNA clusters act as traps that catch actively transposed TEs. Moreover, piRNAs corresponding to one subfamily are predominantly antisense to the TE, whereas piRNAs corresponding to another subfamily are predominantly sense. This is consistent with the sense/antisense bias of corresponding TE insertion to piRNA clusters, suggesting that the direction of TE insertion into the piRNA clusters is one of the major factors for setting the strong bias to the production of these piRNAs. Although a biological role for mouse pachytene piRNA clusters remains unknown, our findings suggest that pachytene piRNA clusters function in both TE silencing and regulation of protein-coding gene expression and may even have functions in meiosis.
RESULTS
Small RNA profiling in the marmoset adult testes
We sequenced total small RNAs of ∼18–35 nt from the testis of an adult marmoset (41 months old; note that marmosets are normally reproductively active from 2 to ∼8 years of age) (Abbott et al. 2003) using Illumina MiSeq, resulting in a total of 18,832,688 reads. We mapped the reads to the marmoset genome, which yielded 13,818,718 perfectly matching reads, mostly in the size range of 21–33 nt (Supplemental Fig. S1A; Supplemental Table S1). Among these, 28.1% mapped to multiple regions within the genome (Supplemental Fig. S1B), suggesting that they originate from repetitive elements like TEs. Small RNA profiling of mapped reads revealed two distinct peaks, one at the 21–23 nt position (6.7% of the total mapped reads), corresponding to miRNA and endo-siRNA in size, and another at the 24–33 nt position, which is the most abundant class of small RNAs (83.5% of the total mapped reads), corresponding to piRNAs in size (Supplemental Fig. S1A). To annotate the genome-mapped small RNA reads, we referred to the UCSC Genome Browser (Meyer et al. 2013) and Ensembl database (Flicek et al. 2013) for the annotation of marmoset genomic regions. We found that 19.28% of small RNAs map to repetitive elements, 9.11% to cellular noncoding (nc) RNAs, including miRNAs (3.60%) and transfer RNAs (tRNAs) (4.65%), 1.68% to coding genes, and 69.96% to unannotated regions (Supplemental Fig. S1C).
miRNAs in the marmoset testis
To precisely analyze the first peak (21–23 nt) observed in marmoset total small RNAs (Supplemental Fig. S1A), the nucleotide bias was checked, and the first nucleotide at the 5′ end was shown to be biased to “U” and “A” (U, 63.2%; A, 24.4%) (Supplemental Fig. S2A), which is a known characteristic of animal miRNAs (Seitz et al. 2011). The 21–23 nt length reads were mapped to the genome and checked for annotation (Supplemental Fig. S2B), revealing that 43.7% match miRNA coding regions registered in the Ensembl database (Flicek et al. 2013). However, more than half were annotated to other regions, such as TEs, repeats, and unannotated intergenic regions, indicating that they may also be capable of producing miRNAs. To clarify this, we identified miRNAs with miRDeep2 pipeline (Friedländer et al. 2012), using total small RNA-seq reads ranging in length from 21–23 nt as input. This search assigned 88.8% of 21–23 nt small RNA reads to 685 miRNAs, of which 332 were identical to miRNAs registered in the Ensembl database and 353 miRNAs were novel (Supplemental Table S2). Of the novel miRNAs, 174 had seed sequences in common with other primate miRNA species. The genomic regions from where these miRNAs originate varied, and some were expressed from protein coding regions (Supplemental Fig. S2C).
A primate-specific X-linked miRNA cluster was of particular interest (Supplemental Fig. S2D; Zhang et al. 2007; Li et al. 2010; Meunier et al. 2013). This cluster is comprised of tandem repeats of the Medium Reiteration frequency (MER) TE, which folds into a palindromic structure similar to the hairpin of the miRNA precursor and appears expressed almost exclusively in the testis (Yuan et al. 2010; Ahn et al. 2013; Meunier et al. 2013). During mammalian male germline development, the X and Y chromosomes are compartmentalized into a perinuclear subdomain called the sex body and become transcriptionally silenced from early pachytene onward, through to the end of spermatogenesis. This process is known as MSCI. X-linked miRNA cluster appears to be expressed during pachytene, suggesting escape from MSCI (Song et al. 2009; Meunier et al. 2013). The expression of two miRNAs from this cluster (miR-MER91C-5-3p and -21-3p) was confirmed by northern blotting (Supplemental Fig. S2E). Consistent with the recent studies that have revealed rapid evolution of this cluster in primates, including frequent tandem duplications and nucleotide substitutions (Zhang et al. 2007; Meunier et al. 2013; Sun et al. 2013), only two of the 23 seed sequences of the X-linked cluster-encoded miRNAs were conserved between marmoset and human, indicating the rapid divergence of this miRNA cluster (Supplemental Tables S3, S4). These findings also suggest that this cluster of miRNAs has a testis-specific function and that a form of genetic conflict may be driving its rapid evolution in primates.
Lack of endo-siRNA in the marmoset testis
To identify endo-siRNAs, we used 21–23-nt long total small RNA-seq reads that mapped to genomic regions other than the location of miRNA genes (476,689 reads). We searched for complementary small RNAs with 2-nt 3′ overhangs corresponding to products excised from long double-stranded precursors by Dicer in endo-siRNA biogenesis, and we also manually searched for genomic regions that can potentially give rise to phased endo-siRNAs by folding into long stem–loop secondary structures (Okamura and Lai 2008). However, we were unable to detect endo-siRNA-like small RNAs with significant reads (data not shown; also see Supplemental Material). This suggests that adult marmoset testes express very few endo-siRNAs, although recent studies have shown that murine adult spermatogenic cells express abundant endo-siRNAs (Song et al. 2011).
Abundant expression of MARWI, a homolog of mouse MIWI, in the marmoset adult testis
As many marmoset small RNAs are derived from unannotated intergenic regions (Supplemental Fig. S1C), which is reminiscent of mouse MIWI- or MILI-associated piRNAs that start to be expressed at the pachytene stage of meiosis (pachytene piRNAs) (Li et al. 2013), we analyzed the expression levels of AGO/PIWI genes using RNA-seq data to determine if marmoset homologs of mouse Miwi and/or Mili are expressed in adult testes. Computational searches of the marmoset genome (UCSC Genome Browser and Ensembl database) revealed eight Argonaute genes: four AGO subfamily genes (AGO1–AGO4) and four PIWI subfamily genes (PIWIL1–4). We named the marmoset PIWI subfamily genes MARWI, MARLI, MARWI2, and MARWI3, which correspond to human HIWI (PIWIL1), HILI (PIWIL2), HIWI2 (PIWIL4), and HIWI3 (PIWIL3) (Sasaki et al. 2003), respectively. We performed directional RNA-seq using total RNA obtained from two adult marmoset testes (41 and 51 months old). Replicates were highly correlated with each other (R2 = 0.50) (Supplemental Fig. S3A); therefore, we merged the reads, resulting in 168,398,076 RNA-seq reads (Supplemental Table S1). Reads were used to calculate the RPKM value for each gene of the Argonaute family. RPKM values indicated that expression levels of MARWI, a marmoset homolog of mouse Miwi, were vastly overrepresented among the gene family in this tissue (Supplemental Fig. S3B), suggesting that most observed piRNA-like reads bind to MARWI. Expression levels of the AGO subfamily in the testis are relatively low, which correlates with the fact that endo-siRNAs were barely detectable in the small RNA library. MARWI3, which does not have a mouse homolog, does not appear to be expressed in the adult testis (Supplemental Fig. S3B).
Comparison of piRNAs in total testis small RNAs with MARWI-associated piRNAs
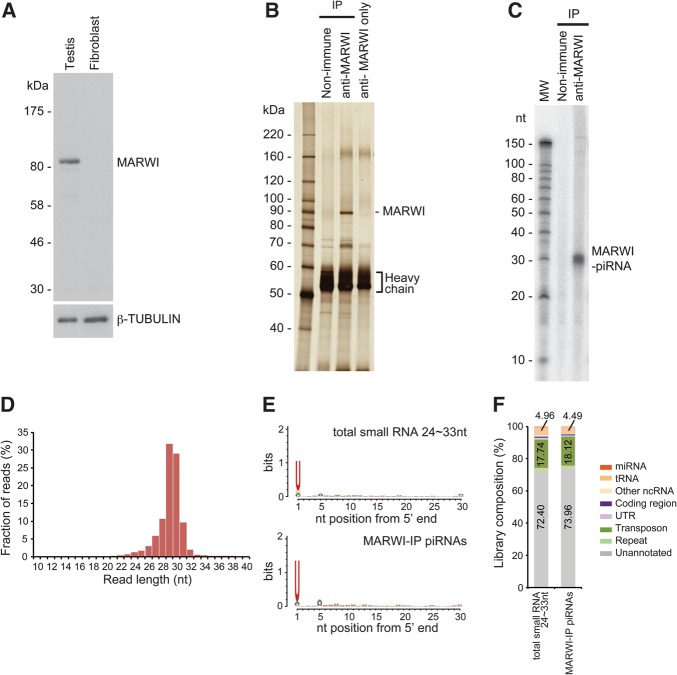
To test whether MARWI binds to the majority of piRNAs in adult testes, we immunopurified MARWI from adult testes using monoclonal antibodies we generated (Fig. 1A,B; Supplemental Fig. S4), in which it is expressed from spermatocytes to elongating spermatids (see below) and examined its associated RNAs (Fig. 1C). piRNAs associated with MARWI (MARWI piRNAs) in testes were ∼30 nt in length (Fig. 1C,D). piRNAs in fly and mouse are known to be modified at the 3′ terminal nucleotide with a 2′-O-methyl marker (Kirino and Mourelatos 2007; Ohara et al. 2007; Saito et al. 2007; Simon et al. 2011; Siomi et al. 2011). The resistance of MARWI piRNAs to periodate oxidation (NaIO4) and β-elimination reactions (Supplemental Fig. S4B) show that they also have the same 3′ modification.
FIGURE 1.
MARWI associates with piRNAs. (A) Western blotting was performed from marmoset testes and fibroblasts with anti-MARWI and anti-β-TUBULIN antibodies. MARWI protein is only detected in the testis. The anti-MARWI antibody (1A5) clearly detects a single band in the testis lysate. (B) Silver staining of protein components in the testis immunoprecipitant with anti-MARWI antibody from testis identifies a 90-kDa band for MARWI protein by LC-MS/MS (data not shown). MARWI protein is exclusively immunoprecipitated without protein contaminants. (C) Isolated RNAs from MARWI immunoprecipitates were 32P-labeled and separated by a denaturing polyacrylamide gel. MARWI protein associates with ∼29–30-nt-long piRNAs. (D) The size distribution of MARWI-associated piRNAs, which peak at 29–30 nt. (E) Nucleotide bias of 24–33 nt total small RNAs (piRNA-like) and MARWI-associated piRNAs. MARWI-piRNAs have a uridine bias at their 5′ end, consistent with the observation in 24–33 nt total small RNA reads. (F) Annotation of 24–33 nt total small RNAs and MARWI-associated piRNAs. Both library compositions resemble each other. This, together with the length distribution and annotation shown in E and F, indicates that MARWI-piRNA is the major component of total small RNA in the marmoset testis.
To further characterize MARWI piRNAs, we performed Illumina HiSeq sequencing using piRNA samples immunopurified with an anti-MARWI antibody from two individual adult marmoset testes (41 and 51 months old). Replicates were highly correlated with each other (R2 = 0.89) (Supplemental Fig. S5); therefore, we merged the reads, resulting in 140,662,017 marmoset MARWI-IP piRNA reads in total (Supplemental Table S1). Expression of highly detected piRNA reads was confirmed by northern blotting (Supplemental Fig. S4C). As in the case of total small RNA reads (using the testis from the 41-month-old marmoset), the length of sequenced reads ranged from 24–33 nt with peaks at 29 and 30 nt (Fig. 1D) and possessed a uracil nucleotide bias at the 5′ end (1U-bias) (Fig. 1E). Annotation of these MARWI piRNA reads was similar to that of the 24–33 nt total small RNA reads (Fig. 1F): Most (∼74.0%) mapped to unannotated intergenic regions and only ∼18.1% of the reads originated from TEs. This profile is similar to that of pachytene piRNAs associated with mouse MIWI, in which ∼70% derive from intergenic regions and ∼24% from TEs (Reuter et al. 2011).
tRNA-derived piRNAs and genic piRNAs
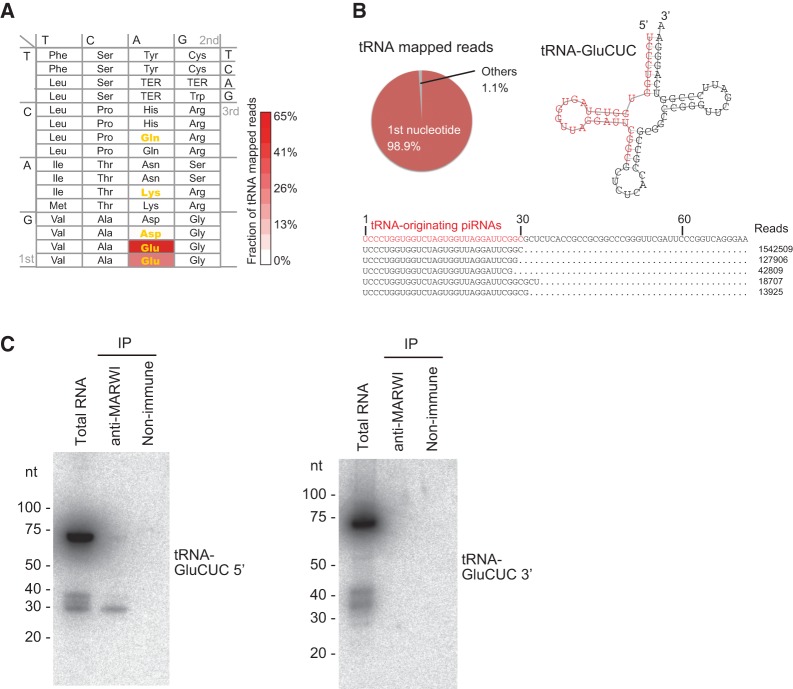
Some (4.5%) piRNAs corresponds to tRNAs (Fig. 1F), which has not been described in previous analyses of mammalian piRNAs. Surprisingly, MARWI piRNAs mapped to the 5′ end of only five tRNA species (tRNA-GluUUC, tRNA-GluCUC, tRNA-AspGUC, tRNA-GlyGCC, and tRNA-ValAAC) account for 99.6% of piRNA reads mapped to tRNAs (Fig. 2A; Supplemental Table S5). The vast majority of these tRNA-derived piRNAs mapped to tRNAs for Glu. Moreover, the tRNAs with the most abundant piRNA species mapped, tRNA-GluCUC, had ∼1.9 million reads mapped to its 5′ end, corresponding to 1.4% of the total number of MARWI-associated piRNAs reads (Fig. 2B). Northern blots of MARWI piRNAs revealed that the 5′, but not the 3′, fragments of the tRNA are loaded onto MARWI (Fig. 2C). We further confirmed that the 5′ fragments of tRNA-AspGUC are also detected by northern blotting analysis but not the 5′ fragments of tRNAs, which hardly match MARWI piRNA reads (Supplemental Fig. S6). These findings together suggest that these tRNAs are specifically processed into MARWI-bound piRNAs. Recent studies have revealed that some tRNA-derived small RNAs associate with AGO/PIWI proteins and function as classical siRNAs or miRNAs, whereas others also inhibit translation in a complementary base-pairing-independent manner or modify functions of associated AGO/PIWI proteins (Sobala and Hutvagner 2011).
FIGURE 2.
MARWI associates with tRNA-derived piRNAs. (A) Heat map showing abundance of MARWI-piRNA reads originating from tRNAs for each codon: (red) higher fraction of reads; (white) lower fraction of reads. Codons for tRNAs with first U at 5′ end are shown in yellow. (B) Pie chart showing the ratio of tRNA-derived piRNA reads mapped from the first nucleotide of mature tRNAs. The secondary structure of tRNA-GluCUC that recognizes the GAG codon is shown together with the region from where piRNAs are derived (in red). The number of reads and piRNA-read sequences are also indicated. (C) Northern blotting with mature tRNA-GluCUC probes targeting either the 5′ or 3′ region of tRNA. Signals were detected in MARWI immunoprecipitant for 5′ but not 3′ probes.
Some mRNAs in flies and vertebrates are known to be processed into piRNAs (Robine et al. 2009; Saito et al. 2009; Siomi et al. 2011). We identified 73 mRNAs that map more than 1000 MARWI piRNAs to their 3′ UTRs (Supplemental Fig. S7; Supplemental Table S6). Marmoset genic piRNAs were also found to originate from irregular positions in host 3′ UTRs forming pronounced peaks (or hot spots) and gaps of piRNA density as seen in flies and mice (Supplemental Fig. S7A). Similarly, some transcripts (seven of 73) exhibited piRNA enrichment in both the 3′ UTR and protein coding sequence (Supplemental Fig. S7B; Supplemental Table S6; Robine et al. 2009). We analyzed the expression levels of these piRNA-generating mRNAs using RNA-seq data and found that their median RPKM value (5.0) was only slightly higher than that of total mRNAs (1.7), which is consistent with previous findings showing that mouse genic piRNA-generating transcripts are only slightly skewed toward highly expressed genes, and house-keeping genes are underrepresented (Robine et al. 2009). These observations suggest a selective mechanism of genic piRNA production, although the mechanism that selects genic piRNA precursors from all cellular RNAs and directs them for piRNA processing remains to be elucidated.
piRNA clusters in the marmoset genome
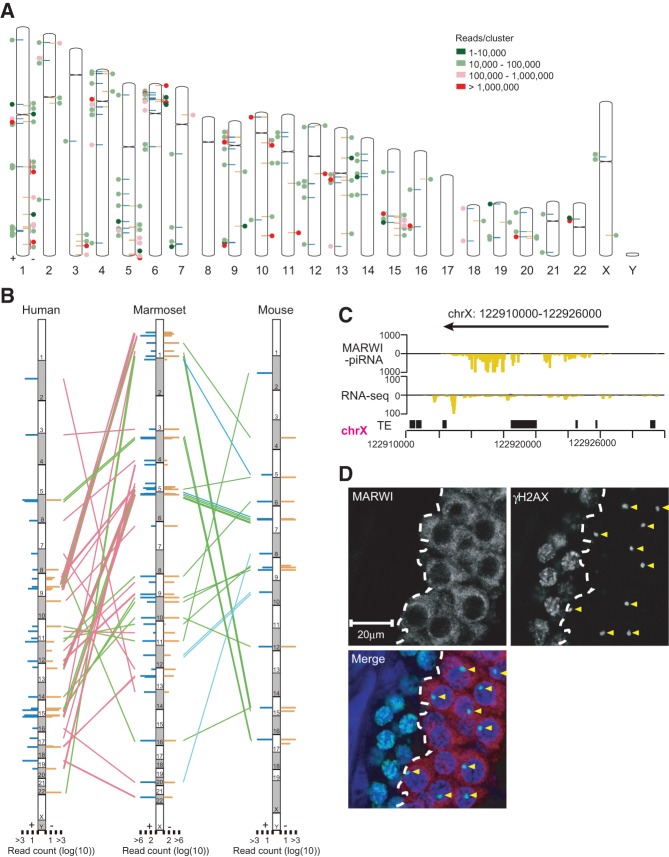
Genomic mapping of piRNAs in flies, zebra fish, mice, and humans has shown that multiple piRNAs map within discrete genomic intervals, forming piRNA clusters that can exceed 100 kb (Aravin et al. 2006; Girard et al. 2006; Brennecke et al. 2007; Houwing et al. 2007). This pronounced clustering also suggests that multiple piRNAs are processed from long primary transcripts. To determine if this occurs in marmosets, we mapped MARWI piRNA reads and found a total of 199 piRNA clusters with an average size of ∼17 kb and the largest at 132 kb (Fig. 3A; Supplemental Table S7; for details to define piRNA clusters, see Supplemental Material). This analysis also indicated that most MARWI piRNAs (83.4% of piRNAs mapped to the genome) are produced in a clustered fashion, although piRNA clusters constitute just 0.1% of the marmoset genome. Both unidirectional clusters, in which piRNAs map to only one strand, and bidirectional clusters, in which the polarity of piRNA production switches between plus and minus strands, were found in the marmoset genome as seen in other species. Analysis of directional RNA-seq data showed that the distribution of RNA-seq reads occurs in regions where piRNA clusters are located, indicating that clustered MARWI piRNAs are likely to be derived from the same single-stranded precursor transcripts.
FIGURE 3.
MARWI-associated piRNA cluster. (A) Location of MARWI-piRNA clusters on marmoset chromosomes. Circles indicate the position of piRNA clusters within each chromosome, and the color of the circles indicates the number of piRNA reads per cluster. Left and right bars on each chromosome show mapped positions of clusters in positive and negative strands, respectively. Of genome-mapped reads, 83.4% were included within piRNA clusters and were distributed randomly within most chromosomes. (B) Genome-wide representation of the synteny of clusters between humans, marmosets, and mice: (green) three species; (red) human and marmoset; (blue) marmoset and mouse. Synteny of piRNA clusters was conserved among species, and the large ratio is conserved only between the human and marmoset. (C) Example of MARWI-piRNA cluster on chromosome X. Distributions of uniquely mapped MARWI-piRNAs and RNA-seq reads on the X chromosome located piRNA cluster are shown. Blue bar indicates sequence mapped to the sense orientation, and yellow bar indicates sequence in antisense orientation. (D) MARWI expresses from pachytene spermatocytes. Coimmunostaining with MARWI and γH2AX are shown: (red) MARWI; (green) γH2AX; (blue) DAPI. γH2AX express mitotic spermatocytes with different forms; some signals are spread around the nucleus, and the others are restricted to the sex bodies (yellow arrowhead). MARWI protein signals are not detected in γH2AX diffused cells but are found in sex body–formed cells, where transcription is silenced on the sex chromosomes. Dashed line indicates MARWI expression boundary.
Although the primary sequences of piRNA clusters are not conserved, their genomic location is highly conserved from rodents to humans (Aravin et al. 2006, 2008; Girard et al. 2006; Beyret et al. 2012). Indeed, only a small fraction of MARWI piRNAs could be mapped to the genomes of human and mouse (7.3% were mapped to the human genome with perfect matches and 4.5% to the mouse genome). To examine if the genomic locations of marmoset piRNA clusters are conserved, we searched for MARWI piRNA syntenic loci in humans and mice, using a previously published data set (Girard et al. 2006). Chromosomal positions of most previously detected piRNA clusters from humans and mice were conserved in the marmoset (Fig. 3B). We also detected a large number of clusters that were likely to be conserved only between marmosets and humans, indicating the existence of primate-specific piRNA clusters. However, we also observed that several clusters are conserved between humans and mice but apparently lost in marmosets, and several others were conserved between marmosets and mice but apparently lost in humans.
Interestingly, we identified three piRNA clusters on X chromosome (Fig. 3A,C). From pachytene spermatocyte onward, X and Y chromosome-linked genes are transcriptionally silenced owing to the MSCI (Turner 2007; Heard and Turner 2011). MIWI, the MARWI ortholog in mice, expresses from pachytene spermatocyte to elongating spermatids during spermatogenensis (Deng and Lin 2002; Di Giacomo et al. 2013). To determine MARWI expression in detail during spermatogenesis, coimmunostaining with meiotic marker γH2AX was performed. During leptotene to zygotene spermatocyte, punctate staining of γH2AX is seen throughout the nucleus. In contrast, at pachytene spermatocyte, γH2AX becomes restricted to the sex body (Mahadevaiah et al. 2001; Fernandez-Capetillo et al. 2003; Di Giacomo et al. 2013). MARWI protein signal is not detected in the early spermatocyte but is observed in the cytoplasm from the pachytene onward, which is similar to ortholog MIWI (Fig. 3D; Supplemental Fig. S8; Di Giacomo et al. 2013). Thus, MARWI and MARWI-associated piRNAs express from pachytene onward, suggesting that the X-linked clusters are transcribed during meiosis in spite of MSCI.
New classes of piRNA clusters
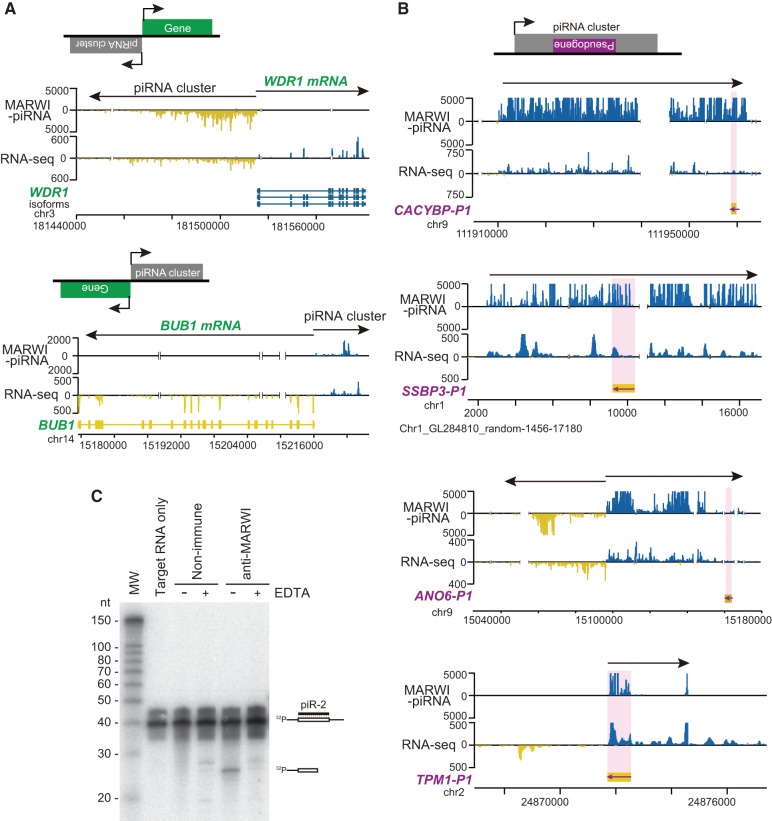
Neither the function of piRNA clusters nor the functional implication of such extensive synteny in mammals are currently understood, so we further characterized the marmoset piRNA clusters identified in the present study. We found two new classes of piRNA clusters: clusters consisting of two segments in which the polarity of piRNA and mRNA production switches between the plus and minus strands (Fig. 4A) and clusters with pseudogenes (Fig. 4B). piRNA mapping, together with directional RNA-seq data, revealed that piRNAs mapped to only one strand but not to the mRNAs (Fig. 4A). In the former class, one of these strands encodes the gene named WD-repeat protein 1 gene (WDR1), also called actin-interacting protein1 (AIP1), which encodes a protein promoting cofilin-mediated actin filament disassembly (Fujibuchi et al. 2005), thereby playing a likely role in round spermatid formation. Budding uninhibited by benzimidazole 1 (BUB1), which encodes a serine/threonine protein kinase that activates spindle check point assembly and is highly expressed in testes, also forms a bidirectional cluster-like transcript (Fig. 4A). Importantly, this gene is essential for spermatogenesis, because the testes of deficient mice showed impaired normal chromosome segregation (Perera et al. 2007). A large number of piRNA clusters in mice appear to be transcribed bidirectionally by the MYB family of transcriptional factors (Li et al. 2013). This suggests that a single transcriptional factor may transcribe both piRNA clusters and protein-coding genes bidirectionally from a single promoter in a coordinated manner.
FIGURE 4.
piRNA clusters contain pseudogenes. (A) Distribution of MARWI-piRNAs and RNA-seq reads where unidirectional piRNA cluster and protein-coding mRNAs are bidirectionally transcribed. WDR1 and BUB1 gene loci are shown. (B) Examples of piRNA clusters including pseudogenes in the opposite orientation. CACYBP-P1 (ENSCJAG00000001398) pseudogene, SSBP3-P1 (ENSCJAG00000010873) pseudogene, ANO6-P1 pseudogene, and TMP1-P1 (ENSCJAG00000015442) pseudogene loci inserted in unidirectional and bidirectional piRNA clusters are shown. (C) MARWI-piR-2 complementary RNA incubation with MARWI immunoprecipitates. Cleavage product was found in the MARWI immunoprecipitates, where the cleavage was inhibited by EDTA. piR-2 is a MARWI piRNA, which occupies the second largest number of reads according to MARWI-piRNA sequencing data.
piRNA clusters with pseudogenes
The second class of clusters contains processed pseudogenes. We found five such piRNA clusters, of which four contain pseudogenes with an antisense orientation relative to the polarity of host clusters (Fig. 4B). piRNAs derived from these pseudogenes are therefore antisense-oriented relative to their parental genes, suggesting that they regulate expression of their parental functional genes. The parental genes of these four pseudogenes in this class of cluster include TROPOMYOSIN-1 (TMP1, also called α-TROPOMYOSIN) encoding a coiled-coil protein that regulates actin filament function (Lange et al. 2007), CACYBP (also called SIP) encoding a S100A6- and Siah-1 binding protein with unknown function (Wasik and Filipek 2013), ANO6 (also called Tmem16F) encoding a protein that forms a Ca2+-activated cation channel required for phospholipid scrambling and linked to Scott syndrome, a rare bleeding disorder caused by a defect in phospholipid scrambling activity (Kunzelmann et al. 2014), and SSBP3 (also called SSDP1) encoding a nuclear protein that binds single-stranded pyrimidine-rich sequences and is involved in transcription (Bayarasaihan et al. 1998). An in vitro cleavage assay confirmed that MARWI-piRNA complexes were capable of specifically cleaving target RNAs containing sequences that are perfectly complementary to piRNAs (Fig. 4C). We also confirmed that MARWI was a cytoplasmic protein (Fig. 3D; Supplemental Fig. S8). These findings together indicate that pseudogene-derived piRNAs play important roles in the regulation of protein-coding gene expression in the cytoplasm, and also suggest a model in which pachytene piRNA clusters regulate protein-coding genes through the processed pseudogenes trapped within.
Strong strand bias of piRNAs derived from each TE subfamily
To further understand TE-derived piRNAs, we first examined the read frequency and strand bias of piRNAs that mapped to each of the TE subfamilies. We aligned MARWI piRNA sequences to a comprehensive set of marmoset TE sequences obtained from RepeatMasker (UCSC Genome Browser track) (Smit et al. 2010), which includes each of the TE sequences originating from different genomic positions. Reads aligned to multiple sequences were normalized by the number of aligned sequences. Vertebrate TEs can be assigned to one of four broad classes according to their mechanism of transposition: long interspersed elements (LINEs), short interspersed elements (SINEs), long terminal repeat (LTR) retrotransposons, and DNA transposons (Mandal and Kazazian 2008); they are further classified into distinct subfamilies according to the degree of deviation from family consensus sequences (Smit et al. 2010; Sookdeo et al. 2013). The MARWI piRNA read counts for each TE sequence were gathered by subfamily classes to integrate TEs originating from multiple regions within the genome as one.
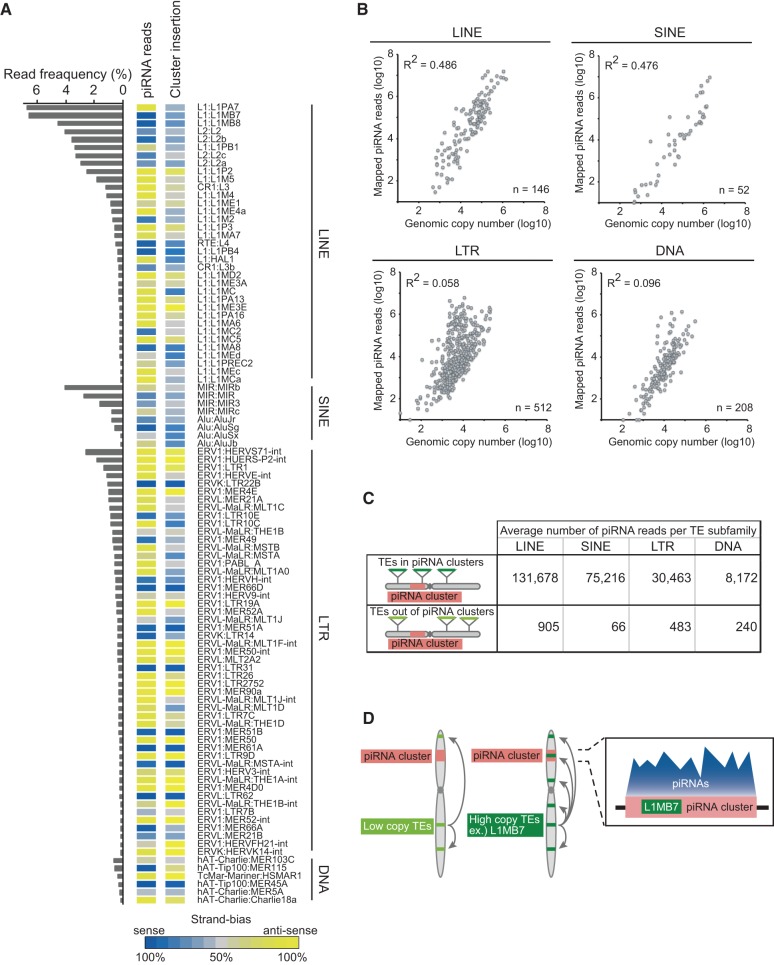
We made a list of the top 100 TE subfamilies with highly mapped piRNA reads and analyzed what fraction of the total TE-mapped piRNA reads mapped to each TE subfamily (Fig. 5A). We found that most piRNA reads were accumulated at specific TE subfamilies. Of 918 analyzed TE subfamilies, 88.2% of TE-derived piRNA reads were mapped to the top 100 TE subfamilies, and the top 20 TE subfamilies occupied more than half (55.8%) of TE-derived piRNA reads. TE-derived piRNAs (∼24%) associated with MIWI in mice have a strand bias skewed to the sense orientation: Two-thirds are sense-oriented (Reuter et al. 2011). To determine whether this is also true for MARWI piRNAs, we examined the strand bias of the mapped piRNA reads (Fig. 5A), but found that 48.0% of TE-mapped piRNAs were sense-oriented, indicating no clear strand bias. However, 80.0% of TE subfamily members were biased to either sense or antisense at a ratio higher than 75%, suggesting that members within the same TE subfamily tend to be transcribed unidirectionally independent of their genomic location (see also below).
FIGURE 5.
Transposable element insertion to piRNA clusters gives rise to the source of piRNA production. (A) Heat maps show strand bias of transposon-derived MARWI-piRNAs for each TE subfamily (left) and the direction of each TE subfamily insertion into piRNA clusters (right). Transposons are grouped into LINEs, SINEs, LTRs, and DNA transposons. Color intensities indicate the degree of strand bias: (blue) sense; (yellow) antisense; (gray) unbiased. The frequencies of piRNAs mapped to each TE subfamily over the total TE-mapped piRNAs are shown in the bar graph. (B) Dot plot shows the relationship between genomic copy number and mapped piRNA reads for LINE, SINE, LTR, and DNA transposons. (C) Table showing average number of piRNA reads obtained from each group of transposon subfamilies. The values are averaged over transposon subfamilies with or without insertions into piRNA clusters. (D) Schematic of marmoset piRNA clusters working as “TE-traps.”
A fraction of piRNAs originating from TEs is generated by the ping-pong cycle, which produces secondary piRNAs with ping-pong signatures (Brennecke et al. 2007; Gunawardane et al. 2007). We found that both MARWI piRNAs and 24–33-nt-long small RNAs were biased for a 5′-terminal uracil (1U) with no 10th adenine (10A) bias, indicating that they harbor characteristic of primary piRNAs and that secondary piRNAs are absent or produced at only low levels (Fig. 1E). To corroborate this, we searched for piRNAs with initial 10-nt complementarity in both MARWI piRNAs and total small RNAs (Supplemental Fig. S9A). However, only 2.5% of total small RNAs and 0.2% of MARWI piRNAs had 10-nt complementary partners, suggesting that piRNA biogenesis in the adult marmoset testis is mostly, if not completely, independent of the ping-pong cycle, as in the case of adult mouse piRNAs (Beyret et al. 2012).
Trans-targeting by TE-derived piRNAs
Because these piRNAs are not amplified by the ping-pong cycle and the production of piRNAs was biased to particular TE subfamilies (Fig. 5A), we hypothesized that these piRNAs may not only be capable of regulating TEs within the same subfamily but also other subfamily members by mediating the cleavage of transcripts from corresponding opposite strands. To test this hypothesis, we searched for TEs potentially targeted by MARWI-piRNA complexes (Supplemental Fig. S9B). Recent studies have revealed that although mismatches at several 3′-terminal nucleotides of piRNAs are tolerated, base-pairing of nucleotides 2–21 is required for efficient target cleavage by MIWI proteins (Reuter et al. 2011). We therefore searched for TE partners with perfect matches at the first 2–21 nt (see Supplemental Material) and found that 9.4% of TE-derived MARWI piRNAs have potential target TEs other than those from the same subfamily. This also implies that MARWI-piRNA complexes, with Slicer activity (Fig. 4C), are capable of regulating TEs in trans by cleaving them. These findings suggest a model in which the mutual cleavage of TE transcripts originating from each subfamily member sets a threshold of expression for the entire TE family.
piRNA clusters as the major source of TE-derived piRNAs
We found that the amount of piRNA reads produced per TE was higher for some members of LINEs and SINEs compared with LTR retrotransposons and DNA transposons (Fig. 5A; Supplemental Fig. S10A; Supplemental Table S8). As this might be caused by the higher activity of recently moved and therefore relatively younger LINEs and SINEs, we checked this for each LINE and SINE with higher piRNA production (Fig. 5A). However, both the evolutionarily younger members (that is, still actively transposing; e.g., L1PA, L1PB, AluS) and older and ancient members (that is, transposition-incompetent; e.g., L2, L1ME, MIR) (Giordano et al. 2007) were equally identified as the source of higher piRNA production. Therefore, we concluded that the amount of piRNAs produced per TE does not rely on recent TE activity.
We next checked for the correlation between genomic copy numbers of TEs and mapped piRNA reads per TEs (Fig. 5B) and observed a positive correlation for LINEs and SINEs. This suggests that the more copies in the genome, the more piRNAs they generate. On the other hand, the correlation was relatively low for LTR retrotransposons and DNA transposons (see below). In addition, we examined the annotation of MARWI piRNA cluster regions and found that up to 33.7% of the regions were occupied by TEs, suggesting that piRNA clusters serve as a major source for TE-derived piRNAs. This is similar to the degree of TE occupancy in mouse pachytene piRNA clusters, although mouse fetal prepachytene clusters are full of TEs (Lakshmi and Agrawal 2008). Thus, we analyzed the correlation between genomic copy numbers of TEs and copy numbers of the same class of TEs in piRNA clusters (Supplemental Fig. S10B) and found that they were highly correlated, especially for LINEs and SINEs. This result together with the observation that piRNA production from TEs is dependent on their genomic copy number, suggest that the number of elements transposed to piRNA clusters might determine the piRNA production from each TE.
In the case of LTR retrotransposons, some of the members with lower copy numbers are capable of producing higher amounts of piRNA (Fig. 5B). We surmised that this might be caused by the genomic position at which LTR retrotransposons have integrated. To test this, we normalized the amounts of piRNAs by the total length of each TE and compared them to the ratio of the TE insertion to piRNA clusters (Supplemental Fig. S10C). Interestingly, we found that LTR retrotransposons can efficiently produce piRNAs compared to the other TE family members. Additionally, although the ratio of LINEs and SINEs integrated into piRNA clusters was similar to the ratio of piRNA clusters within the genome (∼0.1%), LTR retrotransposons were enriched in piRNA clusters. This suggests that LTR retrotransposons might have preferentially integrated into piRNA clusters.
To ask if the TE copy numbers transposed into piRNA clusters are correlated with levels of piRNA production, we analyzed the amounts of piRNAs derived from groups of TE subfamilies with or without copies in piRNA clusters (Fig. 5C; Supplemental Fig. S10D). We divided TEs into two groups: one with at least one copy of elements integrated in a piRNA cluster and the other with no such copies. When the production of piRNAs was compared, the former group was shown to produce many more piRNAs. We therefore hypothesize that actively transposed TEs are more likely to jump into piRNA clusters, and therefore start to produce more piRNAs. Moreover, when we checked the direction of TE insertion into each of the piRNA clusters, we found that the strand bias was similar to that of highly biased piRNAs (R2 = 0.48) (Fig. 5A). Therefore, the direction of TE insertion may be one of the main factors that set the strong sense/antisense bias in piRNA production (Supplemental Fig. S10E). These findings also suggest a model in which TEs that happen to jump into piRNA clusters in the opposite direction regulate other subfamily members expressed from different genomic positions (Fig. 5D; Supplemental Fig. S10F). Although the function of mouse pachytene piRNA clusters is unknown, our findings also suggest that they act as TE-traps, as seen in fly piRNA clusters (Malone and Hannon 2009; Karginov and Hannon 2010).
DISCUSSION
In the present study, we describe the small RNA profile from the adult testes of the common marmoset, a small New World primate, which has been attracting much attention in the research field of biomedical sciences as a model primate species (Sasaki et al. 2009; Orsi et al. 2011; Okano et al. 2012). Our small RNA profiling confirmed the importance of using the common marmoset to analyze primates. Our main findings were (1) marmoset-specific miRNAs, including a primate-specific X-linked cluster; (2) piRNAs may mediate in trans regulation among TE subfamilies; (3) TE-derived piRNAs biased to either sense or antisense strands of each element; (4) abundantly expressed tRNA-derived piRNA species; and (5) piRNA clusters on the X chromosome, suggesting that these clusters are able to evade MSCI, which can be informative for studies of the primate germ line and spermatogenesis. We also revealed some interesting findings that may be common to species other than primates. These include (1) MARWI piRNAs as the most abundant class of small RNAs in the marmoset adult testis, which show characteristics of mouse pachytene piRNAs with unknown functions; (2) bidirectionally transcribed coding genes and piRNA clusters; (3) some pseudogenes integrated into piRNA clusters that may regulate cognate functional genes; and (4) piRNA clusters, mostly localized to intergenic regions, serving as “traps” for TEs (see below).
piRNAs in the adult testis of the common marmoset
Marmosets encode four AGO and four PIWI subfamily members of Argonautes, of which the expression of MARWI is overrepresented in adult testes (Supplemental Fig. S3B). Consistent with this, most piRNAs associate with MARWI (Fig. 1D–F; Supplemental Fig. S1A). Moreover, the predominant class of small RNAs expressed in the marmoset adult testis is likely to be MARWI piRNAs. These have characteristics of mouse pachytene piRNAs that associate with MIWI (Aravin et al. 2006; Girard et al. 2006; Reuter et al. 2011; Beyret et al. 2012), including a higher proportion of intergenic, unannotated sequences (74.0%) with a diminished contribution of TE sequences (18.1%) and very few piRNAs with ping-pong signatures. In addition, mouse piRNA clusters constitute just 0.3% of the genome yet account for 95% of piRNAs in the adult mouse testis (Li et al. 2013). Similarly, marmoset piRNA clusters constitute only 0.1% of the genome yet account for 83.4% of MARWI piRNAs.
We found that within the same TE family (for example, L1), some subfamily members generate larger amounts of piRNAs than others (Fig. 5A). These subfamily-derived piRNAs, regardless of their strand biases, establish an expression threshold for TE families by cleaving homologous TE transcripts. Recently, MIWI was shown to be necessary to maintain the Slicer-dependent silencing of the L1 retrotransposon in the mouse testis after birth (Reuter et al. 2011). This indicates that MIWI Slicer activity directly cleaves TE mRNAs, thereby silencing TE without piRNA amplification. Two-thirds of TE-derived piRNAs in pachytene piRNAs that associate with MIWI derive from the sense strand of TEs, suggesting that the antisense piRNA dosage required for silencing of the L1 is relatively low, probably because of the catalytic nature of Slicer. These findings also suggest that the predominant generation of antisense piRNAs from some TEs observed in the adult marmoset testis contribute to silencing of the other family members in that organ.
piRNA clusters in adult marmoset testes stipulate characteristics of pachytene piRNAs
We found that the TE group with copies in piRNA clusters produces larger amounts of piRNAs (Fig. 5C; Supplemental Fig. S10D). This suggests a model in which TEs that happen to jump into piRNA clusters regulate other elements in the same TEs expressed from different genomic positions (Fig. 5D). As originally proposed by Bergman and colleagues and later elaborated by Hannon and colleagues following Drosophila analysis, piRNA clusters act as “TE traps” that rely on the innate mobility of TEs to passively acquire new content by chance transposition (Bergman et al. 2006; Malone and Hannon 2009; Karginov and Hannon 2010; Sabin et al. 2013). TEs that have jumped into the clusters can then become fixed by evolutionary selection and become part of a silencing program that instructs a small RNA-based immunity to selectively silence homologous elements in germ cells, thereby providing an adaptive immunity to the host genome (Khurana et al. 2011). Although the function of mouse pachytene piRNA clusters is unknown, our findings suggest that pachytene piRNA clusters can also capture and efficiently regulate TEs that express in meiotic or post-meiotic cells during spermatogenesis. Indeed, 33.7% of MARWI piRNA clusters are composed of TEs. Moreover, from the observation that pseudogenes are found in piRNA clusters (Fig. 4B), we hypothesize that piRNA clusters trap not only TEs but also other movable genetic elements. Thus, piRNA clusters may enable the genome to recognize self and non-self and regulate expression of deleterious transposable genetic elements by determining the quality and quantity of piRNAs.
On the other hand, we found that LTR retrotransposons were enriched in piRNA clusters regardless of their abundance in the genome (Fig. 5B; Supplemental Fig. S10C). The reason for this remains unclear, but one hypothesis is that there is a specific preference of the region into which LTR retrotransposons tend to integrate. A previous study indicated that LTR retrotransposons are enriched in open chromatin sites (Jacques et al. 2013), which may include highly transcribed piRNA cluster transcripts. An alternative explanation would be that LTR retrotransposons might have played the role in generation of piRNA clusters. In fact, it has been reported that LTR retrotransposons contribute as a source of long ncRNAs (Kapusta et al. 2013). Further analysis would be necessary to reveal this point.
piRNA clusters as possible regulators for protein-coding genes
Marmoset piRNA clusters harbor not only TEs but also pseudogenes, which tend to be antisense-oriented relative to the polarity of host clusters (Fig. 4B). Thus, these clusters generate antisense piRNAs relative to the pseudogenes and their parental genes. Historically, pseudogenes have been regarded as nonfunctional evolutionary relics (Balakirev and Ayala 2003). Recent evidence indicates that endo-siRNAs arising from pseudogenes in mouse oocytes can adjust the level of the founding source mRNA by mediating mRNA cleavage (Tam et al. 2008; Watanabe et al. 2008). By analogy, piRNAs originating from processed pseudogenes may also regulate the expression of their parental genes post-transcriptionally by cleaving mRNAs in the marmoset adult testis. Processed pseudogenes are generated by the reverse transcription of mRNA transcripts followed by integration of the cDNAs into the genome. It is tempting to speculate that just like TEs, random trapping of the cDNAs into piRNA clusters can then become fixed by evolutionary selection to selectively regulate homologous elements in germ cells. In addition, none of the pseudogenes we identified in marmoset piRNA clusters are found in known mouse piRNA clusters, suggesting that pseudogene-derived piRNAs may have species-specific functions in the testis.
Taken together, the piRNAs and the set of piRNA clusters in marmoset testes defined by our work provide an important insight into the evolutionary origins and functions of pachytene piRNAs. Future work should analyze small RNAs associated with each of the eight marmoset Argonautes and corresponding human proteins in gonads to determine whether primate-specific mechanisms are deployed to regulate gene expression during germ cell development.
MATERIALS AND METHODS
Animals and tissue collection
This study was approved by the animal ethics committee of the Central Institute for Experimental Animals (CIEA) and Keio University, and it was performed in accordance with CIEA and Keio University guidelines. Mice were obtained from CREA Japan, Inc., and Japan SLC, Inc. Information on individual marmosets is described in Supplemental Table S9.
Generation of anti-MARWI/PIWIL1 monoclonal antibodies
Anti-MARWI monoclonal antibodies were produced essentially as described previously (Ishizuka et al. 2002; Nishida et al. 2007). Briefly, monoclonal mouse antibodies against MARWI were generated by injecting mice with GST-MARWI-N (1–204 amino acids) and then fusing lymph node and spleen cells with the myeloma cell line P3U1 to produce hybridomas. Note that the anti-MARWI antibodies produced also recognize mouse MIWI/PIWIL1 protein (Supplemental Fig. S11).
Small RNA-seq and data processing
To prepare total small RNA libraries, total RNAs from the marmoset testis were extracted using the miRNeasy mini kit (QIAGEN, 217004), and gel-purified small RNAs were cloned using the NEBNext Small RNA Library Sample Prep Set (NEB, E7330) with slight modifications. Detailed protocols are described in Supplemental Material. Total small RNA reads were sequenced using MiSeq (illumina), obtaining 18,832,666 reads in total.
To identify MARWI associated piRNAs, coimmunoprecipitated RNA was extracted from MARWI immunoprecipitates with phenol and chloroform, and gel-purified MARWI-associated piRNAs were cloned. MARWI-associated piRNA libraries were analyzed by HiSeq2000 (Illumina) using two replicates from different samples. Replicates were highly correlated (R2 = 0.89), so the reads were merged giving a total of 140,662,017.
Adapter sequences were removed from obtained reads and mapped to the marmoset reference genome (calJac3, WUSTL 3.2) using Bowtie (Langmead et al. 2009), allowing zero mismatches. Annotation of the reads was determined according to UCSC (Meyer et al. 2013) and Ensembl databases (Flicek et al. 2013). Details of further sequence analysis can be found in Supplemental Material.
DATA DEPOSITION
Total small RNA sequences, MARWI-associated small RNA sequences, and total RNA sequences data have been deposited in the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) under accession no. GSE52927.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank members of the Siomi laboratory for helpful discussions and support, especially Kazumichi M. Nishida for technical assistance with monoclonal antibody generation and Soichiro Yamanaka for critical reading of the manuscript. We thank Yukiteru Ono for bioinformatic support and Ikuko Sagawa for mass spectrometry analysis. We are very grateful to Cedric Feschotte for critical reading of the manuscript, to Yasunori Aizawa and Hervé Seitz for useful discussions on TE-derived piRNA sequences and MSCI, and to Satomi Kuramochi-Miyagawa for fruitful discussions on piRNA clusters and reagents. This work was supported by Grants-in-Aid for Scientific Research to H.S. and M.C.S.; a Grant-in-Aid for the Global COE program to Keio University to H.S., T.H., Z.Y.-C.L., E.S., and H.O. from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) of Japan; the Research Fellowships for Young Scientists to T.H.; Grants-in-Aid for Young Scientists to Y.W.I.; the Funding Program for Next-Generation World-Leading Researchers to K.S.; and the “Funding Program for World-leading Innovative R&D on Science and Technology” to H.O. from the Japan Society for the Promotion of Science (JSPS).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.045310.114.
REFERENCES
- Abbott DH, Barnett DK, Colman RJ, Yamamoto ME, Schultz-Darken NJ 2003. Aspects of common marmoset basic biology and life history important for biomedical research. Comp Med 53: 339–350 [PubMed] [Google Scholar]
- Ahn K, Gim JA, Ha HS, Han K, Kim HS 2013. The novel MER transposon-derived miRNAs in human genome. Gene 512: 422–428 [DOI] [PubMed] [Google Scholar]
- Aravin AA, Hannon GJ 2008. Small RNA silencing pathways in germ and stem cells. Cold Spring Harb Symp Quant Biol 73: 283–290 [DOI] [PubMed] [Google Scholar]
- Aravin AA, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, et al. 2006. A novel class of small RNAs bind to MILI protein in mouse testes. Nature 442: 203–207 [DOI] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ 2007. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science 316: 744–747 [DOI] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Bourc'his D, Schaefer C, Pezic D, Toth KF, Bestor T, Hannon GJ 2008. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell 31: 785–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R 2008. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev 22: 2773–2785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balakirev ES, Ayala FJ 2003. Pseudogenes: Are they “junk” or functional DNA? Annu Rev Genet 37: 123–151 [DOI] [PubMed] [Google Scholar]
- Bartel DP 2009. MicroRNAs: target recognition and regulatory functions. Cell 136: 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayarasaihan D, Soto RJ, Lukens LN 1998. Cloning and characterization of a novel sequence-specific single-stranded-DNA-binding protein. Biochem J 331: 447–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman CM, Quesneville H, Anxolabéhère D, Ashburner M 2006. Recurrent insertion and duplication generate networks of transposable element sequences in the Drosophila melanogaster genome. Genome Biol 7: R112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyret E, Liu N, Lin H 2012. piRNA biogenesis during adult spermatogenesis in mice is independent of the ping-pong mechanism. Cell Res 22: 1429–1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ 2007. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128: 1089–1103 [DOI] [PubMed] [Google Scholar]
- Buchon N, Vaury C 2006. RNAi: a defensive RNA-silencing against viruses and transposable elements. Heredity 96: 195–202 [DOI] [PubMed] [Google Scholar]
- Carmell MA, Girard A, van de Kant HJG, Bourc'his D, Bestor TH, de Rooij DG, Hannon GJ 2007. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev Cell 12: 503–514 [DOI] [PubMed] [Google Scholar]
- De Fazio S, Bartonicek N, Di Giacomo M, Abreu-Goodger C, Sankar A, Funaya C, Antony C, Moreira PN, Enright AJ, O'Carroll D 2011. The endonuclease activity of Mili fuels piRNA amplification that silences LINE1 elements. Nature 480: 259–263 [DOI] [PubMed] [Google Scholar]
- Deng W, Lin H 2002. miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Dev Cell 2: 819–830 [DOI] [PubMed] [Google Scholar]
- Di Giacomo M, Comazzetto S, Saini H, De Fazio S, Carrieri C, Morgan M, Vasiliauskaite L, Benes V, Enright AJ, O'Carroll D 2013. Multiple epigenetic mechanisms and the piRNA pathway enforce LINE1 silencing during adult spermatogenesis. Mol Cell 50: 601–608 [DOI] [PubMed] [Google Scholar]
- Dönertas D, Sienski G, Brennecke J 2013. Drosophila Gtsf1 is an essential component of the Piwi-mediated transcriptional silencing complex. Genes Dev 27: 1693–1705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Capetillo O, Mahadevaiah SK, Celeste A, Romanienko PJ, Camerini-Otero RD, Bonner WM, Manova K, Burgoyne P, Nussenzweig A 2003. H2AX is required for chromatin remodeling and inactivation of sex chromosomes in male mouse meiosis. Dev Cell 4: 497–508 [DOI] [PubMed] [Google Scholar]
- Flicek P, Ahmed I, Amode MR, Barrell D, Beal K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fairley S, et al. 2013. Ensembl 2013. Nucleic Acids Res 41: D48–D55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedländer MR, Mackowiak SD, Li N, Chen W, Rajewsky N 2012. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res 40: 37–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujibuchi T, Abe Y, Takeuchi T, Imai Y, Kamei Y, Murase R, Ueda N, Shigemoto K, Yamamoto H, Kito K 2005. AIP1/WDR1 supports mitotic cell rounding. Biochem Biophys Res Commun 327: 268–275 [DOI] [PubMed] [Google Scholar]
- Ghildiyal M, Zamore PD 2009. Small silencing RNAs: an expanding universe. Nat Rev Genet 10: 94–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M, Seitz H, Horwich MD, Li C, Du T, Lee S, Xu J, Kittler ELW, Zapp ML, Weng Z, et al. 2008. Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science 320: 1077–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giordano J, Ge Y, Gelfand Y, Abrusán G, Benson G, Warburton PE 2007. Evolutionary history of mammalian transposons determined by genome-wide defragmentation. PLoS Comput Biol 3: e137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard A, Hannon GJ 2008. Conserved themes in small-RNA-mediated transposon control. Trends Cell Biol 18: 136–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard A, Sachidanandam R, Hannon GJ, Carmell MA 2006. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature 442: 199–202 [DOI] [PubMed] [Google Scholar]
- Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami-Okada TN, Siomi H, Siomi MC 2007. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science 315: 1587–1590 [DOI] [PubMed] [Google Scholar]
- Heard E, Turner J 2011. Function of the sex chromosomes in mammalian fertility. Cold Spring Harb Perspect Biol 3: a002675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houwing S, Kamminga LM, Berezikov E, Cronembold D, Girard A, van den Elst H, Filippov DV, Blaser H, Raz E, Moens CB, et al. 2007. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell 129: 69–82 [DOI] [PubMed] [Google Scholar]
- Huang XA, Yin H, Sweeney S, Raha D, Snyder M, Lin H 2013. A major epigenetic programming mechanism guided by piRNAs. Dev Cell 24: 502–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizu H, Siomi H, Siomi MC 2012. Biology of PIWI-interacting RNAs: new insights into biogenesis and function inside and outside of germlines. Genes Dev 26: 2361–2373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizuka A, Siomi MC, Siomi H 2002. A Drosophila fragile X protein interacts with components of RNAi and ribosomal proteins. Genes Dev 16: 2497–2508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacques P-É, Jeyakani J, Bourque G 2013. The majority of primate-specific regulatory sequences are derived from transposable elements. PLoS Genet 9: e1003504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapusta A, Kronenberg Z, Lynch VJ, Zhuo X, Ramsay L, Bourque G, Yandell M, Feschotte C 2013. Transposable elements are major contributors to the origin, diversification, and regulation of vertebrate long noncoding RNAs. PLoS Genet 9: e1003470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karginov FV, Hannon GJ 2010. The CRISPR system: small RNA-guided defense in bacteria and archaea. Mol Cell 37: 7–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura Y, Saito K, Kin T, Ono Y, Asai K, Sunohara T, Okada TN, Siomi MC, Siomi H 2008. Drosophila endogenous small RNAs bind to Argonaute 2 in somatic cells. Nature 453: 793–797 [DOI] [PubMed] [Google Scholar]
- Ketting RF 2011. The many faces of RNAi. Dev Cell 20: 148–161 [DOI] [PubMed] [Google Scholar]
- Khurana JS, Wang J, Xu J, Koppetsch BS, Thomson TC, Nowosielska A, Li C, Zamore PD, Weng Z, Theurkauf WE 2011. Adaptation to P element transposon invasion in Drosophila melanogaster. Cell 147: 1551–1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN, Han J, Siomi MC 2009. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol 10: 126–139 [DOI] [PubMed] [Google Scholar]
- Kirino Y, Mourelatos Z 2007. Mouse Piwi-interacting RNAs are 2′-O-methylated at their 3′ termini. Nat Struct Mol Biol 14: 347–348 [DOI] [PubMed] [Google Scholar]
- Kunzelmann K, Nilius B, Owsianik G, Schreiber R, Ousingsawat J, Sirianant L, Wanitchakool P, Bevers E, Heemskerk JM 2014. Molecular functions of anoctamin 6 (TMEM16F): a chloride channel, cation channel, or phospholipid scramblase? Pflugers Arch 466: 407–414 [DOI] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Kimura T, Ijiri TW, Isobe T, Asada N, Fujita Y, Ikawa M, Iwai N, Okabe M, Deng W, et al. 2004. Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development 131: 839–849 [DOI] [PubMed] [Google Scholar]
- Lakshmi SS, Agrawal S 2008. piRNABank: a web resource on classified and clustered Piwi-interacting RNAs. Nucleic Acids Res 36: D173–D177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange K, Kammerer M, Hegi ME, Grotegut S, Dittmann A, Huang W, Fluri E, Yip GW, Götte M, Ruiz C, et al. 2007. Endothelin receptor type B counteracts tenascin-C–induced endothelin receptor type A–dependent focal adhesion and actin stress fiber disorganization. Cancer Res 67: 6163–6173 [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Thomas A, Rogers AK, Webster A, Marinov GK, Liao SE, Perkins EM, Hur JK, Aravin AA, Toth KF 2013. Piwi induces piRNA-guided transcriptional silencing and establishment of a repressive chromatin state. Genes Dev 27: 390–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Liu Y, Dong D, Zhang Z 2010. Evolution of an X-linked primate-specific micro RNA cluster. Mol Biol Evol 27: 671–683 [DOI] [PubMed] [Google Scholar]
- Li XZ, Roy CK, Dong X, Bolcun-Filas E, Wang J, Han BW, Xu J, Moore MJ, Schimenti JC, Weng Z, et al. 2013. An ancient transcription factor initiates the burst of piRNA production during early meiosis in mouse testes. Mol Cell 50: 67–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevaiah SK, Turner JM, Baudat F, Rogakou EP, de Boer P, Blanco-Rodríguez J, Jasin M, Keeney S, Bonner WM, Burgoyne PS 2001. Recombinational DNA double-strand breaks in mice precede synapsis. Nat Genet 27: 271–276 [DOI] [PubMed] [Google Scholar]
- Malone CD, Hannon GJ 2009. Small RNAs as guardians of the genome. Cell 136: 656–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal PK, Kazazian H Jr 2008. SnapShot: vertebrate transposons. Cell 135: 192–192.e1 [DOI] [PubMed] [Google Scholar]
- Meunier J, Lemoine F, Soumillon M, Liechti A, Weier M, Guschanski K, Hu H, Khaitovich P, Kaessmann H 2013. Birth and expression evolution of mammalian microRNA genes. Genome Res 23: 34–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer LR, Zweig AS, Hinrichs AS, Karolchik D, Kuhn RM, Wong M, Sloan CA, Rosenbloom KR, Roe G, Rhead B, et al. 2013. The UCSC Genome Browser database: extensions and updates 2013. Nucleic Acids Res 41: D64–D69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi K, Miyoshi T, Hartig JV, Siomi H, Siomi MC 2010. Molecular mechanisms that funnel RNA precursors into endogenous small-interfering RNA and microRNA biogenesis pathways in Drosophila. RNA 16: 506–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muerdter F, Guzzardo PM, Gillis J, Luo Y, Yu Y, Chen C, Fekete R, Hannon GJ 2013. A genome-wide RNAi screen draws a genetic framework for transposon control and primary piRNA biogenesis in Drosophila. Mol Cell 50: 736–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida KM, Saito K, Mori T, Kawamura Y, Nagami-Okada TN, Inagaki S, Siomi H, Siomi MC 2007. Gene silencing mechanisms mediated by Aubergine–piRNA complexes in Drosophila male gonad. RNA 13: 1911–1922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohara T, Sakaguchi Y, Suzuki T, Ueda H, Miyauchi K, Suzuki T 2007. The 3′ termini of mouse Piwi-interacting RNAs are 2′-O-methylated. Nat Struct Mol Biol 14: 349–350 [DOI] [PubMed] [Google Scholar]
- Ohtani H, Iwasaki YW, Shibuya A, Siomi H, Siomi MC, Saito K 2013. DmGTSF1 is necessary for Piwi-piRISC–mediated transcriptional transposon silencing in the Drosophila ovary. Genes Dev 27: 1656–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Lai EC 2008. Endogenous small interfering RNAs in animals. Nat Rev Mol Cell Biol 9: 673–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Chung W-J, Ruby JG, Guo H, Bartel DP, Lai EC 2008. The Drosophila hairpin RNA pathway generates endogenous short interfering RNAs. Nature 453: 803–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano H, Hikishima K, Iriki A, Sasaki E 2012. The common marmoset as a novel animal model system for biomedical and neuroscience research applications. Semin Fetal Neonatal Med 17: 336–340 [DOI] [PubMed] [Google Scholar]
- Orsi A, Rees D, Andreini I, Venturella S, Cinelli S, Oberto G 2011. Overview of the marmoset as a model in nonclinical development of pharmaceutical products. Regul Toxicol Pharmacol 59: 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera D, Tilston V, Hopwood JA, Barchi M, Boot-Handford RP, Taylor SS 2007. Bub1 maintains centromeric cohesion by activation of the spindle checkpoint. Dev Cell 13: 566–579 [DOI] [PubMed] [Google Scholar]
- Pillai RS, Chuma S 2012. piRNAs and their involvement in male germline development in mice. Dev Growth Differ 54: 78–92 [DOI] [PubMed] [Google Scholar]
- Reuter M, Berninger P, Chuma S, Shah H, Hosokawa M, Funaya C, Antony C, Sachidanandam R, Pillai RS 2011. Miwi catalysis is required for piRNA amplification-independent LINE1 transposon silencing. Nature 480: 264–267 [DOI] [PubMed] [Google Scholar]
- Robine N, Lau NC, Balla S, Jin Z, Okamura K, Kuramochi-Miyagawa S, Blower MD, Lai EC 2009. A broadly conserved pathway generates 3′UTR-directed primary piRNAs. Curr Biol 19: 2066–2076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabin LR, Delás MJ, Hannon GJ 2013. Dogma derailed: the many influences of RNA on the genome. Mol Cell 49: 783–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Siomi MC 2010. Small RNA-mediated quiescence of transposable elements in animals. Dev Cell 19: 687–697 [DOI] [PubMed] [Google Scholar]
- Saito K, Sakaguchi Y, Suzuki T, Suzuki T, Siomi H, Siomi MC 2007. Pimet, the Drosophila homolog of HEN1, mediates 2′-O-methylation of Piwi-interacting RNAs at their 3′ ends. Genes Dev 21: 1603–1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Inagaki S, Mituyama T, Kawamura Y, Ono Y, Sakota E, Kotani H, Asai K, Siomi H, Siomi MC 2009. A regulatory circuit for piwi by the large Maf gene traffic jam in Drosophila. Nature 461: 1296–1299 [DOI] [PubMed] [Google Scholar]
- Sasaki T, Shiohama A, Minoshima S, Shimizu N 2003. Identification of eight members of the Argonaute family in the human genome. Genomics 82: 323–330 [DOI] [PubMed] [Google Scholar]
- Sasaki E, Suemizu H, Shimada A, Hanazawa K, Oiwa R, Kamioka M, Tomioka I, Sotomaru Y, Hirakawa R, Eto T, et al. 2009. Generation of transgenic non-human primates with germline transmission. Nature 459: 523–527 [DOI] [PubMed] [Google Scholar]
- Seitz H, Tushir JS, Zamore PD 2011. A 5′-uridine amplifies miRNA/miRNA* asymmetry in Drosophila by promoting RNA-induced silencing complex formation. Silence 2: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sienski G, Dönertas D, Brennecke J 2012. Transcriptional silencing of transposons by Piwi and Maelstrom and its impact on chromatin state and gene expression. Cell 151: 964–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon B, Kirkpatrick JP, Eckhardt S, Reuter M, Rocha EA, Andrade-Navarro MA, Sehr P, Pillai RS, Carlomagno T 2011. Recognition of 2′-O-methylated 3′-end of piRNA by the PAZ domain of a Piwi protein. Structure 19: 172–180 [DOI] [PubMed] [Google Scholar]
- Siomi H, Siomi MC 2009. On the road to reading the RNA-interference code. Nature 457: 396–404 [DOI] [PubMed] [Google Scholar]
- Siomi MC, Sato K, Pezic D, Aravin AA 2011. PIWI-interacting small RNAs: the vanguard of genome defence. Nat Rev Mol Cell Biol 12: 246–258 [DOI] [PubMed] [Google Scholar]
- Smit A, Hubley R, Green P 2010. RepeatMasker. http://www.repeatmasker.org
- Sobala A, Hutvagner G 2011. Transfer RNA-derived fragments: origins, processing, and functions. Wiley Interdiscip Rev RNA 2: 853–862 [DOI] [PubMed] [Google Scholar]
- Song R, Ro S, Michaels JD, Park C, McCarrey JR, Yan W 2009. Many X-linked microRNAs escape meiotic sex chromosome inactivation. Nat Genet 41: 488–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song R, Hennig GW, Wu Q, Jose C, Zheng H, Yan W 2011. Male germ cells express abundant endogenous siRNAs. Proc Natl Acad Sci 108: 13159–13164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sookdeo A, Hepp CM, McClure MA, Boissinot S 2013. Revisiting the evolution of mouse LINE-1 in the genomic era. Mob DNA 4: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Zhang Y, Zhang R, Qi X, Su B 2013. Functional divergence of the rapidly evolving miR-513 subfamily in primates. BMC Evol Biol 13: 255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam OH, Aravin AA, Stein P, Girard A, Murchison EP, Cheloufi S, Hodges E, Anger M, Sachidanandam R, Schultz RM, et al. 2008. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature 453: 534–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson T, Lin H 2009. The biogenesis and function of PIWI proteins and piRNAs: progress and prospect. Annu Rev Cell Dev Biol 25: 355–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner JMA 2007. Meiotic sex chromosome inactivation. Development 134: 1823–1831 [DOI] [PubMed] [Google Scholar]
- Vourekas A, Zheng Q, Alexiou P, Maragkakis M, Kirino Y, Gregory BD, Mourelatos Z 2012. Mili and Miwi target RNA repertoire reveals piRNA biogenesis and function of Miwi in spermiogenesis. Nat Struct Mol Biol 19: 773–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasik U, Filipek A 2013. The CacyBP/SIP protein is sumoylated in neuroblastoma NB2a cells. Neurochem Res 38: 2427–2432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Totoki Y, Toyoda A, Kaneda M, Kuramochi-Miyagawa S, Obata Y, Chiba H, Kohara Y, Kono T, Nakano T, et al. 2008. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature 453: 539–543 [DOI] [PubMed] [Google Scholar]
- Yuan Z, Sun X, Jiang D, Ding Y, Lu Z, Gong L, Liu H, Xie J 2010. Origin and evolution of a placental-specific microRNA family in the human genome. BMC Evol Biol 10: 346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Peng Y, Wang W, Su B 2007. Rapid evolution of an X-linked microRNA cluster in primates. Genome Res 17: 612–617 [DOI] [PMC free article] [PubMed] [Google Scholar]