Pokeweed antiviral protein (PAP) decreases the production of HIV-1 without being toxic for the host cells. This study suggests that the effect of PAP is related to a specific depurination of rev mRNA, preventing the synthesis of active Rev and, consequently, the transport of unspliced and single-spliced viral mRNA to the cytoplasm.
Keywords: HIV-1, Rev, pokeweed antiviral protein, glycosidase, ribosome inactivating protein
Abstract
Processing of HIV-1 transcripts results in three populations in the cytoplasm of infected cells: full-length RNA, singly spliced, and multiply spliced RNAs. Rev, regulator of virion expression, is an essential regulatory protein of HIV-1 required for transporting unspliced and singly spliced viral transcripts from the nucleus to the cytoplasm. Export allows these RNAs to be translated and the full-length RNA to be packaged into virus particles. In our study, we investigate the activity of pokeweed antiviral protein (PAP), a glycosidase isolated from the pokeweed plant Phytolacca americana, on the processing of viral RNAs. We show that coexpression of PAP with a proviral clone alters the splicing ratio of HIV-1 RNAs. Specifically, PAP causes the accumulation of multiply spliced 2-kb RNAs at the expense of full-length 9-kb and singly spliced 4-kb RNAs. The change in splicing ratio is due to a decrease in activity of Rev. We show that PAP depurinates the rev open reading frame and that this damage to the viral RNA inhibits its translation. By decreasing Rev expression, PAP indirectly reduces the availability of full-length 9-kb RNA for packaging and translation of the encoded structural proteins required for synthesis of viral particles. The decline we observe in virus protein expression is not due to cellular toxicity as PAP did not diminish translation rate. Our results describing the reduced activity of a regulatory protein of HIV-1, with resulting change in virus mRNA ratios, provides new insight into the antiviral mechanism of PAP.
INTRODUCTION
The current treatment strategy for HIV-1 includes combined viral inhibitors, referred to as highly active antiretroviral therapy (HAART) (Broder 2010; Doyle and Geretti 2012). A significant challenge for treatment is the mutagenic nature of the virus, which has often led to drug resistance (Kuritzkes 2011; Tang and Shafer 2012). The administration of drug mixtures aims to reduce the chance of resistance by targeting several steps in the virus lifecycle, namely entry, reverse transcription, integration, and maturation (Flexner 2007; Mehellou and De Clercq 2010). Though HAART has significantly extended the lifespan and delayed the onset of AIDS for HIV-1-infected individuals, concerns with this treatment include side effects, resistance, and high cost. Discovery and development of additional ways to inhibit HIV-1 are a constant and essential research effort.
Pokeweed antiviral protein (PAP), a glycosidase isolated from the pokeweed plant (Phytolacca americana), removes purines from various RNA templates (Endo et al. 1988; Karran and Hudak 2008; Mansouri et al. 2009). The enzyme is of research interest because of its broad-range activity against viruses of agricultural and medical relevance. For example, the antiviral activity of PAP against HIV-1 has been observed in mammalian cell cultures and animal models. Addition of picomolar concentrations of PAP to HIV-infected CD4+ T cells substantially inhibited the synthesis of viral proteins without affecting viability or function of the cells (Zarling et al. 1990). Lack of cytotoxicity has been reported in cultured human cells (HEK 293T) (Chan Tung et al. 2008; Mansouri et al. 2009), monkeys (Macaca cynomolgus) (Uckun et al. 1998), and in mouse models (Hu-PBL-SCID, B6C3F1, and CD-1) (Uckun et al. 1998; D'Cruz et al. 2004) at concentrations of PAP that inhibit viral replication. Co-incubation of HIV-1 genomic RNA with PAP resulted in concentration-dependent release of adenines into the media, indicating depurination of the RNA (Rajamohan et al. 1999). These studies have focused primarily on evaluating the clinical potential of PAP as an anti-HIV-1 agent and have established that PAP is a potent, apparently nontoxic, inhibitor of the virus (Uckun et al. 1999).
The goal of this study was to investigate the antiviral mechanism of PAP, specifically to determine what post-transcriptional events in the HIV-1 lifecycle are affected by PAP. Following integration of the HIV-1 provirus and transcription of genomic RNA by cellular RNA polymerase, the full-length viral RNA (∼9 kb) is spliced into transcripts of ∼4 and 2 kb (Purcell and Martin 1993). The unspliced 9-kb transcripts, as well as the singly spliced 4-kb transcripts, contain introns and cis-acting repressive sequences (CRS) that promote retention in the nucleus (Schwartz et al. 1992; Zhang et al. 1996). As a result, only the 2-kb multiply spliced mRNAs exit from the nucleus using common cellular transport pathways (Kang and Cullen 1999). The 2-kb mRNAs encode three proteins, including regulator of virion expression (Rev) (Kim et al. 1989; Pomerantz et al. 1990). Rev facilitates the export of CRS-containing RNAs into the cytoplasm by binding the Rev response element (RRE) found within these transcripts (Sodroski et al. 1986; Felber et al. 1989; Cullen 1998; Pollard and Malim 1998). Although the direct role of Rev is the export of 9- and 4-kb RNAs to the cytoplasm, by virtue of its activity, Rev affects HIV-1 splicing, translation, and packaging (Hope 1999). In our current work, we show that expression of PAP alters the splicing ratio of HIV-1 RNAs by significantly reducing Rev activity. Without the important regulatory function of this protein, virus production declines.
RESULTS
Expression of PAP decreases HIV-1 protein expression without altering cellular translation rate
To characterize the anti-HIV-1 effects of PAP, 293T cells were cotransfected with the noninfectious clone pMenv(-) and the pcPAP plasmid encoding 3X-FLAG-tagged PAP. As a negative control for the catalytic activity of PAP, cells were cotransfected with pcPAPx, an active-site mutant of PAP (E176V) that cannot depurinate RNA (Hur et al. 1995). Immunoblotting confirmed that both PAP and PAPx were effectively expressed in this system (Fig. 1A) and that PAP cotransfection with the proviral clone substantially reduced the levels of viral proteins encoded by 9- and 4-kb mRNAs, including capsid, reverse transcriptase, and Vif (Fig. 1B). Moreover, the release of particles from PAP-expressing cells was diminished, as measured by the amount of capsid protein of particles collected from growth media (Fig. 1B). This decrease agreed with our previous p24 ELISA results illustrating a 450-fold reduction in virus production from cells expressing PAP (Mansouri et al. 2012). To test whether the antiviral effect of PAP was due to cellular toxicity, cells were metabolically radiolabeled with [35S]methionine, harvested over a 6-h time period (time zero = 40 h post-transfection), and assessed for changes in translation rate, with cycloheximide treatment serving as a positive control for translation inhibition. Neither wild-type nor mutant form of PAP inhibited the overall translation rate of the cells relative to cells transfected with empty vector pcDNA3 (Fig. 1C). Mock cells were untransfected and their translation rate was always slightly higher than transfected cells.
FIGURE 1.
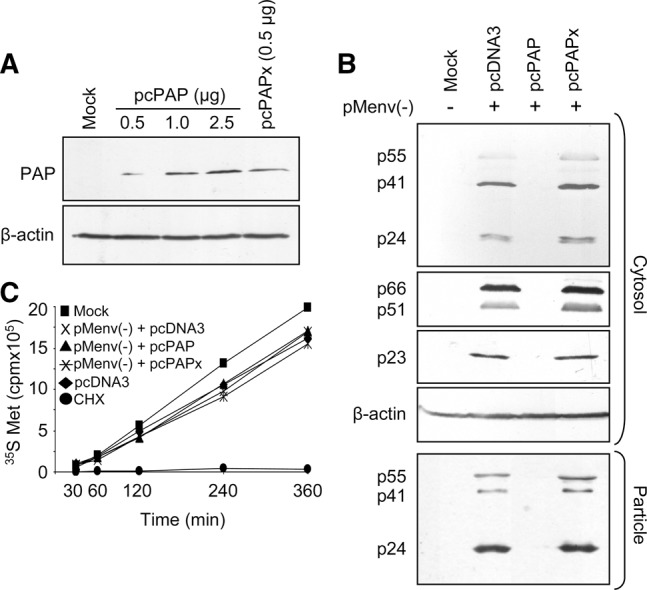
PAP expression in 293T cells inhibits HIV-1 protein expression without decreasing cellular translation rates. (A) 293T cells were transfected with pcPAP (0.5, 1.0, or 2.5 μg) or pcPAPx (0.5 μg). Cells were harvested 40 h post-transfection and total cytoplasmic proteins (120 μg) were separated by 12% SDS-PAGE, followed by immunoblot analysis using a monoclonal FLAG-specific antibody (1:5,000). Blots were also probed with a monoclonal β-actin-specific antibody (1:5,000) for loading control estimates. (B) 293T cells were cotransfected with pMenv(-) proviral clone (5 μg) and pcPAP, pcPAPx, or pcDNA3 (2.5 μg). Cells were harvested 40 h post-transfection and total cytoplasmic proteins (120 μg) were separated by 12% SDS-PAGE, followed by immunoblot analysis using a monoclonal p24 capsid-specific antibody (1:5,000), polyclonal p66, p51 RT-specific antibody (1:2,500), or polyclonal p23 Vif-specific antibody (1:2,500). Blots were also probed with a monoclonal β-actin-specific antibody (1:5,000) as a loading control. Viral particles were collected from 1 mL of growth medium and analyzed by immunoblot for p24 capsid. (C) 293T cells were metabolically labeled with [35S]methionine 40 h post-cotransfection with pMenv(-) proviral clone (5 µg) and pcPAP, pcPAPx, or pcDNA3 (2.5 μg). Mock cells were untransfected, pcDNA3 cells were transfected only with the empty pcDNA3 vector, and CHX cells were treated with 25 μg/mL cycloheximide at the beginning of the time course. Cells were harvested at the indicated time points following addition of [35S]methionine and the incorporated radioactivity was scintillation counted. Points represent means for three independent experiments.
PAP expression increases HIV-1 2-kb mRNA accumulation
To determine the effect of PAP on the level of HIV-1 mRNAs, Northern blotting was carried out using an HIV-1-specific probe that bound a region shared by all three mRNA size classes (Fig. 2A). The normal level of HIV-1 mRNAs was observed in the presence of the empty vector control, with all three mRNA classes readily detected (9, 4, and 2 kb) (Fig. 2B). PAP expression greatly altered the splicing ratio of RRE-free and RRE-containing mRNAs within the cytoplasm (Fig. 2C), evident by the accumulation of 2-kb mRNAs and the reduction of 9- and 4-kb mRNAs (Fig. 2B). The splicing phenotype of HIV-1 remained normal with the expression of PAPx, indicating that the change seen in the presence of PAP was likely due to its catalytic activity.
FIGURE 2.
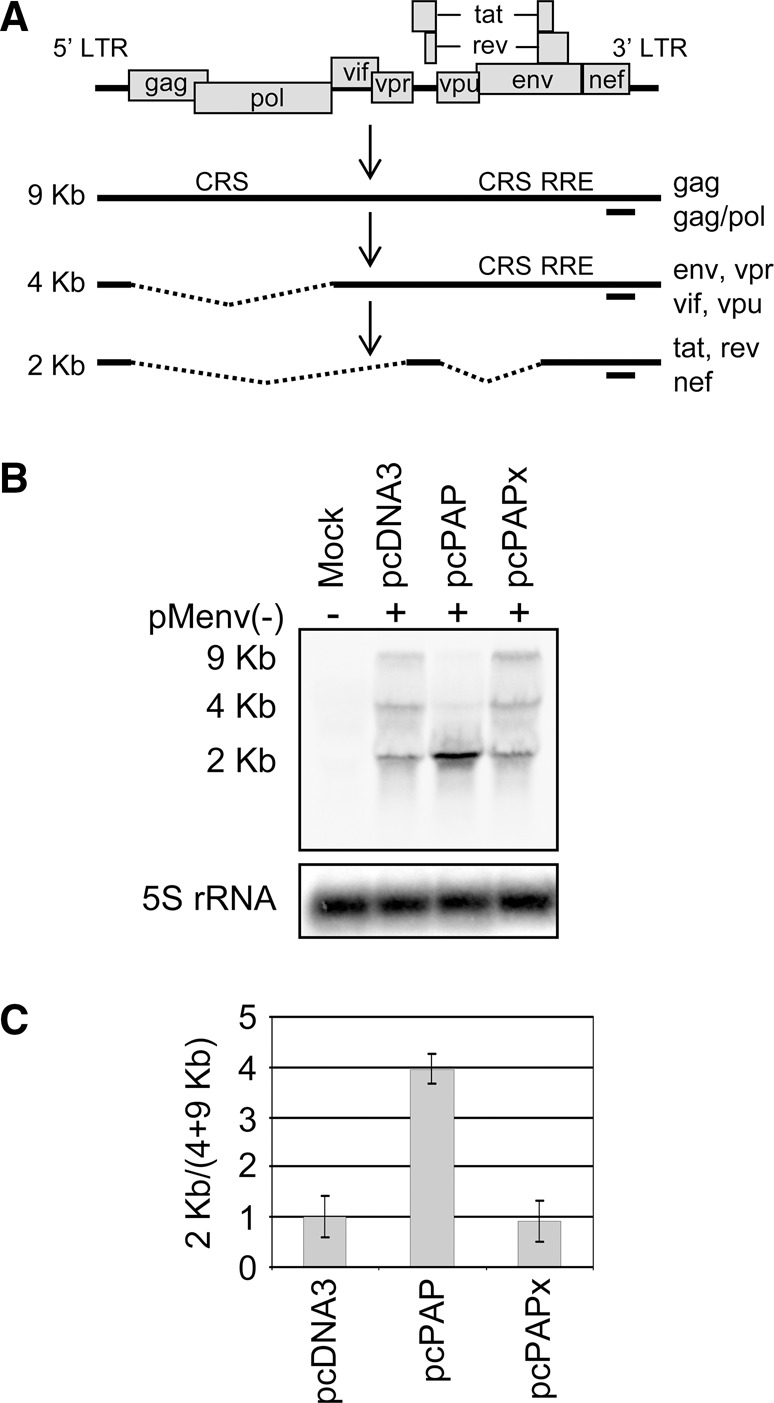
PAP increases HIV-1 splicing and 2-kb mRNA accumulation. (A) Schematic representation of the HIV-1 genome organization, transcription, and splice products. Location of cis-repressive signals (CRS) and Rev responsive element (RRE) are indicated. The annealing site for the HIV-1-specific negative-strand riboprobe is indicated below each mRNA size class as a solid line. (B) 293T cells were cotransfected with pMenv(-) proviral clone (5 μg) and pcPAP, pcPAPx, or pcDNA3 (2.5 μg). Cells were harvested 40 h post transfection and Northern blotting was performed on total cellular RNA (15 μg). Levels of the three HIV-1 mRNA classes were visualized following hybridization with the HIV-1-specific probe. Blots were also hybridized with a 5S rRNA-specific probe as a loading control. (C) The abundance of HIV-1 mRNAs in each size class was measured by quantifying the band intensity using a PhosphorImager. The ratio of RRE-free mRNAs (2 kb) to RRE-containing mRNAs (9 and 4 kb) was plotted relative to 5S rRNA. Error bars represent means ± SE for three independent experiments.
RRE-free mRNAs accumulate on polysomes
The splicing pattern observed in PAP-expressing cells resembled that of Rev-deficient proviral clones since RRE-containing RNAs are retained in the nucleus in the absence of Rev (Pomerantz et al. 1992). The observed alteration in the splicing phenotype in the presence of PAP may be due to lack of Rev translation, even though 2-kb transcripts accumulated. To assess this possibility, polysome analysis was carried out to identify changes in the allocation of HIV-1 mRNAs with respect to ribosome-associating and “free” pools of RNAs (i.e., those RNAs recruited to processing bodies). Northern blot analysis probing for HIV-1 mRNAs within fractions collected from density gradients revealed that there was a significant accumulation of 2-kb mRNAs on polysomes in the presence of PAP when compared with the empty vector control (Fig. 3A,B). Surprisingly, there was no significant difference in the proportion of HIV-1 mRNAs on polysomes relative to total HIV-1 mRNA in the presence or absence of PAP (Fig. 3C). This suggests that the translational profile of HIV-1 was not altered by PAP and the antiviral effect was not likely due to recruitment of viral mRNAs to nontranslating pools of RNA.
FIGURE 3.
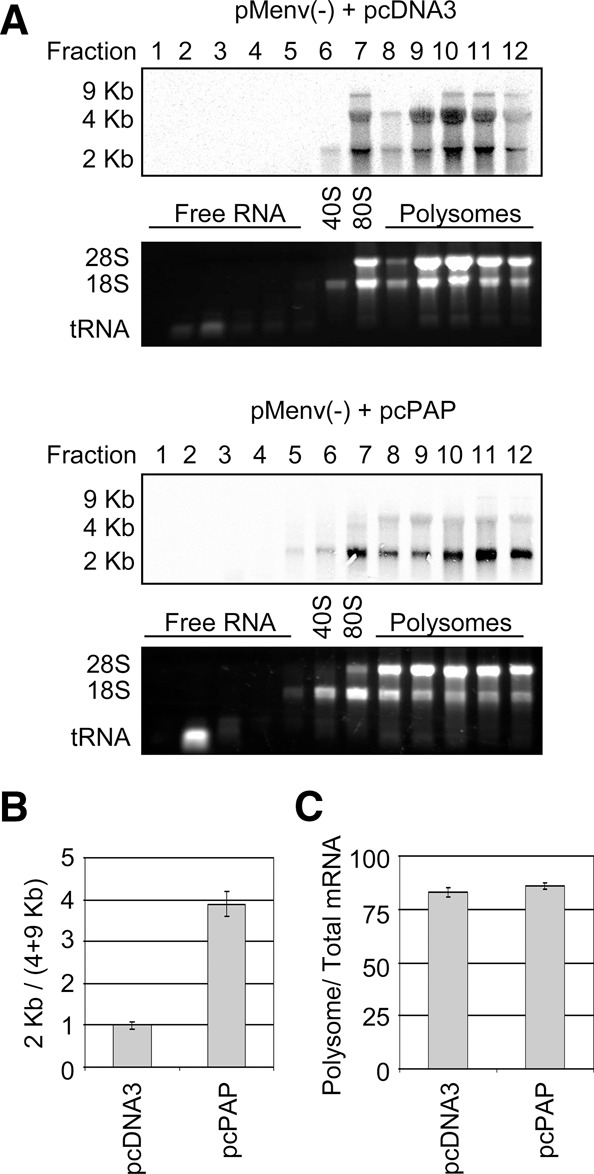
PAP causes RRE-free mRNAs to accumulate on polysomes. (A) 293T cells were cotransfected with pMenv(-) proviral clone (5 μg) and pcPAP or pcDNA3 (2.5 μg). Cells were harvested 40 h post-transfection, and lysates were resolved by density gradient centrifugation and collected in 12 fractions. RNA was isolated from each fraction and levels of the three HIV-1 mRNA classes were visualized by Northern blotting following hybridization with the HIV-1-specific probe. The polysome profile was visualized prior to Northern blotting by ethidium bromide staining of the agarose gel. (B) The abundance of HIV-1 mRNAs in each size class was measured by quantifying the band intensity using a PhosphorImager and the ratio of RRE-free mRNAs (2 kb) to RRE-containing mRNAs (9 and 4 kb) was plotted. (C) The abundance of HIV-1 mRNAs on polysomes was plotted as a percentage of total HIV-1 RNA. Error bars in B and C represent means ± SE for three independent experiments.
Level of rev mRNA increases in the presence of PAP
Since the accumulated 2-kb mRNAs associated with polysomes in the presence of PAP, it was possible that more regulatory proteins (Tat, Rev, and Nef) were being produced. However, the splicing pattern observed in PAP-expressing cells was similar to Rev-deficient clones, suggesting that the level of rev mRNA may be reduced in these cells. RNase protection assay was carried out to identify which 2-kb mRNAs were accumulating in the presence of PAP. Due to overlapping sequences in the three mRNA classes of HIV-1, the ∼2-kb mRNA population was isolated from low-melt agarose gels and equal amounts of RNA were analyzed from cells cotransfected with pcPAP or the empty vector. Figure 4A is a schematic representation of the three 2-kb mRNAs of HIV-1, showing the annealing site of the riboprobe and the expected sizes following the assay. The housekeeping gene glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as a loading control due to its size similarity to the 2-kb mRNAs (∼1.4 kb) and thus could be cut from the gel at the same time. The levels of all three 2-kb mRNAs, tat, rev, and nef, increased approximately twofold with PAP expression (Fig. 4B), relative to cells transfected with empty vector (Fig. 4C). Therefore, rev mRNA was present in cells in which the splicing phenotype resembled a Rev-deficient clone.
FIGURE 4.
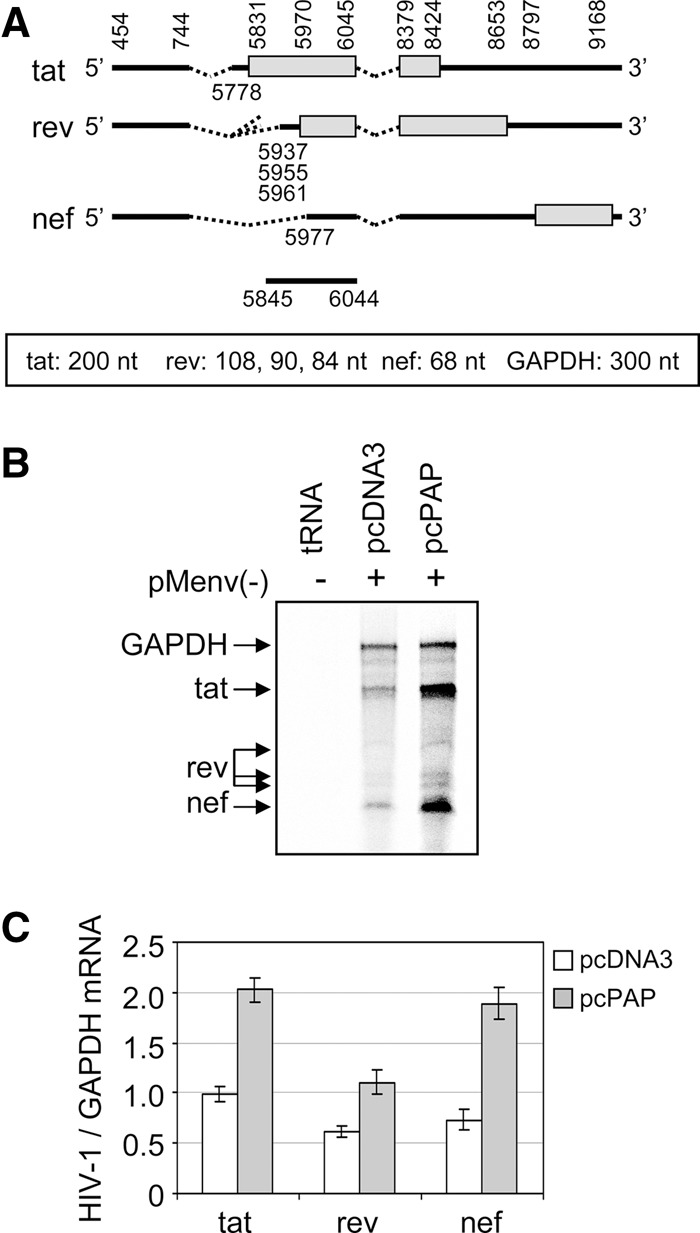
The three HIV-1 2-kb mRNAs accumulate in the presence of PAP. (A) Schematic representation of the HIV-1 2-kb mRNA variants and their corresponding splice junctions (numbered according to the HXB2 genome, accession no. K03455.1). The locations of the open reading frames for each variant are indicated as rectangles and the annealing site for the HIV-1-specific negative-sense radiolabeled riboprobe is indicated below the nef mRNA as a solid line. The expected product sizes from the RNase protection assay are indicated below the schematic for each of the 2-kb mRNA variants, as well as for GAPDH, which served as a loading control. (B) 293T cells were cotransfected with pMenv(-) proviral clone (5 μg) and pcPAP or pcDNA3 (2.5 μg). Cells were harvested 40 h post-transfection and RNase protection assay was performed on the ∼2-kb RNAs (15 μg) or tRNA (15 μg), following hybridization with the 2-kb-specific and GAPDH-specific riboprobes. (C) The abundance of tat, rev, and nef mRNAs was measured by quantifying the band intensity using a PhosphorImager. The ratio of each mRNA variant in the presence or absence of PAP was plotted relative to GAPDH abundance. Error bars represent means ± SE for three independent experiments.
PAP decreases Rev-dependent mRNA transport
The presence of rev mRNA in PAP-expressing cells, taken together with an increase in HIV-1 mRNA splicing ratio, suggested that the rev mRNA was not producing active Rev protein. We attempted analysis of Rev by immunoblot but could only detect Rev when overexpressed, by transfection of cells with a plasmid encoding rev. Therefore, we tested the level of Rev activity from the proviral clone by transfecting cells with the Rev activity reporter plasmid pDM128. This construct contains the chloramphenicol acetyltransferase (CAT) gene upstream of a RRE, between splice donor and acceptor sites (Fig. 5A). The CAT mRNA would only be detected in the cytoplasm if Rev facilitated its transport from the nucleus; otherwise, the CAT open reading frame would be spliced from the mRNA. Northern blot assay using a CAT-specific riboprobe showed that PAP significantly reduced the level of Rev activity relative to the empty vector and PAPx controls (Fig. 5B). In addition, Rev activity was rescued by transfecting cells with a plasmid encoding rev (pcREV) in the presence of PAP. The lack of adequate Rev activity from the proviral clone in the presence of PAP was consistent with the observed increase in the splicing phenotype of HIV-1.
FIGURE 5.
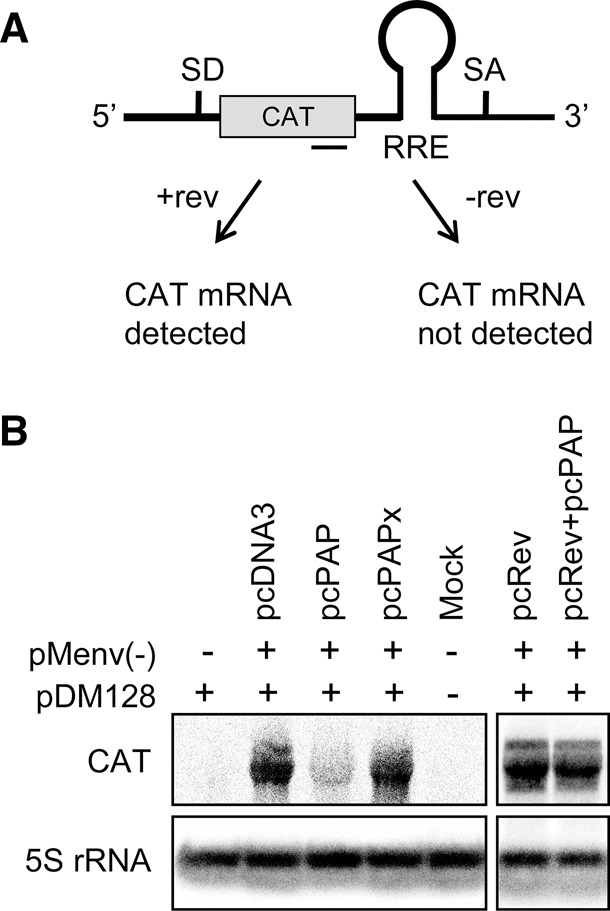
PAP decreases the level of Rev activity. (A) Schematic representation of the pDM128 reporter construct (Hope et al. 1990) and the predicted outcomes with or without Rev activity. The annealing site for the CAT-specific negative-strand riboprobe is indicated below the reporter construct as a solid line. (B) 293T cells were cotransfected with pDM128 reporter construct (5 μg), pMenv(-) proviral clone (5 μg), and pcPAP, pcPAPx, or pcDNA3 (2.5 μg), with or without pcRev (2.5 μg). Cells were harvested 40 h post-transfection and Northern blotting was performed on total cellular RNA (15 μg). Level of the cytoplasmic CAT mRNA was visualized by hybridization with the CAT-specific probe. Blots were also hybridized with a 5S rRNA-specific probe as a loading control.
pcRev rescues the HIV-1 splicing phenotype in the presence of PAP
To assess whether the PAP-induced change to HIV-1 mRNA splicing was due to loss of Rev activity, similar rescue experiments were performed. PAP-expressing cells were supplemented with a plasmid encoding Rev and analyzed for restoration of the splicing phenotype. Addition of pcRev regained the splicing phenotype in a Rev-deficient proviral clone pRev(-) (Fig. 6A, second lane), showing that Rev expressed from this plasmid functioned as expected. Similarly, the rescue of the splicing phenotype was successful for both HIV-1 proviral clones, pRev(-) (Fig. 6A, third lane), and pMenv(-) (Fig. 6B, fifth lane) in the presence of PAP. Therefore, the increase in splicing observed upon expression of PAP (Fig. 2C) was due to loss of Rev, since its addition restored the RRE-containing mRNAs while decreasing the accumulation of 2-kb mRNAs. The addition of pcRev did not detectably affect the normal splicing phenotype of pMenv(-) (Fig. 6B, eighth lane).
FIGURE 6.
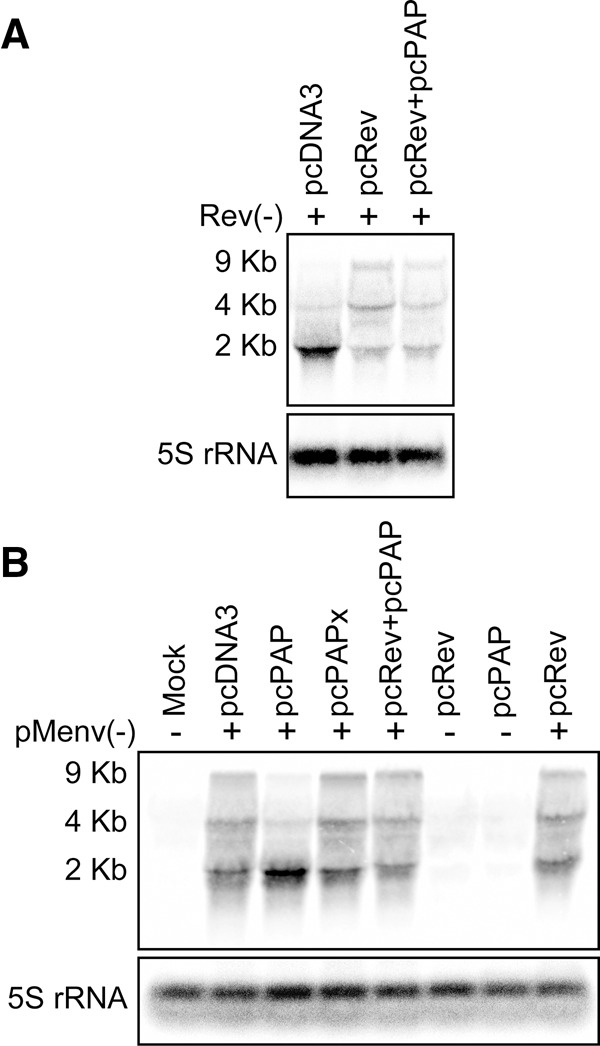
Addition of pcRev rescues the HIV-1 splicing phenotype in the presence of PAP. 293T cells were cotransfected with (A) pRev(-) proviral clone (5 μg) or (B) pMenv(-) proviral clone (5 μg) and pcPAP, pcPAPx, or pcDNA3 (2.5 μg), with or without pcRev (2.5 μg). Cells were harvested 40 h post-transfection and Northern blotting was performed on total cellular RNA (15 μg). Levels of the three HIV-1 mRNA classes were visualized by hybridization with the HIV-1-specific riboprobe. Blots were also hybridized with a 5S rRNA-specific riboprobe as a loading control.
PAP depurinates the rev open reading frame of HIV-1
The rescue of the HIV-1 splicing ratio through the addition of pcRev, taken together with an accumulation of 2-kb mRNAs (including rev) and a substantial reduction in Rev activity, suggested that PAP may exhibit its antiviral effects on HIV-1 by targeting the rev mRNA. To determine whether PAP depurinates the viral RNA, primer extension analysis was conducted on regions of the 2-kb mRNAs. Termination of the reverse transcriptase at site A8597, and to a lesser extent G8594, was reproducibly observed with RNA isolated from PAP expressing cells (Fig. 7A) but not with the empty vector. These same nucleotides were depurinated following incubation of isolated 2-kb RNAs with PAP in vitro. The location of these potential depurination sites (Fig. 7B), numbered according to the first nucleotide of the 5′ U3 region of the proviral DNA (accession no. K03455.1), were identified by sequencing the pMenv(-) proviral DNA with the same reverse primer as used for primer extension. These termination sites were located within the rev ORF, in a region that is unique to rev and does not overlap with any other ORF of the 2-kb population.
FIGURE 7.
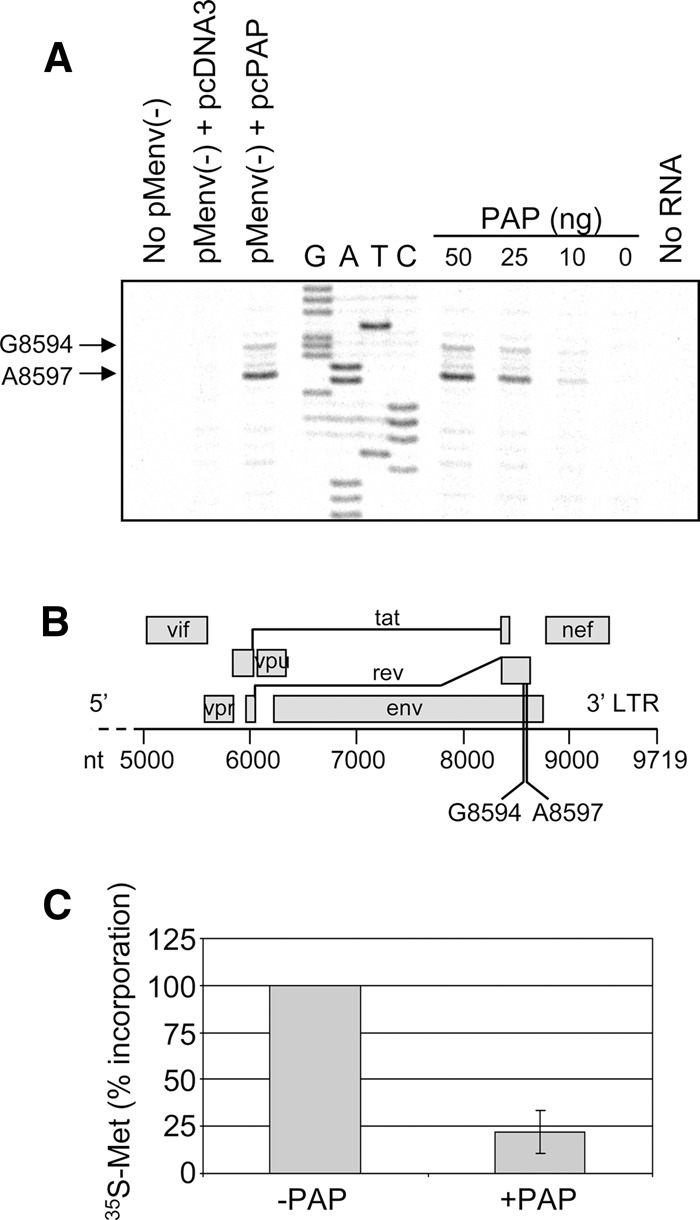
PAP depurinates the rev open reading frame of HIV-1 and inhibits its translation. (A) 2-kb RNAs were isolated from 293T cells cotransfected with pMenv(-) proviral clone (5 μg) and pcPAP or pcDNA3 (2.5 μg) 40 h post-transfection; 2-kb RNAs were also isolated from 293T cells transfected only with pMenv(-) proviral clone (5 μg) and incubated with purified PAP (50, 25, 10 ng) or buffer alone (0 ng). RNAs were analyzed by primer extension using HIV-1-specific radiolabeled reverse primer complementary to region 8648–8668 nt, numbered according to the first nucleotide of the 5′ U3 region of the proviral DNA (HXB2 genome, accession no. K03455.1). No RNA indicates radiolabeled reverse primer without extension template. No pMenv(-) indicates extension from RNAs of cells without transfection of pMenv(-). cDNA products were separated through 7 M urea/8% acrylamide gel and visualized using a PhosphorImager. Manual dideoxynucleotide sequencing was performed on pMenv(-) proviral clone using the same reverse primer. A8597 and G8594, potential depurination sites in the rev ORF. (B) Schematic representation of the 3′ portion of the HIV-1 genome. Open reading frames are indicated as rectangles and the two depurinated nucleotides within rev mRNA are indicated below the nucleotide scale bar. (C) Rev in vitro transcript (5 μg) was incubated with or without purified PAP (25 ng) and extracted from PAP following incubation. The treated RNA was template for in vitro translation in rabbit reticulocyte lysate containing [35S]methionine, and the levels of Rev protein synthesized were measured by trichloroacetic acid precipitation and scintillation counting. The percent incorporation values were normalized to a no-RNA control reaction with the incorporation of the −PAP sample set to 100%. Error bar ± SE for three independent experiments.
Depurinated rev mRNA is translated inefficiently
Given our previous results showing that elongating ribosomes stall at depurinated nucleotides (Gandhi et al. 2008), we hypothesized that depurinated rev mRNA would not translate well. To test this directly, rev ORF was transcribed in vitro and incubated with PAP prior to translation in a cell-free system. Incorporation of radiolabeled methionine during translation of PAP-treated rev mRNA was ∼25% that of untreated mRNA, indicating that PAP directly reduced the synthesis of Rev protein and depurinated mRNA was a poor template for translation (Fig. 7C). The inhibitory effect of PAP on rev mRNA translation supports our initial observation that Rev activity was reduced in PAP-expressing cells, even though rev mRNA accumulated.
DISCUSSION
Our goal was to investigate the post-transcriptional effects of PAP on HIV-1. We transfected 293T cells with a proviral clone of HIV-1 and show that PAP expression altered the splicing ratio of cytosolic HIV-1 mRNAs. Specifically, PAP increased the amount of multiply spliced 2-kb mRNAs relative to unspliced 9-kb and singly spliced 4-kb mRNAs. This accumulation of multiply spliced mRNAs was due to a PAP-dependent decrease in Rev activity. We correlate this decreased activity with depurination of the rev ORF, which inhibited translation of the protein. Depurination of viral RNAs by PAP in mammalian cells presents an interesting model for the mechanism of antiviral activity of PAP. The creation of abasic sites in viral RNAs results in templates with intact sugar phosphate backbones that would be substrates for translation. However, our previous investigations showed that depurinated viral mRNAs stalled elongating ribosomes during translation (Gandhi et al. 2008). The current decline in translational efficiency of rev mRNA treated with PAP agrees with our previous mechanistic study. By decreasing Rev levels, PAP indirectly inhibited the release of full-length genomic RNA into the cytoplasm for packaging and the synthesis of encoded proteins required for particle production.
In its role of facilitating the transport of 9- and 4-kb HIV-1 RNAs from the nucleus to the cytoplasm, Rev is essential for balancing the products of viral gene expression. In addition, Rev promotes the translation of RRE-containing viral messages. Cotransfection of HeLa cells constitutively expressing Tat with Rev and a gag reporter construct increased the amount of Gag protein relative to cells without Rev. This enhanced translation of Gag was only observed with constructs containing a RRE, suggesting that the Rev–RRE interaction was required for increased translation (D'Agostino et al. 1992). A subsequent study using a recombinant vaccinia virus system to synthesize env mRNA directly in the cytoplasm, thereby excluding nucleo-cytoplasmic transport effects, also resulted in increased Env levels in the presence of Rev (Perales et al. 2005). Therefore, Rev regulates the processing and translation of viral RNAs, and any decline in its activity would result in substantial impact on virus production.
Given the importance of Rev as a regulatory protein, cellular factors affecting Rev have been targeted for development of new antiviral agents. For example, the eukaryotic translation initiation factor 5A (eIF-5A) is a Rev cofactor, needed for nuclear export of viral mRNAs. Drugs that block the activity of enzymes required for the hypusine modification of eIF5A have been described (Hauber et al. 2005; Hoque et al. 2009). The host factor Sam68 also promotes Rev activity by functioning synergistically with Rev to increase the association of viral RNA with the cellular translation machinery. A C-terminal deletion mutant of Sam68 altered the cytoplasmic localization of RRE-containing RNA to the nuclear periphery, preventing translation of viral RNA following nuclear export (Soros et al. 2001). Recently, the drug digoxin has been shown to decrease Rev synthesis due to its effect on host factors required for splicing of 2-kb HIV-1 RNAs (Wong et al. 2013). Targeting host cell factors instead of viral proteins may provide antiviral activity against all strains of HIV-1; however, disruption of host factors essential to the cell may also have the potential for toxicity.
As with any anti-HIV-1 drug, there are questions about side effects and this is also true for PAP, a ribosome inactivating protein that is well known to depurinate ribosomal RNA (Dallal and Irvin 1978) in addition to viral targets. PAP expression in 293T cells results in 17% depurination of rRNA (Chan Tung et al. 2008), and consistent with previous results, metabolic labeling of cells showed that this level of depurination is not sufficient to decrease the cellular translation rate (Mansouri et al. 2009). We appreciate that our current study was conducted in a cell line and our future work needs to be confirmed in primary cells over a longer time frame; however, we and others have shown previously that expression or application of PAP is not toxic to cells more relevant to HIV-1 infection, such as MT-2, a lymphoblastoid T cell line, and CD4+ T cells (Zarling et al. 1990; Mansouri et al. 2012). Therefore, we suggest that changes in HIV-1 translation would not be due to a ribotoxic effect of PAP, but rather due to a more specific targeting of viral translation caused by damage to viral RNA. Translation inhibition can be achieved through recruitment of RNAs to nontranslating RNPs, such as processing bodies, which are distinct cytoplasmic foci involved in storage and/or degradation of targeted RNAs (Parker and Sheth 2007). This was not the case for the accumulated 2-kb mRNAs in the presence of PAP; there was no change in the allocation of HIV-1 mRNAs along the polysome profile, with the accumulated mRNAs still associating with polysomes. Therefore, it is not likely that the apparent Rev deficiency was due to the recruitment of 2-kb mRNAs away from ribosomes. Moreover, there did not seem to be accumulation of RNAs in fractions associated with 40S ribosomal subunits or monosomes, suggesting that translation of the 2-kb mRNAs was not inhibited during small subunit scanning or initiation.
Our model for the antiviral activity of PAP is that depurination of viral RNA inhibits its translation, resulting in decreased viral protein levels. We show that PAP, expressed in 293T cells, removes an adenine and a guanine base from the rev ORF. We are currently analyzing how PAP targets viral RNAs and whether particular sequence or conformation of RNA, or presence of host factors, is required. The consistent feature of PAP's enzyme activity is that purines, rather than pyrimidines, are hydrolyzed from RNA. The investigation of PAP as a potential antiviral agent must consider the possibility of the evolution of a resistant strain. The hypermutagenic nature of HIV-1 is a concern for treatment effectiveness and is the primary reason for multi-drug therapies such as HAART (Mehellou and De Clercq 2010). The lack of proofreading capability of the HIV-1 RT results in a range of from two to 10 errors per round of replication of the viral genome in vivo (Preston et al. 1988; Ji and Loeb 1992). In addition, purine to purine (or pyrimidine to pyrimidine) mismatches occur more frequently than those of purine to pyrimidine (or vice versa), likely due to similarity in nucleotide shape (Preston et al. 1988). Other studies reported misincorporation of dAMP as the most frequent mutation produced by HIV-1 RT (Ji and Loeb 1992; Balzarini et al. 2001). This incorporation bias is favorable for maintaining and even creating additional PAP-susceptible sites by preserving purine bases. Furthermore, the misincorporation bias of the HIV-1 RT avoids purine to pyrimidine mutations required for PAP resistance. However, these scenarios do not preclude the possibility that mutations at sites not depurinated by PAP could alter mRNA shape such that certain regions would no longer bind PAP. We are testing the impact of mutation on shape of the 2-kb HIV-1 RNA and whether these mutations inhibit or change location of depurination on the mRNA. Our current results show that PAP targets the mRNA of Rev, a regulatory protein of HIV-1, and that inhibition of Rev activity is not due to cellular toxicity, which support continued investigation of PAP antiviral activity.
MATERIALS AND METHODS
Plasmid constructs, cell culture, and transfection
pMenv(-) is an envelope null mutant of HIV-1 (HXB2) (Sadaie et al. 1992) and was used as a source of proviral DNA in transfections (catalog #2089, NIH AIDS Reference and Reagent Program). The pRev(-) plasmid is a Rev-deficient clone of pMenv(-) generated by the destruction of a BamHI site within the rev ORF. The pcRev plasmid encodes the mature wild-type Rev protein cloned into pcDNA3 mammalian expression vector using KpnI and EcoRI restriction sites. The reporter construct pDM128 was used to assess Rev activity and was kindly provided by Dr. A. Cochrane and has been described previously (Hope et al. 1990). The plasmids encoding 3X-FLAG-tagged mature PAP protein, pcPAP, and the active site mutant (E176V) of PAP, pcPAPx, have been described previously (Mansouri et al. 2009). Human embryonic kidney (HEK) 293T cells were maintained in Dulbecco's Modified Eagle's Medium (DMEM) supplemented with 10% Fetal Bovine Serum (FBS), penicillin (100 U/mL), and streptomycin (100 μg/mL). Cells were grown on 10-cm treated plates in a humidified incubator at 37°C with 5% CO2. Plasmid DNA was transfected into cells (5 × 105 cells/mL) by standard calcium phosphate coprecipitation (Chen and Okayama 1988). Cells were refed 18 h after transfection and were harvested 40 h after transfection. Virus particles released from cells were isolated from 1 mL of medium 40 h after transfection. The medium was centrifuged at 5000g for 5 min at room temperature to remove cell debris, and the supernatant was centrifuged at 16,100g for 90 min at 4°C to pellet virus particles.
Polysome analysis
To determine the effects of PAP on the translational profile of HIV-1, polysome analysis was carried out. HEK 293T cells were transfected, and 40 h following transfection the growth medium was replaced with 10 mL DMEM containing 100 μg/mL cycloheximide and cells were incubated for 20 min at 37°C. Cells were then harvested in 1X PBS containing 100 μg/mL cycloheximide and 1 mM PMSF and centrifuged at 1000g for 5 min at 4°C, washed with 10 mL of ice cold 1X PBS containing 100 μg/mL cycloheximide and centrifuged again. The resulting pellets were resuspended in 0.5 mL Magnesium Buffer (10 mM HEPES-KOH, 10 mM NaCl, 7 mM MgCl2, 3 mM CaCl2, 1 mM DTT, 0.5% NP-40), vortexed for 10 sec and kept for 10 min on ice. Samples were centrifuged at 14,000g for 4 min at 4°C, the supernatant was combined with 1 mg of heparin and quantified at OD260nm. Equal amounts (40 OD260nm) were loaded onto 14%–47% linear sucrose gradients and centrifuged at 240,000g for 2.5 h at 4°C. The gradients were divided into 12 fractions and incubated in 1% SDS and 0.48 mM proteinase K for 30 min at 37°C. RNA was isolated from each fraction for Northern blot analysis.
Protein analyses
Pellets of harvested cells were combined with equal volumes of Lysis Buffer (25 mM HEPES-KOH at pH 7.5, 2 mM EGTA, 1 mM DTT, 10% glycerol, 1% NP-40), vortexed for 20 sec, and incubated on ice for 10 min. Samples were centrifuged at 16,100g for 10 min at 4°C and total protein of the resulting supernatant was quantified by the Bradford Assay. Cell lysates or virus particles were separated through a 12% SDS–polyacrylamide gel, transferred to nitrocellulose, blocked in 5% milk and probed with appropriate primary and secondary antibodies. PAP and PAPx were detected with an anti-FLAG monoclonal antibody (1:1,000; Sigma Aldrich). HIV-1 proteins were detected with antibodies obtained through the NIH AIDS Research and Reference Reagent Program. HIV-1 Gag was detected with an anti-p24 monoclonal antibody (1:5,000; NIH, cat # 3537); RT was detected with an anti-p51 polyclonal antibody (1:2,500; NIH, cat # 6195); and Vif was detected with an anti-Vif polyclonal antibody (1:2,500; NIH, cat # 2221). As a loading control, β-actin was detected with an anti-β-actin monoclonal antibody (1:5,000; Sigma Aldrich). Secondary antibodies were conjugated to horseradish peroxidase and proteins were detected by chemiluminescence.
To assess the rate of cellular translation during PAP expression, cells were metabolically radiolabeled by [35S]methionine incorporation. The day after transfection, cells were seeded in 6-well plates at a density of 1 × 106 and incubated overnight at 37°C with 5% CO2. On the following day, growth medium was replaced with 2 mL of minimum essential medium (MEM) supplemented with 10% FBS and incubated for 30 min at 37°C. Following incubation, the medium was replaced with 1 mL MEM containing 12.5 μCi of [35S]methionine (Perkin Elmer). At this time (40 h post-transfection), the negative control sample was also supplemented with 25 μg/mL cycloheximide. Cells were harvested at the indicated time points (30, 60, 120, 240, and 360 min). RIPA Buffer (60 μL; 150 mM NaCl, 10 mM HEPES-KOH at pH 7.4, 1% Triton-X100, 0.1% SDS), supplemented with 1X complete protease inhibitor cocktail tablets (Roche), was added to cell pellets, vortexed for 20 sec, and incubated on ice for 10 min. Proteins were precipitated in trichloroacetic acid and collected on glass microfiber filters. The incorporated radioactivity was quantified using a scintillation counter.
To determine the direct effect of PAP on viral RNA translation, rev transcript was incubated with purified PAP and translated in a cell-free system. The rev open reading frame was subcloned into pET28a at NcoI and EcoRI sites and the resulting pET-Rev plasmid was linearized with EcoRI. Run-off in vitro transcripts (5 μg) were incubated with or without 25 ng purified PAP in 1X ribosome inactivating protein buffer (60 mM KCl, 10 mM Tris-HCl at pH 7.4, 10 mM MgCl2) for 30 min at 30°C. Reactions were extracted with phenol/chloroform/isoamyl alcohol and ethanol precipitated. The recovered transcripts were supplied as template for in vitro translation with rabbit reticulocyte lysate (Promega) in the presence of [35S]methionine for 90 min at 30°C. Protein product was precipitated with trichloroacetic acid and filtered through glass fiber filters. 35S incorporation was measured by scintillation counting.
RNA analyses
Harvested cell pellets were lysed with an equal volume of Lysis Buffer (25 mM HEPES-KOH at pH 7.5, 2 mM EGTA, 1 mM DTT, 10% glycerol, 1% NP-40) and centrifuged at 16,100g for 10 min at 4°C to pellet cellular debris. Total RNA was extracted with TRI Reagent (Molecular Research Center) following the manufacturer's instructions and precipitated in ethanol. Northern blot analysis was used to detect and quantify levels of HIV-1 mRNAs. Isolated RNA samples (15 μg) were denatured at 85°C in formamide buffer (60% formamide, 20% formaldehyde, 20 mM MOPS at pH 7.0, 2 mM NaOAc, 1 mM EDTA) and separated through a 0.8% agarose denaturing gel. Following separation, RNA was transferred to a positively charged nylon membrane and probed for HIV-1 mRNAs with 1 × 106 cpm [α-33P]-radiolabeled riboprobe specific to nucleotides 8881–9060 of pMenv(-) proviral clone. To detect chloramphenicol acetyl-transferase (CAT) mRNA for Rev activity assay, the blot was probed with a radiolabeled riboprobe specific to nucleotides 220–420 of CAT mRNA. Antisense riboprobe specific for nucleotides 24–96 of 5S rRNA was used to probe the rRNA as a loading control in Northern blotting. Radiolabeled RNA was visualized and quantified with a PhosphorImager.
To isolate 2-kb mRNAs for RNase protection assay and primer extension, total RNA was separated through a 1% low-melt agarose gel. The RNA was stained with ethidium bromide and regions corresponding to 2 kb in size were cut from the gel. Gel pieces were heated for 5 min at 70°C, combined with an equal volume of 2X PK Buffer (300 mM NaCl, 25 mM EDTA, 20 mM Tris-HCl at pH 8.0, 2% SDS) and proteinase K to a final concentration of 0.25 mg/mL, and incubated for 10 min at 37°C. The RNA was extracted from the melted gel with phenol (saturated with 50 mM NaOAc at pH 5.2, 10 mM EDTA), further extracted with phenol:chloroform:isoamyl alcohol (25:24:1), and precipitated in ethanol.
The levels of 2-kb HIV-1 mRNAs were measured by RNAse protection assay using an antisense riboprobe specific to nucleotides 5845–6044 of the pMenv(-) proviral clone. GAPDH mRNA was used as a loading control and was detected with an antisense riboprobe specific for nucleotides 1–341 of pTRI-GAPDH plasmid. Isolated RNA samples (15 μg) were combined with 21 μL Hybridization Buffer (400 mM NaCl, 40 mM PIPES at pH 6.4, 1 mM EDTA, 80% formamide) containing 5 × 105 cpm [α-33P]-radiolabeled probe. Samples were incubated for 30 min at 80°C and then hybridized overnight at 50°C. Each sample was treated with 144 units of RNase T1 and 0.72 units of RNase A in 200 μL of RNase Buffer (300 mM NaCl, 10 mM EDTA, 10 mM Tris-HCl at pH 7.5) for 30 min at room temperature. Samples were then incubated with 17 μL of proteinase K:10% SDS (1:5) for 30 min at 37°C. The RNA was extracted with TRI Reagent (Molecular Research Center) and chloroform and precipitated in ethanol. RNA pellets were resuspended in formamide loading buffer and separated through a 7-M urea/6% polyacrylamide gel. An RNA ladder was visualized by ethidium bromide staining and the sizes of protected radiolabeled RNA probe were detected and quantified with a PhosphorImager.
Primer extension was used to detect depurination sites of rev mRNA by PAP. The HXB28648 (5′-CTTTAGTTCCTGACTCCAATACTGTAGGAG-3′) reverse primer, used to show a depurination site in rev ORF, annealed 51 nucleotides downstream from nucleotide A8597. Isolated 2-kb HIV-1 mRNAs (5 μg) were combined with 5 × 105 cpm of [γ-33P]-end-labeled primer, heat denatured at 90°C for 5 min, then chilled on ice. Samples were brought to 20 μL in 1X buffer (75 mM KCl, 50 mM Tris-HCl at pH 8.3, 10 mM DTT, 3 mM MgCl2, 1 mM dNTPs, 40 units of RNase inhibitor) and incubated for 10 min at room temperature. After annealing, samples were heated to 48°C and 100 units of MMLV Reverse Transcriptase enzyme (New England Biolabs) were added to each sample and incubated for 50 min. The reaction was stopped by the addition of 10 μL of formamide loading buffer and samples were separated through a 7-M urea/8% polyacrylamide gel. To identify the depurinated nucleotides, dideoxynucleotide sequencing of pMenv(-) HIV-1 plasmid was performed with the same reverse primer used for the primer extension assay. Radiolabeled cDNAs were visualized with a PhosphorImager.
ACKNOWLEDGMENTS
We thank Ms. Fatema Fatakdawala for construction of the pcRev plasmid and the pRev(-) proviral clone. We also thank Dr. A. Cochrane (University of Toronto, Canada) for the reporter construct pDM128. The following reagents were obtained through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: proviral clone pMenv(-) (catalog #2089) from Dr. Reza Sadaie; antiserum to p24 (catalog #3537) from Dr. Bruce Chesebro and Kathy Wehrly; antiserum to RT (catalog #6195) from Dr. Stuart Le Grice; antiserum to Vif (catalog #2221) from NIAID (produced by US Biological). This work was funded by a Discovery Grant from the Natural Sciences and Engineering Research Council of Canada to K.A.H.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.043141.113.
REFERENCES
- Balzarini J, Camarasa MJ, Perez-Perez MJ, San-Felix A, Velazquez S, Perno CF, De Clercq E, Anderson JN, Karlsson A 2001. Exploitation of the low fidelity of human immunodeficiency virus type 1 (HIV-1) reverse transcriptase and the nucleotide composition bias in the HIV-1 genome to alter the drug resistance development of HIV. J Virol 75: 5772–5777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broder S 2010. The development of antiretroviral therapy and its impact on the HIV/AIDS pandemic. Antiviral Res 85: 1–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan Tung KW, Mansouri S, Hudak KA 2008. Expression of pokeweed antiviral protein in mammalian cells activates c-Jun NH2-terminal kinase without causing apoptosis. Int J Biochem Cell Biol 40: 2452–2461 [DOI] [PubMed] [Google Scholar]
- Chen CA, Okayama H 1988. Calcium phosphate-mediated gene transfer: a highly efficient transfection system for stably transforming cells with plasmid DNA. Biotechniques 6: 632–638 [PubMed] [Google Scholar]
- Cullen BR 1998. Posttranscriptional regulation by the HIV-1 Rev protein. Semin Virol 8: 327–334 [Google Scholar]
- D'Agostino DM, Felber BK, Harrison JE, Pavlakis GN 1992. The Rev protein of human immunodeficiency virus type 1 promotes polysomal association and translation of gag/pol and vpu/env mRNAs. Mol Cell Biol 12: 1375–1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallal JA, Irvin JD 1978. Enzymatic inactivation of eukaryotic ribosomes by the pokeweed antiviral protein. FEBS Lett 89: 257–259 [DOI] [PubMed] [Google Scholar]
- D'Cruz OJ, Waurzyniakt B, Uckun FM 2004. A 13-week subchronic intravaginal toxicity study of pokeweed antiviral protein in mice. Phytomedicine 11: 342–351 [DOI] [PubMed] [Google Scholar]
- Doyle T, Geretti AM 2012. Low-level viraemia on HAART: significance and management. Curr Opin Infect Dis 25: 17–25 [DOI] [PubMed] [Google Scholar]
- Endo Y, Tsurugi K, Lambert JM 1988. The site of action of six different ribosome-inactivating proteins from plants on eukaryotic ribosomes: the RNA N-glycosidase activity of the proteins. Biochem Biophys Res Commun 150: 1032–1036 [DOI] [PubMed] [Google Scholar]
- Felber BK, Hadzopoulou-Cladaras M, Cladaras C, Copeland T, Pavlakis GN 1989. Rev protein of human immunodeficiency virus type 1 affects the stability and transport of the viral mRNA. Proc Natl Acad Sci 86: 1495–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flexner C 2007. HIV drug development: the next 25 years. Nat Rev Drug Discov 6: 959–966 [DOI] [PubMed] [Google Scholar]
- Gandhi R, Manzoor M, Hudak KA 2008. Depurination of Brome mosaic virus RNA3 in vivo results in translation-dependent accelerated degradation of the viral RNA. J Biol Chem 283: 32218–32228 [DOI] [PubMed] [Google Scholar]
- Hauber I, Bevec D, Heukeshoven J, Kratzer F, Horn F, Choidas A, Harrer T, Hauber J 2005. Identification of cellular deoxyhypusine synthase as a novel target for antiretroviral therapy. J Clin Invest 115: 76–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope TJ 1999. The ins and outs of HIV Rev. Arch Biochem Biophys 365: 186–191 [DOI] [PubMed] [Google Scholar]
- Hope TJ, Huang X, McDonald D, Parslow TG 1990. Steroidreceptor fusion of the human immunodeficiency virus type 1 Rev transactivator: mapping of cryptic functions of the arginine-rich motif. Proc Natl Acad Sci 87: 7787–7791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoque M, Hanauske-Abel HM, Palumbo P, Saxena D, D'Alliessi Gandolfi D, Park MH, Pe'ery T, Mathews MB 2009. Inhibition of HIV-1 gene expression by Ciclopirox and Deferiprone, drugs that prevent hypusination of eukaryotic initiation factor 5A. Retrovirology 13: 60 10.1186/1742-4690-6-90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hur Y, Hwang DJ, Zoubenko O, Coetzer C, Uckun FM, Tumer NE 1995. Isolation and characterization of pokeweed antiviral protein mutations in Saccharomyces cerevisiae: identification of residues important for toxicity. Proc Natl Acad Sci 92: 8448–8452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji JP, Loeb LA 1992. Fidelity of HIV-1 reverse transcriptase copying RNA in vitro. Biochemistry 31: 954–958 [DOI] [PubMed] [Google Scholar]
- Kang Y, Cullen BR 1999. The human Tap protein is a nuclear mRNA export factor that contains novel RNA-binding and nucleocytoplasmic transport sequences. Genes Dev 13: 1126–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karran RA, Hudak KA 2008. Depurination within the intergenic region of Brome mosaic virus RNA3 inhibits viral replication in vitro and in vivo. Nucleic Acids Res 36: 7230–7239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Byrn R, Groopman J, Baltimore D 1989. Temporal aspects of DNA and RNA synthesis during human immunodeficiency virus infection: evidence for differential gene expression. J Virol 63: 3708–3713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuritzkes DR 2011. Drug resistance in HIV-1. Curr Opin Virol 1: 582–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansouri S, Choudhary G, Sarzala PM, Ratner L, Hudak KA 2009. Suppression of human T-cell leukemia virus I gene expression by pokeweed antiviral protein. J Biol Chem 284: 31453–31462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansouri S, Kutky M, Hudak KA 2012. Pokeweed antiviral protein increases HIV-1 particle infectivity by activating the cellular mitogen activated protein kinase pathway. PLoS One 7: 363–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehellou Y, De Clercq E 2010. Twenty-six years of anti-HIV drug discovery: Where do we stand and where do we go? J Med Chem 53: 521–538 [DOI] [PubMed] [Google Scholar]
- Parker R, Sheth U 2007. P bodies and the control of mRNA translation and degradation. Mol Cell 25: 635–646 [DOI] [PubMed] [Google Scholar]
- Perales C, Carrasco L, Gonzalez ME 2005. Regulation of HIV-1 env mRNA translation by Rev protein. Biochim Biophys Acta 1743: 169–175 [DOI] [PubMed] [Google Scholar]
- Pollard VW, Malim MH 1998. The HIV-1 Rev protein. Annu Rev Microbiol 52: 491–532 [DOI] [PubMed] [Google Scholar]
- Pomerantz RJ, Trono D, Feinberg MB, Baltimore D 1990. Cells nonproductively infected with HIV-1 exhibit an aberrant pattern of viral RNA expression: a molecular model for latency. Cell 61: 1271–1276 [DOI] [PubMed] [Google Scholar]
- Pomerantz RJ, Seshamma T, Trono D 1992. Efficient replication of human immunodeficiency virus type 1 requires a threshold level of Rev: potential implications for latency. J Virol 66: 1809–1813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston BD, Poiesz BJ, Loeb LA 1988. Fidelity of HIV-1 reverse transcriptase. Science 242: 1168–1171 [DOI] [PubMed] [Google Scholar]
- Purcell DF, Martin MA 1993. Alternative splicing of human immunodeficiency virus type 1 mRNA modulates viral protein expression, replication, and infectivity. J Virol 67: 6365–6378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajamohan F, Venkatachalam TK, Irvin JD, Uckun FM 1999. Pokeweed antiviral protein isoforms PAP-I, PAP-II, and PAP-III depurinate RNA of human immunodeficiency virus (HIV)-1. Biochem Biophys Res Commun 260: 453–458 [DOI] [PubMed] [Google Scholar]
- Sadaie MR, Kalyanaraman VS, Mukopadhayaya R, Tschachler E, Gallo RC, Wong-Staal F 1992. Biological characterization of noninfectious HIV-1 particles lacking the envelope protein. Virology 187: 604–611 [DOI] [PubMed] [Google Scholar]
- Schwartz S, Felber BK, Pavlakis GN 1992. Distinct RNA sequences in the gag region of human immunodeficiency virus type 1 decrease RNA stability and inhibit expression in the absence of Rev protein. J Virol 66: 150–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sodroski J, Goh WC, Rosen CA, Dayton A, Terwilliger E, Haseltine WA 1986. A second post-transcriptional activator gene required for HTLV-III replication. Nature 321: 412–417 [DOI] [PubMed] [Google Scholar]
- Soros VB, Carvajal HV, Richard S, Cochrane AW 2001. Inhibition of human immunodeficiency virus type 1 Rev function by a dominant-negative mutant of Sam68 through sequestration of unspliced RNA at perinuclear bundles. J Virol 75: 8203–8215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang MW, Shafer RW 2012. HIV-1 antiretroviral resistance: scientific principles and clinical applications. Drugs 72: e1–e25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uckun FM, Chelstrom LM, Tuel-Ahlgren L, Dibirdik I, Irvin JD, Langlie MC, Myers DE 1998. TXU (anti-CD7)-pokeweed antiviral protein as a potent inhibitor of human immunodeficiency virus. Antimicrob Agents Chemother 42: 383–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uckun FM, Bellomy K, O'Neill K, Messinger Y, Johnson T, Chen CL 1999. Toxicity, biological activity, and pharmacokinetics of TXU (anti-CD7)-pokeweed antiviral protein in chimpanzees and adult patients infected with human immunodeficiency virus. J Pharmacol Exp Ther 291: 1301–1307 [PubMed] [Google Scholar]
- Wong RW, Balachandran A, Ostrowski MA, Cochrane A 2013. Digoxin suppresses HIV-1 replication by altering viral RNA processing. PLoS Pathog 9: e1003241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarling JM, Moran PA, Haffar O, Sias J, Richman DD, Spina CA, Myers E, Kuebelbeck V, Ledbetter JA, Uckun FM 1990. Inhibition of HIV replication by pokeweed antiviral protein targeted to CD4+ cells by monoclonal antibodies. Nature 347: 92–95 [DOI] [PubMed] [Google Scholar]
- Zhang G, Zapp ML, Yan G, Green MR 1996. Localization of HIV-1 RNA in mammalian nuclei. J Cell Biol 135: 9–18 [DOI] [PMC free article] [PubMed] [Google Scholar]


