This article describes the identification of two new eIF4E family members in Trypanosomes. Analysis of eIF4E5 showed that it interacts with two of the five eIF4 homologs and its knockdown resulted in loss of productive cell movement.
Keywords: 14-3-3 protein, kinetoplastid, mRNA cap, social motility, translation initiation factor
Abstract
Members of the eIF4E mRNA cap-binding family are involved in translation and the modulation of transcript availability in other systems as part of a three-component complex including eIF4G and eIF4A. The kinetoplastids possess four described eIF4E and five eIF4G homologs. We have identified two new eIF4E family proteins in Trypanosoma brucei, and define distinct complexes associated with the fifth member, TbEIF4E5. The cytosolic TbEIF4E5 protein binds cap 0 in vitro. TbEIF4E5 was found in association with two of the five TbEIF4Gs. TbIF4EG1 bound TbEIF4E5, a 47.5-kDa protein with two RNA-binding domains, and either the regulatory protein 14-3-3 II or a 117.5-kDa protein with guanylyltransferase and methyltransferase domains in a potentially dynamic interaction. The TbEIF4G2/TbEIF4E5 complex was associated with a 17.9-kDa hypothetical protein and both 14-3-3 variants I and II. Knockdown of TbEIF4E5 resulted in the loss of productive cell movement, as evidenced by the inability of the cells to remain in suspension in liquid culture and the loss of social motility on semisolid plating medium, as well as a minor reduction of translation. Cells appeared lethargic, as opposed to compromised in flagellar function per se. The minimal use of transcriptional control in kinetoplastids requires these organisms to implement downstream mechanisms to regulate gene expression, and the TbEIF4E5/TbEIF4G1/117.5-kDa complex in particular may be a key player in that process. We suggest that a pathway involved in cell motility is affected, directly or indirectly, by one of the TbEIF4E5 complexes.
INTRODUCTION
Trypanosoma brucei is a kinetoplastid protozoan belonging to the Excavata, a group that contains free-living and parasitic representatives (Adl et al. 2012). During the digenetic lifecycle, T. brucei is commonly found in the intestinal tract and salivary glands of tsetse flies and in the blood and cerebrospinal fluid of mammalian hosts, in which the parasite causes the tropical diseases human African trypanosomiasis and nagana in cattle. Transmission of the parasite occurs through the bite of the hematophagous tsetse fly vector, which harbors distinct intestinal and salivary forms. The kinetoplastid protozoa share unusual features of gene expression. Most protein-coding genes are arranged in directional gene clusters that are transcribed polycistronically (Alsford et al. 2012). Mature mRNAs possessing 5′ caps and 3′ poly(A) tails are generated by trans-splicing, which is linked mechanistically to polyadenylation (Liang et al. 2003; Preußer et al. 2012). Regulation of transcription initiation by RNA polymerase II is minimal (Martínez-Calvillo et al. 2010; Günzl 2012). The control of protein abundance and function during the parasite lifecycle is mediated predominantly through post-transcriptional mechanisms such as mRNA stability and availability, translational efficiency, post-translational modification, and protein stability (Clayton and Shapira 2007; Fernandez-Moya and Estevez 2010; Kramer 2012).
A key step in post-transcriptional control is the regulation of translation initiation by the tripartite mRNA-binding complex eIF4F (Topisirovic et al. 2011). The core eIF4F complex consists of three proteins: eIF4E, an mRNA m7G cap-binding protein; eIF4G, a large scaffold protein interacting directly with both eIF4E and the poly(A)-binding protein PABP; and eIF4A, a helicase linked to eIF4G that unwinds RNA secondary structure between the 5′ cap and the initiation codon (Gingras et al. 1999). Binding of eIF4F to the mRNA enables recruitment of multiple other translation initiation factors and the two ribosomal subunits to the initiation codon (Aitken and Lorsch 2012). eIF4E function represents a central control point in translational regulation (Jackson et al. 2010). The ability to bind eIF4G can be negated by 4E-binding proteins (Raught and Gingras 1999; Richter and Sonenberg 2005), and binding activity is implicated in the selective translation of RNA subgroups designated “regulons” (Culjkovic et al. 2007; Keene 2007). The existence of coordinated mRNA regulons is supported in the kinetoplastid protozoa (Ouellette and Papadopoulou 2009; Queiroz et al. 2009; Das et al. 2012; De Gaudenzi et al. 2013), however their mechanism of cap-mediated translation regulation remains a mystery.
The complexity of eIF4F components varies among organisms, including the protists (Joshi et al. 2005; Jagus et al. 2012). The yeast Saccharomyces cerevisiae possesses a single eIF4E and two eIF4Gs with distinct functions (Prévôt et al. 2003; Clarkson et al. 2010); humans have four eIF4E isoforms and two eIF4Gs (Prévôt et al. 2003; Joshi et al. 2004). The nematode Caenorhabditis elegans, in contrast, possesses five eIF4E variants (Keiper et al. 2000) that displays distinct preferences for m7G and m2,2,7G cap structures (Jankowska-Anyszka et al. 1998), and a single eIF4G gene that gives rise to two isoforms (Contreras et al. 2008). Variants of eIF4E may show developmental regulation. Drosophila melanogaster has eight isoforms that show distinct patterns of expression during embryogenesis (Hernández et al. 2005). Distinct variant eIF4E–eIF4G interactions are thus expected and observed (Ptushkina et al. 2001). Not all homologs of eIF4E have a role in constitutive translation initiation (Rhoads 2009); they may function as competitive inhibitors of eIF4G recruitment and as scaffolds for interactions with other potential regulatory proteins (Groppo and Richter 2009; Blewett and Goldstrohm 2012; Gosselin et al. 2013). Some eIF4E-binding proteins like 4E-BP repress translation by inhibiting eIF4F formation, whereas other eIF4E-binding proteins, such as Cup and Maskin, use alternative 3′ UTR–protein interactions for selective repression of translation (Groppo and Richter 2009). Further, selective translation of mRNAs can occur via cap binding of an eIF4E homologous protein, 4E-HP (Cho et al. 2005) and discrete protein–3′ UTR interactions (Lasko et al. 2005).
The related pathogens T. brucei and Leishmania spp. have four eIF4E homologs (EIF4E) and five eIF4G homologs (EIF4G) (Dhalia et al. 2005; Freire et al. 2011; Zinoviev and Shapira 2012). In some cases, the respective interacting partners have been identified: EIF4E3 binds to EIF4G4, and EIF4E4 binds to EIF4G3 (Yoffe et al. 2009; Freire et al. 2011; Zinoviev et al. 2012), whereas EIF4E1 appears to lack an EIF4G partner (Zinoviev et al. 2011). The kinetoplastid mRNA cap, termed cap 4, is a complex 5-nt structure comprised of three base methylations and four ribose methylations (Bangs et al. 1992). The Leishmania EIF4E variants display differing affinities for synthetic cap analogs in vitro (Yoffe et al. 2006), suggesting differential roles in cap recognition. EIF4E4 is the best candidate for the workhorse translation initiation factor (Yoffe et al. 2009; Zinoviev et al. 2012).
The functions of the extended family of EIF4E proteins in kinetoplastids remain obscure. Here we extend the T. brucei EIF4E family with the identification of the fifth and sixth members, designated TbEIF4E5 and TbEIF4E6, and characterize the properties and macromolecular composition of TbEIF4E5 (or TbE5). We demonstrate that TbE5 binds to mRNA caps in vitro, is cytosolic, and associates with multiprotein complexes including either the TbEIF4G1 or TbEIF4G2 proteins (also called TbG1 or TbG2, respectively). The TbE5:TbG1 complex contains one protein with two RNA-binding domains and another with both guanylyltransferase and methyltransferase motifs, implicating the complex in cap modification of specific transcripts. The TbE5:TbG2 complex is distinguished by the presence of both 14-3-3 isoforms (Inoue et al. 2005; Benz et al. 2010), hetero- and homodimer forming proteins whose functions are determined by interactions with phosphorylated serine or threonine residues (Mackintosh 2004). The manifestation of a specific phenotype upon knockdown of the TbE5 protein is suggestive of a role for post-transcriptional gene regulation of at least one cellular pathway by a TbE5-containing complex.
RESULTS
Two new members of the kinetoplastid eIF4E-homolog family
In pursuit of our longstanding interest in SL RNA cap function in kinetoplastid gene expression, a search was conducted for potential cap-binding proteins using known entities from other model systems. The eIF4E protein that recognizes the cap 0 structure as a first step in mRNA translation fits this description, and searches using the yeast eIF4E sequence yielded a set of five potential eIF4E family members that are present in T. brucei (Tb4EIF4E), T. cruzi, and Leishmania spp. Of this set, four have been identified and studied to varying degrees by other groups (Yoffe et al. 2004, 2006, 2009; Dhalia et al. 2005; Freire et al. 2011). A sixth homolog was identified by reciprocal BLAST with the uncharacterized TbEIF4E sequence. Here we designate the additional two putative members as TbEIF4E5 (Tb927.10.5020) and TbEIF4E6 (Tb927.7.1670) and present the molecular characterization of TbEIF4E5, a.k.a. TbE5.
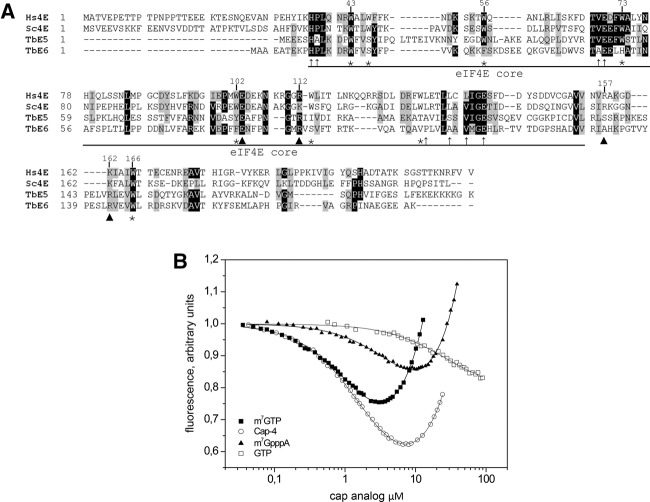
The TbE5 and TbEIF4E6 genes produce the two smallest members of the Tb4EIF4E family at estimated molecular weights of 21.9 and 20.8 kDa, respectively (Fig. 1A). Both proteins carry features conserved generally in eIF4Es, including the eIF4E “core” and amino acids suggestive of RNA cap 0 binding as indicated. Key tryptophan residues are conserved in the trypanosome proteins. The tryptophan at position 73, relative to the human eIF4E-1 numbering, that is key for eIF4G interaction is found in TbE5 and shows a substitution by histidine in TbEIF4E6. Tryptophans at positions 56 and 102 of the human protein act in cap binding through stacking interactions with m7G of the cap structure. Position 56 is conserved in TbE5, while TbEIF4E6 carries a conservative substitution with phenylalanine. Both T. brucei homologs show conservative substitutions to either tyrosine or phenylalanine at position 102. Position 166, which in the human protein is implicated in m7G recognition, shows absolute conservation in the kinetoplastid proteins, as does the tryptophan at position 43. Phylogenetic and BLAST analyses suggest that TbE5 and TbEIF4E6 are more closely related to each other than to other TbEIF4Es (data not shown) and may thus represent a distinct category within this RNA cap-binding protein family.
FIGURE 1.
TbEIF4E5 and TbEIF4E6 are shorter than the eIF4E homologs from yeast and human. (A) Alignment of the sequences was performed using Clustal W. Identical amino acids are indicated by black shading. Amino acids defined as similar, by the BLOSUM 62 Matrix, in >60% of the sequences are shaded gray. Dashes represent spaces that were inserted to allow better alignment. Asterisks represent tryptophan residues conserved in the eIF4E protein family. Arrowheads indicate nontryptophan residues required for the interaction with the cap structure: E103 hydrogen bonded with guanine; and, basic residues at positions 112, 157, and 162 with the phosphate bridge (Marcotrigiano et al. 1997). Thin arrows indicate conserved nontryptophan residues shown to be involved in eIF4G binding (Marcotrigiano et al. 1999). GenBank Accession numbers: Hs (human) eIF4E-1, NP_001959; Sc (yeast) eIF4E, NP_014502. (B) In vitro cap-binding ability of recombinant TbE5. The fluorescence-titration curves with four cap analogs were determined by fluorescence-binding assays. The protein fluorescence was excited at 280 nm and observed at 340 nm. The WT mRNA cap in trypanosomes was represented by hypermethylated cap 4, while the typical eukaryotic cap structure was represented by both the m7GTP and m7GpppA cap 0 structures. The nonmethylated GTP served as a negative control for cap 0-specific binding.
TbEIF4E5 binds cap analogs in vitro
The conservation of aromatic amino acids corresponding to positions 56, 102, and 166 suggests that TbE5 should be capable of binding mRNA caps. Basic arginine residues at positions 112 and 162 could interact with the phosphate chain of the cap via hydrogen bonds. Recombinant TbE5 protein was tested in vitro for binding ability to various cap analogs using fluorescence assays (Fig. 1B). TbE5 bound better to the cap 4 analog corresponding to the structure found on all kinetoplastid nuclear mRNAs (Kas = 0.55 ± 0.01 µM−1) and the m7GTP analog (Kas = 0.65 ± 0.01 µM−1) than to m7GpppA (Kas = 0.12 ± 0.01 µM−1) or GTP (Kas = ∼0.037 µM−1). TbE5 bound the trypanosome-specific substrate 47-fold less than the mouse eIF4E (Yoffe et al. 2006). The association constant for TbE5 with cap 4 was within the range observed for Leishmania major EIF4Es (Yoffe et al. 2006), falling between LeishIF4E-2 and LeishIF4E-4. Generally, TbE5 and all Leishmania EIF4Es show a preference for cap analogs with an N7-methylated guanosine. The energetic gain from methylation of the guanine ring for TbE5 is similar by ▵▵GO of about –1.6 kcal/mol. The presence of additional nucleotides that simulate an mRNA 5′ end showed decreased binding relative to the m7GTP substrate. In all instances, including mouse, the cap 4 substrate was bound better than the cap 0 analog, probably due to the stabilization effect of the phosphate group in the oligonucleotide.
Due to the virtual absence of transcriptional control in kinetoplastids, downstream methods for the modulation of gene expression are of particular interest. The divergent TbE5 protein represents a potential player in the modulation of post-transcriptional control events in the parasite.
TbEIF4E5 interacts directly with two of the five TbEIF4Gs
A hallmark characteristic of the eIF4E proteins is their association with eIF4Gs. In T. brucei five EIF4G family members have been described, thus the potential for specific interactions among the EIF4E and EIF4G homologs could yield a range of combinatorial arrangements leading to an array of biological consequences. To determine if any of these interactions are favored by TbE5 we performed an exhaustive interaction study between the prospective players.
The yeast two-hybrid assay detects interactions between two proteins, termed “bait” and “prey,” as indicated by the activation of a promoter in the yeast cells that permits growth (Fig. 2). The reciprocal interactions were tested, with compatible results (data not shown). The assays revealed either large yeast colonies indicating that TbE5 associated strongly with TbEIF4G2 (Tb09.160.3980; TbG2) or smaller colonies indicating a weaker interaction with TbEIF4G1 (Tb927.5.1490; TbG1). In the yeast two-hybrid assay, binding between TbEIF4E and TbEIF4G proteins in the presence of 5.0 mM 3AT indicates a strong interaction, to the exclusion of other weaker TbEIF4E–TbEIF4G pairings.
FIGURE 2.
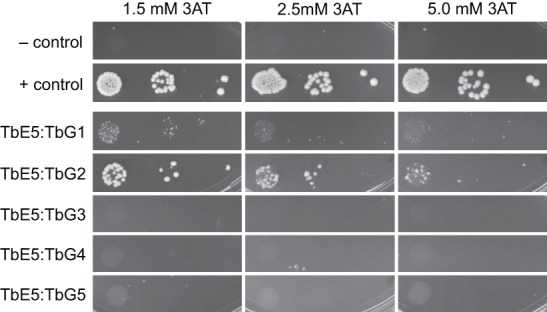
Direct interaction of TbEIF4E5 with two of the five T. brucei EIF4G homologs. Interactions between TbE5 and the five TbEIF4G homologs were challenged using the yeast two-hybrid assay in the presence of increasing amounts of 3AT, increasing the stringency of the assay. Positive controls were pGADT7-T and pGBKT7-53; the negative controls were the empty vectors. Interaction strength is inferred by colony size: ≥2 mm = strong; 1–2 mm = moderate; ≤1 mm = weak.
The interaction of TbE5 with more than one TbEIF4G may be an indication of specific functions associated with either TbG1 or TbG2 pairings. With the exception of the site of EIF4E:EIF4G interaction in Leishmania (Yoffe et al. 2009), no functionality can be predicted for any of the five TbEIF4G sequences outside the core family-defining MIF4G domain (Dhalia et al. 2005). TbG1 and TbG2 have not been observed to interact with other trypanosomatid EIF4E homologs to date. They are the two largest members of the TbEIF4G family and may serve as a scaffold for binding of additional proteins, as observed in humans and yeast (Pestova and Hellen 2003; Jackson et al. 2010). These isolated interactions serve as a guide for the behaviors of proteins in T. brucei, however the incorporation of other proteins or RNAs into the equation may alter the actual complex components.
TbEIF4E5 is present in high molecular weight complexes
The typical eIF4F complex contains three components: eIF4E, eIF4G, and eIF4A (Gingras et al. 1999). Our yeast two-hybrid interaction assays indicate TbE5 associates with two specific TbEIF4G homologs, TbG1 and TbG2. The interaction of TbE5 and the two TbEIF4G subunits observed in yeast was examined in T. brucei first by visualizing the size of complexes containing PTP-tagged proteins by Blue Native gel electrophoresis, followed by MudPIT analysis of tandem-affinity purified complexes.
To validate that TbE5 was in a high molecular weight complex(es), extracts of TbE5−/PTP were resolved by Blue Native gel analysis and visualized by Western blotting using anti-protein A antibody. Two bands of ∼100 kDa (migrating between the 66 and 146 kDa markers) and ∼900 kDa (migrating between the 720 and 1048 kDa markers) were detected in the TbE5 extracts (Fig. 3A), indicating complexes of substantially greater size than predicted for free TbE5 (22 kDa + 19 kDa for the PTP tag), which was either undetectable or not running true to predicted size. The proteins implicated by yeast two-hybrid analysis, TbG1 and TbG2, were PTP-tagged as well to allow reciprocal comparisons. For both TbG1−/PTP and TbG2−/PTP lines multiple discrete and smeared higher molecular weight bands were seen, suggestive of their involvement in multiple distinct complexes, some of which may contain TbE5. The predicted sizes of TbG1 and TbG2 are 122.1 kDa and 97.3 kDa, respectively; thus adding the contribution of the PTP tags, the bottom bands in each lane likely represent the migration of free proteins. The size markers may not represent true complex sizes, as the shape of the complex will play a major role in the migration of species through this nondenaturing gel system. Both of the TbEIF4G proteins show more complex patterns than TbE5.
FIGURE 3.
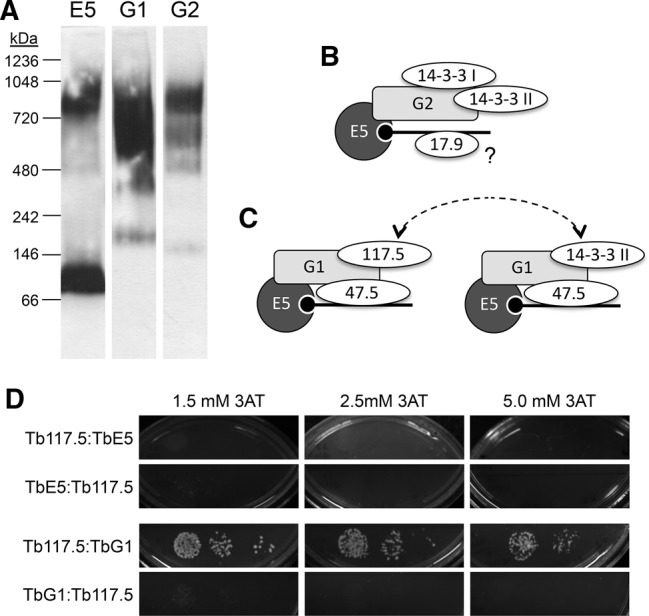
TbEIF4E5 is present in high molecular weight complexes. (A) Blue Native gel electrophoresis of cell extracts from transfected T. brucei containing PTP-tagged proteins. Lysates were run through Blue Native gels, transferred to nitrocellulose membranes and probed with antibody directed against the protein A domain of the PTP tag (TbE5, TbG1, TbG2 = E5, G1, G2, respectively). The migration of size standards is indicated in kDa. (B) The components that co-purified with TbEIF4E5 and TbEIF4G2. The RNA (black line; black circle represents 5′ end cap 4) is recognized by the cap-binding eIF4E component TbE5 (dark gray circle, E5). The eIF4G-like scaffold protein TbG2 (light gray rectangle, G2) interacts directly with TbE5, as indicated by overlap of the respective shapes. Other TbE5-associated proteins are shown as ovals. Overlap of the ovals with other shapes is speculative and based on the assumed scaffold function of TbG2, and shapes do not reflect protein size. The placement of Tb17.9 on the RNA is speculative, as indicated by the “?”. (C) The TbEIF4E5/TbEIF4G1 dynamic complex model. The 117.5-kDa protein in association with the TbG1 scaffold may be regulated by the presence of the 14-3-3 II homodimer, the binding of which is dependent on TbG1 phosphorylation. The dotted arrows highlight the either/or aspect of the complexes. (D) Yeast two-hybrid analysis of Tb117.5 with TbE5 or TbG1 in both bait and prey orientations. Controls and interpretations are shown in Figure 2.
The combined data from these experiments are consistent with the prediction that TbEIF4E/TbEIF4G form high MW complexes in vivo, and indicate the presence of additional components in the complexes formed by TbE5, TbG1, and TbG2 proteins.
TbEIF4E5 associates with two distinct sub-complexes
To isolate the specific complexes and identify their constituents, the TbE5−/PTP line was used for tandem-affinity complex purification with subsequent protein identification by MudPIT. The purification was performed four times under different salt conditions. The comparative results revealed 11 proteins that co-purified with TbE5 in all four preparations (Table 1). Based on low abundance, low numbers of peptides detected, and in some cases the identity of the protein, the bottom four proteins are considered to be contaminants.
TABLE 1.
Proteins that co-purify consistently with TbEIF4E5-PTP
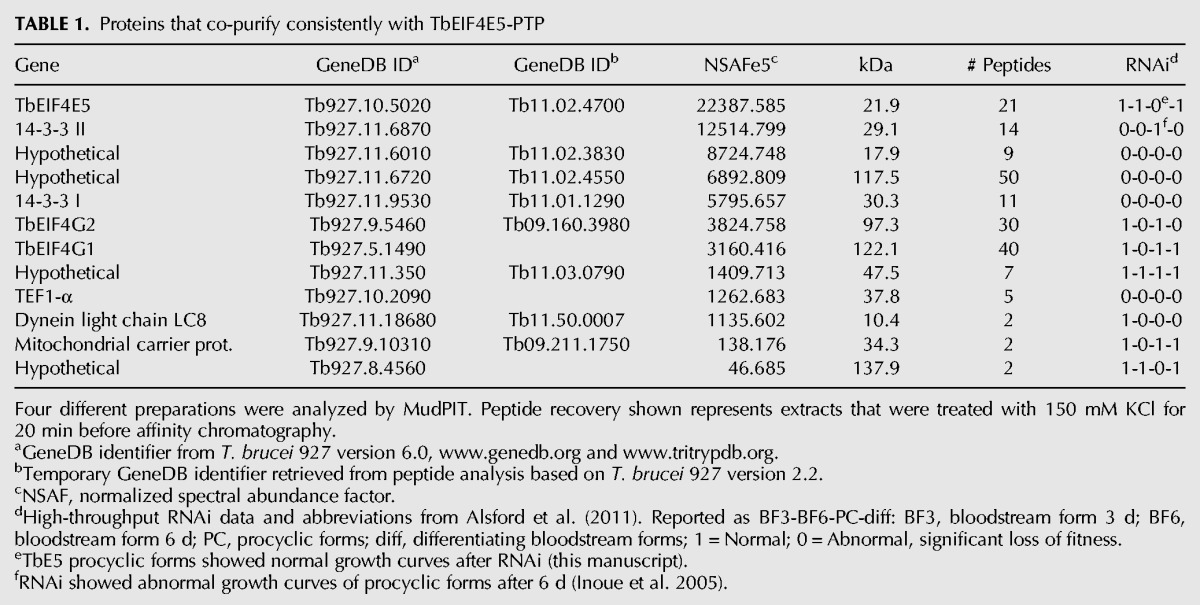
In accordance with the yeast two-hybrid assay both TbG1 and TbG2 co-purified consistently with TbE5. Additional proteins that co-purified with TbE5 complex include both isoforms of the ∼30 kDa phosphoserine/phosphothreonine-binding protein named Tb14-3-3 I and Tb14-3-3 II (Inoue et al. 2005), and proteins of 17.9, 47.5, and 117.5 kDa all designated as “hypothetical” in GeneDB.
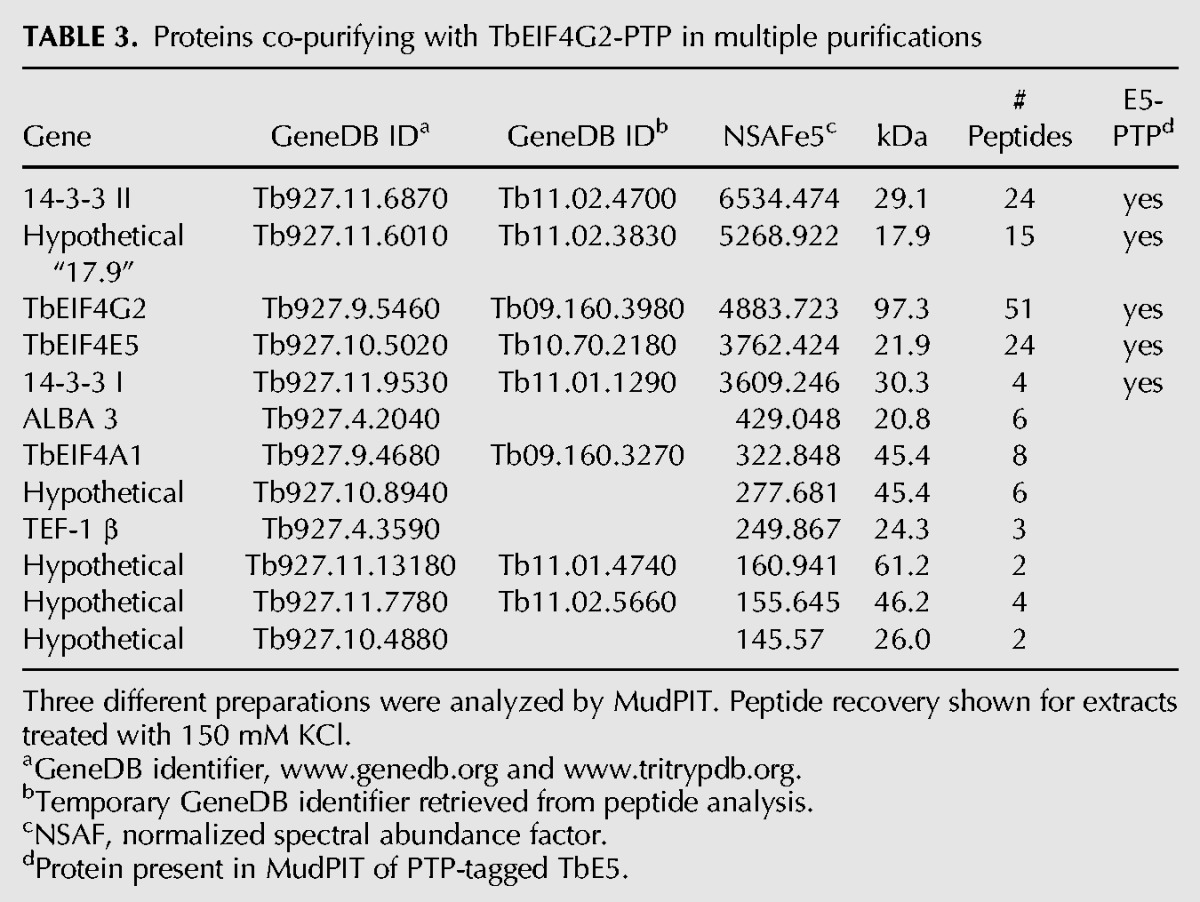
Based on the yeast two-hybrid and TbE5−/PTP MudPIT results, extracts from the TbG1−/PTP and TbG2−/PTP cell lines were subjected to tandem-affinity chromatography and subsequent MudPIT analysis. These purifications partitioned the proteins co-purifying with TbE5 into two distinct subsets. The TbG1-PTP purifications identified 14 proteins that co-purified in two separate preparations (Table 2). Of these, five were present in all TbE5-PTP purifications, supporting the existence of a discrete complex comprised of five members: TbE5, TbG1, Tb14-3-3 II, and the 47.5-kDa and 117.5-kDa proteins. Notably, the first six proteins identified other than TbG1 itself were not present in the TbE5-PTP purification; thus although they may be legitimate associates of TbG1, at this point there is no evidence to include them in the TbE5/TbG1 complex. TbG1 may participate in multiple complexes, not all of which include TbE5, consistent with the wider range of complexes visible for TbG1 in the Blue Native gel analysis (see Fig. 3A). Three tandem-affinity preparations of TbG2-PTP yielded a set of 13 proteins, of which five were present in all TbE5-PTP purifications (Table 3). The second TbE5 complex was thus defined by the presence of TbG2 along with TbE5, Tb14-3-3 I, Tb14-3-3 II, and the 17.9-kDa hypothetical protein (Fig. 3B), yielding a predicted complex mass of 196.5 kDa. In the case of TbG2, the top five identifications in terms of NSAF score are found in the TbE5 purification, and several of the remaining proteins are common to the TbG1 purification, indicating that they may function in a chaperoning role or be contaminants of EIF4G purifications. The TbEIF4A1 protein in particular is interesting, since it is a known homolog of the standard eIF4F translation initiation complex; however, the absence of this protein from the TbE5 purification essentially eliminates it from both the TbG1- and TbG2-containing complexes. Yeast two-hybrid analyses detected no interactions between Tb17.9 and either TbE5 or TbG2 (data not shown), and if this result reflects the behavior of the protein in T. brucei, at least two possibilities may remain: Tb17.9 (1) binds to the 14-3-3 I protein, or (2) interacts with an RNA recognized by the complex. Alternatively, this could be a case of the fusion proteins interfering with the assay. The presence of the PTP tag may interfere with binding of a particular protein, but in this case the TbEIF4A1 protein is predicted to interact directly with the TbEIF4G scaffold; because the PTP construction exists in the absence of the wild-type (WT) allele, PTP tag interference should have resulted in nonviable or impaired cells, and these lines behaved as WT.
TABLE 2.
Proteins co-purifying with TbEIF4G1-PTP in multiple purifications
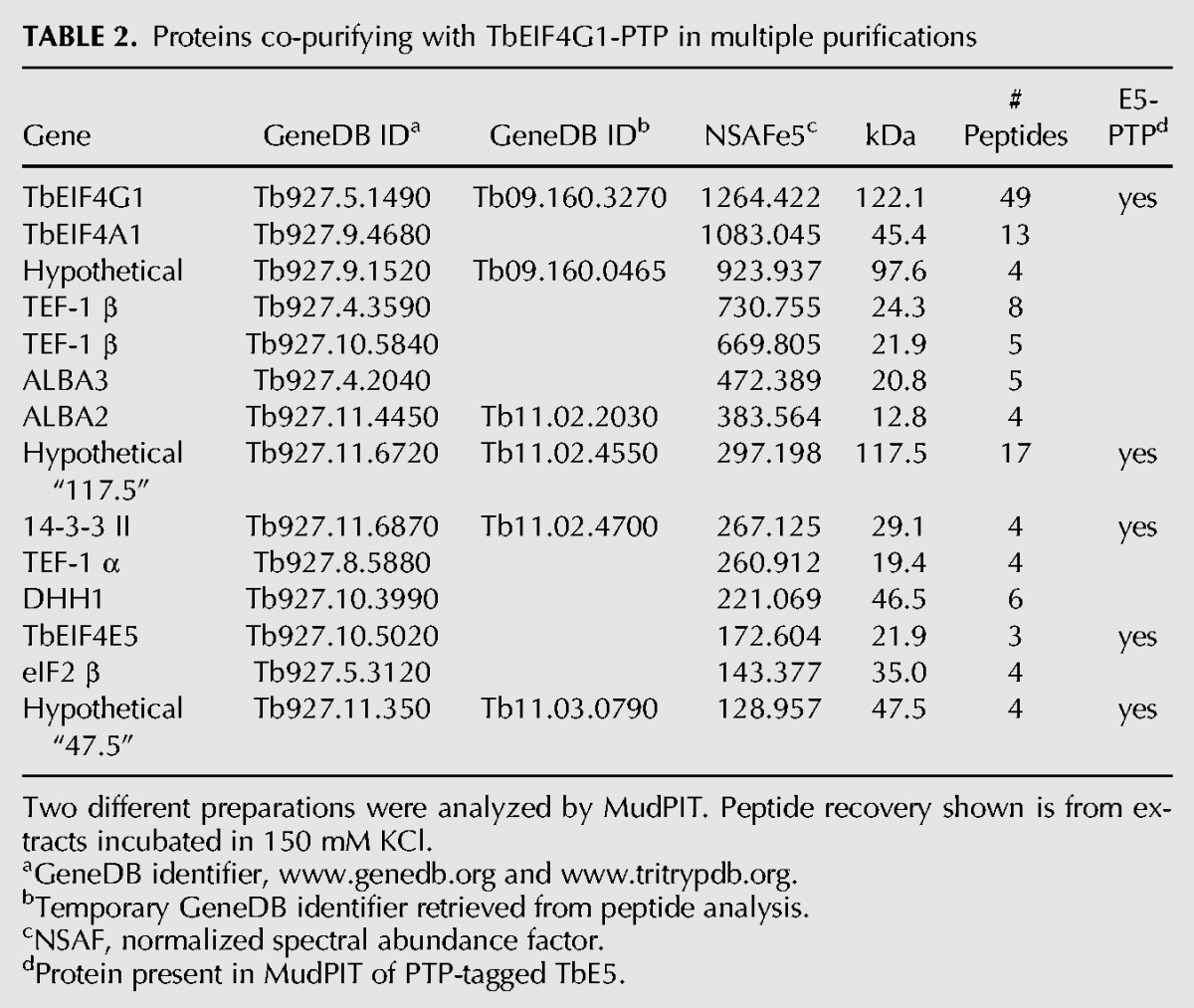
TABLE 3.
Proteins co-purifying with TbEIF4G2-PTP in multiple purifications
These results validate the unusual association of two distinct TbEIF4G proteins with a single TbEIF4E cap-binding protein. While the reciprocal scenario of multiple eIF4E proteins binding a single eIF4G scaffold has been documented (Prévôt et al. 2003; Clarkson et al. 2010), the TbE5 situation is notable. Furthermore, the associates of each complex are distinct, implying that the two complexes could perform unique regulatory functions.
Dynamic associations within the TbEIF4E5/TbEIF4G1 complex implied
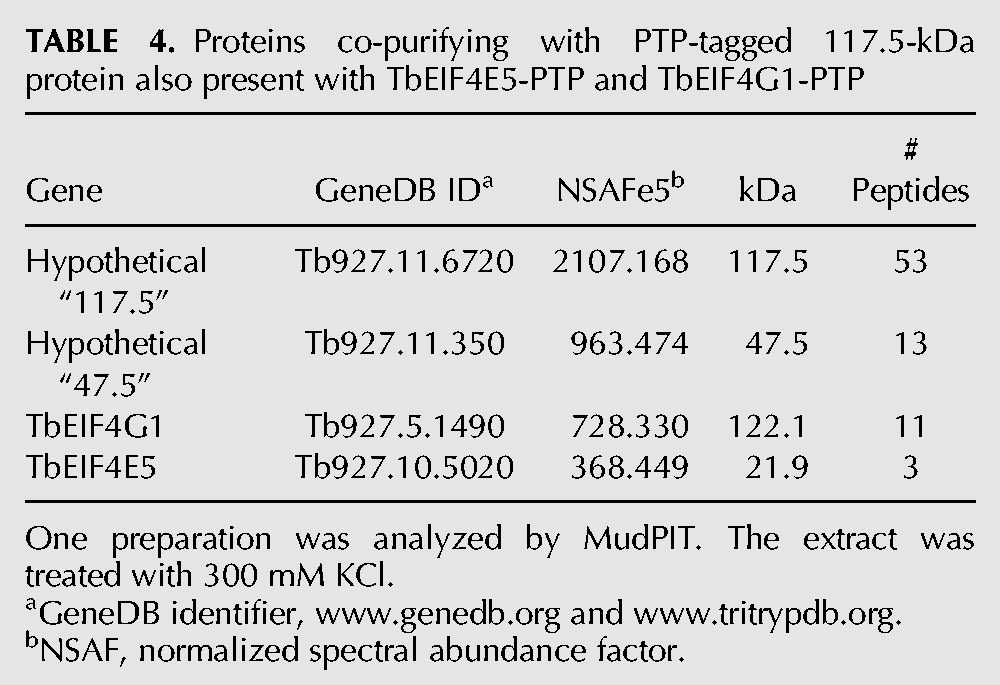
To get a better understanding of the TbE5/TbG1 complex, we performed a PTP purification using a third component. Analysis of a stringent purification using the 117.5-kDa protein tagged with PTP revealed the presence of the 47.5-kDa protein, TbG1, and TbE5 (Table 4), along with other proteins (data not shown). The 14-3-3 II protein was absent from the 117.5-kDa protein-based purification, thus 117.5-kDa protein binding may be modulated by 14-3-3 II interaction with TbG1 (Fig. 3C). Switching between a bound 14-3-3 II dimer and the 117.5-kDa protein could reflect a key dynamic within the TbE5–TbG1 complex. Alternatively, the PTP tag on the 117.5-kDa protein may impair 14-3-3 II binding to the complex. The aggregate molecular weights of these two alternatives are 309 kDa for the 117.5 kDa–protein complex and 249.7 kDa for the 14-3-3 complex, including the dimer form of 14-3-3 II. Examination of 14-3-3 II was considered; however, since the protein is known to interact in numerous cellular pathways and would thus not guarantee a definitive result, we will pursue other experimental avenues to validate this interaction model. Yeast two-hybrid analysis was consistent with direct interaction between TbG1 and the 117.5-kDa protein, with one orientation substantially stronger than the other; results were negative for TbE5–Tb117.5 interaction (Fig. 3D).
TABLE 4.
Proteins co-purifying with PTP-tagged 117.5-kDa protein also present with TbEIF4E5-PTP and TbEIF4G1-PTP
The dynamic interactions within the TbE5/TbG1 complex led to the definition of three TbE5-containing complexes, each of which may have different roles in post-transcriptional gene regulation. Association of the phosphoprotein-binding 14-3-3 components may serve to inactivate the action of the Tb117.5 proteins (see below).
TbEIF4E5 is a cytosolic protein resistant to gene knockout
The four published kinetoplastid EIF4E protein family members are either cytosolic or present in both the nucleus and cytosol (Kramer et al. 2008; Freire et al. 2011). The family member involved in the lion's share of mRNA translation should be critical for cell survival. To assess function, the TbE5 protein was localized and assayed for its importance in cell viability under normal procyclic culture conditions. The absence of strong antibodies for our target proteins and the availability of excellent epitope tagging systems prompted us to generate a cell line designated TbE5−/PTP, with one WT allele replaced by a PHLEO-resistance marker and the other allele tagged in situ by insertion of PTP epitope sequences (Schimanski et al. 2005). The robust behavior of the TbE5−/PTP line indicated that the PTP tag was not interfering with TbE5 protein function.
Immunolocalization of the TbE5 fusion protein in the TbE5−/PTP line via the PTP tag revealed a diffuse cytosolic distribution with numerous foci of greater intensity, and apparent exclusion from the nucleus (Fig. 4), similar to what was seen for homologs TbEIF4E3 and TbEIF4E4 (Kramer et al. 2008; Freire et al. 2011).
FIGURE 4.
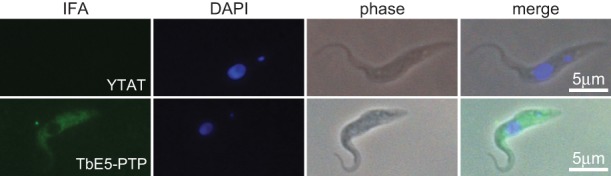
TbEIF4E5 is cytosolic in T. brucei procyclic cells. Subcellular localization was determined by indirect immunofluorescence using antibody against the protein A component of the PTP tag on the TbE5 fusion protein. Nontransfected control YTAT cells served as the negative control. Nuclear and kinetoplast DNA was visualized by counterstaining with DAPI.
In our hands, inducible double-stranded RNA interference (RNAi) directed against the TbE5 transcript in both TbE5+/+ and TbE5+/PTP RNAi procyclic cells showed no catastrophic effect on cell growth in standard growth medium (Fig. 5A). The level of epitope-tagged TbE5 was reduced to <12.5% of uninduced levels by day 2 (Fig. 5B). To provide the cells with ample time to display any effects, we extended our RNAi analysis to 15 d, well beyond our usual 7-d limit, at which point RNAi cell lines are prone to escape the RNAi constraints. The induced TbE5+/PTP RNAi cell lines continued to divide under TbE5 knockdown, albeit at a slightly reduced rate relative to the uninduced culture (data not shown). To quantitate the effect of protein knockdown on general translation, RNAi cells at day 4 and day 7 post-induction were labeled metabolically with 35S-methionine (Fig. 5C). The level of isotope incorporation was similar for both uninduced and induced cultures on day 4. By day 7, isotope incorporation in induced TbE5 cells was reduced by 16%.
FIGURE 5.
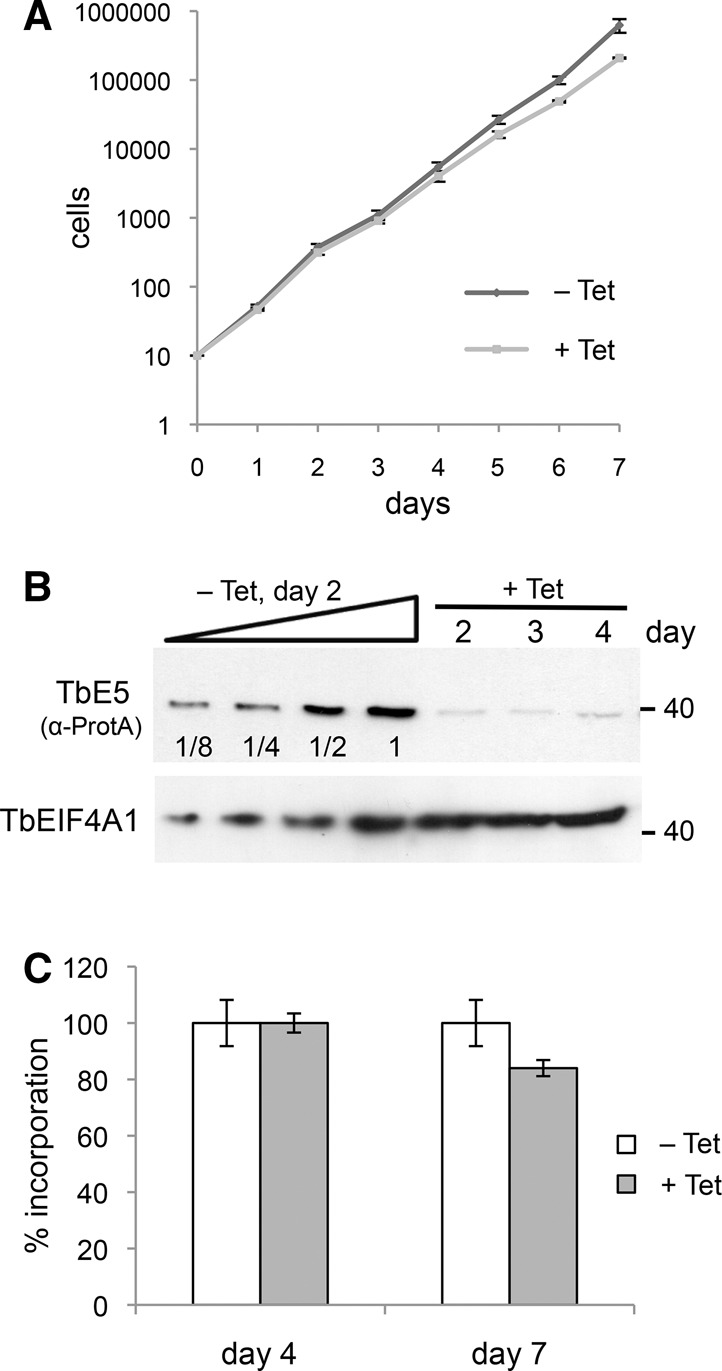
TbEIF4E5 does not have a primary role in translation. (A) Growth of TbE5 RNAi tetracycline (Tet) induced and uninduced cells measured over 7 d. (B) Assessment of protein knockdown by RNAi using the TbE5PTP/+ background cell line. SDS-PAGE analysis of protein levels at days 2, 3, and 4 post-induction. TbE5-PTP was detected by anti-Protein A antibody (α-ProtA). Serial twofold dilutions of uninduced samples at day 2 are shown for comparison. Levels of TbEIF4A1 are included as protein loading controls. (C) Metabolic labeling of cultures induced for TbE5 RNAi knockdown as measured by 35S-methionine incorporation at 4 and 7 d post-induction.
As a general rule, when an RNAi line shows no significant growth defect, the next experimental step is to create a knockout cell line so that the absence of the protein of interest can be assessed in a clean background. No third allele was detected in the construction of the TbE5−/PTP line, thus the diploid nature of these genes was confirmed. Double allele knockout of the TbE5 gene was attempted three times, and all attempts failed in the elimination of the second targeted allele. In both cases a WT allele persisted, despite integration of the second selective marker (data not shown). We interpret this to signify that the TbE5 protein is essential, and the lowered protein levels achieved by our RNAi inductions allowed evaluation at reduced levels of TbE5 function. Discordant results for individual genes and the Alsford study (Alsford et al. 2011) are not unknown (Badjatia et al. 2013). The inability to obtain a knockout TbE5 line may be technical rather than linked to the function of the gene, and alternative approaches (Kim et al. 2013; Merritt and Stuart 2013; Park et al. 2014) are in progress.
Knockdown of the TbEIF4E5 protein affects motility
During the course of the RNAi experiments, the culture phenotype of TbE5 RNAi-induced cells was altered. Initially, after 48 h of stationary incubation, we observed that cells accumulated on the bottom of the culture flasks while noninduced cells remained in suspension. This “settling” phenotype was reproducible, and was measured by OD600 changes over 24 h.
For each tetracycline (Tet) treatment, six equivalent cultures were established. At each time point, three cultures were shaken to resuspend the cells, accommodating for the effect of cell growth during the experimental period. The OD600 relative sedimentation measurements for each time period were determined by dividing the experimental unshaken cuvette reading by the shaken cuvette reading for the same culture condition. The sedimentation assays showed a significant reduction in OD600 at 8 h and 24 h for the TbE5 RNAi-induced culture relative to the uninduced culture (Fig. 6A). At the beginning of the experiment, all cultures showed a 1:1 ratio of suspended cells. After 8 h, WT and the uninduced TbE5 RNAi cultures were not different, but both showed signs of cell settling in the 20%–25% range. In contrast, the induced TbE5 RNAi line displayed a markedly higher level of settling, with about half of the cells settled. This trend continued in the 24-h sample, with the induced sample at ∼50% the level of suspension relative to the WT and uninduced cultures. The settling phenotype is slower than observed for mutants with defects in proteins directly related to motility, e.g., flagellar proteins (Ralston and Hill 2006; Ralston et al. 2006).
FIGURE 6.
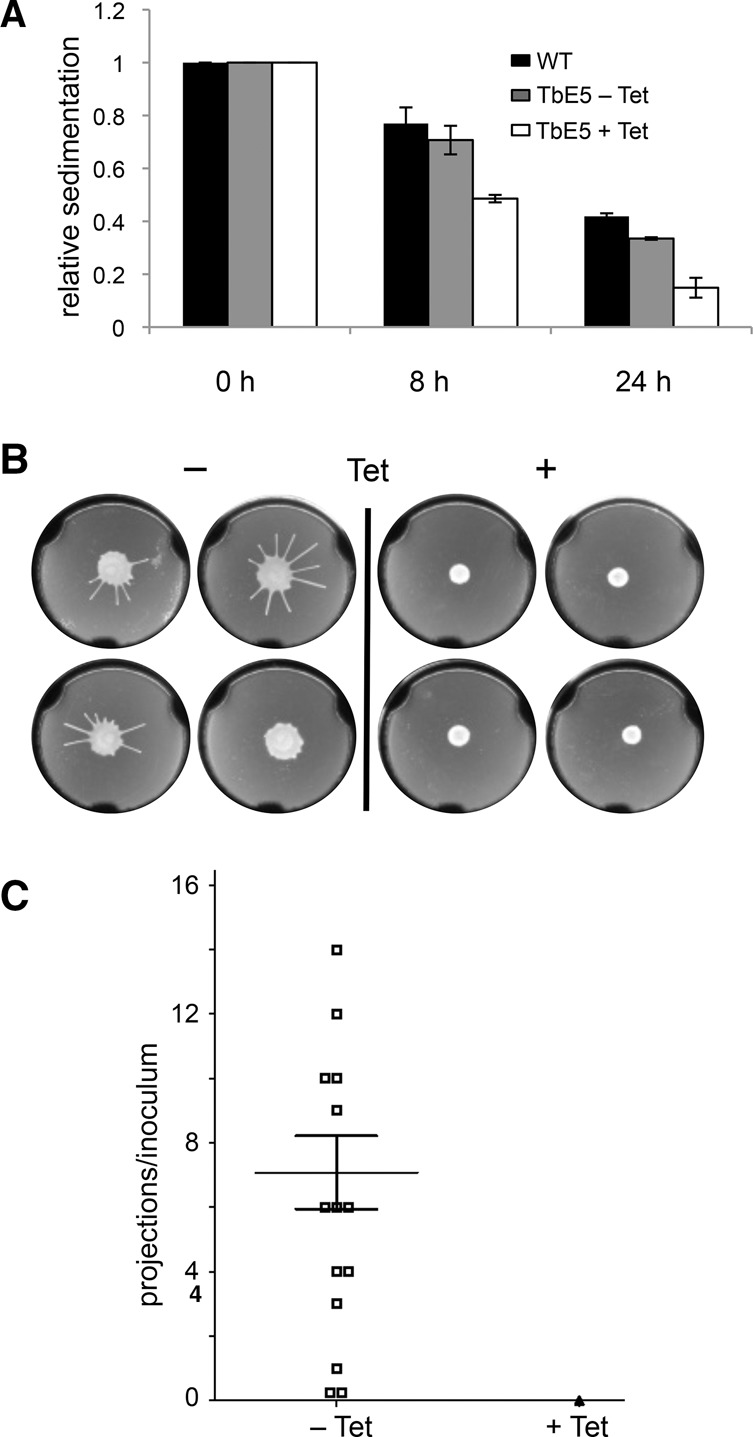
Reduction of TbEIF4E5 results in motility defects. (A) Culture turbidity was measured to assess cell settling in nonshaken cultures. Cells induced for RNAi against TbE5 (white column) were compared with noninduced culture (gray column) and WT cells (black column). Standard error was determined for experiments performed in triplicate. (B) Representative soft agarose plate Social Motility assays for TbE5 RNAi-uninduced (−Tet) or RNAi-induced cells (+Tet) 5 d post-plating. The − plate was scored as showing 10 radial projections, a typical manifestation of social motility in T. brucei. (C) Graphical summary of two sets of SoMo assays indicating means and standard errors. Symbols represent the number of radial projections from the site of inoculation: Open squares represent uninduced (−Tet); the single filled triangle represents all 14 induced samples (+Tet).
The settling phenotype of TbE5 RNAi-induced cells prompted us to examine whether there was a comparable effect on Social Motility (SoMo) on semisolid agar plates (Oberholzer et al. 2010). SoMo is visualized by the formation of massed cell projections from a single inoculum point after 5 d. Under conditions of RNAi knockdown the TbE5 cell line showed a significant defect in SoMo as measured by counting radial projections (Fig. 6B). RNAi induction targeting TbE5 resulted in no SoMo activity on any +Tet plates, while the uninduced TbE5 cultures showed SoMo projections on 12/14 plates (85.7%), with a mean of 7 ± 1 projections (Fig. 6C). Thus the reduction of TbE5 levels resulted in defects in cell motility in both liquid and semisolid media.
Direct light microscopy of cells induced for RNAi against TbE5 revealed no morphological defects that might explain the sedimentation and SoMo phenotypes (data not shown); however, the motility in liquid medium appeared nondirectional and “lethargic.” Motility defects due to knockdown of an mRNA-cap-binding protein were an unexpected finding that may represent downstream or secondary effects.
RNA-binding and cap 0 maturation domains identified in TbEIF4E5 complex components
Phyre2 analysis (Kelley and Sternberg 2009) identified a guanylyltransferase domain (99.4% confidence; positions 409–701), two zinc fingers (positions 711–780), and a m7G methyltransferase domain (100% confidence; positions 774–1039) in the 117.5-kDa protein, while the 47.5-kDa protein carries two RNA-binding motifs (Fig. 7). The 47.5-kDa protein motifs are distinct, with the first containing a ferredoxin-like fold RNA-binding domain (97.7% confidence) and the second a yth domain RNA-binding motif (98.1% confidence) found in splicing factor yt521. The 117.5-kDa protein showed a superficial resemblance to the SL RNA cap 0 formation enzyme TbCGM1 (Ruan et al. 2007; Takagi et al. 2007) in the N7-methyltransferase domain (P-value = 0.54), as well as a bioinformatic kinship with the guanylyltransferase domain (P-value = 0.018) of a 142.1-kDa nucleoside triphosphate hydrolase/guanylyltransferase/zinc finger protein associated with the induction of stumpy bloodstream forms (Fig. 7; Mony et al. 2014). No clues regarding the function of the 17.9-kDa protein were revealed by Phyre2 or BLAST analyses. Other consistently purifying proteins with strong NSAF scores included a TEF1-A isoform and dynein light chain LC8, while the mitochondrial carrier protein and the 137.9-kDa hypothetical protein showed lower scores. With the exception of the 47.5-kDa protein, all the co-purifying proteins are essential in one or more life stages of the trypanosome (Table 1) based on high-throughput RNAi analysis (Alsford et al. 2011). The 17.9-kDa hypothetical protein has a predicted mitochondrial location (Zhang et al. 2010), however it was associated consistently with TbE5 and TbG2 in reciprocal purifications.
FIGURE 7.
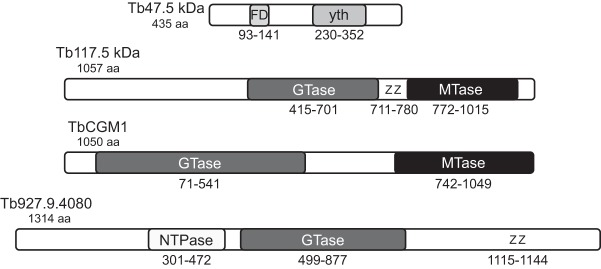
Identified domains in two hypothetical TbEIF4E5/TbEIF4G1 complex members. Schematic locations of the conserved structural domains in the TbE5-associated 47.5-kDa and 117.5-kDa proteins as predicted by Phyre2 analysis, along with two proteins sharing domains with the 117.5-kDa protein. FD, ferredoxin-like fold RNA-binding domain; yth, yth domain RNA-binding motif; GTase, guanylyltransferase; Z, zinc finger; MTase, m7G methyltransferase; NTPase, nucleoside triphosphate hydrolase.
The association of TbE5 in a complex with a protein predicted to have enzymatic activities involved in two of the three steps of mRNA cap formation is a compelling argument for the involvement of TbE5 in mRNA metabolism, and hence the regulation of translation, opening a multitude of new questions. Coupled with the motility phenotype detected upon the depletion of the TbE5 protein, an intriguing pathway for nuanced control of gene expression appears to have been identified.
DISCUSSION
Regulation of translation can be implemented at multiple levels including mRNA degradation, covalent, and noncovalent modification of proteins involved in initiation (Raught and Gingras 1999; Richter and Sonenberg 2005), and mRNA sequestration (Parker and Sheth 2007) and selection (Culjkovic et al. 2007; Keene 2007). The concept of RNA regulons, multiple mRNAs with a shared functional relationship, has been implicated in kinetoplastids (Zick et al. 2005; Estévez 2008; Ouellette and Papadopoulou 2009; Queiroz et al. 2009; Fernandez-Moya et al. 2012). Notably, mammalian eIF4E is a central protein in an RNA regulon related to mRNA export and cell proliferation (Culjkovic et al. 2005; Culjkovic et al. 2006). In this manuscript we characterize a new cytosolic EIF4E family member in T. brucei, TbE5. While not involved primarily in translation, TbE5 can bind cap 4 in vitro and is found in distinct complexes with different eIF4G homologs, TbG1 and TbG2 (Figs. 1B, 3B). Other proteins classified as essential (Alsford et al. 2011) are found in the complexes, including the 117.5-kDa protein, which contains domains consistent with a role in RNA cap formation. Guanylyltransferase and N7-methyltransferase functions are required for the last two steps of eukaryotic mRNA cap formation (Ghosh and Lima 2010), however the enzyme identified here is distinct from that performing the nuclear co-transcriptional capping of the SL RNA, TbCGM1 (Ruan et al. 2007; Takagi et al. 2007; Sturm et al. 2012). The further impact on distinct aspects of cell motility following knockdown of the TbE5 suggests unidentified roles for the complexes in the regulation of protein expression and function in these parasites.
The Alsford genome-wide RNAi study indicates that TbE5 is an essential protein in the cultured insect stage; however, TbE5 is not required for viability in bloodstream forms and differentiating bloodform cells (Alsford et al. 2011). In our hands, procyclic RNAi lines targeting the TbE5 transcript are viable, with protein levels reduced but detectable. Several phenotypic oddities were noted in our TbE5 RNAi knockdowns. TbE5 cells did not remain in suspension in liquid culture and showed “lethargic” activity, and social motility behavior was abolished. The role of an RNA-cap-binding protein in eliciting downstream effects on motility could be direct or indirect depending on whether mRNAs are recognized with specificity or nonspecifically. The absence of a dramatic cell-death or growth-arrest phenotype in the TbE5 knockdown coupled with the lack of a catastrophic effect on 35S-methionine incorporation indicates that TbE5 is not the primary EIF4E responsible for general translation in T. brucei. The lack of effect on translation for TbE5 knockdown on day 4 contrasts the 64% and 72% reduction in translation observed on day 4 RNAi knockdown of TbEIF4E3 and TbEIF4E4, respectively (Freire et al. 2011), the best candidates for a main role in translation initiation. The third component of the eIF4F complex, TbEIF4A, did not co-purify with PTP-tagged TbE5 or the PTP-tagged 117.5-kDa protein, however it was relatively abundant in the TbG1-PTP preparation and of low abundance in the TbG2 preparation. RNAi knockdown of flagellum components often are detrimental to the cell cycle and cell division (Branche et al. 2006; Broadhead et al. 2006; Ralston et al. 2006); no such defects were observed during TbE5 knockdown. The small decrease in translation hints at a minor role in translation or an indirect role in regulation, nevertheless an insufficient level of knockdown or secondary effects cannot be discounted.
TbE5 interacts with proteins associated with the TbEIF4F complex, specifically members of the anticipated TbEIF4G family. As with the TbEIF4E proteins, in kinetoplastids the TbEIF4G family is expanded relative to other eukaryotes, containing five identified members (Dhalia et al. 2006). The direct interaction between the TbE5 and the TbG1 and TbG2 homologs predicted by the initial yeast two-hybrid assay was confirmed in vivo. TbE5 was purified from T. brucei extracts in association with either TbG1 or TbG2 in distinct complexes. In mouse, the eIF4G interaction domain of eIF4E is defined as a pocket that contains three conserved acidic amino acids and four conserved nonpolar residues (Marcotrigiano et al. 1997), thus given the conservation of predicted tertiary structure for TbE5 the same region is expected to determine the specificity of a specific TbEIF4E and specific TbEIF4Gs.
A TbE5 complex containing TbG1 is associated with a predicted 117.5-kDa bifunctional capping enzyme, which contains guanylyltransferase and N7-methyltransferase domains, and is essential in multiple life stages of the parasite (Alsford et al. 2011). The associated 47.5-kDa protein likely possesses RNA-binding activity that may confer transcript selection specificity to the protein aggregate. An alternative complex that swaps the 117.5-kDa protein for a 14-3-3 II protein suggests that association of the 117.5-kDa protein may be modulated by phosphorylations recognized by a 14-3-3 II homodimer, most likely of the scaffold provided by TbG1. The third complex contains TbG2 along with three other proteins that are all essential in the Alsford RNAi analysis (Alsford et al. 2011). The additional members of the third TbE5 complex include a “hypothetical” protein and two related phosphoserine/phosphothreonine-binding proteins, Tb14-3-3 I and Tb14-3-3 II. Tb14-3-3 I/II likely function as homo- or heterodimeric regulators of activity (Mackintosh 2004) and may interact with one of the other protein components to block or facilitate complex formation or RNA binding.
The kinetoplastid 14-3-3 proteins can interact with multiple partners via different motifs (Inoue et al. 2010) so pleiotropic effects are expected in RNAi experiments. Indeed, defects in surface glycoprotein recycling (Benz et al. 2010), motility, cell cycle, and cytokinesis (Inoue et al. 2005) have been observed. The settling phenotype observed for TbE5 in our motility assay is similar to that seen for 14-3-3 knockdowns in procyclic cells (Inoue et al. 2005), suggesting that the phenotype may be the result of a common defect in the protein complex described here. Differential phosphorylation of TbE5 cannot be ruled out as the determinant; TbE5 is predicted to contain four phosphorylation sites (Palmeri et al. 2011), however no phosphorylation was detected in vivo on TbE5 in either procyclic or bloodstream stage (Urbaniak et al. 2013). In TbG1 five phosphorylated serines and one tyrosine have been detected (Urbaniak et al. 2013). TbG2, which has a human eIF4G counterpart with a scaffold function (Prévôt et al. 2003), has three phosphorylated serines and two phosphorylated threonines (Urbaniak et al. 2013), making it the best candidate for 14-3-3 binding.
The presence of a potential 5′-capping enzyme in a TbE5-containing complex raises a paradox, as the TbE5 recognizes m7G-capped RNA 5′ ends, yet the guanylyltransferase acts on 5′ diphosphates, and the N7-methyltransferase requires an unmodified inverted guanosine. What therefore is the substrate for the capping enzyme? The TbE5 component is likely to form a stable interaction with the RNA, while proteins with cap-forming and cap-modifying activities may have a transient association with their substrate. The interplay of the two proteins, with their distinct activities and affinities, will dictate which is the driving force for complex functions. The potential roles of the TbE5 complexes in the manipulation and remodeling of RNA 5′ end structure could have major repercussions for the captured transcripts in terms of stability and translation, providing a vital junction of control for translation initiation in a genetic system believed to be subject to constitutive transcription.
Our working model is that capped mRNAs are recognized and bound by TbE5, which serves a role of recruiting mRNAs, but does not perform any direct catalysis on the transcript. TbE5 is bound by a distinct EIF4G, either TbG1 or TbG2, which provides a scaffold for additional protein interactions. Candidate interactors include the 117.5-kDa protein, which has the potential to modify transcripts released from TbE5. TbE5 complex formation, or action upon a bound RNA, may be regulated by interaction of the Tb14-3-3 II proteins with any of the other associated components, the most direct possibility being the modulation of the 117.5-kDa protein activity upon mRNAs bound to the complex. The presence of a TbCGM1-like protein in an eIF4F-like structure opens a broad range of possibilities for the fate of mRNAs that we can assess. For example, mammalian eIF4E has been implicated in targeting mRNAs to P-bodies (Andrei et al. 2005; Ferraiuolo et al. 2005). The provocative possibility of cytosolic mRNA capping activities highlights the elusive identity of the kinetoplastid mRNA 5′-decapping enzyme (Clayton and Shapira 2007; Banerjee et al. 2009), a lynchpin in the control of translation initiation. Cytosolic 5′ decapping of mRNA is one of two major pathways for eukaryotic mRNA degradation (Coller and Parker 2004), however it is not necessarily the end of life for the mRNA (Topisirovic et al. 2011). Candidate activities, such as a cytosolic complex isolated from human cells, can add a cap structure onto RNA molecules with a 5′ monophosphate (Otsuka et al. 2009). A goal in future studies will be to identify the mRNAs or other capped RNA species associated with each TbE5 complex, and to determine if they change during the lifecycle or in response to select stresses. Other members of the kinetoplastid EIF4E family, TbEIF4E1 through TbEIF4E4, respond to stress by forming cytosolic granules (Kramer et al. 2008, 2012), and comparable behavior has been noted for TbE5 (data not shown). In a group of organisms that predominantly lack regulation of gene/protein expression at the level of transcription initiation (Martínez-Calvillo et al. 2010; Alsford et al. 2012; Kramer 2012), the presence of a putative cytosolic capping enzyme suggests a specific role for RNA processing and/or regulation by the TbE5 complexes. The link between cytosolic mRNA metabolism and the motility-defect phenotypes could lie in TbEIF4E-mediated control of RNA regulons associated with motility.
MATERIALS AND METHODS
Bioinformatics
The two newest T. brucei EIF4E homologs were identified in BLAST searches of the GeneDB database (Hertz-Fowler et al. 2004). TbEIF4E5 was identified using the yeast eIF4E (GenBank number P07260) as a query. Multiple sequence alignment and phylogenetic analysis was performed with Clustal W (http://www.ebi.ac.uk/Tools/msa/clustalw/). Secondary structure predictions were performed using Phyre2 (Kelley and Sternberg 2009).
Plasmid construction
Recombinant TbE5 was expressed from the p2171 plasmid (Dhalia et al. 2006). For interaction assays, the TbE5 and TbEIF4G homolog ORFs were amplified by PCR, and cloned into the yeast two-hybrid vectors pGAD and pGBK. For conditional knockdown by RNAi, gene-internal fragments were PCR-amplified and cloned into the p2T7-177 vector (Wickstead et al. 2002). Gene-specific allele knockout constructions were made by insertion of the respective 5′ and 3′ flanking regions into the pKO vector (Lamb et al. 2001). The 3′ terminal gene fragments required for carboxy-terminal epitope tagging were amplified and the resulting fragments cloned into the pC-PTP-Neo plasmid (Schimanski et al. 2005).
In vitro cap-binding assay
His6-tagged recombinant TbE5 protein was expressed in Escherichia coli Rosetta 2 (DE3) cells. Expression was induced with 0.5 mM IPTG for 3 h at 37°C. Cells were harvested, disrupted by sonication, and centrifuged. The pellet was washed two times in 20 mM HEPES/KOH (pH 7.2) 1 M guanidine hydrochloride, 2 mM DTT, 10% glycerol. Inclusion bodies were dissolved in 50 mM HEPES/KOH (pH 7.2), 6 M guanidine hydrochloride, 10% glycerol, 2 mM DTT. Cell debris was removed by centrifugation (43,000g for 30 min). The protein was diluted to <0.1 mg/mL and refolded by one-step dialysis against 50 mM HEPES/KOH (pH 7.2), 100 mM KCl, 0,5 mM EDTA, 2 mM DTT, and purified by ion exchange chromatography through a HiTrap SP column.
Time-synchronized fluorescence titrations were carried out on a PerkinElmer LS 55 Fluorescence Spectrometer at 20 ± 0.3°C (Niedzwiecka et al. 2002) in 50 mM HEPES/KOH (pH 7.2), 100 mM KCl, 0.5 mM EDTA, 2 mM DTT. During the time-course titration, aliquots of 1 μL cap analog solution (Yoffe et al. 2006) were added to 1400 μL protein solution (0.1 μM, 0.2 μM, or 0.3 μM protein concentration). Changes in fluorescence intensities were measured at 325 nm or 340 nm with excitation at 280 nm and corrected for sample dilution and inner-filter effects. Equilibrium association constants (Kas) were determined by fitting the theoretical curve of fluorescence intensity for total cap analog concentration to the experimental data points (Niedzwiecka et al. 2002). The final Kas was calculated as a weighted average of three to five independent titrations. The fitting procedure utilized nonlinear, least-squared regression analysis and was performed using ORIGIN 6.0 (Microcal Software). The Gibbs free energy of binding was calculated from the Kas value according to the standard equation ▵G° = −RTlnKas.
T. brucei cell culture and RNAi
YTAT procyclic T. brucei was grown at 27°C in SM medium (Cunningham 1977) supplemented with 10% fetal bovine serum, and was used for PTP-targeted integrations for MudPIT analyses. Procyclic forms of T. brucei Lister 427 strain 29-13 were used for RNAi. Transfection was performed as described (Hill et al. 1999). Selection was performed with G418 (15 μg/mL), puromycin (10 μg/mL), or phleomycin (2.5 μg/mL), and clonal lines of selected cultures were obtained by limiting dilution in 96-well plates. To induce RNAi, 1 μg/mL tetracycline (Tet) was added to mid-log phase cultures and the growth measured daily. Single knockout/PTP-tagged lines in YTAT were constructed as described (Schimanski et al. 2005). The 29-13 cell line was transfected with the plasmid cPTP-puro-TbE5, selected with puromycin and checked for PTP-tagged protein expression, then transfected with the RNAi plasmid p2T7-177-TbE5 and selected with phleomycin, generating the TbE5+/PTPRNAi cell line. PTP tagging of TbE5 to monitor knockdown in the 29-13 RNAi cell line affected one allele, leaving the second as WT.
Fluorescence microscopy
T. brucei cultures in mid-log phase (5 × 106– 107 cells/mL) were used for immunofluorescence imaging as described (Oberholzer et al. 2011). Aliquots of 1 mL were washed twice in 1 mL PBS, resuspended in 1 mL PBS/0.01% paraformaldehyde, and incubated on ice for 5 min. The cells were centrifuged and resuspended in 0.5 mL PBS. Approximately 20 µL of the cell suspension was spread on a microscope slide and dried at room temperature (RT), followed by fixation at −20°C in acetone for 5 min and −20°C in methanol for 5 min. The slides were dried at RT, the cells were rehydrated with 1 mL PBS for 15 min and blocked for 1.5 h at RT with blocking solution (PBS/5% Normal Goat Serum/5% BSA). Blocked cells were incubated for 1.5 h in a 1:3000 Anti-Protein A antibody produced in rabbit (Sigma) in blocking solution, washed three times with PBS-T (PBS/0.05% Tween 20), incubated in 1:3000 anti-rabbit IgG Alexa 488 (Invitrogen), washed three times with PBS-T, once with PBS and mounted on slides with Vectashield (Vector Laboratories) containing DAPI, and viewed by fluorescence microscopy.
Metabolic labeling assay
[35S]-methionine incorporation was determined as described (Freire et al. 2011). RNAi-induced and uninduced TbE5+/PTPRNAi mid-log phase cultures were centrifuged at 3000 RPM at RT, washed once in methionine-free SM medium, and resuspended to a concentration of 1 × 107 cells/mL in the methionine-free SM medium supplemented with 50 μCi/mL [35S]-methionine. After 1-h incubation at 28°C, 50 μL aliquots were lysed (5 μL 10% SDS, 2.5 μL 1 M NaOH), 10 μL of these lysates were spotted in triplicate to Whatman filter papers, and dried at RT. The filters were then incubated on ice in 10% TCA for 15 min followed by boiling in 5% TCA for 10 min. After one methanol wash and one acetone wash the filters were dried at RT. The radiolabel incorporated into proteins was measured with a Beckman LS 6500 Scintillation Counter. Experiments were performed three times in triplicate. Standard error was calculated and plotted in Microsoft Excel.
Sedimentation assay
Motility in a liquid environment was quantified by spectrophotometry (Bastin et al. 1999). Cells with integrated RNAi constructions were incubated +/–Tet for 72 h, then resuspended at 5 × 106 cells/mL in fresh medium with or without drug. Six replicates (1 mL) were transferred to cuvettes and incubated without shaking under standard conditions. The optical density at 600 nm (OD600) was measured in triplicate every 8 h, with three cuvettes left undisturbed to measure sedimentation and three cuvettes mixed prior to measurement. The ΔOD600 for each sample was calculated by dividing the OD600 of the resuspended samples by those of the undisturbed samples.
Motility assay
Social motility assays on semi-solid 0.4% agarose plates were performed as described (Oberholzer et al. 2010). Plates contained either methanol (diluent) or Tet (final 1 μg/mL) for “–Tet” and “+Tet” conditions. The plates were inoculated using 5.5 µL cells from suspension cultures at ∼1.0 × 107 cells/mL. The +Tet cells were induced for 72 h prior to plating. Following inoculation, the plates were closed, left to sit for 20 min, then sealed with Parafilm and incubated at 27°C with 5% CO2. Plates were photographed at 120 h using a white light box and a velvet cloth to provide background contrast. Images were acquired using a Pentax Optio M30 camera, and cropped in Adobe Photoshop.
Yeast two-hybrid assays
Yeast strain PJ69-4A was cultivated overnight at 30°C in YPD medium (Ammerman et al. 2012). Each transformation used 1 mL cell suspension washed and resuspended in 100 µL TE/100 mM lithium acetate buffer and incubated at RT for 15 min. The cells were centrifuged and resuspended in 360 µL transformation buffer (1 × TE/1 mM LiOAc; 50% PEG 8000 and 2 mg/mL boiled salmon sperm DNA), simultaneously transformed with GBK (tryp+) and GAD (leu+) plasmids expressing TbE5 and individual T. brucei 4G homologs, and incubated for 30 min at RT. Subsequently, the cells were incubated at 42°C for 20 min, and then spun down. The pellet was resuspended in 2 mL dropout medium (minimal medium minus tryptophan and leucine) and incubated overnight at 30°C. After dropout incubation the OD600 was checked and all cultures centrifuged, diluted to OD600 0.5 in dropout medium (minimal medium minus tryptophan, leucine, and histidine), applied on to 2% agar dropout medium plates containing 3-amino-1,2,4-triazole (3AT) in serial dilutions, and incubated at 30°C for 5 d. The positive control plates used plasmids pGADT7-T and pGBKT7-53 (CLONTECH Laboratories Inc.) for transformation.
Native gel electrophoresis for protein complex detection and sizing
Blue Native gel analysis was performed according to manufacturer instructions (Novex). Samples were prepared as follows: Mid-Log phase culture cells were washed twice in PBS, resuspended in 24 μL extraction buffer (25 mM HEPES, 150 mM sucrose, 20 mM potassium glutamate, 3 mM MgCl2, 0.5% NP40, 150 mM KCl, 0.5 mM DTT, and SIGMAFAST Protease Inhibitor Cocktail, EDTA-free [Sigma-Aldrich]), incubated on ice for 20 min, and centrifuged at full speed for 10 min/4°C. Eighteen microliters of the supernatant was added to 6.25 μL of 4× Native PAGE buffer and 1 μL of G-250 Coomassie sample buffer. The samples were electrophoresed through precast 4%–16% NativePAGE Novex Bis-Tris gels following the manufacturer specifications (Life Technologies). The NativeMark Unstained Protein standard (Life Technologies) was used to estimate complex sizes. Proteins were transferred to Immun-Blot PVDF 0.2 μm membranes (BioRad). Membranes were fixed in 8% acetic acid for 15 min, rinsed with water, and incubated with primary or secondary antibody. The size marker lane was removed prior to antibody incubation, air-dried, equilibrated with methanol, and stained with Coomassie dye for visualization.
Tandem-affinity purification
Purification was performed from 500 mL of culture (5 × 106 cell/mL). For tandem-affinity purification, the PTP tag was utilized and purification performed as described (Schimanski et al. 2005). Total elution from the protein C column was resolved either by SDS-PAGE and visualized by silver staining (BioRad Silver Staining plus), or TCA precipitation and subjected to tandem mass spectrometry (MudPIT).
MudPIT
The TCA precipitated proteins were digested by trypsin and subjected to mass spectrometry as described (Zamudio et al. 2009). The proteomic data were analyzed using the SEQUEST and DTASelect2 algorithms against the T. brucei genome database (Berriman et al. 2005), filtering by a peptide-level false-positive rate of 5%, and a minimum of two peptides per protein (Peng et al. 2002).
ACKNOWLEDGMENTS
We thank Bidyottam Mittra, Jun Urano, Laurie Read, and Fuyu Tamanoi for strains, reagents, and help with the Y2H assay; and Arthur Günzl for the PTP-immunofluorescence protocol. This work was supported by a UCLA Stein-Oppenheimer award; the National Institute of Allergy and Infectious Diseases, the National Institutes of Health, and the National Institute of General Medical Sciences (grant numbers AI056034, AI073806, TW009035 to D.A.C. and N.R.S., grant number GM089778 to J.A.W., and grant number AI052348 to K.H.); and the National Science Centre, Poland (grant numbers UMO-2012/07/B/NZ1/00118 and UMO-2013/08/A/NZ1/00866 to E.D. and J.Z.). Scholarship support for A.M.M. and J.F.N. was provided by Fundação de Amparo à Ciência e Tecnologia de Pernambuco; Coordenação de Aperfeiçoamento de Pessoal de Nível Superior; and Conselho Nacional de Desenvolvimento Científico e Tecnológico.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.045534.114.
REFERENCES
- Adl SM, Simpson AG, Lane CE, Lukes J, Bass D, Bowser SS, Brown MW, Burki F, Dunthorn M, Hampl V, et al. 2012. The revised classification of eukaryotes. J Eukaryot Microbiol 59: 429–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aitken CE, Lorsch JR 2012. A mechanistic overview of translation initiation in eukaryotes. Nat Struct Mol Biol 19: 568–576 [DOI] [PubMed] [Google Scholar]
- Alsford S, Turner DJ, Obado SO, Sanchez-Flores A, Glover L, Berriman M, Hertz-Fowler C, Horn D 2011. High-throughput phenotyping using parallel sequencing of RNA interference targets in the African trypanosome. Genome Res 21: 915–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsford S, duBois K, Horn D, Field MC 2012. Epigenetic mechanisms, nuclear architecture and the control of gene expression in trypanosomes. Expert Rev Mol Med 14: e13. [DOI] [PubMed] [Google Scholar]
- Ammerman ML, Downey KM, Hashimi H, Fisk JC, Tomasello DL, Faktorova D, Kafkova L, King T, Lukes J, Read LK 2012. Architecture of the trypanosome RNA editing accessory complex, MRB1. Nucleic Acids Res 40: 5637–5650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrei MA, Ingelfinger D, Heintzmann R, Achsel T, Rivera-Pomar R, Luhrmann R 2005. A role for eIF4E and eIF4E-transporter in targeting mRNPs to mammalian processing bodies. RNA 11: 717–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badjatia N, Ambrosio DL, Lee JH, Gunzl A 2013. Trypanosome cdc2-related kinase 9 controls spliced leader RNA cap4 methylation and phosphorylation of RNA polymerase II subunit RPB1. Mol Cell Biol 33: 1965–1975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee H, Palenchar JB, Lukaszewicz M, Bojarska E, Stepinski J, Jemielity J, Guranowski A, Ng S, Wah DA, Darzynkiewicz E, et al. 2009. Identification of the HIT-45 protein from Trypanosoma brucei as an FHIT protein/dinucleoside triphosphatase: substrate specificity studies on the recombinant and endogenous proteins. RNA 15: 1554–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bangs JD, Crain PF, Hashizume T, McCloskey JA, Boothroyd JC 1992. Mass spectrometry of mRNA cap 4 from trypanosomatids reveals two novel nucleosides. J Biol Chem 267: 9805–9815 [PubMed] [Google Scholar]
- Bastin P, Pullen TJ, Sherwin T, Gull K 1999. Protein transport and flagellum assembly dynamics revealed by analysis of the paralysed trypanosome mutant snl-1. J Cell Sci 112: 3769–3777 [DOI] [PubMed] [Google Scholar]
- Benz C, Engstler M, Hillmer S, Clayton C 2010. Depletion of 14-3-3 proteins in bloodstream-form Trypanosoma brucei inhibits variant surface glycoprotein recycling. Int J Parasitol 40: 629–634 [DOI] [PubMed] [Google Scholar]
- Berriman M, Ghedin E, Hertz-Fowler C, Blandin G, Renauld H, Bartholomeu DC, Lennard NJ, Caler E, Hamlin NE, Haas B, et al. 2005. The genome of the African trypanosome Trypanosoma brucei. Science 309: 416–422 [DOI] [PubMed] [Google Scholar]
- Blewett NH, Goldstrohm AC 2012. A eukaryotic translation initiation factor 4E-binding protein promotes mRNA decapping and is required for PUF repression. Mol Cell Biol 32: 4181–4194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branche C, Kohl L, Toutirais G, Buisson J, Cosson J, Bastin P 2006. Conserved and specific functions of axoneme components in trypanosome motility. J Cell Sci 119: 3443–3455 [DOI] [PubMed] [Google Scholar]
- Broadhead R, Dawe HR, Farr H, Griffiths S, Hart SR, Portman N, Shaw MK, Ginger ML, Gaskell SJ, McKean PG, et al. 2006. Flagellar motility is required for the viability of the bloodstream trypanosome. Nature 440: 224–227 [DOI] [PubMed] [Google Scholar]
- Cho PF, Poulin F, Cho-Park YA, Cho-Park IB, Chicoine JD, Lasko P, Sonenberg N 2005. A new paradigm for translational control: inhibition via 5′–3′ mRNA tethering by Bicoid and the eIF4E cognate 4EHP. Cell 121: 411–423 [DOI] [PubMed] [Google Scholar]
- Clarkson BK, Gilbert WV, Doudna JA 2010. Functional overlap between eIF4G isoforms in Saccharomyces cerevisiae. PLoS One 5: e9114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton C, Shapira M 2007. Post-transcriptional regulation of gene expression in trypanosomes and leishmanias. Mol Biochem Parasitol 156: 93–101 [DOI] [PubMed] [Google Scholar]
- Coller J, Parker R 2004. Eukaryotic mRNA decapping. Ann Rev Biochem 73: 861–890 [DOI] [PubMed] [Google Scholar]
- Contreras V, Richardson MA, Hao E, Keiper BD 2008. Depletion of the cap-associated isoform of translation factor eIF4G induces germline apoptosis in C. elegans. Cell Death Differ 15: 1232–1242 [DOI] [PubMed] [Google Scholar]
- Culjkovic B, Topisirovic I, Skrabanek L, Ruiz-Gutierrez M, Borden KL 2005. eIF4E promotes nuclear export of cyclin D1 mRNAs via an element in the 3′UTR. J Cell Biol 169: 245–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culjkovic B, Topisirovic I, Skrabanek L, Ruiz-Gutierrez M, Borden KL 2006. eIF4E is a central node of an RNA regulon that governs cellular proliferation. J Cell Biol 175: 415–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culjkovic B, Topisirovic I, Borden KL 2007. Controlling gene expression through RNA regulons: the role of the eukaryotic translation initiation factor eIF4E. Cell Cycle 6: 65–69 [DOI] [PubMed] [Google Scholar]
- Cunningham I 1977. New culture medium for maintenance of tsetse tissues and growth of trypanosomatids. J Protozool 24: 325–329 [DOI] [PubMed] [Google Scholar]
- Das A, Morales R, Banday M, Garcia S, Hao L, Cross GA, Estevez AM, Bellofatto V 2012. The essential polysome-associated RNA-binding protein RBP42 targets mRNAs involved in Trypanosoma brucei energy metabolism. RNA 18: 1968–1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Gaudenzi JG, Carmona SJ, Aguero F, Frasch AC 2013. Genome-wide analysis of 3′-untranslated regions supports the existence of post-transcriptional regulons controlling gene expression in trypanosomes. PeerJ 1: e118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhalia R, Reis CR, Freire ER, Rocha PO, Katz R, Muniz JR, Standart N, de Melo Neto OP 2005. Translation initiation in Leishmania major: characterisation of multiple eIF4F subunit homologues. Mol Biochem Parasitol 140: 23–41 [DOI] [PubMed] [Google Scholar]
- Dhalia R, Marinsek N, Reis CR, Katz R, Muniz JR, Standart N, Carrington M, de Melo Neto OP 2006. The two eIF4A helicases in Trypanosoma brucei are functionally distinct. Nucleic Acids Res 34: 2495–2507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estévez AM 2008. The RNA-binding protein TbDRBD3 regulates the stability of a specific subset of mRNAs in trypanosomes. Nucleic Acids Res 36: 4573–4586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Moya SM, Estevez AM 2010. Posttranscriptional control and the role of RNA-binding proteins in gene regulation in trypanosomatid protozoan parasites. Wiley Interdiscip Rev RNA 1: 34–46 [DOI] [PubMed] [Google Scholar]
- Fernandez-Moya SM, Garcia-Perez A, Kramer S, Carrington M, Estevez AM 2012. Alterations in DRBD3 ribonucleoprotein complexes in response to stress in Trypanosoma brucei. PLoS One 7: e48870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferraiuolo MA, Basak S, Dostie J, Murray EL, Schoenberg DR, Sonenberg N 2005. A role for the eIF4E-binding protein 4E-T in P-body formation and mRNA decay. J Cell Biol 170: 913–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freire ER, Dhalia R, Moura DM, da Costa Lima TD, Lima RP, Reis CR, Hughes K, Figueiredo RC, Standart N, Carrington M, et al. 2011. The four trypanosomatid eIF4E homologues fall into two separate groups, with distinct features in primary sequence and biological properties. Mol Biochem Parasitol 176: 25–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh A, Lima CD 2010. Enzymology of RNA cap synthesis. Wiley Interdiscip Rev RNA 1: 152–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras AC, Raught B, Sonenberg N 1999. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem 68: 913–963 [DOI] [PubMed] [Google Scholar]
- Gosselin P, Martineau Y, Morales J, Czjzek M, Glippa V, Gauffeny I, Morin E, Le Corguille G, Pyronnet S, Cormier P, et al. 2013. Tracking a refined eIF4E-binding motif reveals Angel1 as a new partner of eIF4E. Nucleic Acids Res 41: 7783–7792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groppo R, Richter JD 2009. Translational control from head to tail. Curr Opin Cell Biol 21: 444–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Günzl A 2012. RNA polymerases and transcription factors of trypanosomes. In RNA metabolism in trypanosomes (ed. Bindereif A), Vol. 28, pp. 1–27 Springer, Berlin [Google Scholar]
- Hernández G, Altmann M, Sierra JM, Urlaub H, Diez del Corral R, Schwartz P, Rivera-Pomar R 2005. Functional analysis of seven genes encoding eight translation initiation factor 4E (eIF4E) isoforms in Drosophila. Mech Dev 122: 529–543 [DOI] [PubMed] [Google Scholar]
- Hertz-Fowler C, Peacock CS, Wood V, Aslett M, Kerhornou A, Mooney P, Tivey A, Berriman M, Hall N, Rutherford K, et al. 2004. GeneDB: a resource for prokaryotic and eukaryotic organisms. Nucleic Acids Res 32: D339–D343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill KL, Hutchings NR, Russell DG, Donelson JE 1999. A novel protein targeting domain directs proteins to the anterior cytoplasmic face of the flagellar pocket in African trypanosomes. J Cell Sci 112: 3091–3101 [DOI] [PubMed] [Google Scholar]
- Inoue M, Nakamura Y, Yasuda K, Yasaka N, Hara T, Schnaufer A, Stuart K, Fukuma T 2005. The 14-3-3 proteins of Trypanosoma brucei function in motility, cytokinesis, and cell cycle. J Biol Chem 280: 14085–14096 [DOI] [PubMed] [Google Scholar]
- Inoue M, Yasuda K, Uemura H, Yasaka N, Inoue H, Sei Y, Horikoshi N, Fukuma T 2010. Phosphorylation-dependent protein interaction with Trypanosoma brucei 14-3-3 proteins that display atypical target recognition. PLoS One 5: e15566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RJ, Hellen CU, Pestova TV 2010. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol 11: 113–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagus R, Bachvaroff TR, Joshi B, Place AR 2012. Diversity of eukaryotic translational initiation factor eIF4E in protists. Comp Funct Genomics 2012: 134839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankowska-Anyszka M, Lamphear BJ, Aamodt EJ, Harrington T, Darzynkiewicz E, Stolarski R, Rhoads RE 1998. Multiple isoforms of eukaryotic protein synthesis initiation factor 4E in Caenorhabditis elegans can distinguish between mono- and trimethylated cap structures. J Biol Chem 273: 10538–10542 [DOI] [PubMed] [Google Scholar]
- Joshi B, Cameron A, Jagus R 2004. Characterization of mammalian eIF4E-family members. Eur J Biochem 271: 2189–2203 [DOI] [PubMed] [Google Scholar]
- Joshi B, Lee K, Maeder DL, Jagus R 2005. Phylogenetic analysis of eIF4E-family members. BMC Evol Biol 5: 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene JD 2007. RNA regulons: coordination of post-transcriptional events. Nat Rev Genet 8: 533–543 [DOI] [PubMed] [Google Scholar]
- Keiper BD, Lamphear BJ, Deshpande AM, Jankowska-Anyszka M, Aamodt EJ, Blumenthal T, Rhoads RE 2000. Functional characterization of five eIF4E isoforms in Caenorhabditis elegans. J Biol Chem 275: 10590–10596 [DOI] [PubMed] [Google Scholar]
- Kelley LA, Sternberg MJ 2009. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc 4: 363–371 [DOI] [PubMed] [Google Scholar]
- Kim HS, Li Z, Boothroyd C, Cross GA 2013. Strategies to construct null and conditional null Trypanosoma brucei mutants using Cre-recombinase and loxP. Mol Biochem Parasitol 191: 16–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer S 2012. Developmental regulation of gene expression in the absence of transcriptional control: the case of kinetoplastids. Mol Biochem Parasitol 181: 61–72 [DOI] [PubMed] [Google Scholar]
- Kramer S, Queiroz R, Ellis L, Webb H, Hoheisel JD, Clayton C, Carrington M 2008. Heat shock causes a decrease in polysomes and the appearance of stress granules in trypanosomes independently of eIF2α phosphorylation at Thr169. J Cell Sci 121: 3002–3014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer S, Marnef A, Standart N, Carrington M 2012. Inhibition of mRNA maturation in trypanosomes causes the formation of novel foci at the nuclear periphery containing cytoplasmic regulators of mRNA fate. J Cell Sci 125: 2896–2909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb JR, Fu V, Wirtz E, Bangs JD 2001. Functional analysis of the trypanosomal AAA protein TbVCP with trans-dominant ATP hydrolysis mutants. J Biol Chem 276: 21512–21520 [DOI] [PubMed] [Google Scholar]
- Lasko P, Cho P, Poulin F, Sonenberg N 2005. Contrasting mechanisms of regulating translation of specific Drosophila germline mRNAs at the level of 5′-cap structure binding. Biochem Soc Trans 33: 1544–1546 [DOI] [PubMed] [Google Scholar]
- Liang X-h, Haritan A, Uliel S, Michaeli S 2003. trans and cis splicing in trypanosomatids: mechanisms, factors, and regulation. Eukaryot Cell 2: 830–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackintosh C 2004. Dynamic interactions between 14-3-3 proteins and phosphoproteins regulate diverse cellular processes. Biochem J 381: 329–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcotrigiano J, Gingras AC, Sonenberg N, Burley SK 1997. Cocrystal structure of the messenger RNA 5′ cap-binding protein (eIF4E) bound to 7-methyl-GDP. Cell 89: 951–961 [DOI] [PubMed] [Google Scholar]
- Marcotrigiano J, Gingras AC, Sonenberg N, Burley SK 1999. Cap-dependent translation initiation in eukaryotes is regulated by a molecular mimic of eIF4G. Mol Cell 3: 707–716 [DOI] [PubMed] [Google Scholar]
- Martínez-Calvillo S, Vizuet-de-Rueda JC, Florencio-Martínez LE, Manning-Cela RG, Figueroa-Angulo EE 2010. Gene expression in trypanosomatid parasites. J Biomed Biotechnol 2010: 525241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt C, Stuart K 2013. Identification of essential and non-essential protein kinases by a fusion PCR method for efficient production of transgenic Trypanosoma brucei. Mol Biochem Parasitol 190: 44–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mony BM, MacGregor P, Ivens A, Rojas F, Cowton A, Young J, Horn D, Matthews K 2014. Genome-wide dissection of the quorum sensing signalling pathway in Trypanosoma brucei. Nature 505: 681–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedzwiecka A, Marcotrigiano J, Stepinski J, Jankowska-Anyszka M, Wyslouch-Cieszynska A, Dadlez M, Gingras AC, Mak P, Darzynkiewicz E, Sonenberg N, et al. 2002. Biophysical studies of eIF4E cap-binding protein: recognition of mRNA 5′ cap structure and synthetic fragments of eIF4G and 4E-BP1 proteins. J Mol Biol 319: 615–635 [DOI] [PubMed] [Google Scholar]
- Oberholzer M, Lopez MA, McLelland BT, Hill KL 2010. Social motility in african trypanosomes. PLoS Pathogens 6: e1000739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberholzer M, Langousis G, Nguyen HT, Saada EA, Shimogawa MM, Jonsson ZO, Nguyen SM, Wohlschlegel JA, Hill KL 2011. Independent analysis of the flagellum surface and matrix proteomes provides insight into flagellum signaling in mammalian-infectious Trypanosoma brucei. Mol Cell Proteomics 10: M111.010538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka Y, Kedersha NL, Schoenberg DR 2009. Identification of a cytoplasmic complex that adds a cap onto 5′-monophosphate RNA. Mol Cell Biol 29: 2155–2167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouellette M, Papadopoulou B 2009. Coordinated gene expression by post-transcriptional regulons in African trypanosomes. J Biol 8: 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmeri A, Gherardini PF, Tsigankov P, Ausiello G, Spath GF, Zilberstein D, Helmer-Citterich M 2011. PhosTryp: a phosphorylation site predictor specific for parasitic protozoa of the family trypanosomatidae. BMC Genomics 12: 614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SH, Nguyen BN, Kirkham JK, Nguyen TN, Gunzl A 2014. A new strategy of RNA interference that targets heterologous sequences reveals CITFA1 as an essential component of class I transcription factor A in Trypanosoma brucei. Eukaryot Cell 13: 785–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R, Sheth U 2007. P bodies and the control of mRNA translation and degradation. Mol Cell 25: 635–646 [DOI] [PubMed] [Google Scholar]
- Peng J, Elias JE, Thoreen CC, Licklider LJ, Gygi SP 2002. Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: the yeast proteome. J Proteome Res 2: 43–50 [DOI] [PubMed] [Google Scholar]
- Pestova TV, Hellen CU 2003. Coupled folding during translation initiation. Cell 115: 650–652 [DOI] [PubMed] [Google Scholar]
- Preußer C, Jaé N, Günzl A, Bindereif A 2012. Pre-mRNA splicing in Trypanosoma brucei: factors, mechanisms, and regulation. In RNA metabolism in trypanosomes (ed. Bindereif A), pp. 49–78 Springer, Berlin [Google Scholar]
- Prévôt D, Darlix JL, Ohlmann T 2003. Conducting the initiation of protein synthesis: the role of eIF4G. Biol Cell 95: 141–156 [DOI] [PubMed] [Google Scholar]
- Ptushkina M, Berthelot K, von der Haar T, Geffers L, Warwicker J, McCarthy JE 2001. A second eIF4E protein in Schizosaccharomyces pombe has distinct eIF4G-binding properties. Nucleic Acids Res 29: 4561–4569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queiroz R, Benz C, Fellenberg K, Hoheisel JD, Clayton C 2009. Transcriptome analysis of differentiating trypanosomes reveals the existence of multiple post-transcriptional regulons. BMC Genomics 10: 495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralston KS, Hill KL 2006. Trypanin, a component of the flagellar Dynein regulatory complex, is essential in bloodstream form African trypanosomes. PLoS Pathogens 2: e101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralston KS, Lerner AG, Diener DR, Hill KL 2006. Flagellar motility contributes to cytokinesis in Trypanosoma brucei and is modulated by an evolutionarily conserved dynein regulatory system. Eukaryot Cell 5: 696–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raught B, Gingras AC 1999. eIF4E activity is regulated at multiple levels. Int J Biochem Cell Biol 31: 43–57 [DOI] [PubMed] [Google Scholar]
- Rhoads RE 2009. eIF4E: new family members, new binding partners, new roles. J Biol Chem 284: 16711–16715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter JD, Sonenberg N 2005. Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature 433: 477–480 [DOI] [PubMed] [Google Scholar]
- Ruan J-p, Shen S, Ullu E, Tschudi C 2007. Evidence for a capping enzyme with specificity for the trypanosome spliced leader RNA. Mol Biochem Parasitol 156: 246–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimanski B, Nguyen TN, Günzl A 2005. Highly efficient tandem affinity purification of trypanosome protein complexes based on a novel epitope combination. Eukaryot Cell 4: 1942–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm NR, Zamudio JR, Campbell DA 2012. SL RNA biogenesis in kinetoplastids: a long and winding road. In RNA metabolism in trypanosomes (ed. Bindereif A), pp. 29–47 Springer, Berlin [Google Scholar]
- Takagi Y, Sindkar S, Ekonomidis D, Hall MP, Ho CK 2007. Trypanosoma brucei encodes a bifunctional capping enzyme essential for cap 4 formation on the spliced leader RNA. J Biol Chem 282: 15995–16005 [DOI] [PubMed] [Google Scholar]
- Topisirovic I, Svitkin YV, Sonenberg N, Shatkin AJ 2011. Cap and cap-binding proteins in the control of gene expression. Wiley Interdiscip Rev RNA 2: 277–298 [DOI] [PubMed] [Google Scholar]
- Urbaniak MD, Martin DM, Ferguson MA 2013. Global quantitative SILAC phosphoproteomics reveals differential phosphorylation is widespread between the procyclic and bloodstream form lifecycle stages of Trypanosoma brucei. J Proteome Res 12: 2233–2244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickstead B, Ersfeld K, Gull K 2002. Targeting of a tetracycline-inducible expression system to the transcriptionally silent minichromosomes of Trypanosoma brucei. Mol Biochem Parasitol 125: 211–216 [DOI] [PubMed] [Google Scholar]
- Yoffe Y, Zuberek J, Lewdorowicz M, Zeira Z, Keasar C, Orr-Dahan I, Jankowska-Anyszka M, Stepinski J, Darzynkiewicz E, Shapira M 2004. Cap-binding activity of an eIF4E homolog from Leishmania. RNA 10: 1764–1775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoffe Y, Zuberek J, Lerer A, Lewdorowicz M, Stepinski J, Altmann M, Darzynkiewicz E, Shapira M 2006. Binding specificities and potential roles of isoforms of eukaryotic initiation factor eIF4E in Leishmania. Eukaryot Cell 5: 1969–1979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoffe Y, Léger M, Zinoviev A, Zuberek J, Darzynkiewicz E, Wagner G, Shapira M 2009. Evolutionary changes in the Leishmania eIF4F complex involve variations in the eIF4E-eIF4G interactions. Nucleic Acids Res 10: 3243–3253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamudio JR, Mittra B, Chattopadhyay A, Wohlschlegel JA, Sturm NR, Campbell DA 2009. Trypanosoma brucei spliced leader RNA maturation by the cap 1 2′-O-ribose methyltransferase and SLA1 H/ACA snoRNA pseudouridine synthase complex. Mol Cell Biol 29: 1202–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Cui J, Nilsson D, Gunasekera K, Chanfon A, Song X, Wang H, Xu Y, Ochsenreiter T 2010. The Trypanosoma brucei MitoCarta and its regulation and splicing pattern during development. Nucleic Acids Res 38: 7378–7387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zick A, Onn I, Bezalel R, Margalit H, Shlomai J 2005. Assigning functions to genes: identification of S-phase expressed genes in Leishmania major based on post-transcriptional control elements. Nucleic Acids Res 33: 4235–4242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinoviev A, Shapira M 2012. Evolutionary conservation and diversification of the translation initiation apparatus in trypanosomatids. Comp Funct Genomics 2012: 813718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinoviev A, Leger M, Wagner G, Shapira M 2011. A novel 4E-interacting protein in Leishmania is involved in stage-specific translation pathways. Nucleic Acids Res 39: 8404–8415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinoviev A, Manor S, Shapira M 2012. Nutritional stress affects an atypical cap-binding protein in Leishmania. RNA Biol 9: 1450–1460 [DOI] [PubMed] [Google Scholar]