In this study, the authors present a series of in vitro and in vivo experiments that demonstrate that miR-190 is able to significantly suppress angiogenesis. They show that miR-190 directly regulates a subset of pro-angiogenic genes including VEGF.
Keywords: miR-190, VEGF, angiogenesis, synergistic effect, metastasis
Abstract
VEGF is a major contributor to angiogenesis, a vital process in normal growth and development and tumor transition. However, the current clinical efficacy of VEGF inhibitors is limited, and the molecular mechanism underlying VEGF regulation remains to be elucidated. Here we show that miR-190 directly targets a group of angiogenic effectors besides VEGF per se. Noted, these effectors can transcriptionally regulate VEGF expression in an intracellular or intercellular manner, thus demonstrating that miR-190 modulates the VEGF-mediated tumor angiogenesis at three levels. The synergistic effect of miR-190 and its target genes demonstrates a complex but apparently more stable system, allowing for the tight control of the level of VEGF. Finally, we showed that miR-190 significantly suppresses tumor metastasis, especially angiogenesis. Together, these results indicate that miR-190 is a promising antitumor target in clinical applications.
INTRODUCTION
Angiogenesis is essential for tumors to grow and progress. The formation of a neovascular is a multistep process which includes endothelial cells (ECs) proliferation, migration, vascular tubule formation, and cell survival (Coultas et al. 2005). During this process, VEGF has been demonstrated to be a major modulator (Folkman 2007; Birk et al. 2008). Inhibiting VEGF signal pathway has shown promise for tumor antiangiogenesis therapy.
The expression of VEGF is regulated at many levels. A large number of transcription factors, such as Stat3, AP-1, TCF4, and Smad2/4, participate in the transcriptional regulation of VEGF (Clifford et al. 2008). Additionally, the expression of VEGF is affected by a variety of microenvironmental factors, such as hypoxia, acidosis, and oxidative stress, as well as growth factors and cytokines (Samani et al. 2007; Sullivan and Graham 2007). A dozen VEGF pathway inhibitors have been already approved or are being clinically evaluated for treating patients with many different types of aggressive tumors. However, these VEGF pathway inhibitors elicit only transitory improvements or even no objective benefit in many cases. Therefore, the detailed characterization of the key molecules in the VEGF pathway is critical to the understanding of this apparent conundrum in the activity of angiogenic inhibitors.
As a new class of signaling modulators, miRNAs have attracted great interest due to certain unique features they possess, including multi-target regulation, tissue specificity, and widespread conservation across species (Flynt and Lai 2008). A few miRNAs, including miR-92a, miR-126, and miR-132, have been demonstrated to play a critical role in the regulation of angiogenesis (Fish et al. 2008; Kuehbacher et al. 2008; Wurdinger et al. 2008; Anand et al. 2010; Nicoli et al. 2010). Our recently reported results quantitatively examining the impact of all of the known human miRNAs on cell migration, referred to as “migration functional analysis,” found that one fifth of all the miRNAs are involved in the regulation of cell migration in both epithelial cells and human umbilical vein endothelial cells (HUVECs) (Zhang et al. 2011). On the basis of those results, we further found that four new miRNAs, miR-190, miR-23b, miR-215, and miR-451, which have such cell migratory regulation capacity, had distinct expression patterns in human colon cancer samples compared with normal ones (Supplemental Fig. S1). In this study, we characterized the molecular mechanism of miR-190 in the regulation of colon tumor, especially angiogenesis. It was demonstrated that miR-190 modulates a group of target genes centralized on the VEGF biogenesis, thus highlighting the therapeutic potential of this miRNA in the treatment of cancers.
RESULTS
miR-190 suppresses the angiogenic traits of HUVECs
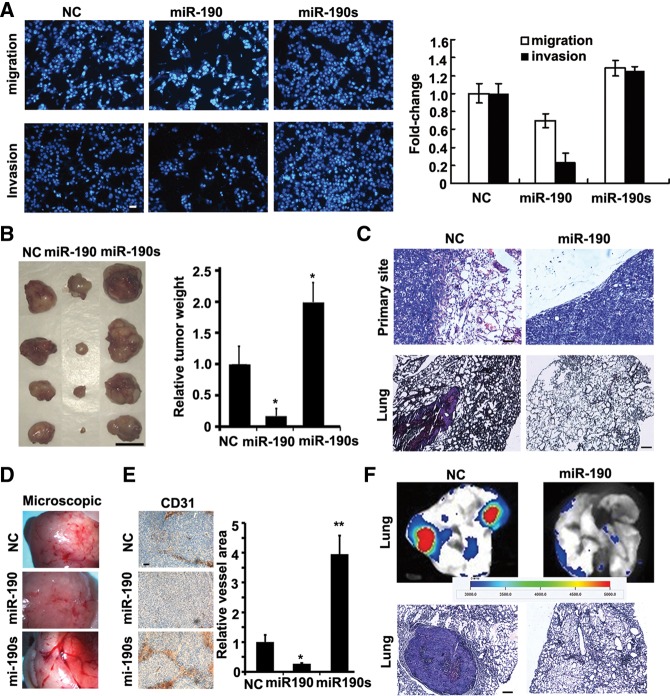
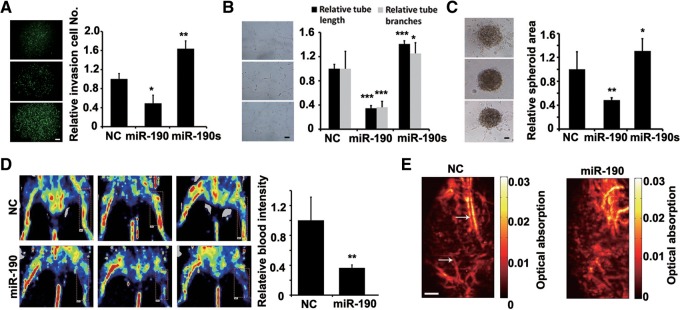
To investigate whether miR-190 acts as a dominant determinant of angiogenesis, we first studied the capacity of miR-190 to regulate the main angiogenic traits of HUVECs, i.e., cell migration, tubule formation, and sprouting. To confirm the transfection efficiency, a lentiviral construct expressing miR-190, an miR-190 sponge, or negative control (NC) as well as enhanced green fluorescent protein (eGFP) was transfected into HUVECs (Supplemental Fig. S2). First, the Boyden chamber-based transwell migration assay showed that miR-190 induced a potent inhibition of the cell migration, while its sponge increased the migratory capability by >60% (Fig. 1A). The tube formation assay was then used to measure the capacity of endothelial cells to form capillary-like structures. At 24 h after transfection with the lentiviral constructs, three different kinds of HUVECs were individually plated on Matrigel for another 12-h culture. It was shown that miR-190 significantly suppressed both tubule elongation and branching, while its sponge stimulated tubule formation, in contrast to scramble siRNA (Fig. 1B). Sprouting angiogenesis is the process by which the growth of new capillary vessels out of pre-existing ones takes place. We found that forced overexpression of miR-190 in human HUVECs inhibited sprout formation in a three-dimensional model of angiogenesis, whereas overexpression of the miR-190 sponge enhanced sprout formation (Fig. 1C).
FIGURE 1.
miR-190 suppresses angiogenesis in vitro and in vivo. (A) Migration and invasion assays after infection of HUVECs with miR-190, miR-190 sponge, or NC vectors. NC, a scramble miRNA. N = 6. (*) P < 0.05, (**) P < 0.01 (t-test). Scale bar, 100 μm. (B) Tubule formation of HUVECs infected as indicated. Representative images are shown at 12 h after plating. Quantitation of tubule branching and length. N = 5. (*) P < 0.05, (***) P < 0.005 (t-test). Scale bar, 50 μm. (C) Spheroid-based angiogenesis assay of HUVECs infected as indicated. Quantitation of cumulative length of the sprouts. N = 10. (*) P < 0.05, (**) P < 0.01 (t-test). Scale bar, 50 μm. (D) Mouse hind limb ischemia model assay. Mice were injected with miR-190 or NC duplex four times after surgery (right leg). Quantitation of laser Doppler-derived blood flow in the 14th day after surgery. Three representative experiments are shown. N = 6. (**) P < 0.01 (t-test). (E) Photoacoustic microscopy imaging results of ischemic limbs treated with NC or miR-190 in the 14th day after surgery. Scale bar, 1 mm. Arrows indicate new vessels.
Next, we used a mouse hind limb ischemia model to characterize the role of miR-190 in angiogenesis in vivo. miR-190 or scramble siRNA (NC) was packaged with nanoparticles (Lin et al. 2011) and was locally administrated at injured sites on day 0, 3, 7, or 12 after the induction of hind limb ischemia. On day 14, laser Doppler-derived blood flow was examined. The results showed that the local injection of miR-190 significantly inhibited the angiogenesis in ischemic limbs, as evidenced by <40% of blood intensity in comparison with limbs treated with a control scramble siRNA (Fig. 1D). Furthermore, the vessel densities were quantified by using photoacoustic microscopy (PAM). Less neovascularization has been generated in ischemic limbs treated with miR-190 (Fig. 1E). Together, these results demonstrated that miR-190 suppresses the angiogenic traits of HUVECs both in vitro and in vivo.
miR-190 inhibits tumor metastasis and angiogenesis in vivo
Another key question was whether miR-190 would inhibit tumor metastasis, especially angiogenesis, in tumor development. Ectopic expression of miR-190 produced a potent inhibition to the cell migration and invasion, but its sponge increased the migratory and invasive capability by >30% (Fig. 2A).
FIGURE 2.
miR-190 inhibits the tumor angiogenesis in vivo. (A) Transwell migration assay and Matrigel invasion assay of HCT116 cells when transfected with miR-190-expressing plasmid or its sponge, respectively (quantified right). N = 3. Scale bar, 50 μm. (B) Images of tumors formed by indicated HCT116 cells harvested from nude mice. Quantification of tumor weight. N = 4. (*) P < 0.05 (t-test). Scale bar, 1 cm. (C) H&E stain of HCT116 primary tumors or lungs isolated from mice that received subcutaneous injection of miR-190-transduced or mock-infected HCT116 cells at 60 d after transplantation. Eight mice were implanted with NC or miR-190 stable-expressing HCT116 cells subcutaneously for each group. H&E staining of lungs indicated tumors could be found in all the eight mice implanted with NC-expressing HCT116 cells, but not in those implanted with miR-190-expressing HCT116 cells. Scale bar, 50 μm. (D) Images of blood vessels on the surface tumors formed by indicated HCT116 cells harvested from nude mice. N = 4. (E) CD31 staining sections of primary tumors formed by indicated HCT116 cells. Quantification of blood vessel area. N = 10. (*) P < 0.05, (**) P < 0.01 (t-test). Scale bar, 50 μm. (F) Images of lungs isolated from mice that received a tail vein injection of miR-190-transduced or mock-infected GFP-labeled HCT116 cells. Metastatic nodules were measured within a period of 5 wk after injection. N = 3. H&E stain of lungs isolated from mice that received a tail vein injection of miR-190-transduced or mock-infected HCT116 cells. Eight mice were injected with NC or miR-190 stable-expressing HCT116 cells from the tail vein for each group. H&E staining of lungs indicated tumors could be found in the seven mice implanted with NC HCT116 cells, but not in those implanted with miR-190-expressing HCT116 cells. Scale bar, 50 μm.
We then generated a series of xenograft mouse models and characterized the molecular role of miR-190 in tumorigenesis in vivo. We delivered a lentiviral construct into HCT116 cells to generate a stable cell line that stably expressed eGFP as well as miR-190 or its sponge. These stable cell lines were subcutaneously injected into mice. The tumor weights obtained on day 60 indicated that miR-190 suppressed primary tumor growth by 80% and its sponge promoted tumor growth over twofold (Fig. 2B). We found that miR-190-expressing tumors were well encapsulated and noninvasive (Fig. 2C). Examination of lungs revealed metastatic nodules developed in control mice, but not in mice implanted with miR-190-expressing cells (Fig. 2C).
Bright field microscopic images demonstrated that the miR-190 sponge dramatically increased the intensity of blood vessels (Fig. 2D). The evidence of CD31 staining showed that miR-190-expressing tumors generated fivefold fewer blood vessels and that the sponge-expressing tumors generated fourfold more blood vessels (Fig. 2E).
We next determined whether miR-190's impact on metastasis was also attributable to effects on later steps of the extravasation-initiation at distant sites, independent of its influence on local invasion. We then injected miR-190-expressing HCT116 directly into the circulation of severe combined immunodeficiency (SCID) mice. Metastatic nodules developed in the skull, liver, or lungs in a period of 5 wk after injection in control mice. Examination of lung revealed 14-fold fewer lung metastases in mice implanted with miR-190-expressing HCT116 cells (Fig. 2F).
Finally, we collected and analyzed the expression patterns of miR-190 in 50 pairs of primary colon carcinoma and normal adjunct samples. Expression analysis confirmed a twofold lower miR-190 level in the carcinoma samples (P = 1.10 × 10−5) (Fig. 3A). We then examined the expression level of miR-190 in 10 pairs of matched samples of primary colon cancer and lymph node metastasis tumor, each from the same patient. It was found that the mean expression of miR-190 exhibited a significant inverse relationship with metastasis in human colon patients (P = 0.00697) (Fig. 3B). The in situ hybridization data for miR-190 showed that the proportion of cells expressing miR-190 was reduced threefold in the colon tumor samples relative to the adjacent normal ones (Fig. 3C). On the basis of these observations in vivo, we conclude that miR-190 played a critical role in tumor development, especially in both the early and late stages of metastasis.
FIGURE 3.
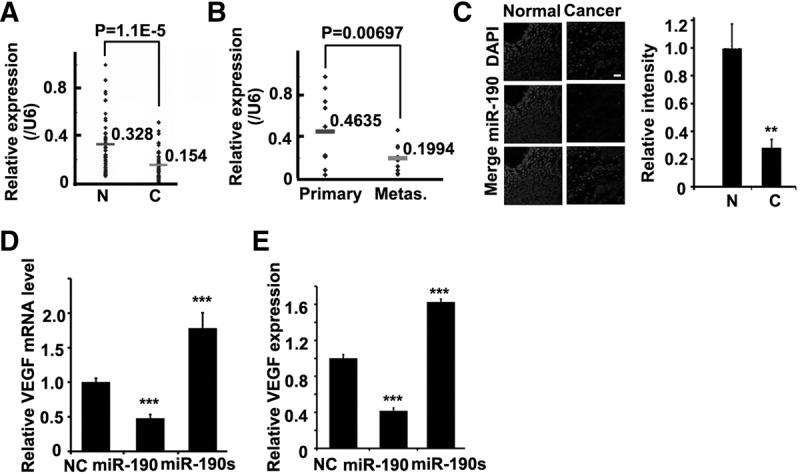
Expression patterns of miR-190 in human colon samples. (A) Real-time PCR for miR-190 in 50 pairs of human colon tumors or normal adjunct samples. There was a significant difference in miR-190 expression between these two groups. N, normal; C, cancer. Each sample is repeated three times. (B) The same samples as in A were divided into two groups: primary colon cancer and lymph node metastasis tumor samples, each from the same patient. There was a significant difference in miR-190 expression between these two groups. Each sample is repeated three times. (C) The in situ hybridization for miR-190 (middle) in the clinical samples (quantification right); DAPI counterstain (top). N, Normal; C, Cancer. N = 3. (**) P < 0.01 (t-test). Scale bar, 200 μm. (D) Relative VEGF mRNA levels in HCT116 cells infected as indicated by real-time PCR. GAPDH was used as the loading control in real-time PCR assay. N = 3. (***) P < 0.005 (t-test). (E) Expression of VEGF in HCT116 cells infected as indicated by ELISA assay for secreted VEGF. N = 3. (***) P < 0.005 (t-test).
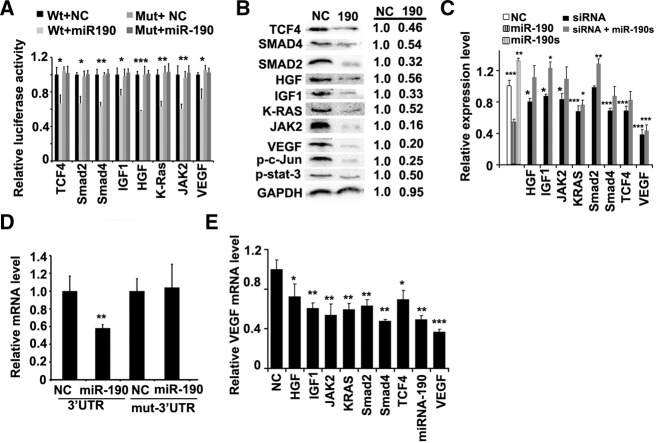
We hypothesized that miR-190 inhibitory effect on the angiogenesis resulted from the regulation of VEGF expression by this miRNA. To investigate this hypothesis, we then measured the transcriptional and translational levels of VEGF after transfection with miR-190 or its sponge in HCT116s. Clearly, the overexpression of miR-190 resulted in a decrease of VEGF mRNA and protein by 40%–50%, while its sponge resulted in an increase of VEGF mRNA and protein by 60%–80%, as evidenced by real-time PCR and ELISA (Fig. 3D,E).
miR-190 regulates a group of angiogenic target genes
To determine the molecular mechanism of miR-190, we employed three algorithms that predict the targets of a miRNA, PicTar (Krek et al. 2005), TargetScan (Grimson et al. 2007), and miRDB (Wang 2008). More than 100 genes appeared to be regulated by miR-190 on the basis of at least two of these algorithms (Supplemental Fig. S3). First, we investigated whether miR-190 directly targets VEGF, since the 3′ untranslated region (3′ UTR) of the VEGF mRNA contains one conserved predicted binding site for miR-190. We cloned the VEGF 3′ UTR sequence into a luciferase construct and examined luciferase activity after cotransfection with miR-190 or NC in 293T cells (Supplemental Table S1). Indeed, overexpression of miR-190 reduced the luciferase activity, while the down-regulation of luciferase activity was prevented by mutating the target sequence (Fig. 4A; Supplemental Fig. S4A).
FIGURE 4.
miR-190 regulates a group of target genes centralizing VEGF. (A) Luciferase activity in the 293T cells upon transfection with different reporter constructs as indicated. NC stands for an empty vector. N = 3. (*) P < 0.05, (**) P < 0.01, (***) P < 0.005 (t-test). (B) Immunoblots for endogenous TCF4, Smad2, Smad4, RAS2, Jak2, HGF, IGF1, VEGF, phosphorylated c-Jun, or phosphorylated Stat3 in the indicated HCT116 cells. Representative images are shown at left and the average value for each gene is calculated at right (N = 4). GAPDH was used as the loading control. (C) ELISA assay of secreted VEGF in HCT116 cells as indicated. N = 3. (*) P < 0.05, (**) P < 0.01, (***) P < 0.005 (t-test). (D) Relative mRNA levels of luciferase reporters containing VEGF 3′ UTR or mutant 3′ UTR in HCT116 cells infected as indicated by real-time PCR. GAPDH was used as the loading control in real-time PCR assay. N = 3. (**) P < 0.01 (t-test). (E) Relative VEGF mRNA levels in HCT116 cells infected as indicated by the real-time PCR. GAPDH was used as the loading control in real-time PCR assay. N = 3. (*) P < 0.05, (**) P < 0.01, (***) P < 0.005 (t-test).
Second, we cloned the 3′ UTR of another 20 putative miR-190 targets into a luciferase construct and performed quantitative analysis of luciferase activity (Supplemental Fig. S4B). Among them, eight reporters fused to the 3′ UTR of these putative targets resulted in a silencing of at least 80% by miR-190. Consistent with this, the luciferase activity remained unchanged when these reporters were fused to their mutant 3′ UTRs (Fig. 4A). We then assayed the endogenous protein levels of the eight candidate genes, including TCF4, Smad2, Smad4, RAS2, Jak2, HGF, IGF1, and VEGF when miR-190 was overexpressed in HCT116 cells. We found that miR-190 reduced the levels of these proteins by 46%–84% (Fig. 4B). Furthermore, we investigated whether the downstream modulators of the target genes were altered accordingly when miR-190 was overexpressed in HCT116 cells. The phosphorylated c-Jun or Stat3 proteins, which are downstream modulators of RAS or Jak2, were decreased by 75% and 50%, respectively (Fig. 4B).
Third, we synthesized a group of siRNAs, each of which silenced an individual gene. These siRNAs, similar to miRNA-190, inhibited the expression level of VEGF with the exception of Smad2 siRNA (Fig. 4C). What is noted, the silence effect of the individual gene on VEGF expression, with the exception of VEGF per se, is smaller than the effect of miR-190. Consistently, in the presence of the miR-190 sponge, the siRNA inhibitory effects were compromised, except for VEGF (Fig. 4C). Taken together, these algorithmic and experimental results indicate that miR-190 regulates a group of angiogenic target genes, including TCF4, Smad2, Smad4, RAS2, Jak2, HGF, IGF1, and VEGF. These target genes have been reported to regulate VEGF at the transcriptional or intracellular level.
To characterize the role of this additional-layer regulation, we examined the contribution of miR-190 to VEGF transcription or translation levels, respectively. We first quantitatively examined the direct effect of miR-190 on VEGF mRNA or protein by use of the luciferase reporter containing VEGF 3′ UTR. The real-time PCR results and the luciferase assays demonstrated the consistent inhibition, with the 30%–40% decrease (Fig. 4A,D). Second, the total inhibition effect of miR-190 on VEGF transcription, ∼50%, was bigger than the silence effect of the individual gene, with the exception of VEGF per se (Fig. 4E). However, the total inhibition of miR-190 on VEGF endogenous protein level was close to 80% (Fig. 4B). Therefore, the addition of miR-190-mediated regulation in the canonical VEGF pathways affords a simple but efficient approach to tuning the expression of VEGF.
miR-190 inhibits the tumor microenvironment angiogenesis
The tumor microenvironment affects angiogenesis by modulating the signaling pathways required for vascular construction. We then studied whether the supernatants from the different HCT116 cells would regulate the angiogenic features of HUVECs in vitro. At 24 h culture after transfection, HUVECs were treated with the supernatants from the different HCT116 cells. It was found that, in contrast to scramble siRNA, the supernatant from the HCT116 cells expressing miR-190 significantly suppressed both tubule and sprout formation, while the supernatant from HCT116 cells expressing the miR-190 sponge stimulated them (Fig. 5A,B). To confirm that the expression level of miR-190 in colon cancer cell lines affects the angiogenic status of HUVECs by regulating the expression of VEGF, we then measured the transcriptional and translational levels of VEGF in HCT116 cells transfected with miR-190 or its sponge. The overexpression of miR-190 resulted in a decrease of the VEGF mRNA and protein by 50%, while its sponge induced an increase of the VEGF mRNA and protein by 30%–40% (Fig. 5C). We also examined the levels of p-VEGFR2 in HCT116, and found that the level of p-VEGFR2 was decreased by 20% when treated with miR-190 (Supplemental Fig. S5). When HUVECs were treated with the supernatants from the HCT116 cells expressing miR-190, the level of p-VEGFR2 was decreased by 60%.
FIGURE 5.
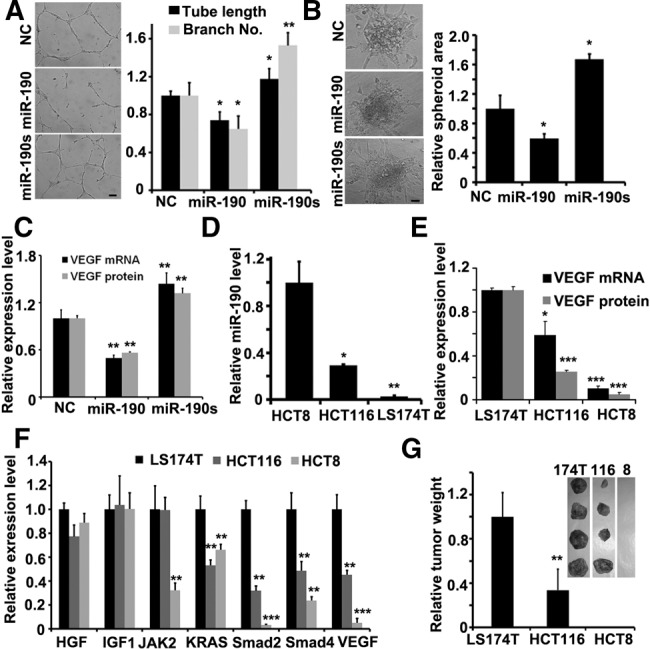
miR-190 inhibits the tumor microenvironment angiogenesis. (A) Tubule formation of HUVECs treated with supernatant as indicated. Shown is a representative image at 24 h after plating. Quantitation of tubule branching and tubule length. N = 5. (*) P < 0.05, (**) P < 0.01 (t-test). Scale bar, 50 μm. (B) Spheroid-based angiogenesis assay of HUVECs treated with supernatant as indicated. Quantitation of cumulative length of the sprouts. N = 10. (*) P < 0.05 (t-test). Scale bar, 50 μm. (C) Expression of VEGF in HCT116 cells transfected with different vectors as indicated by real-time PCR or ELISA assay. N = 3. (**) P < 0.01 (t-test). (D) Expression of miR-190 in different colon cancer cell lines as indicated by real-time PCR. N = 3. (*) P < 0.05, (**) P < 0.01 (t-test). (E) Expression of VEGF mRNA or secreted protein in different colon cancer cell lines as indicated by real-time PCR or ELISA assay. N = 3. (*) P < 0.05, (***) P < 0.005 (t-test). (F) The mRNA level of HGF, IGF1, Jak2, KRAS, Smad2, Smad4, or VEGF in HCT116, HCT8, and LS174T cell lines. (G) Images of tumors formed by different colon cancer cell lines as indicated harvested from nude mice. No solid tumor found in nude mice after injection of HCT8 cells. Quantification of tumor weight. N = 4. (**) P < 0.01 (t-test).
Finally, we characterized the correlation of miR-190 and VEGF in the colon cancer cell lines HCT116, HCT8, and LS174T. As expected, miR-190 expression level exhibited a reverse pattern compared with the transcriptional or secreted levels of VEGF in these three types of cells (Fig. 5D,E). The results showed that LS174T exhibited the lowest expression level of miR-190, along with the highest levels of VEGF mRNA and secreted protein. We also examined the transcriptional levels of HGF, IGF1, Jak2, KRAS, Smad2, or Smad4 in three cells, and demonstrated the similar pattern of Smad2 or Smad4 to one of VEGF (Fig. 5F). Furthermore, we subcutaneously injected these three cell lines into mice, respectively. The tumor weight on day 40 indicated that LS174T cells had the highest capacity for tumorigenesis, while HCT8 cells had the lowest capacity (Fig. 5G). In sum, these in vitro and in vivo experiments demonstrated that miR-190 plays a critical role in the tumor microenvironment, especially in terms of angiogenesis.
DISCUSSION
To date, only a few miRNAs, such as miR-20 and miR-93, have been experimentally shown to directly target the 3′ UTR region of VEGF mRNA (Dews et al. 2006; Lei et al. 2009). The results of this study reveal an miR-190-mediated regulation network that is centralized on VEGF (Fig. 6). It is well known that miRNA regulates a group of functionally related genes that modulate the activities of a specific cell process (Hobert 2007). However, to the best of our knowledge, this study is the first demonstration of one miRNA governing a large group of angiogenic effectors, including TCF4, Smad2, Smad4, RAS2, Jak2, IGF1, and HGF besides VEGF. In addition, these target genes could also mitigate the effect of miR-190 and buffer the expression level of VEGF on the whole. The synergistic effect of miR-190 and its target genes demonstrates a complex but seemingly more stable system for stabilizing the concentration of VEGF. It seems likely that the implementation of this type of switch would be beneficial to the robustness of regulation and/or reduction of the variability incurred by ever-changing environmental conditions.
FIGURE 6.
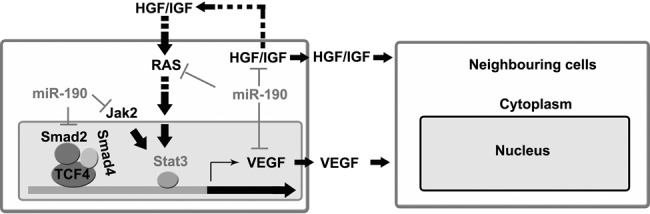
miR-190 regulates the expression of VEGF at three levels. Schematic diagram showing the synergistic effect of miR-190 and its target genes on VEGF-mediated angiogenesis.
miR-190 is conserved in mice, rats, and humans, and is localized in the intronic regions of the talin2 gene. Its expression was dependent on the phosphorylated state of YinYang1 (YY1), a highly conserved and ubiquitously expressed protein (Zheng et al. 2010). However, the expression of YY1 was in turn up-regulated by insulin-like growth factor-1 (IGF1) (Flanagan 1995). Given that miR-190 directly targets IGF1, as described in this report, we believe that there exists a complex feedback network between miR-190 and YY1. In addition, the down-expression of miR-190 was also found in melanoma cancer samples (Mueller et al. 2009). The impaired miRNA biogenesis was observed in cancers, which may result in the global repression of miRNAs (Thomson et al. 2006; Kumar et al. 2007). Actually, we tried to determine whether the effect of miR-190 is attributed to endothelial cells or cancer cells, which is still a challenging task. First, we quantified the expression level of miR-190 in different cells including HUVECs or a few of cancer cell lines (Supplemental Fig. S6). Thus, miR-190 might make a different effect in cancer cells relative to endothelial cells, and the mechanism of decreased miR-190 levels in colon cancer tumor samples and cell lines remains to be characterized in the future. Second, although we did not discriminate the role of miR-190 in endothelial cells or cancer cells, the inhibition effect of this miRNA on VEGF or angiogenesis is consistent in either type of cells. One miRNA could target hundreds of genes and behave as a functional molecule, thus indicating that a miRNA regulates a certain gene in a cellular context-dependent manner. Therefore, we concentrated on the molecular mechanism underling the interaction between miR-190 and its target genes, and would put more effort into the investigation of the cellular context-dependent regulation in the future. We demonstrated that miR-190 directly regulates a group of angiogenic effectors besides VEGF per se, which modulates the expression of VEGF through either canonical transcription regulation or sponging the effect miR-190, thus buffering the expression level of VEGF on the whole.
Our results showed that miR-190 targets the two diffusible growth factors HGF and IGF1, thus altering the local microenvironment and subsequently regulating the expression of VEGF in neighboring endothelial cells. A series of in vitro and in vivo experiments showed that miR-190 plays a critical role in both the early and late stages of colon cancer metastasis (Fig. 2). Therefore, the result of miR-190 having the dual capacity of antitumor and anti-angiogenesis shows that miR-190 is a high potential in cancer therapy target.
Two challenging issues need to be overcome prior to the employment of these strategies in preclinical as well as clinical studies. First, an accurate but high-throughput method is still urgently needed to identify all the target genes a miRNA regulates (Thomas et al. 2010), even though many prediction-based algorithmic and experimental approaches have already been developed. Second, the tissue-specific delivery of miRNA inhibitors or mimics is still challenging. This is the case despite the fact that pioneering studies have demonstrated the potential of miRNA-based strategies in clinical applications (Ma et al. 2007; Kota et al. 2009). However, the finding that miR-190 regulates angiogenesis via a modulation of various effectors centralized on VEGF points to a new way of understanding the molecular mechanism of this type of small RNA, which will finally realize the potential of miRNAs against a variety of diseases, including cancer.
MATERIALS AND METHODS
Ethics statement
All human colon samples were obtained from Beijing Cancer Hospital. Before surgery at the center, all patients provided written informed consent to allow any excess tissue to be used for research studies. All research involving animals complied with protocols approved by the Laboratory Animal Center, Peking University. All the BALB/C nude mice were purchased from the animal center of Peking University.
Cell culture
Cell lines including HCT116, HCT8, and LS174T were cultured in Dulbecco's Modified Eagle Medium (DMEM) containing 10% fetal bovine serum (FBS), 100 units/mL penicillin and 0.1 mg/mL streptomycin under humidified conditions in 95% air and 5% CO2 at 37°C. HUVECs were cultured for three passages in ECM medium (ScienCell) containing 1% ECGS, 5% FBS, and 1% P/S. Cells were washed with PBS and incubated in 0.25% trypsin containing 5 mM EDTA. DMEM containing 10% FBS were added to terminate trypsin. After centrifugation, the pallet was diluted with the medium and counted via haemocytometer.
Image capture and analysis
Images for cells and tissues were taken by Nikon TE2000-E (CCD: Regita 2000R, Qimaging), Nikon TE2000-S (CCD: QHY IMG2P, Xinminke), or ImageXpress Micro (MD). Quantization was calculated by the software of ImageJ.
Synthesis of siRNA duplex and miRNA mimics
siRNA duplexes, scramble siRNA, and all human miRNA mimics were obtained from the Viewsolid. The silence efficiency of these RNAi constructs for their targeted mRNAs was tested by quantitative real-time PCR at 24–48 h after transfection. All reactions were run in triplicate.
Transwell migration assay
Cells were seeded into the upper chamber of a Transwell insert (24-well insert; pore size, 8 µm; Costar) in 100 μL serum-free medium per well. Six-hundred-microliter medium containing 10% serum was placed in the lower chamber to act as a chemoattractant. Nonmigratory cells were removed from the upper chamber by scraping with a cotton bud. The cells remaining on the lower surface of the insert were fixed with 2% formaldehyde (Sigma) and stained by DAPI (Roche). Cells were quantified as the number of cells found in 15 random microscope fields in three independent inserts. Error bars represent the SD from three separate experiments.
Luciferase assays
4.0 × 104 293T cells were cotransfected with 200 ng of the indicated pGL3 firefly luciferase construct and 20 ng of a pGL3 Renilla luciferase normalization control. The media was changed after 6 h, and luciferase activity was measured after 48 h using the dual luciferase reporter assay system (Promega). The 3′ UTRs of all genes cloned are shown in Supplemental Table S1.
Immunoblotting
Lysates were resolved by electrophoresis, transferred to a polyvinylidene difluoride membrane (Millipore Corporation), and probed with antibodies against GAPDH (Santa Cruz), VEGF (Santa Cruz), SMAD2 (CST), IGF1 (Santa Cruz), SMAD4 (CST), JAK2 (CST), HGF (Abcam), Phospho-STAT3 (CST), Phospho-c-Jun (CST).
miRNA in situ hybridization
miRNA expression was assessed from paraffin sections. In brief, after 2-h prehybridization, a 5′ FITC-labeled miRCURY LNA probe targeting miR-190 (Exiqon) was hybridized to proteinase K-treated 10-mm sections at 37°C for 12 h.
In vitro Matrigel angiogenesis assays
Briefly, 24 h after infection of miR-190, miR-190 sponge, or NC, HUVECs were seeded on Matrigel (BD Biosciences) in 24-well plate (Sigma). Tube lengths and branches were measured and quantified by ImageJ software.
Quantitative real-time PCR
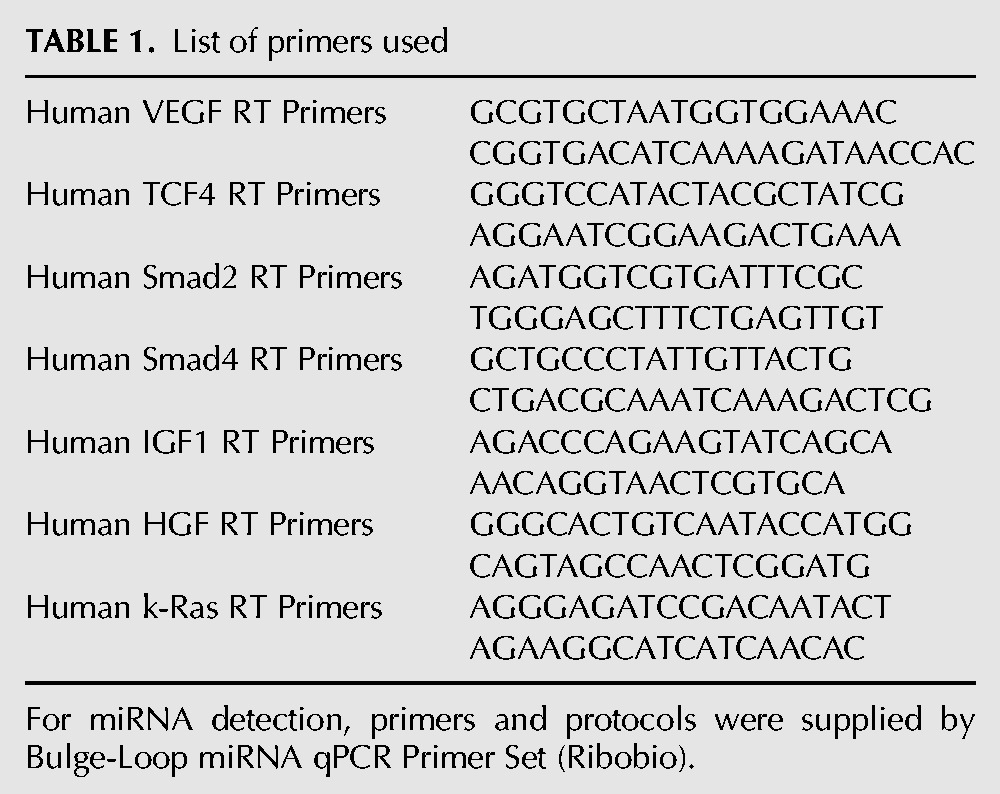
Quantitative real-time PCR (qRT-PCR) was processed by Real Master Mix (Tiangen Biotech, Co. Ltd) using Bio-Rad Opticon2 Real-Time PCR Detector (Bio-Rad Laboratories). The primers are shown in Table 1.
TABLE 1.
List of primers used
Enzyme-linked immunosorbent assay
The concentration of VEGF in culture medium supernatants was determined by enzyme-linked immunosorbent assay (ELISA). The human VEGF-A ELISA kits were purchased from R&D Systems. The assay was performed according to the manufacturer's protocol.
Spheroid-based angiogenesis assay
Endothelial cell spheroids of a defined cell number were generated as described previously (Korff and Augustin 1998; Diehl et al. 2007). In vitro angiogenesis was quantified by measuring the cumulative length of the sprouts that had grown out of each spheroid using ImageJ software analyzing 10 spheroids per group and experiment.
Animal studies and Immunohistochemistry
Mice were injected into the dorsum or tail vein with indicated HCT116, HCT8, and LS174T cells in PBS. Immunohistochemistry was accomplished at Chinese Academy of Medical Sciences. Tissue samples were fixed in 10% buffered formalin for 12 h, followed by a wash with PBS and transfer to 70% ethanol, and then embedded in paraffin, sectioned, and stained with haematoxylin and eosin. The immunohistochemistry detection with anti-CD-31 antibodies (BD Biosciences) was performed on paraffin sections.
Murine model of hind limb ischemia and evaluation of blood flow, collateralization, and neovascularization
Male C57 mice were subjected to unilateral hind limb surgery under anesthesia with sodium pentobarbital (50 mg/kg). The left femoral artery was separated from the vein and nerve, ligated proximally, and excised. The right hind limb served as control. Blood flow was measured by using a laser Doppler perfusion imager (PeriScan PIM3; Perimed). Ischemic and nonischemic limb perfusion was measured before and after surgery and 1 wk later. Semiquantitative assessment of the severity of limb ischemia was performed according to a clinical score standard. The neovascularization was quantified by using PAM (laser pulses, 532 nm; repetition rate: 10 Hz, pulse width: 10 nsec).
Preparation of nanoparticles/microRNA complexes
NPs/microRNA complexes were prepared via electrostatic interactions. For example, to prepare PEGb–PAEM/microRNA complexes at N/P = 10 (the charge ratio [N/P] of polymers and microRNA was calculated as the mole ratio of moles of the amino/guanidino groups [N] on polymer to the phosphate groups [P] on microRNA), PEGb–PAEM was diluted to 4.2 mg/10 mL in 10 mm DEPC water (pH = 7.3), microRNA was diluted to 0.665 mg/10 mL in 10 mm DEPC water (pH = 7.3). Complexes were prepared by adding 50 μL NPs suspension to an equal volume of microRNA solution by pipetting and then incubated for 30 min before characterization and other experiments. A given formulation was administered to each mouse via orthotopic injection at 2.5 mg/kg.
Human colon tumor samples
Samples were snap frozen in OCT and stored at −80°C until use. The pathologic status of the specimens was provided by the hospital. The quantitative examination of miRNA expression in those samples follows to previous description.
Statistical analysis
For statistical analysis, two-sided paired Student's t-tests were processed by Excel spreadsheet. P-values were given, but in general, P < 0.05 was considered as statistically significant. Error bars represented SDs of at least three independent experiments, unless indicated otherwise.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank Jingyan Han from Peking University for providing the Doppler perfusion imager. This work was supported by projects of MOST (Grant No. 2011CB809106, 2013CB917803) and NSFC (Grant No. 81030040, 31371443, 30600142).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.044651.114.
REFERENCES
- Anand S, Majeti BK, Acevedo LM, Murphy EA, Mukthavaram R, Scheppke L, Huang M, Shields DJ, Lindquist JN, Lapinski PE, et al. 2010. MicroRNA-132-mediated loss of p120RasGAP activates the endothelium to facilitate pathological angiogenesis. Nat Med 16: 909–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birk DM, Barbato J, Mureebe L, Chaer RA 2008. Current insights on the biology and clinical aspects of VEGF regulation. Vasc Endovascular Surg 42: 517–530 [DOI] [PubMed] [Google Scholar]
- Clifford RL, Deacon K, Knox AJ 2008. Novel regulation of vascular endothelial growth factor-A (VEGF-A) by transforming growth factor β1: requirement for Smads, β-catenin, and GSK3β. J Biol Chem 283: 35337–35353 [DOI] [PubMed] [Google Scholar]
- Coultas L, Chawengsaksophak K, Rossant J 2005. Endothelial cells and VEGF in vascular development. Nature 438: 937–945 [DOI] [PubMed] [Google Scholar]
- Dews M, Homayouni A, Yu D, Murphy D, Sevignani C, Wentzel E, Furth EE, Lee WM, Enders GH, Mendell JT, et al. 2006. Augmentation of tumor angiogenesis by a Myc-activated microRNA cluster. Nat Genet 38: 1060–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl F, Rossig L, Zeiher AM, Dimmeler S, Urbich C 2007. The histone methyltransferase MLL is an upstream regulator of endothelial-cell sprout formation. Blood 109: 1472–1478 [DOI] [PubMed] [Google Scholar]
- Fish JE, Santoro MM, Morton SU, Yu S, Yeh RF, Wythe JD, Ivey KN, Bruneau BG, Stainier DY, Srivastava D 2008. miR-126 regulates angiogenic signaling and vascular integrity. Dev Cell 15: 272–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan JR 1995. Autologous stimulation of YY1 transcription factor expression: role of an insulin-like growth factor. Cell Growth Differ 6: 185–190 [PubMed] [Google Scholar]
- Flynt AS, Lai EC 2008. Biological principles of microRNA-mediated regulation: shared themes amid diversity. Nat Rev 9: 831–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folkman J 2007. Angiogenesis: an organizing principle for drug discovery? Nat Rev Drug Discov 6: 273–286 [DOI] [PubMed] [Google Scholar]
- Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP 2007. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell 27: 91–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert O 2007. miRNAs play a tune. Cell 131: 22–24 [DOI] [PubMed] [Google Scholar]
- Korff T, Augustin HG 1998. Integration of endothelial cells in multicellular spheroids prevents apoptosis and induces differentiation. J Cell Biol 143: 1341–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kota J, Chivukula RR, O'Donnell KA, Wentzel EA, Montgomery CL, Hwang HW, Chang TC, Vivekanandan P, Torbenson M, Clark KR, et al. 2009. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell 137: 1005–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krek A, Grün D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, et al. 2005. Combinatorial microRNA target predictions. Nat Genet 37: 495–500 [DOI] [PubMed] [Google Scholar]
- Kuehbacher A, Urbich C, Dimmeler S 2008. Targeting microRNA expression to regulate angiogenesis. Trends Pharmacol Sci 29: 12–15 [DOI] [PubMed] [Google Scholar]
- Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T 2007. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet 39: 673–677 [DOI] [PubMed] [Google Scholar]
- Lei Z, Li B, Yang Z, Fang H, Zhang GM, Feng ZH, Huang B 2009. Regulation of HIF-1α and VEGF by miR-20b tunes tumor cells to adapt to the alteration of oxygen concentration. PLoS One 4: e7629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin D, Huang Y, Jiang Q, Zhang W, Yue X, Guo S, Xiao P, Du Q, Xing J, Deng L, et al. 2011. Structural contributions of blocked or grafted poly(2-dimethylaminoethyl methacrylate) on PEGylated polycaprolactone nanoparticles in siRNA delivery. Biomaterials 32: 8730–8742 [DOI] [PubMed] [Google Scholar]
- Ma L, Teruya-Feldstein J, Weinberg RA 2007. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature 449: 682–688 [DOI] [PubMed] [Google Scholar]
- Mueller DW, Rehli M, Bosserhoff AK 2009. miRNA expression profiling in melanocytes and melanoma cell lines reveals miRNAs associated with formation and progression of malignant melanoma. J Invest Dermatol 129: 1740–1751 [DOI] [PubMed] [Google Scholar]
- Nicoli S, Standley C, Walker P, Hurlstone A, Fogarty KE, Lawson ND 2010. MicroRNA-mediated integration of haemodynamics and Vegf signalling during angiogenesis. Nature 464: 1196–1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samani AA, Yakar S, LeRoith D, Brodt P 2007. The role of the IGF system in cancer growth and metastasis: overview and recent insights. Endocr Rev 28: 20–47 [DOI] [PubMed] [Google Scholar]
- Sullivan R, Graham CH 2007. Hypoxia-driven selection of the metastatic phenotype. Cancer Metastasis Rev 26: 319–331 [DOI] [PubMed] [Google Scholar]
- Thomas M, Lieberman J, Lal A 2010. Desperately seeking microRNA targets. Nat Struct Mol Biol 17: 1169–1174 [DOI] [PubMed] [Google Scholar]
- Thomson JM, Newman M, Parker JS, Morin-Kensicki EM, Wright T, Hammond SM 2006. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev 20: 2202–2207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X 2008. miRDB: a microRNA target prediction and functional annotation database with a wiki interface. RNA 14: 1012–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurdinger T, Tannous BA, Saydam O, Skog J, Grau S, Soutschek J, Weissleder R, Breakefield XO, Krichevsky AM 2008. miR-296 regulates growth factor receptor overexpression in angiogenic endothelial cells. Cancer Cell 14: 382–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Hao Y, Yang J, Zhou Y, Li J, Yin S, Sun C, Ma M, Huang Y, Xi JJ 2011. Genome-wide functional screening of miR-23b as a pleiotropic modulator suppressing cancer metastasis. Nat Commun 2: 554. [DOI] [PubMed] [Google Scholar]
- Zheng H, Chu J, Zeng Y, Loh HH, Law PY 2010. Yin Yang 1 phosphorylation contributes to the differential effects of μ-opioid receptor agonists on microRNA-190 expression. J Biol Chem 285: 21994–22002 [DOI] [PMC free article] [PubMed] [Google Scholar]