Abstract
Cancer-associated IDH mutations are characterized by neomorphic enzyme activity and resultant 2 hydroxyglutarate (2HG) production. Mutational and epigenetic profiling of a large AML patient cohort revealed that IDH1/2-mutant AMLs display global DNA hypermethylation and a specific hypermethylation signature. Furthermore, expression of 2HG-producing IDH alleles in cells induced global DNA hypermethylation. In the AML cohort, IDH1/2 mutations were mutually exclusive with mutations in the α-ketoglutarate-dependent enzyme TET2, and TET2 loss-of-function mutations associated with similar epigenetic defects as IDH1/2 mutants. Consistent with these genetic and epigenetic data, expression of IDH mutants impaired TET2 catalytic function in cells. Finally, either expression of mutant IDH1/2 or Tet2 depletion impaired hematopoietic differentiation and increased stem/progenitor cell marker expression, suggesting a shared proleukemogenic effect.
Introduction
Acute myeloid leukemia (AML) pathogenesis is characterized by recurrent chromosomal translocations and somatic mutations that can define biologically distinct disease subtypes. Among these abnormalities are mutations or rearrangement of genes encoding aberrant transcription factors or cofactors that directly perturb gene expression and disrupt cell differentiation and survival. Others include gain of function mutations of kinases involved in transduction of growth and proliferation signals. However, in many AML cases the identity and function of pathogenetic mutations remains obscure. Recent genomic sequencing efforts in AML and in other malignancies have identified new classes of oncogenic disease alleles. One recently identified class of genes mutated in cancer are those coding for enzymes involved in citrate metabolism (Mardis et al., 2009; Parsons et al., 2008). The most prevalent such mutations identified to date affect the genes for cytosolic isocitrate dehydrogenase 1 (IDH1) and its mitochondrial homolog IDH2. IDH1 and IDH2 lesions occur in ~70% of patients with lower grade (grade II-III) brain tumors such as astrocytomas, as well as in secondary glioblastomas derived from lower grade glial tumors (Hartmann et al., 2009; Parsons et al., 2008; van den Bent et al., 2010; Yan et al., 2009). IDH1 and IDH2 mutations were subsequently observed in myeloid malignancies including de novo and secondary AML (~15–30%), and pre-leukemic clonal malignancies including myelodysplasia and myeloproliferative neoplasms (~5% of chronic phase and ~20% of transformed cases) (Marcucci et al., 2010; Mardis et al., 2009; Paschka et al., 2010; Tefferi et al., 2010; Wagner et al., 2010; Ward et al., 2010). The mutational data in malignant gliomas and in myeloid malignancies suggest that IDH1/2 mutations can occur early in disease pathogenesis and may drive tumorigenesis.
The precise genetic context in which IDH1/2 mutations occur is not known, nor is the mechanism through which they contribute to the malignant phenotype. IDH1/2 mutations are heterozygous, with tumors retaining one wild-type copy of the relevant IDH1 or IDH2 allele, suggesting that the mutations are selected for an enzymatic gain of function rather than a loss of function (Dang et al., 2009; Ward et al., 2010), and that retention of the wild-type allele may be required for normal cellular metabolism. IDH1 and IDH2 are NADP+-dependent enzymes that normally catalyze the interconversion of isocitrate and alpha-ketoglutarate (αKG - also known as 2-oxoglutarate). The most common IDH1/2 mutations in AML and brain tumors, affecting R132 of IDH1 or R140 and R172 of IDH2, have the common feature of acquiring a neomorphic enzymatic activity catalyzing the NADPH-dependent reduction of α KG to R(−)-2-hydroxyglutarate (2HG) (Dang et al., 2009; Ward et al., 2010). Presumably it is the production of 2HG that provides a biological advantage that contributes to malignant transformation. As 2HG is a structural analogue of aKG, differing only in the substitution of the alphaketo group on aKG for a hydroxyl group, it is plausible that generation of 2HG by mutant IDH1 and IDH2 might impair the function of enzymes that require αKG as a substrate.
Recent epigenetic studies of large AML patient cohorts have demonstrated that aberrant DNA methylation is a hallmark of AML (Figueroa et al., 2010). Importantly, promoter methylation data can be used to classify AML in distinct clusters defined by specific patterns of methylation. Although a subset of DNA methylation signatures have been found to be associated with known genetic mutations, specific AML subsets were recognized solely based on their DNA methylation profiles (Figueroa et al., 2010). It is not currently known whether there are somatic genetic events that define any of the 5 epigenetically defined clusters in AML. The recent finding that glioblastomas with a methylator phenotype are associated with cytosolic IDH1 mutations suggested a potential causative link between these features, although mutations of the mitochondrial IDH2 enzyme were not detected in this study and as such the relationship between IDH2 mutations and epigenetic state could not be assessed (Noushmehr et al., 2010). In order to explore the mechanism of action of both IDH1 and IDH2 mutations in malignant transformation we performed a large scale genetic, epigenetic and transcriptional profiling study in a cohort of 385 de novo AML patients of <60yrs of age enrolled in a phase III, multicenter Eastern Cooperative Oncology Group (ECOG) clinical trial (Fernandez et al., 2009). This is a different patient population than reported previously(Figueroa et al., 2010), although the methylation data from 344 patients in the Erasmus cohort was used to validate the current results. In this study we test the hypothesis that increased cellular 2HG levels induced by mutant IDH isoforms might contribute to malignant transformation by interfering with the normal cycle of DNA methylation and demethylation through inhibiting α KG-dependent enzymes such as TET2.
Results
Mutations in IDH1 and IDH2 are frequent and mutually exclusive in de novo AML
385 specimens from a total cohort of 398 patients with de novo AML younger than 60 years of age, enrolled in the ECOG E1900 clinical trial (Fernandez et al., 2009) were subjected to DNA sequence analysis for AML-associated recurrent mutations, gene expression microarray profiling and DNA methylation microarray profiling. Patient characteristics are summarized in Table 1. High throughput resequencing of IDH1 and IDH2 revealed IDH1 R132 mutations in 6.2% of patients and IDH2 mutations in 8.6% of patients (6.3% R140Q and 2.3% R172K). Patients with mutations in IDH1 or IDH2 did not differ from IDH1/2-wild-type patients in terms of age, sex, or percentage of bone marrow blasts at diagnosis (Table 1). No additional somatic IDH1/2 mutations were found. All IDH1 and IDH2 mutations were heterozygous, consistent with retention of the wild-type allele as previously reported (Dang et al., 2009; Ward et al., 2010). Although mutations in IDH1 and IDH2 are thought to be mutually exclusive based on mutational studies in malignant gliomas (Hartmann et al., 2009; Yan et al., 2009), occasional rare AML patients have been reported with concurrent mutations in both IDH1 and IDH2 (Paschka et al., 2010). In this large cohort of patients with de novo AML, mutations in IDH1 and IDH2 were mutually exclusive.
Table 1.
Clinical and Genetic Parameters of IDH1/2- and TET2-wild-type and mutant AML samples from the ECOG E1900 Cohort.
|
IDH & TET2 status |
Median Age (Range) |
Gender (M/F) |
Cytogenetic Risk Class (Favorable/Intermediate/ Unfavorable/ Indeterminate) |
% FLT-3 mutant (ITD/TKD) |
NPM1- mutant (%) |
CEBPA mutant (%) |
% bone marrow blast at sample acquisition Median (range) |
|---|---|---|---|---|---|---|---|
|
TET2 and IDH1/2 Wildtype (n=300) |
45.5 (18–60) | 160/140 | 61/133/51/55 | 32%/8% | 11% | 11% | 65% (3–100) |
|
TET2 mutant (n=28) |
55 (30–60) | 17/11 | 2/10/4/12 | 35.7%/3.6% | 21.4% | 10.7% | 69.5% (20–99) |
|
IDH1 or 2 mutant (n=57) |
46.5 (18–60) | 25/32 | 1/35/5/15 | 19.3%/3.5% | 24.6% | 1.8% | 79% (11–100) |
|
IDH1 mutant (n=24) |
46 (18–58) | 10/14 | 1/18/0/5 | 16.7%/0% | 25% | 4.2% | 79% (30–96) |
|
IDH2 mutant (n=33) |
46.5 (24–60) | 15/18 | 0/18/5/10 | 21.2%/6.1% | 24.2% | 0% | 78% (11–100) |
| All patients (n=385) |
46.5 (18–60) | 202/183 | 64/179/60/82 | 31.7%/7% | 14% | 9.9% | 68% (3–100%) |
We then asked if IDH1 and IDH2 mutations were associated with distinct clinical subsets of AML. We found that mutations in IDH1 and IDH2 were significantly enriched in AML patients with intermediate-risk cytogenetics. Specifically, 27.1% of patients with intermediate-risk cytogenetics harbored a mutation in IDH1 or IDH2 vs. only 12.6% of samples from patients who presented with cytogenetic favorable or poor risk AML (p=0.009). We next determined the coincidence of IDH1 or IDH2 with other commonly mutated genes in AML, namely FMS-like tyrosine kinase 3 (FLT3), NPM1, and CEBPA, in the same cohort. IDH1/2 mutations were significantly enriched in patients with cytoplasmic NPM1 mutations (p=0.01), as was recently reported (Abbas et al., 2010; Paschka et al., 2010). In contrast IDH1/2 mutations were not significantly correlated with or exclusive of CEBPA or FLT3-ITD mutations.
IDH1/2 mutations associate with specific cytosine methylation distribution profiles
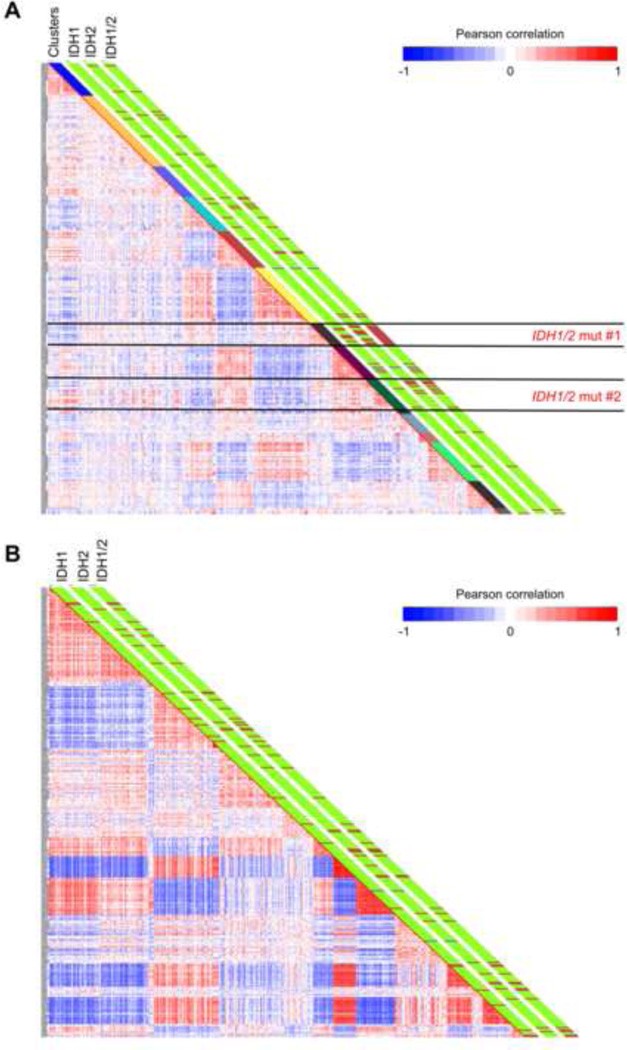
We next performed DNA methylation profiling of ~14,000 promoters (~50,000 CpG sites) in 398 samples in the cohort analyzed for IDH1/2 mutations using the HELP assay (HpaII tiny fragment enrichment by ligation-mediated PCR) (Figueroa et al., 2009). 15 samples did not pass our microarray quality control criteria and were excluded from subsequent analysis. The accuracy of the HELP assay in detecting cytosine methylation levels was validated in these patients using MassArray Epityping, a gold standard quantitative single locus assay (correlation coefficient r= −0.87 in these AML patients, Figure S1A). 51 AML patient samples analyzed by the HELP assay were positive for mutations in either IDH1 (n=22) or IDH2 (n=29). Unsupervised analysis of informative probesets (see supplementary methods) using hierarchical clustering (1 - Pearson correlation distance and Ward’s clustering method) revealed that AMLs harboring IDH1/2 mutations clustered together into two groups based on their common DNA methylation profile, as can be observed in the graphical representation of correlation between samples shown in Figure 1A.
Figure 1. IDH1 and IDH2 mutant AML cases tend to cluster based on their DNA methylation profiles.
(A) Heatmap representation of a correlation matrix in which each patient’s DNA methylation profile is correlated with that of the other patients in the dataset. Patients are ordered according to the unsupervised analysis (hierarchical clustering) results, so that highly correlated patients are located next to each other. Parallel bars on the right of the heatmap have been used to indicate, from left to right: cluster membership, IDH1 mutational status (Green: WT, Dark red: Mutant), IDH2 mutational status (Green: WT, Dark red: Mutant) and combined IDH1/2 mutational status (Green: WT, Dark red: Mutant). (B) Heatmap representation of a correlation matrix in which each patient’s gene expression profile is correlated with that of the other patients in the dataset. Patients are ordered according to the unsupervised analysis (hierarchical clustering) results, so that highly correlated patients are located next to each other. Parallel bars on the right of the heatmap have been used to indicate, from left to right: IDH1 mutational status (Green: WT, Dark red: Mutant), IDH2 mutational status (Green: WT, Dark red: Mutant) and combined IDH1/2 mutational status (Green: WT, Dark red: Mutant). (See also Figure S1 and Table S1).
Hypermethylation is the dominant feature of IDH1/2 mutant AMLs
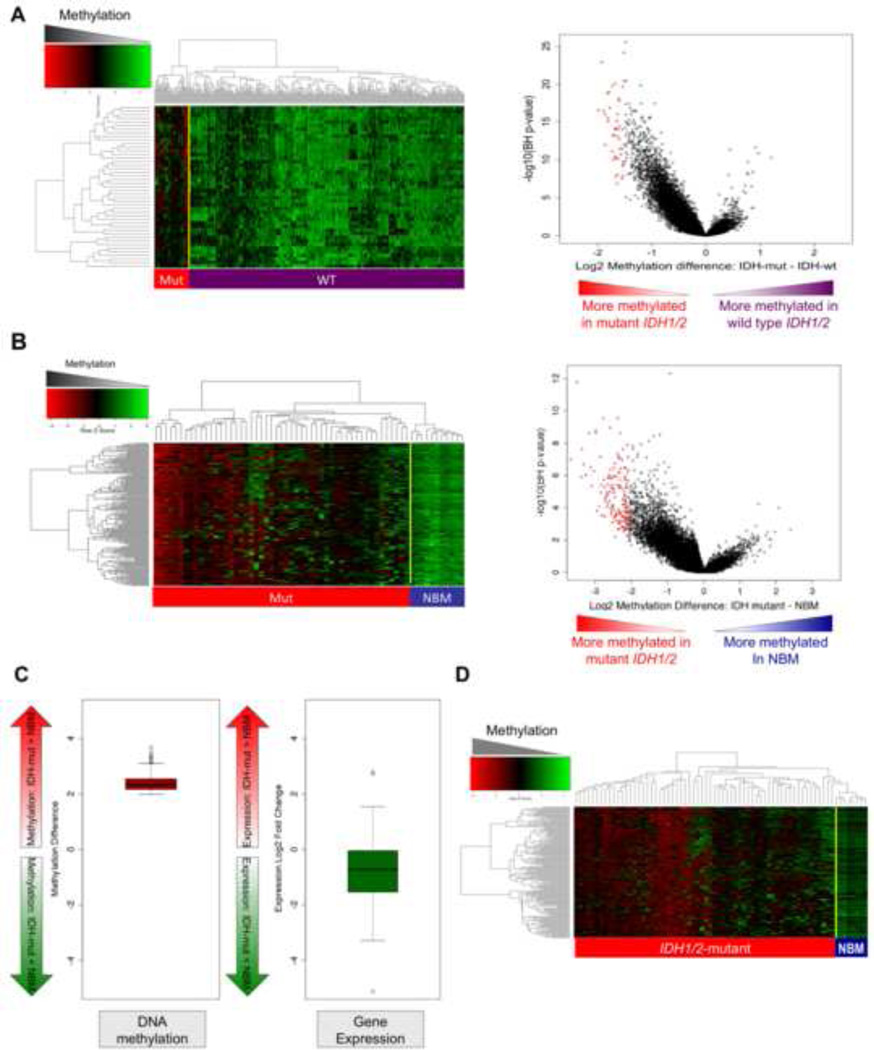
In order to more specifically characterize DNA methylation in IDH1/2-mutant patient samples we next carried out a supervised analysis comparing IDH1/2 wild-type vs. IDH1/2 mutant AMLs. Using stringent criteria (absolute log2 difference in methylation > 1.5, p<0.05 T-test with Benjamini-Hochberg correction for multiple testing (T+BH)) we identified 45 differentially methylated regions (DMRs), all of which were universally hypermethylated in IDH1/2 mutant AMLs (Figure 2A, and Table S1A). Thirty of these genes could be assessed for gene expression (i.e. they were present on the expression arrays used in this study), and 23/30 (77%) were repressed, consistent with the expected effect of hypermethylation. Although many these genes were also expressed at relatively low abundance in a subset of IDH1/2 wild type AMLs, they are uniquely epigenetically silenced through DNA methylation in IDH1/2 mutant AMLs. Furthermore, as evidenced from the asymmetry observed in the volcano dot plot in Figure 2A, in which methylation difference between IDH1/2 mutant and wild-type AMLs is plotted against statistical significance, we also observed a significant increase in overall promoter DNA methylation levels in IDH1/2 mutant AMLs vs. other AML patients, consistent with a methylator phenotype.
Figure 2. IDH1/2 mutant AMLs have a markedly aberrant hypermethylated DNA profile.
(A)Left: Heatmap representation of a two-dimensional hierarchical clustering of genes identified as differentially methylated between IDH1/2 mutant primary AML cases (indicated by the red bar) and IDH1/2 wild-type cases (indicated by the purple bar). Each row represents a probe set and each column represents a patient. Right: Dot plot of methylation difference between IDH-mutant and IDH-wild-type AMLs (biological significance) vs. statistical significance (-log10 (T+BH p value)). Red points indicate probe sets identified as differentially methylated between the two types of AML. (B)Left: Heatmap representation of a two-dimensional hierarchical clustering of genes identified as differentially methylated between IDH1/2 mutant primary AML cases (Mut; red bar) and normal CD34+ bone marrow cells (NBM; blue bar). Each row represents a probe set and each column represents a patient. Right: Dot plot of methylation difference between IDH1/2 mutant AMLs and normal CD34+ bone marrow cells (biological significance) vs. statistical significance (-log10 (T+BH p value)). Red points indicate probe sets identified as differentially methylated between the two groups. (C) Boxplot illustrating average methylation difference between IDH1/2 mutant AMLs vs. normal CD34+ cells (left) and average gene expression difference between IDH1/2 mutant AMLs vs. normal CD34+ cells (right) of genes aberrantly methylated in IDH1/2 mutant AMLs. (D) Heatmap illustrating the validation of the IDH1/2 mutant methylation signature in an independent cohort of 344 AMLs (IDH1/2 mutant AML = Mut; red bar; normal CD34+ bone marrow cells = NBM; blue bar). (See also Figures S2 and S3 and Table S2A–B).
IDH1/2 mutant AMLs feature an aberrant DNA hypermethylation signature vs. normal bone marrow
We next characterized the specific epigenetic alterations associated with IDH1/2 mutant malignant transformation by performing a supervised analysis of HELP profiles between a cohort of normal CD34+ cells obtained from the bone marrows of 11 healthy individuals (NBM) and the 51 IDH1/2-mutant AML. We again observed a marked increase in DNA methylation levels in IDH1/2-mutant AMLs, as evidenced by the asymmetry of the dot plot of overall methylation levels (Figure 2B). Moreover, 154 genes were consistently and robustly differentially methylated in IDH1/2 mutant AMLs vs. normal bone marrow (> 2 log2 methylation difference and p<0.05 T+BH). All of these were aberrantly hypermethylated (Figure 2B, and Table S1B), and we observed an overall trend to an inverse correlation between methylation of these specific genes and their gene expression, since a large proportion of these genes were > 2 fold repressed compared to normal CD34+ cells (Figure 2C). We next investigated whether genes aberrantly methylated in IDH1/2 mutant AMLs could be validated in an independent cohort of AMLs carrying these mutations. The methylation status of genes included in the IDH1/2 epigenetic signature was examined in a series of 344 AMLs from Erasmus University Medical Center, which were previously analyzed by HELP assays using the same microarray design (Figueroa et al., 2010). IDH1/2 mutations were identified in 68 of these cases (Abbas et al., 2010). We observed that the aberrantly methylated signature identified in the E1900 cohort was comparably hypermethylated in this independent cohort of IDH1/2 mutant AMLs (Fig 2D). Moreover, unsupervised analysis of the 344 Erasmus cases again revealed that the IDH1/2 mutant cases clustered together into two epigenetically defined methylation clusters we had identified previously (clusters five and seven) (Figueroa et al., 2010) (Figure S1B).
IDH1 and IDH2 mutations display similar DNA methylation profiles
The fact that IDH1 and IDH2 mutations are mutually exclusive suggests that their biological effect is similar and that their impact on cytosine methylation distribution would overlap. We first carried out principal component analysis (PCA) in order to determine if any amount of the dataset’s variance could be explained by which IDH gene was affected or by the type of mutation. PCA showed that neither the specific IDH gene affected nor the specific allele (IDH1 R132, IDH2 R172, IDH2 R140) constituted a significant source of variance (Figure S2A, S2B). We then performed supervised analyses separately comparing DNA methylation in IDH1 or IDH2 mutant AML samples to NBM samples. Consistent with IDH1 and IDH2 mutations having a common enzymatic gain of function, we observed that methylation changes were quite similar for both groups, although a greater number of genes reached statistical significance (log2 ratio >2 and p< 0.05 T+BH) in IDH1-mutant compared to IDH2-mutant AML samples (268 DMRs for IDH1-mut vs. 168 DMRs for IDH2-mut) (Tables S1C and S1D). Nonetheless, 140 DMRs (83.3% of the hypermethylated genes in IDH2 AML, Fisher p value < 0.0001) were common to both signatures (Figure S2C), and the extent of overall promoter hypermethylation was similar (data not shown). Furthermore, when the IDH1 and IDH2 signatures are compared using a less stringent threshold (p<0.05 T+BH), only four genes in each group failed to reach statistical significance in the other group. The data demonstrate that IDH1 and IDH2 neomorphic mutations have similar effects on DNA methylation in AML cells.
Integrative analysis reveals gene networks perturbed in IDH1/2 mutant AMLs
Next an unsupervised analysis of gene expression profiling data was performed. Unlike DNA methylation profiling, gene expression profiling did not define specific transcriptional cluster(s) based on IDH1/2 mutational status in this AML patient cohort (Figure 1B). These data indicate that the biochemical effects of IDH1/2 mutations affect the epigenome more extensively and specifically than the transcriptome. Similar results were observed in the analysis of gene expression in the Erasmus University Medical Center cohort (not shown). However, a supervised analysis comparing IDH1/2 mutant AML patients to normal CD34+ cells revealed a robust gene expression signature associated with IDH1/2 mutations (fold difference >2 and p<0.001 T+BH) (Figure S2D and Table S1E). The fact that unsupervised analysis based on gene expression does not segregate IDH1/2 mutant AML cases suggests that subsets of the genes deregulated in IDH1/2 mutant AMLs may be shared with other leukemia subtypes, or that gene expression in AML is affected by additional disease alleles which do not impact DNA methylation signatures. We then performed supervised analyses comparing the gene expression profiles of IDH1 and IDH2 mutant AMLs to normal CD34+ cells (>2 fold change, p<0.001 T+BH). There was a significant degree of overlap (Fisher p value<0.0001) between the gene expression signatures defined by IDH1 and IDH2 mutants, respectively (Figure S3A, S3B and Tables S1F, S1G). At a statistical cut off of p<0.05 (T+BH) only 1.3% and 2.5% of IDH1 and IDH2 signature genes did not overlap respectively, further underlining the functional similarity of these alleles. We next combined the genes contained within the aberrant IDH1/2 mutant gene expression and DNA methylation signatures, and integrated them in order to identify biological pathways and functions that are perturbed in these specific leukemias. This integrative pathway analysis of aberrantly methylated and expressed genes revealed a significant over-representation of genes involved in pathways known to contribute to hematopoietic malignant transformation (Figure S3C and S3D and Table S2A).
Expression of 2HG-producing IDH enzymes in cells induces a global increase in 5-methylcytosine
Based on the observed association of IDH1/2 mutations with hypermethylation in AML patient samples, we next sought to determine if expression of mutant IDH1 or IDH2 was sufficient to increase global levels of 5-methylcytosine in cells. As shown previously (Ward et al., 2010), expression of R132H mutant IDH1 or R172K mutant IDH2 in 293T cells led to a marked elevation of 2HG levels compared with expression of the corresponding wild-type enzymes (Figure 3A and 3B). We next determined the level of 5-methylcytosine in these cells by immunofluorescence staining with 5-methylcytosine-specific antibody (Figure S4A) and quantified the fluorescence intensity in > 200 cells using the count nuclei application module in MetaMorph software. Mutant IDH1 or IDH2 expression resulted in a statistically significant increase in 5-methylcytosine compared to cells expressing wild-type IDH1 or IDH2 (Figure 3C). Overexpression of wild-type IDH2 also led to a small but significant increase in 5-methylcytosine levels compared to vector control, consistent with the small 2HG elevation observed in these cells. The increase in 5-methylcytosine correlated well with the levels of 2HG measured in the cells. To extend these findings to hematopoietic cells, wild-type or R172K mutant IDH2 were stably expressed in mouse myeloid progenitor 32D cells, which normally express wild-type forms of IDH1, IDH2 and TET2. Expression of wild-type and mutant IDH2 protein was confirmed by western blot. 2HG levels were measured by gas chromatography-mass spectrometry (GC-MS) and were only induced in cells stably expressing mutant IDH2 (Figure 3D and 3E). The levels of 5-methylcytosine were assessed by slot blot of DNA extracted from the cells using 5-methylcytosine-specific antibody. Quantification of band intensity showed that expression of R172K mutant IDH2 led to a significant increase in global 5-methylcytosine levels (Figure 3F). These results were also confirmed using liquid chromatography-electrospray ionization tandem mass spectrometry (LC-ESI-MS/MS) and showed a similar increase in the 5-methylcytosine level in cells expressing R172K mutant IDH2 (Figure S4B). We also assessed 5-methylcytosine levels using LC-ESI-MS/MS in murine primary bone marrow cells expressing wild-type and mutant IDH1 or IDH2, and observed an increase in 5-methylcytosine levels in cells expressing IDH1 R132H or IDH2 R172K, but not wild-type IDH1 or IDH2 (Figure S4C). The increase in DNA methylation we observed in hematopoietic cells after mutant IDH1/2 expression suggests that the IDH1/2 mutants may specifically alter DNA methylation in AML cells.
Figure 3. Expression of 2HG-producing IDH proteins increases global 5-methylcytosine levels.
(A) 293T cells were transiently transfected with empty vector, wild-type or R132H mutant IDH1, or wild-type or R172K mutant IDH2. After 3 days, cells were lysed and assessed for IDH1 expression levels by Western blot, and then re-probed for IDH2. β-actin antibody was used as a control. (B) Cells transfected in parallel to those lysed in (A) were extracted for intracellular metabolites. Metabolites were then derivatized with MTBSTFA and analyzed by GC-MS. Shown is the quantitation of 2HG signal intensities relative to the intrasample glutamate signals for a representative experiment. (C) Global DNA methylation levels in cells were analyzed 3 days following transfection by immunofluorescence using antibody against 5-methylcytosine. Quantification of fluorescence intensities from one experiment is shown. Data is representative of three independent experiments. (D) 32D cells were transduced with empty retroviral vector or with wild-type or R172K mutant IDH2, selected in 2.5 µg/ml puromycin for 7 days, and then lysed to confirm stable expression of IDH2. Tubulin antibody was used as a control. (E) Cells were extracted for their intracellular metabolites which were then derivatized with MTBSTFA and analyzed by GC-MS. Shown are representative gas chromatographs from wild-type and mutant IDH2 expressing cells depicting the derivatized metabolites eluting between 31.3 and 33.5 min, including 4-oxoproline (4-oxo Pro), glutamate (Glu), and 2HG. Metabolite abundance refers to GC-MS signal intensity. (F) DNA was extracted from cells with stable wild-type or mutant IDH2 expression, and global DNA methylation levels were measured by slot blot using antibody against 5-methylcytosine. Relative intensity of signals of three independent experiments was quantified. Error bars: +/− SD for triplicate experiments. (See also Figure S4)
Mutations in IDH1/2 are mutually exclusive with mutations in TET2
One potential test of whether I DH1/2 mutants contribute to leukemogenesis through the impairment of DNA demethylation is through assessment of IDH1/2 mutational status in tumors with or without somatic mutations in an enzyme involved in the removal of DNA methylation. TET2 is a Fe(II) and αKG-dependent enzyme known to display loss of function mutations in AML and other myeloid malignancies (Abdel-Wahab et al., 2009; Delhommeau et al., 2009; Langemeijer et al., 2009). Recently all TET family members including TET2 were shown to catalyze the conversion of 5-methylcytosine to 5-hydroxymethylcytosine (5-OH-MeC) (Ito et al., 2010; Tahiliani et al., 2009). The reaction depends on αKG, iron and oxygen. Although the physiological significance of 5-OH-MeC remains to be determined, it is likely to be the intermediate in the pathway that actively demethylates 5-methylcytosine (Wu and Zhang, 2010). Importantly, expression of TET1 or TET2 in cellular systems results in a reduction in 5-methylcytosine levels (Ito et al., 2010). Unlike other DNA demethylases identified so far, the hydroxylation reaction does not involve any DNA damage, suggesting an important physiological function of TET2 in removing methylation marks (Ito et al., 2010; Tahiliani et al., 2009).
We first examined the genetic status of TET2 in AML samples wild-type or mutant for IDH1/2. As controls in analyzing for association with TET2 mutations, we also examined the mutational status of 11 other genes known to be recurrently mutated in AML (including PHF6, WT1, TP53, ASXL1, PTEN, RUNX1, KIT, NPM1, FLT3, CEBPA and RAS) in this same cohort. There were 40 somatic TET2 mutations identified in 28/385 patients (7.3% of the ECOG patient cohort). Although only 1 homozygous TET2 mutation was observed, 11 of the 28 TET2-mutant patients had >1 TET2 mutation, consistent with previous studies reporting biallelic TET2 mutations. Of the 40 TET2 mutations, 40% were insertions/deletions resulting in a frameshift mutation, 30% were nonsense mutations, and 30% were somatic missense mutations. AML patients with TET2 mutations did not differ from TET2 wild-type patients in terms of age, sex, cytogenetic risk, or percentage of bone marrow blasts at diagnosis. TET2 mutant patients also did not differ from TET2 wild-type AML patients in frequency of mutations in FLT3, NPM1, or CEBPA.
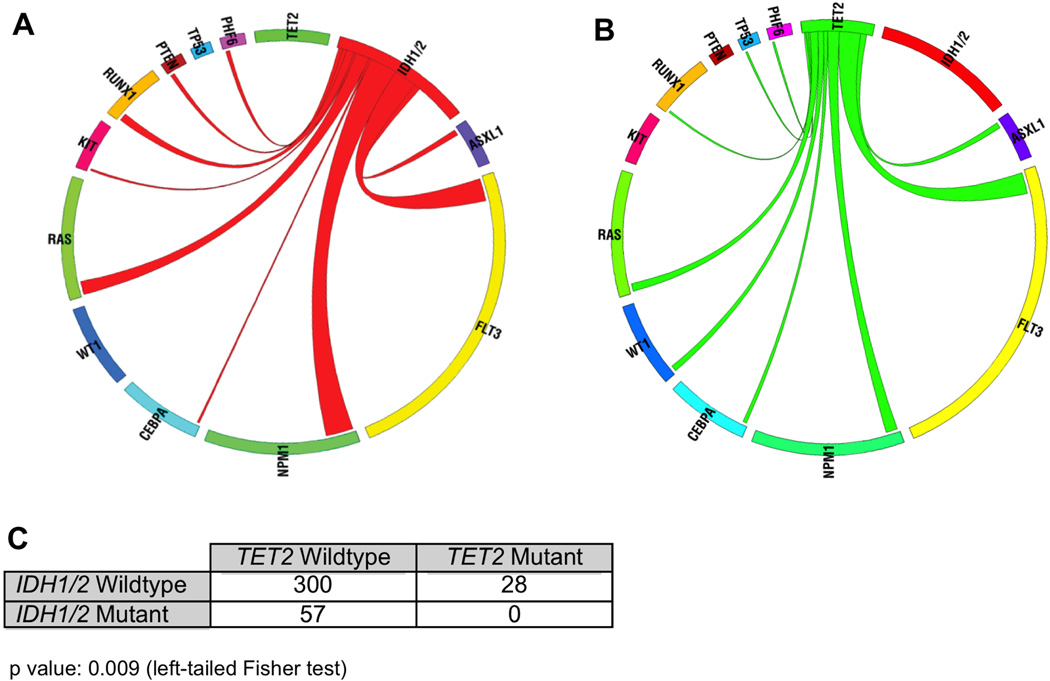
In contrast to the other genes analyzed, the relationship between IDH1/2 and TET2 mutations was striking in this AML cohort. Although IDH1, IDH2, and TET2 mutations were each identified in a significant proportion of patients in our study, there was a statistically significant inverse correlation between IDH1/2 and TET2 mutations in AML (p=0.009, left-tailed Fisher’s exact test). Specifically, we did not identify a single patient with concurrent mutations in TET2 and in IDH1 or IDH2; 0/57 IDH1/2 mutant cases were TET2 mutant versus 28/300 IDH1/2 wild-type AML cases (Figure 4). These data suggest that TET2 and IDH1/2 mutations form a distinct mutational class in AML, and that IDH1/2 mutations and TET2 mutations have overlapping roles in AML pathogenesis.
Figure 4. IDH1/2 mutations are mutually exclusive with mutations in TET2 in de novo AML.
(A) Circos diagram revealing relative frequency and pairwise co-occurrences of mutations in IDH1 and IDH2 in de novo AML. (B) Circos diagram revealing relative frequency and pairwise co-occurrences of mutations in TET2 in de novo AML. (C) Two-by-two table showing that mutations in IDH1/2 and TET2 were mutually exclusive in AML (Left-tailed Fisher p value: 0.009).
IDH mutation inhibits the hydroxylation of 5-methylcytosine by TET2
TET2 has the classical features of an αKG -Fe(II) dioxygenase, and requires αKG in order to mediate 5-methylcystosine hydroxylation (Ito et al., 2010; Tahiliani et al., 2009). The five carbon dicarboxylic acids 2HG and α-KG are chemically analogous. The substitution of the keto (oxo) group on α-KG to hydroxyl group on 2HG could potentially interfere with Fe(II) binding and stabilization of the reaction intermediate. We therefore hypothesized that 2HG produced by mutant IDH might inhibit the hydroxylation reaction of 5-methylcytosine by TET2. To test this hypothesis, we expressed FLAG-tagged TET2 in 293T cells. Consistent with previous findings (Ito et al., 2010; Tahiliani et al., 2009), cells expressing TET2 showed higher levels of 5-OH-MeC in the nucleus as detected by immunofluorescence staining with 5-OH-MeC specific antibody. Co-transfection of TET2 with IDH1 R132H, but not wild-type IDH1, reversed this increase in 5-OH-MeC (Figure 5A). To obtain a more quantitative assessment, we also performed flow cytometric analysis of 5-OH-MeC fluorescence intensity. Staining with an anti-FLAG antibody allowed us to discriminate between TET2-positive and TET2-negative populations; importantly the TET2-positive population showed a ~4-fold increase in the 5-OH-MeC fluorescence intensity. Co-transfection with R132H mutant IDH1 led to a ~40% decrease in the fluorescence intensity, while wild-type IDH1 had no significant impact on 5-OH-MeC fluorescence intensity (Figure 5B). Consistent with its proposed role in DNA demethylation, TET2 transfection also caused a decrease in 5-methylcytosine, which can be reversed by co-expression of IDH1 R132H mutant but not wild-type IDH1 (Figure S5). Together these data suggest that expression of mutant IDH is able to inhibit the hydroxylation of 5-methylcytosine by TET2 and subsequent DNA demethylation.
Figure 5. Mutant IDH1 expression inhibits hydroxylation of 5-methylcytosine by TET2.
(A) 293T cells were transiently transfected with FLAG-tagged TET2 in the absence or presence of wild-type or R132H mutant IDH1. Three days following transfection, global levels of 5-methylcytosine hydroxylation were analyzed by immunofluorescence using antibody against 5-hydroxy-methylcytosine (5-OH-MeC). Representative images from mock-transfected, TET2-transfected, TET2 + IDH1 WT co-transfected, and TET2 + IDH1 R132H co-transfected cells are shown. Scale bar: 100 µM. (B) Transfected cells were analyzed by flow cytometry and gated as TET2 positive or negative by FLAG antibody. Representative gating is shown. Intensities of 5-OH-methylcytosine staining within the TET2 positive and negative populations are shown as histogram overlays. Data in (A) and (B) are representative of three independent experiments. (See also Figure S5)
TET2 mutant AMLs display an overlapping hypermethylation signature with IDH1/2 mutant AMLs
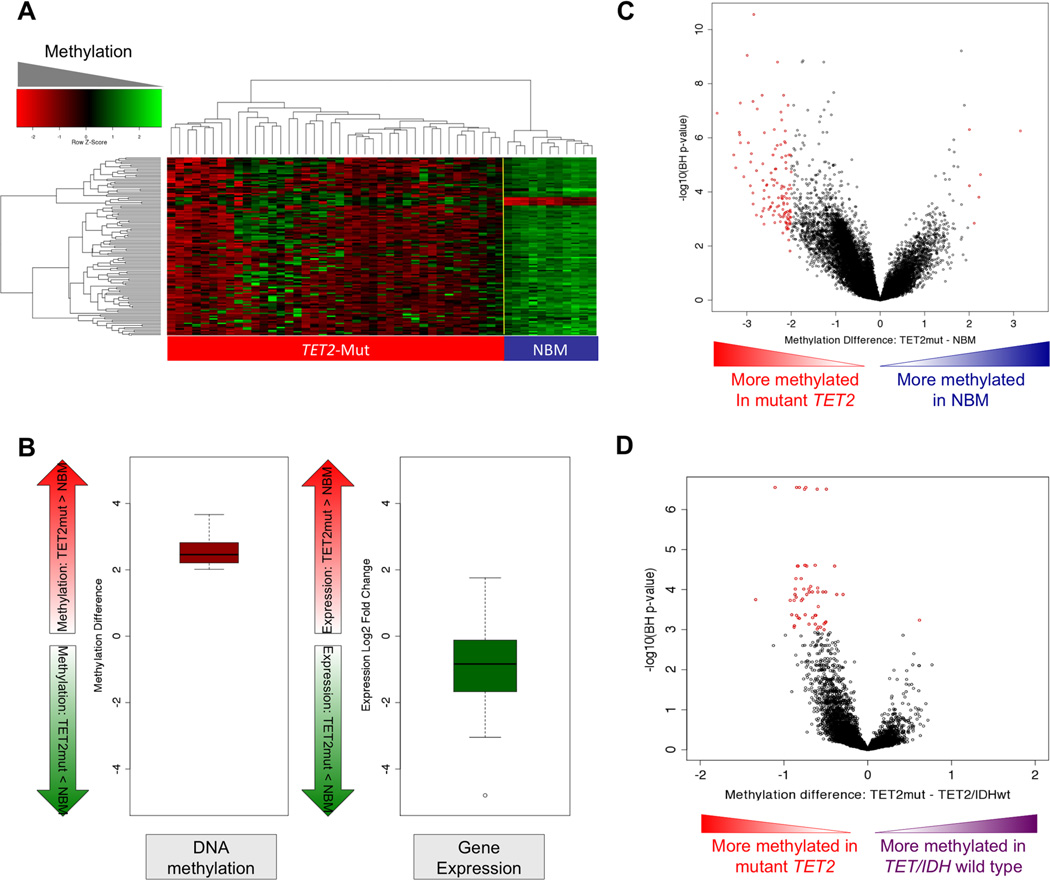
The fact that IDH1/2 and TET2 mutations are mutually exclusive in AML, and that IDH1/2 mutations induce hypermethylation and inhibit TET2 mediated 5-hydroxymethylation suggested that both mutations may function by altering the cytosine methylation profiles of hematopoietic cells. To determine whether this is the case we performed a supervised analysis comparing DNA methylation profiles of TET2 mutant AMLs to NBM. This allowed us to identify a set of 129 DMRs (> 2 log2 methylation difference and p<0.05 T+BH), all of which were hypermethylated in TET2 patients (Figure 6A and Table S3A). As in the case of the IDH1/2 mutant signature, there was a strong tendency for transcript abundance of these genes to be repressed (Figure 6B) Notably, 79 (61%) of these genes overlapped with the IDH1/2-mutant epigenetic signature (Fisher test p value <0.0001) (Table S3B). We then compared the methylation signature of TET2 mutant AML cases to AMLs that were wild-type for the IDH1/2 and TET2 loci. This analysis allowed us to determine that TET2 mutant AMLs displayed a unique methylation signature consisting of 57 DMRs (> 2 log2 methylation difference and p<0.05 T+BH). IDH1/2 mutant AMLs also displayed a 57 DMR signature when compared to IDH1/2 wild-type and TET2 wild-type AMLs (> 2 log2 methylation difference and p<0.05 T+BH). Note that this IDH1/2 signature is slightly different than the one reported in figure 2 since it excludes TET2 mutant AMLs from the control group in order to make a fair comparison. Remarkably, there was an eight gene overlap between this unique IDH1/2 mutant signature and the TET2 mutant signature, which is greater than expected by chance (Fisher exact test p value =5.45e−13), again linking the DNA methylation defect induced by IDH1/2 and TET2 mutations (Table S3C).
Figure 6. TET2-mutant AML is associated with a hypermethylation phenotype.
(A) Heatmap representation of a two-dimensional hierarchical clustering of genes identified as differentially methylated between TET2 mutant primary AML cases (Mut; red bar) and normal CD34+ bone marrow cells (NBM; blue bar). Each row represents a probe set and each column represents a patient. (B) Boxplot illustrating average methylation difference between TET2 mutant AMLs vs. normal CD34+ cells (left) and average gene expression difference between TET2 mutant AMLs vs. normal CD34+ cells (right) of genes aberrantly methylated in TET2 mutant AMLs. (C) Dot plot of methylation difference between TET2-mutant AMLs and normal CD34+ bone marrow cells (biological significance) vs. statistical significance (-log10 (BH p value)). Red points indicate probe sets identified as differentially methylated between the two groups. (D) Dot plot of methylation difference between TET2-mutant AMLs and TET2 and IDH1/2- wild-type AMLs (biological significance) vs. statistical significance (-log10 (T+BH)). Red points indicate probe sets statistically significant between the two groups. (See also table S3A–C).
Ninety-three percent of genes aberrantly expressed in TET2 mutant AMLs (Fold difference >2 and P<0.001 T+BH) were also aberrantly expressed in IDH1/2 mutant AML (Fisher test p value <0.0001). Furthermore, when we re-examined the two epigenetic clusters enriched in patients having IDH1/2 mutations, we found that six IDH1/2 negative cases had TET2 mutations.
Similar to the case of IDH1/2, there was a marked overall trend towards promoter hypermethylation in TET2-mutant AMLs compared to TET2 and IDH1/2-wild-type AMLs and to NBM CD34+ cells (Figure 6C, D). In order to confirm that this hypermethylation phenomenon was specific to IDH1/2 and TET2 mutants, we carried out a similar analysis of AML1-ETO AMLs vs. AMLs negative for this fusion gene. Despite the fact that AML1-ETO positive AMLs have a robust hypermethylation signature when compared to normal CD34+ cells (Figueroa et al., 2010), they did not display a global hypermethylation profile when compared to other AMLs (Figure S2E).
We also examined whether loss of TET2 in cells resulted in a similar increase in 5-methylcytosine levels as did expression of mutant IDH1/2. Two independent short-hairpin RNAs (shRNAs) were used to stably knock down TET2 in murine primary BM cells. LC-ESI-MS/MS analysis revealed that expression of validated TET2 shRNA constructs or IDH1/2 mutations in murine primary bone marrow cells led to a ~20% increase in 5-methylcytosine levels (Figure S4C). These data suggest that lossof-function mutations in TET2 and IDH1/2 mutations contribute to leukemogenesis through a shared mechanism that disrupts DNA demethylation.
GATA and EVI1 binding sites are enriched in aberrantly methylated loci of IDH1/2 mutant AMLs
DNA motif analysis in the promoter regions of genes associated with the identified DMRs in IDH1/2 mutant AMLs revealed a statistically significant enrichment of the GATA and EVI1 consensus DNA binding sequences (Figure S3E), two transcription factor classes known to recruit chromatin modifying enzymes and to direct myeloid differentiation (e.g. (Lugthart et al., 2010; Spensberger and Delwel, 2008; Stopka et al., 2005)). Notably, out of the 164 genes in the IDH1/2 mutant methylation signature, 65 (40%) are known GATA2 direct target genes, and 31 (19%) are known to be GATA1 direct targets (Table S2B) (Fujiwara et al., 2009). Additionally, in our patient cohort, GATA1 was aberrantly hypermethylated and silenced specifically in IDH1 mutant AMLs (not shown). These data suggest that aberrant methylation in IDH1/2 AMLs may perturb the function of transcription factors which control myeloid differentiation.
Expression of IDH2 mutants as well as loss of TET2 increase expression of stem cell markers and impair myeloid differentiation
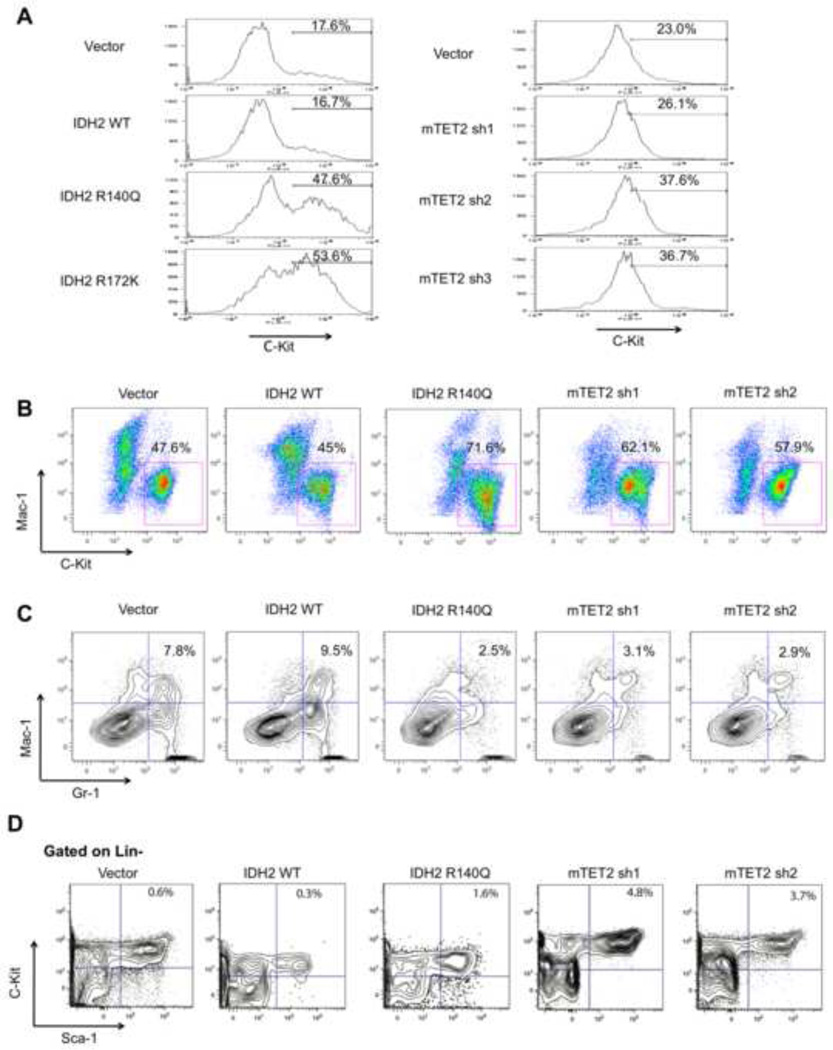
Analysis of the aberrant DNA methylation and gene expression signatures of IDH1/2 mutant AMLs suggested a role for these genetic lesions in impairing myeloid differentiation. To investigate the biological significance of IDH1/2 mutations, the effects of stable expression of wild-type, R172K mutant or R140Q mutant IDH2 were examined in 32D myeloid cells and primary mouse bone marrow (BM) cells. Expression of mutant IDH2, but not of wild-type IDH2 resulted in an average of ~40% increase in C-Kit expression in 32D cells stably expressing IDH2 constructs (p=0.04; Figure 7A). Likewise, shRNA-mediated stable knockdown (KD) of Tet2 expression by 40–80% with 3 different shRNAs resulted in a significant increase in C-Kit expression (p=0.02; Figure 7A and Figure S6A). We next assessed the effects of wild-type and mutant IDH2 expression in murine primary bone marrow cells, and demonstrated that expression of mutant, but not wild-type IDH2 resulted in an increase in C-Kit expression in murine primary BM cells after 14 days of growth in methylcellulose (Figure 7B) as well as after 5 days of growth in ex-vivo liquid culture under myeloid differentiation conditions (Figures S6B, S6C). After 14 days in methylcellulose, 71.6% of IDH2 R140Q–expressing cells were C-Kit positive compared with 45% of IDH2 wild-type-expressing cells. Likewise, stable shRNA-mediated knockdown of Tet2 (50–70% Tet2 mRNA expression levels) in murine BM cells resulted in a >50% increase in C-Kit expression after 14 days of growth in methylcellulose such that 57.9%–62.1% of cells with Tet2 knockdown expressed C-Kit compared with 47.6% of empty vector treated cells (Figures 7B and Figure S6B). We also assessed the effects of loss of Tet2 as well as expression of IDH2 R140Q and wild-type IDH2 on the differentiation of murine bone marrow cells.
Figure 7. IDH2 mutant expression and TET2 knockdown in hematopoietic cells impairs differentiation.
(A) 32D cells retrovirally transduced with empty vector, IDH2 WT, IDH2 R140Q, IDH2 R172K or three independent shRNAs against mouse TET2 were analyzed for C-Kit expression by flow cytometry. Intensities of fluorescence signals are depicted as histograms. (B) Primary mouse bone marrow cells were retrovirally transduced with MIGR1 vector, IDH2 WT, IDH2 R140Q, or two shRNAs against mouse TET2. GFP-positive cells were sorted and expanded in methylcellulose media for 14 days Cells were analyzed for Mac-1 and C-Kit expression by flow cytometry. (C) Cells treated as in (B) were analyzed for Mac-1 and Gr-1 expression by flow cytometry. (D) Murine primary bone marrow cells were retrovirally transduced with MIGR1 vector, IDH2 WT, IDH2 R140Q, or two shRNAs against mouse TET2. Cells were grown in liquid culture for 5 days ex-vivo and assessed for the percentage of LSK cells out of the total lineage-negative, GFP-positive cell population. (See also Figure S6)
Importantly, we found that mutant IDH2 expression as well as Tet2 loss, but not wild-type IDH2 expression results in a reduced expression of the mature myeloid markers Gr-1 and Mac-1 (Figure 7C). After 14 days in methylcellulose culture, 7.8% of empty vector treated cells and 9.5% of IDH2 wild-type-expressing cells were Mac-1/Gr-1 double-positive. In contrast BM cells expressing IDH2 R140Q (2.5% of cells double-positive for Mac-1/Gr-1) or with stable Tet2 KD (2.9–3.1% of cells double-positive for Mac-1/Gr-1) had a >3-fold reduction the proportion of cells co-expressing these mature myeloid markers. CFU plating did not reveal any significant differences in types or numbers of colonies formed amongst groups (Figure S6D). We then assessed the effects of TET2 knockdown or mutant IDH expression on the proportion of Lin−/Sca-1+/C-Kit+ (LSK) cells, which includes hematopoietic stem cells (HSC). We found that overexpression of IDH2 R140Q or knockdown of Tet2 in murine primary BM cells grown in liquid culture for 5 days ex-vivo demonstrated an increase in the percentage of LSK cells compared to control cells expressing empty vector or wild-type IDH2 (Figure 7D). Analysis of cell morphology with Wright-Giemsa staining also revealed an increase in the proportion of immature myeloid progenitors (Figure S6E). These data suggest that expression of IDH2 mutants or reduction in Tet2 expression result in similar effects on hematopoietic differentiation with a concordant increase in the proportion of hematopoietic stem/progenitor cells and in inhibition of normal myeloid differentiation.
Discussion
The combination of DNA methylation, DNA sequencing and gene expression profiling offers the possibility of defining new subsets of AML and gaining additional insight into AML pathogenesis. Although we have previously shown that aberrant DNA methylation patterning is a characteristic feature of AML (Figueroa et al., 2010), here we demonstrate that IDH1/2 mutant AML is associated with more extensive promoter hypermethylation compared to other AML subtypes. In addition to the global increase in promoter methylation, IDH1/2-mutant AMLs display a robust and unique epigenetic signature, consisting almost entirely of hypermethylated genes, indicating that aberrant cytosine methylation in IDH1/2-mutant AML affects specific loci with known and putative roles in leukemogenesis. Importantly, we validated these findings in a second cohort of 344 patients in which we had previously defined five AML clusters based on distinct epigenetic signatures without known genetic or molecular features (Figueroa et al., 2010). We now demonstrate that two of those five epigenetically distinct clusters of AML patients are defined by IDH1/2 mutations.
Leukemias harboring translocations or mutations in transcriptional regulators such as AML1-ETO, CBFB-MYH11, PML-RARA, MLL and CEBPA each display their own specific and distinct epigenetic signatures in our previously reported cohort (Figueroa et al., 2010) and in the ECOG cohort which is the focus of our current study (data not shown). This is not surprising since it is expected that mutations which disrupt hematopoietic transcription factors would have an impact on epigenetic gene regulation. In contrast, FLT3 mutations, which activate signaling pathways without a known role in affecting the epigenetic state, are not associated with a mutation-specific epigenetic signature in AML (Figueroa et al., 2010). It would not necessarily be anticipated that mutant IDH1/2 proteins, which are metabolic enzymes normally involved in citrate metabolism, would specify an epigenetically defined disease. In previous studies in glioma, it was suggested that IDH-mutant brain tumors were characterized by a methylator phenotype which favors the acquisition of IDH1/2 mutations (Noushmehr et al., 2010). Alternatively IDH1/2 mutations could be early events in the process of hematopoietic transformation and precede the stochastic acquisition and accumulation of epigenetic changes that eventually culminate in overt AML. However, our data suggest a more direct mechanistic link between IDH1/2 mutations and dysregulated epigenetic programming in leukemia cells. Specifically, our data suggest that DNA hypermethylation is a consequence of mutant IDH protein expression and that IDH1/2-mutant mediated epigenetic effects contribute to AML pathogenesis. Several lines of evidence support this hypothesis. Firstly, there was an inverse correlation between gene expression and methylation among the IDH1/2 methylation signature genes and many of these genes have known and suspected roles in oncogenesis. Second, we show that expression of IDH 1/2 mutant proteins resulted in marked increases in DNA methylation. Third, we found that IDH1/2 mutations were mutually exclusive of TET2 mutations in AML and that expression of mutant, but not wild-type IDH proteins disrupted TET2 function in cells.
We observed that IDH1/2 and TET2 mutations were mutually exclusive in a large, genetically annotated de novo AML cohort, suggesting that these lesions may be biologically redundant. TET2 is a member of a family of α KG-dependent enzymes that catalyze cytosine 5-hydroxymethylation and induce subsequent demethylation of DNA (Ito et al., 2010; Tahiliani et al., 2009). TET2 loss of function would be anticipated to result in hypermethylation, and the data reported here support that. Furthermore, IDH1/2 mutations, which lead to elevated levels of 2HG, an αKG analogue, might be anticipated to inhibit the function of TET2. In support of this possibility we observed that TET2 and IDH1/2 mutant AMLs presented with more pronounced hypermethylation profiles than other AMLs and shared an overlapping epigenetic signature. Most importantly, expression of IDH 1/2 mutants induced an increase in global DNA hypermethylation, and inhibited TET2 induced cytosine 5-hydroxymethylation. These data suggest that TET2 and IDH1/2 mutations constitute a distinct mutational class in AML, which affects the epigenetic state; we would predict that additional mutations will be identified which will be classified into this “epigenetic regulator” mutational class in AML and in other malignancies.
The present data do not exclude the possibility that IDH1/2 mutant-mediated transformation is in part mediated by TET2 independent effects. Other families of enzymes that also require αKG for their catalytic activity, including Jumonji-C domain histone demethylases, could be affected by accumulation of 2HG in leukemic cells. Jumonji-C domain proteins can impair the spread of DNA methylation, alter the function of transcription factors and demethylate various chromatin marks including gene activation marks such as H3K4me3 and H3K36me3 (Tsukada et al., 2006) and gene repressive marks such as H3K9me3 (Yamane et al., 2006). It will therefore be important in subsequent studies to assess the effects of mutant IDH protein expression and 2HG accumulation on the function of other αKG-dependent enzymes expressed in hematopoietic cells and on the chromatin state of normal and leukemic stem/progenitor cells. Moreover, it is important to note that other somatic lesions may influence the IDH1/2 and TET2 mutant DNA methylation profiles, which likely explains why a number of these cases did not distribute to the two main IDH clusters. Ongoing studies of AML genetic and epigenetic lesions will help to explain these observations.
Although in vivo studies will be needed to specifically delineate the effects of IDH 1/2 mutant protein expression on stem/progenitor cell function and on hematopoietic differentiation, the data suggest that expression of IDH 1/2 proteins in hematopoietic cells results in impairment of myeloid differentiation and increased expression of immature cell surface markers. Consonant with these findings, we found that many of the genes hypermethylated in the context of IDH1/2-mutant AML contained DNA binding motifs for GATA1, GATA2, and EVI1, transcription factors known play a role in leukemogenesis and in normal myeloid differentiation. These data are supported by recent ChIP-seq data demonstrating many of these loci are in fact GATA1 and GATA2 direct targets (Fujiwara et al., 2009). We also found that GATA1 is aberrantly methylated and silenced in IDH1-mutant AMLs, implicating GATA1 loss of function might contribute to the ability of IDH1/2 mutant enzymes to perturb differentiation in myeloid cells. By contrast, EVI1 is known to bind to DNA methyltransferases 3A and 3B and may directly methylate target genes in IDH1/2-mutant leukemias (Lugthart et al., 2010).
Collectively, the data suggest a paradigm whereby oncogenic alterations in core cellular metabolic pathways can lead to leukemic transformation by dysregulating the epigenetic machinery in hematopoietic progenitors. Additionally, the data support the notion that IDH1/2 mutants are a powerful leukemia driver with a well-defined phenotype distinct from other AMLs and we propose that IDH1/2-mutant AML be considered a separate subtype of AML. Moreover, since mutant IDH1/2 are associated with a neomorphic function not characteristic of the wild-type protein, targeted therapies that inhibit the neomorphic function of mutant IDH enzymes might reverse epigenetic patterning, promote myeloid differentiation, and improve outcomes in IDH1/2-mutant AML.
Experimental procedures
Patient samples
398 AML samples were obtained at diagnosis from patients enrolled in the Eastern Cooperative Oncology Group’s (ECOG) E1900 clinical trial (Fernandez et al., 2009). Samples were de-identified at the time of inclusion in this study. IRB approval was obtained at Weill Cornell Medical College and Memorial Sloan Kettering Cancer Center. Sixteen specimens of human CD34+ bone marrow cells were provided by the Stem Cell and Xenograft Core Facility of the University of Pennsylvania. These studies were performed in accordance with the Helsinki protocols. Table 1 summarizes patients’ characteristics.
Statistical Analysis
Statistical analysis of demographic factors, disease characteristics, and mutational frequencies was performed using Fisher’s exact test and Chi-square tests for categorical variables and t-tests and Mann-Whitney U test for continuous variables. Student’s t test was used for analysis of fluorescence intensities.
DNA methylation microarrays
High-molecular-weight DNA was isolated from mononuclear cell fractions consisting of >90% blasts using the PureGene kit from Qiagen (Valencia, CA). The HELP (HpaII tiny fragment enrichment by ligation-mediated PCR) assay was carried out as previously described (Figueroa et al., 2009; Khulan et al., 2006) and samples were hybridized onto a custom human promoter array covering 25,626 HpaII amplifiable fragments (~50,000 CpGs), annotated to ~14,000 genes (Roche NimbleGen, Design name: 2006-10–26_HG17_HELP_Promoter, Design ID: 4802).
Mouse bone marrow infections, ex-vivo liquid culture and CFU assay
Bone marrow was harvested from the femurs of wild-type Black/6 mice immediately after sacrifice. Animal care was in strict compliance with institutional guidelines established by the Memorial Sloan-Kettering Cancer Center, the National Academy of Sciences Guide for the Care and Use of Laboratory Animals, and the Association for Assessment and Accreditation of Laboratory Animal Care International. After red cell lysis in RBC lysis buffer (Purgene), the bone marrow were cultured in media containing RPMI/10% FBS and murine IL-3, IL-6, and SCF. 24h later bone marrow cells were infected with retroviral vectors expressing MIGR1 empty vector or MIGR1 vector containing IDH2 wild-type, IDH2 R140Q, or at least 2 different shRNAs targeting mTET2. Cells were infected twice with 24 h between infections and then sorted for GFP+ cells 48 h following second infection using a FACS Aria. IDH1/2 over-expression and TET2 KD were confirmed by qRTPCR using SYBR green quantification in an ABI 7500 sequence detection system. Primer sequences for IDH1/2 qRTPCR were as follows: IDH1F 5’-GGGTTGGCCTTTGTATCTGA-3’; IDH1R 5’-TTTACAGGCCCAGATGAAGC-3’; IDH2F 5’-CGGCACTTTCAAAATGGTCT-3’; IDH2R 5’GCATACTGGAAGCAGCTGTG-3’. After sorting, GFP+ bone marrow cells were placed in liquid culture or methylcellulose for colony-forming assays. For ex-vivo liquid culture, GFP+ cells expressing empty vector or IDH1/2 constructs were plated at 2 × 105/ml and grown for 4 days in StemSpan SFEM (StemCell Technologies) supplemented with rmIL3 (20ng/mL), rhIL6 (10ng/mL), rmSCF (10ng/mL), rmGMCSF (10ng/mL), rmTPO (50ng/mL), and rhFlt3L (100ng/mL). For CFU assays cells 1 × 104 GFP-positive cells were plated in duplicate in methylcellulose with complete cytokine mix (MethoCult GF M3434 (Stem Cell Technologies)). Colonies were scored at 14 days after seeding and followed by flow cytometric analysis of the population of cells plated in methylcellulose.
Supplementary Material
Significance.
Aberrant epigenetic programming is a hallmark of cancer and yet very little is known concerning the mechanisms through which this occurs. Here we demonstrate that leukemic neomorphic mutations of the citrate metabolism genes IDH1 and IDH2 that generate the aberrant metabolite 2HG, induce DNA hypermethylation, and impair differentiation in hematopoietic cells. These effects are caused in part through inhibition of TET2, a DNA demethylase enzyme also mutated in leukemia. IDH1/2 and TET2 mutant primary AML cells both displayed a similar defect in epigenetic programming consisting of global hypermethylation and a gene-specific methylation signature. This work identifies IDH1/2 and TET2 mutant leukemias as a biologically distinct disease subtype, and links cancer metabolism with epigenetic control of gene expression.
Acknowledgements
MEF is funded by Leukemia and Lymphoma Society Special Fellow award. AM is funded by a Leukemia and Lymphoma Society SCOR and TRP award, and is a Burroughs Wellcome Clinical Translational Scholar and Scholar of the Leukemia and Lymphoma Society. AM and MEF are also supported by the Sackler Center for Biomedical and Physical Sciences. JDL is funded by RO1 HL082950. CBT, CL, and PSW are supported in part by grants from the NCI and NIH. PSW is also supported in part by the University of Pennsylvania Medical Scientist Training Program. This work was supported in part by U54CA143798-01 from the National Cancer Institute to RLL and to a grant from the Starr Cancer Consortium to KF, RLL, and AM. RLL is an Early Career Award Recipient of the Howard Hughes Medical Institute and is a Geoffrey Beene Junior Faculty Chair at Memorial Sloan Kettering Cancer Center. EP is supported by NCI CA21115 and CA 114737. We would like to thank Adriana Heguy and Igor Dolgalev for assistance with DNA resequencing, and also thank Hans-Guido Wendel and Dino Mavrakis for assistance with TET2 shRNA construction. We thank Justin Cross and Dan Zlotoff for technical help and Sachie Marubayashi for assistance with murine bone marrow studies.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Additional methods are explained in the supplementary materials section.
Accession numbers. All arrays were submitted to the GEO repository (http://www.ncbi.nlm.nih.gov/geo), accession number GSE24505.
Authors’ contributions:
M.E.F., P.S.W., O.A.W., C.L., J.P., C.B.T., R.L.L., and A.M. designed the study. M.E.F., P.S.W., O.A.W., C.L., J.P., Y.L. N.B., K.W., J.D.L., W.L., A.S., A.V. and S.C. performed research; M.E.F., P.S.W., O.A.W., C.L., J.K.P., V.F., A.V., L.A.G., C.B.T., R.L.L. and A.M. analyzed the data; J.D.L., J.P., E.P. and P.J.V. contributed research material; R.D., B.L., P.J.V., Z.S., M.T. and H.F. contributed to data interpretation; M.E.F., O.A.W, P.S.W., C.L., C.B.T, R.L.L. and A.M. wrote the manuscript.
WL, SC, VF, and CBT are employees or consultants of Agios Pharmaceuticals and have financial interest in Agios.
References
- Abbas S, Lugthart S, Kavelaars FG, Schelen A, Koenders J, Zeilemaker A, van Putten WJ, Rijneveld A, Lowenberg B, Valk PJ. Acquired mutations in the genes encoding IDH1 and IDH2 both are recurrent aberrations in acute myeloid leukemia (AML): prevalence and prognostic value. Blood. 2010 doi: 10.1182/blood-2009-11-250878. [DOI] [PubMed] [Google Scholar]
- Abdel-Wahab O, Mullally A, Hedvat C, Garcia-Manero G, Patel J, Wadleigh M, Malinge S, Yao J, Kilpivaara O, Bhat R, et al. Genetic characterization of TET1, TET2, and TET3 alterations in myeloid malignancies. Blood. 2009;114:144–147. doi: 10.1182/blood-2009-03-210039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhommeau F, Dupont S, Della Valle V, James C, Trannoy S, Masse A, Kosmider O, Le Couedic JP, Robert F, Alberdi A, et al. Mutation in TET2 in myeloid cancers. N Engl J Med. 2009;360:2289–2301. doi: 10.1056/NEJMoa0810069. [DOI] [PubMed] [Google Scholar]
- Dickins RA, Hemann MT, Zilfou JT, Simpson DR, Ibarra I, Hannon GJ, Lowe SW. Probing tumor phenotypes using stable and regulated synthetic microRNA precursors. Nat Genet. 2005;37:1289–1295. doi: 10.1038/ng1651. [DOI] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez HF, Sun Z, Yao X, Litzow MR, Luger SM, Paietta EM, Racevskis J, Dewald GW, Ketterling RP, Bennett JM, et al. Anthracycline dose intensification in acute myeloid leukemia. N Engl J Med. 2009;361:1249–1259. doi: 10.1056/NEJMoa0904544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa ME, Lugthart S, Li Y, Erpelinck-Verschueren C, Deng X, Christos PJ, Schifano E, Booth J, van Putten W, Skrabanek L, et al. DNA methylation signatures identify biologically distinct subtypes in acute myeloid leukemia. Cancer Cell. 2010;17:13–27. doi: 10.1016/j.ccr.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa ME, Melnick A, Greally JM. Genome-wide determination of DNA methylation by Hpa II tiny fragment enrichment by ligation-mediated PCR (HELP) for the study of acute leukemias. Methods Mol Biol. 2009;538:395–407. doi: 10.1007/978-1-59745-418-6_20. [DOI] [PubMed] [Google Scholar]
- Fujiwara T, O’Geen H, Keles S, Blahnik K, Linnemann AK, Kang YA, Choi K, Farnham PJ, Bresnick EH. Discovering hematopoietic mechanisms through genome-wide analysis of GATA factor chromatin occupancy. Mol Cell. 2009;36:667–681. doi: 10.1016/j.molcel.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann C, Meyer J, Balss J, Capper D, Mueller W, Christians A, Felsberg J, Wolter M, Mawrin C, Wick W, et al. Type and frequency of IDH1 and IDH2 mutations are related to astrocytic and oligodendroglial differentiation and age: a study of 1,010 diffuse gliomas. Acta Neuropathol. 2009;118:469–474. doi: 10.1007/s00401-009-0561-9. [DOI] [PubMed] [Google Scholar]
- Huesken D, Lange J, Mickanin C, Weiler J, Asselbergs F, Warner J, Meloon B, Engel S, Rosenberg A, Cohen D, et al. Design of a genome-wide siRNA library using an artificial neural network. Nat Biotechnol. 2005;23:995–1001. doi: 10.1038/nbt1118. [DOI] [PubMed] [Google Scholar]
- Ito S, D’Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H, Ehrlich LI, Seita J, Murakami P, Doi A, Lindau P, Lee H, Aryee MJ, Irizarry RA, Kim K, et al. Comprehensive methylome map of lineage commitment from haematopoietic progenitors. Nature. 2010;467:338–342. doi: 10.1038/nature09367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khulan B, Thompson RF, Ye K, Fazzari MJ, Suzuki M, Stasiek E, Figueroa ME, Glass JL, Chen Q, Montagna C, et al. Comparative isoschizomer profiling of cytosine methylation: the HELP assay. Genome Res. 2006;16:1046–1055. doi: 10.1101/gr.5273806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langemeijer SM, Kuiper RP, Berends M, Knops R, Aslanyan MG, Massop M, Stevens-Linders E, van Hoogen P, van Kessel AG, Raymakers RA, et al. Acquired mutations in TET2 are common in myelodysplastic syndromes. Nat Genet. 2009;41:838–842. doi: 10.1038/ng.391. [DOI] [PubMed] [Google Scholar]
- Lugthart S, Figueroa ME, Bindels E, Skrabanek L, Valk PJ, Li Y, Meyer S, Erpelinck-Verschueren C, Greally J, Lowenberg B, et al. Aberrant DNA hypermethylation signature in acute myeloid leukemia directed by EVI1. Blood. 2010 doi: 10.1182/blood-2010-04-281337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcucci G, Maharry K, Wu YZ, Radmacher MD, Mrozek K, Margeson D, Holland KB, Whitman SP, Becker H, Schwind S, et al. IDH1 and IDH2 gene mutations identify novel molecular subsets within de novo cytogenetically normal acute myeloid leukemia: a Cancer and Leukemia Group B study. J Clin Oncol. 2010;28:2348–2355. doi: 10.1200/JCO.2009.27.3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardis ER, Ding L, Dooling DJ, Larson DE, McLellan MD, Chen K, Koboldt DC, Fulton RS, Delehaunty KD, McGrath SD, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009;361:1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, Berman BP, Pan F, Pelloski CE, Sulman EP, Bhat KP, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17:510–522. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschka P, Schlenk RF, Gaidzik VI, Habdank M, Kronke J, Bullinger L, Spath D, Kayser S, Zucknick M, Gotze K, et al. IDH1 and IDH2 mutations are frequent genetic alterations in acute myeloid leukemia and confer adverse prognosis in cytogenetically normal acute myeloid leukemia with NPM1 mutation without FLT3 internal tandem duplication. J Clin Oncol. 2010;28:3636–3643. doi: 10.1200/JCO.2010.28.3762. [DOI] [PubMed] [Google Scholar]
- Song L, Prey JD, Xue J, Kanter P, Manzotti C, Bombardelli E, Morazzoni P, Pendyala L. Pharmacokinetic measurements of IDN 5390 using electrospray ionization tandem mass spectrometry: structure characterization and quantification in dog plasma. Rapid Commun Mass Spectrom. 2005;19:3617–3625. doi: 10.1002/rcm.2235. [DOI] [PubMed] [Google Scholar]
- Spensberger D, Delwel R. A novel interaction between the proto-oncogene Evi1 and histone methyltransferases, SUV39H1 and G9a. FEBS Lett. 2008;582:2761–2767. doi: 10.1016/j.febslet.2008.06.056. [DOI] [PubMed] [Google Scholar]
- Stopka T, Amanatullah DF, Papetti M, Skoultchi AI. PU.1 inhibits the erythroid program by binding to GATA-1 on DNA and creating a repressive chromatin structure. Embo J. 2005;24:3712–3723. doi: 10.1038/sj.emboj.7600834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tefferi A, Lasho TL, Abdel-Wahab O, Guglielmelli P, Patel J, Caramazza D, Pieri L, Finke CM, Kilpivaara O, Wadleigh M, et al. IDH1 and IDH2 mutation studies in 1473 patients with chronic-, fibrotic- or blast-phase essential thrombocythemia, polycythemia vera or myelofibrosis. Leukemia. 2010;24:1302–1309. doi: 10.1038/leu.2010.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, Zhang Y. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439:811–816. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- van den Bent MJ, Dubbink HJ, Marie Y, Brandes AA, Taphoorn MJ, Wesseling P, Frenay M, Tijssen CC, Lacombe D, Idbaih A, et al. IDH1 and IDH2 mutations are prognostic but not predictive for outcome in anaplastic oligodendroglial tumors: a report of the European Organization for Research and Treatment of Cancer Brain Tumor Group. Clin Cancer Res. 2010;16:1597–1604. doi: 10.1158/1078-0432.CCR-09-2902. [DOI] [PubMed] [Google Scholar]
- Wagner K, Damm F, Gohring G, Gorlich K, Heuser M, Schafer I, Ottmann O, Lubbert M, Heit W, Kanz L, et al. Impact of IDH1 R132 mutations and an IDH1 single nucleotide polymorphism in cytogenetically normal acute myeloid leukemia: SNP rs11554137 is an adverse prognostic factor. J Clin Oncol. 2010;28:2356–2364. doi: 10.1200/JCO.2009.27.6899. [DOI] [PubMed] [Google Scholar]
- Ward PS, Patel J, Wise DR, Abdel-Wahab O, Bennett BD, Coller HA, Cross JR, Fantin VR, Hedvat CV, Perl AE, et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell. 2010;17:225–234. doi: 10.1016/j.ccr.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SC, Zhang Y. Active DNA demethylation: many roads lead to Rome. Nat Rev Mol Cell Biol. 2010;11:607–620. doi: 10.1038/nrm2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane K, Toumazou C, Tsukada Y, Erdjument-Bromage H, Tempst P, Wong J, Zhang Y. JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell. 2006;125:483–495. doi: 10.1016/j.cell.2006.03.027. [DOI] [PubMed] [Google Scholar]
- Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.