Abstract
Background
Identification of diagnostic and prognostic biomarkers is a research priority for the improved management of pancreatic ductal adenocarcinoma (PDAC). Insulin-like growth factor binding protein 2 (IGFBP2) and mesothelin (MSLN) have shown potential as serum biomarkers in other cancers, but have not been adequately studied in PDAC.
Methods
Serum IGFBP2 and MSLN levels were quantified by enzyme-linked immunosorbent assay (ELISA) in a cohort of 84 PDAC patients, 84 healthy control subjects and 40 chronic pancreatitis (ChPT) patients. Regression models related IGFBP2 and MSLN levels to diagnosis, gender, age, stage and survival.
Results
IGFPB2 and MSLN serum levels were diagnostic for PDAC in age-adjusted models (P = 0.032 and P = 0.002, respectively) when compared with ChPT and healthy control samples. At a 95% specificity threshold, the sensitivity for IGFBP2 was 22% and the sensitivity for MSLN was 17%. Neither protein approached the diagnostic accuracy of CA 19-9. However, IGFBP2 or MSLN or both correctly identified 18 of the 28 samples misidentified by CA 19-9. In age-adjusted models, neither serum IGFBP2 (P = 0.36) nor MSLN (P = 0.29) were significant predictors of survival.
Discussion
Serum IGFBP2 and MSLN are weak diagnostic classifiers individually, but may be useful in a diagnostic biomarker panel.
Introduction
Pancreatic cancer accounts for 3% of new cancer diagnoses,1 but in spite of this low incidence rate, it ranks as the fourth most common cause of cancer mortality for both men and women annually in the United States. The problem is only made more troubling by the fact that the number of deaths per 100 000 caused by pancreatic cancer per has not decreased significantly in 15 years.1 These distressing statistics are in large part as a result of the difficulty in detecting and diagnosing pancreatic cancer at a treatable stage. Pancreatic ductal adenocarcinoma (PDAC), if detected before metastasis, has a survival rate of approximately 20%. If detection does not occur until after metastasis, the point at which symptoms begin to be noticeable, the survival rate plummets to 5%.2 These statistics highlight the need for better detection methods.
Of the secreted proteins that are commonly over expressed in PDAC,3 we identified insulin-like growth factor binding protein 2 (IGFBP2) and mesothelin (MSLN) for further analysis as potential PDAC diagnostic and prognostic biomarkers. IGFBP2 has not been evaluated as a potential diagnostic or prognostic biomarker for PDAC. The gene and protein exhibited an average over expression of 4.8-fold change in PDAC and has been studied as a circulating biomarker in lung and liver cancers.3–5 In pancreatic tissue, IGFPB2 was elevated in PDAC samples, but not in chronic pancreatitis samples.6 IGFBP2 was also elevated in pancreatic juice from pancreatic cancer patients.7 MSLN RNA demonstrated an average over expression of 6.4-fold change in PDAC3,8–10 and has shown diagnostic and prognostic potential in lung, ovarian and breast cancers.3,11–13 Owing to its role in cancer progression,12–14 including PDAC,13–18 MSLN has become a target for intervention in mesothelin-expressing cancers.14,15,19–23 Serum MSLN has been studied as a diagnostic marker in PDAC,24,25 however, the number of samples evaluated was small and the protein was not assessed as a prognostic biomarker. At the level of the gene MSLN is thought to be co-regulated with TIMP-1,26 which has previously been identified as an effective PDAC biomarker.27
Methods
Levels of IGFBP2 and MSLN were determined in serum samples from 84 patients with histologically or cytologically confirmed PDAC, 84 gender-matched and age-approximated healthy control subjects (CON), and 40 patients with chronic pancreatitis (ChPT). Serum samples from healthy control subjects were obtained from two sources: healthy adults accompanying index patients and excess sera obtained from a reference library (ARUP Laboratories) managed by the University of Utah Department of Pathology. All blood samples were collected prior to treatment, separated into the serum component and frozen for later analysis. Subject characteristics are detailed in Table 1. Patient treatment and outcomes information were obtained from a curated research database maintained by the Pancreas Cancer Research Program at the University of Utah. Staging information was abstracted from patient records. Survival information was determined from the last contact date for censored cases or patient records, local and regional cancer registries, the Social Security Death Index or national obituary records for deceased cases. Nine of the 84 PDAC cases evaluated in this study were censored at the time of analysis. Of those, five were actively followed and four were lost to follow-up, but had not appeared in death registries.
Table 1.
Subject demographics
| Diagnosis | Subject group | No. cases | Median age (Range), years |
|---|---|---|---|
| Pancreatic adenocarcinoma | Total | 84 | 65.5 (44–87) |
| Female | 40 | 69 (44–87) | |
| Male | 44 | 61.5 (47–83) | |
| Class N0 | 18 | 64 (48–83) | |
| Class N1 | 39 | 66 (47–87) | |
| Class M1 | 25 | 66 (44–81) | |
| Healthy control | Total | 84 | 63.5 (29–94) |
| Female | 40 | 63 (49–94) | |
| Male | 44 | 64 (29–85) | |
| Chronic pancreatitis | Total | 40 | 48 (29–80) |
| Female | 20 | 46.5 (30–70) | |
| Male | 20 | 50 (29–80) | |
Serum protein levels were determined by enzyme-linked immunosorbent assay (ELISA) (Human IGFBP2 ELISA Kit; RayBiotech, Inc., Norcross, GA, Human Mesothelin Quantikine ELISA Kit; R&D Systems, Inc. Minneapolis, MN, Gastrointestinal Cancer Antigen CA 19-9 EIA Test Kit; Diagnostic Automation, Inc., Calabasas, CA, USA). The serum samples were diluted 1:200 for IGFBP2 and 1:25 for MSLN and then assayed according to the manufacturer's recommended protocol. CA 19-9 levels were measured according to the manufacturer's recommendations, although assays were performed using 25 μl of serum. Protein levels were calculated by comparing absorbance readings against a calibration curve generated from standards of known concentration. Samples that yielded readings outside the linear range of detection were further diluted and re-evaluated.
Linear models were used to relate IGFBP2 and MSLN levels to gender, age, stage and class. Univariate and multivariate Cox models were employed for survival analyses. Correlation analyses were performed to compare MSLN and TIMP-1 levels. TIMP-1 levels were previously determined in these samples.27 Receiver-operating characteristic (ROC) curves were determined and the area under the curve (AUC) was calculated as a comparative measure of diagnostic accuracy. All statistical analyses were performed using ‘R’ statistical computing software, version 2.15.0.28 P-values < 0.05 were considered significant.
All studies were performed with the approval of the Institutional Review Board at the University of Utah.
Results
IGFBP2 and MSLN as diagnostic biomarkers
To assess the diagnostic potential of IGFBP2 and MSLN, serum levels were determined in CON cases, ChPT cases and patients PDAC. Serum IGFBP2 was significantly elevated in PDAC cases (450.7 ± 301.7 ng/ml, mean ± standard deviation) when compared with serum from ChPT cases (258 ± 145.1 ng/ml, P = 0.0002), but did not reach the level of significance relative to serum from healthy control subjects (321.2 ± 285.0 ng/ml, P = 0.069). Serum IGFBP2 was significantly elevated in PDAC cases compared with the combined CON and ChPT group (P = 0.002) (Fig. 1a). Similarly, serum MSLN was significantly elevated in PDAC cases (32.87 ± 17.50 ng/ml) when compared with serum from ChPT cases (25.81 ± 12.36 ng/ml, P = 0.042) as well as healthy control subjects (24.31 ± 13.25 ng/ml, P = 0.0003). MSLN was significantly elevated in PDAC cases when compared with the combined controls (P = 0.0002) (Fig. 1b). Linear models indicated that age was also significantly related to serum protein levels when assessed in healthy control subjects, with an increase of 8.94 ng/ml for IGFBP2 and 0.4239 ng/ml for MSLN per year (P = 0.0017 and P = 0.00029 for slope, respectively). The direct relationship between protein levels and age was also evident in PDAC cases for MSLN (0.53 ng/ml/year, P = 0.003), although the relationship did not reach the level of significance for IGFBP2 (6.01 ng/ml/year, P = 0.058). In spite of this age dependence, serum IGFBP2 and MSLN remained significant predictors of PDAC diagnosis when compared with combined controls after adjusting for age (P = 0.032 and P = 0.002, respectively). Serum protein levels of IGFBP2 (P = 0.95) and MSLN (P = 0.56) were not significantly related to gender.
Figure 1.
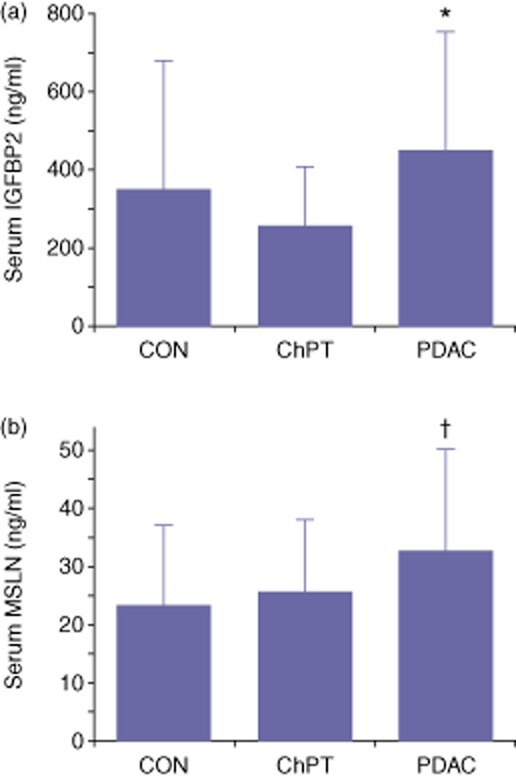
Mean serum protein levels of insulin-like growth factor binding protein 2 (IGFBP2) and mesothelin (MSLN). IGFBP2 (a) and MSLN (b) were measured in serum samples obtained from healthy control subjects (CON), patients with chronic pancreatitis (ChPT) and pre-treatment patients with confirmed pancreatic ductal adenocarcinoma (PDAC). Error bars represent a standard deviation. *P = 0.069 versus CON, P = 0.0002 versus ChPT and P = 0.0022 versus CON + ChPT. †P = 0.0003 versus CON, P = 0.042 versus ChPT and P = 0.0002 versus CON + ChPT
Although aggregate data demonstrated elevated IGFBP2 and MSLN in serum from PDAC cases compared with controls, there was substantial overlap between subject groups when individual cases were considered (Fig. 2a,b) suggesting a limited diagnostic utility of the individual biomarkers. At a threshold value corresponding to a specificity of 95%, the sensitivity for IGFBP2 was 22% (Fig. 2a) and the sensitivity for MSLN was 17% (Fig. 2b). Demonstrating the accuracy to discriminate PDAC from both CON and ChPT over the full range of threshold values, the diagnostic strength of each protein serum level can be summarized by the area under the ROC curve (Fig. 3). The AUC for IGFBP2 was 0.655 (Fig. 3a) and for MSLN was 0.668 (Fig. 3b). For comparison, CA 19-9, the most commonly used biomarker for PDAC, yielded an AUC of 0.921 in this cohort (Fig. 3c). The 95% specificity threshold yielded a sensitivity of 76% for CA 19-9. At the commonly used clinical threshold of 37 U/ml, CA 19-9 produced a sensitivity of 84% and a specificity of 88%. However, of the 28 samples misidentified by CA 19-9 at the 37 U/ml threshold, 18 were correctly identified by either IGFBP2 or MSLN or both using their respective 95% specificity threshold.
Figure 2.
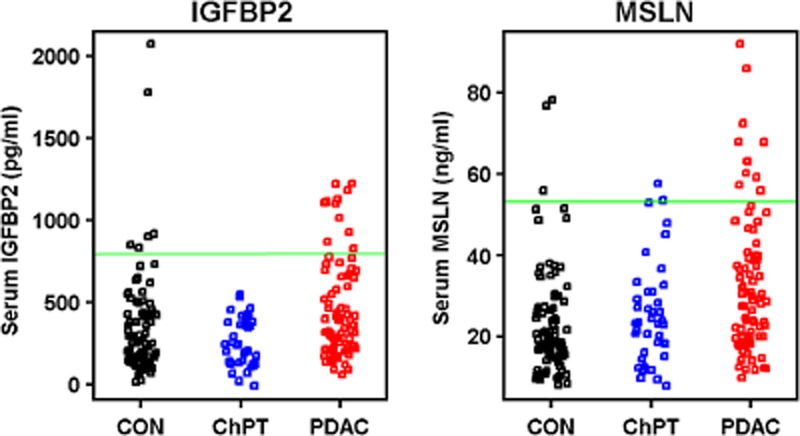
Distribution of serum insulin-like growth factor binding protein 2 (IGFBP2) and mesothelin (MSLN) in individual samples. Each data point represents the serum level for IGFBP2 (a) and MSLN (B) in individual healthy control subjects (CON), patients with chronic pancreatitis (ChPT) and patients with pancreatic ductal adenocarcinoma (PDAC). A threshold value corresponding to a specificity of 95% is represented by a green line
Figure 3.
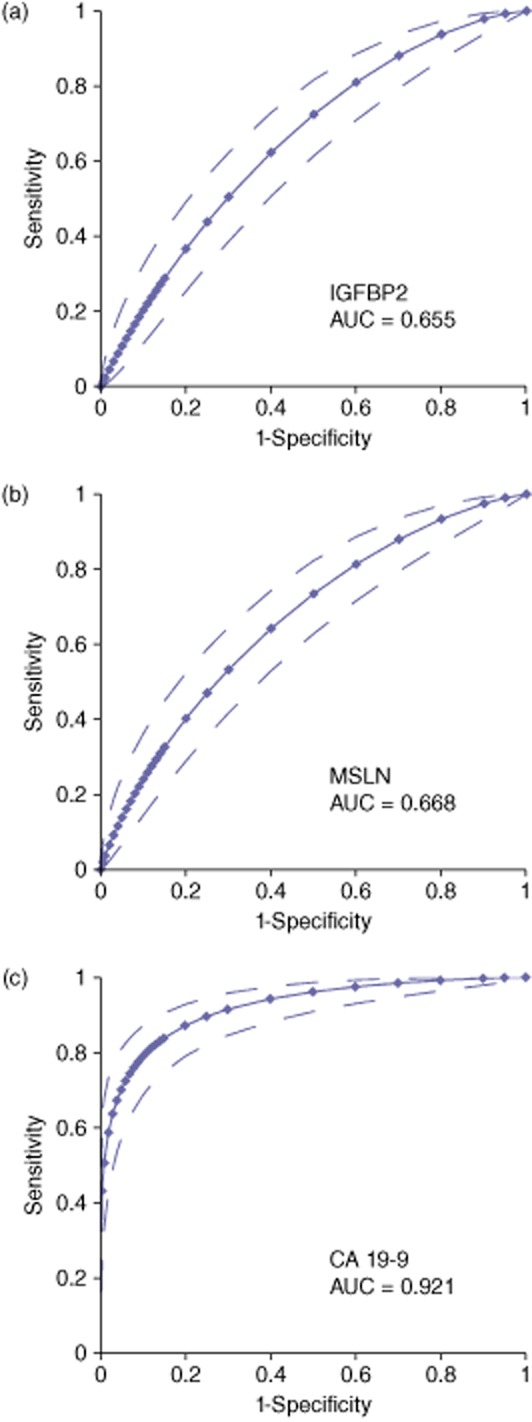
Receiver-operating characteristics (ROC) curves for IGFBP2 (a), MSLN (b) and CA 19-9 (c). Dotted lines indicate the upper and lower 95% confidence levels. AUC = area under the curve
Relationship between TIMP-1 and MSLN in serum
The potential co-regulation of MSLN and TIMP-1 was examined by comparing serum levels of each protein using univariate linear modelling. Consistent with previous findings, both MSLN and TIMP-1 were significantly related to diagnosis. There is no significant relationship between serum MSLN and serum TIMP-1 values (P = 0.22, data not shown) after adjusting for diagnosis in multivariate models.
IGFBP2 and MSLN as disease progression biomarkers
To examine the correlation of biomarker with extent of disease, serum IGFBP2 and MSLN levels were compared at different PDAC stages (Fig. 4). Mean IGFBP2 levels increased as the stage increased (P < 0.0001, Fig. 4a) indicating that serum IGFBP2 levels correlated with tumour burden. MSLN levels did not change significantly as the stage advanced (P = 0.59, Fig. 4b). Neither IGFBP2 (P = 0.44) nor MSLN (P = 0.26) serum levels were significantly different when compared by tumour grade (data not shown).
Figure 4.
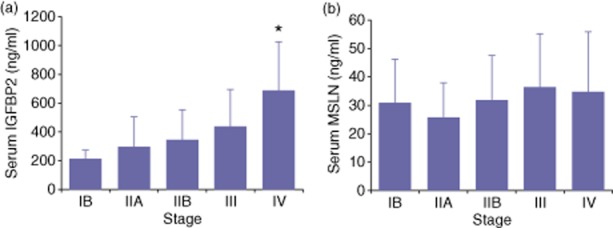
Stage distribution of serum insulin-like growth factor binding protein 2 (IGFBP2) and mesothelin (MSLN) in pancreatic ductal adenocarcinoma (PDAC) samples. Mean levels of IGFBP2 (a) and MSLN (b) at each stage are represented by the solid bars. Error bars indicate a standard deviation. *P < 0.03 versus all other stages by ANOVA and Tukey–Kramer post-hoc test
IGFBP2 and MSLN as prognostic biomarkers
Linear models were developed to investigate the relationship of IGFBP2 and MSLN levels to PDAC patient survival. In univariate analyses, serum IGFBP2, serum MSLN, age, gender and class were all significant predictors of survival (Table 2). In multivariate analysis, age in IGFBP2 models, and M1 status in both IGFBP2 and MSLN models remained significant predictors of survival. Adjusting for age, gender and class, serum IGFBP2 (P = 0.36) and MSLN (P = 0.29) were no longer significant predictors of survival (Table 2). Although adequately powered to detect the observed 2.0 hazard ratios in univariate interquartile range differences in the data, the study was underpowered to detect the observed 1.2 hazard ratios in mutivariate interquartile range differences. Thus, a larger study might reveal a prognostic potential for serum IGFBP2 and MSLN.
Table 2.
Univariate and multivariate analyses for factors affecting survival in pancreatic ductal adenocarcinoma (PDAC) cases (N = 84)
| Predictor | Level | Hazard ratio | Univariate P-value (log-rank or Wald) | Multivariate P-value (IGFBP2 model) | Multivariate P-value (MSLN model) |
|---|---|---|---|---|---|
| IGFBP2 | – | 1.0015b | < 0.0001 | 0.36a | |
| MSLN | – | 1.015b | 0.022 | 0.29a | |
| Age at Sample | – | 1.031c | 0.0057 | 0.039 | 0.053 |
| Gender | Female | 1.00 | (reference) | (reference) | (reference) |
| Male | 0.5456 | 0.0090 | 0.30 | 0.23 | |
| Class | N0 | (reference) | (reference) | (reference) | |
| N1 | 1.578 | 0.17 | 0.27 | 0.30 | |
| M1 | 6.436 | <0.0001 | <0.0001 | <0.0001 | |
Adjusted for age, gender and class.
Per unit change.
Per year.
Bold numbers indicate statistically significant comparisons.
Discussion
The diagnosis and management of PDAC is hampered by the lack of effective biomarkers. The conventionally used PDAC biomarker, CA 19-9, has limited diagnostic accuracy, is not synthesized by about 10% of the population and is not specific for PDAC as it is elevated in several gastrointestinal diseases.29 For these reasons, CA 19-9 is not used for screening but is commonly employed to monitor disease progression and response to therapy. In spite of extensive research, no single diagnostic biomarker has emerged that is more effective than CA 19-9. Numerous combinations of genetic lesions lead to PDAC development30 indicating a high potential for variability in the molecular signatures in different patients. This heterogeneity suggests that a panel of biomarkers may be a more effective approach. It is in such a panel that serum levels of IGFBP2 and MSLN, although independently weak, may prove useful as a subset of cases showed elevated serum levels. This idea is supported by the observation that IGFBP2 and MSLN levels correctly identified samples that were misidentified by CA 19-9.
In the context of a biomarker panel, co-expressed proteins would provide redundant diagnostic information. Based on the presence of a cancer-related promoter element, MSLN and TIMP-1 have been proposed to be transcriptionally co-regulated.26 However, in our sample set, MSLN and TIMP-1 were not correlated suggesting that any transcriptional co-regulation did not extend to circulating serum levels. Thus, TIMP-1, which we previously showed to be diagnostically useful,27 may provide information supplementary to MSLN in a biomarker panel.
Circulating MSLN was prognostic for survival in both gastric and ovarian cancers,31,32 but MSLN serum levels were unrelated to stage or survival in our PDAC sample set. The result that serum MSLN was independent of PDAC stage is consistent with a previous report.25 When measured in tumour samples via tissue microarray, MSLN levels in PDAC were predictive of early cancer-caused mortality.33 Poor outcomes would be expected in highly invasive disease and MSLN levels in PDAC tumours have been clearly linked to increased invasiveness.14,15 Our results suggest that, in PDAC, circulating MSLN is probably not related to tumor levels. In contrast, IGFBP2 serum levels increased with increasing stage in our sample set indicating a direct relationship with tumour burden. As advanced stage disease correlates with poor outcomes, it is unclear why IGFBP2 levels were not related to survival, but it remains possible that a larger sample set will reveal a significant relationship.
In summary, our study found that serum levels of IGFBP2 and MSLN were both significantly elevated in PDAC relative to non-cancer controls. Although individually weak diagnostic biomarkers, serum levels of the proteins may prove useful when combined with other biomarkers in a panel. Neither IGFBP2 nor MSLN demonstrated prognostic accuracy in this sample population although a study with a larger sample population may be warranted.
Conflicts of interest
None declared.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Compton CC, Mulvihill SJ. Prognostic factors in pancreatic carcinoma. Surg Oncol Clin N Am. 1997;6:533–554. [PubMed] [Google Scholar]
- 3.Harsha HC, Kandasamy K, Ranganathan P, Rani S, Ramabadran S, Gollapudi S, et al. A compendium of potential biomarkers of pancreatic cancer. PLoS Med. 2009;6:e1000046. doi: 10.1371/journal.pmed.1000046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang Y, Ying X, Han S, Wang J, Zhou X, Bai E, et al. Autoantibodies against insulin-like growth factorbinding protein-2 as a serological biomarker in the diagnosis of lung cancer. Int J Oncol. 2013;42:93–100. doi: 10.3892/ijo.2012.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou Q, Mao YQ, Jiang WD, Chen YR, Huang RY, Zhou XB, et al. Development of IGF signaling antibody arrays for the identification of hepatocellular carcinoma biomarkers. PLoS One. 2012;7:e46851. doi: 10.1371/journal.pone.0046851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen R, Brentnall TA, Pan S, Cooke K, Moyes KW, Lane Z, et al. Quantitative proteomics analysis reveals that proteins differentially expressed in chronic pancreatitis are also frequently involved in pancreatic cancer. Mol Cell Proteomics. 2007;6:1331–1342. doi: 10.1074/mcp.M700072-MCP200. [DOI] [PubMed] [Google Scholar]
- 7.Chen R, Pan S, Yi EC, Donohoe S, Bronner MP, Potter JD, et al. Quantitative proteomic profiling of pancreatic cancer juice. Proteomics. 2006;6:3871–3879. doi: 10.1002/pmic.200500702. [DOI] [PubMed] [Google Scholar]
- 8.Argani P, Iacobuzio-Donahue C, Ryu B, Rosty C, Goggins M, Wilentz RE, et al. Mesothelin is overexpressed in the vast majority of ductal adenocarcinomas of the pancreas: identification of a new pancreatic cancer marker by serial analysis of gene expression (SAGE) Clin Cancer Res. 2001;7:3862–3868. [PubMed] [Google Scholar]
- 9.Hassan R, Laszik ZG, Lerner M, Raffeld M, Postier R, Brackett D. Mesothelin is overexpressed in pancreaticobiliary adenocarcinomas but not in normal pancreas and chronic pancreatitis. Am J Clin Pathol. 2005;124:838–845. [PubMed] [Google Scholar]
- 10.Iacobuzio-Donahue CA, Ashfaq R, Maitra A, Adsay NV, Shen-Ong GL, Berg K, et al. Highly expressed genes in pancreatic ductal adenocarcinomas: a comprehensive characterization and comparison of the transcription profiles obtained from three major technologies. Cancer Res. 2003;63:8614–8622. [PubMed] [Google Scholar]
- 11.O'Shannessy DJ, Somers EB, Palmer LM, Thiel RP, Oberoi P, Heath R, et al. Serum folate receptor alpha, mesothelin and megakaryocyte potentiating factor in ovarian cancer: association to disease stage and grade and comparison to CA125 and HE4. J Ovarian Res. 2013;6:29. doi: 10.1186/1757-2215-6-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Servais EL, Colovos C, Rodriguez L, Bograd AJ, Nitadori J, Sima C, et al. Mesothelin overexpression promotes mesothelioma cell invasion and MMP-9 secretion in an orthotopic mouse model and in epithelioid pleural mesothelioma patients. Clin Cancer Res. 2012;18:2478–2489. doi: 10.1158/1078-0432.CCR-11-2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y, Wang L, Li D, Wang HB, Chen QF. Mesothelin promotes invasion and metastasis in breast cancer cells. J Int Med Res. 2012;40:2109–2116. doi: 10.1177/030006051204000608. [DOI] [PubMed] [Google Scholar]
- 14.Wang K, Bodempudi V, Liu Z, Borrego-Diaz E, Yamoutpoor F, Meyer A, et al. Inhibition of mesothelin as a novel strategy for targeting cancer cells. PLoS One. 2012;7:e33214. doi: 10.1371/journal.pone.0033214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shimizu A, Hirono S, Tani M, Kawai M, Okada K, Miyazawa M, et al. Coexpression of MUC16 and mesothelin is related to the invasion process in pancreatic ductal adenocarcinoma. Cancer Sci. 2012;103:739–746. doi: 10.1111/j.1349-7006.2012.02214.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bharadwaj U, Li M, Chen C, Yao Q. Mesothelin-induced pancreatic cancer cell proliferation involves alteration of cyclin E via activation of signal transducer and activator of transcription protein 3. Mol Cancer Res. 2008;6:1755–1765. doi: 10.1158/1541-7786.MCR-08-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen SH, Hung WC, Wang P, Paul C, Konstantopoulos K. Mesothelin binding to CA125/MUC16 promotes pancreatic cancer cell motility and invasion via MMP-7 activation. Sci Rep. 2013;3:1870. doi: 10.1038/srep01870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zheng C, Jia W, Tang Y, Zhao H, Jiang Y, Sun S. Mesothelin regulates growth and apoptosis in pancreatic cancer cells through p53-dependent and -independent signal pathway. J Exp Clin Cancer Res. 2012;31:84. doi: 10.1186/1756-9966-31-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deng L, Ke X, He Z, Yang D, Gong H, Zhang Y, et al. A MSLN-targeted multifunctional nanoimmunoliposome for MRI and targeting therapy in pancreatic cancer. Int J Nanomedicine. 2012;7:5053–5065. doi: 10.2147/IJN.S34801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hassan R, Cohen SJ, Phillips M, Pastan I, Sharon E, Kelly RJ, et al. Phase I clinical trial of the chimeric anti-mesothelin monoclonal antibody MORAb-009 in patients with mesothelin-expressing cancers. Clin Cancer Res. 2010;16:6132–6138. doi: 10.1158/1078-0432.CCR-10-2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li M, Bharadwaj U, Zhang R, Zhang S, Mu H, Fisher WE, et al. Mesothelin is a malignant factor and therapeutic vaccine target for pancreatic cancer. Mol Cancer Ther. 2008;7:286–296. doi: 10.1158/1535-7163.MCT-07-0483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Showalter SL, Huang YH, Witkiewicz A, Costantino CL, Yeo CJ, Green JJ, et al. Nanoparticulate delivery of diphtheria toxin DNA effectively kills Mesothelin expressing pancreatic cancer cells. Cancer Biol Ther. 2008;7:1584–1590. doi: 10.4161/cbt.7.10.6562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamasaki S, Miura Y, Davydova J, Vickers SM, Yamamoto M. Intravenous genetic mesothelin vaccine based on human adenovirus 40 inhibits growth and metastasis of pancreatic cancer. Int J Cancer. 2013;133:88–97. doi: 10.1002/ijc.27983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnston FM, Tan MC, Tan BR, Jr, Porembka MR, Brunt EM, Linehan DC, et al. Circulating mesothelin protein and cellular antimesothelin immunity in patients with pancreatic cancer. Clin Cancer Res. 2009;15:6511–6518. doi: 10.1158/1078-0432.CCR-09-0565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sharon E, Zhang J, Hollevoet K, Steinberg SM, Pastan I, Onda M, et al. Serum mesothelin and megakaryocyte potentiating factor in pancreatic and biliary cancers. Clin Chem Lab Med. 2012;50:721–725. doi: 10.1515/CCLM.2011.816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ren YR, Patel K, Paun BC, Kern SE. Structural analysis of the cancer-specific promoter in mesothelin and in other genes overexpressed in cancers. J Biol Chem. 2011;286:11960–11969. doi: 10.1074/jbc.M110.193458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poruk KE, Firpo MA, Scaife CL, Adler DG, Emerson LL, Boucher KM, et al. Serum osteopontin and tissue inhibitor of metalloproteinase 1 as diagnostic and prognostic biomarkers for pancreatic adenocarcinoma. Pancreas. 2013;42:193–197. doi: 10.1097/MPA.0b013e31825e354d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.The R Project for Statistical Computing. [updated 12 May 2013] Available at http://www.r-project.org (last accessed 15 July 2012)
- 29.Poruk KE, Gay DZ, Brown K, Mulvihill JD, Boucher KM, Scaife CL, et al. The clinical utility of CA 19-9 in pancreatic adenocarcinoma: diagnostic and prognostic updates. Curr Mol Med. 2013;13:340–351. doi: 10.2174/1566524011313030003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aktas B, Kasimir-Bauer S, Wimberger P, Kimmig R, Heubner M. Utility of mesothelin, L1CAM and Afamin as biomarkers in primary ovarian cancer. Anticancer Res. 2013;33:329–336. [PubMed] [Google Scholar]
- 32.Einama T, Homma S, Kamachi H, Kawamata F, Takahashi K, Takahashi N, et al. Luminal membrane expression of mesothelin is a prominent poor prognostic factor for gastric cancer. Br J Cancer. 2012;107:137–142. doi: 10.1038/bjc.2012.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Winter JM, Tang LH, Klimstra DS, Brennan MF, Brody JR, Rocha FG, et al. A novel survival-based tissue microarray of pancreatic cancer validates MUC1 and mesothelin as biomarkers. PLoS One. 2012;7:e40157. doi: 10.1371/journal.pone.0040157. [DOI] [PMC free article] [PubMed] [Google Scholar]


