Summary
Eukaryotic cells compartmentalize biochemical processes in different organelles, often relying on metabolic cycles to shuttle reducing equivalents across intracellular membranes. NADPH serves as the electron carrier for the maintenance of redox homeostasis and reductive biosynthesis, with separate cytosolic and mitochondrial pools providing reducing power in each respective location. This cellular organization is critical for numerous functions but complicates analysis of metabolic pathways using available methods. Here we develop an approach to resolve NADP(H)-dependent pathways present within both the cytosol and the mitochondria. By tracing hydrogen in compartmentalized reactions that use NADPH as a cofactor, including the production of 2-hydroxyglutarate by mutant isocitrate dehydrogenase enzymes, we can observe metabolic pathway activity in these distinct cellular compartments. Using this system we determine the direction of serine/glycine interconversion within the mitochondria and cytosol, highlighting the ability of this approach to resolve compartmentalized reactions in intact cells.
Introduction
One of the defining characteristics of eukaryotic cell metabolism is the compartmentalization of reactions in different organelles. Although coordination of metabolic flux across organelles is critical for cell physiology, the inability to distinctly observe identical reactions present in more than one subcellular location has been a major barrier to understanding cell metabolism. Many of these compartmentalized reactions are oxidation/reduction (redox) reactions that utilize pyridine nucleotide-based cofactors to transfer electrons between metabolites to support biosynthesis, redox homeostasis, signal transduction, and ATP generation (Pollak et al., 2007a). For instance, reduction of NAD+ to NADH captures energy from catabolic reactions to drive ATP synthesis through mitochondrial oxidative phosphorylation, whilst NADPH is regenerated via a different set of reactions to maintain reduced glutathione (GSH) pools and support reductive biosynthesis (Lunt and Vander Heiden, 2011). As such, NADPH has been hypothesized to be limiting for proliferation, lipid biosynthesis, and survival in response to cell stress (Diehn et al., 2009; Jeon et al., 2012; Jiang et al., 2013; Schafer et al., 2009). These compartmentalized metabolic processes impact numerous cell and tissue functions; therefore, understanding how biochemical networks function across compartments is necessary to determine how metabolism contributes to disease pathologies.
The pool of NADP(H) in cells is small relative to flux through pathways that utilize this cofactor (Pollak et al., 2007a). Thus, interconversion between the oxidized and reduced states must be coupled across all reactions involving this cofactor, and changes in abundance may not be informative for assessing the use of NADPH in a particular pathway. Neither NAD(H) nor NADP(H) are known to be transported across intracellular membranes (Nikiforov et al., 2011; Pollak et al., 2007b), and multistep shuttles involving compartmentalized redox reactions are used to transfer electrons between the mitochondria and cytosol (Bissell et al., 1976; LaNoue et al., 1974; LaNoue and Schoolwerth, 1979). This organization facilitates the maintenance of different NADPH/NADP+ ratios in each subcellular location and allows for the execution of compartment-specific metabolic processes. Classically, cytosolic NADPH is thought to be regenerated primarily via the oxidative pentose phosphate pathway (PPP) (Lunt and Vander Heiden, 2011; Pollak et al., 2007a). Other potential sources of cytoplasmic NADPH exist in mammalian cells, including reactions catalyzed by specific isozymes of isocitrate dehydrogenase (IDH), malic enzyme (ME), aldehyde dehydrogenase (ALDH), and methylene tetrahydrofolate dehydrogenase (MTHFD) (Pollak et al., 2007a; Tibbetts and Appling, 2010). However, isoforms of several of these enzymes also catalyze identical reactions in the mitochondria and can potentially transfer reducing equivalents between the mitochondria and the cytosol. For example, the reductive carboxylation of alpha-ketoglutarate (αKG) to isocitrate by IDH2 consumes mitochondrial NADPH, with citrate/isocitrate subsequently transported to the cytosol where it can be oxidized by IDH1 to produce cytosolic NADPH (Sazanov and Jackson, 1994; Wise et al., 2011). Theoretically, the reverse cycle may be used to produce mitochondrial NADPH. Metabolic cycles such as this utilize compartment-specific enzymes, and existing methods for tracing metabolism rely on breaking apart cells and pooling metabolites from all compartments, making it impossible to reliably distinguish the net reaction flux through each enzyme or pathway.
Results
Tracing NADPH with 2H-labeled glucose
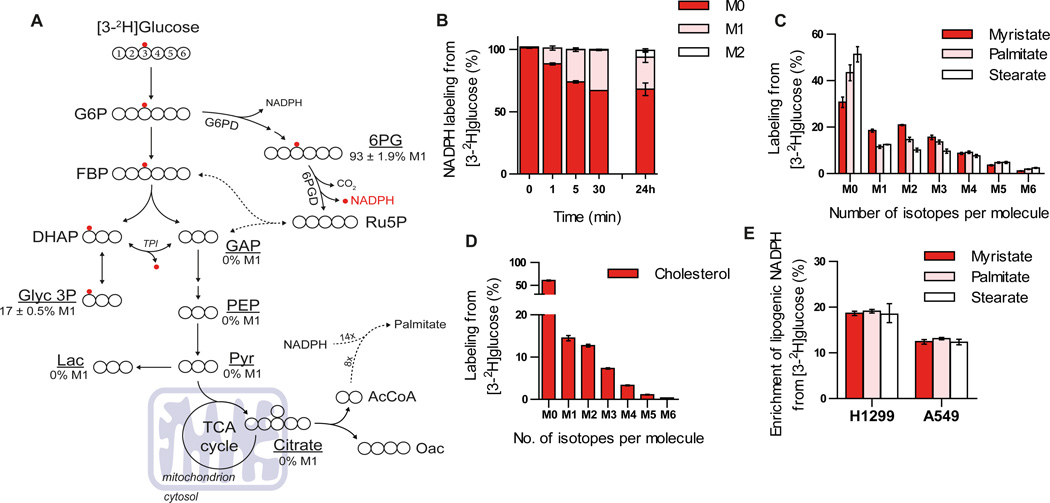
Because reaction mechanisms involving pyridine nucleotides transfer electrons as a hydride (H−) ion, isotope-labeled hydrogen atoms can be used to follow electron movement in these reactions (Katz et al., 1965; Rendina et al., 1984). The transfer of 2H and 3H can also be used to observe redox reactions in central carbon metabolism, an approach that has been used to generate insight into NAD(P)H metabolism in eukaryotic cells (Ben-Yoseph et al., 1994; Ruhl et al., 2012). Glucose is the primary carbon source for glycolysis and the oxidative PPP in mammalian cells, with the latter pathway representing an important source of cytosolic NADPH. Non-labile hydrogen atoms on specific glucose carbons (the 1 and 3 positions, respectively) are transferred to NADPH by the oxidative PPP enzymes glucose-6-phosphate dehydrogenase (G6PD) and 6-phosphogluconate dehydrogenase (6PGD). The hydrogen atom on carbon-3 of glucose (which becomes carbon-1 of dihydroxyacetone phosphate in glycolysis) can exchange with water during isomerization to glyceraldehyde-3-phosphate (GAP) by triose phosphate isomerase (TPI) (Katz et al., 1965). This prevents confounding labeling of downstream metabolites including TCA cycle intermediates, suggesting that tracing this hydrogen atom could provide a means of quantifying the contribution of 6PGD to the cellular NADPH pool (Figure 1A) (Katz et al., 1966; Katz and Rognstad, 1978). To test this possibility, we cultured H1299 non-small cell lung cancer cells in the presence of [3-2H]glucose and observed labeling of NADPH using LC/MS-MS (Figure 1B). The rapid turnover of NADPH allows labeling from [3-2H]glucose to reach isotopic steady state within 30 minutes, as evidenced by the lack of any increased label incorporated into NADPH after culturing cells in the presence of [3-2H]glucose for 24 hours (Figure 1B). NADPH has two hydrogens that can be transferred when it acts as an electron donor. Once labeled, either the labeled or unlabeled hydrogen atom can be transferred depending on the stereospecificity of downstream NADPH-utilizing enzymes (You, 1985). Transfer of the unlabeled hydride from labeled NADPH generates labeled NADP+ (Figure S1A), and subsequent labeling of the second hydrogen on NADP+ yields NADPH heavy by two mass units (M2) at later time points (Figure 1B). Some labeling of ribose-5-phosphate and ribulose-5-phosphate was also observed at late time points, presumably via flux through the nonoxidative PPP (Figure S1B). This label incorporation into the ribose moieties of NADP(H) could account for a minor portion of the isotope enrichment observed at 24 hours as suggested by the small amount of M+2 labeling of NADP+ (Figure S1A), but these atoms would not be subject to transfer in downstream reactions utilizing NADPH as a cofactor. [3-2H]glucose does not contribute to NAD(H) except by incorporation into the ribose moieties, and only a small amount of NAD(H) is labeled at late time points (Figure S1C), arguing that the majority of NADPH labeling from [3-2H]glucose reflects hydride transfer. Importantly, metabolites in lower glycolysis such as pyruvate and lactate are not labeled from [3-2H]glucose (see % values in Figure 1A) implying that the presence of label on downstream metabolites must arise as a result of hydride transfer from labeled NADPH. [1-2H]glucose labels NADPH via G6PD in the oxidative PPP (Figures S1D–E), and similar to [3-2H]glucose this label is detected on NADP+ and pentose phosphate pathway intermediates (Figures S1E–F). However, deuterium present on carbon-1 of [1-2H]glucose can be lost due to reversibility of phosphoglucose isomerase, resulting in less glucose-6-phosphate labeling from [1-2H]glucose in cells compared to labeling from [3-2H]glucose (Figure S1G) (Ben-Yoseph et al., 1994; Hellerstein et al., 1986; Katz and Rognstad, 1976). In contrast to [3-2H]glucose, the deuterium isotope from carbon-1 of glucose does not exchange with water in any of the reactions of glycolysis and is retained on carbon entering the TCA cycle (Figure S1D). Therefore, deuterium from [1-2H]glucose has the potential to label downstream metabolites by either hydride transfer from NADPH or label retention on the carbon from glucose. As a result, use of [3-2H]glucose is preferable for tracing NADPH produced by the oxidative PPP.
Figure 1. Use of 2H glucose to label cytosolic NADPH.
A) Atom-transition map depicting a model of deuterium transfer from [3-2H]glucose through glycolysis and the pentose phosphate pathway. Open large circles represent carbon and small red circles indicate deuterium label from [3-2H]glucose. Where measured, enrichment of M1 isotopomer (%) for glycolytic intermediates in parental H1299 cells is shown. B) Labeling of NADPH from [3-2H]glucose in parental H1299 cells over time. C) Saturated fatty acid labeling (myristate; C14:0, palmitate; C16:0 and stearate C18:0) from [3-2H]glucose in parental H1299 cells following incubation for 72 hours. D) Cholesterol labeling from [3-2H]glucose in parental H1299 cells cultured for 72 hours. E) Enrichment of lipogenic [2H]-NADPH by [3-2H]glucose estimated by a model for saturated fatty acid synthesis (ISA) in parental H1299 and A549 cells following incubation with tracer for 72 hours. Data plotted in a-d represent mean ± SD of at least three biological replicates. For e, data presented are mean ± 95% confidence interval of at least three biological replicates.
The NADPH labeling we observe is a subset of the total cellular pool. To gain insights into compartment-specific redox reactions we next quantified 2H enrichment in specific metabolites downstream of NADPH-dependent reactions. For example, fatty acid and cholesterol synthesis occur specifically in the cytosol and require NADPH. When H1299 cells are cultured with [3-2H] glucose for 72 hours to allow for accumulation of new lipid molecules, we detected significant label on newly synthesized fatty acids; including myristate (C14:0), palmitate (C16:0), and stearate (C18:0); as well as cholesterol (Figures 1C–D). Importantly, no label from [3-2H]glucose was detected on citrate (see % value in Figure 1A), suggesting that isotope enrichment on lipids was from the NADPH pool that was labeled by the oxidative PPP. We also detected labeling of fatty acids from [1-2H]-glucose (Figure S1H); however, tracing of G6PD-derived NADPH is complicated by deuterium from [1-2H]glucose being retained on citrate and lipogenic acetyl-CoA (Figure S1D). We next applied isotopomer spectral analysis (ISA) to estimate the contribution of [3-2H]glucose to the lipogenic NADPH pool used for palmitate synthesis, as fourteen NADPH molecules are required during the production of one palmitate molecule (Kharroubi et al., 1992; Metallo et al., 2012). The ISA model includes two parameters representing the deuterium enrichment of the NADPH pool and the percentage of palmitate that was synthesized de novo (Figure S2). Using this method we estimated the enrichment of lipogenic NADPH from [3-2H]glucose ranged from 12–20% in A549 and H1299 cells (Figure 1E). ISA modelling of other saturated fatty acids (e.g. myristate and stearate) yielded similar estimations for the enrichment of lipogenic NADPH from [3-2H]glucose (Figure 1E).
Use of 2H glucose to trace NADH metabolism
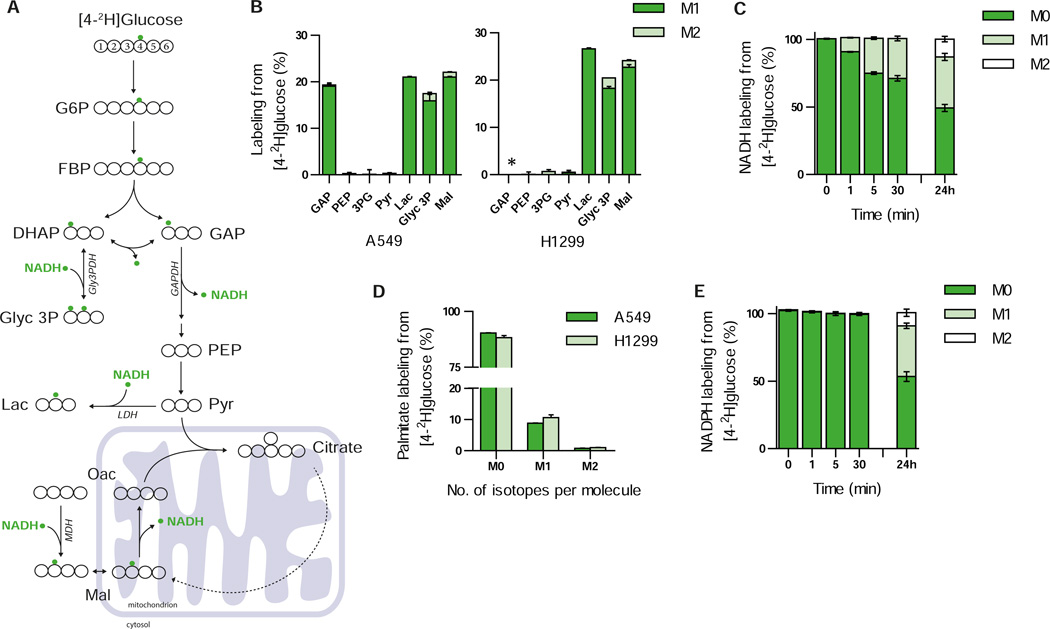
To maintain flux through glycolysis cytosolic NAD+ pools are regenerated primarily by three enzymes: lactate dehydrogenase (LDH), malate dehydrogenase (MDH), and/or the glycerol phosphate shuttle (Glyc3PDH) (Lunt and Vander Heiden, 2011; Metallo and Vander Heiden, 2013). Distinct hydrogen atoms on glucose are transferred to NAD+ during glycolysis via glyceraldehyde phosphate dehydrogenase (GAPDH). In theory, up to half of the hydrogen transferred to NADH via GAPDH comes from carbon four of glucose; however, exchange with water in the aldolase and TPI reactions decreases the net contribution of this hydrogen atom to NADH (Go et al., 2009) (Figure 2A). Upon culturing A549 and H1299 cells with [4-2H]glucose, significant labeling of lactate, malate, and glycerol 3-phosphate was observed (Figure 2B). Label was detected on GAP in A549 cells; however, the level of GAP in H1299 cells was below the limit of detection. In addition, no label was detected on metabolites in lower glycolysis including PEP, 3PG, and pyruvate. This pattern fits with known reactions using NADH in central carbon metabolism (Figure 2A) and suggests that [4-2H]glucose can be used to label the NADH pool produced by glycolysis in cells. Consistent with these findings, we observed rapid labeling from [4-2H]glucose on NADH (Figure 2C), as well as label on NAD+ (Figure S3A) arising from donation of the unlabeled hydride from M+1 labeled NADH.
Figure 2. Use of 2H glucose to label NADH.
A) Atom-transition map depicting a model of deuterium transfer from [4-2H]glucose through glycolysis and NAD+-dependent shuttle systems (malate dehydrogenase MDH, glycerol 3-phosphate dehydrogenase Gly3PDH, and lactate dehydrogenase LDH). Open large circles represent carbon and small green circles indicate deuterium label from [4-2H]glucose. B) Labeling of glycolytic intermediates from [4-2H]glucose in A549 (left panel) and H1299 (right panel) cells. Glyceraldehyde-3-phosphate (GAP) was below the limit of detection in H1299 cells, indicated by *. C) Labeling of NADH from [4-2H]glucose in parental H1299 cells over time. D) Palmitate labeling from [4-2H]glucose in A549 and H1299 cells following incubation with tracer for 72 hours. E) Time course labeling of NADPH from [4-2H]glucose in parental H1299. Data presented are mean ± SD of at least three biological replicates.
Interestingly, we observed isotope incorporation into fatty acid pools from [4-2H]glucose (Figure 2D), suggesting that some label from NADH transfers to cytosolic NADPH through an unknown mechanism (Figure S3F). Whilst deuterium label from [4-2H]glucose was detected on aspartate, citrate, and isocitrate due to the symmetry of fumarate (Figures S3B–D), carbons labeled in this manner do not contribute to lipogenic acetyl-CoA, demonstrating that the observed fatty acid labeling is derived from hydride transferring from NAD(H) to cytosolic NADP(H). We also observed some isotope enrichment on NADP+ and NADPH from [4-2H]glucose (Figure 2E and Figure S3A); however, ribose 5-phosphate and ribulose 5-phosphate are also labeled from [4-2H]glucose (Figure S3E). Furthermore, these direct measurements of total cellular NADP(H) cannot distinguish between cytosolic and mitochondrial pools, highlighting the need for methods to elucidate compartment-specific NADP(H) pools.
A reporter system to trace compartmentalized sources of NADPH
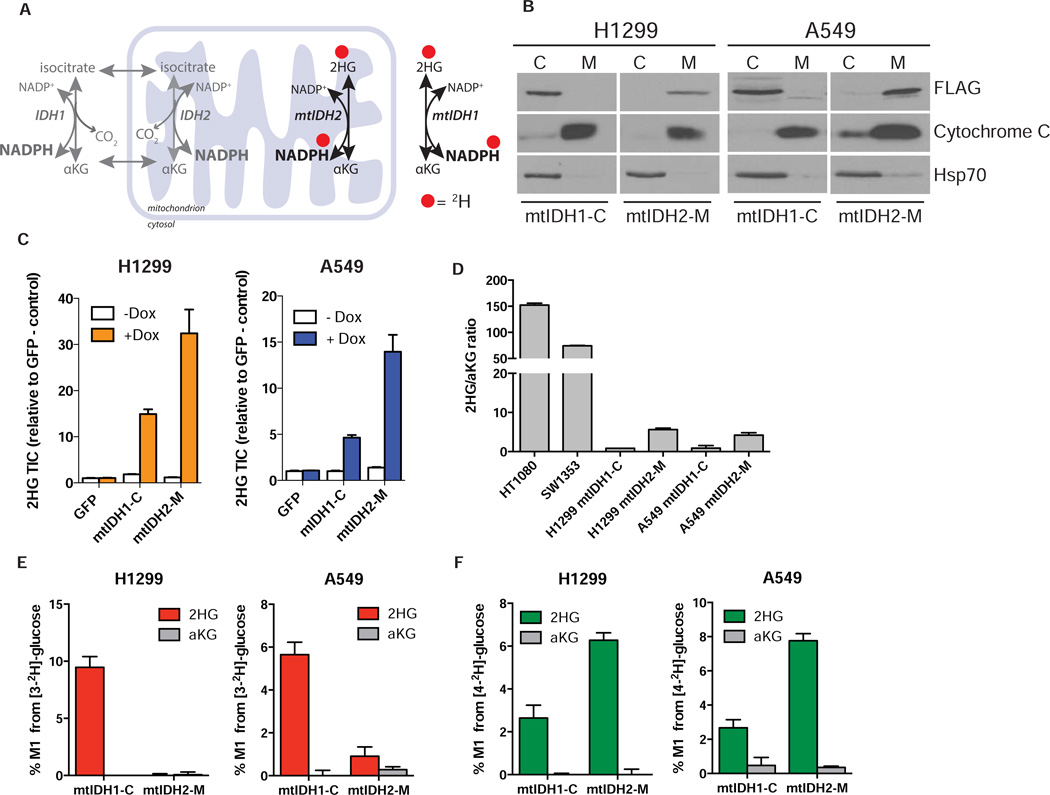
The above data demonstrate that we can observe cytosolic production of NADPH and NADH in intact cells. Although we were able to quantify the contribution of oxidative PPP enzymes to the lipogenic NADPH pool, deuterium tracing alone cannot distinguish other compartmentalized sources of NADPH. Therefore, we sought to develop a reporter system that can detect pathway-specific NADPH production in different subcellular compartments. To accomplish this, we took advantage of the neomorphic mutant IDH enzymes that produce (D)2-hydroxyglutarate (2HG) from αKG. This reaction reduces αKG by transferring a hydride from NADPH to form 2HG. As 2HG is a xenometabolite that is only present at very low levels in most cells (Matsunaga et al., 2012), it can be used as an end-product readout. By applying specific metabolic 2H-tracers to cells and measuring enrichment of 2HG produced by ectopically expressed mutant IDH1 (cytosol) or IDH2 (mitochondria), we reasoned that pathway-specific information on NADPH metabolism in each compartments could be obtained (Figure 3A).
Figure 3. Generation and characterization of cell lines expressing inducible mutant IDH.
A) Schematic demonstrating the transfer of deuterium (red dots) from NADPH to 2HG via the reaction catalyzed by mutant IDH enzymes. mtIDH1 is localized to the cytoplasm and mtIDH2 is localized to the mitochondria. By expressing compartment specific mutant enzymes and combining this with deuterated glucose tracer, it is possible to track 2HG production, and therefore the source of the NADPH used to make the 2HG in the cytosol or the mitochondria. B) Mutant IDH1-R132H is localized to the cytosol (mtIDH1-C) and mutant IDH2-R172K is localized to the mitochondria (mtIDH2-M) in H1299 and A549 cells. Cells were transduced with lentiviral constructs containing cDNA encoding C-terminal FLAG-tagged IDH1-R132H or IDH2-R172K under the control of a doxycycline-inducible promoter. Once stable cell lines were established, cells were treated with 0.1 µg/mL doxycycline for 24 hrs. Protein expression was analyzed by cellular fractionation and Western blotting using antibodies against FLAG, Cytochrome C (mitochondrial-specific marker) and Hsp70 (cytoplasmic-specific marker). C: Supernatant-100 fraction (cytoplasm); M: Mitochondria. White lines between blots in the horizontal direction indicate separate gels. C) Cell lines expressing inducible IDH mutants produce 2HG in a doxycycline-dependent manner. H1299 and A549 cells stably expressing inducible mtIDH1-C or mtIDH2-M constructs were treated with doxycycline (0.1 µg/mL) for 24 hrs. Amounts of 2HG (total ion counts: TIC) are shown relative to GFP control cells treated with vehicle. D) 2HG production, as measured by 2HG/αKG ratio, is much higher in cell lines harbouring endogenous mutations for IDH1 (R132C/+, HT1080) and IDH2 (R172S/+, SW1353) than cells expressing mtIDH1-C and mtIDH2-M. E) NADPH produced by the pentose phosphate pathway (6PGD) is cytosolic. Cells were cultured in [3-2H]-glucose (10mM) for 24 hrs before adding doxycycline (0.1µg/mL) for 24 hours to induce mutant IDH expression and amount of M1 label (%) from [3-2H]-glucose incorporated into 2HG and αKG was measured. F) NADH supports NADPH production in the mitochondria. Cells were incubated with 10mM [4-2H]-glucose for 24 hours and treated and analyzed as in E. Data represent mean ± SEM of at least three biological replicates.
We generated H1299 and A549 cell lines that express epitope-tagged mutant IDH1-R132H (mtIDH1-C) or mutant IDH2-R172K (mtIDH2-M) in a doxycycline-dependent manner (Figure S4A). We employed a weak promoter to minimize effects on endogenous IDH metabolism. Indeed, flag-tagged mtIDH1-C was expressed at levels that were not detectable with an antibody recognizing wild-type IDH1 enzyme in these cells (Figure S4A). mtIDH1-C is expected to be expressed in the cytoplasm and mtIDH2-M in the mitochondria, and we confirmed the localization of each using cell fractionation and Western blotting (Figure 3B). We also confirmed that the flag-tagged mutant IDH enzymes produce 2HG in a doxycycline-dependent manner in both H1299 and A549 cell lines (Figure 3C). Interestingly, mtIDH1-C produced less 2HG than mtIDH2-M in cells, consistent with observations that ectopically expressed IDH2 mutants produce more 2HG than IDH1 mutants due to their mitochondrial localization (Ward et al., 2013). In all cases 2HG levels were far below those observed in tumor cell lines expressing endogenous mutations in IDH1 (R132C/+, HT1080) or IDH2 (R172S/+, SW1353) (Figure 3D). Introduction of mutant IDH enzymes could impact NADPH or TCA metabolism; however, 2HG production flux observed in cell lines expressing IDH1 mutations at higher levels is small relative to other αKG-dependent reaction fluxes, suggesting mutant IDH expression has a minimal direct impact on αKG pools (Grassian et al., 2014). Furthermore, no significant change in [3-2H]glucose contribution to cytosolic NADPH was observed in A549 cells following mtIDH1-C or mtIDH2-M expression (Figure S4G).
Although doxycycline can affect the metabolism and proliferation of some mammalian cancer cell lines in culture (Ahler et al., 2013), we saw no dox-dependent changes in the abundance of central carbon metabolites (Figure S4B) or in the proliferation rate (Figure S4C) of A549 or H1299 cells. Importantly, cells expressing dox-inducible GFP also showed no changes in metabolite pool sizes or proliferation rates (Figures S4B–C), indicating that when added at this concentration doxycycline does not significantly affect metabolism in this system. In addition, we observed no significant differences in pool sizes of NAD+, NADH, NADP+ or NADPH in H1299 mtIDH1-C and mtIDH2-M cells following the addition of doxycycline for 24 hours, suggesting that dox-dependent production of 2HG was not altering the availability of these cofactors for use in other redox reactions (Figure S4D). This is supported by the reported kcat of mutant IDH1 enzymes being small relative to wild type IDH1 (Dang et al., 2009) and suggests that any direct effects of these enzymes on cellular redox state are minimal.
Validation of compartment-specific cofactor tracing
To validate the ability of this system to trace compartment-specific NADPH metabolism we induced expression of the mutant IDH enzymes in cells cultured in the presence of [3-2H]glucose and measured enrichment of 2H in the 2HG pool. In order to ensure that the cells were at or near isotopic steady-state prior to induction of mutant IDH expression, the cells were incubated with tracer for 24 hours prior to the addition of doxycycline. Consistent with [3-2H]glucose producing cytosolic NADPH via the oxidative PPP, 2HG was only significantly labeled from [3-2H]glucose in the mtIDH1-C cell lines and not in the mtIDH2-M cell lines (Figure 3E). Importantly, little to no label was observed on αKG under these conditions, ensuring that label on 2HG was a direct result of hydride ion transfer from NADPH by the mutant enzyme. We next asked whether label from [4-2H]glucose was incorporated into 2HG by either mtIDH1-C or mtIDH2-M. Notably, more 2HG was labeled from [4-2H]glucose in mtIDH2-M cells than in mtIDH1-C cells (Figure 3F). These data suggest that transfer of H- from NADH to NADPH occurs through a mitochondrial intermediate (e.g. malate) or via nicotinamide nucleotide transhydrogenase (NNT), and transfer of reducing equivalents from NADH to NADPH mostly supports the mitochondrial NADPH pool. Similar results were observed in cell lines with endogenous, heterozygous IDH1 and IDH2 mutations (Figures S4E–F).
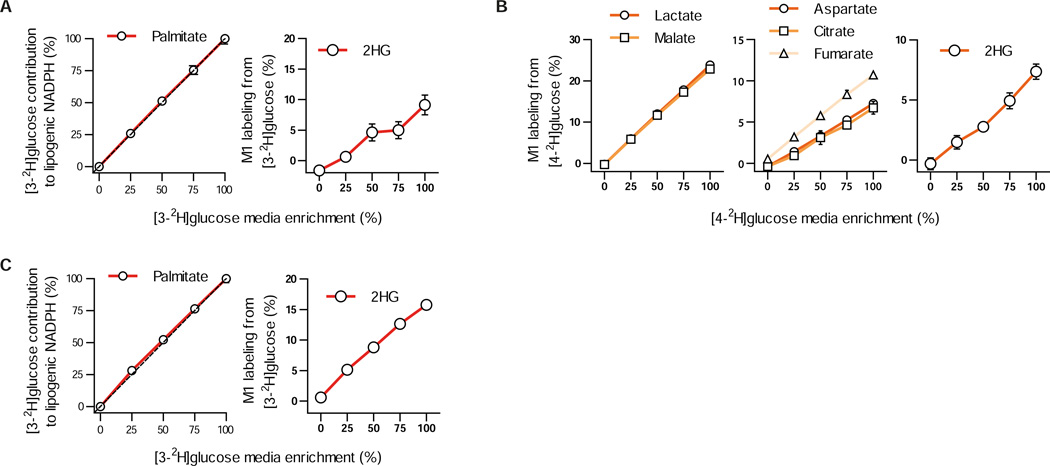
In vitro steady state enzyme kinetics experiments have demonstrated that rate constants for reactions involving 2H transfer can be lower compared to studies conducted with unlabeled substrate (Rendina et al., 1984). This phenomenon has been an invaluable tool for elucidating biochemical reaction mechanisms, including many of the reactions responsible for label transfer in our system. However, given the diverse means through which metabolism is regulated and the challenges associated with understanding which enzymatic steps are rate limiting for pathways in intact cells, the relevance of isotope effects to intracellular metabolic fluxes is not clear. To determine the significance of this phenomenon in our system we cultured cells in different ratios of [3-2H]glucose and unlabeled glucose and measured downstream labeling of lipogenic NADPH and 2HG in H1299 mtIDH1-C cells (Figure 4A). Similarly, we titrated [4-2H]glucose with unlabeled substrate in H1299 mtIDH2-M cells and observed whether label transfer to lactate, malate, aspartate, fumarate, citrate, and 2HG was affected by different amounts of labeled substrate (Figure 4B). We reasoned that if reaction rates are affected by the presence of 2H isotopes, the use of unlabeled substrates would be favored and less relative transfer of label would be observed as unlabeled substrate is titrated into the medium. However, in all cases transfer of label from either tracer decreased linearly as the tracer was diluted with unlabeled substrate, suggesting that kinetic isotope effects minimally impact the results of these experiments (Figures 4A–B). A linear decrease in lipogenic NADPH and 2HG labeling was also observed in endogenous mtIDH1 cells (HT1080) that exhibit much higher rates of 2HG production when we titrated [3-2H]glucose, further supporting the notion that isotope effects minimally affect substrate fluxes through the reactions we traced in intact cells (Figure 4C).
Figure 4. Kinetic isotope effect minimally affects [3-2H]glucose and [4-2H]glucose metabolism.
A) [3-2H]glucose was titrated with unlabeled glucose and added to H1299 cells expressing mtIDH1-C. Contribution from [3-2H]glucose to lipogenic NADPH (left panel, normalized to contribution at 100% [3-2H]glucose media enrichment) and 2HG (right panel) was measured. Dashed lined (left panel) represents 1:1 contribution of [3-2H]glucose to lipogenic NADPH to enrichment of [3-2H]glucose in media. B) [4-2H]glucose was titrated with unlabeled glucose and added to H1299 cells expressing mtIDH2-M. Labeling from [4-2H]glucose on lactate and malate (left panel); aspartate, citrate, and fumarate (middle panel); and 2HG (right panel) was measured. C) [3-2H]glucose was titrated with unlabeled glucose in HT1080 cells harbouring endogenous IDH1 mutations (R132C/+). Contribution from [3-2H]glucose to lipogenic NADPH (left panel, normalized to contribution at 100% [3-2H]glucose media enrichment) and M1 labeling on 2HG (right panel) was quantified at 0, 25, 50, 75, and 100 percent dilution with unlabeled glucose (left panel) in HT1080 cells cultured with [3-2H]glucose diluted with unlabeled glucose at 0, 25, 50, 75, and 100 percent enrichment. Data presented are shown as mean ± SD of three biological replicates for panels A (right panel), B, and C (right panel). Data represent mean ± 95% confidence interval for at least three biological replicates for panels A (left panel) and C (left panel)
Characterizing serine/glycine metabolism in the cytosol and mitochondria
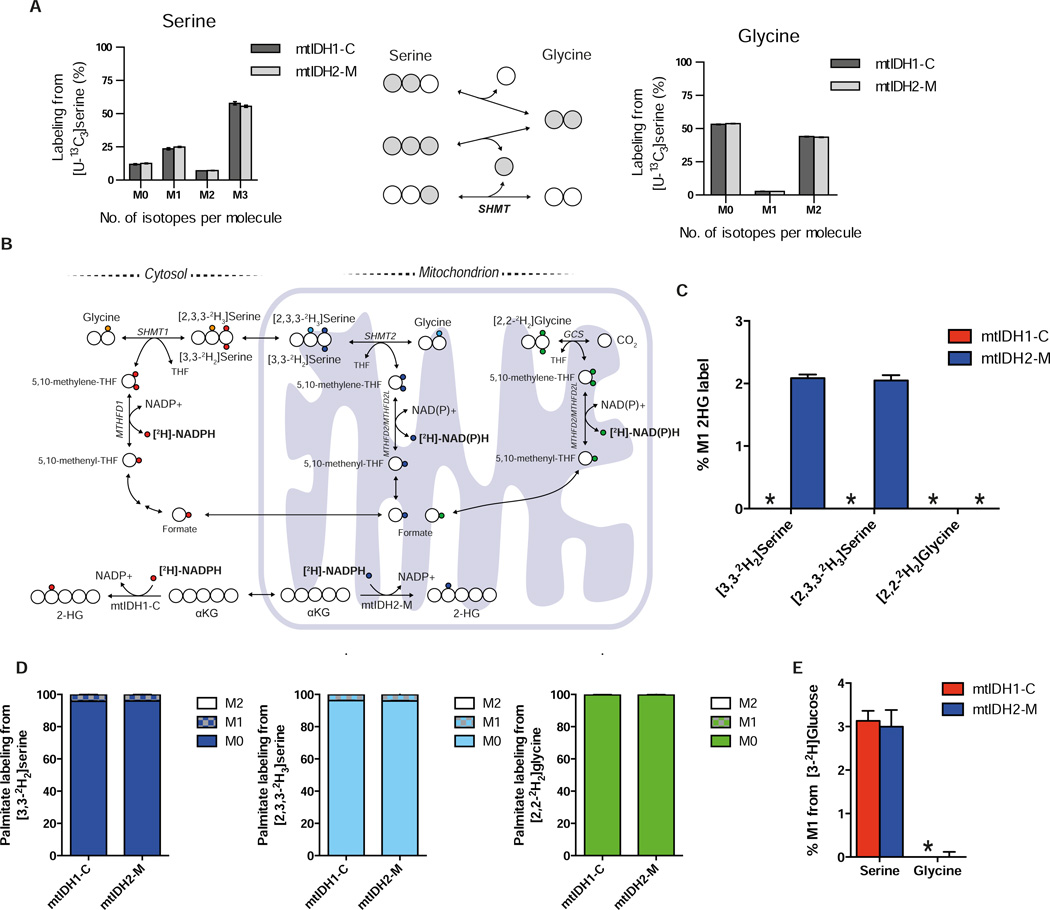
We next sought to use our reporter system to examine a compartmentalized metabolic cycle. Reactions that make up folate-mediated one carbon metabolism exist in both the cytosol and the mitochondria, although it is not clear from current literature whether these are linked in a cycle and/or in which direction the reactions proceed (Anderson et al., 2011; Nilsson et al., 2014; Tedeschi et al., 2013; Tibbetts and Appling, 2010). Serine and glycine interconversion via serine hydroxymethyltransferase (SHMT) has been observed in cultured cells (Jain et al., 2012; Levintow and Eagle, 1961; Perry et al., 2007) but 13C tracing is unable to ascertain the directionality, compartmentalization, and interconnectivity of this process. Indeed, upon culture with [U-13C3]serine we observed significant interconversion of serine and glycine in A549 mtIDH1-C and mtIDH2-M cells (Figure 5A). The reactions catalyzed by MTHFD1 (cytosolic) and MTHFD2/MTHFD2L (mitochondrial) utilize NAD(P)H (Figure 5B); therefore, we hypothesized that 2H serine and glycine tracing in combination with our compartment reporter system would enable us to experimentally determine the direction of serine-glycine exchange reactions in the cytosol and mitochondria. Hydrogens on carbon-3 of serine are transferred to 5,10-methylenetetrahydrofolate (5,10-methyleneTHF) and subsequently to NADPH via MTHFD1/2 (Figure 5B). Additionally, the glycine cleavage system (GCS) exists in the mitochondria and could transfer hydrogen from carbon two of glycine to 5,10-methylene-THF and generate NADPH (Figure 5B) (Kikuchi et al., 2008).
Figure 5. Characterizing serine/glycine metabolism in the cytosol/mitochondria.
A) Serine (left panel) and glycine (right panel) labeling in A549 mtIDH1-C and mtIDH2-M cells cultured with [U-13C3]serine. Cells were incubated with [U-13C3]serine for 24 hours prior to dox-induction (0.1 µg/mL) for an additional 48 hours. Middle panel demonstrates interconversion of serine and glycine by SHMT. Data plotted represent mean ± SD for three biological replicates. B) A schematic of folate-mediate one carbon metabolism in cytosolic and mitochondrial compartments catalyzed via SHMT and MTHFD. Deuterium transfer from [3,3-2H2]serine is shown for pathways containing SHMT and MTHFD and is indicated by small red or blue circles for cytosolic and mitochondrial isozymes, respectively. The extra deuterium on [2,3,3-2H3]serine is indicated by an orange (cytosolic) or a turquoise (mitochondrial) circle. Deuterium transfer from [2,2-2H2]glycine is shown for the glycine cleavage system (GCS) pathway indicated by small green circles. C) 2HG labeling from [3,3-2H2]serine, [2,3,3-2H3]serine or [2,2-2H2]glycine in A549 mtIDH1-C and mtIDH2-M cells. Cells were incubated with either tracer for 24 hours prior to dox-induction (0.1 µg/mL) for an additional 48 hours. No label was detected on 2HG in mtIDH1-C cells from either [3,3-2H2]serine or [2,3,3-2H3]serine, nor was label detected on 2HG from [2,2-2H2]glycine in mtIDH1-C and mtIDH2-M cells (indicated by *). D) Fatty acid labeling from A549 mtIDH1-C and mtIDH2-M cells cultured with either [3,3-2H2]serine, [2,3,3-2H3]serine, or [2,2-2H2]glycine. Cells were incubated with tracer for 24 hours prior to dox-induction (0.1 µg/mL) for an additional 48 hours. E) Serine and glycine labeling in A549 mtIDH1-C and mtIDH2-M cells cultured with [3-2H]glucose. Cells were incubated with tracer for 24 hours prior to dox-induction (0.1 µg/mL) for 48 hours. Data represent mean ± SEM of at least three biological replicates for panels C–E.
To study these compartment-specific pathways, we cultured A549 mtIDH1-C and mtIDH2-M cells with either [3,3-2H2]serine or [2,3,3-2H3]serine and unlabeled glycine or [2,2-2H2]glycine and unlabeled serine and measured incorporation of 2H in cytosolic or mitochondrial 2HG, respectively. Strikingly, we detected label from [3,3-2H2]serine and [2,3,3-2H3]serine on 2HG only in mtIDH2-M cells, strongly suggesting that serine to glycine conversion occurs primarily in the mitochondria in these cells with the MTHFD2/MTHFD2L reaction operating oxidatively (Figures 5C and Figure S5A–C). We did not observe labeling of 2HG from [2,2-2H2]glycine in cells expressing either mutant IDH (Figure 5C), indicating that either the majority of mitochondrial glycine is generated by SHMT2 (rather than glycine import) or the label is lost in the GCS. Consistent with the lack of label transfer from 2H-labeled serine or glycine to 2HG in mtIDH1-C cells, we detected minimal contribution of these tracers in the lipogenic NADPH pool (Figure 5D). To further confirm the direction of MTHFD1, we cultured A549 mtIDH1-C and mtIDH2-M cells with [3-2H]glucose, which specifically labels cytosolic NADPH. We observed transfer of 2H from [3-2H]glucose onto serine suggesting that cytosolic MTHFD1 can operate in the reductive direction in these cells (Figure 5E). The lack of glycine labeling from [3-2H]glucose confirms label transfer to serine was obtained from 5,10-methyleneTHF (Figure 5E, S5D). The lack of labeling from either [2,3,3-2H3]serine or [3,3-2H2]serine on either fatty acids or 2HG produced by mtIDH1-C is also consistent with minimal contribution of MTHFD1 to the cytoplasmic NADPH pool in these cells, although channeling to other reactions cannot be ruled out by these methods. Shuttling of serine labeled by [3-2H]glucose by MTHFD1/SHMT1 into the mitochondria for catabolism by MTHFD2/SHMT2 may account for the small amount of label (<1%) we observe on mitochondrial 2HG in A549 mtIDH2-M cells cultured with [3-2H]glucose (Figure 3E). Collectively, these data provide direct evidence that serine metabolism can contribute to regenerating mitochondrial NADPH in cells.
Discussion
We have developed a system that can distinguish compartmentalized pools of NADPH, demonstrating the directionality and interconnectivity of serine/glycine metabolism in the cytosol and mitochondria of intact cells. In the cells studied, conversion of serine to glycine occurs primarily in the mitochondria with the reaction catalyzed by MTHFD2/MTHFD2L contributing to NADPH production in this compartment. Interestingly, label transfer from both [2,3,3-2H3]serine and [3,3-2H2]serine to mitochondrial 2HG was observed, suggesting that serine metabolism by SHMT2 is a contributor to the mitochondrial NADPH and glycine pools. These data also provide direct experimental support for the hypothesis that the cytoplasmic source of formate used for purine synthesis can be mitochondrially-derived in some cells. Previous efforts to ascertain directionality of folate-mediated one carbon metabolism have been unable to distinguish between compartments, relying on expression data (Nilsson et al., 2014), mathematical modeling (Scotti et al., 2013; Tedeschi et al., 2013), or isolated mitochondrial preparations (Barlowe and Appling, 1988). The importance of distinguishing compartmentalized redox pathways is highlighted by the large number of potential pathways that have been implicated in the shuttling of reducing equivalents between the cytosol and mitochondria (Tibbetts and Appling, 2010). For instance, compartment-specific metabolic cycling through citrate/αKG (Sazanov and Jackson, 1994; Ward et al., 2010), malate/pyruvate (Jiang et al., 2013; Son et al., 2013), proline (Hagedorn and Phang, 1983; Nilsson et al., 2014), and serine have been suggested to be important for mammalian cell physiology. Although carbon tracing is increasingly combined with genetic approaches to implicate a role for compartment-specific isozymes in such processes, adaptation to genetic depletion strategies that break these cycles can confound interpretation. The reporter system described here circumvents these issues for reactions involving NAPDH by providing direct visualization of compartmentalized reaction activity and direction in intact cells.
Other subcellular compartments also have distinct metabolic needs in eukaryotic cells and could be probed with an analogous approach by engineering the localization of mutant IDH enzymes and/or monitoring 2H transfer between other metabolites. The endoplasmic reticulum (ER) is an important site for protein folding, disulfide bond formation, long chain fatty acid extension, and sterol reduction; as such, the NADP+/NADPH ratio within the ER can influence diverse cellular and physiological processes (Banhegyi et al., 2009; Kardon et al., 2008; Szaraz et al., 2010). This approach may also be adapted to quantify the NADP+/NADPH ratio in particular organelles and the turnover rate of NADPH in specific compartments. Altering the cofactor selectivity of the mutant IDH enzyme, or relying on NADH-dependent production of another xenometabolite could similarly be used to visualize compartmentalized reactions that utilize NADH. Finally, these data may be integrated with compartmentalized 13C metabolic flux analysis (MFA) models to better understand the role of cofactor metabolism in metabolic engineering applications or disease models (Ruhl et al., 2012). Thus, this approach opens up new avenues to observe metabolic processes in complex cells and improve our understanding of metabolism in normal and disease states.
Experimental Procedures
Cell Culture and Isotopic Labeling
All cell lines were maintained in DMEM supplemented with 10% FBS, 100 U/mL penicillin/streptomycin and 4 mM L-glutamine. The pSLIK-mtIDH cell lines (see below) were maintained as above, but FBS was substituted for Tet-free FBS (Clontech). Cell number was determined using an automated cell counter (Nexcelom) or by haemocytometer. For isotopic labeling experiments in the pSLIK-mtIDH cell lines, cells were cultured in 6-well plates in glucose- and glutamine- free DMEM, supplemented with 10% dialyzed Tet-free FBS, 100 U/mL penicillin/streptomycin, 4 mM L-glutamine and 10 mM or 15 mM of the appropriate deuterated glucose tracer ([3-2H, 95% or 98%]glucose or [4-2H, 94% or 98%]glucose) for 24 hour or 48–72 hour incubation, respectively (Omicron and Cambridge Isotope Laboratories, Inc.). For cholesterol labeling experiments, parental H1299 cells were cultured in DMEM supplemented with 1% FBS for two passages prior to 72 hour incubation with [3-2H]glucose. Cells were cultured in tracer medium for 24 hours prior to the addition of doxycycline hyclate (0.1 µg/mL in water: Sigma) for 24 to 48 hours in order to induce mutant IDH expression and accumulate 2HG. Isotope-labeled glycine and serine tracer medium was prepared from custom phenol red-, glucose-, sodium pyruvate-, amino acid- and sodium bicarbonate-free DMEM (Hyclone Laboratories, Inc.) supplemented with 10% dialyzed Tet-free FBS, 3.7 g/L sodium bicarbonate, and DMEM-levels of L-arginine, L-cystine, L-glutamine, L-histidine, L-isoleucine, L-leucine, L-lysine, L-methionine, L-phenylalanine, L-threonine, L-tryptophan, L-tyrosine, and L-valine prepared as a 100× stock in aqueous acid (pH 2.0). A549 pSLIK mtIDH1-C and mtIDH2-M cells were cultured in serine- and glycine- free DMEM supplemented with either [2,3,3-2H3, 98%]serine or [3,3-2H2, 98%]serine and unlabeled glycine (0.4 mM) (Cambridge Isotope Laboratories, Inc.), or [2,2-2H2, 98%]glycine and unlabeled serine (0.4 mM) (Cambridge Isotope Laboratories, Inc.). Cells were cultured in the presence of tracer medium for 24 hours prior to doxycycline addition (0.1 µg/mL) for a further 48 hours.
Generation of cell lines stably expressing inducible forms of flag-tagged mutant IDH
To generate the doxycycline-inducible mutant IDH (mtIDH) cell lines, full-length cDNA for IDH1-R132H and IDH2-R172K was amplified by PCR and cloned into the p3xFLAG-CMV14 vector (Sigma) to generate C-terminal Flag-tagged constructs. cDNA for IDH1-R132H–FLAG and IDH2-R172K–FLAG was then amplified by PCR and cloned into the pEN_TTmcs entry vector for recombination into the pSLIK-hygro lentiviral vector (Both vectors from Addgene (Shin et al., 2006)). Lentiviruses were produced by transfecting HEK-293T cells with the pSLIK-hygro-IDH1-R132H or pSLIK-hygro-IDH2-R172K plasmids along with the lentiviral packaging plasmids pMDLg/pRRE and pRSV-Rev and the envelope plasmid pMD2.G (all from Addgene). Supernatants containing lentiviral particles were collected 48 hours after transfection and used to infect sub-confluent H1299 and A549 cells. Infected cells were allowed to recover for 24 hours before being placed under selection with 350 µg/mL hygromycin (Invitrogen) for ten days. Protein expression was induced using 0.1 µg/mL doxycycline hyclate (Sigma) for 24–48 hours.
Protein expression analysis and cellular fractionation
For whole cell extracts, cells were lysed in RIPA buffer. Mitochondrial and cytoplasmic fractions were prepared as previously described (Vander Heiden et al., 1997). Briefly, cells were harvested in buffer A (250 mM sucrose, 20 mM HEPES, 10 mM KCl, 1.5 mM MgCl2, 1 mM EDTA, 1 mM EGTA, 1 mM DTT, 1× EDTA-free protease inhibitor cocktail tablet (Roche)[pH 7.4]) and broken apart using a mechanical homogenizer (H & Y Enterprise, Redwood City, CA). Following centrifugation at 750 xg to remove unlysed ells and nuclei, mitochondria were isolated by centrifuging at 10,000 xg for 25 mins. The resulting pellet was resuspended in buffer A and represents the mitochondrial fraction. The remaining supernatant containing cytoplasmic and membrane proteins was centrifuged for 1 hour at 100,000 xg. The supernatant from this final spin represents the S100 fraction. Protein expression was analyzed by Western blotting using antibodies against FLAG (DYKDDDDK Tag: Cell Signaling), IDH1 (Santa Cruz), IDH2 (Abcam), Cytochrome C (clone 7H8.2C12, Abcam), Hsp 70 (Cell Signaling).
Metabolite extraction and GC-MS analysis
Polar metabolites and fatty acids were extracted using methanol/water/chloroform as previously described (Metallo et al., 2012). Parental and pSLIK-mtIDH cells were cultured in 6-well or 12-well plates and volumes of tracer media and extraction buffers were adjusted accordingly. Derivatization of both polar metabolites and fatty acids has been described previously (Metallo et al., 2012). Briefly, polar metabolites were derivatized to form methoxime-tBDMS derivatives by incubation with 2% methoxylamine hydrochloride (MP Biomedicals) in pyridine (or MOX reagent (Thermo Scientific) followed by addition of N-tert-butyldimethylsilyl-N-methyltrifluoroacetamide (MTBSTFA) with 1% tert-butyldimethylchlorosilane (t-BDMCS) (Regis Technologies). Non-polar fractions, including triacylglycerides and phospholipids were saponified to free fatty acids and esterified to form fatty acid methyl esters either by incubation with 2% H2SO4 in methanol or by using Methyl-8 reagent (Thermo Scientific). Derivatized samples were analysed by GC-MS using a DB-35MS column (30m × 0.25mm i.d. × 0.25 µm, Agilent J&W Scientific) installed in an Agilent 7890A gas chromatograph (GC) interfaced with an Agilent 5975C mass spectrometer (MS). Mass isotopomer distributions were determined by integrating metabolite ion fragments (Table S1) and corrected for natural abundance using in-house algorithms adapted from Fernandez et al. (Fernandez et al., 1996).
Extraction of NAD+, NADH, NADP+ and NADPH and analysis by LC-MS/MS
The extraction protocol for NADPH was based on one previously described by Fendt et al. (Fendt et al., 2013), and was optimized for analysis by LC-MS/MS. Briefly, cells were cultured in 6-well plates over the course of 30 minutes, washed once in ice cold water and immediately quenched in liquid nitrogen. 200 µL ice cold extraction buffer (40:40:20 acetonitrile/methanol/200 mM NaCl, 10 mM Tris-HCl, pH 9.2) was added directly to the cells. Cells were scraped on ice and cleared by centrifugation at 4°C. 50 µL of supernatant was transferred to a polypropylene vial and samples were analysed using a Q Exactive Benchtop LC-MS/MS (Thermo Fisher Scientific).
For measurement of NAD(P)H at 24 hours, cells were cultured in 10 cm plates. After 24 hour incubation with tracer, approximately 1×107 cells were washed in ice cold 0.9% saline and immediately quenched in 1 mL of 80% methanol at −80°C. Cells were scraped on dry ice and cleared by centrifugation at 4°C. Cleared supernatant was transferred to Eppendorf tube, dried under vacuum using a CentriVap (Labconco), resuspended in water and immediately loaded onto a XSELECT HSS XP 150 mm × 2.1 mm × 2.5 µm (Waters®, Milford, MA) with an UFLC XR HPLC (Shimadzu, Columbia, MD) coupled to an AB SCIEX Qtrap® 5500 mass spectrometer (AB SCIEX, Framingham, MA) operating in negative ion mode. Mass isotopomer distributions were corrected for natural abundance using in-house software adapted from Fernandez et al. (Fernandez et al., 1996). Additional information regarding chromatographic separation, mass spectrometry, and data acquisition can be found in the Supplemental Experimental Procedures.
Isotopomer Spectral Analysis (ISA)
The ISA method compares a measured palmitate mass isotopomer distribution to one that is simulated using a reaction network for palmitate synthesis whereby 14 NADPH molecules are consumed to form one palmitate molecule. Models were also generated for myristate and stearate synthesis whereby 12 or 16 NADPH molecules are consumed to form one myristate or stearate molecule, respectively. Parameters for the relative enrichment of the lipogenic NADPH pool from a given [2H] tracer and the percentage of fatty acids that are de novo synthesized are extracted from a best fit model using the INCA metabolic flux analysis software package (Figure S2) (Young, 2014). The 95% confidence intervals for both parameters were estimated by evaluating the sensitivity of the sum of squared residuals between measured and simulated palmitate mass isotopomer distributions to small flux variations (Antoniewicz et al., 2006).
Cell Proliferation Assays
On day −1, 1/10 of a confluent 10cm dish of cells were seeded in six six-wells of a six-well plate. 24 hours later, cells were counted on an automated cell counter (Nexcelom) and this time-point was considered T0. At T0, all other time-points were media changed to −/+ dox media (0.1 µg/mL doxycycline hyclate (Sigma) in water). Cells were counted every 24 hours in technical duplicate and biological triplicate. Media was changed every 48 hours to prevent degradation of doxycycline in the media.
Supplementary Material
Highlights.
Use of 2H stable isotopes to trace NAD(P)H metabolism in cells
Quantification of pentose phosphate pathway contribution to cytosolic NADPH
Reporter system to distinguish cytosolic and mitochondrial NADPH
Resolve direction of otherwise identical compartmentalized redox reactions
Acknowledgements
We thank Michael Pacold, Elizaveta Freinkman, David Sabatini, and Bernhard Palsson for providing access to equipment. We are grateful to Patrick Ward and Craig B. Thompson for providing the cDNA for IDH1-R132H and IDH2-R172K. Eric L. Bell and Sarah-Maria Fendt provided advice and reagents. This work was supported, in part, by NIH grants P30CA147882, U54-CA121852-09, and R01CA168653, as well as the Koch Institute/DFHCC Bridge Project, the Koch Institute Frontier Research Fund, the Burroughs Wellcome Fund, the Damon Runyon Cancer Research Foundation, American Cancer Society grant IRG #70-002, DOD grant W81XWH-13-1-0105, a University of California Cancer Research Coordinating Committee grant, and a Searle Scholar Award.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
C.A.L. and S.J.P. conducted stable isotope tracing experiments and prepared samples for GC-MS and LC-MS analysis. N.I.V. and C.R.G. conducted stable isotope tracing experiments and analyzed samples by GC-MS. C.R.G. assisted in serine and glycine tracing experiments for GC-MS analysis. C.A.L. and S.J.P. ran and analyzed samples on GC-MS. B.P.F, D.Y.G., D.M., and A.M.F. developed methods and ran samples on LC-MS. C.A.L. and S.J.P. analyzed data obtained by LC-MS assisted by B.P.F. and D.M. S.J.P. built the isotopomer spectral analysis method and carried out lipid modeling. C.A.L. generated inducible mutant IDH expression system. C.M.M., M.G.V.H., C.A.L., and S.J.P. designed the experiments. C.A.L., S.J.P, C.M.M., and M.G.V.H. designed the study, analyzed all data, and wrote the manuscript.
The authors declare no conflict of interest.
References
- Ahler E, Sullivan WJ, Cass A, Braas D, York AG, Bensinger SJ, Graeber TG, Christofk HR. Doxycycline alters metabolism and proliferation of human cell lines. PLoS One. 2013;8:e64561. doi: 10.1371/journal.pone.0064561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson DD, Quintero CM, Stover PJ. Identification of a de novo thymidylate biosynthesis pathway in mammalian mitochondria. Proc Natl Acad Sci U S A. 2011;108:15163–15168. doi: 10.1073/pnas.1103623108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoniewicz MR, Kelleher JK, Stephanopoulos G. Determination of confidence intervals of metabolic fluxes estimated from stable isotope measurements. Metab Eng. 2006;8:324–337. doi: 10.1016/j.ymben.2006.01.004. [DOI] [PubMed] [Google Scholar]
- Banhegyi G, Csala M, Benedetti A. Hexose-6-phosphate dehydrogenase: linking endocrinology and metabolism in the endoplasmic reticulum. J Mol Endocrinol. 2009;42:283–289. doi: 10.1677/JME-08-0156. [DOI] [PubMed] [Google Scholar]
- Barlowe CK, Appling DR. In vitro evidence for the involvement of mitochondrial folate metabolism in the supply of cytoplasmic one-carbon units. Biofactors. 1988;1:171–176. [PubMed] [Google Scholar]
- Ben-Yoseph O, Kingsley PB, Ross BD. Metabolic loss of deuterium from isotopically labeled glucose. Magn Reson Med. 1994;32:405–409. doi: 10.1002/mrm.1910320317. [DOI] [PubMed] [Google Scholar]
- Bissell MJ, Rambeck WA, White RC, Bassham JA. Glycerol phosphate shuttle in virus-transformed cells in culture. Science. 1976;191:856–858. doi: 10.1126/science.175441. [DOI] [PubMed] [Google Scholar]
- Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehn M, Cho RW, Lobo NA, Kalisky T, Dorie MJ, Kulp AN, Qian D, Lam JS, Ailles LE, Wong M, et al. Association of reactive oxygen species levels and radioresistance in cancer stem cells. Nature. 2009;458:780–783. doi: 10.1038/nature07733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fendt SM, Bell EL, Keibler MA, Olenchock BA, Mayers JR, Wasylenko TM, Vokes NI, Guarente L, Vander Heiden MG, Stephanopoulos G. Reductive glutamine metabolism is a function of the alpha-ketoglutarate to citrate ratio in cells. Nat Commun. 2013;4:2236. doi: 10.1038/ncomms3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez CA, Des Rosiers C, Previs SF, David F, Brunengraber H. Correction of 13C mass isotopomer distributions for natural stable isotope abundance. J Mass Spectrom. 1996;31:255–262. doi: 10.1002/(SICI)1096-9888(199603)31:3<255::AID-JMS290>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Go MK, Amyes TL, Richard JP. Hydron transfer catalyzed by triosephosphate isomerase. Products of the direct and phosphite-activated isomerization of [1-(13)C]-glycolaldehyde in D(2)O. Biochemistry. 2009;48:5769–5778. doi: 10.1021/bi900636c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grassian AR, Parker SJ, Davidson SM, Divakruni AS, Green CR, Zhang X, Slocam KL, Pu M, Lin F, Vickers C, et al. IDH1 mutations alter citric acid cycle metabolism and increase dependence on oxidative mitochondrial metabolism. Cancer Res Published online April 22nd 2014. 2014 doi: 10.1158/0008-5472.CAN-14-0772-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagedorn CH, Phang JM. Transfer of reducing equivalents into mitochondria by the interconversions of proline and delta 1-pyrroline-5-carboxylate. Arch Biochem Biophys. 1983;225:95–101. doi: 10.1016/0003-9861(83)90010-3. [DOI] [PubMed] [Google Scholar]
- Hellerstein MK, Greenblatt DJ, Munro HN. Glycoconjugates as noninvasive probes of intrahepatic metabolism: pathways of glucose entry into compartmentalized hepatic UDP-glucose pools during glycogen accumulation. Proc Natl Acad Sci U S A. 1986;83:7044–7048. doi: 10.1073/pnas.83.18.7044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M, Nilsson R, Sharma S, Madhusudhan N, Kitami T, Souza AL, Kafri R, Kirschner MW, Clish CB, Mootha VK. Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation. Science. 2012;336:1040–1044. doi: 10.1126/science.1218595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon SM, Chandel NS, Hay N. AMPK regulates NADPH homeostasis to promote tumour cell survival during energy stress. Nature. 2012;485:661–665. doi: 10.1038/nature11066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang P, Du W, Mancuso A, Wellen KE, Yang X. Reciprocal regulation of p53 and malic enzymes modulates metabolism and senescence. Nature. 2013;493:689–693. doi: 10.1038/nature11776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardon T, Senesi S, Marcolongo P, Legeza B, Banhegyi G, Mandl J, Fulceri R, Benedetti A. Maintenance of luminal NADPH in the endoplasmic reticulum promotes the survival of human neutrophil granulocytes. FEBS Lett. 2008;582:1809–1815. doi: 10.1016/j.febslet.2008.04.045. [DOI] [PubMed] [Google Scholar]
- Katz J, Landau BR, Bartsch GE. The pentose cycle, triose phosphate isomerization, and lipogenesis in rat adipose tissue. J Biol Chem. 1966;241:727–740. [PubMed] [Google Scholar]
- Katz J, Rognstad R. Futile cycles in the metabolism of glucose. Curr Top Cell Regul. 1976;10:237–289. doi: 10.1016/b978-0-12-152810-2.50013-9. [DOI] [PubMed] [Google Scholar]
- Katz J, Rognstad R. Futile Cycling in Glucose-Metabolism. Trends Biochem Sci. 1978;3:171–174. [Google Scholar]
- Katz J, Rognstad R, Kemp RG. Isotope Discrimination Effects in the Metabolism of Tritiated Glucose. J Biol Chem. 1965;240:PC1484–PC1486. [PubMed] [Google Scholar]
- Kharroubi AT, Masterson TM, Aldaghlas TA, Kennedy KA, Kelleher JK. Isotopomer spectral analysis of triglyceride fatty acid synthesis in 3T3-L1 cells. Am J Physiol. 1992;263:E667–E675. doi: 10.1152/ajpendo.1992.263.4.E667. [DOI] [PubMed] [Google Scholar]
- Kikuchi G, Motokawa Y, Yoshida T, Hiraga K. Glycine cleavage system: reaction mechanism, physiological significance, and hyperglycinemia. Proc Jpn Acad Ser B Phys Biol Sci. 2008;84:246–263. doi: 10.2183/pjab/84.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaNoue KF, Meijer AJ, Brouwer A. Evidence for electrogenic aspartate transport in rat liver mitochondria. Arch Biochem Biophys. 1974;161:544–550. doi: 10.1016/0003-9861(74)90337-3. [DOI] [PubMed] [Google Scholar]
- LaNoue KF, Schoolwerth AC. Metabolite transport in mitochondria. Annu Rev Biochem. 1979;48:871–922. doi: 10.1146/annurev.bi.48.070179.004255. [DOI] [PubMed] [Google Scholar]
- Levintow L, Eagle H. Biochemistry of Cultured Mammalian Cells. Annu Rev Biochem. 1961;30 605-&. [Google Scholar]
- Lunt SY, Vander Heiden MG. Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Annu Rev Cell Dev Biol. 2011;27:441–464. doi: 10.1146/annurev-cellbio-092910-154237. [DOI] [PubMed] [Google Scholar]
- Matsunaga H, Futakuchi-Tsuchida A, Takahashi M, Ishikawa T, Tsuji M, Ando O. IDH1 and IDH2 have critical roles in 2-hydroxyglutarate production in D-2-hydroxyglutarate dehydrogenase depleted cells. Biochem Biophys Res Commun. 2012;423:553–556. doi: 10.1016/j.bbrc.2012.06.002. [DOI] [PubMed] [Google Scholar]
- Metallo CM, Gameiro PA, Bell EL, Mattaini KR, Yang J, Hiller K, Jewell CM, Johnson ZR, Irvine DJ, Guarente L, et al. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature. 2012;481:380–384. doi: 10.1038/nature10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metallo CM, Vander Heiden MG. Understanding metabolic regulation and its influence on cell physiology. Mol Cell. 2013;49:388–398. doi: 10.1016/j.molcel.2013.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikiforov A, Dolle C, Niere M, Ziegler M. Pathways and subcellular compartmentation of NAD biosynthesis in human cells: from entry of extracellular precursors to mitochondrial NAD generation. J Biol Chem. 2011;286:21767–21778. doi: 10.1074/jbc.M110.213298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson R, Jain M, Madhusudhan N, Sheppard NG, Strittmatter L, Kampf C, Huang J, Asplund A, Mootha VK. Metabolic enzyme expression highlights a key role for MTHFD2 and the mitochondrial folate pathway in cancer. Nat Commun. 2014;5:3128. doi: 10.1038/ncomms4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry C, Yu S, Chen J, Matharu KS, Stover PJ. Effect of vitamin B6 availability on serine hydroxymethyltransferase in MCF-7 cells. Arch Biochem Biophys. 2007;462:21–27. doi: 10.1016/j.abb.2007.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollak N, Dolle C, Ziegler M. The power to reduce: pyridine nucleotides--small molecules with a multitude of functions. Biochem J. 2007a;402:205–218. doi: 10.1042/BJ20061638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollak N, Niere M, Ziegler M. NAD kinase levels control the NADPH concentration in human cells. J Biol Chem. 2007b;282:33562–33571. doi: 10.1074/jbc.M704442200. [DOI] [PubMed] [Google Scholar]
- Rendina AR, Hermes JD, Cleland WW. Use of multiple isotope effects to study the mechanism of 6-phosphogluconate dehydrogenase. Biochemistry. 1984;23:6257–6262. doi: 10.1021/bi00320a056. [DOI] [PubMed] [Google Scholar]
- Ruhl M, Le Coq D, Aymerich S, Sauer U. 13C–flux analysis reveals NADPH-balancing transhydrogenation cycles in stationary phase of nitrogen-starving Bacillus subtilis. J Biol Chem. 2012;287:27959–27970. doi: 10.1074/jbc.M112.366492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sazanov LA, Jackson JB. Proton-translocating transhydrogenase and NAD- and NADP-linked isocitrate dehydrogenases operate in a substrate cycle which contributes to fine regulation of the tricarboxylic acid cycle activity in mitochondria. FEBS Lett. 1994;344:109–116. doi: 10.1016/0014-5793(94)00370-x. [DOI] [PubMed] [Google Scholar]
- Schafer ZT, Grassian AR, Song L, Jiang Z, Gerhart-Hines Z, Irie HY, Gao S, Puigserver P, Brugge JS. Antioxidant and oncogene rescue of metabolic defects caused by loss of matrix attachment. Nature. 2009;461:109–113. doi: 10.1038/nature08268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scotti M, Stella L, Shearer EJ, Stover PJ. Modeling cellular compartmentation in one-carbon metabolism. Wiley Interdiscip Rev Syst Biol Med. 2013;5:343–365. doi: 10.1002/wsbm.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin KJ, Wall EA, Zavzavadjian JR, Santat LA, Liu J, Hwang JI, Rebres R, Roach T, Seaman W, Simon MI, et al. A single lentiviral vector platform for microRNA-based conditional RNA interference and coordinated transgene expression. Proc Natl Acad Sci U S A. 2006;103:13759–13764. doi: 10.1073/pnas.0606179103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son J, Lyssiotis CA, Ying H, Wang X, Hua S, Ligorio M, Perera RM, Ferrone CR, Mullarky E, Shyh-Chang N, et al. Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway. Nature. 2013;496:101–105. doi: 10.1038/nature12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szaraz P, Banhegyi G, Benedetti A. Altered redox state of luminal pyridine nucleotides facilitates the sensitivity towards oxidative injury and leads to endoplasmic reticulum stress dependent autophagy in HepG2 cells. Int J Biochem Cell Biol. 2010;42:157–166. doi: 10.1016/j.biocel.2009.10.004. [DOI] [PubMed] [Google Scholar]
- Tedeschi PM, Markert EK, Gounder M, Lin H, Dvorzhinski D, Dolfi SC, Chan LL, Qiu J, Dipaola RS, Hirshfield KM, et al. Contribution of serine, folate and glycine metabolism to the ATP, NADPH and purine requirements of cancer cells. Cell Death Dis. 2013;4:e877. doi: 10.1038/cddis.2013.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibbetts AS, Appling DR. Compartmentalization of Mammalian folate-mediated one-carbon metabolism. Annu Rev Nutr. 2010;30:57–81. doi: 10.1146/annurev.nutr.012809.104810. [DOI] [PubMed] [Google Scholar]
- Vander Heiden MG, Chandel NS, Williamson EK, Schumacker PT, Thompson CB. Bcl-xL regulates the membrane potential and volume homeostasis of mitochondria. Cell. 1997;91:627–637. doi: 10.1016/s0092-8674(00)80450-x. [DOI] [PubMed] [Google Scholar]
- Ward PS, Lu C, Cross JR, Abdel-Wahab O, Levine RL, Schwartz GK, Thompson CB. The potential for isocitrate dehydrogenase mutations to produce 2-hydroxyglutarate depends on allele specificity and subcellular compartmentalization. J Biol Chem. 2013;288:3804–3815. doi: 10.1074/jbc.M112.435495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward PS, Patel J, Wise DR, Abdel-Wahab O, Bennett BD, Coller HA, Cross JR, Fantin VR, Hedvat CV, Perl AE, et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell. 2010;17:225–234. doi: 10.1016/j.ccr.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise DR, Ward PS, Shay JE, Cross JR, Gruber JJ, Sachdeva UM, Platt JM, DeMatteo RG, Simon MC, Thompson CB. Hypoxia promotes isocitrate dehydrogenase-dependent carboxylation of alpha-ketoglutarate to citrate to support cell growth and viability. Proc Natl Acad Sci U S A. 2011;108:19611–19616. doi: 10.1073/pnas.1117773108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You KS. Stereospecificity for nicotinamide nucleotides in enzymatic and chemical hydride transfer reactions. CRC Crit Rev Biochem. 1985;17:313–451. doi: 10.3109/10409238509113625. [DOI] [PubMed] [Google Scholar]
- Young JD. INCA: a computational platform for isotopically non-stationary metabolic flux analysis. Bioinformatics. 2014 doi: 10.1093/bioinformatics/btu015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.