Abstract
The role of Group X secreted phospholipase A2 (GX-sPLA2) during influenza infection has not been previously investigated. We examined the role of GX-sPLA2 during H1N1 pandemic influenza infection in a GX-sPLA2 gene targeted mouse (GX−/−) model and found that survival after infection was significantly greater in GX−/− mice than in GX+/+ mice. Downstream products of GX-sPLA2 activity, PGD2, PGE2, LTB4, cysteinyl leukotrienes and Lipoxin A4 were significantly lower in GX−/− mice BAL fluid. Lung microarray analysis identified an earlier and more robust induction of T and B cell associated genes in GX−/− mice. Based on the central role of sPLA2 enzymes as key initiators of inflammatory processes, we propose that activation of GX-sPLA2 during H1N1pdm infection is an early step of pulmonary inflammation and its inhibition increases adaptive immunity and improves survival. Our findings suggest that GX-sPLA2 may be a potential therapeutic target during influenza.
Keywords: Secreted phospholipase A2, Influenza, Host response, Phospholipids, H1N1 pandemic influenza, Leukotrienes, Prostaglandins, Lipoxin A4, Pathogenesis, Inflammation
Highlights
-
•
We investigated GX-sPLA2 −/− mice infected with pandemic H1N1.
-
•
GX−/− mice had increased survival and significantly lower eicosanoids in BAL fluid.
-
•
Lung RNA analysis identified earlier and increased induction of T and B cell responses in GX−/− mice.
-
•
GX may initiate influenza inflammation; inhibition promotes adaptive immunity and survival.
-
•
Our findings suggest that GX-sPLA2 may be a potential therapeutic target during influenza.
Introduction
Influenza is a leading source of morbidity and mortality worldwide that is caused by ever changing and newly emerging influenza viruses including the introduction of the 2009 A/H1N1/2009 (H1N1pdm) and novel avian H7N9 viruses (Fisher et al., 2005, Groom and Luster, 2011, Widegren et al., 2011). While effective vaccines and antiviral drugs have been developed for circulating strains of human influenza (Santone et al., 2008), continued antigenic drift and shift generate novel virus strains that pose a threat to immunologically naïve populations. The emergence of pandemic influenza H1N1pdm in the spring of 2009 led to hundreds of thousands of hospitalizations with significant numbers of fatalities in North America (Update: Influenza Activity – United States, 2009–10). Severe cases were characterized by viral pneumonia and uncontrollable pulmonary inflammation, and were similar to the inflammation observed in severe cases of SARS, H5N1 and Spanish Influenza patients (Baillie and Digard, 2013, Bermejo-Martin et al., 2010, Cameron et al., 2012, Curfs et al., 2008) Importantly there is little understood regarding the pathways driving the pulmonary inflammatory process for these diseases.
Host-defenses against influenza include anatomic barriers, mucociliary clearance, anti-microbial secretions and innate and adaptive immune responses. Early host responses are characterized by the mobilization of leukocytes, such as alveolar and circulating macrophages, polymorphonuclear leukocytes (PMN) (World Health Organization, 2012) and NK cells which continues into the activation of adaptive immune cells such as T-cells, B-cells and dendritic cells. Importantly, factors that lead to the activation of these cells and cell networks are increased after H1N1pdm Infection (Influenza Activity – United States and Worldwide,, Ohtsuki et al., 2006, Ohtsuki et al., 2006, Paquette et al., 2014). These include inflammatory mediators such as chemokines, cytokines and lipid mediators like eicosanoids.
The first step in the generation of eicosanoids, inflammatory mediators that participate in the regulation of the inflammatory response, is catalyzed by PLA2 enzymes, which release arachidonic acid (AA) from phospholipids (Del et al., 2007). To date, 11 secreted PLA2 enzymes (sPLA2; IB, IIA, IIC, IID, IIE, IIF, III, V, X, XIIA and XIIB) (Murakami et al., 1999), six cytosolic PLA2 enzymes (cPLA2s; α, β, γ, δ, ε, and ξ) (Kim et al., 2007), nine Ca2+-independent PLA2 enzymes (iPLA2s; α, β, γ, δ2, δ, ε, ε2 ξ, θ, and η) (Murakami et al., 1999), and two lysosomal PLA2 enzymes (Femling et al., 2005) have been described. The sPLA2 enzymes are structurally related, Ca2+-dependent proteins with unique biological properties, enzymatic activities against membrane phospholipids and tissue and cellular locations, suggest distinct roles for these enzymes in various pathophysiological events. sPLA2 enzymes are implicated in lipid mediator release, degranulation, cellular proliferation, destruction of invading bacteria (Murakami et al., 1999), viruses (Kennedy et al., 1995, Mazur et al., 2007) and activation of intracellular signaling cascades (Kim et al., 2007). GX-sPLA2 is expressed in alveolar macrophages and epithelial cells in the lungs of patients with pneumonia (Marshall et al., 2000), neuronal cells (Gaudreault and Gosselin, 2008), male reproductive organs (Dennis, 1994) and atherosclerotic plaques (Huang et al., 2011), and is cleaved to its active form in inflamed tissues (Lu et al., 2006).
Arachidonic acid (AA) is the precursor of prostaglandins, thromboxanes, leukotrienes and lipoxins, eicosanoids that regulate pulmonary vascular and bronchial responses, leukocyte activation, adhesion and emigration (Guan et al., 2013, Henderson et al., 1995). Eicosanoids also regulate antigen presenting cell [APC] function (Degousee et al., 2001, Murakami et al., 2011, Truchetet et al., 2012), T cell maturation (Myers et al., 2012) and Th17 expansion (Stephenson et al., 1988, Zhao et al., 2011). We found that IL-17 and Th17 cells are dysregulated during human H1N1pdm infection (Baillie and Digard, 2013, Bermejo-Martin et al., 2010, Ohtsuki et al., 2006). Therefore, sPLA2 enzymes and eicosanoids may play a central role in determining the outcome of pulmonary viral infection. The role of sPLA2 enzymes in the immune responses to H1N1pdm infection in vivo has not been evaluated.
GX-sPLA2 has been highly implicated in the inflammatory response including pattern recognition receptor function, and displays the highest activity among all mammalian sPLA2s on phosphatidylcholine-rich liposomes in vitro (Crooks and Stockley, 1998, Henderson et al., 1995). Recently, GX-sPLA2 has been suggested as a signal amplifier in TLR4 stimulation which further suggests a role for GX-sPLA2 in the regulation of the inflammatory response (Schultz-Cherry and Jones, 2010). Considering the potential of GX-sPLA2 in the inflammatory response, sPLA2 enzymes may play a central role in determining the outcome of pulmonary viral infections, which cause uncontrolled inflammatory destruction of the respiratory tract (Henderson et al., 1995).
We have developed a robust lethal mouse model of H1N1pdm infection to study innate host defense mechanisms and antiviral compound activity (Paquette et al., 2012). We have shown that H1N1pdm infection in this mouse model leads to pulmonary inflammation, a histopathological picture similar to what is observed in fatal human cases, and over 90% lethality within 5–8 days (Paquette et al., 2012). In this study, we document a marked increase in GX-sPLA2 expression in lung following infection in GX+/+ mice. To specifically evaluate the pathophysiological role of GX-sPLA2 in our lethal influenza mouse model, we subjected GX+/+ and GX−/− mice (Henderson et al., 1995) to H1N1pdm infection in vivo. Our results showed that, in two distinct mouse strains, targeted deletion of GX significantly increased survival following H1N1pdm infection in comparison with GX+/+ mice. In addition, eicosanoid accumulation in BAL fluid was attenuated and induction of T cell and B cell associated genes was higher in GX−/− than GX+/+ mice after H1N1pdm infection. Taken together, our data suggests a negative role for GX-sPLA2 in the immune response to pulmonary infection with H1N1pdm influenza in vivo. Furthermore, these findings implicate GX-sPLA2 as a potential therapeutic target during severe influenza infection.
Results
GX-sPLA2 is increased in the lungs during H1N1pdm infection
Airway epithelial cells and myeloid cells can both express GX-sPLA2 (Marshall et al., 2000). Previously, we have investigated the host immune responses to pulmonary viral infections, including infection with the influenza viruses H5N1 and H1N1pdm (Baillie and Digard, 2013, Bermejo-Martin et al., 2010, Bhavsar et al., 2010, Cameron et al., 2008, Cameron et al., 2012, Escoffier et al., 2010, Huang et al., 2009, Huang et al., 2012, Kudo and Murakami, 1999). Furthermore, we have also delineated the biology and molecular regulation of many of the enzymes that catalyze eicosanoid biosynthesis in vitro and in vivo (De et al., 2012, Degousee et al., 2006, Degousee et al., 2008, Degousee et al., 2002, Degousee et al., 2003, Leon et al., 2012, Lu et al., 2006, Rowe et al., 2010, Saez de et al., 2011). To begin to investigate the role of GX-sPLA2 during H1N1pdm infection, we evaluated the pulmonary expression of GX-sPLA2 in our H1N1 pandemic influenza mouse model.
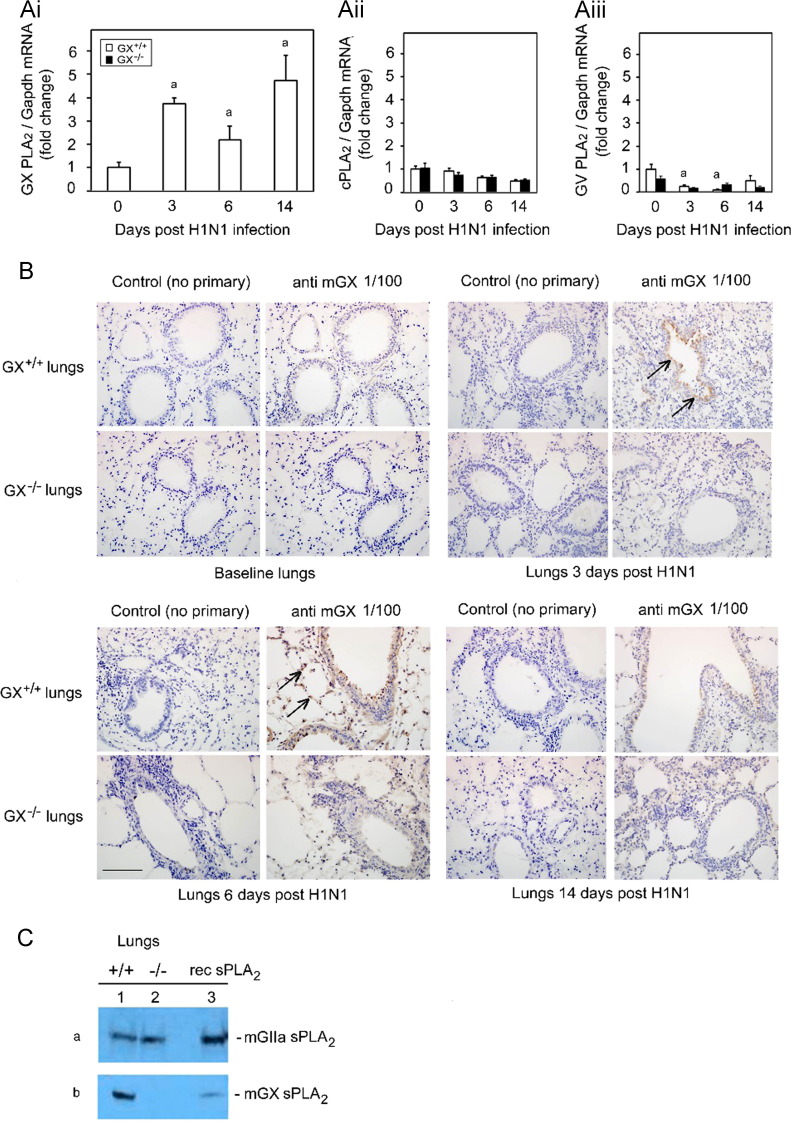
GX+/+ mice were infected intranasally with A/Mexico/4108/2009 (H1N1pdm) and lung tissues were harvested at baseline and on day 3, 6 and 14 post infection (pi). Real-time PCR was performed on the extracted RNA and identified a significant increase in the ratio of GX-sPLA2 to GAPDH mRNA on day 3 and day 14, but not day 6 pi (Fig. 1Ai). GX-sPLA2/GAPDH mRNA increased approximately four fold on day 3 and three fold on day 14 compared to baseline. Furthermore, we also determined the regulation of cytosolic PLA2 (cPLA2) (Fig. 1Aii) and the sPLA2 family member GV-PLA2 (Fig. 1Aiii) in both GX+/+ and GX−/− mice. There was negligible total protein upregulation of cPLA2 and GV-PLA2 was not regulated throughout the infection time course. The absence of the change of total protein of cPLA2 was also confirmed by immunohistochemistry (data not shown). No statistical differences in mRNA transcripts for cPLA2 and GV-PLA2 were noted between the GX+/+ or GX−/− mice. These results demonstrated that H1N1pdm influenza infection stimulated a bimodal increase in pulmonary GX-sPLA2 mRNA expression which was specific for this sPLA2 since neither cPLA2 nor GV-sPLA2 was up-regulated.
Fig. 1.
Infection with H1N1pdm influenza stimulates the expression of GX-sPLA2 in bronchial epithelial cells and inflammatory cells. Mice were infected with H1N1pdm (A/Mexico/4108/2009) and the lungs were assessed for the mRNA and protein expression and localization of PLAs during a 14 day time course. GX-sPLA2 mRNA (Ai), cPLA2 mRNA (Aii) and GV-sPLA2 mRNA (Aiii) expression quantified by Real-Time RT-PCR was normalized to GAPDH, GX+/+ (open bars) and GX−/− (filled bars) mice (C3H/HeN background mice). GX+/+ and GX−/− mouse lungs were perfusion fixed in situ with 4% paraformaldehyde, sectioned and subject to immunohistochemical analysis with the IgG fraction of rabbit anti-mouse GX-sPLA2 antiserum (1/100 dilution) (B). GIIA and GX-sPLA2 protein expression determined by immunoblot analysis of lung tissue homogenates of wild type GX+/+ (lane 1) and knockout GX−/− (lane 2) mice (C). For each blot, the corresponding recombinant sPLA2 enzyme (rec sPLA2) was run alone (lane 3) as a control. Representative results for five separate experiments are shown. All the mice used in these experiments were genotyped littermates and grouped and analyzed by their genotype. a, p<0.05 GX+/+ or GX−/− vs. base; b, p<0.05 GX+/+ vs. GX−/− at any time point, ANOVA followed by paired t-test, two tailed, assuming unequal variance. n≥8 per group; 400×; scale bar: 50 μm for immunohistochemistry.
Since GX-sPLA2 mRNA levels increased in response to H1N1pdm infection, we investigated the spatial and temporal expression of GX-sPLA2 protein in mouse lungs after influenza infection. Lungs from GX+/+ and GX−/− mice infected with H1N1pdm were harvested at baseline, 3, 6 and 14 days pi and subjected to immunohistochemical analysis with anti-mouse GX-sPLA2 antiserum. Visualization by light microscopy revealed GX-sPLA2 protein accumulation in the lungs of infected mice compared to baseline (Fig. 1B). GX-sPLA2 protein was identified in inflammatory cells that had infiltrated in the alveolar space on day 3 and 6 in GX+/+ mice infected with H1N1pdm (shown by arrows). GX-sPLA2 protein was also clearly identified in epithelial cells lining the bronchioles on day 3, 6 and 14 in GX+/+ mice infected with H1N1pdm (Fig. 1B, upper right panels). No staining for GX-sPLA2 protein was observed in GX−/− mice at baseline or at any time point after infection with H1N1pdm (Fig. 1B, lower panel rows). Similarly, no proteins cross reacting with the secondary antibody alone was identified in GX+/+ or GX−/− mice (Fig. 1B, left hand panels). We confirmed the loss of GX-sPLA2 in the GX−/− mice by immunoblot analysis. We indeed observed a specific depletion of GX-sPLA2 but no change in the expression of GIIA-sPLA2 in the GX−/− mice compared to GX+/+ mice (Fig. 1C). Taken together, these results show that intranasal infection with H1N1pdm increases GX-sPLA2 RNA and protein expression in the lung that corresponds to the increase in lung inflammation associated with influenza infection. This suggests a possible role for GX-sPLA2 in the pathogenesis of pulmonary H1N1pdm influenza infection.
Depletion of GX-sPLA2 increases host survival following H1N1pdm infection
Since GX-sPLA2 was upregulated in the lung during H1N1pdm infection, we explored its role in the host response to pulmonary infection with H1N1pdm influenza. We first examined the clinical outcome of GX-sPLA2 deletion by assessing weight loss and survival of G+/+ and GX-sPLA2 gene targeted mice−/− mice on two different genetic backgrounds following infection and assessed weight loss and survival.
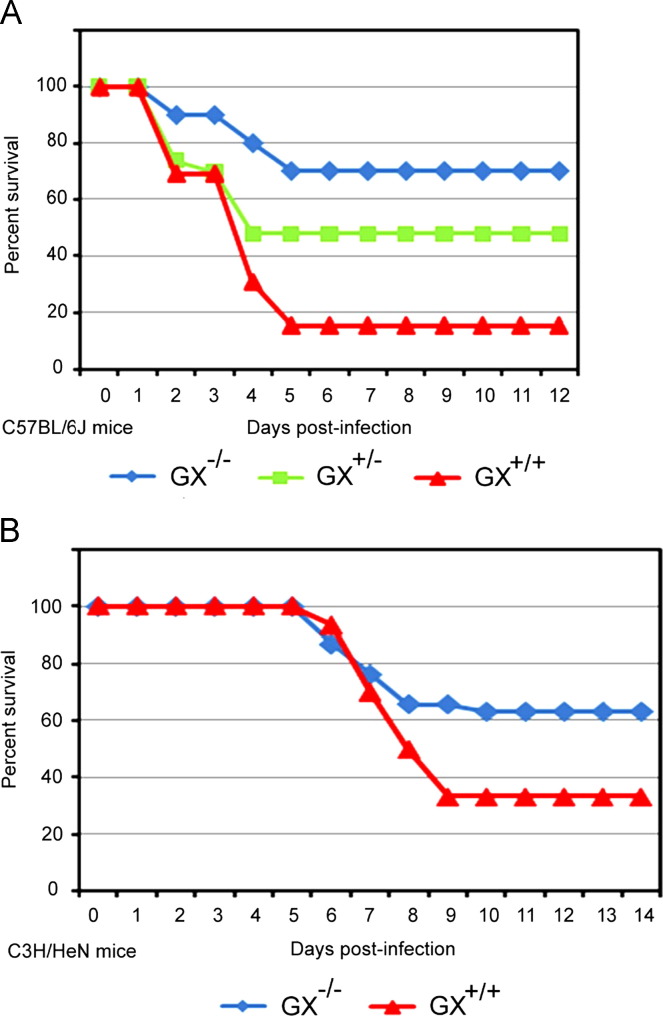
In the first series of infections, GX+/+ (n=25), GX+/– (n=32), and GX−/− (n=24) mice on a C57BL/6J background were infected intranasally with H1N1pdm influenza A/Mexico/4108/2009 ( Fig. 2A). Mice on this background lack the GIIA-sPLA2 gene (Karabina et al., 2006). Animals were euthanized if their body weight decreased to less than 80% of baseline weight, or if the 14-day duration of the study was completed. Survival 14 days after H1N1pdm influenza infection was 70% in GX−/− mice (blue line), 48% in GX+/– mice (green line) and 15% in GX+/+ mice (red line). The difference in survival between GX−/− and GX+/– mice, and between GX−/− and GX+/+ mice after H1N1pdm infection was statistically significant, p≤0.01.
Fig. 2.
Increased survival of GX−/− vs. GX+/− or GX+/+ mice following A/Mexico/4108/2009 infection. GX+/+ (n=25), GX+/− (n=32), and GX−/− (n=24) mice (C57BL/6J background, lacks GIIA-sPLA2) were infected intranasally with A/Mexico/4108/2009 and survival was assessed for a 14 day period (A). GX+/+ (n=71) and GX−/− (n=57) mice (C3H/HeN background, expresses GIIA-sPLA2) were infected intranasally with A/Mexico/4108/2009 and survival was assessed for a 14 day period (B). Animals were sacrificed if their body weight decreased to less than 80% of baseline weight, or if the 14-day duration of the study was completed. Log rank test, p<0.05 GX−/− vs. GX+/+ and GX+/− mice or p<0.05, GX−/− vs. GX+/+ mice. All the mice used in these experiments were genotyped littermates and grouped and analyzed by their genotype.
To independently confirm these findings, we evaluated the survival of GX+/+ (n=71) and GX−/− (n=57) mice on a C3H/HeN background (Fig. 2B) which have a functional GIIA-sPLA2 gene (Karabina et al., 2006). As with the studies with the C57BL/6J mice, animals were infected intranasally with A/Mexico/4108/2009 and euthanized if their body weight decreased to less than 80% of baseline weight, or at the end of the study. Survival of GX−/− mice on the C3H/HeN background was again significantly higher (62%, blue line) following H1N1pdm infection than survival of GX+/+ mice on a C3H/HeN background (36%, red line). Together, these studies showed that targeted deletion of GX-sPLA2 in two different mouse models led to increased survival following H1N1pdm infection in vivo. Furthermore, since the C3H/HeN mice expressed endogenous GIIA-sPLA2, these results demonstrate that the ability to express GIIA-sPLA2 does not compensate for the loss of GX-sPLA2 during host immune responses to pulmonary H1N1pdm influenza infection.
Depletion of GX-sPLA2 during H1N1pdm infection leads to a decrease in downstream phospholipid catalysis (AA) products but no difference in innate cell recruitment
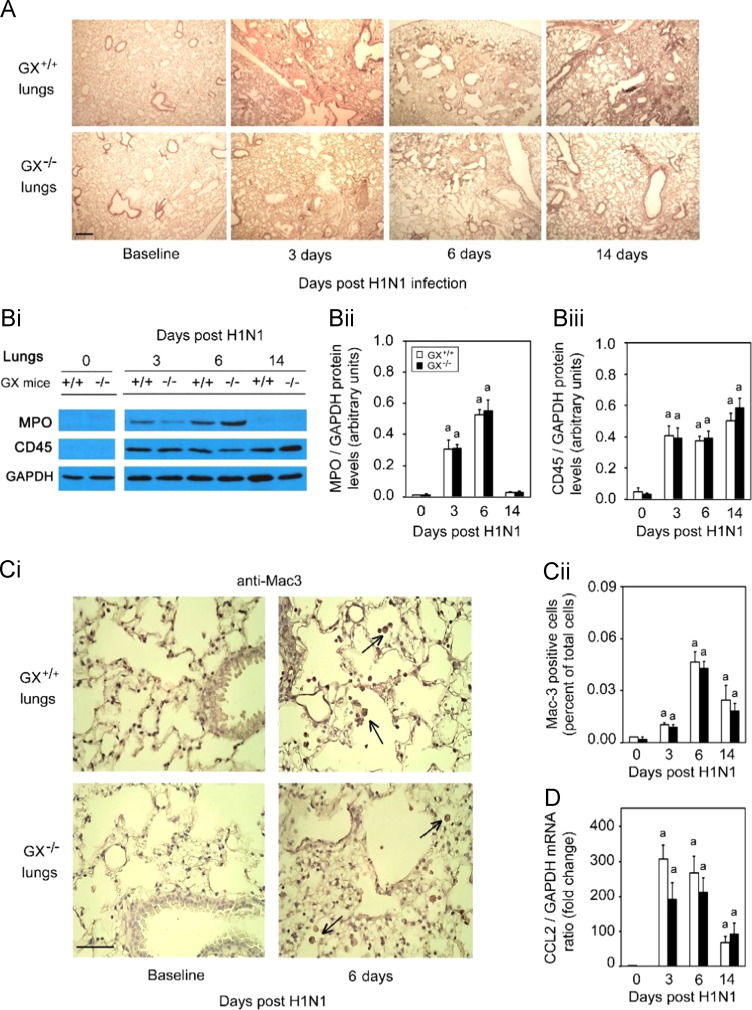
GX+/+ and GX-sPLA2 gene targeted mice−/− on a C3H/HeN background were infected with A/Mexico/4108/2009, and BAL fluid was harvested 3 or 6 days post H1N1pdm infection. To assess the general inflammatory response and lung tissue destruction that typically occurs during H1N1pdm infection (Ohtsuki et al., 2006, Paquette et al., 2012), we investigated the histopathology by H&E staining of lungs isolated from both GX−/− and GX+/+ mice at baseline, day 3, 6 and 14 pi ( Fig. 3A). The pulmonary pathology peaked quickly by day 3 pi in infected GX+/+ and GX−/− animals. Bronchiolitis and alveolitis with mononuclear cell and neutrophil infiltration were observed in several loci of the infected lungs of both groups. Hemorrhage, edema, and necrotizing respiratory epithelia were also observed with similar severity among both groups. Pathology persisted until day 7 pi where mononuclear cell and neutrophil infiltration were still profound and caused patches of consolidation in the lung tissue in both groups. It seemed by day 14 pi that the pulmonary pathology was slightly more minimal in the GX−/− mice with reduced level of leukocyte infiltration. In contrast, multi foci cell infiltration and tissue consolidation was still prominent in the GX+/+ lungs by day 14 pi.
Fig. 3.
Infection with H1N1pdm influenza induces similar pulmonary inflammation and recruitment of inflammatory cells in GX+/+ and GX−/−mice. GX+/+ and GX−/− mice (C3H/HeN background mice) were infected with H1N1pdm (A/Mexico/4108/2009) influenza and the lungs were perfusion fixed in situ with 4% paraformaldehyde on specific time points following infection, sectioned and subject to hematoxylin and eosin staining (A). MPO protein (neutrophil marker), CD45 protein (leukocyte marker) and GAPDH protein (loading control) expression levels were determined by immunoblot analysis from lung tissue homogenates of GX+/+ and GX−/− mice over a 14 day time course of H1N1pdm influenza infection (Bi). Densitometric analysis of MPO (Bii) and CD45 (Biii) protein levels normalized to GAPDH levels in lung tissue of GX+/+ (open bars) and GX−/− (filled bars) mice after H1N1pdm influenza infection, are presented. Immunohistochemical analysis with specific rabbit primary antibody against mouse Mac-3 antigen (marker for macrophages) is shown (Ci). Assessment of Mac-3 positive cells (indicated by →) per high power field before, 3, 6 or 14 days after infection with H1N1pdm influenza is shown (Cii). CCL2 mRNA expression normalized to GAPDH was determined by quantitative real-time PCR in lung tissue of GX+/+ and GX−/− mice after H1N1pdm influenza infection (D). Representative images (×200) from five independent experiments are shown. Scale bar: 100 μm (A) or 50 μm (C). a, p<0.05 GX+/+ or GX−/− vs. base; b, p<0.05 GX+/+ vs. GX−/− at any time point, ANOVA followed by paired t-test, two tailed, assuming unequal variance. n≥8 per group.
To further examine the inflammatory cell types that may be recruited to the lung during H1N1pdm infection we analyzed lung homogenates from day 0, 3, 6 and 14 pi from both GX+/+ and GX−/− mice by immunoblot for neutrophil and leukocyte cell markers, MPO and CD45 respectively. MPO was induced on day 3 and day 6 pi in both the mouse genotypes and returned to baseline on day 14 and CD45 was induced from baseline for all time points measured. Neither MPO nor CD45 showed any variation in the lungs between GX−/− or GX+/+ throughout the infection time course (Fig. 3Bi); densitometry did not reveal any statistical differences (Fig. 3Bii). Furthermore, we also analyzed GX−/− and GX+/+ lungs for the presence and activation of macrophages by immunohistochemistry and Real-Time RT-PCR (Fig. 3Ci, Cii and D). Here we found that the macrophage marker Mac-3 was significantly increased and peaked at day 6 following infection as determined by immunohistochemistry staining which was further confirmed by quantifying the staining and quantification of the signal (Fig. 3Ci and Cii). Furthermore the mRNA for the inflammatory chemokine CCL2 was also significantly increased following infection (days 3 and 6) and decreased by day 14 (Fig. 3D). No difference was determined for Mac-3 or CCL2 expression between GX−/− or GX+/+ mice. Taken together, these results suggest a similar inflammatory and innate response in both the GX+/+ and GX−/− mice.
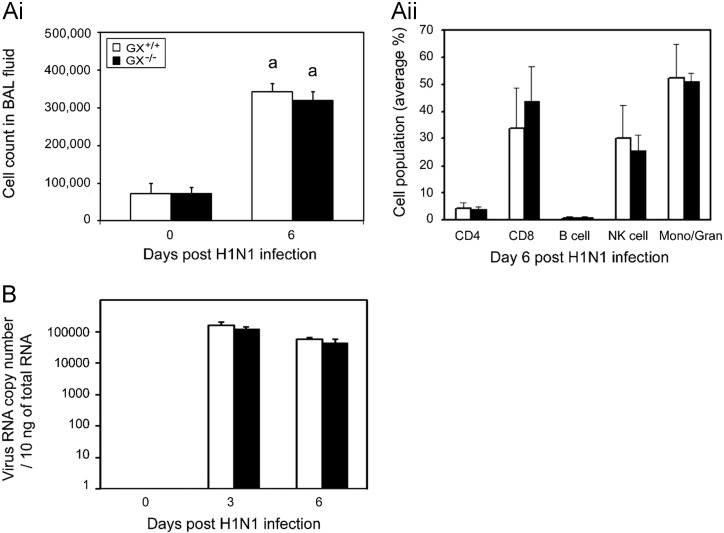
To assess the role of GX-sPLA2 in leukocyte infiltration into the bronchoalveolar space after H1N1pdm infection, we measured total leukocyte cell counts and the levels of different leukocyte cell types in the BAL fluid of H1N1pdm infected in GX+/+ and GX−/− mice ( Fig. 4A). No significant difference in total cell counts (Fig. 4Ai) or the percentage of CD4+, CD8+, B or natural killer cells, or granulocytes were identified in the BAL fluid of GX+/+ and GX−/− mice 6 days after H1N1pdm infection (Fig. 4Aii). In addition, targeted deletion of GX-sPLA2 had no effect on lung viral titers 3 or 6 days pi (Fig. 4B).
Fig. 4.
Lung viral titers and cell counts in BAL fluid show similar cell numbers and cell population distributions following infection in GX+/+ and GX−/− mice. GX−/− and GX+/+ mice infected with A/Mexico/4108/2009 were investigated for lung cell numbers, populations and viral load. BAL fluid was harvested from infected GX+/+ (open bars) and GX−/− (filled bars) mice (C3H/HeN background mice) on day 0 and 6 and the cell numbers (Ai) and cell population distributions (Aii) were assessed by FACS. Viral load was determined on day 0, 3 and 6 pi of GX+/+ (open bars) and GX−/− (filled bars) mice by Real-Time RT-PCR vRNA quantification (B). All the mice used in these experiments were genotyped littermates and grouped and analyzed by their genotype. a, p<0.05 GX+/+ vs. base, ANOVA followed by paired t-test, two tailed, assuming unequal variance. n≥7 per group.
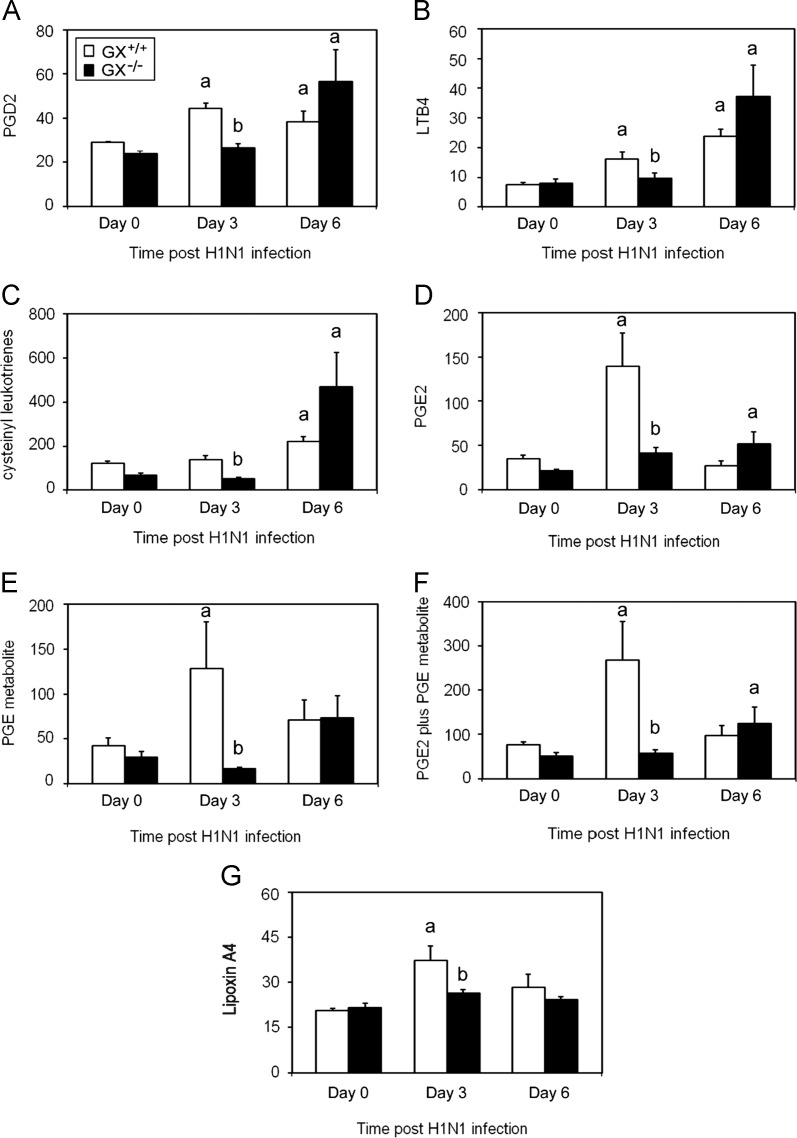
We next determined by ELISA the levels of diferent AA metabolites including PGD2, LTB4, cysteinyl leukotrienes, PGE2, a stable PGE metabolite, and Lipoxin A4 which are known to regulate bronchiolar reactivity and inflammatory cell adhesion, migration and activation (Henderson et al., 1995) were determined by ELISA. Levels of PGD2 ( Fig. 5A), LTB4 (Fig. 5B), cysteinyl leukotrienes (Fig. 5C), PGE2 (Fig. 5D), the stable PGE metabolite (Fig. 5E), PGE2 plus PGE metabolite (Fig. 5F) and Lipoxin A4 (Fig. 5G) were all significantly lower in the BAL fluid from GX−/− mice (solid bars) on day 3 pi compared to GX+/+ mice. Conversely, on day 6 pi, the levels of these metabolites were similar in GX+/+ and GX−/− mice (Fig. 5A–F). In summary, these results show that deletion of GX-sPLA2 in mice led to a transient but significant decrease in the levels of a panel of AA metabolites when mice were infected with a lethal H1N1pdm influenza virus that was not associated with alterations in inflammatory cell infiltration or viral clearance.
Fig. 5.
Decreased eicosanoid levels in the BAL fluid 3 but not 6 days after infection with A/Mexico/4108/2009 in GX−/− vs. GX+/+ mice. The eicosanoid levels in the BAL fluid of GX−/− and GX+/+ mice (C3H/HeN background) were investigated at baseline and following infection with A/Mexico/4108/2009. GX+/+ (open bars) and GX−/− (filled bars) mice (C3H/HeN background mice) BAL fluid was harvested by instilling ice cold NaCl (1 ml) five times and pooled. Levels of PGD2 MOX (A), LTB4 (B), cysteinyl leukotrienes (C), PGE2 (D), stable PGE metabolite (E), PGE2 plus PGE metabolite (F) and Lipoxin A4 (G) were assessed by ELISA. These results are the mean of 5 independent studies. All the mice used in these experiments were genotyped littermates and grouped and analyzed by their genotype. GX+/+ (open bars) and GX−/− (filled bars). Results are expressed in pg/mL. a, p<0.05 GX+/+ or GX−/− vs. base; b, p<0.05 GX+/+ vs. GX−/− at any time point, ANOVA followed by paired t-test, two tailed, assuming unequal variance. n≥7 per group.
Increased expression of immunoglobulin chain, lymphocyte differentiation, antigen processing genes and presence of CD3+ T cells in the lungs of mice lacking GX-sPLA2 after H1N1pdm infection
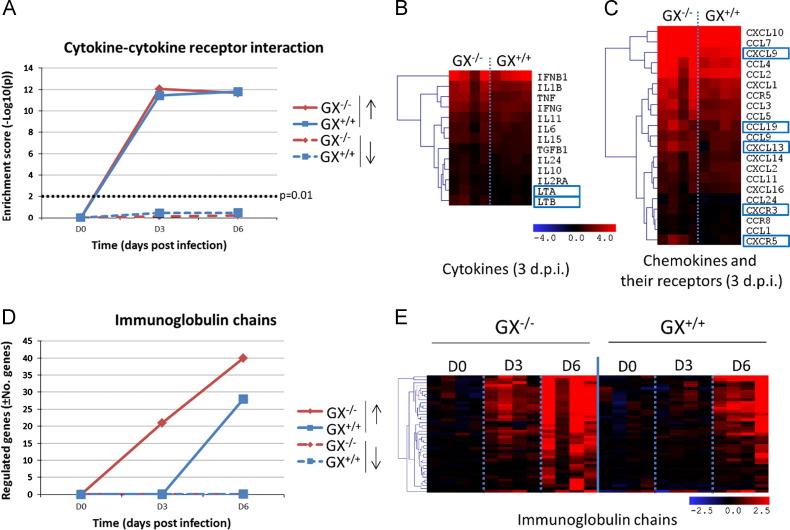
To increase our understanding of the molecular events leading to increased survival following H1N1pdm infection in GX−/− mice, we conducted microarray analysis of RNA extracted from the lungs of GX+/+ and GX−/− influenza infected animals. As previously reported by our group (Kudo and Murakami, 1999, Paquette et al., 2014, Rowe et al., 2010), influenza infection caused a progressive increase in the total number of upregulated genes in the lung tissue of GX+/+ mice (1246 genes at 3 days pi and 2469 genes at 6 days pi). Genes that belonged to different functional groups, such as immune response, inflammatory response and prostaglandin signaling pathways ( Fig. 6, Fig. 7) showed a progressive increase that was parallel to the global evolution of gene expression. Conversely, the expression of cytokine-related genes reached maximal levels 3 days pi and were maintained thereafter (Fig. 6A).
Fig. 6.
Effect of GX-sPLA2 deficiency in the mRNA expression levels of cytokines, chemokines and their receptors and immunoglobulin chains in the lung tissue of mice during influenza infection. GX+/+ and GX−/− mice (C3H/HeN background mice) were infected with influenza A/Mexico/4108/2009 and the gene expression profiles were analyzed in the lung tissue at day 0, 3, and 6 days after infection by microarray analysis (n=4 per group). Evolution of gene enrichment (Fisher׳s exact test) of the KEGG category “cytokine-cytokine receptor interaction” (A). Differences in the expression levels of cytokines (B) and chemokines (C) and their receptors at 3 days pi. The heatmaps show the genes that are significantly upregulated with respect to the control group and the blue boxes indicate that the expression levels of those genes are significantly higher in the GX−/− than in the wild type mice at the same time-point. Evolution in the expression levels of immunoglobulin genes: total number of regulated genes (D) and overview of different experimental groups and time-points (E). All the mice used in these experiments were genotyped littermates and grouped and analyzed by their genotype.
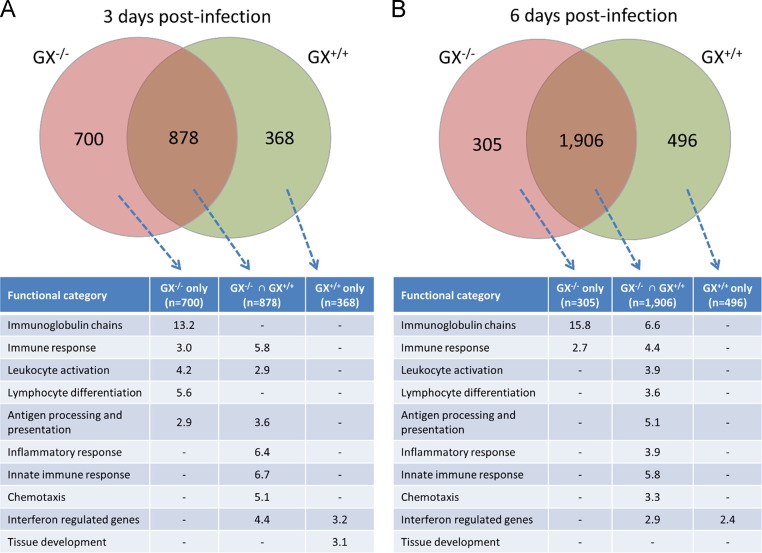
Fig. 7.
Intersect analysis of the genes up-regulated in the lung tissue of GX−/− and GX+/+ mice during influenza infection and functional classification of the resulting gene subsets. Venn diagrams are representative of the total number of genes that are significantly up-regulated with respected to the uninfected mice. David Annotation tool was used to classify the genes of each subset, and the fold enrichment is shown for each category. All the mice used in these experiments were genotyped littermates and grouped and analyzed by their genotype. * The “Immunoglobulin chains” category was manually curated and contains 84 genes. ⁎⁎ The “interferon responses category”.
At first sight, lack of GX-sPLA2 did not modify the global evolution of gene expression in the lungs. Similarly to GX+/+ mice, GX−/− mice showed a progressive increase in the number of upregulated genes (1578 at 3 days pi and 2469 at 6 days pi). Further analysis demonstrated that on day 3 pi, GX−/− mice showed significantly higher levels of the cytokines LTA and LTB, the chemokines CCL19, CXCL9 and CXCL13 and the chemokine receptors CXCR3 and CXCR5 (Fig. 6B and C). In contrast, the expression pattern of cytokines and chemokines showed no differences between GX+/+ and GX−/− mice 6 days pi (data not shown). Interestingly, expression of 21 immunoglobulin chains, including heavy and light chains, was identified in GX−/− mice 3 days pi, while no expression of immunoglobulin chain genes was identified in GX+/+ mice at this time point. In addition, the number of immunoglobulin chain related genes was higher in GX−/− than GX+/+ mice 6 days after H1N1pdm infection (Fig. 6B). No differences were observed in the patterns of interferon regulated genes between GX+/+ and GX−/− mice after H1N1pdm infection (data not shown).
To determine which functional pathways are differentially enriched between GX+/+ and GX−/− mice after H1N1pdm infection, we performed intersect analysis of the respective sets of upregulated genes (Fig. 7). At 3 days pi, expression of interferon regulated, inflammatory response and innate immune response genes were common to both GX+/+ and GX−/− mice. A number of genes related with eicosanoid synthesis and their receptors were found to be regulated during influenza infection; however, GX-sPLA2 deficiency did not cause any major alterations in their expression profiles (Fig. S1).
The set of genes specifically enriched in the GX−/− mice at 3 day pi were those related to adaptive immune responses, such as immunoglobulin chains, lymphocyte differentiation and antigen processing and presentation. On the other hand, the set of genes more enriched in the GX+/+ mice were genes involved in the tissue development category at 3 days pi (Fig. 7A). At 6 days pi (Fig. 7B), the enrichment profiles of upregulated genes in GX+/+ and GX−/− mice were nearly identical. While immunoglobulin chain gene expression was identified in both GX+/+ and GX−/− mice, expression of immunoglobulin chain genes remained elevated only in the GX−/− set of genes 6 days pi, while the GX+/+ set of genes continued to show enrichment in the tissue development category at day 6 pi.
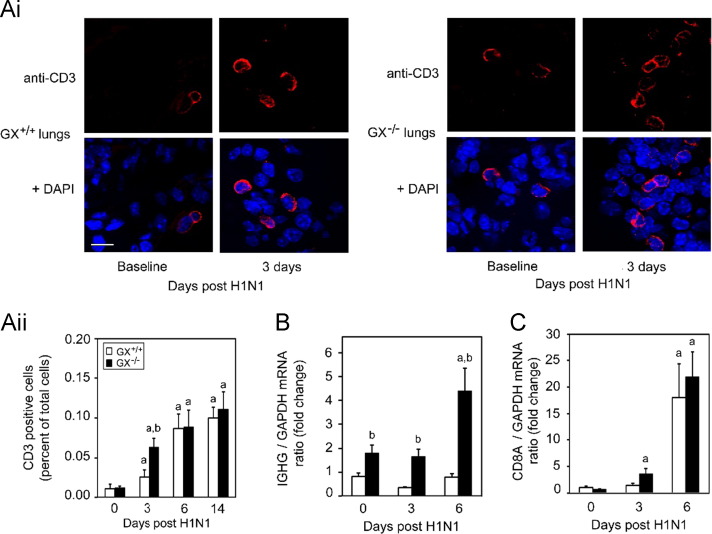
To further evaluate the adaptive immune system of the GX−/− mice infected with H1N1pdm we investigated the T and B cell responses within the lung during infection. Here we stained lung sections with anti-CD3 to assess infiltration of T cells using immunocytochemistry ( Fig. 8A and B). We found a significant increase of CD3 positive T cells in the lung on day 3 pi in the GX−/− mice compared to GX+/+ mice (Fig. 8A, upper right panels) by approximately 2 fold (Fig. 8B). Interestingly, CD3 staining of the GX+/+ animals had increased to similar levels seen in the GX−/− mice by day 6 and both genotypes had sustained levels of CD3 on day 14. Moreover, we also investigated CD8A and IgG (IGHG) mRNA levels in the lungs of the GX−/− mice throughout the time course and there was a slight trend for increase CD8A levels. Taken together, the results from the CD3 and IgG analysis supported the microarray studies where the adaptive immune system of the GX−/− had a faster and more robust initiation.
Fig. 8.
GX-sPLA2 deficiency increases T cell recruitment and immunoglobulin heavy chain mRNA expression in the lung tissue of mice during influenza infection. Day 0 and 3 pi with H1N1pdm (A/Mexico/4108/2009) influenza the lungs from GX+/+ and GX−/− mice (C3H/HeN background mice) were perfusion fixed in situ with 4% paraformaldehyde, sectioned and subject to immunofluorescence analysis with specific rabbit primary antibody against mouse CD3 antigen (marker for T-cell) (Ai). Representative images (at ×400 with 3.4 zoom factor) from five independent experiments are shown. Assessment of CD3 positive cells per high power field for day 0, 3, 6 and 14 days following infection with H1N1pdm influenza is shown (Aii). IgG (B) and CD8A (C) mRNA expression normalized to GAPDH were determined by quantitative real-time PCR in lung tissue of GX+/+ and GX−/− mice after H1N1pdm influenza infection. Scale bar: 10 μm. a, p<0.05 GX+/+ or GX−/− vs. base; b, p<0.05 GX+/+ vs. GX−/− at any time point, ANOVA followed by paired t-test, two tailed, assuming unequal variance. n≥8 per group. All the mice used in these experiments were genotyped littermates and grouped and analyzed by their genotype.
Discussion
GX-sPLA2 has been highly implicated in various inflammatory diseases of the respiratory tract, including Th2 cytokine-driven asthma (de Jong et al., 2006, Henderson et al., 2007) and lung injury (Napolitani et al., 2009), but its role during influenza infection has not been previously investigated. Here we evaluated the pathophysiological role of GX-sPLA2 during severe influenza A H1N1pdm infection in the mouse. We found that GX-sPLA2 expression was increased following infection, and that targeted deletion of GX-sPLA2 led to increased survival in mice. Lack of GX-sPLA2 resulted in decreased levels of PGD2, LTB4, cysteinyl leukotrienes, PGE2 and Lipoxin A4 and increased adaptive immune responses at 3 but not 6 days following H1N1pdm infection. This demonstrates that GX-sPLA2 plays an important role in the production of several biologically active inflammatory lipid mediators during the early phase of the inflammatory response that follows H1N1pdm influenza infection. Human patients with a severe respiratory disease caused by influenza infection have a dysregulated inflammatory response that leads to lung pathogenesis associated with hypercytokinemia in most cases (Bermejo-Martin et al., 2010, Curfs et al., 2008). Taken together with the previous findings showing a role of GX-sPLA2 in inflammatory lung diseases, our work supports the further investigation of the therapeutic potential of attenuating GX-sPLA2 during severe influenza infection as well as the interplay between eicosanoids and adaptive immunity.
sPLA2 has previously been implicated in pulmonary disease onset and progression putting it forth as a potential biomarker for severe respiratory diseases (Henderson et al., 1995, Henderson et al., 2007). We show that GX-sPLA2 protein and mRNA expression increased in the lungs of GX+/+ mice following H1N1pdm infection, suggesting that GX-sPLA2 may be used as a possible biomarker of severe influenza infection. This is the first report of increased GX-sPLA2 expression following influenza virus infection. Both epithelial cells and leukocytes were found to be sources of GX-sPLA2 during infection, and GX-sPLA2 expression was detected in epithelial cells 3 days prior to the infiltration of leukocytes. In will be interesting to determine in future experiments whether the specific deletion of GX-sPLA2 expression in epithelial cells vs. infiltrating leukocytes or both is responsible for the increased survival. Here we observed a bimodal expression pattern of GX-sPLA2 during the 14 day time course of infection. It is possible that this occurred due to the protein stability as it is used to regulate bioactive lipid mediator synthesis. If the protein does not remain stable throughout the course of infection and recovery, it may be important to have a second increase in GX-sPLA2 in the later stages of infection to compensate for the loss of protein. It is in fact possible that the protein exerts distinct roles in the clearance of the virus and tissue remodeling in addition to the regulation of immune cells. Such a scenario would require inductions at specific time points during infection. It will be important to further explore the local expression in the virus niche and the stability of protein GX-sPLA2 during influenza infection to better understand how GX-sPLA2 stability may influence influenza severity in the initiation of the innate immune response, adaptive maintenance, and recovery. Furthermore, it would also be of value to investigate the source of GX-sPLA2 by expression analysis of each cell type and also by investigating the role of hematopoietic GX-sPLA2 compared to epithelial GX-sPLA2. The latter could be studied by employing bone marrow transplantation experiments from GX−/− mice into GX+/+ and the reverse. While the association between GX-sPLA2 and influenza related complications has not previously been investigated, LTB4, a downstream product of GX-sPLA2 has been suggested to be a biomarker for pulmonary disease and respiratory complications following trauma (Influenza Activity – United States and Worldwide,, Henderson et al., 2011, Shridas et al., 2011).
Multiple studies have implicated GX-sPLA2 in the pathophysiology of pulmonary diseases onset and progression, suggesting GX-sPLA2 might be a suitable therapeutic target in lung (Henderson et al., 1995, Morita et al., 2013). Deletion of GX-sPLA2 in a Th2 cytokine-driven mouse asthma model significantly impairs development of asthma (Henderson et al., 1995) and accordingly, administration of a human GX-sPLA2 selective inhibitor in a human GX-sPLA2 knock-in mouse model led to a significant reduction in airway inflammation, mucus hypersecretion and airway hyperresponsiveness (Henderson et al., 2007). Furthermore, although not specific for human GX-sPLA2, the indole-based sPLA2 inhibitor varespladib has been shown to significantly inhibit sPLA2 activity in the BAL fluid of infants with post-neonatal ARDS (de Jong et al., 2006) during induced asthma, suggesting the involvement of sPLA2 among other sPLA2s. Our results showing increased survival of the GX−/− mice after infection with H1N1pdm further support the notion that GX-sPLA2 is a therapeutic target in pulmonary diseases due to viral infection and that infection with H1N1 might be better controlled by inhibiting this sPLA2.
One of the main functions of GX-sPLA2 is likely the generation of bioactive lipid mediators which play important roles in lung inflammatory diseases (Gao et al., 2013, Gaudreault and Gosselin, 2007, Van Elssen et al., 2011). Although we did not see any major differences in the mRNA analysis of the eicosanoid pathways between the GX−/− and GX+/+ mice, measuring the mRNA levels of these genes may have limited value to determine the level of activation of their signaling pathways. Conversely, we observed decreased levels of PGD2, LTB4, cysteinyl leukotrienes, PGE2 and Lipoxin A4 in BAL fluid 3 day pi in the GX−/− mice, indicating that GX-sPLA2 acts upstream of these bioactive lipid mediators during influenza infection and thereby suggests a possible role of these bioactive mediators in pulmonary pathogenesis after influenza infection. In agreement with our findings, PGD2 has been implicated during influenza A infection as PGD2 expression in the lungs of older animals inhibits regulatory dendritic cells activity and T cell responses (Zhang et al., 2000). Other eicosanoids have been implicated in different lung diseases, and the dysregulation of leukotrienes and lipoxins have been reported as contributing factors to the pathogenesis and severity of other respiratory diseases (Bermejo-Martin et al., 2009). LTB4 has been suggested to play a destructive inflammatory role in the lung by priming neutrophils for adhesion, chemotaxis and stimulation of granule release (Cameron et al., 2007). As well PGD2, PGD receptor, lipocalin-type PGD synthase and LTB4 have been implicated in asthma pathogenesis (Arima and Fukuda, 2011, Masuda et al., 2005, Rusinova et al., 2012). Although asthma and pulmonary disease due to influenza infection differ in derivation, both are characterized by hyper-inflammation of the respiratory tract. Taken together, our data supports a role of GX-sPLA2 signaling and bioactive mediator production in the regulation of the pulmonary response to H1N1pdm infection. In the future it would be important to specifically determine whether PGD2, LTB4, cysteinly leukotrienes, PGE2, Lipoxin A4 or another AA metabolite specifically modulates the response to H1N1pdm infection.
The inflammatory response may be simultaneously beneficial and destructive during lung infection (Baillie and Digard, 2013, Bermejo-Martin et al., 2010). Although destructive killing of foreign pathogens is imperative for eradication and microbe clearing, the over production of inflammatory mediators leading to an overt inflammatory response may accentuate disease pathology, as is the case during severe influenza H5N1 and H1N1 infection (Bermejo-Martin et al., 2010, Curfs et al., 2008, Huang et al., 2009). This illustrates the dual role of proinflammatory mediators, which has also been suggested for some GX-sPLA2 downstream lipid mediators. For instance, LTB4 has been shown to increase the activity of nasal neutrophil killing of human coronavirus, RSV, and influenza B virus (Van Elssen et al., 2011) and to induce the release of antimicrobial peptides in vivo in the lungs of mice infected with viruses (Gao et al., 2013, Gaudreault and Gosselin, 2007). The lipid product protectin D1 has been implicated in influenza therapeutics (Arima and Fukuda, 2011, Mitsuishi et al., 2006). Although these previous reports seem to suggest a conflicting role for GX-sPLA2 in consideration of our data, it may be possible that LTB4 and GX-sPLA2 promote antiviral activity and are significant during a viral response but only at moderate levels. Alternatively, it is possible that GX-sPLA2 prevents H1N1 infection but also triggers excessive inflammation that is associated with lipid surfactant destruction. More work is needed to understand how the function of GX-sPLA2 mediates both beneficial and deleterious roles during influenza infection.
Our survival data from GX gene targeted mice indicated that the loss of GX-sPLA2 was beneficial to the host during influenza infection. The microarray mRNA data from lungs of GX+/+ infected mice were in agreement with our previously published data on pandemic H1N1 2009 virus infection, in mice including the progressive increase of immune and inflammatory responses and of the prostaglandin signaling pathway (Paquette et al., 2014). Together, the results of microarray analysis, gene expression by Real-Time RT PCR and immunocytochemistry of the lungs suggested that GX−/− mice exhibited a more robust adaptive immune response than GX+/+ mice. Indeed, we observed significant differences in lymphocyte gene profiles at day 3 pi, associated with differences in the levels of lymphotoxin alpha and beta, B cell chemokines, T cell chemokine receptors and B cell immunoglobulin chains as measured which by immunofluorescence and RT-PCR. Expression of B cell immunoglobulin chain genes were substantially increased on day 3 pi in the GX−/− mice but not in the GX+/+. B cell immunoglobulin gene expression was significantly greater on day 6 pi. and the expression of the T cell, B cell and dendritic cell chemokines and chemokine receptors, i.e., CCL19, CXCR3, etc., were significantly higher in the GX−/− samples than GX+/+. These results suggest that the downstream products of GX-sPLA2, such as PGD2, PGE2, LTB4 may inhibit the early adaptive immune responses of T and B cells during viral infection and this fits with the fact that aspirin, which attenuates eicosanoid production, can be an effective therapy for patients with influenza infection (Matsuoka et al., 2000). It would be of value in future studies to further investigate the effect of GX-sPLA2 on the proliferation, activation and differentiation of T and B lymphocytes. Consistent with this notion, the chemokines and chemokine receptors found to be upregulated in the H1N1pdm infected GX−/− mice are known to play significant roles in T and B cell migration and localization to the lymph nodes (Goracci et al., 2010, Muthuswamy et al., 2010). CXCL13/CXCR5 signaling has been shown to activate B cells (Sadik and Luster, 2012), which may explain the increased immunoglobulin chain gene expression observed in GX−/− mice. Our data supports previous findings implicating PGD2 in the inhibition of cell migration to lymph nodes (Zhang et al., 2000), and PGE2 in the inhibition of adaptive immune cellular events such as chemokine production by DCs and the attraction of naïve T cells (Murakami et al., 2011, Truchetet et al., 2012).
In conclusion, our findings provide new insights into the molecular pathophysiology of lethal influenza infection, highlighting a new role for GX-sPLA2 during H1N1pdm infection. Overall, the sPLA2 appears as a negative effector but it may act at several steps during infection. We found that GX-sPLA2 and its downstream products may have a role in the inhibition of adaptive immunity during viral infection in mice thereby contributing to pathogenesis. Within this mechanism, it is in fact possible that T and B cell maturation and activation are initiated in mice lacking GX-sPLA2 prior to virus infection, and that a more robust and earlier adaptive immune response increased the survival of GX−/− mice after H1N1pdm infection. Since GX-sPLA2 may contribute to inflammatory response dysregulation during influenza infection and contribute to the morbidity and mortality associated with hospitalized influenza patients, this work may shed important insight into the molecular mechanisms of severe influenza infection. Our findings further support the notion that GX-sPLA2 is an interesting therapeutic target in lung inflammatory diseases. Whether inhibition or attenuation of GX-sPLA2 activity during severe influenza infection has a therapeutic effect remains to be demonstrated.
Materials and methods
Generation of GX-sPLA2 KO mice
To dissect the role of GX-sPLA2 in the molecular regulation of pulmonary infection with H1N1pdm influenza, mice that lack GX-sPLA2 (GX−/− mice) on the C57BL/6J background previously described were used (Matsuoka et al., 2000). This mixed background strain has a naturally occurring mutation in the gene encoding GIIA-sPLA2 (Karabina et al., 2006), which has been implicated in bacterial phospholipid hydrolysis (Fang et al., 2011). Furthermore, we generated GX−/− mice on the C3H/HeN background ( Fig. 1C) by backcrossing the C57BL/6J GX−/− mice for 10 generations which had functional GIIA-sPLA2.
Animals maintenance
Mice were maintained on standard animal feed and water ad libitum in the conventional environmental conditions and controlled temperature and humidity with a 12 h light and dark cycle. For infection studies, animals were housed in HEPA-filtered cage racks adherent to ABSL2+ conditions (Toronto General Hospital, Animal Resource Centre, Toronto, Canada). All animal procedures were performed in a certified class II biosafety cabinet (Baker Company, Sanford, NC, USA). Housing and experimental procedures were approved by the Animal Care Committee of the University Health Network, and were in accordance with the Guide for the Care and Use of Laboratory Animals Research Statutes, Ontario (1980).
Viral infection
All infection experiments were conducted with H1N1pdm strain, A/Mexico/4108/2009 (H1N1pdm), provided by the Centers for Disease Control and Prevention (Atlanta, GA, USA). Virus was propagated and titrated in embryonated eggs and titrated prior to animal challenge. Viral stocks were stored in liquid nitrogen and thawed prior to use. Mice were weighed and randomly assigned for sample collection, and were infected through intranasal instillation with 50 µL phosphate-buffered saline (mock infection) or 50 µL A/Mexico/4108/2009 (H1N1pdm) at 1×105 or 1×104 50% egg infectious dose (EID)50. Virus dosage were 1×104 EID50 and ×105 EID50 for host response profiling in C57BL/6J mice and 1×104 EID50 for comparing disease severity between GX+/+ and GX−/− mice. Throughout infection experiments, animal survival, clinical signs, and weights were recorded daily. In accordance with Animal Care Committee recommendation, mice were euthanized when recorded body weight fell below 80% of original body weight.
Viral load measurement
At day 0, 3 and 6 pi, 3 GX+/+ and 3 GX−/− mice were euthanized and lung homogenates collected for viral load determination by either Madin–Darby Canin Kidney (MDCK) cell growth determination or Real-time RT-PCR (RNA Analysis methods and Table S1). For MDCK determination lungs were homogenized (10% w/v) in High Glucose (4.5 g/L) Dulbecco׳s Modified Eagle Medium (DMEM), supplemented with 1% bovine serum albumin, 50 µg/mL Gentamycin, 100 U/mL Penicillin, 100 µg/mL Streptomycin, and 1 µg/mL TPCK-Trypsin (vDMEM). Homogenates were then serially diluted (0.5 log10) in quadruplicate over Madin–Darby Canine Kidney cells, cultured at 2.0×104 cells/well in 96-well plates. Cells were incubated for 2 h at 37 °C and 5% CO2. Homogenates were then removed and replaced with fresh vDMEM. Cells infected were incubated for 6 days at 37 °C and 5% CO2, after which cell culture supernatants were tested for the presence of virus by hemagglutination assay using 0.5% (v/v) turkey red blood cells (LAMPIRE Biological Laboratories, Pipersville, PA, USA). Viral loads were determined as the reciprocal of the dilution at which 50% of wells were positive for viral infection. Viral loads were reported as TCID50 per gram of lung tissue. Limit of detection of 101 TCID50/g.
Host gene expression and viral load measurement by Real Time RT-PCR
Lung tissues from both GX−/− and GX+/+ mice were collected at 3 and 6 days post infection (pi) and from uninfected controls (four mice per group). RNA was purified from lung tissue using TriPure (Roche, Indianapolis, IN, USA). Purified RNA was then reverse transcribed using ImProm-II Reverse Transcription System (Promega, Madison, WI, USA). Real Time RT-PCR was performed using the ABI-Prism 7900HT Sequence Detection Systems (Applied Biosystems, Foster City, CA, USA). Data was collected with Applied Biosystems Sequence Detection Systems Version 2.3 software. Each reaction well contained 4 μL of 0.625 ng/μL cDNA, 0.5 μL each of forward and reverse primers (final concentration of 200 nM), and 5 μL SYBR Green Master Mix, for a total reaction volume of 10 μL and run in quadruplicate. Host gene expression was normalized to the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) housekeeping gene, and quantified by relative standard curve method. Viral load was quantified by the absolute standard curve method, normalized to GAPDH housekeeping. Primer sequences are listed in Table S1.
Histology, immunohistochemistry, immunofluorescence and immunoblotting
GX+/+ and GX−/− mice were euthanized at baseline, day 3, day 6 and day 14 pi (n>5 mice) and the mouse whole body was vascular perfused by cardiac puncture in situ with a fixative solution of 10% buffered formalin by a continuous release pump under pressure and volume-controlled conditions. Fixed lung tissues were paraffin wax embedded for histology and immunohistochemistry. For H&E, tissue slides were then stained with hematoxylin–eosin for histopathology assessment. Rabbit anti-murine GX-sPLA2 (Degousee et al., 2008) was used to assess the tissue distribution of the GX-sPLA2 protein. Sections were counter-stained with hematoxylin and eosin and observed under light microscope (Accu-scope®, Commack, NY, USA). Images were captured using a digital camera and SE Premium software (MicrometricsTM, Londonderry, NH, USA) (Degousee et al., 2006). For Mac-3 tissue expression, rat anti-mouse Mac-3 was used (BD Biosciences, Mississauga, ON).
For CD3 immunofluorescence, following heat-induced antigen retrieval, lung tissue sections were blocked with donkey serum and stained with primary antibodies rabbit anti-CD3 (Dako, Burlington, ON). Donkey anti-rabbit Cy3 was used as secondary antibodies (Millipore, Billerica, MA) and DAPI (Sigma) for nuclear counterstain. Images were recorded with an Olympus Fluo View 1000 confocal laser scanning microscope (Olympus, Tokoyo, Japan).
Immunoblots for GX-sPLA2 were carried out as described by our group (Degousee et al., 2008). For Immunoblotting detection antibodies against MPO (Upstate, Lake Placid), CD45 (BD Biosciences, Mississauga, ON), and GAPDH (Santa Cruz Biotechnology, Dallas, TX).
Eicosanoid analysis
BAL fluid collection
Lungs were lavaged at 0 and 6 days post H1N1 infection with 5 ml of normal saline. The BAL fluid was centrifuged at 250g for 10 min and the supernatant was used for estimation of PGD2, PGE2, LTB4, cysteinyl leukotriens and Lipoxin A4 content.
Analysis of PGD2 in BAL fluid
0.5 ml of BAL fluid was mixed with 0.5 ml of ice-cold acetone, incubated on ice for 5 min and centrifuged for 10 min at 3000g at 4 °C. After supernatant aspiration, the pellet was extracted with 1 ml of ice-cold acetone and centrifuged again. The acetone extracts were combined and the acetone evaporated under nitrogen. All the samples were then methoximated (PGD2-MOX EIA kit (Cayman Chemical), and purified on Oasis HLB columns (Waters Corporation) equilibrated with methanol/0.2% formic acid. Methanol eluants were evaporated in a Savant Speed Vac concentrator and samples dissolved in Cayman EIA buffer before EIA analysis, according to the manufacturer׳s instructions.
Analysis of PGE2, LTB4 and cysteinyl leukotriens in BAL fluid
1.2 ml of BAL fluid was mixed with 2.4 ml of methanol containing 0.2% formic acid, incubated on ice for 5 min and centrifuged at 3000g for 10 min at 4 °C. After adjustment of methanol to 15%, supernatants were loaded on Oasis HLB column equilibrated with methanol/0.2% formic acid (Waters Corporation). Columns were processed in a vacuum manifold (Waters Corporation). After wash with water/0.03% formic acid, the samples were eluted with methanol/0.2% formic acid, methanol eluants were evaporated in a Savant Speed Vac concentrator and samples dissolved in Cayman EIA buffer before EIA analysis for PGE2, LTB4 and cysteinyl leukotrienes (EIA kits, Cayman Chemical) according to the manufacturer׳s instructions.
Analysis of Lipoxin A4 in BAL fluid
0.6 ml of BAL fluid was extracted with 1.2 ml of ice-cold methanol, incubated on ice for 5 min and centrifuged at 3000g for 10 min at 4 °C. The supernatants were diluted with water to achieve 11% methanol concentration and adjusted to pH 3.5 with 1N HCl. Samples were purified on C18 Sep-Pak columns (Waters Corporation) preconditioned with methanol. After column wash with water followed by hexane, samples were eluted with methyl formate. The eluants were evaporated under nitrogen and the samples reconstituted in EIA buffer and assayed for Lipoxin A4 content (Neogen Corporation) according to the manufacturer׳s protocol.
Microarray analysis
Lung tissues from both GX−/− and GX+/+ mice were collected at 3 and 6 days pi and from uninfected controls (four mice per group) as with the Real Time RT-PCR. RNA was purified from lung tissue using TriPure (Roche, Indianapolis, IN, USA) and amplified with Illumina TotalPrep RNA Amplification Kit (Ambion, Austin, TX, USA). 1.5 µg of cRNA was labeled and hybridized to MouseWG-6 v2.0 Expression BeadChip (Illumina, San Diego, CA, USA) and scanned on Illumina BeadStation 500GX. Raw data was processed with Illumina GenomeStudio V2010.3 software. The data sets were subjected to quantile normalization, variance stabilization and log 2 transformation. Genes were considered significantly regulated if the expression levels with respect to the uninfected controls were ≥1.5-fold different and the Student t-test׳s p value was <0.05. DAVID Bioinformatics Resource v6.7 (http://david.abcc.ncifcrf.gov/home.jsp) (Hicks et al., 2007) was used to perform functional classification of differentially expressed genes. Additionally, interferon regulated genes were selected by using the Interferome (v2) database (http://interferome.its.monash.edu.au/interferome/home.jspx) (Rubin et al., 2005). Immunoglobulin chains and prostaglandin-related gene categories were defined by searching for relevant keywords in the annotated microarray datasets. MultiExperiment Viewer v4.7.2 (http://www.tm4.org/mev/) was used to perform complete Hierarchical clustering and generate heatmap representations of selected genes.
Statistical analysis
Data are presented as mean±SEM. Analyses of data recorded at one time point were performed by 2-tailed, unpaired, Student t-tests. Analyses of data recorded at several time points for two groups (GX+/+ and GX−/− mice) were performed by 2-way ANOVA (to evaluate the effect of group, time and group–time interactions); if significant, a Bonferroni correction for multiple comparisons was applied for post-hoc analysis between different time points or between different groups at the same time point. Survival after H1N1pdm influenza infection was assessed by a log-rank test. A value of p<0.05 was accepted as statistically significant. The authors had full access to and take full responsibility for the integrity of the data. All authors have read and agree to the manuscript as written.
Acknowledgments
A/Mexico/4108/2009 was obtained through the Influenza Reagent Resource, Influenza Division, WHO Collaborating Center for Surveillance, Epidemiology and Control of Influenza, Centers for Disease Control and Prevention, Atlanta, GA, USA. We thank the Li Ka-Shing Foundation of Canada, Immune Diagnostics & Research, Shantou University Medical College, NIH (Grant R37 HL36235), NIH 1U01AI11598-01 Subaward no. 0038591(123721-3) to the support of this study and the Canadian Institutes of Health Research (CIHR) (Grant MOP 126205 (Dr. Rubin)) for the support of this study. We thank the staff from the Animal Resource Center of University Health Network for their help with the animal experiments.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.virol.2014.01.030.
Appendix A. Supplementary materials
Supplementary Data
Supplementary Data
References
- Arima M., Fukuda T. Prostaglandin D(2) and T(H)2 inflammation in the pathogenesis of bronchial asthma. Korean J. Intern. Med. 2011;28:8–18. doi: 10.3904/kjim.2011.26.1.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baillie J.K., Digard P. Influenza – time to target the host? N. Engl. J. Med. 2013;369:191–193. doi: 10.1056/NEJMcibr1304414. [DOI] [PubMed] [Google Scholar]
- Bermejo-Martin J.F., Ortiz de L.R., Pumarola T., Rello J., Almansa R., Ramirez P., Martin-Loeches I., Varillas D., Gallegos M.C., Seron C., Micheloud D., Gomez J.M., Tenorio-Abreu A., Ramos M.J., Molina M.L., Huidobro S., Sanchez E., Gordon M., Fernandez V., Del C.A., Marcos M.A., Villanueva B., Lopez C.J., Rodriguez-Dominguez M., Galan J.C., Canton R., Lietor A., Rojo S., Eiros J.M., Hinojosa C., Gonzalez I., Torner N., Banner D., Leon A., Cuesta P., Rowe T., Kelvin D.J. Th1 and Th17 hypercytokinemia as early host response signature in severe pandemic influenza. Crit. Care. 2009;13:R201. doi: 10.1186/cc8208. (doi:cc8208 [pii];10.1186/cc8208 [doi]) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermejo-Martin J.F., Martin-Loeches I., Rello J., Anton A., Almansa R., Xu L., Lopez-Campos G., Pumarola T., Ran L., Ramirez P., Banner D., Ng D.C., Socias L., Loza A., Andaluz D., Maravi E., Gomez-Sanchez M.J., Gordon M., Gallegos M.C., Fernandez V., Aldunate S., Leon C., Merino P., Blanco J., Martin-Sanchez F., Rico L., Varillas D., Iglesias V., Marcos M.A., Gandia F., Bobillo F., Nogueira B., Rojo S., Resino S., Castro C., Ortiz de L.R., Kelvin D. Host adaptive immunity deficiency in severe pandemic influenza. Crit. Care. 2010;14:R167. doi: 10.1186/cc9259. (doi:cc9259 [pii];10.1186/cc9259 [doi]) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhavsar P.K., Levy B.D., Hew M.J., Pfeffer M.A., Kazani S., Israel E., Chung K.F. Corticosteroid suppression of lipoxin A4 and leukotriene B4 from alveolar macrophages in severe asthma 12. Respir. Res. 2010;11:71. doi: 10.1186/1465-9921-11-71. (doi:1465-9921-11-71 [pii];10.1186/1465-9921-11-71 [doi]) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron C.M., Cameron M.J., Bermejo-Martin J.F., Ran L., Xu L., Turner P.V., Ran R., Danesh A., Fang Y., Chan P.K., Mytle N., Sullivan T.J., Collins T.L., Johnson M.G., Medina J.C., Rowe T., Kelvin D.J. Gene expression analysis of host innate immune responses during Lethal H5N1 infection in ferrets. J. Virol. 2008;82:11308–11317. doi: 10.1128/JVI.00691-08. (doi:JVI.00691-08 [pii];10.1128/JVI.00691-08 [doi]) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron M.J., Ran L., Xu L., Danesh A., Bermejo-Martin J.F., Cameron C.M., Muller M.P., Gold W.L., Richardson S.E., Poutanen S.M., Willey B.M., Devries M.E., Fang Y., Seneviratne C., Bosinger S.E., Persad D., Wilkinson P., Greller L.D., Somogyi R., Humar A., Keshavjee S., Louie M., Loeb M.B., Brunton J., McGeer A.J., Kelvin D.J. Interferon-mediated immunopathological events are associated with atypical innate and adaptive immune responses in patients with severe acute respiratory syndrome. J. Virol. 2007;81:8692–8706. doi: 10.1128/JVI.00527-07. (doi:JVI.00527-07 [pii];10.1128/JVI.00527-07 [doi]) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron M.J., Kelvin A.A., Leon A.J., Cameron C.M., Ran L., Xu L., Chu Y.K., Danesh A., Fang Y., Li Q., Anderson A., Couch R.C., Paquette S.G., Fomukong N.G., Kistner O., Lauchart M., Rowe T., Harrod K.S., Jonsson C.B., Kelvin D.J. Lack of innate interferon responses during SARS coronavirus infection in a vaccination and reinfection Ferret Model2. PLoS One. 2012;7:e45842. doi: 10.1371/journal.pone.0045842. (doi:10.1371/journal.pone.0045842 [doi];PONE-D-12-09714 [pii]) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks S.W., Stockley R.A. Leukotriene B42. Int. J. Biochem. Cell Biol. 1998;1998;3030:173–178. 173–178. doi: 10.1016/s1357-2725(97)00123-4. (doi:S1357-2725(97)00123-4 [pii]) (doi:S1357-2725(97)00123-4 [pii]) [DOI] [PubMed] [Google Scholar]
- Curfs D.M., Ghesquiere S.A., Vergouwe M.N., van d.M., Gijbels I.M.J., Greaves D.R., Verbeek J.S., Hofker M.H., de Winther M.P. Macrophage secretory phospholipase A2 group X enhances anti-inflammatory responses, promotes lipid accumulation, and contributes to aberrant lung pathology. J. Biol. Chem. 2008;283:21640–21648. doi: 10.1074/jbc.M710584200. (doi:M710584200 [pii];10.1074/jbc.M710584200 [doi]) [DOI] [PubMed] [Google Scholar]
- De L.D., Minucci A., Piastra M., Cogo P.E., Vendittelli F., Marzano L., Gentile L., Giardina B., Conti G., Capoluongo E.D. Ex vivo effect of varespladib on secretory phospholipase A2 alveolar activity in infants with ARDS. PLoS One. 2012;7:e47066. doi: 10.1371/journal.pone.0047066. (doi:10.1371/journal.pone.0047066 [doi];PONE-D-12-14582 [pii]) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degousee N., Stefanski E., Lindsay T.F., Ford D.A., Shahani R., Andrews C.A., Thuerauf D.J., Glembotski C.C., Nevalainen T.J., Tischfield J., Rubin B.B. p38 MAPK regulates group IIa phospholipase A2 expression in interleukin-1b stimulated rat neonatal cardiomyocytes. J. Biol. Chem. 2001;276:43842–43849. doi: 10.1074/jbc.M101516200. [DOI] [PubMed] [Google Scholar]
- Degousee N., Ghomashchi F., Stefanski E., Singer A., Smart B.P., Borregaard N., Reithmeier R., Lindsay T.F., Lichtenberger C., Reinisch W., Lambeau G., Arm J., Tischfield J., Gelb M.H., Rubin B.B. Groups IV, V, and X phospholipases A2s in human neutrophils: role in eicosanoid production and gram-negative bacterial phospholipid hydrolysis. J. Biol. Chem. 2002;277:5061–5073. doi: 10.1074/jbc.M109083200. [DOI] [PubMed] [Google Scholar]
- Degousee N., Martindale J., Stefanski E., Cieslak M., Lindsay T.F., Fish J.E., Marsden P.A., Thuerauf D.J., Glembotski C.C., Rubin B.B. MAP kinase kinase 6-p38 MAP kinase signaling cascade regulates cyclooxygenase-2 expression in cardiac myocytes in vitro and in vivo. Circ. Res. 2003;92:757–764. doi: 10.1161/01.RES.0000067929.01404.03. [DOI] [PubMed] [Google Scholar]
- Degousee N., Angoulvant D., Fazel S., Stefanski E., Saha S., Iliescu K., Lindsay T.F., Fish J.E., Marsden P.A., Li R.K., Audoly L.P., Jakobsson P.J., Rubin B.B. c-Jun N-terminal kinase-mediated stabilization of microsomal prostaglandin E2 synthase-1 mRNA regulates delayed microsomal prostaglandin E2 synthase-1 expression and prostaglandin E2 biosynthesis by cardiomyocytes. J. Biol. Chem. 2006;281:16443–16452. doi: 10.1074/jbc.M602815200. [DOI] [PubMed] [Google Scholar]
- Degousee N., Fazel S., Angoulvant D., Stefanski E., Pawelzik S.C., Korotkova M., Arab S., Liu P., Lindsay T.F., Zhuo S., Butany J., Li R.K., Audoly L., Schmidt R., Angioni C., Geisslinger G., Jakobsson P.J., Rubin B.B. Microsomal prostaglandin E2 synthase-1 deletion leads to adverse left ventricular remodeling after myocardial infarction. Circulation. 2008;117:1701–1710. doi: 10.1161/CIRCULATIONAHA.107.749739. [DOI] [PubMed] [Google Scholar]
- Del P.A., Shao W.H., Mitola S., Santoro G., Sozzani S., Haribabu B. Regulation of dendritic cell migration and adaptive immune response by leukotriene B4 receptors: a role for LTB4 in up-regulation of CCR7 expression and function1. Blood. 2007;109:626–631. doi: 10.1182/blood-2006-02-003665. (doi:blood-2006-02-003665 [pii];10.1182/blood-2006-02-003665 [doi]) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis E.A. Diversity of group types, regulation, and function of phospholipase A2. J. Biol. Chem. 1994;269:13057–13060. [PubMed] [Google Scholar]
- de Jong M.D., Simmons C.P., Thanh T.T., Hien V.M., Smith G.J., Chau T.N., Hoang D.M., Chau N.V., Khanh T.H., Dong V.C., Qui P.T., Cam B.V., Ha d.Q., Guan Y., Peiris J.S., Chinh N.T., Hien T.T., Farrar J. Fatal outcome of human influenza A (H5N1) is associated with high viral load and hypercytokinemia. Nat. Med. 2006;12:1203–1207. doi: 10.1038/nm1477. (doi:nm1477 [pii];10.1038/nm1477 [doi]) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escoffier J., Jemel I., Tanemoto A., Taketomi Y., Payre C., Coatrieux C., Sato H., Yamamoto K., Masuda S., Pernet-Gallay K., Pierre V., Hara S., Murakami M., De W.M., Lambeau G., Arnoult C. Group X phospholipase A2 is released during sperm acrosome reaction and controls fertility outcome in mice. J. Clin. Invest. 2010;120:1415–1428. doi: 10.1172/JCI40494. (doi:40494 [pii];10.1172/JCI40494 [doi]) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y., Banner D., Kelvin A.A., Huang S.S., Paige C.J., Corfe S.A., Kane K.P., Bleackley R.C., Rowe T., Leon A.J., Kelvin D.J. Seasonal H1N1 infection induces cross protective pandemic H1N1 immunity through a CD8 independent, B cell dependent mechanism. J. Virol. 2011;86:2229–2238. doi: 10.1128/JVI.05540-11. (doi:JVI.05540-11 [pii];10.1128/JVI.05540-11 [doi]) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Femling J.K., Nauseef W.M., Weiss J.P. Synergy between extracellular group IIA phospholipase A2 and phagocyte NADPH oxidase in digestion of phospholipids of Staphylococcus aureus ingested by human neutrophils. J. Immunol. 2005;175:4653–4661. doi: 10.4049/jimmunol.175.7.4653. [DOI] [PubMed] [Google Scholar]
- Fisher A.B., Dodia C., Feinstein S.I., Ho Y.S. Altered lung phospholipid metabolism in mice with targeted deletion of lysosomal-type phospholipase A2. J. Lipid Res. 2005;46:1248–1256. doi: 10.1194/jlr.M400499-JLR200. (doi:M400499-JLR200 [pii];10.1194/jlr.M400499-JLR200 [doi]) [DOI] [PubMed] [Google Scholar]
- Gao R., Cao B., Hu Y., Feng Z., Wang D., Hu W., Chen J., Jie Z., Qiu H., Xu K., Xu X., Lu H., Zhu W., Gao Z., Xiang N., Shen Y., He Z., Gu Y., Zhang Z., Yang Y., Zhao X., Zhou L., Li X., Zou S., Zhang Y., Li X., Yang L., Guo J., Dong J., Li Q., Dong L., Zhu Y., Bai T., Wang S., Hao P., Yang W., Zhang Y., Han J., Yu H., Li D., Gao G.F., Wu G., Wang Y., Yuan Z., Shu Y. Human infection with a novel avian-origin influenza A (H7N9) virus. N. Engl. J. Med. 2013;368:1888–1897. doi: 10.1056/NEJMoa1304459. (doi:10.1056/NEJMoa1304459 [doi]) [DOI] [PubMed] [Google Scholar]
- Gaudreault E., Gosselin J. Leukotriene B4-mediated release of antimicrobial peptides against cytomegalovirus is BLT1 dependent. Viral Immunol. 2007;20:407–420. doi: 10.1089/vim.2006.0099. (doi:10.1089/vim.2006.0099 [doi]) [DOI] [PubMed] [Google Scholar]
- Gaudreault E., Gosselin J. Leukotriene B4 induces release of antimicrobial peptides in lungs of virally infected mice. J. Immunol. 2008;180:6211–6221. doi: 10.4049/jimmunol.180.9.6211. (doi:180/9/6211 [pii]) [DOI] [PubMed] [Google Scholar]
- Goracci G., Ferrini M., Nardicchi V. Low molecular weight phospholipases A2 in mammalian brain and neural cells: roles in functions and dysfunctions4. Mol. Neurobiol. 2010;41:274–289. doi: 10.1007/s12035-010-8108-6. (doi:10.1007/s12035-010-8108-6 [doi]) [DOI] [PubMed] [Google Scholar]
- Groom J.R., Luster A.D. CXCR3 ligands: redundant, collaborative and antagonistic functions4. Immunol. Cell Biol. 2011;89:207–215. doi: 10.1038/icb.2010.158. (doi:icb2010158 [pii];10.1038/icb.2010.158 [doi]) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y., Farooqui A., Zhu H., Dong W., Wang J., Kelvin D.J. H7N9 incident, immune status, the elderly and a warning of an influenza pandemic. J. Infect. Dev. Ctries. 2013;7:302–307. doi: 10.3855/jidc.3675. [DOI] [PubMed] [Google Scholar]
- Henderson L.M., Banting G., Chappell J.B. The arachidonate-activable, NADPH oxidase-associated H+ channel. Evidence that gp91-phox functions as an essential part of the channel. J. Biol. Chem. 1995;270:5909–5916. [PubMed] [Google Scholar]
- Henderson W.R., Jr., Chi E.Y., Bollinger J.G., Tien Y.T., Ye X., Castelli L., Rubtsov Y.P., Singer A.G., Chiang G.K., Nevalainen T., Rudensky A.Y., Gelb M.H. Importance of group X-secreted phospholipase A2 in allergen-induced airway inflammation and remodeling in a mouse asthma model. J. Exp. Med. 2007;204:865–877. doi: 10.1084/jem.20070029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson W.R., Jr., Oslund R.C., Bollinger J.G., Ye X., Tien Y.T., Xue J., Gelb M.H. Blockade of human group X secreted phospholipase A2 (GX-sPLA2)-induced airway inflammation and hyperresponsiveness in a mouse asthma model by a selective GX-sPLA2 inhibitor. J. Biol. Chem. 2011;286:28049–28055. doi: 10.1074/jbc.M111.235812. (doi:M111.235812 [pii];10.1074/jbc.M111.235812 [doi]) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks A., Monkarsh S.P., Hoffman A.F., Goodnow R., Jr. Leukotriene B4 receptor antagonists as therapeutics for inflammatory disease: preclinical and clinical developments. Expert Opin. Investig. Drugs. 2007;16:1909–1920. doi: 10.1517/13543784.16.12.1909. (doi:10.1517/13543784.16.12.1909 [doi]) [DOI] [PubMed] [Google Scholar]
- Huang S.S., Banner D., Fang Y., Ng D.C., Kanagasabai T., Kelvin D.J., Kelvin A.A. Comparative analyses of pandemic H1N1 and seasonal H1N1, H3N2, and influenza B infections depict distinct clinical pictures in ferrets. PLoS One. 2011;6:e27512. doi: 10.1371/journal.pone.0027512. (doi:10.1371/journal.pone.0027512 [doi];PONE-D-11-09202 [pii]) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S.S., Banner D., Degousee N., Leon A.J., Xu L., Paquette S.G., Kanagasabai T., Fang Y., Rubino S., Rubin B., Kelvin D.J., Kelvin A.A. Differential pathological and immune responses in newly weaned ferrets are associated with mild clinical outcome of pandemic 2009 H1N1 Infection1. J. Virol. 2012;86:13187–13701. doi: 10.1128/JVI.01456-12. (doi:JVI.01456-12 [pii];10.1128/JVI.01456-12 [doi]) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang d.W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. (doi:nprot.2008.211 [pii];10.1038/nprot.2008.211 [doi]) [DOI] [PubMed] [Google Scholar]
- Influenza Activity – United States and Worldwide, June 13–September 25, 2010. Morbidity and Mortality Weekly Report (MMWR). Vol. 59, pp. 1270–1273. (doi:mm5939a3 [pii]). [PubMed]
- Karabina S.A., Brocheriou I., Le N.G., Agrapart M., Durand H., Gelb M., Lambeau G., Ninio E. Atherogenic properties of LDL particles modified by human group X secreted phospholipase A2 on human endothelial cell function. FASEB J. 2006;20:2547–2549. doi: 10.1096/fj.06-6018fje. (doi:fj.06-6018fje [pii];10.1096/fj.06-6018fje [doi]) [DOI] [PubMed] [Google Scholar]
- Kennedy B.P., Payette P., Mudgett J., Vadas P., Pruzanski W., Kwan M., Tang C., Rancourt D.E., Cromlish W.A. A natural disruption of the secretory group II phospholipase A2 gene in inbred mouse strains. J. Biol. Chem. 1995;270:22378–22385. doi: 10.1074/jbc.270.38.22378. [DOI] [PubMed] [Google Scholar]
- Kim J.O., Chakrabarti B.K., Guha-Niyogi A., Louder M.K., Mascola J.R., Ganesh L., Nabel G.J. Lysis of human immunodeficiency virus type 1 by a specific secreted human phospholipase A2. J. Virol. 2007;81:1444–1450. doi: 10.1128/JVI.01790-06. (doi:JVI.01790-06 [pii];10.1128/JVI.01790-06 [doi]) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudo I., Murakami M. Diverse functional coupling of prostanoid biosynthetic enzymes in various cell types. Adv. Exp. Med. Biol. 1999;469:29–35. doi: 10.1007/978-1-4615-4793-8_5. [DOI] [PubMed] [Google Scholar]
- Leon A.J., Banner D., Xu L., Ran L., Peng Z., Yi K., Chen C., Xu F., Huang J., Zhao Z., Lin Z., Huang S.H., Fang Y., Kelvin A.A., Ross T.M., Farooqui A., Kelvin D.J. Sequencing, annotation and characterization of the influenza ferret infectome1. J. Virol. 2012;87:1957–1966. doi: 10.1128/JVI.02476-12. (doi:JVI.02476-12 [pii];10.1128/JVI.02476-12 [doi]) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z., Serghides L., Patel S.N., Degousee N., Rubin B.B., Krishnegowda G., Gowda D.C., Karin M., Kain K.C. Disruption of JNK2 decreases the cytokine response to Plasmodium falciparum glycosylphosphatidylinositol in vitro and confers protection in a cerebral malaria model. J. Immunol. 2006;177:6344–6352. doi: 10.4049/jimmunol.177.9.6344. [DOI] [PubMed] [Google Scholar]
- Marshall J., Krump E., Lindsay T., Downey G., Ford D.A., Zhu P., Walker P., Rubin B. Involvement of cytosolic phospholipase A2 and secretory phospholipase A2 in arachidonic acid release from human neutrophils. J. Immunol. 2000;164:2084–2091. doi: 10.4049/jimmunol.164.4.2084. [DOI] [PubMed] [Google Scholar]
- Masuda S., Murakami M., Mitsuishi M., Komiyama K., Ishikawa Y., Ishii T., Kudo I. Expression of secretory phospholipase A2 enzymes in lungs of humans with pneumonia and their potential prostaglandin-synthetic function in human lung-derived cells. Biochem. J. 2005;387:27–38. doi: 10.1042/BJ20041307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka T., Hirata M., Tanaka H., Takahashi Y., Murata T., Kabashima K., Sugimoto Y., Kobayashi T., Ushikubi F., Aze Y., Eguchi N., Urade Y., Yoshida N., Kimura K., Mizoguchi A., Honda Y., Nagai H., Narumiya S. Prostaglandin D2 as a mediator of allergic asthma5. Science. 2000;287:2013–2017. doi: 10.1126/science.287.5460.2013. (doi:8366 [pii]) [DOI] [PubMed] [Google Scholar]
- Mazur I., Wurzer W.J., Ehrhardt C., Pleschka S., Puthavathana P., Silberzahn T., Wolff T., Planz O., Ludwig S. Acetylsalicylic acid (ASA) blocks influenza virus propagation via its NF-kappaB-inhibiting activity. Cell Microbiol. 2007;9:1683–1694. doi: 10.1111/j.1462-5822.2007.00902.x. (doi:CMI902 [pii];10.1111/j.1462-5822.2007.00902.x [doi]) [DOI] [PubMed] [Google Scholar]
- Mitsuishi M., Masuda S., Kudo I., Murakami M. Group V and X secretory phospholipase A2 prevents adenoviral infection in mammalian cells. Biochem. J. 2006;393:97–106. doi: 10.1042/BJ20050781. (doi:BJ20050781 [pii];10.1042/BJ20050781 [doi]) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita M., Kuba K., Ichikawa A., Nakayama M., Katahira J., Iwamoto R., Watanebe T., Sakabe S., Daidoji T., Nakamura S., Kadowaki A., Ohto T., Nakanishi H., Taguchi R., Nakaya T., Murakami M., Yoneda Y., Arai H., Kawaoka Y., Penninger J.M., Arita M., Imai Y. The lipid mediator protectin D1 inhibits influenza virus replication and improves severe influenza. Cell. 2013;153:112–125. doi: 10.1016/j.cell.2013.02.027. (doi:S0092-8674(13)00216-X [pii];10.1016/j.cell.2013.02.027 [doi]) [DOI] [PubMed] [Google Scholar]
- Murakami M., Kambe T., Shimbara S., Higashino K., Hanasaki K., Arita H., Horiguchi M., Arita M., Arai H., Inoue K., Kudo I. Different functional aspects of the group II subfamily (Types IIA and V) and type X secretory phospholipase A(2)s in regulating arachidonic acid release and prostaglandin generation. Implications of cyclooxygenase-2 induction and phospholipid scramblase-mediated cellular membrane perturbation57. J. Biol. Chem. 1999;274:31435–31444. doi: 10.1074/jbc.274.44.31435. [DOI] [PubMed] [Google Scholar]
- Murakami M., Taketomi Y., Miki Y., Sato H., Hirabayashi T., Yamamoto K. Recent progress in phospholipase A(2) research: from cells to animals to humans. Prog. Lipid Res. 2011;50:152–192. doi: 10.1016/j.plipres.2010.12.001. (doi:S0163-7827(10)00065-2 [pii];10.1016/j.plipres.2010.12.001 [doi]) [DOI] [PubMed] [Google Scholar]
- Muthuswamy R., Mueller-Berghaus J., Haberkorn U., Reinhart T.A., Schadendorf D., Kalinski P. PGE(2) transiently enhances DC expression of CCR7 but inhibits the ability of DCs to produce CCL19 and attract naive T cells11. Blood. 2010;116:1454–1459. doi: 10.1182/blood-2009-12-258038. (doi:blood-2009-12-258038 [pii];10.1182/blood-2009-12-258038 [doi]) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers R.C., King R.G., Carter R.H., Justement L.B. Lymphotoxin alpha(1) beta(2) expression on B cells is required for follicular dendritic cell activation during the germinal center response1. Eur. J. Immunol. 2012;43:348–359. doi: 10.1002/eji.201242471. ([doi]) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napolitani G., Acosta-Rodriguez E.V., Lanzavecchia A., Sallusto F. Prostaglandin E2 enhances Th17 responses via modulation of IL-17 and IFN-gamma production by memory CD4+ T cells3. Eur. J. Immunol. 2009;39:1301–1312. doi: 10.1002/eji.200838969. ([doi]) [DOI] [PubMed] [Google Scholar]
- Ohtsuki M., Taketomi Y., Arata S., Masuda S., Ishikawa Y., Ishii T., Takanezawa Y., Aoki J., Arai H., Yamamoto K., Kudo I., Murakami M. Transgenic expression of group V, but not group X, secreted phospholipase A2 in mice leads to neonatal lethality because of lung dysfunction. J. Biol. Chem. 2006;281:36420–36433. doi: 10.1074/jbc.M607975200. (doi:M607975200 [pii];10.1074/jbc.M607975200 [doi]) [DOI] [PubMed] [Google Scholar]
- Paquette S.G., Banner D., Zhao Z., Fang Y., Huang S.S., Leomicronn A.J., Ng D.C., Almansa R., Martin-Loeches I., Ramirez P., Socias L., Loza A., Blanco J., Sansonetti P., Rello J., Andaluz D., Shum B., Rubino S., de Lejarazu R.O., Tran D., Delogu G., Fadda G., Krajden S., Rubin B.B., Bermejo-Martin J.F., Kelvin A.A., Kelvin D.J. Interleukin-6 is a potential biomarker for severe pandemic H1N1 influenza A infection2. PLoS One. 2012;7:e38214. doi: 10.1371/journal.pone.0038214. (doi:10.1371/journal.pone.0038214 [doi];PONE-D-11-22886 [pii]) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquette S.G., Banner D., Chi T.B., le A.J., Leomicronn L., Xu L., Ran S.S., Huang A., Farooqui D.J., Kelvin, Kelvin A.A. Pandemic H1N1 influenza A directly induces a robust and acute inflammatory gene signature in primary human bronchial epithelial cells downstream of membrane fusion. Virology. 2014;448:91–103. doi: 10.1016/j.virol.2013.09.022. (doi:S0042-6822(13)00555-2 [pii];10.1016/j.virol.2013.09.022 [doi]) [DOI] [PubMed] [Google Scholar]
- Rowe T., Banner D., Farooqui A., Ng D.C.K., Kelvin A.A., Rubino S., Huang S.S.H., Fang Y., Kelvin D.J. In vivo ribavirin activity against severe pandemic H1N1 influenza A/Mexico/4108/2009. J. Gen. Virol. 2010 doi: 10.1099/vir.0.024323-0. [DOI] [PubMed] [Google Scholar]
- Rowe T., Leon A.J., Crevar C.J., Carter D.M., Xu L., Ran L., Fang Y., Cameron C.M., Cameron M.J., Banner D., Ng D.C., Ran R., Weirback H.K., Wiley C.A., Kelvin D.J., Ross T.M. Modeling host responses in ferrets during A/California/07/2009 influenza infection. Virology. 2010;401:257–265. doi: 10.1016/j.virol.2010.02.020. (doi:S0042-6822(10)00129-7 [pii];10.1016/j.virol.2010.02.020 [doi]) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin B.B., Downey G.P., Koh A., Degousee N., Ghomashchi F., Nallan L., Stefanski E., Harkin D.W., Sun C., Smart B.P., Lindsay T.F., Cherepanov V., Vachon E., Kelvin D., Sadilek M., Brown G.E., Yaffe M.B., Plumb J., Grinstein S., Glogauer M., Gelb M.H. Cytosolic phospholipase A2-a is necessary for platelet-activating factor biosynthesis, efficient neutrophil-mediated bacterial killing, and the innate immune response to pulmonary infection: cPLA2-a does not regulate neutrophil NADPH oxidase activity. J. Biol. Chem. 2005;280:7519–7529. doi: 10.1074/jbc.M407438200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusinova I., Forster S., Yu S., Kannan A., Masse M., Cumming H., Chapman R., Hertzog P.J. INTERFEROME v2.0: an updated database of annotated interferon-regulated genes1. Nucl. Acids Res. 2012;41:D1040–D1046. doi: 10.1093/nar/gks1215. (doi:gks1215 [pii];10.1093/nar/gks1215 [doi]) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadik C.D., Luster A.D. Lipid-cytokine-chemokine cascades orchestrate leukocyte recruitment in inflammation2. J. Leukoc. Biol. 2012;91:207–215. doi: 10.1189/jlb.0811402. (doi:jlb.0811402 [pii];10.1189/jlb.0811402 [doi]) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saez de G.J., Barrio L., Mellado M., Carrasco Y.R. CXCL13/CXCR5 signaling enhances BCR-triggered B-cell activation by shaping cell dynamics1. Blood. 2011;118:1560–1569. doi: 10.1182/blood-2011-01-332106. (doi:blood-2011-01-332106 [pii];10.1182/blood-2011-01-332106 [doi]) [DOI] [PubMed] [Google Scholar]
- Santone D.J., Shahani R., Rubin B.B., Romaschin A.D., Lindsay T.F. Mast cell stabilization improves cardiac contractile function following hemorrhagic shock and resuscitation. Am. J. Physiol. Heart Circ. Physiol. 2008;294:H2456–H2464. doi: 10.1152/ajpheart.00925.2007. [DOI] [PubMed] [Google Scholar]
- Schultz-Cherry S., Jones J.C. Influenza vaccines: the good, the bad, and the eggs. Adv. Virus Res. 2010;77:63–84. doi: 10.1016/B978-0-12-385034-8.00003-X. (doi:B978-0-12-385034-8.00003-X [pii];10.1016/B978-0-12-385034-8.00003-X [doi]) [DOI] [PubMed] [Google Scholar]
- Shridas P., Bailey W.M., Talbott K.R., Oslund R.C., Gelb M.H., Webb N.R. Group X secretory phospholipase A2 enhances TLR4 signaling in macrophages1. J. Immunol. 2011;187:482–489. doi: 10.4049/jimmunol.1003552. (doi:jimmunol.1003552 [pii];10.4049/jimmunol.1003552 [doi]) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson A.H., Lonigro A.J., Hyers T.M., Webster R.O., Fowler A.A. Increased concentrations of leukotrienes in bronchoalveolar lavage fluid of patients with ARDS or at risk for ARDS. Am. Rev. Respir. Dis. 1988;138:714–719. doi: 10.1164/ajrccm/138.3.714. [DOI] [PubMed] [Google Scholar]
- Truchetet M.E., Allanore Y., Montanari E., Chizzolini C., Brembilla N.C. Prostaglandin I(2) analogues enhance already exuberant Th17 cell responses in systemic sclerosis3. Ann. Rheum. Dis. 2012;71:2044–2050. doi: 10.1136/annrheumdis-2012-201400. (doi:annrheumdis-2012-201400 [pii];10.1136/annrheumdis-2012-201400 [doi].) [DOI] [PubMed] [Google Scholar]
- Update: Influenza Activity – United States, 2009–10 season. Morbidity and Mortality Weekly Report (MMWR). Vol. 59, pp. 901–908. (doi:mm5929a2 [pii]). [PubMed]
- Van Elssen C.H., Vanderlocht J., Oth T., Senden-Gijsbers B.L., Germeraad W.T., Bos G.M. Inflammation-restraining effects of prostaglandin E2 on natural killer-dendritic cell (NK-DC) interaction are imprinted during DC maturation1. Blood. 2011;118:2473–2482. doi: 10.1182/blood-2010-09-307835. (doi:blood-2010-09-307835 [pii];10.1182/blood-2010-09-307835 [doi]) [DOI] [PubMed] [Google Scholar]
- Widegren H., Andersson M., Borgeat P., Flamand L., Johnston S., Greiff L. LTB4 increases nasal neutrophil activity and conditions neutrophils to exert antiviral effects2. Respir. Med. doi:S0954-6111(10)00566-4 [pii];10.1016/j.rmed.2010.12.021 [doi] 2011;2011;105:997–1006. doi: 10.1016/j.rmed.2010.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization, 2012. Global Influenza Programme. (Ref Type: Online Source).
- Zhang P., Summer W.R., Bagby G.J., Nelson S. Innate immunity and pulmonary host defense. Immunol. Rev. 2000;173:39–51. doi: 10.1034/j.1600-065x.2000.917306.x. [DOI] [PubMed] [Google Scholar]
- Zhao J., Zhao J., Legge K., Perlman S. Age-related increases in PGD(2) expression impair respiratory DC migration, resulting in diminished T cell responses upon respiratory virus infection in mice1. J. Clin. Invest. 2011;121:4921–4930. doi: 10.1172/JCI59777. (doi:59777 [pii];10.1172/JCI59777 [doi]) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data
Supplementary Data