Abstract
The International Mouse Phenotyping Consortium (IMPC) (http://www.mousephenotype.org) will reveal the pleiotropic functions of every gene in the mouse genome and uncover the wider role of genetic loci within diverse biological systems. Comprehensive informatics solutions are vital to ensuring that this vast array of data is captured in a standardised manner and made accessible to the scientific community for interrogation and analysis. Here we review the existing EuroPhenome and WTSI phenotype informatics systems and the IKMC portal, and present plans for extending these systems and lessons learned to the development of a robust IMPC informatics infrastructure.
Introduction
The availability of thousands of targeted mutations in C57BL/6N embryonic stem (ES) cells from the International Mouse Knockout Consortium (IKMC, http://www.knockoutmouse.org) is accelerating efforts worldwide to understand mammalian gene function (Collins et al. 2007; Skarnes et al. 2011). Gene-based, phenotype-driven screens on a genome-wide scale are now possible in mice, facilitating both hypothesis-driven and unbiased screens to address the role of individual genes in normal mouse development and physiology. To date, a number of programs have utilised the IKMC resource in broad-based systematic phenotyping pipelines. The European Mouse Disease Clinic (EUMODIC) comprises four European centres [MRC Harwell (UK), Wellcome Trust Sanger Institute (WTSI) (UK), Helmholtz Zentrum München (Germany), and Institut Clinique de la Souris in Stras-bourg-Illkirch (France)] and employs the EMPReSSlim pipelines (phenotyping pipeline of 20 platforms) to phenotype lines from the IKMC resource. To date, EUMODIC has phenotyped 423 mutant lines. In addition to EUMODIC, the Wellcome Trust Sanger Institute has phenotyped 651 lines through the Mouse Genetics Project (MGP). The KOMP312 project is a pilot study funded at the Children’s Hospital Oakland Research Institute in California and the Mouse Biology Program, University of California, Davis to phenotype 312 mutant lines evaluating LacZ reporter expression and additional phenotyping on 100 homozygous mutants. Building upon these efforts, a number of newly funded initiatives have begun under the International Mouse Phenotyping Consortium (IMPC) umbrella to phenotype 5,000 knockout mouse lines over the next 5 years. The KOMP2 program, funded by the National Institutes of Health (NIH), will deliver 2,500 of these lines (Brown and Moore 2012).
In general, the international multicentre effort, spanning years of activity, mouse production, and phenotyping, necessitates careful tracking of mutant mouse colonies as they move from production stage to phenotyping. Tracking ensures that production targets are maintained, allows users to be notified when a mouse is available, and identifies colonies with potential problems such as those that stay in some production stage for longer than expected. Distributed high-throughput mouse mutagenesis and phenotyping requires that the data generated be comparable and quality controlled regardless of where they were created and that the results are rapidly available to all through a standard statistical analysis pipeline.
For IMPC, the interpretation of the differences and similarities in phenotypes seen between different alleles can be done only if the phenotype data generated uses robust, common semantics and rigorous quality control (QC) is applied. The development and use of comprehensive and standardised phenotyping procedures (SOPs) are vital. Effective SOPs ensure that results are comparable within and between different laboratories and over time, and are also essential in relating phenotypic data to ontological descriptions in any automatic annotation pipeline (Brown et al. 2005, 2006). A data collection challenge is the requirement to interact with diverse Laboratory Information Management Systems (LIMS), instrumentation, and animal husbandry conditions. The inherent high complexity of phenotype data requires standardisation and semantics for all data, including images. Moreover, the analysis and annotation of the phenotypes identified by imaging techniques in an automatic or manually curated manner is important to ensure that the data can be integrated with the text-based and numerical data. The capture, integration, and dissemination of mouse production and phenotyping data from current projects are reviewed here as we have begun to address the informatics challenges of providing access to this complex data resource. This review addresses the effort to consolidate and build upon these existing experiences and database resources to deliver an integrated web portal that will allow the mouse and clinical research communities to have access to the raw and analysed data.
Mouse production data
IKMC portal and data management
The IKMC resource of mutant mouse ES cells is the foundation for both large-scale phenotyping programs and investigator-driven studies. The IKMC portal provides a central point of access for vectors, ES cells, and mice available to researchers from designated repositories (Ringwald et al. 2011). The site was developed and is maintained jointly between the NIH-funded KOMP Data Coordination Centre and the EU-funded International Data Coordination Centre. The IKMC portal displays detailed information on targeting vectors for 16,000 genes, mutant ES cells for 17,000 genes, and 1,600 mutant mouse strains developed from the resource. Because several distinct pipelines were set up to develop these reagents, each using different mutagenesis strategies and ES cell lines, the resource is not uniform. Thus, an important function of this site is to provide summary information for each production pipeline as well as nucleotide-level descriptions of each allele.
Underlying the portal are several key databases containing contributions from all IKMC members: each database reflects a different aspect of the production process. In particular, the annotated master gene list serves to record the production status of every targeting project initiated by IKMC members and the availability of any generated products. Project status data are fed from each production centre to the Mouse Genome Informatics Group (MGI, http://www.informatics.jax.org/) at the Jackson Laboratory, which updates the status against a unified catalogue of mouse genes and gene models. The targeting repository holds annotated molecular descriptions in the form of GenBank files for every IKMC targeting vector and mutant allele, as well as summary results of the genotyping assays used for quality control of each ES cell clone. Data are placed into the targeting repository nightly by the production centres, and the repository in turn supplies the IKMC portal with Gen-Bank files and images of vectors and mutant alleles. Together these two systems form the core of the IKMC portal.
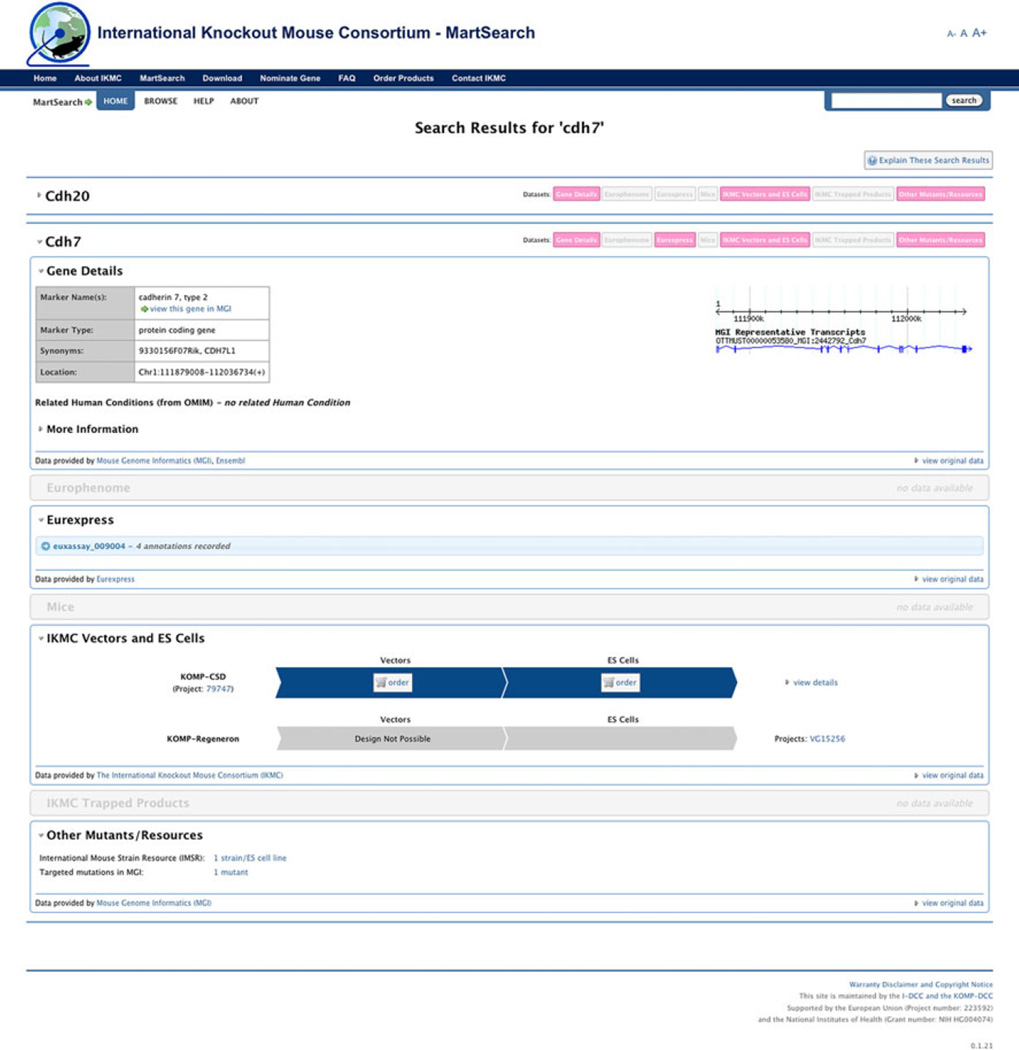
The IKMC portal also incorporates BioMart technology (Kasprzyk 2011) to integrate other biological information such as gene expression and mouse phenotypes with the existing IKMC alleles. The Martsearch section of the portal (http://www.knockoutmouse.org/martsearch) integrates IKMC mutant allele information with embryonic gene expression data from the EurExpress BioMart and mouse phenotype data from EuroPhenome BioMart (Fig. 1). Users can search this site using anatomy or phenotype terms and view summaries of expression and phenotype results for IKMC mutants alongside detailed mutant allele descriptions. All information supplied by the IKMC portal is distributed to other projects and computer systems via BioMart, which permits advanced queries of the data and bulk data downloads. The use of BioMart technology serves as a model for future integration and display of IKMC resources within the IMPC web portal.
Fig. 1.
IKMC BioMart search. Resulting gene details page for the gene Chd7 displaying a number of data panels that have returned data from the BioMart query. This shows the different genes that are found with the specific query and the various data that are returned
Mouse phenotyping data
Phenotyping procedures and pipelines
The standardisation of phenotyping procedures and pipelines is vital to ensure that the data generated from laboratories performing high-throughput phenotyping can be integrated and shared. The EUMORPHIA consortium, comprising 18 laboratories across Europe, has defined and standardised a comprehensive reference set of procedures for high-throughput phenotyping. The resulting collection called EMPReSS defines over 150 SOPs covering a broad spectrum of body systems (Brown et al. 2005). The EMPReSS database provides access to these SOPs through a web portal and via programmatic access (Mallon et al. 2008). To ensure that these data and results from this diverse set of SOPs is in context, a community-accepted set of minimum-information guidelines [Minimal Information for Mouse Phenotyping Procedures (MIMPP)] (Taylor et al. 2008) was developed in a collaboration between the SDOP-DB at RIKEN (Tanaka et al. 2010), the Mouse Phenome Database (MPD) at JAX (Bogue and Grubb 2004), and the EMPReSS database at MRC Harwell. This set of guidelines describes the methods, data, and metadata required to define a mouse phenotyping procedure.
LIMS development
The scale and complexity of the data generated from highthroughput mouse breeding and phenotyping require that it is captured in a robust LIMS that can effectively manage and track this information from the point at which it is generated to upload into a public data repository. The primary phenotyping centres in IMPC capture breeding and phenotyping data in centre-specific LIMS which were developed to serve the requirements of the individual animal facility. These systems use a range of relational database implementations from Sybase to Oracle to Access and are typically web-based implementations that run locally at each site.
Standardised data capture in a central database
EUMODIC implements standardised data capture with a common XML data format to exchange data from the diverse spectrum of local LIMS described above into a centralised database called EuroPhenome (Morgan et al. 2010). The data format adopted defines the required data for each individual mouse (e.g., sex, litter ID, mouse ID, zygosity, strain background, and date of birth) and the phenotyping data generated from it (e.g., glucose concentration, bone mineral density). The LIMS systems developed automated export procedures to generate the XML files and place them on a local FTP server. The EuroPhenome database retrieves these files automatically and loads them into the database, triggering a series of validation methods to ensure accuracy and completeness, with feedback presented to the data-generating centres so they could resolve identified issues. This data validation utilises the data definition information included in the SOP records such as data range information, e.g., body weight cannot be <0, and has significantly reduced the occurrence of data errors. EuroPhenome is now also uploading data from the Centre for Modelling Human Disease (CMHD) in Toronto.
Data analysis and annotation
The phenotyping procedures adopted in each pipeline measure primary phenotype parameters such as bone mineral density, blood glucose concentration, and oxygen consumption. Once collected, these primary data are analysed by an annotation pipeline that compares data from the mutant line to the baseline inbred strain to identify statistically significant phenodeviants. The current EuroPhenome annotation pipeline utilises the Wilcoxon rank-sum test for numerical parameters and Fisher’s exact test or the χ2 test for categorical parameters. The Wilcoxon rank-sum test is used to calculate a p value under the null hypothesis that the mutant and control groups have the same population distribution. Fisher’s exact test is used in the case of contingency tables with one degree of freedom (df) (i.e., 2 × 2 tables), and a χ2 test is used for tables more than 1 df to calculate this p value. In each case the p value is stored, and by default a line is called phenodeviant in that test if p < 0.0001. A nominal significance level of 10−4 was chosen because it implied a Bonferroni-controlled familywise error rate of 0.040 (from testing 398 parameters at the 10−4 level), which can be interpreted as allowing only 4 % of tested mutant lines to have one or more false-positive hits. Identified phenodeviants are annotated automatically with the Mammalian Phenotype (MP) ontological terms stored at the level of the SOP in the EMPReSS database and all this resultant data are stored in the EuroPhenome database (Table 1).
Table 1.
Various projects that contribute to the global mouse phenotyping effort, including links to their online resources
| Acronym | Project | URL |
|---|---|---|
| IMPC | International Mouse Phenotyping Consortium | http://www.mousephenotype.org |
| IKMC | International Mouse Knockout Consortium | http://www.knockoutmouse.org/ |
| EUMODIC | European Mouse Disease Clinic | http://www.eumodic.org/ |
| MGP | Mouse Genetics Project | http://www.sanger.ac.uk/mouseportal/ |
| KOMP312 | Knockout Mouse Phenotyping Pilot | http://www.kompphenotype.org/ |
| KOMP2 | Knockout Mouse Phenotyping Project | http://commonfund.nih.gov/KOMP2/ |
| NIH | National Institutes of Health | http://www.nih.gov |
| MPD | Mouse Phenome Database | http://phenome.jax.org/ |
| MGI | Mouse Genome Informatics | http://www.informatics.jax.org/ |
| SDOP-DB | Standardised Description of Operating Procedures Database | http://www.brc.riken.jp/lab/bpmp/SDOP/index.html |
| EMPReSS | European Mouse Phenotyping Resource of Standardised Screens | http://empress.har.mrc.ac.uk/ |
| EUMORPHIA | European Union Mouse Research for Public Health and Industrial Applications | http://www.eumorphia.org/ |
| CMHD | Centre for Modelling Human Disease | http://www.cmhd.ca/ |
| EMMA | European Mouse Mutant Archive | http://www.emmanet.org/ |
| IMPReSS | International Mouse Phenotyping Resource of Standardised Screens | http://www.mousephenotype.org/impress |
In the WTSI program, a different approach to data analysis was implemented, where numerical parameters were analysed via a reference range approach and a line was called a phenodeviant if more than 60 % of the mutant results are outside of a 95 % reference range derived by interpolation from all appropriate control data. For categorical parameters Fisher’s exact test is used, and a line is called phenodeviant if the resulting p value is <0.05 and the total change in frequency of any one value is >60 % (e.g., if the baseline rate of abnormality is 5 %, the mutant rate must be 65 % or greater for the line to be called phenodeviant). In addition, the decision tree adopted enabled the statistical calls to be overridden by a human expert.
Phenotyping portals
A number of distributed phenotyping portals generated to support the pilot projects described in the Introduction share primary phenotyping data and analysed phenodeviant calls with the community. Here we review these portals and summarise the key features for identifying new mutant lines of interest by the community.
EuroPhenome
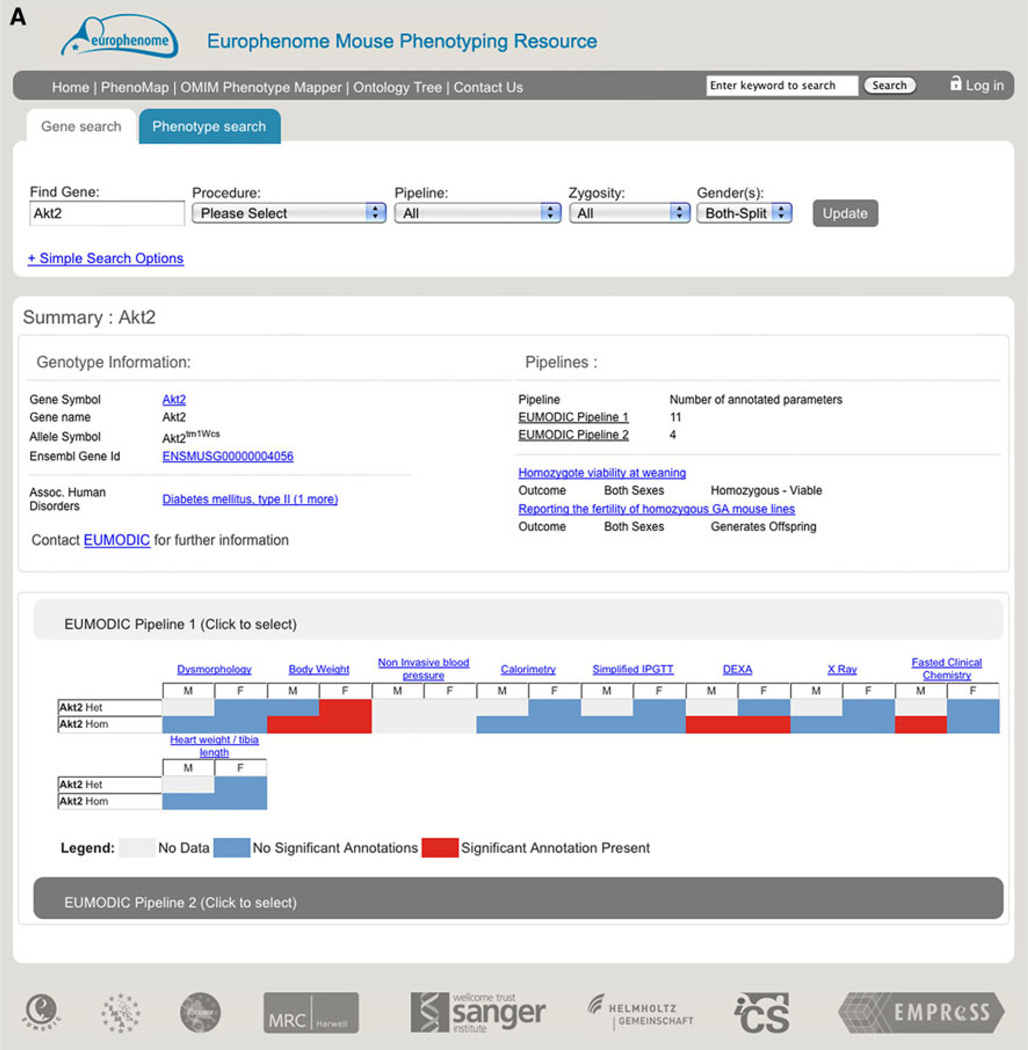
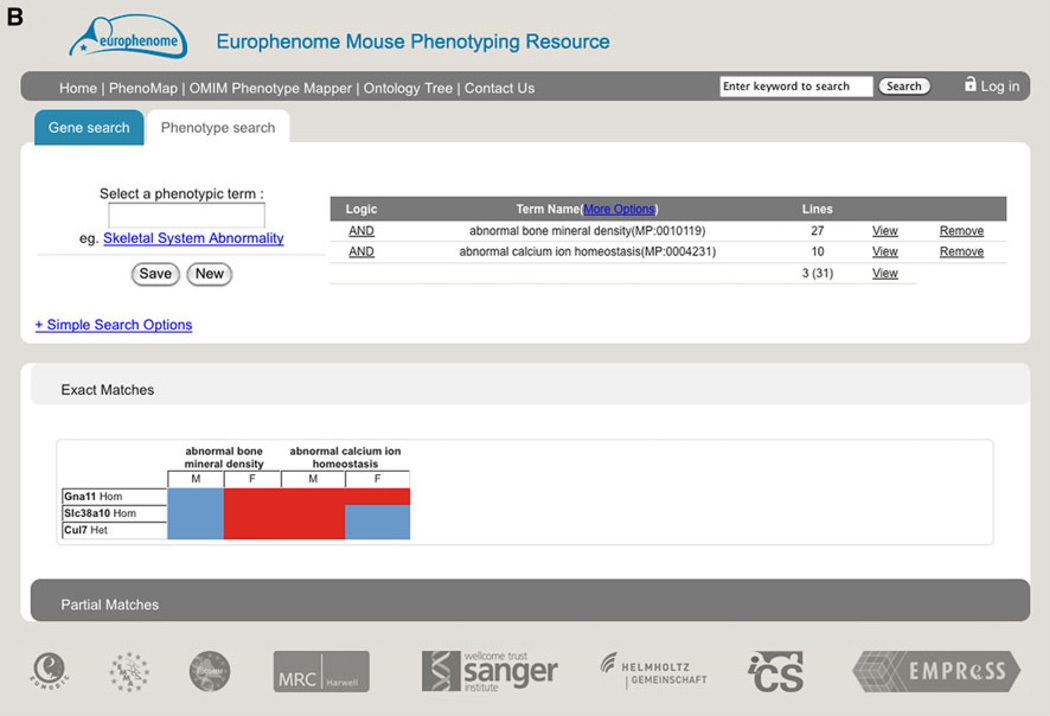
The EuroPhenomeweb portal (http://www.europhenome.org) provides tools for the comparative analysis of summary and detailed phenotype data aggregated from different EUMODIC mouse lines. The EuroPhenome portal provides a number of data access and analysis tools, with two primary search methods enabling users to access the data by a gene or phenotype query. Querying the portal via the “gene search,” e.g., Akt2, returns a summary page for that allele which displays both key information about the gene and a visual “heatmap” summary of the phenotype calls resulting from the annotation pipeline (Fig 2a). The second key search method is to mine with phenotype terms, e.g., abnormal glucose homeostasis, which will return a list of all alleles that have a significant phenodeviant hit for that ontology term or any of its children. Key feedback from the user community and secondary partners in EUMODIC drove the development of a “complex logical phenotype search” allowing users to identify alleles by defining a “combination of phenotypes of interest” search, e.g., abnormal bone mineral density AND abnormal calcium ion homeostasis (Fig. 2b). In addition to categorical and numerical data, EuroPhenome captures images from X-ray, ophthalmoscope, and slit lamp procedures. X-ray images are captured as DICOM files, rendered as png thumbnails in the web portal in a display that allows comparison of both mutant and baseline images together. As of 15 May 2012, the EuroPhenome portal contains data for 459 mutant strains, 44 inbred strains, 27,873 mice, 8,795,125 data points, and 3,412 significant annotations.
Fig. 2.
EuroPhenome gene and phenotype queries. a Summary page for the Akt2 allele displaying key information about the gene and a visual heatmap summary of the phenotype calls resulting from the annotation pipeline. At the top are links and search tools to find the relevant data and below this is information about the line, including genotype information, an overall summary of the results of the phenotyping, and specific results in a heatmap format. This is available from http://www.europhenome.org/databrowser/viewer.jsp?set=true&m=true&l=10035. b Phenotype overview page from the “complex logical phenotype search” showing the alleles that have a significant annotation to both abnormal bone mineral density AND abnormal calcium ion homeostasis. Below the primary navigation is the phenotype selection interface that allows the selection of a number of phenotype terms and the customisation of the search. Below this are the results of the search in a heatmap format
The Mouse Genetics Projects portal
The Mouse Genetics Project portal (http://www.sanger.ac.uk/mouseportal/) combines MGP-specific phenotype data and mutant mouse availability with the details of the mutation structure of IKMC mutant ES cells. Users can search for mouse lines by gene and the phenotypic associations observed at WTSI based on Mammalian Phenotype (MP) ontology terms, as well as functional associations based on Gene Ontology (GO) terms and Interpro structures. The phenotyping data presented on the portal reflects the standard broad-based phenotype assays developed for the IMPC and available on the EuroPhenome website. Additional assays of interest to WTSI researchers and their collaborators are performed on mice, including infection challenge, skin histopathology, and brain development. The data can be interrogated by using either a traditional assay-by-assay format or root Mammalian Phenotype (MP) ontology terms that have been associated with the phenotypic observations made at WTSI. The portal also aims to direct users to the appropriate archive to obtain the appropriate mouse or ES cell resource for their research.
The portal is implemented using a highly extensible architecture based on BioMart and SOLR indexing and is easily extended to integrate new sources of relevant information (generated by WTSI and others) as they become available.
KOMP312 portal
Data from the KOMP312 project are available from the KOMP phenotyping pilot web page (http://www.kompphenotype.org). Users can explore the phenotyping data by selecting a phenotype category, e.g., LacZ, and the genes that have a positive phenotype hit are displayed. Clicking on a gene with a positive hit will display a summary page on which users can view the data. In addition, users can search on genes and mouse anatomical structures of interest. The results from the anatomy search display LacZ stained whole-mount and frozen-section images.
Integrating mouse phenotyping data
The wider context of mouse phenotype data includes information on normal and mutant gene expression, e.g., expression array experiments, availability of mouse lines for biologists to order for experimental use, e.g., from the EMMA database (Wilkinson et al. 2010), and genetic variation data. High-complexity, high-dimensionality data from array-based and sequencing technologies are stored in the European Bioinformatics Institute’s (EBI) gene expression atlas (Kapushesky et al. 2012) and are available as a series of meta-analysed experiments coanalysed with human orthologues of mouse genes (Zheng-Bradley et al. 2010). Integration of these data is challenging because often the allele and/or strain background is under- or unreported in the gene expression data submission, meaning that these data need to be retrofitted. Nevertheless, it is possible to identify differentially expressed genes at the level of tissues and make comparisons with comparable human data sets. For mouse phenotypic data to be useful to the clinical community it must be summarised at the gene or phenotype level and integrated with resources that are used by this community. These include genome-wide association studies (GWAS) and other variation data held in the Ensembl variation database (Flicek et al. 2012), as well as gene expression data.
Future perspectives: Mouse Phenotyping Informatics Infrastructure (MPI2)
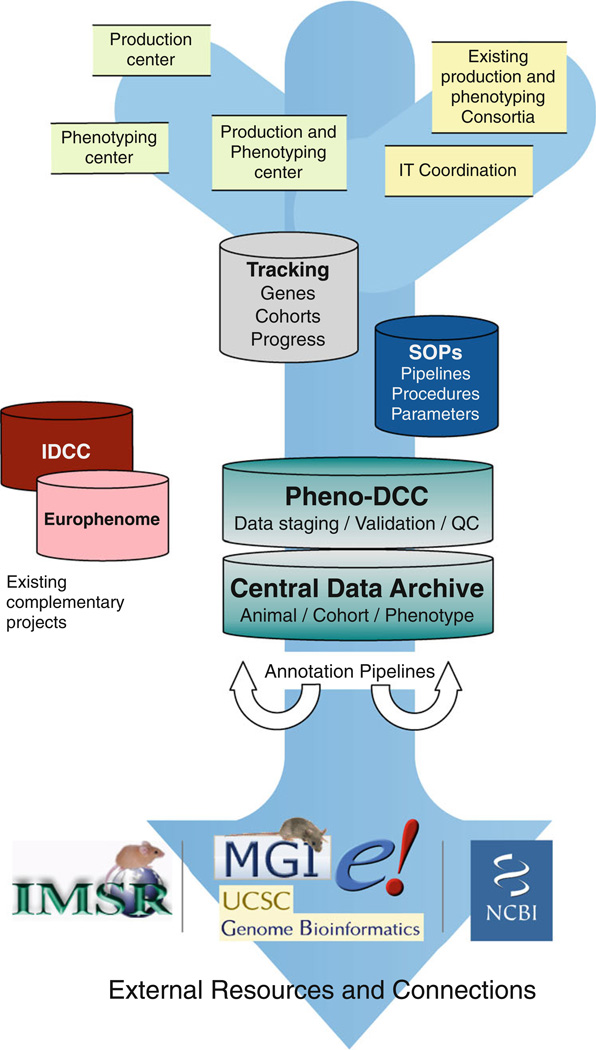
The goal of the Mouse Phenotyping Informatics Infrastructure (MPI2) is to develop and deploy the IT infrastructure, database, and web portal required to efficiently capture, manage, annotate, integrate, and disseminate the phenotyping data from KOMP2 and wider IMPC programmes to the scientific and biomedical communities in an accurate, timely, and intuitive manner. We have established a consortium comprising the EBI, MRC Harwell, and the Wellcome Trust Sanger Institute to develop the components of the MPI2 infrastructure that will build on previous experiences described in this review. The primary components shown in Fig. 3 are described here.
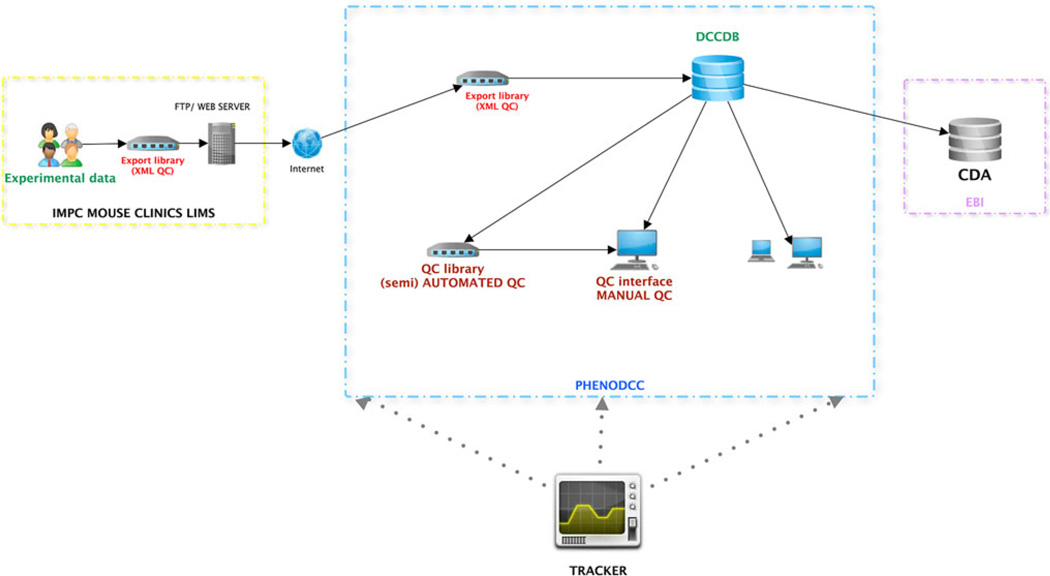
Fig. 3.
Overview of the data flow components of the MPI2 Consortium, from the production and phenotyping centres to the DCC and the CDA
The data coordination centre (DCC)
The DCC acts as a staging area to ensure that the data generated from the production and phenotyping centres are captured, validated, and quality controlled before deposition into the publically accessible data centre. The DCC builds on the knowledge and code developed in the EuroPhenome and IKMC projects to deliver a data management system that supports high-level summaries and detailed reports for data-generating centres and funding bodies via the public web portal. The key components of the DCC are as follows.
Data tracking: iMITS
The iMITS database (http://www.mousephenotype.org/imits) coordinates and provides summary reports on the production and phenotyping of mice from all IMPC members. The key contents of this database are the gene, allele, production and phenotyping plans of IMPC members; time-stamped records of their progress towards those plans; and essential metadata (e.g., genotyping QC results). The system is flexible. It is designed to allow IMPC members to enter data manually via a web interface or automatically via computer services. iMITS has four key outputs: First, iMITS reports potential duplication of production between different consortia. Second, iMITS creates summary reports of total production and pipeline efficiencies for each IMPC member and monthly reports of activity. Third, iMITS summary data are used to actively inform participants registered at the IMPC website of progress on their genes of interest. Finally, iMITS provides publicly accessible data via a BioMart to the IMPC portal, current IKMC portals, mouse repositories, and all other end users. The iMITS database extends previous versions, is in use by KOMP2 centres, and is a component of the IMPC website through the gene search as shown in Fig. 4.
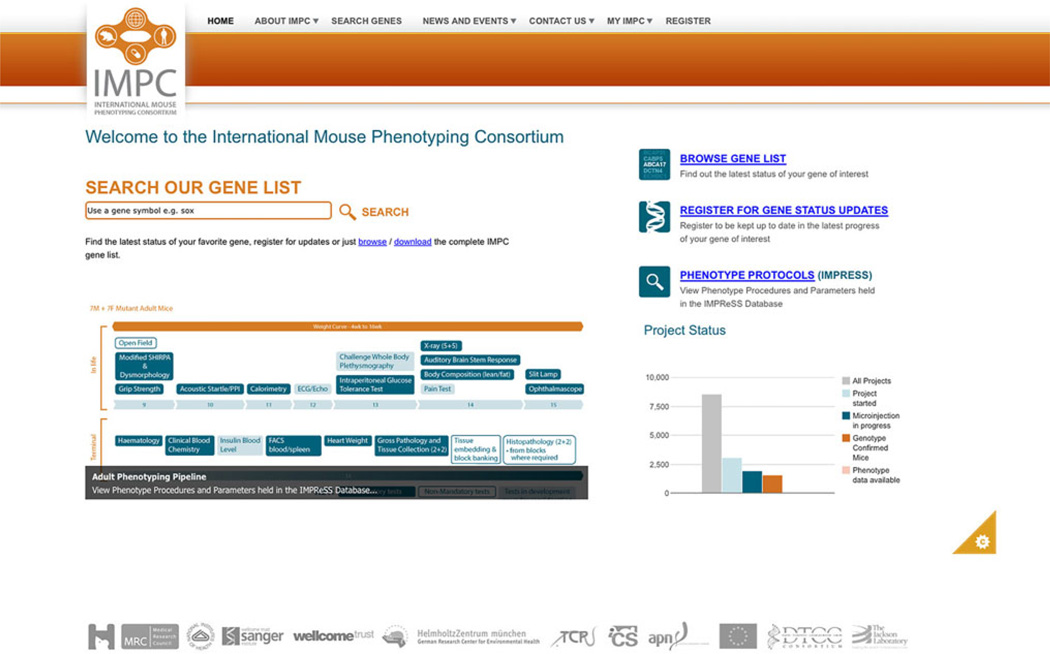
Fig. 4.
The current version of the IMPC web portal illustrating that summary data from the iMITS tracking database can be retrieved by searching your gene of choice, the search function that provides access to the gene list, and links to the various parts of the site
SOP data management: IMPReSS
IMPReSS (http://www.mousephenotype.org/impress) is a database and web portal developed to manage and track the phenotyping procedures implemented in IMPC and is an extended and enhanced version of the EMPReSS website (Fig. 5). IMPReSS enables users to view and download the procedures in the IMPC pipeline, e.g., IPGTT, and users can search for procedures that measure a phenotype of interest, e.g., abnormal glucose homeostasis. IMPReSS will provide a system for tracking changes to the procedures throughout the IMPC project as they are reviewed and improved, as well as integrate additional richer ontological annotations and assess consequences on linked data.
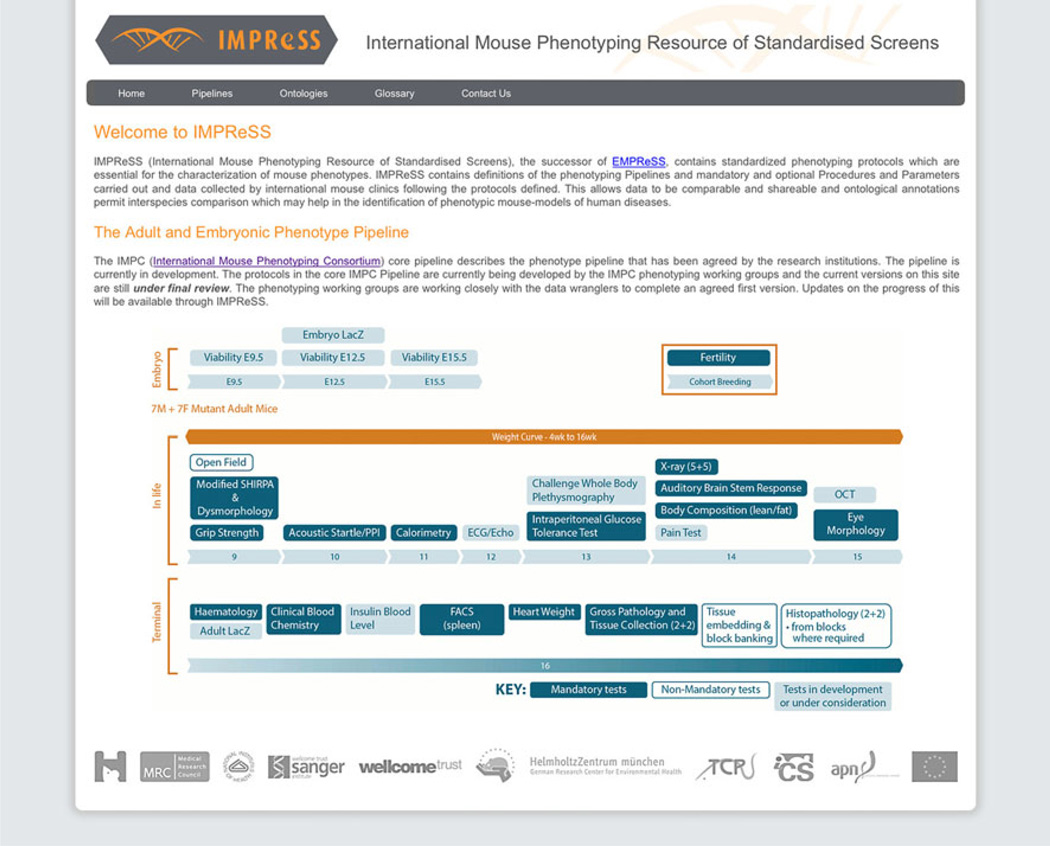
Fig. 5.
The IMPReSS web portal, which can be searched to retrieve phenotyping protocols of interest in the IMPC pipeline. This shows the phenotyping pipeline, including embryonic, in-life, and terminal tests and has links to the various tools
Data upload, validation, and quality control: pheno-DCC
The data upload process developed in EuroPhenome is being extended and improved within MPI2 to adapt to an increasing number of phenotyping centres and an expanded number of procedures. The initial version of the new XML schemas (https://github.com/mpi2) and data export library has been revised by all the IMPC centres and is in the process of being implemented to ensure IMPC data can be exported to the Pheno-DCC. New features are additional data validation modules which will prevent erroneous data entry and automated quality control modules which will identify data for further investigation by expert data wranglers and/or phenotyping centres. Expert data wranglers in the Pheno-DCC will manage the QC and validation processes and interact with the phenotyping centres to ensure that the data exported from the DCC to the core data archive is accurate and valid. The phenotyping data will be uploaded into the Pheno-DCC from the centres’ LIMS as it is generated and will be displayed on the IMPC portal in a timely manner to enable the scientific community access to the data and the mutant lines as they progress through the pipelines. The data served to the IMPC portal from the Pheno-DCC will be flagged as “incomplete QC data” to make sure users are aware that they must take caution when interpreting the data as it may be partial or include QC errors. Once the data for a mutant line is complete and QC approved, the data will be exported to the core data archive for further analysis and at this point the data on the portal will be flagged as “complete and QC approved” (Fig. 6).
Fig. 6.
The data flow of phenotyping data from the centres through the DCC, highlighting the QC process before being exported to the CDA
Data annotation and statistical analysis
The automated annotation pipeline (AAP), which assigns phenotype ontology terms (based on the phenotyping procedure definitions) to the statistically significant phenodeviants, relies on the reliability and reproducibility of calling significant phenodeviants, which is impacted by the experimental design of each procedure. The experimental design and statistical data analysis for each procedure are being reviewed by an expert statistical working group and the outcomes will define the statistical tests chosen in the pipeline. The recommendations of this expert group will be utilised to extensively extend and redevelop the existing AAP to scale for IMPC. The choice of the modular architecture and the specific modules in the AAP incorporates lessons learned from the EUMODIC project. The design includes a “Data Selection Layer,” a “Statistical Analysis Layer,” and an “Annotation Generator Layer.” These layers will enable the annotation pipeline infrastructure to be utilised at phenotyping centres, the Pheno-DCC, and the data centre, with appropriate modifications to each layer for the required tasks of each location.
Effective annotation and use of the resulting data are impossible without the assignment of the appropriate ontological phenotype term to a parameter or derived parameter when the mutant data are deemed to be statistically different from the control data (e.g., parameter: glucose; MP terms: increased or decreased glucose concentration). The definition of these ontology terms is captured in IMPReSS at the level of each parameter and is developed collaboratively between the data wranglers, the phenotyping centres, and domain experts. The annotation of the IMPC data with additional ontological descriptions will be critical to ensure cross-species integration, so additional ontologies from the community, such as the phenotypic quality ontology (PATO) (Gkoutos et al. 2005) and the experimental factor ontology (EFO) (Malone et al. 2010), will be adopted. Annotations from image-based phenotyping procedures will be incorporated into the pipeline as will the addition of value from other mouse gene function databases (e.g., MGI) or human GWAS projects. Statistical tools utilised in the annotation pipeline will be made generally available through the web portal and R packages to give expert users of the data flexibility to define the methods they would like to use in their own data analysis.
Core data archive (CDA)
The CDA is the archive for the IMPC data and is coordinated with other resources in the EBI such as Ensembl and the EB-Eye query system (Valentin et al. 2010). Data are transferred from the DCC data staging area after the completion of QC and annotation. Centralisation of the IMPC data in a single resource ensures that the data are preserved and available at a single location for bulk download and that coanalysis can be performed across the entire growing data set as it appears from the phenotyping centres. The CDA architecture contains components for storing or accessing ontologies and genomic and genome variation level, as well as a tracking component for the authoritative source and version for each category of information. This is critical as these sources, e.g., gene models, are updated with successive Ensembl builds. SOLR technology indexes the CDA content and serves this information back to the IMPC portal for complex query. DAS technology (Prlic et al. 2007) is used to access the mouse genome and existing DAS Tracks showing allele and cassette details from IKMC. Ensembl is used as a gene/genome-level integration strategy for EBI users, and phenotype information will be projected as a DAS track and for query via EB-Eye.
IMPC portal
The IMPC portal (http://www.mousephenotype.org) provides a single point of access to all IMPC data for the biomedical and scientific communities and will integrate data from the complete MPI2 infrastructure. This portal is extensible to include data sets from past and future IMPC projects; interested data owners should contact info@mousephenotype.org for discussion. The current implementation already allows users to participate in forums for SOPs, register for genes of interest, perform gene queries to track progress of mouse production and phenotyping, and in the future will include identification of mouse models of interest from phenotypes and models of human disease. The web portal will include tools to access and view the primary data, tools to search the IMPC data integrated with an array of third-party data on mouse gene function, pathway data, data display, and analysis tools. In addition, the data will be made available through a number of programmatic routes such as web services and database dumps. The current functionalities on the site are designed to ensure that mouse biologists can search and view the data because they are envisaged as the primary users. Future plans are to extend the tools, as described above, to widen the user base to clinicians and bioinformaticians as key users of these data.
Contributor Information
Ann-Marie Mallon, Email: a.mallon@har.mrc.ac.uk, Mammalian Genetics Unit, Medical Research Council Harwell, Harwell, Oxfordshire OX11 0RD, UK.
Vivek Iyer, The Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge CB10 1HH, UK.
David Melvin, The Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge CB10 1HH, UK.
Hugh Morgan, Mammalian Genetics Unit, Medical Research Council Harwell, Harwell, Oxfordshire OX11 0RD, UK.
Helen Parkinson, European Bioinformatics Institute, Hinxton, Cambridge CB10 1ST, UK.
Steve D. M. Brown, Mammalian Genetics Unit, Medical Research Council Harwell, Harwell, Oxfordshire OX11 0RD, UK
Paul Flicek, European Bioinformatics Institute, Hinxton, Cambridge CB10 1ST, UK.
William C. Skarnes, The Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge CB10 1HH, UK
References
- Bogue MA, Grubb SC. The Mouse Phenome Project. Genetica. 2004;122:71–74. doi: 10.1007/s10709-004-1438-4. [DOI] [PubMed] [Google Scholar]
- Brown SD, Moore MW. Towards an encyclopaedia of mammalian gene function: the International Mouse Phenotyping Consortium. Dis Model Mech. 2012;5:289–292. doi: 10.1242/dmm.009878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SD, Chambon P, de Angelis MH. EMPReSS: standardized phenotype screens for functional annotation of the mouse genome. Nat Genet. 2005;37:1155. doi: 10.1038/ng1105-1155. [DOI] [PubMed] [Google Scholar]
- Brown SD, Hancock JM, Gates H. Understanding mammalian genetic systems: the challenge of phenotyping in the mouse. PLoS Genet. 2006;2:e118. doi: 10.1371/journal.pgen.0020118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins FS, Rossant J, Wurst W. A mouse for all reasons. Cell. 2007;128:9–13. doi: 10.1016/j.cell.2006.12.018. [DOI] [PubMed] [Google Scholar]
- Flicek P, Amode MR, Barrell D, Beal K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fairley S, Fitzgerald S, Gil L, Gordon L, Hendrix M, Hourlier T, Johnson N, Kahari AK, Keefe D, Keenan S, Kinsella R, Komorowska M, Koscielny G, Kulesha E, Larsson P, Longden I, McLaren W, Muffato M, Overduin B, Pignatelli M, Pritchard B, Riat HS, Ritchie GR, Ruffier M, Schuster M, Sobral D, Tang YA, Taylor K, Trevanion S, Vandrovcova J, White S, Wilson M, Wilder SP, Aken BL, Birney E, Cunningham F, Dunham I, Durbin R, Fernandez-Suarez XM, Harrow J, Herrero J, Hubbard TJ, Parker A, Proctor G, Spudich G, Vogel J, Yates A, Zadissa A, Searle SM. Ensembl 2012. Nucleic Acids Res. 2012;40:D84–D90. doi: 10.1093/nar/gkr991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gkoutos GV, Green EC, Mallon AM, Hancock JM, Davidson D. Using ontologies to describe mouse phenotypes. Genome Biol. 2005;6:R8. doi: 10.1186/gb-2004-6-1-r8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapushesky M, Adamusiak T, Burdett T, Culhane A, Farne A, Filippov A, Holloway E, Klebanov A, Kryvych N, Kurbatova N, Kurnosov P, Malone J, Melnichuk O, Petryszak R, Pultsin N, Rustici G, Tikhonov A, Travillian RS, Williams E, Zorin A, Parkinson H, Brazma A. Gene Expression Atlas update— a value-added database of microarray and sequencing-based functional genomics experiments. Nucleic Acids Res. 2012;40:D1077–D1081. doi: 10.1093/nar/gkr913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasprzyk A. BioMart: driving a paradigm change in biological data management. Database (Oxford) 2011;2011 doi: 10.1093/database/bar049. bar049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallon AM, Blake A, Hancock JM. EuroPhenome and EMPReSS: online mouse phenotyping resource. Nucleic Acids Res. 2008;36:D715–D718. doi: 10.1093/nar/gkm728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone J, Holloway E, Adamusiak T, Kapushesky M, Zheng J, Kolesnikov N, Zhukova A, Brazma A, Parkinson H. Modeling sample variables with an Experimental Factor Ontology. Bioinformatics. 2010;26:1112–1118. doi: 10.1093/bioinformatics/btq099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan H, Beck T, Blake A, Gates H, Adams N, Debouzy G, Leblanc S, Lengger C, Maier H, Melvin D, Meziane H, Richardson D, Wells S, White J, Wood J, de Angelis MH, Brown SD, Hancock JM, Mallon AM. EuroPhenome: a repository for high-throughput mouse phenotyping data. Nucleic Acids Res. 2010;38:D577–D585. doi: 10.1093/nar/gkp1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prlic A, Down TA, Kulesha E, Finn RD, Kahari A, Hubbard TJ. Integrating sequence and structural biology with DAS. BMC Bioinformatics. 2007;8:333. doi: 10.1186/1471-2105-8-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringwald M, Iyer V, Mason JC, Stone KR, Tadepally HD, Kadin JA, Bult CJ, Eppig JT, Oakley DJ, Briois S, Stupka E, Maselli V, Smedley D, Liu S, Hansen J, Baldock R, Hicks GG, Skarnes WC. The IKMC web portal: a central point of entry to data and resources from the International Knockout Mouse Consortium. Nucleic Acids Res. 2011;39:D849–D855. doi: 10.1093/nar/gkq879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skarnes WC, Rosen B, West AP, Koutsourakis M, Bushell W, Iyer V, Mujica AO, Thomas M, Harrow J, Cox T, Jackson D, Severin J, Biggs P, Fu J, Nefedov M, de Jong PJ, Stewart AF, Bradley A. A conditional knockout resource for the genome-wide study of mouse gene function. Nature. 2011;474:337–342. doi: 10.1038/nature10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka N, Waki K, Kaneda H, Suzuki T, Yamada I, Furuse T, Kobayashi K, Motegi H, Toki H, Inoue M, Minowa O, Noda T, Takao K, Miyakawa T, Takahashi A, Koide T, Wakana S, Masuya H. SDOP-DB: a comparative standardized-protocol database for mouse phenotypic analyses. Bioinformatics. 2010;26:1133–1134. doi: 10.1093/bioinformatics/btq095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor CF, Field D, Sansone SA, Aerts J, Apweiler R, Ashburner M, Ball CA, Binz PA, Bogue M, Booth T, Brazma A, Brinkman RR, Michael Clark A, Deutsch EW, Fiehn O, Fostel J, Ghazal P, Gibson F, Gray T, Grimes G, Hancock JM, Hardy NW, Hermjakob H, Julian RK, Jr, Kane M, Kettner C, Kinsinger C, Kolker E, Kuiper M, Le Novere N, Leebens-Mack J, Lewis SE, Lord P, Mallon AM, Marthandan N, Masuya H, McNally R, Mehrle A, Morrison N, Orchard S, Quackenbush J, Reecy JM, Robertson DG, Rocca-Serra P, Rodriguez H, Rosenfelder H, Santoyo-Lopez J, Scheuermann RH, Schober D, Smith B, Snape J, Stoeckert CJ, Jr, Tipton K, Sterk P, Untergasser A, Vandesompele J, Wiemann S. Promoting coherent minimum reporting guidelines for biological and biomedical investigations: the MIBBI project. Nat Biotechnol. 2008;26:889–896. doi: 10.1038/nbt.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentin F, Squizzato S, Goujon M, McWilliam H, Paern J, Lopez R. Fast and efficient searching of biological data resources— using EB-eye. Brief Bioinform. 2010;11:375–384. doi: 10.1093/bib/bbp065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson P, Sengerova J, Matteoni R, Chen CK, Soulat G, Ureta-Vidal A, Fessele S, Hagn M, Massimi M, Pickford K, Butler RH, Marschall S, Mallon AM, Pickard A, Raspa M, Scavizzi F, Fray M, Larrigaldie V, Leyritz J, Birney E, Tocchini-Valentini GP, Brown S, Herault Y, Montoliu L, de Angelis MH, Smedley D. EMMA—mouse mutant resources for the international scientific community. Nucleic Acids Res. 2010;38:D570–D576. doi: 10.1093/nar/gkp799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng-Bradley X, Rung J, Parkinson H, Brazma A. Large scale comparison of global gene expression patterns in human and mouse. Genome Biol. 2010;11 doi: 10.1186/gb-2010-11-12-r124. R124. [DOI] [PMC free article] [PubMed] [Google Scholar]