Abstract
Amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD) have distinct clinical features but a common pathology—cytoplasmic inclusions rich in TDP43. Rare TDP43 mutations cause ALS or FTD, but abnormal TDP43 levels and localization may cause disease even if TDP43 lacks a mutation. Here we showed that individual neurons vary in their ability to clear TDP43 and are exquisitely sensitive to TDP43 levels. To measure TDP43 clearance, we developed and validated a single-cell optical method that overcomes the confounding effects of aggregation and toxicity, and discovered that pathogenic mutations significantly shorten TDP43 half-life. Novel compounds that stimulate autophagy improved TDP43 clearance and localization, and enhanced survival in primary murine neurons and in human stem cell–derived neurons and astrocytes harboring mutant TDP43. These findings indicate that the levels and localization of TDP43 critically determine neurotoxicity and show that autophagy induction mitigates neurodegeneration by acting directly on TDP43 clearance.
INTRODUCTION
ALS and FTD are fundamentally connected1. Neuronal and glial cytoplasmic accumulations of TDP43 (transactive response element DNA-binding protein of 43 kDa) occur in most ALS cases and the most common form of FTD (FTLD-TDP)2, and mutations in several genes cause ALS, FTD, or both1. TDP43 is a nuclear RNA-binding protein involved in several aspects of RNA processing3 that actively shuttles between the nucleus and cytoplasm4. In ALS and FTLD-TDP, TDP43 is excluded from the nucleus2, but such cytoplasmic mislocalization is common in neuronal injury or stress5,6, and TDP43-positive inclusions may represent secondary pathology in some neurodegenerative disorders7.
Over 30 mutations in the TDP43 gene (TARDBP) cause familial ALS (fALS) and FTLD-TDP8. We previously demonstrated that TARDBP mutations promote cytoplasmic TDP43 mislocalization and that mutant TDP43-induced neurodegeneration could be explained by cytoplasmic TDP43 toxicity9. Cytoplasmic accumulations of wild-type (WT) TDP43 are pathognomonic for sporadic ALS (sALS) and FTLD-TDP2, and elevated WT TDP43 levels are neurotoxic in multiple systems10,11, linking abnormal protein homeostasis with sALS and fALS. Despite the potential importance of TDP43 metabolism to neuronal survival, prior studies were limited to cell lines or non-neuronal cell types12,13. Could neuron-specific differences in protein metabolism14,15 underlie their susceptibility to TDP43-dependent pathology?
TDP43 accumulates when the ubiquitin-proteasome system (UPS) or autophagy is inhibited16,17, but it is unclear which is primarily responsible for clearing TDP43. Conflicting data suggest that autophagy accelerates or slows disease progression: although autophagy induction with rapamycin exacerbated neurodegeneration in transgenic ALS mice18, genetically upregulating autophagy enhanced survival in the same animals19. Rapamycin was beneficial in FTLD-TDP transgenic mice20, but despite the effects of this and other mTOR (mammalian target of rapamycin) inhibitors in non-neuronal cells, their ability to induce neuronal autophagy per se is limited14,21,22, and their off-target effects might affect disease activity in an autophagy-independent manner23. Careful pharmacodynamic studies confirming the autophagy inducing effects of rapamycin and its derivatives in the central nervous system (CNS) are often lacking or, when performed, fail to demonstrate autophagy induction21. In the few cases where autophagy has been clearly induced, the mechanism by which this pathway affects disease pathogenesis remains obscure, since autophagy has broad effects on mitochondrial turnover, energy balance, and global protein catabolism23.
Using a structure-activity assay, we assembled a neuronal autophagy-inducing pharmacophore, used it to discover novel autophagy inducers and then investigated the therapeutic potential of autophagy induction in a neuronal model of ALS. Stimulating neuronal autophagy enhanced TDP43 clearance and improved neuronal survival, and the changes in TDP43 turnover explained much of the survival benefit of autophagy induction, providing mechanistic insight into how autophagy mitigates TDP43-induced neurodegeneration.
RESULTS
TDP43 toxicity is dose-dependent
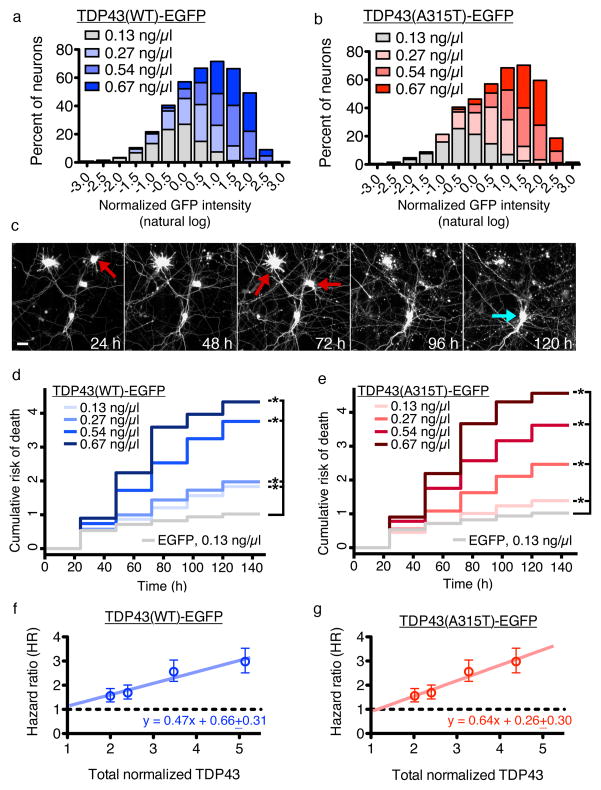
We fused WT and ALS-associated mutant (A315T) TDP4324 to enhanced green fluorescent protein (EGFP) and transfected primary rodent cortical neurons with different amounts of construct and mApple, a survival and morphology marker (Supplementary Results, Supplementary Fig. 1). To determine TDP43-EGFP levels for each neuron, we measured single-cell fluorescence by automated fluorescence microscopy (AFM)9,25. Transient transfection with 0.13 ng/μl DNA gave a broad distribution of expression levels (Fig. 1a, b). Increasing the amount of DNA right-shifted the distribution, with more cells exhibiting higher TDP43-EGFP levels. We thus created specific populations of neurons expressing WT or mutant TDP43-EGFP at defined “doses.”
Figure 1. The toxicity of TDP43 depends strongly on dose.
(a, b) Histograms of TDP43(WT)-EGFP and TDP43(A315T)-EGFP levels in individual neurons. Increasing amounts of DNA significantly shifted the distribution of log-transformed expression levels towards higher values (p < 0.01 for all comparisons, two-sided Kolmogorov-Smirnov test). TDP43(WT)-EGFP, n=1287 neurons, with 290–342 cells per group; TDP43(A315T)-EGFP, n=1290, with 315–332 neurons per group. Values pooled from 12 wells per condition, performed in duplicate. (c) Automated fluorescence microscopy. Primary rodent cortical neurons were transfected with mApple and TDP43-EGFP, and survival was determined by repeated imaging at regular intervals. The last time at which the cell was noted to be alive (red arrows) was used as the time of death. Cells that survive the entire length of the experiment (blue arrow) were censored. Scale bar = 25 μm. (d, e) Cumulative risk of death over time for neurons transfected with EGFP, TDP43(WT)-EGFP or TDP43(A315T)-EGFP. Supplementary Table 1 lists the number of neurons, hazard ratios, 95% CI, and p values for each condition, as determined by Cox proportional hazards analysis. Results pooled from 12 wells per condition, performed in duplicate. (f, g) Linear regression of calculated hazard ratio (± SEM) as a function of total TDP43(WT)-EGFP (R2 = 0.9353) or TDP43(A315T)-EGFP (R2 = 0.9574) level. The total TDP43 level was determined by quantitative immunocytochemistry using TDP43-specific antibodies (Supplementary Fig. 3) and normalized to the amount of TDP43 in non-transfected neurons. Dotted lines, reference (HR = 1).
What is the relationship between TDP43 dose and survival? To answer this, we determined the risk of death for neurons expressing WT and mutant TDP43-EGFP by longitudinal AFM (Fig. 1c). AFM measures the lifespan of individual neurons and gauges the impact of pre-specified factors on the risk of death. Criteria for cell death in these assays—loss of mApple fluorescence or disruption of cell integrity—are more sensitive than conventional measures25. We calculated risk of death for each population of neurons through Cox hazards analysis, resulting in a hazard ratio (HR) analogous to relative risk in human clinical trials. Risk of death increased steadily with levels of TDP43(WT)-EGFP and TDP43(A315T)-EGFP (Fig. 1d, e, Supplementary Table 1). WT and mutant TDP43 increased risk of death by 25–50% at low doses and ~300% at higher levels. For a given expression level, TDP43 was more toxic than other disease-associated proteins (data not shown), including fused-in-sarcoma (FUS) in fALS1 or mutant huntingtin (Htt) in Huntington’s disease (HD)25. In contrast, we noted no relationship between neuronal survival and EGFP levels (Supplementary Fig. 2a), suggesting a specific dose-dependent toxicity of TDP43.
We next used immunocytochemistry to determine total TDP43 levels in transfected neurons (Supplementary Fig. 3a–e). Neurons receiving 0.13 ng/μl DNA exhibited twice the endogenous TDP43 (en-TDP43) level, and those given 0.67 ng/μl DNA fivefold more. We observed downregulation of en-TDP43 in several neurons (Supplementary Fig. 3f), but downregulation and survival were unrelated. For instance, downregulation was most efficient at the lowest “dose” of TDP43(WT)-EGFP (0.13 ng/μl, Supplementary Fig. 3f), but toxicity was maximal at the highest (0.67 ng/μl, Fig. 1d), consistent with in vivo studies showing no relation between en-TDP43 levels and disease26. In contrast, HR and TDP43 levels were linearly related (Fig. 1f, g): risk of death increased 47% for every fold-change in TDP43(WT)-EGFP levels, and 64% for every fold-change in TDP43(A315T)-EGFP levels. Thus, even small increases in WT and mutant TDP43 can drive toxicity.
Measuring TDP43 clearance in situ
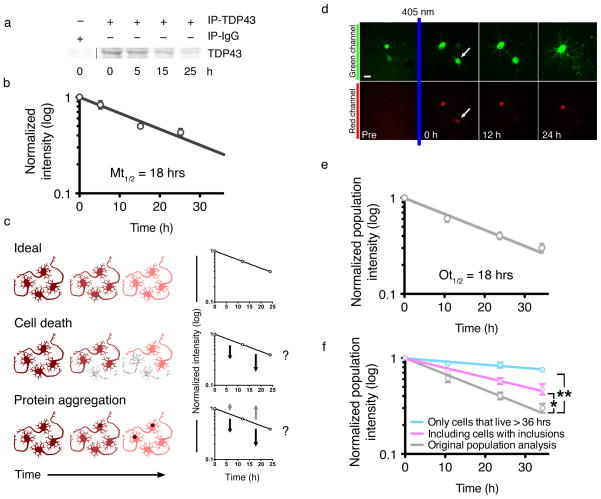
If TDP43 levels are directly proportional to neurodegeneration, augmenting TDP43 clearance might have therapeutic potential. The half-life of en-TDP43 in primary rodent cortical neurons by metabolic pulse chase (MPC) was 18 h (95% CI, 13.4–29.3 h; Fig. 2a, b; Supplementary Fig. 4a), significantly longer than in fibroblasts or immortalized cell lines (4–12 h)12,13. Is this discrepancy due to cell-specific differences in TDP43 stability or an artifact of the method used to measure half-life?
Figure 2. TDP43 turnover in primary neurons.
(a) Primary neurons were labeled with 35S-labeled methionine, and endogenous TDP43 was immunoprecipitated using anti-TDP43 antibodies (IP-TDP43) or non-specific IgG antibodies (IP-IgG). (b) Log-normal plot of normalized TDP43 levels. TDP43 half-life was determined by fitting a first-order exponential curve to the data (R2 = 0.8984). Values pooled from five independent experiments. (c) Potential confounders in half-life analyses. Ideally (top), total protein decreases over time in a predictable manner (top right panel). Toxicity (middle) reduces the amount of measured protein, potentially shortening the calculated half-life (middle right panel). Protein aggregation (bottom) might also shorten half-life estimations if the protein cannot be immunoprecipitated (dark arrows, bottom right panel), or lengthen it if the protein is more stable and successfully immunoprecipitated (grey arrows, bottom right panel). (d) Optical pulse labeling. Primary cortical neurons were transfected with EGFP and TDP43(WT)-Dendra2, photoconverted then imaged by AFM. Dead cells (arrow) were excluded at the time of death. (e) The half-life of TDP43(WT)-Dendra2 was determined by fitting a first-order exponential curve to the data (R2 0.9558). (f) Including cells with aggregates prolonged TDP43(WT)-Dendra2 half-life (purple line, R2 = 0.9003; * p < 0.0001, F 15.34, extra sum-of-squares F-test). Excluding cells that died over the 36 h experiment also prolonged half-life (blue line, R2 = 0.7171; ** p < 0.0001, extra sum-of-squares F-test), suggesting that toxicity reduces measured half-life. Error bars represent ± SEM. Values pooled from eight wells per condition, performed in quadruplicate.
MPC is a population-based assay and therefore susceptible to several potential confounds, including protein aggregation and cell death (Fig. 2c). To circumvent these, we developed a complementary in situ method for measuring TDP43 half-life (Fig. 2d; Supplementary Fig. 4b). Optical pulse labeling (OPL) uses photoconvertible proteins, such as Dendra227, that irreversibly switch their spectral properties when exposed to short-wavelength light. We tagged WT TDP43 with Dendra2 and expressed the construct in rodent primary cortical neurons, then photoconverted with 405-nm light. Neurons were imaged by AFM, and the data analyzed in a population-based manner, as with MPC. Neurons were prospectively tracked and excluded after an aggregate was identified, and single-cell red fluorescence intensities were summed to generate a population total and normalized to the total immediately after photoconversion. TDP43(WT)-Dendra2 half-life by OPL (18 h, 95% CI 15.1–23.4 h; Fig. 2e) was similar to en-TDP43 half-life measured by MPC (Fig. 2b). These results have two important implications. First, overexpression and Dendra2 fusion do not affect TDP43 turnover. Second, OPL and MPC provided the same estimate of TDP43 half-life when the data are analyzed in an equivalent fashion, suggesting both techniques measure the same biological processes acting on TDP43.
But do the results accurately reflect TDP43 clearance? At least two potential confounds are relevant to these measurements—cellular toxicity and protein aggregation (Fig. 2c). However, their respective contributions have previously been impossible to determine. With OPL, for the first time, we can directly measure the contributions of each to half-life estimations. For protein aggregation, we analyzed all cells, regardless of whether they exhibited visible inclusions. In doing so, TDP43(WT)-Dendra2 half-life increased from 18 to 31 h (Fig. 2f), indicating that TDP43 half-life in aggregates is different from that of diffuse TDP43, which would confound population-based estimates of protein half-life. We next removed from the analysis cells that died and therefore could have biased half-life calculations. When we did so, TDP43(WT)-Dendra2 half-life was significantly longer (Fig. 2f). Thus, toxicity also affects measurements of protein half-life, artificially shortening values by eliminating cells before half-life can be measured. Alternatively, neurons displaying prolonged TDP43(WT)-Dendra2 half-life might live longer. In single-cell analyses, however, we detected no relationship between TDP43(WT)-Dendra2 half-life and survival (data not shown). Therefore, cell death most likely confounds population-based estimates of protein clearance by prematurely excluding cells from analysis; this is particularly evident with proteins, such as TDP43, whose levels are closely tied to survival.
Pathogenic mutations enhance TDP43 turnover
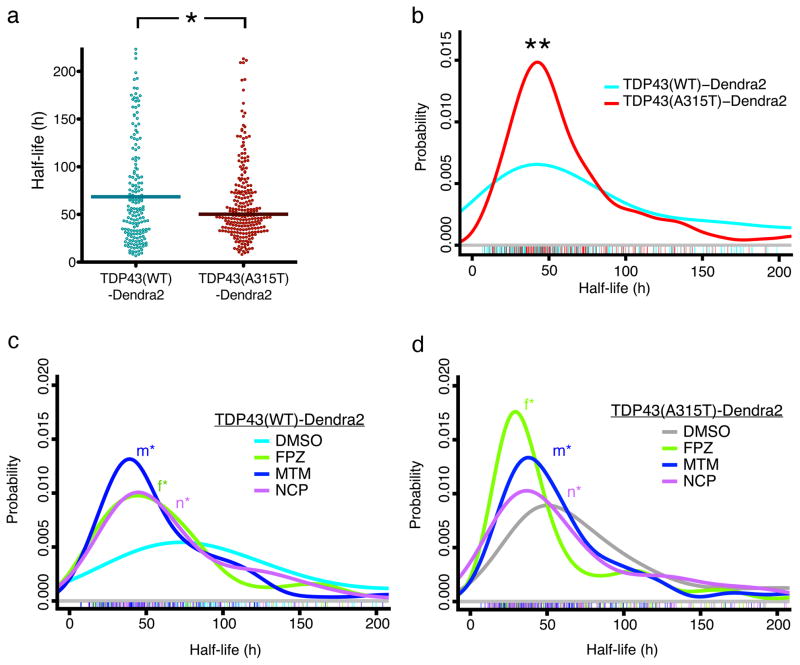
We next used OPL to visualize neuronal TDP43 clearance. Neurons expressing TDP43(WT)-Dendra2 were photoconverted and tracked until an inclusion developed or cell death. For each neuron, we measured the red fluorescence of TDP43(WT)-Dendra2 immediately after photoconversion and used each cell as its own control to normalize subsequent measurements. Individual neurons exhibited marked variability in TDP43(WT)-Dendra2 half-life (Fig. 3a) that was independent of expression (Supplementary Fig. 4c–e). TDP43(WT)-Dendra2 half-life was significantly shorter by single-cell analyses, in comparison to population-based methods (68 vs 18 h; p < 0.0001, F = 96.75, extra sum-of-squares F-test). Therefore, in the absence of confounding from toxicity and aggregation, TDP43(WT)-Dendra2 half-life is substantially greater than previously estimated.
Figure 3. Optical pulse labeling of TDP43.
(a) Scatter plot depicting half-life measurements for individual cells expressing TDP43(WT)-Dendra2 (cyan, n=206) and TDP43(A315T)-Dendra2 (red, n=251). * p=0.0029, U = 11797, Mann-Whitney test. (b) Probability density plot for single-cell half-life measurements, demonstrating a reduced half-life of TDP43(A315T)-Dendra2 (red, n=251), compared to TDP43(WT)-Dendra2 (cyan, n=206; ** p=0.0007, two-sided Kolmogorov-Smirnov test). Values were pooled from eight wells per condition, performed in quadruplicate. Probability density plots of single-cell half-life measurements for TDP43(WT)-Dendra2 (c) and TDP43(A315T)-Dendra2 (d). At 0.5 μM, all three compounds significantly reduced TDP43(WT)-Dendra2 half-life, compared to vehicle control (DMSO, n=110; FPZ, n=96, *f p=1×10−4; MTM, n=129, *m p=2×10−7; NCP, n=111, *n p=2×10−5, two-sided Kolmogorov-Smirnov test). The compounds also reduced TDP43(A315T)-Dendra2 half-life, in comparison to vehicle control (DMSO, n=120; FPZ, n=155, *f p=2×10−9; MTM, n=152, *m p=0.004; NCP, n=111, *n p=0.005, two-sided Kolmogorov-Smirnov test). Colored hash marks represent values from individual neurons in (b), (c) and (d). Values were obtained from eight wells per condition, performed in triplicate.
We next used OPL to determine if pathogenic mutations affect TDP43 clearance. The median TDP43(A315T)-Dendra2 half-life (50 h) was shorter than that of TDP43(WT)-Dendra2 (Fig. 3a), suggesting more rapid clearance of the mutant protein. We also analyzed single-cell half-life using a probability density function, confirming the effect of the A315T mutation on TDP43-Dendra2 half-life (Fig. 3b–d). We also used OPL to measure the half-life of TDP43-Dendra2 carrying a separate ALS-associated mutation, M337V8 (Supplementary Fig. 5a) and found that this mutation also reduced TDP43-Dendra2 half-life. Expression of mutant TDP43 might trigger global changes in protein catabolism, but we found no difference in the half-life of unrelated, Dendra2-fused proteins in neurons expressing WT or mutant TDP43 (data not shown). Therefore, our observations strongly suggest that pathogenic TARDBP mutations enhance TDP43 turnover.
Induction of autophagy increases TDP43 clearance
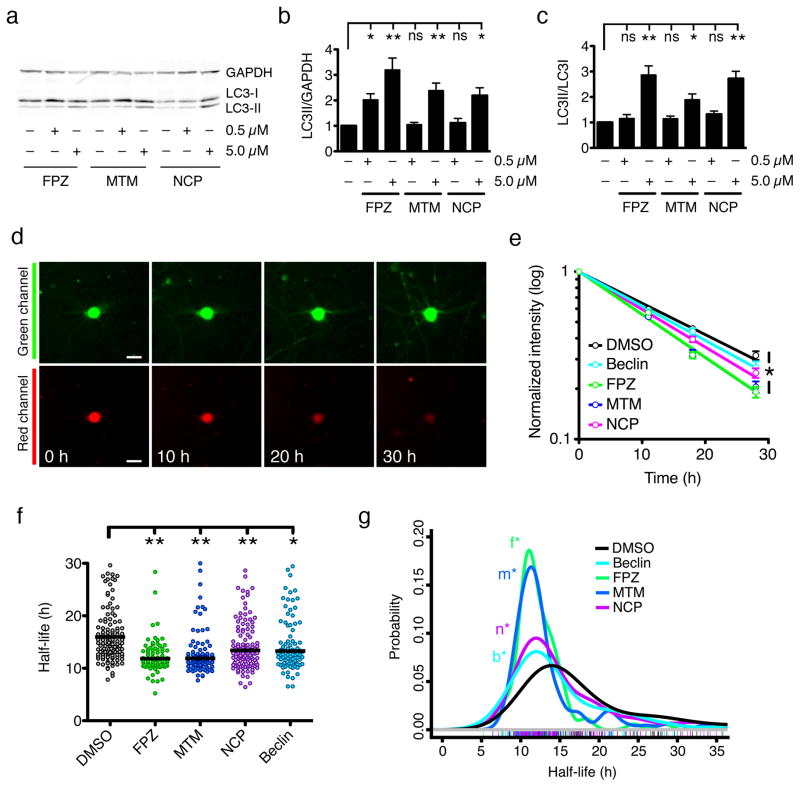
We chose to stimulate TDP43 clearance through autophagy because this pathway degrades long-lived cytoplasmic proteins and organelles28, and cyotoplasmic accumulation of TDP43 is toxic9. Using a neuronal autophagy-inducing pharmacophore22, we screened an in silico library of over 1 million compounds for more potent autophagy stimulators (see Online Methods). The top two candidates from this screen—fluphenazine (FPZ, 1) and methotrimeprazine (MTM, 2)—and the lead compound 10-(4′-(N-diethylamino)butyl)-2-chlorophenoxazine (NCP, 3) were selected for study. We previously demonstrated that NCP effectively induces neuronal autophagy22. Here, all three compounds significantly increased levels of LC3-II, the lipidated isoform of LC3-I (light chain 3 isoform I) that is incorporated into developing autophagosomes29, and the ratio of LC3-II to LC3-I in primary rodent cortical neurons (Fig. 4a–c; Supplementary Fig. 6a), confirming their efficacy in stimulating autophagy. All compounds also stimulated LC3-positive puncta formation and enhanced LC3-II/LC3-I ratios in the presence of NH4Cl, an alkalinizing agent that prevents autophagosome maturation29 (Supplementary Fig. 6b–i), verifying autophagy induction. We reliably detected such effects at 5 μM, but not 0.5 μM, suggesting a dose-dependent response, limited assay sensitivity, or both.
Figure 4. Induction of autophagy by a family of small molecules.
(a). Each compound significantly increased the LC3-II/GAPDH (b) and LC3-II/LC3-I (c) ratios at 5 μM, but not 0.5 μM [ns, p > 0.05; * 0.05 < p < 0.01, ** p < 0.01; F = 13.74 (b) and 9.74 (c), one-way ANOVA with Dunnett’s post-test]. Error bars represent ± SEM. Values pooled from five independent experiments. (d) Optical pulse-labeling of LC3-Dendra2 in rodent primary cortical neurons. Scale bar = 25 μm. (e) The change in red fluorescence intensity over time was used to calculate LC3-Dendra2 half-life, as in Fig. 2 (DMSO, R2 = 0.7333; Beclin, R2 = 0.6968, FPZ, R2 = 0.8752; MTM, R2 = 0.8447; NCP, R2 = 0.8357; * p < 0.0001 for global trend, F = 12.6, extra sum-of-squares F-test). Error bars represent ± SEM. (f) The median single-cell half-life of LC3-Dendra2 was significantly reduced by beclin expression or treatment with 0.5 μM NCP, MTM, and FPZ (* p < 0.01, ** p < 0.001, Kruskal Wallis statistic = 52.56, Kruskal-Wallis with Dunn’s test). (g) Probability density plot of LC3-Dendra2 half-life measurements from individual neurons treated with vehicle control (DMSO, n=140), NCP (n=128), MTM (n=91), or FPZ (n=77), or those expressing beclin (n=101). *n, p=6×10−4; *m, p=1×10−10; *f, p=5×10−9; *b, p=2×10−3; two-sided Kolmogorov-Smirnov test). Colored hash marks represent values from individual neurons. Values pooled from eight wells per condition, performed in triplicate.
The limitations of these techniques for demonstrating autophagic induction are myriad29. Fundamentally, they depend on inferring autophagic flux from changes in levels of intermediates under specific conditions. To accurately measure flux, the rate of protein breakdown must be measured directly for an autophagy substrate. We therefore developed an in situ assay of autophagy flux by fusing LC3 to Dendra2, expressing LC3-Dendra2 in primary rodent cortical neurons (Fig. 4d) and measuring the turnover of LC3-Dendra2 through OPL. First, we used this assay to investigate autophagic flux in response to NCP, MTM, and FPZ. As a positive control, we cotransfected neurons with beclin, an autophagy upregulator30. Beclin significantly reduced median LC3-Dendra2 half-life, as did NCP, MTM, and FPZ (Fig. 4e, f). In single-cell analyses (Fig. 4g), we noted that 0.5 μM of each compound, 1/10 the dose required in conventional assays (Fig. 4a–c), effectively shortened LC3-Dendra2 half-life, suggesting OPL is substantially more sensitive than traditional measures. To ensure the compounds are stimulating autophagy and not acting through off-target pathways, we measured the effects of the compounds in cells depleted of the essential autophagy protein Atg731 (Supplementary Fig. 6j). As expected, Atg7 knockdown prolonged LC3-Dendra2 half-life. No compound reduced LC3-Dendra2 half-life in Atg7-knockdown neurons, indicating that their effects on LC3-Dendra2 flux are mediated specifically through autophagy.
In population-based approaches (Fig. 4e, f), FPZ and MTM appeared to affect autophagic flux similarly. However, in single-cell analyses (Fig. 4g), subtle differences were evident: we noted a slightly higher “peak” in FPZ-treated neurons, and less of a “shoulder” representing neurons with relatively long LC3-Dendra2 half-lives. These observations suggest that FPZ is a more potent autophagy inducer, and illustrate the advantages of single-cell approaches over traditional methods for evaluating target efficacy.
We next asked whether the compounds enhance TDP43 clearance. Primary neurons expressing WT and mutant TDP43-Dendra2 were treated with vehicle (DMSO) or 0.5 μM of each compound, and half-life was determined through OPL. Probability density plots demonstrated effective reduction in TDP43-Dendra2 half-life with each compound (Fig. 3c, d). Thus, all three compounds significantly augment WT and mutant TDP43-Dendra2 clearance in primary neurons by inducing autophagy.
Autophagy induction improves TDP43 metabolism
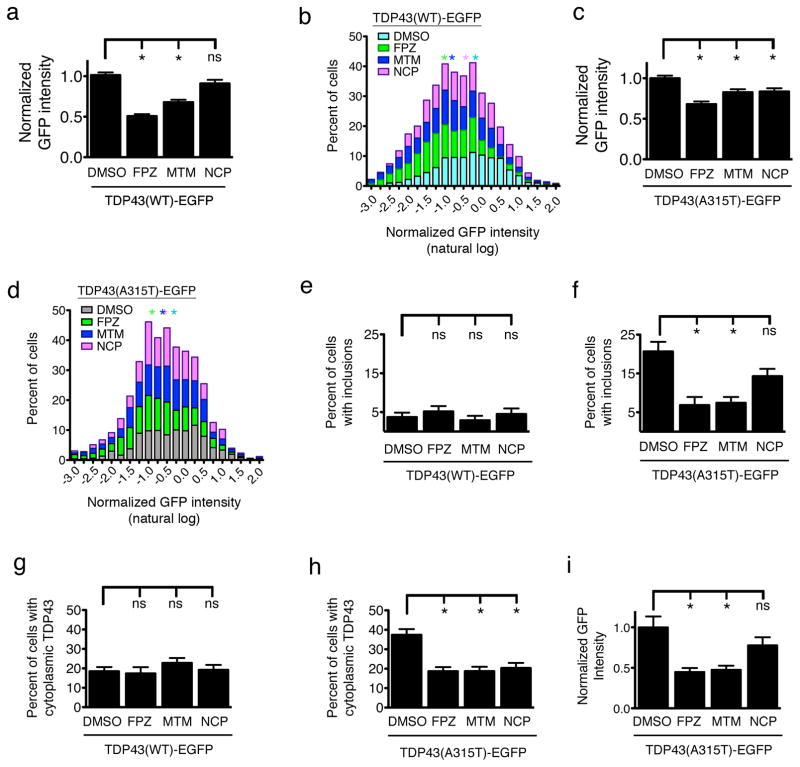
Does enhancing TDP43 turnover affect TDP43 levels, subcellular localization, or aggregation? To answer this question, we expressed WT or mutant TDP43-EGFP in primary neurons, treated the cultures with vehicle or 0.5 μM of each compound, and imaged the neurons by AFM. Because TDP43 shuttles between the nucleus and cytoplasm4, augmenting TDP43 turnover within the cytoplasm through autophagy should lower whole-cell TDP43 levels. FPZ and MTM, but not NCP, significantly decreased TDP43(WT)-EGFP levels (Fig. 5a) and shifted the distribution of expression to the left (Fig. 5b), consistent with their effects on protein half-life (Fig. 3c). All three also lowered whole-cell TDP43(A315T)-EGFP levels (Fig. 5c) and left-shifted the TDP43(A315T)-EGFP expression distribution (Fig. 5d). Very few neurons displayed TDP43(WT)-EGFP inclusions (Fig. 5e; Supplementary Fig. 1). However, almost 20% of neurons expressing TDP43(A315T)-EGFP developed inclusions by 48 h (Fig. 5f), a figure significantly reduced by FPZ and MTM, but not NCP. FPZ and MTM were more effective than NCP in stimulating autophagy (Fig. 4), implying that the observed reductions in TDP43-EGFP levels and aggregation are proportional to autophagy induction.
Figure 5. Autophagy induction reduces TDP43 levels, aggregation, and cytoplasmic mislocalization.
(a) FPZ (n=490) and MTM (n=580), but not NCP (n=523), lowered total TDP43(WT)-EGFP levels in comparison to vehicle (DMSO, n=1007). (b) Histogram of normalized, log-transformed TDP43(WT)-EGFP levels. (c) Each compound also reduced TDP43(A315T)-EGFP levels (DMSO, n=983; FPZ, n=550; MTM, n=542; NCP, n=493). (d) Histogram of single-cell TDP43(A315T)-EGFP levels. Asterisks in (b) and (d) denote median values for cells exposed to vehicle (cyan), FPZ (green), MTM (blue) or NPZ (purple). For FPZ, MTM, NCP vs. DMSO in (a) and (d), p < 1×10−3 by two-sided Kolmogorov-Smirnov test. (e) No compound reduced TDP43(WT)-EGFP inclusions 48 h after transfection (DMSO, n=360; FPZ, n=194; MTM, n=234; NCP, n=203). (f) FPZ and MTM, but not NCP, reduced TDP43(A315T)-EGFP inclusions at 48 h (DMSO, n=327; FPZ, n=226; MTM, n=262; NCP, n=253). (g) No compound affected TDP43(WT)-EGFP localization 24 h after transfection (DMSO, n=256; FPZ, n=212; MTM, n=250; NCP, n=241). Each compound (h) prevented TDP43(A315T)-EGFP cytoplasmic mislocalization (DMSO, n=233; FPZ, n=268; MTM, n=263; NCP, n=270) and (i) reduced cytoplasmic TDP43(A315T)-EGFP levels (DMSO, n=125; FPZ, n=102; MTM, n=121; NCP, n=144. * p < 0.05; ns, p > 0.05). For (a), (c), and (e–i), * p < 0.05; ns, p > 0.05, one-way ANOVA with Dunnett’s test. Error bars represent ± SEM. Values in (a–d) pooled from 8–12 wells per condition, from six experiments; values in (e–h) pooled from eight wells per condition, in triplicate; values in (i) pooled from five wells per condition, in duplicate.
In agreement with our prior observations9, the A315T mutation increased the proportion of neurons with cytoplasmic TDP43-EGFP (Fig. 5g, h). We surmised that stimulating autophagy might also reduce cytoplasmic TDP43-EGFP mislocalization. Autophagy induction effectively reduced the proportion of neurons with cytoplasmic TDP43(A315T)-EGFP, without changing the localization of TDP43(WT)-EGFP. We also noted a reduction of cytoplasmic TDP43(A315T)-EGFP levels with FPZ and MTM, but not NCP (Fig. 5i), mirroring the compounds’ respective abilities to shorten TDP43(A315T)-EGFP half-life (Fig. 3d). These data provide strong evidence that autophagic stimulation lowers WT and mutant TDP43 levels, and reduces mutant TDP43 cytoplasmic mislocalization and aggregation.
Autophagy induction prevents TDP43-mediated cell death
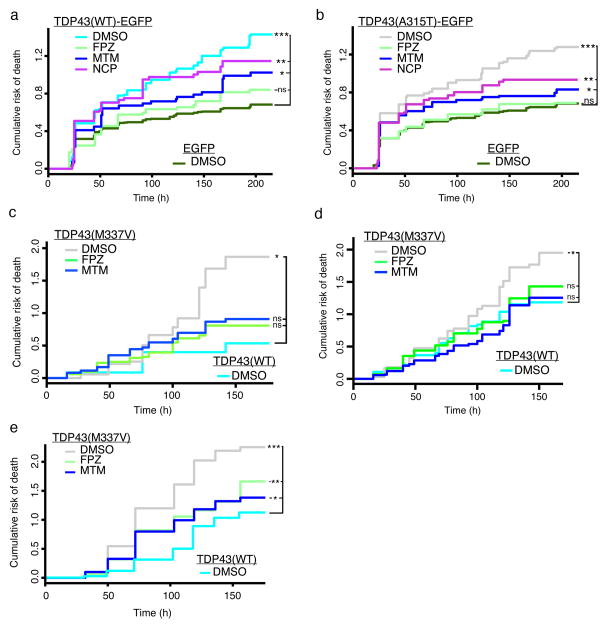
If TDP43 levels and mislocalization are proximal drivers of toxicity, and autophagy induction affects both factors, autophagy induction should improve survival. To test this, we expressed WT and mutant TDP43-EGFP at two- to threefold endogenous levels in primary neurons and imaged cells by AFM (Fig. 6a, b). At 0.5 μM, FPZ and MTM but not NCP significantly improved survival (Supplementary Table 2). Neither FPZ nor MTM affected the survival of control neurons expressing EGFP alone (Supplementary Fig. 2b–e). FPZ also improved survival in neurons expressing a different TDP43 mutant, TDP43(M337V)-EGFP (Supplementary Fig. 5b). We attribute the ineffectiveness of NCP to off-target, compound-specific toxicity that likely counteracts any protective effects mediated by autophagy induction (Supplementary Fig. 2c).
Figure 6. Autophagic stimulation improves survival in neuronal and astrocyte models of ALS.
Primary neurons transfected with TDP43(WT)-EGFP (a) or TDP43(A315T)-EGFP (b) were treated with autophagy inducers and survival determined by AFM. Data obtained from eight wells per condition, performed in triplicate. See Supplementary Table 2 for the number of neurons per condition, hazard ratios, 95% CI, and p values as determined by Cox hazards analysis. (c) Autophagic induction improved survival in TDP43 M337V HB9-positive human iPSC-derived MNs. (d) FPZ and MTM also reduced the risk of death in MAP2-positive human iPSC-derived MNs Values in (c) and (d) pooled from 18 wells per condition, performed in triplicate. (e) Astrocytes differentiated from human iPSCs exhibit mutant TDP43-related toxicity that is mitigated by FPZ and MTM. Values pooled from 18 wells per condition, performed in triplicate. See Supplementary Table 3 for the number of human iPSC-derived neurons and astrocytes per condition, hazard ratios, 95% CI, and p values as determined by Cox hazards analysis.
Can the benefits of autophagy induction be explained through its effects on TDP43 levels or localization? FPZ lowered TDP43(WT)-EGFP levels by 50% and TDP43(A315T)-EGFP levels by 32% (Fig. 5a–d). According to dose-response plots (Fig. 1f, g), a 50% reduction in WT TDP43 levels should lower the experimental HR from 1.9 to 1.28, for a predicted HR of 0.67. The actual HR for the FPZ-treated group was 0.60 (Fig. 6a), suggesting the beneficial effect of autophagic induction can be explained by proportional reduction in WT TDP43 levels. For mutant TDP43, a 32% decrease in levels should lower the experimental HR from 1.8 to 1.31, resulting in a predicted HR of 0.73. Since the actual HR (0.60) for neurons treated with FPZ is better than predicted (0.73; Fig. 6b), stimulating autophagy likely has additional beneficial effects; for instance, autophagy induction also prevents mutant TDP43 mislocalization and reduces cytoplasmic TDP43 levels (Fig. 5h,i), which we reported to be neurotoxic9.
For autophagy inducers to be valid ALS therapies, they must mitigate neurodegeneration in human neurons and astrocytes. We therefore tested the two most potent autophagy inducers, MTM and FPZ, in human stem cell–based ALS models. In prior studies, motor neurons (MNs) derived from human induced pluripotent stem cells (iPSCs) carrying a pathogenic TARDBP mutation (M337V) exhibited a greater risk of death than WT neurons32. Does stimulating autophagy prevent mutant TDP43-mediated toxicity in human MNs? To answer this question, we differentiated MNs from WT and M337V iPSCs32 and introduced a MN-specific survival marker driven by the HB9 promoter33. Cultures were treated with vehicle or 0.5 μM of each compound, and neuronal survival was determined by AFM (Fig. 6c). As expected, M337V-TDP43 MNs displayed a significantly greater risk of death than WT TDP43 MNs (Supplementary Table 3). MTM and FPZ reduced the risk of death for M337V-TDP43 MNs, without affecting the survival of WT TDP43 MNs (Supplementary Fig. 7a). Although ALS typically begins with MN dysfunction and loss, it evolves into a multisystem disorder affecting several different neuronal subtypes34. Therefore, we also evaluated the compounds in iPSC-derived neurons expressing a pan-neuronal MAP2 survival marker35 (Fig. 6d). As with HB9-positive MNs, MTM and FPZ reduced risk of death in M337V-TDP43 MAP2-positive neurons without causing toxicity in WT iPSC-derived neurons (Supplementary Fig. 7b).
Astrocytes are integrally involved in ALS onset and progression36. We reported an increased risk of death in human astrocytes harboring TDP43(M337V)37, indicative of intrinsic toxicity related to pathogenic TARDBP mutations. To determine if autophagy induction is beneficial in human M337V-TDP43 astrocytes, we differentiated WT and mutant iPSCs into astrocytes, transfected them with mApple, and determined time of death by AFM (Fig. 6e, Supplementary Table 3). M337V-TDP43 astrocytes displayed an elevated risk of death that was effectively diminished by 0.5 μM FPZ and MTM. Neither compound affected the survival of WT TDP43 astrocytes (Supplementary Fig. 7c). Inducing autophagy is thus an effective and safe strategy for reducing mutant TDP43-associated toxicity in human neurons and astrocytes.
DISCUSSION
Here, we measured protein turnover in situ by OPL, and showed that, in comparison to conventional methods, OPL is particularly advantageous for investigations of neurodegenerative diseases, where the implicated proteins are toxic and prone to misfolding. We also established a novel autophagic flux assay that can be applied on a single-cell basis, and through an in silico screen identified compounds that effectively stimulate autophagy in neurons, enhance TDP43 clearance and reduce its mislocalization. Finally, and perhaps most importantly, we demonstrated that autophagy induction improves survival in human iPSC-derived neurons and astrocytes from patients with familial ALS, the most genetically precise disease models currently available.
Fueled by autophagy’s broad therapeutic potential, interest in autophagy has grown exponentially. Still, the lack of high-throughput screening assays limits the discovery and development of autophagy inducers as therapies29. Here, we designed and validated a live-cell autophagic flux assay that is uniquely suited to high-throughput and high-content screening: it is robust, quantitiative, and adaptable to any cell type. LC3 clearance in this assay was tightly correlated with autophagic flux, and although definitive proof of this will require equally sensitive measures of autophagic maturation, the assay represents a significant advance over prior screening methods, and we expect it to facilitate the identification of novel autophagy-inducing agents.
Regardless of the model system, the most significant factor in determining neuronal survival is the steady-state level of TDP43—elevated levels are invariably associated with more aggressive neurodegeneration10,11. In sALS, WT TDP43 accumulates within the neuronal cytoplasm2, indicating that protein levels and localization may be primary drivers of neurodegeneration not only in fALS—when they are precipitated by TARDBP mutations—but also in sALS. Our data show that both WT and mutant TDP43 levels are linearly related to toxicity, with nearly identical slopes, suggesting that ALS-associated TARDBP mutations might cause toxicity by increasing TDP43 levels32,38 or mislocalization9,37. Strategies that reduce TDP43 levels and mislocalization, including autophagy induction as shown here, may therefore be effective in both sALS and fALS.
We sought to improve neuronal survival by enhancing TDP43 clearance via small-molecule autophagy inducers. With these compounds, we tested the utility of autophagy induction for desired on-target effects (e.g., TDP43 clearance) and for efficacy against disease-related phenotypes (e.g., survival). The compounds represent well-validated tools for inducing autophagy in neurons, but they and their derivatives must be evaluated in vivo to assess for therapeutic effectiveness or unanticipated toxicities. Still, phenothiazine derivatives, including FPZ, have been used for decades to treat patients with psychoses. These compounds appear to be neuroprotective39, and one phenothiazine derivative, pimozide40, slowed disease progression in ALS patients. Because of its structural similarity to NCP, we tested pimozide in our initial screen22, and found that it does in fact stimulate autophagy, providing indirect support for the beneficial effects of autophagy induction in ALS.
We modeled disease mechanisms over a short time by capitalizing upon a striking and conserved pathologic signature among neurodegenerative diseases: the accumulation of misfolded proteins, such as TDP43. Overexpressing TDP43 in primary neurons reproduces key hallmarks of ALS pathology, including neurodegeneration accompanied by TDP43 accumulation and mislocalization9. Coping responses in acute overexpression models may be different from those occurring in ALS patients, representing a potential caveat of the current system. Still, human iPSC-derived neurons and astrocytes carrying ALS-associated mutations in TARDBP also accumulate mutant TDP43 protein32,37, lending additional support to the notion that excess TDP43 is central to disease pathogenesis. Autophagy induction effectively improved survival in rodent neurons and in human iPSC-derived neurons and astrocytes, supporting the validity of the rodent model system utilized here, and more importantly, showing that enhancing TDP43 clearance is a reasonable neuroprotective strategy for humans with disease.
In a prior study20, rapamycin exhibited neuroprotective effects in TDP43 transgenic animals, but there was little evidence that the benefit was tied to autophagy induction. Rapamycin and other mTOR inhibitors exert multiple effects independent of autophagy23, including some (e.g., inhibition of protein synthesis41) that account for their benefits in another disease model42. The concentrations typically used to mitigate disease are much higher than the EC50 associated with mTOR inhibition, raising serious concerns about their mechanisms of action. In contrast, we identified compounds that effectively stimulate neuronal autophagy and enhance TDP43 turnover. This, in turn, reduced the amount of TDP43, explaining much of the survival benefit observed in primary neurons and human iPSC-derived neurons and astrocytes.
Why is it important to specifically stimulate autophagy within neurons? Increasing evidence suggests that non-neuronal phenomena contribute to ALS pathogenesis36. For instance, mutant TDP43 exhibits intrinsic toxicity in human astrocytes, and these cells enhance the survival of iPSC-derived neurons37. Previous investigations of autophagy stimulation in ALS20 failed to determine if neuronal or glial induction was responsible for the observed benefit, an essential question relating to the drugs’ targets and mechanism of action. We measured autophagic activation at the single-cell level, within living neurons, leaving no doubt as to the targeted cell type.
If mutant TDP43 is cleared rapidly, why does it preferentially accumulate in fALS? In previous work9, we showed that TDP43(A315T) forms detergent-resistant inclusions more readily than TDP43(WT). If mutant TDP43 misfolding and deposition outpace degradation, kinetics will favor the formation of inclusions despite mutant TDP43’s relatively short half-life. According to this model, stabilization of mutant TDP43 through aggregation can be prevented by blocking TDP43 misfolding or enhancing its degradation before inclusions form. Autophagy inducers likely accomplish the latter, effectively augmenting mutant TDP43 clearance and inhibiting the formation of visible TDP43 inclusions. Stimulating autophagy may also improve proteostasis more generally, which could also indirectly improve protein misfolding.
One of the biggest obstacles to therapy development is the unreliable translation from preclinical disease models to humans43. iPSC-derived neurons and astrocytes have the potential to transform therapeutics development for ALS and other disorders44. Here, we observed neuroprotection in iPSC-based ALS models, without the use of artificial stressors to elicit phenotypes38. Astrocytes prevent37,45 or stimulate46 neuronal loss under different circumstances, and are directly affected by disease-associated mutations37. Our results, demonstrating efficacy of small-molecule autophagy inducers in human iPSC-derived neurons and astrocytes, suggests that a common therapeutic strategy might work in more than one cell type.
In prior work we showed that visible TDP43 inclusions do not impact neuronal survival9, and multiple in vivo studies have confirmed that TDP43 inclusions are not required for neurotoxicity10,11. Strong precedence exists for the idea that protein inclusions themselves are not necessarily pathogenic25,47,48. Thus, although autophagy induction reduced visible inclusions in TDP43(A315T)-expressing neurons, we do not believe this directly affected survival. Instead, the observed reduction in inclusions likely followed the drop in TDP43 levels or cytoplasmic localization, since inclusions develop preferentially in cells with high TDP43 levels9, and they are predominantly cytoplasmic.
Neuronal inclusions rich in misfolded proteins are characteristic of HD and ALS, consistent with underlying protein dyshomeostasis in both disorders. Our current and previous49 results suggest that neurons cope with aggregation-prone proteins by targeting them for accelerated clearance. Instead of a non-specific reaction to overexpressed proteins, this may represent a conserved coping response directed against toxic, misfolded proteins. When the cell’s ability to degrade these proteins is insufficient, the proteins accumulate and cause toxicity. Thus, enhancing protein clearance should prevent accumulation and improve survival. We tested this by inducing autophagy and found that stimulating TDP43 clearance via autophagy reduced TDP43 levels and diminished risk of death. These data show that protein dyshomeostasis is a common thread in neurodegenerative diseases, and offer the exciting hope that therapies targeting this thread might have broad therapeutic potential.
ONLINE METHODS
Ethics Statement
Rat (Rattus norvegicus) colonies were maintained in accordance with the University of California, San Francisco, “Assurance of Compliance with PHS Policy on Humane Care and Use of Laboratory Animals by Awardee Institutions” number A3400-01. The animal care facility at The J. David Gladstone Institutes is fully approved and under the supervision of full-time veterinarians. There were no manipulations that might have produced discomfort, distress, pain or injury. The animals were euthanized using CO2, resulting in rapid and painless death, consistent with the recommendations of the Panel on Euthanasia of the American Veterinary Medical Association.
Plasmids
Human TDP43 fused at its C-terminus to EGFP (TDP43-EGFP) was amplified from pcDNA-TDP43-EGFP9 by PCR using the following primers: 5′-ATA TAT AAG CTT CCA TGT CTG AAT ATA TTC GGG TAA CCGA-3′, 5′-ATA TAT GGT ACC GGA GAT CTC TTG TAC AGC TCG TCC ATGC-3′. Using KpnI and HindIII, the insert was then cloned into pGW1-CMV. Dendra2 was cloned into pGW1-CMV from pDendra2-N (Evrogen) using HindIII and MfeI. To create pGW1-TDP43-Dendra2, TDP43 was amplified by PCR using the following primers: 5′-TAT ATA AAG CTT CCA TGT CTG AAT ATA TTC GGG TAA CCGA-3′, 5′-ATA TAT GGT ACC CCT CCC CAG CCA GAA GAC TTA GAA TCC-3′. The PCR product was digested with KpnI and HindIII, then inserted into pGW1-Dendra2 that had been cut with the same enzymes. The A315T mutation was established by site-directed mutagenesis using the following primers: 5′-GTG GTG GGA TGA ACT TTG GTA CGT TCA GCA TTA ATC CAG CC-3′ and 5′-GGC TGG ATT AAT GCT GAA CGT ACC AAA GTT CAT CCC ACC AC-3′. The M337V mutation was also created by site-directed mutagenesis using the following primers: 5′-GCA GTT GGG GTA TGG TGG GCA TGT TAG CC-3′ and 5′-GGC TAA CAT GCC CAC CAT ACC CCA ACT GC-3′. mApple was amplified by PCR from pmApple-C1 by the following primers: 5′-ATA TAA GCT TCG CCA CCA TGG TGA GCAA-3′ and 5′-ATA TGA ATT CTT ATC TAG ATC CGG TGG ATC CCG-3′. The insert was then ligated into pGW1-CMV after digestion with HindIII and EcoRI, creating pGW1-mApple. pGW1-EGFP and pGW1-Dendra2-LC3 were created as described25,49. To make the MAP2:mApple survival marker used in longitudinal microscopy of human iPSC-derived neurons, the human MAP2 promoter (Systems Biosciences) was cloned into pGW1-mApple using BamHI and XhoI. The HB9::GFP reporter was generated as described33. Sequence information for all plasmids is available at http://gind-db.ucsf.edu:8000/cgi-bin/Plasmid/main_menu2.cgi.
Cell culture and transfection
Cortical neurons were dissected from embryonic day 20–21 rat pups and cultured at 0.6×106 cells/ml for 4 days in vitro, as described50. Euthanasia for these experiments is entirely consistent with the recommendations of the Guidelines on Euthanasia of the American Veterinary Medical Association. Transfection of primary neurons was accomplished using Lipofectamine 2000 (Invitrogen). All transfections involved 0.2 μg DNA (total) and 0.5 μl Lipofectamine 2000 per well, unless otherwise noted. Cells were incubated with Lipofectamine/DNA complexes for no more than 20 min at 37°C before rinsing. The remainder of the transfection protocol was per the manufacturer’s suggestions, resulting in an overall transfection efficiency of < 1%. For co-transfections, plasmids were kept at a 1:1 molar ratio, unless otherwise noted in the text. For live-cell nuclear staining, neurons were incubated in PBS supplemented with calcium, magnesium, and Hoechst dye (10 μM final concentration) for 10 min at 37°C, then transferred back to serum-free media for imaging.
Metabolic pulse-chase
Primary rodent cortical neurons were isolated from embryonic day 21 rat pups as described above and cultured in 6-well plates at a density of 0.6×106 cells/ml. On in vitro day 4, cultures were rinsed twice with methionine- and cysteine-free DME (Invitrogen), then placed in the same media for 1 h at 37°C. Proteins were labeled by adding 35S-labeled methionine and cysteine (EasyTag Express, PerkinElmer; final concentration 0.3 mCi/ml), and incubating for 4 h at 37°C. Afterwards, neurons were rinsed twice with methionine- and cysteine-free DME, then cultured in Neurobasal media supplemented with 2% B27, 1% GlutaMAX, and 1% penicillin/streptomycin (all from Invitrogen) for the indicated time. For collection, neurons were rinsed twice with ice-cold PBS then lysed in ice-cold RIPA (0.5% deoxycholate, 1% Triton X-100) supplemented with MiniProtean protease inhibitors (Roche). Samples were kept on ice for 5 min, then centrifuged at 1000 x g for 5 min at room temperature. Supernatants were stored at −80°C until all samples were collected. TDP43 was immunoprecipitated using rabbit polyclonal anti-TDP43 antibodies (G. Yu and J. Herz, University of Texas, Southwestern; ThermoScientific, PA5-17011) diluted 1:100, for 1 h at room temperature. As a control, non-specific rabbit anti-IgG antibodies (Santa Cruz) were added at a similar concentration. Antibody-protein complexes were isolated using protein-A agarose beads (Invitrogen) diluted 1:20, by rotating for 30 min at room temperature. The complexes were rinsed twice with RIPA, and twice more with PBS, prior to elution by boiling in Laemmli sample buffer. Proteins were separated by SDS-PAGE, and gels were fixed in 10% glacial acetic acid, 20% methanol, 3% glycerol (v/v/v) in water for 1 h at room temperature. Gels were then dried by heating to 80°C for 2 h, under vacuum for 3 h. Dried gels were exposed to film (Kodak BioMax MR) with an intensifying screen overnight at −80°C before developing. The amount of immunoprecipitated TDP43 at each time point was determined by densitometry of scanned films using ImageJ software, and normalized to the amount of TDP43 at the initial time point.
Western blotting
Primary rodent cortical neurons were isolated and cultured from embryonic day 21 rat pups as described above in 6-well plates at a density of 0.6×106 cells/ml. On in vitro day 5, cultures were treated with vehicle control (DMSO), 0.5 or 5 μM of compound for 24 h. Cells were rinsed twice in ice-cold PBS and lysed in RIPA (0.5% deoxycholate, 1% Triton X-100) supplemented with MiniProtean protease inhibitors (Roche). Lysates were separated by SDS-PAGE, and proteins detected via western blotting. Primary antibodies included rabbit polyclonal anti-LC3 antibodies raised against a polypeptide modeled after the N-terminus of LC3 (EKTFKQRRSFEQRVEDVR), diluted 1:1000 in 1% non-fat dry milk (BioRad) in TBS-T (Tris-buffered saline with 0.1% tween-20), and mouse monoclonal anti-GAPDH antibodies (AbCam, 6C5) diluted 1:5000 in 1% non-fat dry milk in TBS-T. Proteins were visualized using a Licor digital imager after incubation with Licor anti-rabbit and anti-mouse fluorescently-conjugated antibodies diluted 1:10,000 in TBS-T supplemented with 1% non-fat dry milk. The amounts of LC3-II, LC3-I, and GAPDH were determined by densitometry of scanned films using ImageJ and Fiji.
Automated fluorescence microscopy
Experiments involving neuronal survival analysis and optical pulse imaging utilized an automated microscopy platform as described9.25. Briefly, images were obtained with an inverted microscope (Nikon TE2000) equipped with the PerfectFocus system, a high-numerical aperture 20× objective lens and a 16-bit Andor Clara digital camera with a cooled charge-coupled device. Illumination was provided by a Lambda XL Xenon lamp (Sutter) with a liquid light guide. The ASI 2000 stage was controlled by rotary encoders in all three planes of movement. All components were encased in a custom-designed, climate-controlled environmental chamber (In Vivo Scientific) kept at 37°C and 5% CO2. The Semrock BrightLine full-multiband filter set (DAPI, FITC, TRITC, Cy5) was used for fluorophore photoactivation (DAPI), excitation and detection (FITC, TRITC). The illumination, filter wheels, focusing, stage movements, and image acquisitions were fully automated and coordinated with a mix of proprietary (ImagePro) and publicly available (ImageJ, μManager) software.
Image analysis
Relevant data were extracted from the raw, digital images in a sequential process using an original script developed in PipelinePilot (Accelrys, San Diego, CA). Briefly, the median background fluorescence from a portion of all images was calculated and subtracted from each individual image. The images were then assembled into montages representing each well at each time point. The montages were sequenced and aligned automatically, and neuron cell bodies segmented based on intensity and morphology. Among the variables recorded for each neuron were the fluorescence intensity and the time of death, marked by the loss of cellular fluorescence, rounding or dissolution of the cell body. Neurons with TDP43-Dendra2 intensity values below 20 AU were excluded from half-life analyses. Statistical analyses and the generation of cumulative hazard plots were accomplished using custom-designed algorithms and the survival package within R, while scatter plots and bar graphs were created using Prism (GraphPad).
Motor neuron culturing and differentiation
iPSC were generated and characterized as described32. Control fibroblasts were obtained from a 56-year-old man and a 40-year-old woman (ATCC), while mutant TDP43 fibroblasts were donated from a 56-year-old man. iPSCs were cultured on CF-1 irradiated mouse embryonic fibroblasts with KO-DMEM (Invitrogen) supplemented with 20% knockout serum replacement (Invitrogen), 10 ng/ml basic FGF2 (PeproTech), 1 mM L-glutamine (Invitrogen), 100 mM 2-mercaptoethanol (Invitrogen), 1% penicillin/streptomycin (Invitrogen) and 1% nonessential amino acids. Before differentiation, cells were resuspended in MTESR1 media (Stem Cell Technologies) and passaged three times. Neural precursor cells (NPCs) were generated by dual-SMAD inhibition in medium supplemented with SB431542 (10 μM; Tocris), dorsomorphin (2.5 μM; Calbiochem), and N-acetylcysteine (0.5 μM; Sigma) for 5–7 days. Cells were dissociated with Accutase (Sigma) and caudalized using retinoic acid (0.1 μM; Sigma) for 7–12 days, then dissociated again and plated onto coverslips coated with laminin (Sigma) and fibronectin (Sigma) in Neurobasal medium (Invitrogen) supplemented with 0.1 μM retinoic acid, 1 μM purmorphamine (Calbiochem), 1% N-2 supplement (Invitrogen), 1% nonessential amino acids (Invitrogen), 1% penicillin/streptomycin(Invitrogen), 1% GlutaMAX (Invitrogen), and 5 ng/mL basic FGF. After 7–10 days, MN precursors were transferred to maturation medium, consisting of Neurobasal medium (Invitrogen) supplemented with 0.5% N-2 supplement (Invitrogen), 1% nonessential amino acids (Invitrogen), 1% penicillin/streptomycin (Invitrogen), 0.5% GlutaMAX (Invitrogen), 10 ng/mL brain-derived neurotrophic factor (BDNF; PeproTech), 10 ng/mL glial cell-derived neurotrophic factor (PeproTech), and 10 μM forskolin (Tocris). At 2–4 weeks later, neurospheres were dissociated with papain (Worthington) and immediately electroporated using Neon (Invitrogen) with HB9::GFP or pGW1-MAP2::mApple before plating at 75×103 cells per well of a 96-well plate that had been coated with polyornithine (Sigma) and laminin (Sigma).
Astrocyte culturing and differentiation
Astrocytes were differentiated from iPSCs as described37. Briefly, NPCs were cultured in DMEM/F12 (Invitrogen) supplemented with 1% N2 (Invitrogen), 0.1% B27 (Invitrogen), 1% nonessential amino acids (Invitrogen), 1% penicillin/streptomycin (Invitrogen), 1% GlutaMAX solution (Invitrogen), 20 ng/mL LIF (Sigma) and 20 ng/mL EGF (R&D Systems) for 4–6 weeks. Spheres were then transferred to DMEM/F12 supplemented with 1% N2, 0.1, % B27, 1% nonessential amino acids, 1% penicillin/streptomycin, 1% GlutaMAX, 20 ng/mL FGF-2 (PeproTech) and 20 ng/mL EGF (R&D Systems), and mechanically passaged every 2 weeks. To generate monolayer cultures, spheres were dissociated with papain (Worthington Biochemical) and plated in wells coated with Matrigel (BD Biosciences, 1:80). Monolayers were split with Accutase (Sigma) approximately every 3–4 d, or when confluent. Differentiated astrocytes were created by culturing the cells in Neurobasal medium (Invitrogen) supplemented with 0.2% B27, 1% nonessential amino acids, 1% penicillin/streptomycin, 1% GlutaMAX solution, and 10 ng/mL CNTF (R&D Systems) for 2 weeks. Before survival analysis, astrocytes were dissociated with Accutase and cultured at a density of 2×104 cells per well of a 96-well plate. 2–3 days later, differentiated astrocyte cultures were transfected with pGW1-mApple using Lipofectamine 2000, as described above.
Immunocytochemistry
Cortical neurons from rats were isolated as described50 and plated onto 12-mm glass coverslips coated with laminin (BD Biosciences) and poly-D-lysine (Millipore), then transfected at 4 days in vitro with pGW1-mApple and pGW1-TDP43(WT)-EGFP or pGW1-TDP43(A315T)-EGFP. Immunocytochemistry was performed as described50 using rabbit anti-TDP43 polyclonal antibodies (Proteintech; 1:1000) and anti-rabbit Cy5-labeled secondary antibodies (Jackson Immunochemical; 1:250). For LC3 immunocytochemistry, neurons were treated for 24 h at 37°C with the indicated compound or DMSO, then fixed and probed with rabbit anti-LC3 primary antibodies (Novus Biologicals, 1:200), and anti-rabbit Alexa 488-labeled secondary antibodies (Invitrogen, 1:500). Nuclei were stained by a brief incubation in PBS supplemented with Hoechst dye (10 μM final concentration). Immunocytochemical samples were imaged using a Zeiss LSM510 confocal microscope equipped with Meta. The total normalized TDP43 level was determined by comparing the intensity of TDP43 immunolabeling in transfected cells to that in non-transfected cells within the same high-powered (63×) field.
In silico drug screen
The Discovery Studio 2.1 (Accelrys, San Diego, CA) suite of molecular modeling software was used to assemble and screen a three-dimensional database (3DDB) of commercially available and well-characterized compounds. This collection, totaling over one million compounds, represents a broad cross section of chemical space, including all marketed drugs and drugs in development. It includes compounds from several drug libraries, including Maybridge (http://www.maybridge.com), ChemBridge (http://www.chembridge.com), MicroSource Discovery Systems (http://www.msdiscovery.com), Sigma-Aldrich (http://www.sigmaaldrich.com), TimTec Drug Discovery (http://www.timtec.com), Specs (http://www.specs.com), ZINC (http://zinc.docking.org), the World Drug Index (http://scientificthomsonreuters.com/products/wdi) and Interbioscreen (http://www.ibscreen.com). Pharmacophore hypotheses were generated utilizing the two most active compounds in a previously-performed screen22 utilizing the HipHop algorithm within the Pharmacophore Protocol of Discovery Studio to automatically build the common-features pharmacophore models using hydrophobe, hydrophobe aromatic, acceptor, donor and charged positive features. Up to 255 low energy conformations of each of the compounds (based on the initial low energy conformation) were included in this analysis. Briefly, the program evaluates chemical features that the two compounds have in common based on common conformations. The algorithm starts with a small set of features and extends them until no further common conformation of the input compounds is found. Multiple hypotheses are generated and scored. These hypotheses are spatial dispositions of chemical features the input compounds share in common. All three and four feature hypotheses were evaluated utilizing the Ligand Pharmacophore Mapping function and scoring module within the Pharmacophore Protocol of Discovery Studio. The relative concordance of the scoring function was compared to the neuronal autophagy stimulation ranking for each compound, as previously determined22. The best three-feature pharmacophore model (two hydrophobic aromatic features and one charged positive feature) was selected and systematically modified with exclusion spheres (ligand inaccessible areas) to properly rank each compound with the computed scoring function. Fourteen exclusion spheres were added to the three-feature pharmacophore model, and this model was utilized in conjunction with the 3DDB to conduct the in silico HTS. Scoring was assessed utilizing a fit function that determined the goodness of fit of each compound to the features of the model. In addition, high scoring hits from the initial screen were also fit to the ‘shape’ feature of the model, and the highest scoring compounds from this filter (MTM and FPZ) were evaluated as described in the text.
Compounds
MTM and FPZ were acquired from Sigma (M4406 and F4765, respectively). The purity of MTM was ≥95%, while that of FPZ was ≥98%, determined by thin-liquid chromatography and HClO4 titration. NCP was purchased from EMD Millipore (#124020), with a purity of ≥95% as measured by high-performance liquid chromatography. All compounds were solubilized in DMSO at 10 mM, and stored at −20°C until just prior to use.
Statistical analysis
One-way ANOVA with Dunnett’s test was used for comparison of multiple means in Fig. 4b, c; Fig. 5a, c, e, f, g, h, i; and Supplementary Fig. 6a. The two-sided Kolomogorov-Smirnov test was used to determine differences between non-parametric probability distributions in Fig. 1a, b; Fig. 3b, c, d; Fig. 4g; Fig. 5b, d; Supplementary Fig. 5a and Supplementary Fig. 6j. The Mann-Whitney test was used to compare medians from non-parametric data in Fig. 3a. The Kruskal-Wallis test with Dunn’s post-test was used for comparisons of medians from multiple non-parametric data groups in Fig. 4f. Linear regression was employed in Fig. 1f, g; Supplementary Fig. 3e; and Supplementary Fig. 4c–e. First-order exponential decay curve fitting was used to model half-life in Fig. 2b, e, f; and Fig. 4e. Cox proportional hazards analysis was used to determine differences in survival among populations in Fig. 1d, e; Fig. 6a–e; Supplementary Fig. 2a–e; Supplementary Fig. 5b; and Supplementary Fig. 7a–c.
Supplementary Material
Acknowledgments
We would like to thank Drs. B. Miller, B. Seeley, C. Lomen-Hoerth, and the members of the Finkbeiner lab for all their generous support and advice. H. Zahed and J. Margulis deserve special acknowledgement for their assistance. We thank G. Howard for editorial assistance and K. Nelson for administrative assistance. We also thank G. Yu and J. Herz (University of Texas, Southwestern) for anti-TDP43 antibody. This work was supported by National Institutes of Neurological Disorders and Stroke grants 1K08NS072233-01A1 (to S.B.) 3R01 NS039074, R01 NS083390, R43 NS081844, and U24 NS078370 (to S.F.), the ALS Association (S.F.), the Robert Packard Center for ALS Research and the William H. Adams Foundation (S.F.) and from Target ALS (S.F.). Additional support was provided by the Roddenberry Stem Cell Program (to S.F.), The Taube/Koret Center for Neurodegenerative Disease (S.F.), the Hellman Family Foundation Alzheimer’s Disease Research Program (S.F.), the Protein Folding Diseases Initiative at the University of Michigan (S.B.), California Institute of Regenerative Medicine TR4-06693 (S.F) and U01 MH1050135 (S.F.). The animal care facility was partly supported by an NIH Extramural Research Facilities Improvement Program Project (C06 RR018928).
Footnotes
AUTHOR CONTRIBUTIONS
S.B., A.S., A.A., A.T., S.C. and S.F. designed the study. S.B., A.A., X.L. and A.S cultured cells, performed microscopy and conducted survival analyses. A.T., B.B., C.S., and S.C. provided compounds and iPSCs. M.P. performed in silico drug screening. D.P., D.M.A. and A.D. wrote original scripts for AFM. S.B. and A.A. analyzed data, constructed vectors, performed MPC and OPL, and transfected neurons. S.B. and S.F. wrote and edited the manuscript.
STATEMENT OF COMPETING FINANCIAL INTERESTS
We declare that the authors have no competing interests as defined by Nature Publishing Group, or other interests that might be perceived to influence the results and/or discussion reported in this article.
References
- 1.Robberecht W, Philips T. The changing scene of amyotrophic lateral sclerosis. Nat Rev Neurosci. 2013;14:248–264. doi: 10.1038/nrn3430. [DOI] [PubMed] [Google Scholar]
- 2.Neumann M. Molecular Neuropathology of TDP-43 Proteinopathies. Int J Mol Sci. 2009;10:232–246. doi: 10.3390/ijms10010232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Blitterswijk M, Landers JE. RNA processing pathways in amyotrophic lateral sclerosis. Neurogenetics. 2010 doi: 10.1007/s10048-010-0239-4. [DOI] [PubMed] [Google Scholar]
- 4.Ayala YM, et al. Structural determinants of the cellular localization and shuttling of TDP-43. J Cell Sci. 2008;121:3778–3785. doi: 10.1242/jcs.038950. [DOI] [PubMed] [Google Scholar]
- 5.Moisse K, et al. Cytosolic TDP-43 expression following axotomy is associated with caspase 3 activation in NFL−/− mice: support for a role for TDP-43 in the physiological response to neuronal injury. Brain Res. 2009;1296:176–186. doi: 10.1016/j.brainres.2009.07.023. [DOI] [PubMed] [Google Scholar]
- 6.Dewey CM, et al. TDP-43 is directed to stress granules by sorbitol, a novel physiological osmotic and oxidative stressor. Mol Cell Biol. 2010 doi: 10.1128/MCB.01279-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davidson YS, et al. TDP-43 pathological changes in early onset familial and sporadic Alzheimer’s disease, late onset Alzheimer’s disease and Down’s Syndrome: association with age, hippocampal sclerosis and clinical phenotype. Acta Neuropathol. 2011;122:703–713. doi: 10.1007/s00401-011-0879-y. [DOI] [PubMed] [Google Scholar]
- 8.Barmada SJ, Finkbeiner S. Pathogenic TARDBP mutations in amyotrophic lateral sclerosis and frontotemporal dementia: disease-associated pathways. Rev Neurosci. 2010;21:251–272. doi: 10.1515/revneuro.2010.21.4.251. [DOI] [PubMed] [Google Scholar]
- 9.Barmada SJ, et al. Cytoplasmic mislocalization of TDP-43 is toxic to neurons and enhanced by a mutation associated with familial amyotrophic lateral sclerosis. J Neurosci. 2010;30:639–649. doi: 10.1523/JNEUROSCI.4988-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsao W, et al. Rodent models of TDP-43: Recent advances. Brain Res. 2012:1–14. doi: 10.1016/j.brainres.2012.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roberson ED. Mouse models of frontotemporal dementia. Ann Neurol. 2012;72:837–849. doi: 10.1002/ana.23722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ling SC, et al. ALS-associated mutations in TDP-43 increase its stability and promote TDP-43 complexes with FUS/TLS. Proc Natl Acad Sci USA. 2010;107:13318–13323. doi: 10.1073/pnas.1008227107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Watanabe S, Kaneko K, Yamanaka K. Accelerated disease onset with stabilized familial Amyotrophic Lateral Sclerosis (ALS)-linked TDP-43 mutations. J Biol Chem. 2013;288:3641–3654. doi: 10.1074/jbc.M112.433615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roscic A, Baldo B, Crochemore C, Marcellin D, Paganetti P. Induction of autophagy with catalytic mTOR inhibitors reduces huntingtin aggregates in a neuronal cell model. J Neurochem. 2011;119:398–407. doi: 10.1111/j.1471-4159.2011.07435.x. [DOI] [PubMed] [Google Scholar]
- 15.Prakash A, Levy DE. Regulation of IRF7 through cell type-specific protein stability. Biochem Biophys Res Comm. 2006;342:50–56. doi: 10.1016/j.bbrc.2006.01.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang X, et al. Degradation of TDP-43 and its pathogenic form by autophagy and the ubiquitin-proteasome system. Neurosci Lett. 2010;469:112–116. doi: 10.1016/j.neulet.2009.11.055. [DOI] [PubMed] [Google Scholar]
- 17.Urushitani M, Sato T, Bamba H, Hisa Y, Tooyama I. Synergistic effect between proteasome and autophagosome in the clearance of polyubiquitinated TDP-43. J Neurosci Res. 2010;88:784–797. doi: 10.1002/jnr.22243. [DOI] [PubMed] [Google Scholar]
- 18.Zhang X, et al. Rapamycin treatment augments motor neuron degeneration in SOD1(G93A) mouse model of amyotrophic lateral sclerosis. Autophagy. 2011;7:412–425. doi: 10.4161/auto.7.4.14541. [DOI] [PubMed] [Google Scholar]
- 19.Hetz C, et al. XBP-1 deficiency in the nervous system protects against amyotrophic lateral sclerosis by increasing autophagy. Genes Dev. 2009;23:2294–2306. doi: 10.1101/gad.1830709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang IF, et al. Autophagy activators rescue and alleviate pathogenesis of a mouse model with proteinopathies of the TAR DNA-binding protein 43. Proc Natl Acad Sci USA. 2012;109:15024–15029. doi: 10.1073/pnas.1206362109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fox JH, et al. The mTOR kinase inhibitor Everolimus decreases S6 kinase phosphorylation but fails to reduce mutant huntingtin levels in brain and is not neuroprotective in the R6/2 mouse model of Huntington’s disease. Mol Neurodegen. 2010;5:26. doi: 10.1186/1750-1326-5-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsvetkov AS, et al. A small-molecule scaffold induces autophagy in primary neurons and protects against toxicity in a Huntington disease model. Proc Natl Acad Sci USA. 2010;107:16982–16987. doi: 10.1073/pnas.1004498107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gitcho MA, et al. TDP-43 A315T mutation in familial motor neuron disease. Ann Neurol. 2008;63:535–538. doi: 10.1002/ana.21344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arrasate M, Mitra S, Schweitzer ES, Segal MR, Finkbeiner S. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature. 2004;431:805–810. doi: 10.1038/nature02998. [DOI] [PubMed] [Google Scholar]
- 26.Xu YF, et al. The Pathological Phenotypes of Human TDP-43 Transgenic Mouse Models Are Independent of Downregulation of Mouse Tdp-43. PLoS ONE. 2013;8:e69864. doi: 10.1371/journal.pone.0069864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chudakov DM, Lukyanov S, Lukyanov KA. Tracking intracellular protein movements using photoswitchable fluorescent proteins PS-CFP2 and Dendra2. Nat Protoc. 2007;2:2024–2032. doi: 10.1038/nprot.2007.291. [DOI] [PubMed] [Google Scholar]
- 28.Wong E, Cuervo AM. Integration of clearance mechanisms: the proteasome and autophagy. Cold Spring Harb Perspect Biol. 2010;2:a006734. doi: 10.1101/cshperspect.a006734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klionsky DJ, et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang J. Beclin 1 bridges autophagy, apoptosis and differentiation. Autophagy. 2008;4:947–948. doi: 10.4161/auto.6787. [DOI] [PubMed] [Google Scholar]
- 31.Kim J, Dalton VM, Eggerton KP, Scott SV, Klionsky DJ. Apg7p/Cvt2p is required for the cytoplasm-to-vacuole targeting, macroautophagy, and peroxisome degradation pathways. Mol Biol Cell. 1999;10:1337–1351. doi: 10.1091/mbc.10.5.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bilican B, et al. Mutant induced pluripotent stem cell lines recapitulate aspects of TDP-43 proteinopathies and reveal cell-specific vulnerability. Proc Natl Acad Sci USA. 2012;109:5803–5808. doi: 10.1073/pnas.1202922109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singh Roy N, et al. Enhancer-specified GFP-based FACS purification of human spinal motor neurons from embryonic stem cells. Exp Neurol. 2005;196:224–234. doi: 10.1016/j.expneurol.2005.06.021. [DOI] [PubMed] [Google Scholar]
- 34.Geser F, et al. Clinical and pathological continuum of multisystem TDP-43 proteinopathies. Arch Neurol. 2009;66:180–189. doi: 10.1001/archneurol.2008.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kosik KS, Finch EA. MAP2 and tau segregate into dendritic and axonal domains after the elaboration of morphologically distinct neurites: an immunocytochemical study of cultured rat cerebrum. J Neurosci. 2001;7:3142–3153. doi: 10.1523/JNEUROSCI.07-10-03142.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boillée S, Vande Velde C, Cleveland DW. ALS: a disease of motor neurons and their nonneuronal neighbors. Neuron. 2006;52:39–59. doi: 10.1016/j.neuron.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 37.Serio A, et al. Astrocyte pathology and the absence of non-cell autonomy in an induced pluripotent stem cell model of TDP-43 proteinopathy. Proc Natl Acad Sci USA. 2013 doi: 10.1073/pnas.1300398110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Egawa N, et al. Drug Screening for ALS Using Patient-Specific Induced Pluripotent Stem Cells. Sci Transl Med. 2012;4:145ra104–145ra104. doi: 10.1126/scitranslmed.3004052. [DOI] [PubMed] [Google Scholar]
- 39.Stavrovskaya IG. Clinically Approved Heterocyclics Act on a Mitochondrial Target and Reduce Stroke-induced Pathology. J Exp Med. 2004;200:211–222. doi: 10.1084/jem.20032053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Szczudlik A, Tomik B, Słowik A, Kasprzyk K. Assessment of the efficacy of treatment with pimozide in patients with amyotrophic lateral sclerosis. Introductory notes. Neurol Neurochir Pol. 1998;32:821–829. [PubMed] [Google Scholar]
- 41.Krauss S, et al. Translation of HTT mRNA with expanded CAG repeats is regulated by the MID1–PP2A protein complex. Nat Comm. 1AD;4:1511–9. doi: 10.1038/ncomms2514. [DOI] [PubMed] [Google Scholar]
- 42.Miller J, et al. Quantitative Relationships between Huntingtin Levels, Polyglutamine Length, Inclusion Body Formation, and Neuronal Death Provide Novel Insight into Huntington’s Disease Molecular Pathogenesis. J Neurosci. 2010;30:10541–10550. doi: 10.1523/JNEUROSCI.0146-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Finkbeiner S. Bridging the Valley of Death of therapeutics for neurodegeneration. Nat Med. 2010;16:1227–1232. doi: 10.1038/nm.2222. [DOI] [PubMed] [Google Scholar]
- 44.Dolmetsch R, Geschwind DH. The Human Brain in a Dish: The Promise of iPSC-Derived Neurons. Cell. 2011;145:831–834. doi: 10.1016/j.cell.2011.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jacquier A, Bellouze S, Blanchard S, Bohl D, Haase G. Astrocytic protection of spinal motor neurons but not cortical neurons against loss of Als2/alsin function. Hum Mol Genet. 2009;18:2127–2139. doi: 10.1093/hmg/ddp136. [DOI] [PubMed] [Google Scholar]
- 46.Haidet-Phillips AM, et al. Astrocytes from familial and sporadic ALS patients are toxic to motor neurons. Nat Biotechnol. 2011;29:824–828. doi: 10.1038/nbt.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ko LW, Ko HHC, Lin WL, Kulathingal JG, Yen SHC. Aggregates assembled from overexpression of wild-type alpha-synuclein are not toxic to human neuronal cells. J Neuropathol Exp Neurol. 2008;67:1084–1096. doi: 10.1097/NEN.0b013e31818c3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mucke L, et al. High-level neuronal expression of abeta 1–42 in wild-type human amyloid protein precursor transgenic mice: synaptotoxicity without plaque formation. J Neurosci. 2000;20:4050–4058. doi: 10.1523/JNEUROSCI.20-11-04050.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tsvetkov AS, et al. Proteostasis of polyglutamine varies among neurons and predicts neurodegeneration. Nat Chem Biol. 2013;9:586–592. doi: 10.1038/nchembio.1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saudou F, Finkbeiner S, Devys D, Greenberg ME. Huntingtin acts in the nucleus to induce apoptosis but death does not correlate with the formation of intranuclear inclusions. Cell. 1998;95:55–66. doi: 10.1016/s0092-8674(00)81782-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.