Abstract
Introduction
X-ray crystallography plays an important role in structure-based drug design (SBDD), and accurate analysis of crystal structures of target macromolecules and macromolecule–ligand complexes is critical at all stages. However, whereas there has been significant progress in improving methods of structural biology, particularly in X-ray crystallography, corresponding progress in the development of computational methods (such as in silico high-throughput screening) is still on the horizon. Crystal structures can be overinterpreted and thus bias hypotheses and follow-up experiments. As in any experimental science, the models of macromolecular structures derived from X-ray diffraction data have their limitations, which need to be critically evaluated and well understood for structure-based drug discovery.
Areas covered
This review describes how the validity, accuracy and precision of a protein or nucleic acid structure determined by X-ray crystallography can be evaluated from three different perspectives: i) the nature of the diffraction experiment; ii) the interpretation of an electron density map; and iii) the interpretation of the structural model in terms of function and mechanism. The strategies to optimally exploit a macromolecular structure are also discussed in the context of ‘Big Data’ analysis, biochemical experimental design and structure-based drug discovery.
Expert opinion
Although X-ray crystallography is one of the most detailed ‘microscopes’ available today for examining macromolecular structures, the authors would like to re-emphasize that such structures are only simplified models of the target macromolecules. The authors also wish to reinforce the idea that a structure should not be thought of as a set of precise coordinates but rather as a framework for generating hypotheses to be explored. Numerous biochemical and biophysical experiments, including new diffraction experiments, can and should be performed to verify or falsify these hypotheses. X-ray crystallography will find its future application in drug discovery by the development of specific tools that would allow realistic interpretation of the outcome coordinates and/or support testing of these hypotheses.
Keywords: Big Data, crystallographic data interpretation, functional annotation, protein crystallography, target-based drug discovery, validation, virtual screening
1. Using structural information for structure-based drug discovery
The three-dimensional structures of biological macromolecules, particularly those determined by X-ray crystallography, are often considered as the ‘gold standard’ of data describing the molecular architecture of important proteins and nucleic acids. When the structures of interest have been determined, macromolecular models can yield a wealth of information necessary for modern drug discovery efforts that utilize computer-aided drug design (CADD) [1,2]. We note that sometimes the term ‘rational drug design’ is used instead of CADD or ‘structure-based drug design’. We feel that this is a misnomer – all drug design processes are always rational, albeit not necessarily optimal. Advancements of various software packages has made use of structural information in CADD both readily accessible and automated [3–5]. Developments in fragment-based drug discovery may provide further avenues for the use of structural information in CADD [6].
In addition, there has been an explosion in the number of macromolecular structures that are available. The rate of deposition of such structures to the Protein Data Bank (PDB) has continued to accelerate since its inception in 1971, and the number of deposits in the PDB will likely exceed 100,000 in 2014 [7]. The average data content of each deposit has also increased over time, in terms of the average molecular weight per structure, resolution and the associated experimental data such as structure factors [8]. Fully utilizing the rapid growth of macromolecular structure data, integrated with the wealth of other kinds of biological data available (amino acid or nucleotide sequence, metabolic and signaling pathways, expression patterns, etc.) is a significant challenge for data mining in medical applications such as drug discovery. The ‘Big Data’ paradigm usually refers to techniques for dealing with very large and complex datasets that reach or exceed the effective capabilities of traditional data processing tools or relational database management systems. This approach could be useful to process and analyze these structure data [9].
Whereas there are a number of techniques for determining macromolecular structure, X-ray crystallography is particularly well suited for drug discovery. First, X-ray crystallography is capable of producing structures of high (potentially atomic) resolution. Second, X-ray crystallography can be used to determine the structures of large heteromeric complexes (e.g., ribosome). Third, and perhaps most usefully, X-ray crystallography can provide detailed experimental evidence of the binding mode of small molecule ligands found in the crystal. Macromolecular crystal structures provide a platform for intuitive visualization of the architecture [10] and facilitate the understanding of mechanisms, and ultimately drug activity, at a molecular level. Moreover, crystal structures inspire new hypotheses and experiments to probe biological macromolecules regarding molecular mechanisms, plausible binding modes, and the feasibility of small molecule agents to serve as scaffolds for lead compounds. During the past two decades, we have witnessed an unprecedented success in the development of highly potent and selective drugs or lead compounds based on information obtained from the crystal structures of target proteins. Prominent examples include transition-state analog inhibitors for influenza virus neuraminidase [11], adenosine-derived inhibitors of Leishmania mexicana glyceraldehyde-3-phosphate dehydrogenase [12], and, perhaps most notably, inhibitors of HIV protease [13,14].
Emerging technologies in CADD tend to emphasize the small molecule aspects [2], including virtual screening [15], ‘click chemistry’ (the use of small modular building blocks to generate new compounds) [16], cheminformatics [17] and peptide-based drug discovery [18]. However, accurate understanding of the target macromolecule deserves and requires no less attention and has been the subject of considerable discussion in CADD in the past decade [19]. The major pitfall in structure-based drug design – apart, of course, from the fact that a structure of the target may not be known – is a lack of understanding of the limitations of structural models, both of the apo-forms of macromolecules and of the macromolecule– ligand complexes [20]. Sometimes the atomic coordinates of protein structure models taken from the PDB are used ‘as is’ with potential caveats [19–22], as there is no single parameter, measure or standard that fully describe both the overall and local quality of a structure. In fact, to reliably estimate how trustworthy a given model is for the purpose of its use in CADD, knowing the details of how a particular structure was determined is crucial.
Although most three-dimensional macromolecular structures are determined from experimental data, it is important to recall that the structural model of a given protein or nucleic acid in the PDB is exactly that – a model. In other words, it is a simplified interpretation of the raw experimental data, in much the same way that a linear regression fit models data of two variables. In both cases, many statistics may be calculated to determine how accurately and precisely the model fits (or does not fit) the data, and these statistics need to be carefully examined before making use of the models for further inference. One would not infer that two variables are linearly correlated, for example, without first examining the correlation coefficient. Further, there are limits to the validity of models, largely due to the quality of data used to produce them. For example, a linear regression model is not necessarily valid when one extrapolates beyond the range of the data used to infer the fit. Similarly, macromolecular models are also limited by the scope of the diffraction data (due to data resolution, limited completeness and/or redundancy, missing regions of density, etc.). Whereas the properties of accuracy, precision and validity of linear regression models are fairly well known and understood, the analogous properties of crystallographic models are not as straightforward. However, these properties need to be fully understood if those structures are to be used for drug design [22,23].
Although there is a standard set of validation checks performed on structures deposited to the PDB, many of these checks are informational in nature and deposits may be accepted even if some ‘fail.’ Other aspects of a PDB deposition, most notably the parameters of sample preparation and data collection contained in the PDB header, are only minimally validated, if at all. As a result, the completeness and accuracy of PDB deposits vary significantly (although it should be noted that the PDB has made steady improvement in this area through improved deposition procedures and other curation efforts). In addition to the validity of atomic coordinates, the parameters and experimental details of a crystal structure play a role in its reliability as well. Clearly, for example, a structure determined at 1.5 Å resolution is much better suited for use in CADD than a 3.5 Å structure, although this fact may not be readily apparent looking at the atomic models alone.
There is always a danger that crystal structures may be overinterpreted, or even worse, blindly trusted and used in further computational and experimental drug discovery procedures without a critical assessment. In the evaluation of high-throughput virtual screening methods [24], the accuracy of scoring functions has been widely assessed and criticized [25]. However, there has been little discussion about evaluation of the quality of the structural models used in deriving the parameters of these scoring functions. Quite often, when a set of structures is established as a reference set for derivation of an algorithm or a scoring function, only the resolution of each structure is used as a criterion to address its reliability.
The structures deposited to the PDB are usually of high quality; however, this fact gives a false sense of security that all structural details are optimal. In reality, despite steady improvement in the quality of deposited structures, the spatial coordinates of each atom can only be unambiguously derived from the experimental map for high-resolution structures. For moderate or low resolution macromolecular structures, additional prior information, such as chemical identity, stereochemistry, and the like, must be used to infer atomic positions, which can introduce bias. In fact, a recent validation study highlighted that a considerable number of functional ligands reported in the PDB were not supported by electron density maps [26,27]. There have been some efforts to improve this, through the development of projects such as PDB-REDO [28], which re-refine previously deposited protein structures using the state-of-the-art automated refinement programs. Projects such as PDB-REDO suffer from the fact that they produce not primary databases but rather contain derived data. This shortcoming may limit the acceptance of these secondary databases as a source of reliable structural information, even though the vast majority of the models that they contain fit quantitatively better to the original experimental data. However, automatic re-refinement has its limitations and sometimes does not eliminate even simple errors [22].
The suboptimal quality of some models is especially visible for structures of protein– ligand complexes, whereas exactly these structures are critical for testing and application of CADD methods. For example, recent publications show that a significant number of metal ions reported in the PDB are misassigned and/or incorrectly refined [22,29–31]. Moreover, a significant number of small molecule ligands reported in the PDB do not have sufficient continuous electron density to support their presence and location [26,27]. Many crystallographic deposits in the PDB for which structure factors are available contain significant regions of unidentified density. In some cases, these unidentified regions are modeled as either sets of ‘unknown’ atoms or as solvent waters. A brief analysis of the PDB shows that there are around 2000 structures which have unexplained continuous densities not modeled by either UNK or UNL ‘atoms’. Whereas they represent only 2% of the contents of the PDB as a whole, they include nearly 6% of all unique structures (with ‘uniqueness’ of a macromolecular chain defined as < 90% sequence identity compared to any other structure in the PDB). In these cases, the ‘extra’ information about unknown ligands – which may be very useful to a biochemist or chemist knowledgeable about a target – is completely absent from the PDB model. Identifying such chemical information requires a close examination of the electron density map calculated using the deposited structure factors. Unfortunately, the use of structure factors to generate electron density requires some rudimentary crystallographic knowledge not possessed by all ‘consumers’ of the PDB structures, as well as access to methods for dealing in subsequent analyses with unidentified ligands. Even if a consumer of the structure possesses the crystallographic skills to verify a model, large-scale validation of thousands of PDB models (e.g., by calculating the agreement of a model with its electron density map) is a task very difficult, if not impossible, for individual researchers or even for small research centers.
Although the average quality of macromolecular structures has improved steadily due to advancements in both X-ray crystallography techniques and validation programs [32], the goal of structural validation is not just ensuring that structural models deposited in the PDB are of the best quality. Validation is also important for objectively assessing the models as valid and useful for subsequent research. This is a challenging task, as at present there is no single parameter that fully describes the usefulness of a structure for a particular purpose. However, there is a set of relatively few parameters – resolution, volume of unexplained density, the agreement of stereochemical parameters with the known ideal values and reasonable ligand assignment – which, in our opinion, can reasonably well quantify the usefulness of a structure for structure-based drug discovery.
2. How may a protein crystal structure mislead drug discovery?
Often, both the in silico and in vitro/in vivo work of pharmaceutical researchers is driven by either direct or indirect availability of structural information. Advancements in high-throughput structural biology and structural genomics projects [33], together with the availability of highly automated software packages for processing X-ray diffraction data [34,35], have made structure determination itself a process that no longer mandates fundamental understanding of the underlying crystallographic theory. Moreover, a significant number of crystal structures are used in various stages of the drug discovery pipeline by researchers not necessarily involved in their determination. Unlike many experiments where a positive or a negative outcome can be judged by an established confidence level or expectation value, evaluating the reliability of an X-ray diffraction model as being suitable for drug discovery studies is much harder to quantify and requires consideration of measurement inaccuracies ranging across multiple levels. The sources of these inaccuracies include i) conformational changes and the flexible nature of the proteins and nucleic acids themselves; ii) the physical setup of the diffraction data collection process; iii) temporal and spatial averaging of the crystal lattice; iv) the level of experience of the person who interprets it; and v) the functional interpretation of ‘active sites’ and intermolecular interfaces. Moreover, different structures may require specific treatment on a case-by-case basis.
2.1 Nature of macromolecular X-ray crystallographic experiments
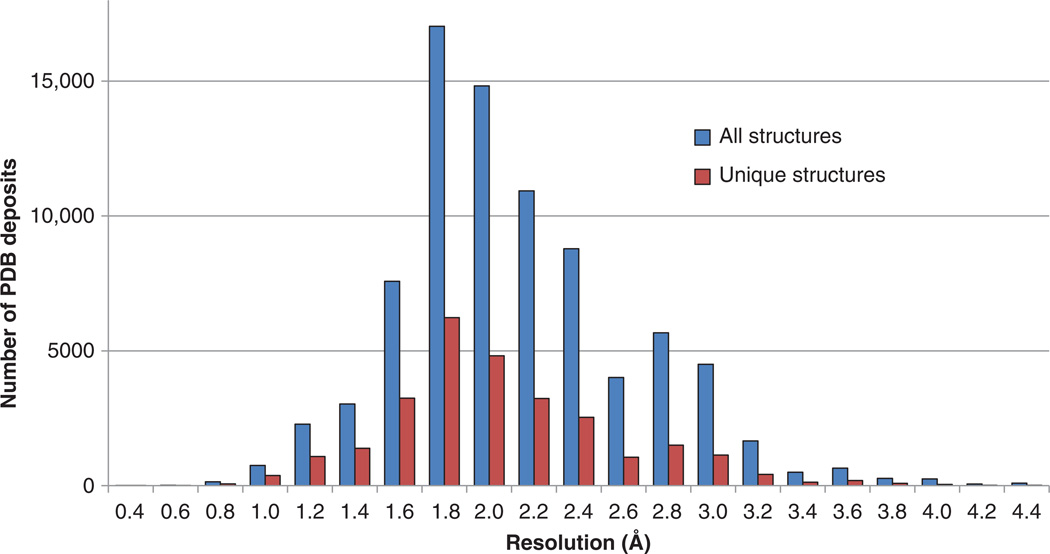
The most often cited parameter related to the quality of macromolecular crystallography data is the diffraction limit – often just called resolution – which reflects the long-range order of a crystal and hence the degree to which it diffracts X-rays. Resolution is more or less a measure of the degree of detail of the electron density maps. At high resolutions (better than 1 Å), peaks indicating the positions of individual atoms can be clearly distinguished. Conversely, at low resolution (around 3 – 4 Å), only the basic contours of a macromolecule backbone are observed in the density and it may be impossible to produce an atomic model with any degree of certainty. The overwhelming majority of macromolecular crystal structures are at moderate resolution, which falls in between these two extremes (Figure 1). When the resolution is moderate or low, it is worse than typical covalent bond distances (1.2 – 1.3 Å), and, thus, there are no separated peaks in the electron density that could be used to determine the atomic coordinates directly (as is the case in small molecule crystallography). At such a resolution, covalently linked atoms are instead represented by contiguous regions of electron density, but atomic positions can still be determined with a reasonable accuracy using both the electron density and prior chemical information. In any case, the uncertainty of the atomic positions strongly depends on the resolution of the diffraction data.
Figure 1. Distribution of resolution of macromolecular structures determined by X-ray crystallography deposited to and released by the PDB prior to October 2013 is shown.
The number of all structures and unique structures (as defined by < 90% sequence identity) are shown for each resolution bin between 0.4 Å and 4.4 Å.
However, the quality of diffraction data also depends on the accuracy and precision of the experimental setup and the skill and experience of the person who performs the diffraction experiment. Inexperienced crystallographers often underestimate the importance of optimizing diffraction protocols [23,29,36,37]. Due to variations in experimental setup limitations, crystal quality and the types of experiments, choosing appropriate diffraction protocols and optimizing them plays a major role in producing the best quality diffraction data, and subsequently, the best models. In addition, crystals can only absorb a limited dose of X-ray radiation before they begin to degrade, which effectively limits the completeness and redundancy of diffraction data that could be collected from a single crystal. In these circumstances, only very experienced experimenters can perform optimal experiments without sophisticated data collection strategy programs. Unfortunately, many strategy programs focus on minimizing the length of an experiment rather than maximizing the amount of information than one can get from a single or multiple crystal(s).
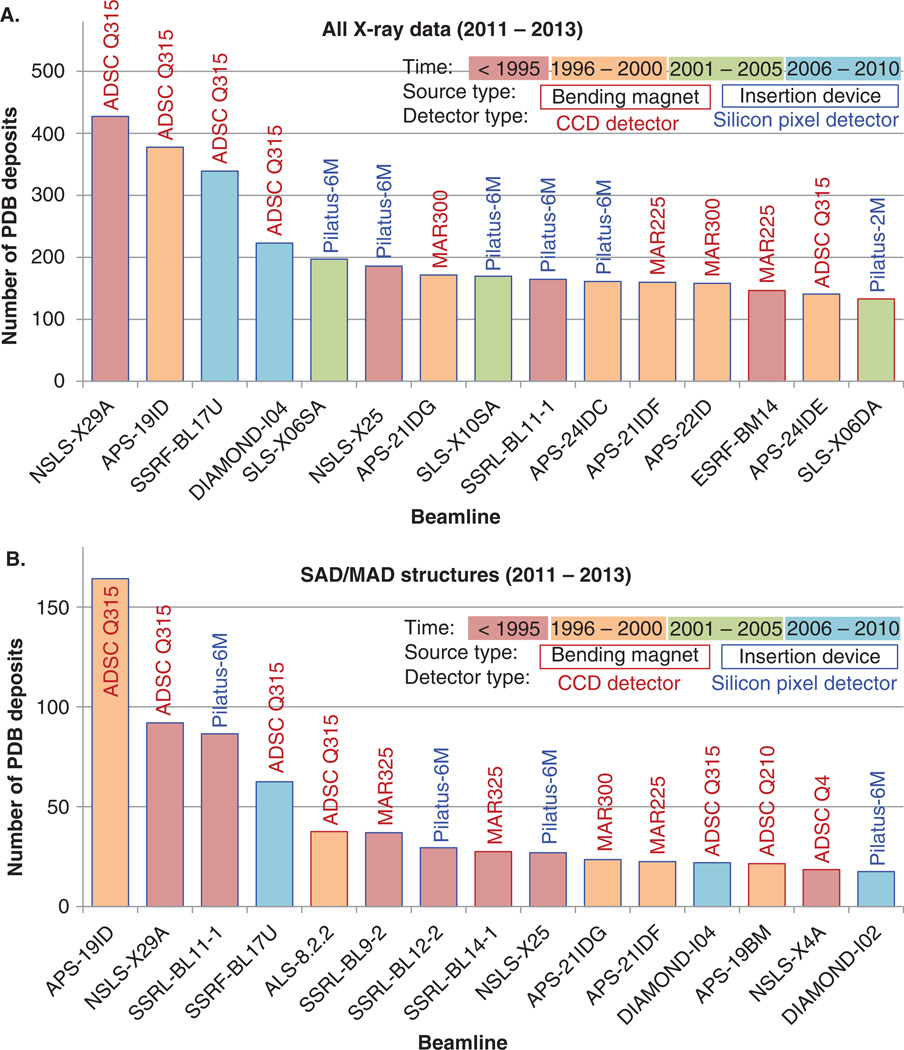
The importance of optimizing the experimental protocols can be illustrated by the productivity of synchrotron beamlines, as measured by the number of depositions to the PDB. Figure 2 shows the number of PDB deposits between January 2011 and September 2013 (including all deposits and single wavelength anomalous dispersion (SAD)/multiple wavelength anomalous dispersion (MAD) deposits) for the 15 most productive synchrotron beamlines in the world. There is no clear correlation between beamline productivity and any aspect of physical setup of the data collection hardware. These 15 beamlines are located at different synchrotrons and vary widely in terms of beam brightness, X-ray optics, detector model, and the like. In our opinion, experimental protocols and customized software that take into account various beamline limitations are critical for ensuring high productivity of a given beamline. It should be noted as well that the best beamlines still average less than one structure a day (when the yearly total is divided by 365). While this calculation has some caveats – synchrotrons typically operate far fewer than 365 days a year, for instance – it is in contrast to frequent reports that only 1 – 5 min of data collection time are needed to generate an entire data set sufficient for structure determination [38,39]. In other words, high throughput is not necessarily correlated with high output.
Figure 2. Recent PDB deposits generated by the 15 most productive synchrotron beamlines, as determined by the number of (A) all X-ray structures with data collected at a synchrotron facility during 2011 to 2013, or (B) SAD/MAD structures with data collected during 2011 to 2013 are shown. The setup of each beamline is shown by the time of synchrotron construction, detector and detector type used, and beamline source type.
APS: Advanced Photon Source, Argonne, IL, USA; NSLS: National Synchrotron Light Source, Brookhaven, NY, USA; Diamond – Harwell, UK; SSRF: Shanghai Synchrotron Radiation Facility, Shanghai, China; SLS: Swiss Light Source, Villigen, Switzerland; ESRF: European Synchrotron Radiation Facility, Grenoble, France; SSRL: Stanford Synchrotron Radiation Laboratory, Palo Alto, CA, USA.
Another factor to consider is how accurately the electron density of macromolecules that are located in a crystal lattice corresponds to the structure of those molecules in solution. In solution, macromolecules are quite flexible and dynamic and typically accommodate a relatively wide spectrum of conformations, including both large-scale motions and local conformational changes (e.g., oscillations of a protein side chain). Ideally, a macromolecular structure should be represented as an animated video showing the trajectory of multiple possible conformations transforming into each other. However, an electron density map (and the atomic model built from it) actually represents an averaged ‘snapshot’ of an ensemble of macromolecules, both temporally over the period of data collection and spatially over all individual copies of the macromolecules in the crystal lattice. Thus, motions on a time scale faster than the period of data collection and changes in conformation across different copies of the macromolecule are ‘smeared out’ and generally cannot be characterized. (One exception is the local vibrations of individual atoms, which are quantified by atomic displacement parameters [40].) This also results in regions of the electron density map where either a superposition of multiple conformations of atoms is observed, or no ordered density is seen at all. Moreover, the range of averaged structural fluctuations observed in the crystalline state is unlikely to represent the full magnitude of fluctuations in the physiological solution state [41]. In this aspect, a combination of X-ray diffraction with other methods that provide more information about the solution dynamics of the target of interest, such as nuclear magnetic resonance, could be extremely beneficial.
The sources of these inaccuracies are rarely evaluated in structure-based drug design, mainly due to the lack of a comprehensive and feasible methodology for quick and routine consideration of coordinate errors, the effects of structure averaging and structural fluctuations. Although subsequent molecular dynamics simulation of a structure before further analysis can partially alleviate some problems through computational means, it is generally not possible to compensate for all types of inaccuracies resulting from the experimental measurements.
2.2 Art of electron density map interpretation
Unlike small molecule crystallography, macromolecular crystallography is notorious for its poor data-to-parameter ratio due to the limited number of diffraction spots, especially for lower resolution structures. In practice, an effective data-to-parameter ratio can be achieved only by applying additional restraints based on prior knowledge of the general properties of macromolecules. The most notable feature of both proteins and nucleic acids is that they are linear polymers composed almost exclusively of a limited set of common subunits (20 amino acids or 5 nucleotides). For proteins, this leads to well-investigated stereochemistry of amino acids [42] and peptide bonds [43], along with the unambiguous chemical identity of the polymer represented by the polypeptide sequence(s). Similar type of information is used to restrain structures of DNA and RNA. In general, the basic stereochemistry of the protein, RNA and DNA represented by an electron density is well known, and thus model building of the macromolecular part of a crystal structure can, in most cases, be fully (or nearly fully) automated. In other words, the procedures used in building the macromolecular components of a structure are usually highly reproducible, regardless of the person performing them.
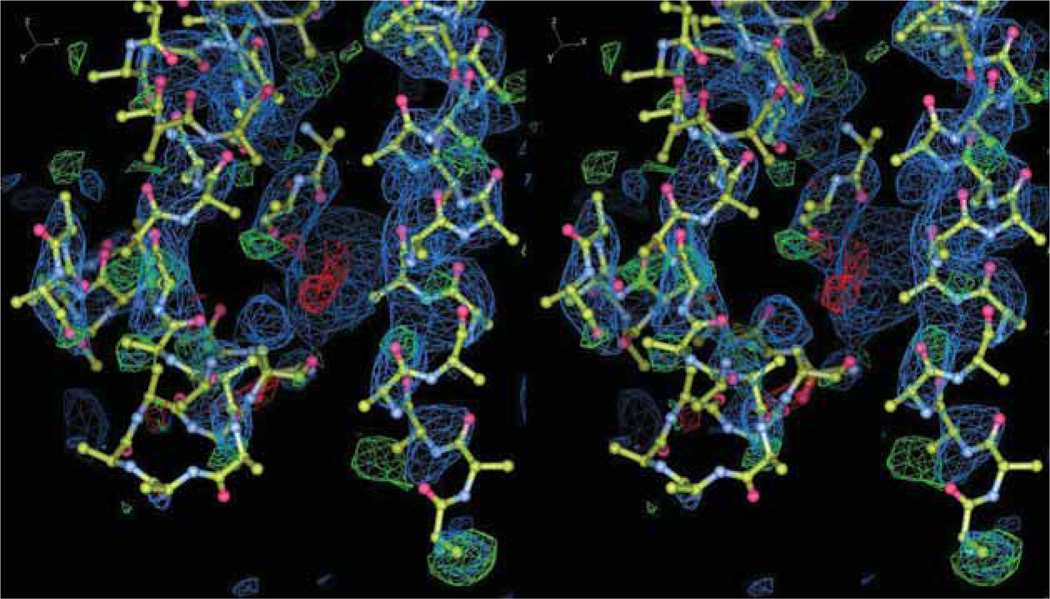
However, the models of macromolecules deposited in the PDB do not always accurately interpret their electron density. A practical example is the crystal structure (PDB deposit 2FHS) of enoyl reductase (FabI) complexed to acyl carrier protein (ACP), in which helix α2 of ACP was predicted to form an interaction with helix α8 of FabI. Both proteins are actively pursued as targets for new antibiotics that would disrupt the fatty acid synthesis pathway in bacteria [44]. When the electron density map for the ACP is investigated in detail, even the main chain of the model fits the density poorly (Figure 3). This result makes conclusions deduced from the crystal structure (i.e., that the intermolecular interaction seen in the complex structure is functionally relevant) and the follow-up computational analysis highly uncertain. Nevertheless, the paper describing this complex structure has been cited > 40 times and the deposit has been downloaded > 28,000 times.
Figure 3.
Sigma-A weighted 2Fo-Fc and Fo-Fc electron density maps with the corresponding modeled coordinates of the ACP component of the FabI– ACP complex from Escherichia coli (PDB code: 2FHS), shown in wall-eyed stereo. The maps were calculated using the deposited experimental structure factors and deposited model. The difference map (Fo-Fc) is shown in green and red at the ± 3.0 σ contour levels, whereas the 2Fo-Fc density map is contoured at 1.0 σ.
Apart from the electron density that corresponds to the main macromolecule(s), the interpretation of residual ‘blobs’ remaining after the macromolecules have been built suffers from ambiguities in chemical identity. Such interpretation can be highly subjective depending on the experience and knowledge of the crystallographer, even for high-resolution structures [45]. Owing to the vast chemical vocabulary that these residual electron densities may represent, the chemical knowledge and crystallographic experience of the researcher may have a significant effect on the final steps of the macromolecular crystal structure determination process.
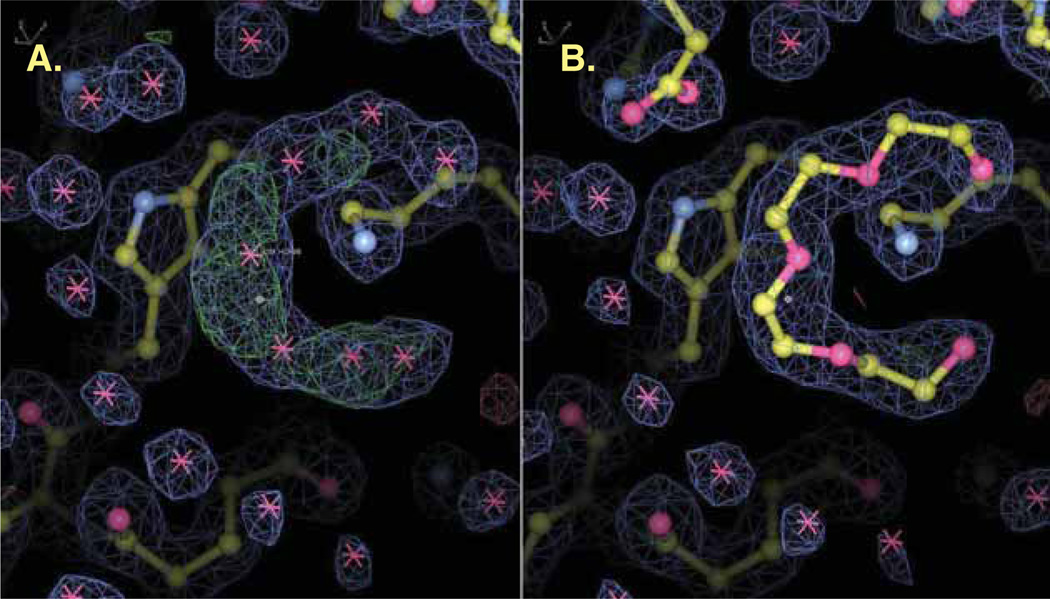
Faithful and precise interpretation of residual density requires familiarity with both the target macromolecule and the structure determination and refinement process. (Note: we use the term ‘residual density’ to refer to non-modeled, non-solvent density remaining after the macromolecule chains comprising the 20 common amino acids and/or the 5 common nucleotides have been built.) In this sense, a residual density may, in fact, represent a moiety that is covalently linked to a macromolecule. The first step of analysis is to tentatively reveal the chemical identity of residual densities by considering the chemicals or the products of chemical reactions introduced by experimental conditions used in sample production, crystallization, soaking or cryoprotection. Such an analysis requires detailed and accurate recordkeeping of the experimental procedures that are used in producing the crystals, ideally through the means of sophisticated laboratory information management systems that are capable of harvesting data automatically during experiments (such as LabDB [46]). The chemicals used during sample preparation are usually present in the experimental environment at high concentration, which can easily result in nonspecific binding. For that reason, the detection and modeling of small molecules in a crystal structure cannot prove their physiological relevance alone without corresponding enzymatic or binding assays. There are also many other circumstances where one may observe residual electron densities that cannot be explained by any chemicals introduced during sample preparation, but rather by endogenous chemical modifications and/or small molecules (such as metabolites) internally bound by the macromolecule prior to its purification. Continuous electron density usually hints at the presence of chemical bonds stronger than the typical hydrogen bonds formed by crystallized water, which might be of great biological significance. However, it is not uncommon to see a PDB deposit with a large ‘blob’ of electron density modeled as multiple water molecules or just annotated as unknown atoms (Figure 4). Quite often there are differing opinions as to the optimal way to interpret these residual densities, depending on the views, knowledge and experience of individual crystallographers.
Figure 4.
The unmodeled blobs in electron density maps should be critically evaluated. The unmodeled blobs (A) in the structure of crystal structure of aq_1716 from Aquifex aeolicus vt5 (PDB code: 2P68) should be more appropriately interpreted as PEG, rebuilt and refined as shown in (B). In both images, 2Fo-Fc electron density contoured at 1.0 σ is shown in blue, and Fo-Fc difference density contoured at ± 3.0 σ is shown in green and red. Ninety minutes of model building and re-refinement with HKL-3000 was sufficient to complete chain B, build other ligands and decrease R from 18.5 to 14.6% and Rfree from 22.3 to 18.2%.
Besides strong peaks that would hint at the presence of heavy atoms, a general strategy is to inspect the distances between each pair of peaks in the electron density maps to figure out the type of each bond to build the heterogeneous component in a ‘bottom-up’ manner. Residual density may be connected to the macromolecular structure through five major types of bonds: i) covalent bonds that chemically modify common amino acid and nucleic acid residues to produce nonstandard residues or other chemical modifications, such as acetylation or glycosylation; ii) bonds that coordinate metal cations; iii) ionic bonds between cations and anions; iv) hydrogen bonds; and v) hydrophobic (van der Waals) interactions. Common small molecules to be modeled include metal ions, anions, organic solvent molecules, common cofactors, glycans, and the like. A good candidate solution should be one that is chemically sensible, satisfies known restraints, and, if possible, is biologically relevant.
However, the number of different organic compounds that may be found is huge, and building a comprehensive library to cover the chemical space is a daunting task. Biological macromolecules (even recombinant ones) prepared for crystallization (or any other in vitro analyses) are almost always purified from in vivo sources. Therefore, by the time a crystal has formed, a given macromolecule may have been exposed to tens of thousands of different small molecule compounds endogenous to the expression organism, not to mention the dozens of compounds and reaction products found in the purification and crystallization buffers. Efforts to build tools to screen a chemical library to find and fit compounds into electron densities are under active exploration [47]; however, undocumented chemicals and unexpected binding modes are frequently encountered [48]. Under such circumstances, the experience of a crystallographer, especially in organic chemistry and bioinorganic coordination chemistry, plays an important role in the faithful and precise interpretation of the residual electron densities. Fortunately, there has been recent demonstrated increase of interest in the issue of quality assessment in ligand modeling [27,49,50].
2.3 The knowns and unknowns during structure-based functional exploration
Besides the considerations of structure imprecision originating from sample quality, experimental setup and human input (both in data collection and interpretation), the lack of precision in the structure– function relationship is the major bottleneck/obstacle for structure-based drug discovery. Compared with the mature macromolecular structure determination pipelines, large-scale Big Data approaches to the exploration of structure– function relationships are rather preliminary.
Understanding the molecular mechanism of a drug target is clearly critical for better utilization of a crystal structure in CADD [51]. While the general chemical, physiological and pathological properties of a drug target have usually been characterized previously, the mechanisms of action that may explain these properties are not necessarily known. Structure-based functional exploration typically includes aspects ranging from a local structural feature to global configurations. Local structural features may include conformations of active site residues, related to a specific biological function, while global configurations may include tertiary and quaternary structures which are responsible for higher level activity regulation and/or creation of binding interfaces of macromolecular complexes. On the tertiary structure level, when the target is an enzyme or a small molecule receptor, the structure-based functional exploration usually involves identification and location of the active site(s), ligands (substrates, cofactors, products, etc.) and their binding modes, and, in the case of enzymes, the catalytic residues and waters involved in the chemical reaction [52]. On the quaternary structure level, it usually involves investigation of the biological units, including complex formation, oligomerization state and allosteric regulation, and the like.
General strategies to characterize an active site and explore the efficacy of inhibitors include methods that are structure-based (e.g., virtual screening [24]) and non-structure-based (e.g., high-throughput screening [53]). Here, we describe two other, more specific structure-based strategies. One of them involves a series of assumptions derived from the ‘key’ residues or ‘hot spots’ [54] in the atomic coordinates, followed by biological assays. Such a strategy for activity identification, guided by structural information, may be successful under certain circumstances [55] but might not be generally applicable. Another strategy is to perform customized cocktail soaking of metabolite libraries in order to obtain structures of macromolecules in complex with specifically bound small molecules [56]. The latter strategy is more flexible since it may or may not need to utilize hints from the structures of the apo-form up front in order to guide the selection of the metabolite library. In many cases, non-structural information (e.g., physiological, biological, metabolic, gene loci, etc.) is needed to select the contents of the initial metabolite library. Nevertheless, the flexibility of this method may require a search of a large chemical space to detect an effectively bound compound.
Inter-macromolecular interactions observed in a crystal may allow structure-based functional exploration on the quaternary structure level, but the determination of whether the intermolecular interfaces are biologically relevant is also challenging. Usually mutagenesis or other studies are needed to verify that intermolecular contacts observed in the crystal structures are biologically relevant [57,58]. Crystallization experiments – even though they are carried out in solution– are influenced by the ubiquitous presence of crystal-packing forces [59], so it would be more precise to consider crystallization experiments separately as the in crystallo state to be differentiated from the in vitro state. Since the crystalline state differs significantly from the physiological solution state, the inter-macromolecular interfaces observed in the crystal need to be critically evaluated to determine if they are potentially crystal packing artifacts. In the case of stable oligomers or strong interactions with tight binding interfaces (e.g., antibody– antigen interactions), the likely biologically relevant interfaces can typically be distinguished from the crystal packing interfaces by structure analysis methods, such as PDBe-PISA [60]. However, in many cases, transient interactions between macromolecules are actually extremely difficult to capture in the crystallized state. In such cases, the crystal packing energy is likely to be of magnitude comparable to the energy involved in the formation of a transient complex [61]. Therefore, it may not be possible to conclusively differentiate true, biologically relevant interfaces from crystal packing interfaces using only crystal structure data in the absence of other in vivo or in vitro evidence, let alone if there are inaccuracies in interpretation of the relevant electron density map regions (Figure 3).
3. Expert opinion
The objective and realistic utilization of a crystal structure is full of pitfalls. Any analyses that use atomic coordinates should always consider not only the resolution but also the methods used for structure determination, refinement and the statistics that describes both. All drug discovery researchers using a crystal structure as a starting point for subsequent studies should i) pay special attention to the technical statistics and parameters described in the PDB file header; ii) verify the geometry and electron density agreement of all heterogeneous residues, which could be highly subjective; and, iii) trace the evidence for all functional exploration, including biological unit assignment. That being said, even for crystal structures of the highest quality possible, critical assessment of the structure should still address the extent of experimental errors and how well the structure represents the target protein in solution. This could be used as a basis to determine to what degree and in what contexts the crystal structure should be considered as reliable for various structure-based drug discovery analyses.
As X-ray crystallography is still one of the best tools for rational drug discovery, the authors would like to emphasize the importance of not only individual efforts but also community-wide efforts for validation of crystal structures. There is a need to either expand the PDB or create a new organization to maintain a library of validated and representative structures along with a checklist of the latest structure validation tools. Since validation techniques are constantly evolving, new validation tools should be applied not only to newly deposited structures but also to the previously deposited structures that should be revisited periodically. Moreover, the exploration of structural ensembles of a single experimentally determined crystal structure is one way to account for protein flexibility in virtual screening and subsequent structure-based drug discovery analyses [62–66].
While macromolecular structures may provide hints about molecular function, typically the information revealed is context-dependent and further biochemical or biophysical experiments are needed to verify these functions. However, in many cases, crystallography should be used to verify aspects of functional experiments: for example, by analysis of reaction mechanisms using transition state analogs or identification of confounding factors, such as binding artifacts derived from protein production buffers [67].
Due to the presence of uncertainty even in validated structural models, the authors also suggest that a crystal structure should not be thought of as a collection of precise atomic coordinates but rather as a framework of hypotheses to be explored and verified (or falsified) experimentally. A static set of atomic coordinates represented by a macromolecular structural model is the closest approximation of macromolecular structures that crystallography can determine and represent. Crystal structures cannot accurately capture all of the structural fluctuations that a macromolecule adopts under physiological conditions and this may unnecessarily confine experimental design in the search of inhibitors. Considered as a hypothesis framework, a crystal structure may be used as the basis to propose or prioritize biological assays in experimental design but should not be used to prove or rule out any hypotheses without further experimental evidence.
3.1 Future challenges
Future challenges for crystallography in structure-based drug discovery are in the fields of data validation, data mining and data management. State-of-the-art validation methodologies in protein crystallography have been broadly documented [32]. However, successful application of these validation principles requires continuous efforts. Easy-to-use and sophisticated tools for the critical assessment and realistic interpretation of macromolecular model coordinates are still in short supply. Advanced tools designed to tackle the aforementioned pitfalls should be of particular interest. These include tools for the visualization and analysis of structure determination statistics, atomic displacement and translation libration screw motions (TLS) parameters [68], and structural fluctuations, as well as validation protocols for verifying stereochemistry and agreement with the electron density of all heterogeneous regions of macromolecular models [26].
Another important issue is preserving and making generally available as much raw data as possible. The PDB has already made large strides in this area, by requiring deposition of structure factors along with a refined model. The availability of diffraction data permits others in the community to evaluate a deposit versus its electron density map and possibly identify over- and/or under-modeled regions of the structure. However, in some cases even structure factor data may be insufficient for structure verification; in these cases, raw diffraction images are needed. A prominent example is reinterpretation of crystallographic data collected on mammalian 15S–lipoxygenase. The originally deposited structure of this protein was solved and modeled in the space group R32 [69]. However, when the structure factors were re-indexed in another space group (R3) and the structure was rebuilt, the shape and size of the substrate-binding cavity changed significantly and the cavity was no longer able to accommodate the ligands proposed to bind in it [70]. The controversy is still unresolved, since any drug discovery researcher would not know which interpretation is correct, as only the structure factors were retained and there were no follow-up experiments. The original deposit 1LOX, as well as the reinterpreted model 2P0M, was each downloaded around 34,000 times.
There has already been a call for organized efforts to collect diffraction images [71]. A number of structural genomics centers, including the Center for Structural Genomics of Infectious Diseases, the Midwest Center for Structural Genomics and Joint Center for Structural Genomics, already make diffraction images for their structural deposits available. After all, electron density maps and structure factor files are derived data, which may not be optimally abstracted from the raw data, that is, the diffraction images. Another indicator of the benefits of preserving raw diffraction images comes from the recent introduction of the CC/2 statistic [72]. This measure provides an alternative method for determining which reflections should be included in crystallographic data reduction. It is possible to reprocess previously deposited structures to higher resolution only if raw diffraction images are available. In addition, new versions of some data reduction programs allow correcting data for radiation decay or for anisotropic diffraction [35,73,74].
Moreover, an important challenge to face is the development of intelligent tools to help establish hypotheses, design experiments and prioritize the order of experiments based on the ‘hypotheses framework’ to facilitate the data mining process. One example of a next-generation intelligent tool in pharmaceutical research is related to a recent breakthrough of the IBM Watson artificial intelligence system developed by the DeepQA research team [75]. However, as tools continue to increase in their level of sophistication, it is still wise to consider the reliability and quality of the data they mine. Any statistical inference is only as reliable as the corpus of data used to generate it, regardless of the level of ‘intelligence’ of the analysis tool. In other words, the principle of ‘garbage in, garbage out’ equally applies to artificial intelligence systems.
Processing of structural information is rapidly becoming more sophisticated, particularly when combined with functional and evolutionary data, and in the context of interaction networks with other biomacromolecules or bioactive chemical compounds. This increasingly requires the use of Big Data paradigms for effective data management [76], as well as for checking data integrity and accuracy. Big Data traditionally refers to the analysis of very large datasets (on the scale of tera- or peta-bytes), but the scale of what it constitutes varies significantly based on the application domain. With the availability of cloud computing and Big Data technologies in genomics [9], new technologies must be developed (or existing technologies adopted) to handle and retrieve structural data and its connections to other data, such as structural flexibility measurements and functional exploration. For example, large-scale macromolecular structure data may benefit from the effective usage of map-reduce paradigms implemented by tools such as Hadoop [77]. This would be most beneficial for establishing strategies to limit the scope of experimental screening and to keep track of all evidence for functional exploration.
However, implementation of Big Data paradigms does not necessarily require massive computation clusters and Google-sized data storage. The most important Big Data paradigm can be summarized by Clifford Stoll’s quotation, ‘Data is not information, information is not knowledge, knowledge is not understanding, understanding is not wisdom’ [78]. Big Data tools, techniques and technologies may be productively applied to working with data at any scale, large or small. The tools to support data harvesting, data mining, computations and sharing data with collaborators should all be available in a straightforward way. The systems should generate reports and dashboards that present information (not data) that helps to manage a project (scientific or not), identify bottlenecks and opportunities, eliminate and suggest scientific experiments, verify experimental protocols, and the like. This is easy to say but extremely difficult to implement, as development of these tools takes time, effort, and most of all, creativity of the leaders and developers of the projects. This is more expensive than just purchase of supercomputers with petabyte storage. A combination of advancements in high-quality data validation, data mining and data management tools would make it possible to convert high-throughput pipelines into high-output pipelines in target-based drug discovery.
Article highlights.
The use of public and private structural information is critical for structure-based drug discovery. The assessment of the quality of private structural data is impossible. The quality of public structural data overall is very good but a small number of poor quality structures need to be excluded from subsequent analyses. The deposition of structure factors allows for re-refinement of suspicious models and validation of structures.
The direct measurement of a macromolecular X-ray crystallographic experiment is the diffraction pattern and not the electron density map that is produced from it. Bias can be introduced into the map by the methodology used to obtain phases.
The macromolecular model is an interpretation of the electron density map. This interpretation may also be biased, especially when experimenters identify and place small molecular compounds adjacent to macromolecules.
The integration of structural, functional and bioinformatics data leads to better information and understanding of mechanisms of action. Experiments that provide contradictory results should not be disregarded but rather carefully analyzed.
Preservation and general availability of as much raw data as possible is critical for clarification of disputable interpretations and/or contradictory results.
The future challenges for crystallography in structure-based drug discovery are in the underappreciated fields of data validation, mining and management. Processing of structural information, combined with functional and other experimental and bioinformatics data, requires the use of Big Data paradigms.
This box summarizes key points contained in the article.
Acknowledgments
The authors’ research was supported by federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under Contract No. HHSN272201200026C, by NIH grants GM094662, GM093342 and GM094585, and in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Footnotes
Declaration of interest
The authors state no conflict of interest in preparation of this manuscript.
Bibliography
Papers of special note have been highlighted as either of interest (•) or of considerable interest (••) to readers.
- 1.Song CM, Lim SJ, Tong JC. Recent advances in computer-aided drug design. Brief Bioinform. 2009;10(5):579–591. doi: 10.1093/bib/bbp023. [DOI] [PubMed] [Google Scholar]
- 2.Kortagere S, Lill M, Kerrigan J. Role of computational methods in pharmaceutical sciences. Methods Mol Biol. 2012;929:21–48. doi: 10.1007/978-1-62703-050-2_3. [DOI] [PubMed] [Google Scholar]
- 3.SYBYL-X. St. Louis, MO: Tripos International; 2013. [Google Scholar]
- 4.Discovery Studio. San Diego, CA: Accelrys, Inc.; 2013. [Google Scholar]
- 5.Maestro. Schrödinger: 2013. [Google Scholar]
- 6.Scott DE, Coyne AG, Hudson SA, Abell C. Fragment-based approaches in drug discovery and chemical biology. Biochemistry. 2012;51(25):4990–5003. doi: 10.1021/bi3005126. [DOI] [PubMed] [Google Scholar]
- 7.Berman HM, Coimbatore Narayanan B, Di Costanzo L, et al. Trendspotting in the protein data bank. FEBS Lett. 2013;587(8):1036–1045. doi: 10.1016/j.febslet.2012.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berman HM, Kleywegt GJ, Nakamura H, Markley JL. How community has shaped the protein data bank. Structure. 2013;21(9):1485–1491. doi: 10.1016/j.str.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. O'Driscoll A, Daugelaite J, Sleator RD. ‘Big data’, hadoop and cloud computing in genomics. J Biomed Inform. 2013;46(5):774–781. doi: 10.1016/j.jbi.2013.07.001. •• The application of the Big Data paradigm to biological data.
- 10.Schrödinger L. The PyMOL molecular graphics system. Version~1. 2010;3r1 In press. [Google Scholar]
- 11.Feng E, Ye D, Li J, et al. Recent advances in neuraminidase inhibitor development as anti-influenza drugs. ChemMedChem. 2012;7(9):1527–1536. doi: 10.1002/cmdc.201200155. [DOI] [PubMed] [Google Scholar]
- 12.Callens M, Hannaert V. The rational design of trypanocidal drugs: selective inhibition of the glyceraldehyde-3-phosphate dehydrogenase in trypanosomatidae. Ann Trop Med Parasitol. 1995;89(Suppl 1):23–30. doi: 10.1080/00034983.1995.11813011. [DOI] [PubMed] [Google Scholar]
- 13.Srivastava HK, Bohari MH, Sastry GN. Modeling anti-HIV compounds: the role of analogue-based approaches. Curr Comput Aided Drug Des. 2012;8(3):224–248. doi: 10.2174/157340912801619085. [DOI] [PubMed] [Google Scholar]
- 14.Wlodawer A. Rational approach to AIDS drug design through structural biology. Annu Rev Med. 2002;53:595–614. doi: 10.1146/annurev.med.53.052901.131947. [DOI] [PubMed] [Google Scholar]
- 15.Heikamp K, Bajorath J. The future of virtual compound screening. Chem Biol Drug Des. 2013;81(1):33–40. doi: 10.1111/cbdd.12054. [DOI] [PubMed] [Google Scholar]
- 16.Durrant JD, McCammon JA. AutoClickChem: click chemistry in silico. PLoS Comput Biol. 2012;8(3):e1002397. doi: 10.1371/journal.pcbi.1002397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vogt M, Bajorath J. Chemoinformatics: a view of the field and current trends in method development. Bioorg Med Chem. 2012;20(18):5317–5323. doi: 10.1016/j.bmc.2012.03.030. [DOI] [PubMed] [Google Scholar]
- 18.Audie J, Swanson J. Advances in the prediction of protein-peptide binding affinities: implications for peptide-based drug discovery. Chem Biol Drug Des. 2013;81(1):50–60. doi: 10.1111/cbdd.12076. [DOI] [PubMed] [Google Scholar]
- 19. Davis AM, Teague SJ, Kleywegt GJ. Application and limitations of X-ray crystallographic data in structure-based ligand and drug design. Angew Chem Int Ed Engl. 2003;42(24):2718–2736. doi: 10.1002/anie.200200539. • A review presenting the use of X-ray crystallography data for drug discovery.
- 20.Davis AM, St-Gallay SA, Kleywegt GJ. Limitations and lessons in the use of X-ray structural information in drug design. Drug Discov Today. 2008;13(19–20):831–841. doi: 10.1016/j.drudis.2008.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Acharya KR, Lloyd MD. The advantages and limitations of protein crystal structures. Trends Pharmacol Sci. 2005;26(1):10–14. doi: 10.1016/j.tips.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 22. Chruszcz M, Domagalski M, Osinski T, et al. Unmet challenges of structural genomics. Curr Opin Struct Biol. 2010;20(5):587–597. doi: 10.1016/j.sbi.2010.08.001. •• An overview of current status and possible future benefits of structural genomics for drug discovery. The study describes the importance of data management and its impact on the accuracy of PDB deposits and provides a discussion of standards that should be used to make structures more easily accessible for the wider biomedical community.
- 23. Wlodawer A, Minor W, Dauter Z, Jaskolski M. Protein crystallography for aspiring crystallographers or how to avoid pitfalls and traps in macromolecular structure determination. FEBS J. 2013;280(22):5705–5736. doi: 10.1111/febs.12495. •• A paper discussing all aspects of protein crystallography, including structure quality and limitations of the technique. A ‘must read’ not only for protein crystallographers but also, most of all, for scientists who apply structural biology results in biomedical research.
- 24.Sukumar N, Das S. Current trends in virtual high throughput screening using ligand-based and structure-based methods. Comb Chem High Throughput Screen. 2011;14(10):872–888. doi: 10.2174/138620711797537120. [DOI] [PubMed] [Google Scholar]
- 25.Scior T, Bender A, Tresadern G, et al. Recognizing pitfalls in virtual screening: a critical review. J Chem Inf Model. 2012;52(4):867–881. doi: 10.1021/ci200528d. [DOI] [PubMed] [Google Scholar]
- 26. Pozharski E, Weichenberger CX, Rupp B. Techniques, tools and best practices for ligand electron-density analysis and results from their application to deposited crystal structures. Acta Crystallogr D. 2013;69:150–167. doi: 10.1107/S0907444912044423. •• Recent advances in validation techniques used in assessing the validity of ligand placement and identification in protein structures. Required reading for practicing structural biologists.
- 27. Weichenberger CX, Pozharski E, Rupp B. Visualizing ligand molecules in twilight electron density. Acta Crystallogr F. 2013;69(Pt 2):195–200. doi: 10.1107/S1744309112044387. • Recent advances in validation techniques used in assessing the validity of ligand placement and identification in protein structures. Required reading for practicing structural biologists.
- 28. Joosten RP, Joosten K, Murshudov GN, Perrakis A. PDB_REDO: constructive validation, more than just looking for errors. Acta Crystallogr D. 2012;68(Pt 4):484–496. doi: 10.1107/S0907444911054515. • A description of a project to provide an improved version of PDB by automatically and systematically re-refining PDB deposits. The automated procedure has its limitations, but comparison of the results of PDB_REDO with the original deposits in the PDB can be useful for structure evaluation.
- 29. Wlodawer A, Minor W, Dauter Z, Jaskolski M. Protein crystallography for non-crystallographers, or how to get the best (but not more) from published macromolecular structures. FEBS J. 2008;275(1):1–21. doi: 10.1111/j.1742-4658.2007.06178.x. • A paper discussing all aspects of protein crystallography, including structure quality and limitations of X-ray crystallography.
- 30. Zheng H, Chruszcz M, Lasota P, et al. Data mining of metal ion environments present in protein structures. J Inorg Biochem. 2008;102(9):1765–1776. doi: 10.1016/j.jinorgbio.2008.05.006. • A paper presenting the analysis of metal– protein interaction distances, coordination numbers, B-factors (displacement parameters) and occupancies of metal-binding sites in protein structures determined by X-ray crystallography.
- 31. Zheng H, Chordia MD, Cooper DR, et al. Validating metal-binding sites in macromolecular structures with the CheckMyMetal web server. Nat Protoc. 2014;9:156–170. doi: 10.1038/nprot.2013.172. •• A description of an excellent protein model validation tool. Provides a discussion of the difficulties related to placement and identification of metal ions in protein structures. The server is available from http://csgid.org/csgid/metal_sites/.
- 32. Cooper DR, Porebski PJ, Chruszcz M, Minor W. X-ray crystallography: assessment and validation of protein-small molecule complexes for drug discovery. Expert Opin Drug Discov. 2011;6(8):771–782. doi: 10.1517/17460441.2011.585154. • An overview of recent approaches for validation of results obtained by X-ray crystallography as used in drug discovery.
- 33. Grabowski M, Chruszcz M, Zimmerman MD, et al. Benefits of structural genomics for drug discovery research. Infect Disord Drug Targets. 2009;9(5):459–474. doi: 10.2174/187152609789105704. • A review of the possible impact of structural genomics on drug discovery research. The review provides a discussion of the weaknesses and strengths of structural genomics programs and how these programs impact research related to drug discovery.
- 34. Adams PD, Baker D, Brunger AT, et al. Advances, interactions, and future developments in the CNS, phenix, and rosetta structural biology software systems. Annu Rev Biophys. 2013;42:265–287. doi: 10.1146/annurev-biophys-083012-130253. • Paper that in part describes the application of the Rosetta macromolecular modeling suite to protein crystallography.
- 35. Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. HKL-3000: the integration of data reduction and structure solution - from diffraction images to an initial model in minutes. Acta Crystallogr D. 2006;62:859–866. doi: 10.1107/S0907444906019949. •• A paper describing HKL-3000, a semi-automatic system for structure determination.
- 36.Chruszcz M, Wlodawer A, Minor W. Determination of protein structures- a series of fortunate events. Biophys J. 2008;95(1):1–9. doi: 10.1529/biophysj.108.131789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Domagalski MJ, Zheng H, Zimmerman MD, et al. The quality and validation of structures from structural genomics. Methods Mol Biol. 2014;1091:297–314. doi: 10.1007/978-1-62703-691-7_21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Joachimiak A. High-throughput crystallography for structural genomics. Curr Opin Struct Biol. 2009;19(5):573–584. doi: 10.1016/j.sbi.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Walsh MA, Evans G, Sanishvili R, et al. MAD data collection - current trends. Acta Crystallogr D Biol Crystallogr. 1999;55(Pt 10):1726–1732. doi: 10.1107/s0907444999008392. [DOI] [PubMed] [Google Scholar]
- 40. Merritt EA. To B or not to B: a question of resolution? Acta Crystallogr D Biol Crystallogr. 2012;68(Pt 4):468–477. doi: 10.1107/S0907444911028320. • A guide that describes the experimental factors that should be considered in deciding whether and how to refine atomic displacement B factors.
- 41.Makowski L, Rodi DJ, Mandava S, et al. Molecular crowding inhibits intramolecular breathing motions in proteins. J Mol Biol. 2008;375(2):529–546. doi: 10.1016/j.jmb.2007.07.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Engh RA, Huber R. Accurate bond and angle parameters for X-ray protein-structure refinement. Acta Crystallogr A. 1991;47:392–400. [Google Scholar]
- 43.Ramachandran GN, Ramakrishnan C, Sasisekharan V. Stereochemistry of polypeptide chain configurations. J Mol Biol. 1963;7:95–99. doi: 10.1016/s0022-2836(63)80023-6. [DOI] [PubMed] [Google Scholar]
- 44.Rafi S, Novichenok P, Kolappan S, et al. Structure of acyl carrier protein bound to FabI, the FASII enoyl reductase from escherichia coli. J Biol Chem. 2006;281(51):39285–39293. doi: 10.1074/jbc.M608758200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Branden C, Jones T. Between objectivity and subjectivity. Nature. 1990;343(6260):687–689. [Google Scholar]
- 46.Zimmerman MD, Grabowski M, Domagalski MJ, et al. Data management in the modern structural biology and biomedical research environment. Methods Mol Biol. 2014 doi: 10.1007/978-1-4939-0354-2_1. In print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. LigSearch identification of possible ligands from 3D protein structure or sequence [Internet] [Cited 30 July 2013];EMBL-EBI. 2013 Available from: http://www.ebi.ac.uk/thornton-srv/databases/LigSearch/index.html. •• An excellent server for the identification of ligands based on crystal structure.
- 48.Malde AK, Mark AE. Challenges in the determination of the binding modes of non-standard ligands in X-ray crystal complexes. J Comput Aided Mol Des. 2011;25(1):1–12. doi: 10.1007/s10822-010-9397-6. [DOI] [PubMed] [Google Scholar]
- 49.Cereto-Massague A, Ojeda MJ, Joosten RP, et al. The good, the bad and the dubious: VHELIBS, a validation helper for ligands and binding sites. J Cheminform. 2013;5(1):36. doi: 10.1186/1758-2946-5-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dauter Z, Weiss MS, Einspahr H, Baker EN. Expectation bias and information content. Acta Crystallogr F. 2013;69(Pt 2):83. doi: 10.1107/S1744309113001486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee D, Redfern O, Orengo C. Predicting protein function from sequence and structure. Nat Rev Mol Cell Biol. 2007;8(12):995–1005. doi: 10.1038/nrm2281. [DOI] [PubMed] [Google Scholar]
- 52.Brylinski M. Unleashing the power of meta-threading for evolution/structure-based function inference of proteins. Front Genet. 2013;4:118. doi: 10.3389/fgene.2013.00118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hann MM, Oprea TI. Pursuing the leadlikeness concept in pharmaceutical research. Curr Opin Chem Biol. 2004;8(3):255–263. doi: 10.1016/j.cbpa.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 54. Vishveshwara S, Ghosh A, Hansia P. Intra and inter-molecular communications through protein structure network. Curr Protein Pept Sci. 2009;10(2):146–160. doi: 10.2174/138920309787847590. • The usage of protein structure networks to infer functional ‘hot spots’ in crystal structures.
- 55.Hermann JC, Marti-Arbona R, Fedorov AA, et al. Structure-based activity prediction for an enzyme of unknown function. Nature. 2007;448(7155):775–779. doi: 10.1038/nature05981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shumilin IA, Cymborowski M, Chertihin O, et al. Identification of unknown protein function using metabolite cocktail screening. Structure. 2012;20(10):1715–1725. doi: 10.1016/j.str.2012.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Abdurahman S, Hoglund S, Hoglund A, Vahlne A. Mutation in the loop C-terminal to the cyclophilin A binding site of HIV-1 capsid protein disrupts proper virus assembly and infectivity. Retrovirology. 2007;4:19. doi: 10.1186/1742-4690-4-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kovacs JM, Hannan JP, Eisenmesser EZ, Holers VM. Biophysical investigations of complement receptor 2 (CD21 and CR2)-ligand interactions reveal amino acid contacts unique to each receptor-ligand pair. J Biol Chem. 2010;285(35):27251–27258. doi: 10.1074/jbc.M110.106617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Krissinel E. Crystal contacts as nature’s docking solutions. J Comput Chem. 2010;31(1):133–143. doi: 10.1002/jcc.21303. [DOI] [PubMed] [Google Scholar]
- 60. Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372(3):774–797. doi: 10.1016/j.jmb.2007.05.022. •• An algorithm to identify intermolecular interfaces in a crystal structure that are potentially biologically relevant.
- 61.Ramakrishnan B, Shah PS, Qasba PK. Alpha-lactalbumin (LA) stimulates milk beta-1 4-galactosyltransferase I (beta 4Gal-T1) to transfer glucose from UDP-glucose to N-acetylglucosamine crystal structure of beta 4Gal-T1 x LA complex with UDP-glc. J Biol Chem. 2001;276(40):37665–37671. doi: 10.1074/jbc.M102458200. [DOI] [PubMed] [Google Scholar]
- 62.Osterberg F, Morris GM, Sanner MF, et al. Automated docking to multiple target structures: incorporation of protein mobility and structural water heterogeneity in AutoDock. Proteins. 2002;46(1):34–40. doi: 10.1002/prot.10028. [DOI] [PubMed] [Google Scholar]
- 63.Cavasotto CN, Abagyan RA. Protein flexibility in ligand docking and virtual screening to protein kinases. J Mol Biol. 2004;337(1):209–225. doi: 10.1016/j.jmb.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 64.Trellet M, Melquiond AS, Bonvin AM. A unified conformational selection and induced fit approach to protein-peptide docking. PLoS One. 2013;8(3):e58769. doi: 10.1371/journal.pone.0058769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Okamoto Y, Kokubo H, Tanaka T. Ligand docking simulations by generalized-ensemble algorithms. Adv Protein Chem Struct Biol. 2013;92:63–91. doi: 10.1016/B978-0-12-411636-8.00002-X. [DOI] [PubMed] [Google Scholar]
- 66.Sinko W, Lindert S, McCammon JA. Accounting for receptor flexibility and enhanced sampling methods in computer-aided drug design. Chem Biol Drug Des. 2013;81(1):41–49. doi: 10.1111/cbdd.12051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Majorek KA, Kuhn ML, Chruszcz M, et al. Structural, functional, and inhibition studies of a Gcn5-related N-acetyltransferase (GNAT) superfamily protein PA4794: a new C-terminal lysine protein acetyltransferase from pseudomonas aeruginosa. J Biol Chem. 2013;288(42):30223–30235. doi: 10.1074/jbc.M113.501353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Painter J, Merritt EA. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr D Biol Crystallogr. 2006;62(Pt 4):439–450. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- 69.Gillmor SA, Villasenor A, Fletterick R, et al. The structure of mammalian 15-lipoxygenase reveals similarity to the lipases and the determinants of substrate specificity. Nat Struct Biol. 1997;4(12):1003–1009. doi: 10.1038/nsb1297-1003. [DOI] [PubMed] [Google Scholar]
- 70.Choi J, Chon JK, Kim S, Shin W. Conformational flexibility in mammalian 15S–lipoxygenase: reinterpretation of the crystallographic data. Proteins. 2008;70(3):1023–1032. doi: 10.1002/prot.21590. [DOI] [PubMed] [Google Scholar]
- 71.Jovine L, Morgunova E, Ladenstein R. Of crystals, structure factors and diffraction images. J Appl Cryst. 2008;41:659. [Google Scholar]
- 72. Karplus PA, Diederichs K. Linking crystallographic model and data quality. Science. 2012;336(6084):1030–1033. doi: 10.1126/science.1218231. • Definition of a single statistically valid guide for determination of the resolution of diffraction data.
- 73. Borek D, Cymborowski M, Machius M, et al. Diffraction data analysis in the presence of radiation damage. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):426–436. doi: 10.1107/S0907444909040177. • A discussion of radiation damage and its influence on diffraction data and structure determination and refinement. The paper also discusses the impact of radiation decay on the choice of data collection strategy.
- 74.Borek D, Minor W, Otwinowski Z. Measurement errors and their consequences in protein crystallography. Acta Crystallogr D Biol Crystallogr. 2003;59(Pt 11):2031–2038. doi: 10.1107/s0907444903020924. [DOI] [PubMed] [Google Scholar]
- 75.Ferrucci D, Levas A, Bagchi S, et al. Watson: beyond jeopardy! Artif Intell. 2013;199:93–105. [Google Scholar]
- 76. Howe D, Costanzo M, Fey P, et al. Big data: the future of biocuration. Nature. 2008;455(7209):47–50. doi: 10.1038/455047a. •• The role of Big Data techniques, as applied to the curation of biological data.
- 77.How hadoop makes short work of big data [Internet] [Cited 30 July 2013];Forbes. 2012 Available from: http://www.forbes.com/sites/netapp/2012/09/24/hadoop-big-data/
- 78. Data is not information, information is not knowledge, knowledge is not understanding, understanding is not wisdom [Internet] [Cited 15 October 2013];Brainy Quote. Available from: http://www.brainyquote.com/quotes/quotes/c/cliffordst212166.html. •• A quotation from Clifford Stoll regarding different hierarchies of intelligence.