SUMMARY
Periodontitis is a polymicrobial oral infection characterized by the destruction of tooth-supporting structures that can be linked to systemic diseases such as cardiovascular disease, diabetes or rheumatoid arthritis. Porphyromonas gingivalis, a bacterium implicated in the etiology of periodontitis, has shown variation in inducing T-cell responses among different strains. Therefore, in this study we investigated the strain-specific immune response using a murine experimental model of periodontitis. Periodontitis was induced by P. gingivalis strains A7A1-28, W83 and W50, and later confirmed by the presence of P. gingivalis in the oral microflora and by alveolar bone resorption. Splenocytes were evaluated for gene expression, cellular proteins and cytokine expression. Dendritic cells were stimulated in vitro for T helper cell–cytokine profiling. Results showed that P. gingivalis had the ability to alter the systemic immune response after bacterial exposure. Strains W50 and W83 were shown to induce alveolar bone loss, whereas the A7A1-28 strain did not significantly promote bone resorption in mice. Splenocytes derived from mice infected with strains W50 and W83 induced expression of high levels of interleukin-4 (IL-4) but A7A1-28 stimulated increased IL-10. Stimulation of dendritic cells in vitro showed a similar pattern of cytokine expression of IL-12p40, IL-6 and transforming growth factor-β among strains. A distinct systemic response in vivo was observed among different strains of P. gingivalis, with IL-10 associated with the least amount of alveolar bone loss. Evaluation of pathogen-driven systemic immune responses associated with periodontal disease pathogenesis may assist in defining how periodontitis may impact other diseases.
Keywords: bone loss, interleukin-10, oral infection, periodontal disease, periodontitis, systemic response
INTRODUCTION
Periodontal disease is an immuno-inflammatory infection of the tooth-supporting structures and a major cause of tooth loss among the adult population. Increasing evidence shows an association between periodontal disease and other diseases, including cardiovascular disease (Friedewald et al., 2009), diabetes mellitus (Lamster et al., 2008) and rheumatoid arthritis (De Pablo et al., 2009). Although this association seems to be bi-directional, studies show that periodontitis alone can be considered a modifying factor for human diseases (Garlet, 2010). Still, the mechanism causing this effect is not well defined and the systemic immunological influence of periodontal disease is not clearly understood.
The acquired immune response is known to be important for periodontal disease development (Garlet, 2010). Specific microbial components that activate antigen-presenting cells, such as dendritic cells, will lead to production of a set of cytokines. The pattern of cytokines expressed determines subsequent polarization of a distinct antigen-specific lymphocyte response. Characteristic markers of T helper type 1 (Th1), Th2, Th17 and regulatory T (Treg) cell subsets have all been described in diseased periodontal tissues (Garlet et al., 2003). Therefore, the exact crosstalk that occurs among Th cytokines in periodontal disease and its impact on disease outcome is still to be determined. However, it is clear that Th cells are essential for periodontal destruction. The absence of B cells does not impede lipopolysaccharide-induced bone resorption (Yamaguchi et al., 2008). But B cells also seem to contribute to periodontal disease development; B-cell deletion prevented alveolar bone loss in mice after infection with Porphyromonas gingivalis (Baker et al., 2009).
Porphyromonas gingivalis is a pathogenic bacterium associated with increased risk to periodontal breakdown, disease recurrence (Socransky et al., 1998, 2002; Byrne et al., 2009). Colonization by P. gingivalis can cause changes in the amount and composition of the oral commensal microbiota by perturbing the host immune response (Hajishengallis et al., 2011). Several strains of P. gingivalis have been isolated from individuals with oral infections and are used to understand periodontal disease pathogenesis. Some studies have directly compared the biological differences induced among P. gingivalis strains in animal models. Classically, P. gingivalis strains are classified as invasive or non-invasive strains based on differences in the ability to cause soft-tissue abscesses following subcutaneous injections at distant or local sites (Neiders et al., 1989). In addition to soft-tissue abscesses, different strains vary in their ability to cause periodontal bone loss (Baker et al., 2000), serum and saliva antibody expression (Katz et al., 1996) and death in mice (Ebersole et al., 1995) and Drosophila (Igboin et al., 2011). Only 36% of mice gavaged with strain A7A1-28 died but W50 strain caused death in 100% of the animals in a 48-h period (Katz et al., 1996), demonstrating the different pathogenic potential of these strains. In addition, the induced pattern of cytokines driving the development of Th cells in vitro has also shown differences among strains (Gemmell et al., 2002; Vernal et al., 2009). Therefore, we speculate that the systemic effect following an oral infection with different P. gingivalis strains is also distinct. In this study, we established periodontal infection in mice with the strains A7A1-28, W83 and W50, and hypothesized that, along with the differences found in the amount of alveolar bone loss established, P. gingivalis would induce a distinct pattern of systemic immune responses among strains. The P. gingivalis strains chosen in this study are among the predominant ones isolated from clinical samples and are used as inducers of periodontitis in preclinical models of disease (Igboin et al., 2009).
METHODS
To evaluate the systemic immune response caused by different P. gingivalis strains, the strains A7A1-28, W83 and W50 were tested both in vivo and in vitro. All animal experiments were approved by the Institutional Animal Care and Use Committee of the University of Michigan and conformity to ARRIVE guidelines for preclinical studies. In vivo, DBA/1J mice (The Jackson Laboratory, Bar Harbor, ME) were infected with the three different strains and followed during 42 days for periodontal disease development (Fig. 1). Periodontal disease was confirmed by the presence of P. gingivalis in the oral cavity by polymerase chain reaction (PCR) and the stimulation of bone loss assessed by micro-computed tomography (micro-CT) and histomorphometric analysis. In addition, splenocytes were collected and evaluated by flow cytometry and gene expression, as well as being treated with phorbol 12-myristate 13-acetate (PMA)/ionomycin and evaluated for protein expression by enzyme-linked immunosorbent assay (ELISA). In vitro, bone-marrow-derived dendritic cells from non-infected mice were isolated and activated with different P. gingivalis strains, and later evaluated for protein expression by ELISA.
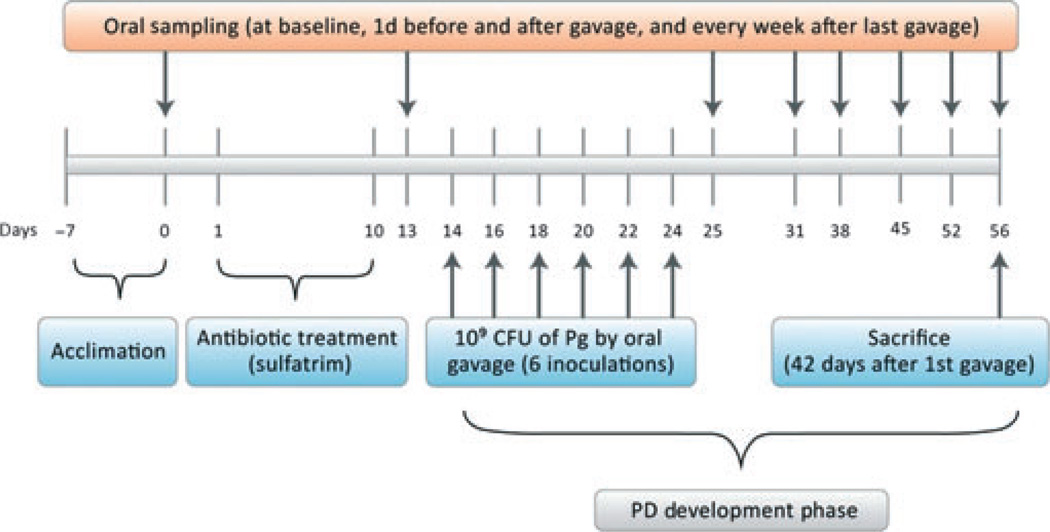
Figure 1.
Schematic diagram illustrating the study experimental design. This timeline displays the in vivo experiment, including mice acclimation, antibiotic treatment, Porphyromonas gingivalis (Pg) infection period and periodontitis developmental (PD) phase, oral microflora sampling, spleen harvest and sacrifice.
Bacterial strains
Porphyromonas gingivalis strains A7A1-28, W83 and W50 were grown under anaerobic conditions (85% N2, 5% CO2 and 10% H2) on pre-reduced brucella broth agar (BD Biosciences, San Jose, CA) plates enriched with 5% sheep blood (Remel, Lenexa, KS), 5 mg ml−1 hemin and 1 µg ml−1 menadione (Sigma-Aldrich, St Louis, MO). The P. gingivalis colonies were confirmed by PCR of Arg-gingipain (201 base pairs) and DNA sequencing before experiments were performed. Before PCR analysis was performed, the samples were heated at 100°C for 30 min. The following primer was used: 5′-CAAAATTACTCCGGGG ATCA-3′ (forward) and 5′-GCCGAAGCAATACAAA GAGC-3′ (reverse). New bacterial passages were further confirmed by the presence of homogeneous black-pigmented colonies or BANA test positivity (Loesche et al., 1992).
Murine periodontitis model
Thirty-two DBA/1J male mice (4 weeks old) were divided into four groups (eight mice/group) that were infected with A7A1-28, W83, W50 or vehicle. Mice were given sulphamethoxazole at 0.87 mg ml−1 and trimethoprim at 0.17 mg−1ml (Hi-Tech Pharmacal Co. Inc., Armityville, NY) in mili-Q water ad libitum for 10 days, followed by 3 days without antibiotics. For infection, P. gingivalis was suspended at 1010 colony-forming units (CFU) ml−1 as determined by optical density at 600 nm. Mice were inoculated with 109 CFU of bacteria in 100 µl phosphate-buffered saline with 2% carboxymethylcellulose (Sigma-Aldrich, St Louis, MO) by oral gavage on six separate occasions, with 2-day intervals as previously described (Baker et al., 1999; Sasaki et al., 2008). The vehicle group received carboxymethylcellulose alone. Mice were weighed at study initiation and at sacrifice (Fig. 1).
Oral microflora analysis
For P. gingivalis colonization analysis, the oral microflora was collected at baseline and 1, 7, 14, 21, 28 and 42 days after the last gavage. A sterile swab was placed in the oral cavity of mice and swirled for 15 s before being placed in 1 ml phosphate-buffered saline. Bacterial infection was confirmed by PCR of Arg-gingipain. To identify the minimum amount of bacteria detectable in the oral microflora of mice, PCR analysis was performed with P. gingivalis at the following concentrations: 0, 102, 103, 104 CFU ml−1. The PCR products were evaluated by electrophoresis and densitometry by ImageJ software (http://rsb.info.nih.gov/ij/download.html).
Micro-computed tomography
Mice were sacrificed at 42 days following gavage and subsequently maxillae were harvested and immediately fixed in 10% formalin for 24 h, and transferred to 70% alcohol. Fixed, non-demineralized mouse maxillae were scanned using a Pxs5-928EA cone-beam micro-CT system for the generation of three-dimensional images (GE Healthcare, Little Chalfont, UK). Analysis was performed by a calibrated masked examiner (SL). All micro-CT maxillary images were reoriented with morphological landmarks identified as previously described (Park et al., 2007). All three planes were identified by morphological landmarks to ensure consistent orientation. The region of interest (ROI) was then chosen to include as much interproximal alveolar bone while excluding tooth structures. Two transverse plane boundaries of ROI were chosen between the M1 root furcation and the root apex of M1 and M3. Tissue mineral content, bone volume, bone volume fraction and bone mineral content were obtained using bone analysis command of GEHC microview analysis plus software (GE Healthcare), as previously described (Park et al., 2007).
Histological preparation and analysis
After analysis by micro-CT, maxillae were decalcified in EDTA (Acros Organics, Fair Lawn, NJ) for a period of 14 days and then embedded in paraffin. Sagittal sections (4–5 µm thick) were obtained from each maxilla at the molar region of M1, M2 and M3 for two-dimensional evaluation. Coded sections were stained with hematoxylin & eosin for histomorphometric analysis, further performed by a masked and calibrated examiner (JM). The distance between the cementum–enamel junction and the alveolar bone crest in the mesial and distal region of M1 with mesial of M2 were measured using NIS elements basic research imaging software (Nikon Inc., Melville, NY) (Park et al., 2011).
Spleen harvest and processing
After mice were sacrificed, spleens were removed, segmented and placed in 10 ml serum-free RPMI −1640 medium (Gibco, Life Technologies, Grand Island, NY). Cells were dispersed through a 70-µm cell strainer into a 50-ml conical tube and centrifuged at 480 g for 3 min. The pellet was suspended in 5 ml ACK lysis buffer (155 mm NH4CI, 10 mm KHCO3, 0.1 mm EDTA) for 5 min to allow for lysis of red blood cells. Serum-free medium was added to halt lysis; supernatant was collected and centrifuged at 480 RCF for 5 min to pellet the leukocytes. The total number of splenocytes was calculated and further experiments were performed.
Quantification of splenocyte messenger RNA expression
Processed splenocytes derived from mice 42 days after gavage were treated with 1 ml TRIzol reagent (Invitrogen, Rockville, MD) and stored at - 80°C for messenger RNA (mRNA) expression and further analysis. The RNA was isolated via the TRIzol method, and total RNA was quantified by spectrophotometry. Double-stranded complementary DNA (cDNA) was synthesized from 0.5 µg RNA using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Carlsbad, CA). The cDNA was amplified via TaqMan Universal PCR Master Mix (Applied Biosystems). The following transcription factors were evaluated: T-bet for a Th1-driven response, GATA-3 for a Th2-driven response, Ror-γ for a Th17-driven response, Foxp3 for a Treg-cell-driven cell response, and FasL for a killer cell response. Amplification was performed using the ABI Prism 7700 Sequence Detection System (Applied Biosystems). Gene expression was normalized with the housekeeping gene GAPDH and relative quantification of the data generated was carried out using the comparative CT method (Schmittgen & Livak, 2008).
Flow cytometric analysis
Splenocytes were isolated and labeled with either anti-mouse CD3-phycoerythrin (PE)-cychrome 7 (Cy7) conjugate (BD Biosciences) and anti-mouse CD4-fluorescein isothiocyanate (Biolegend, San Diego, CA) for T cells or B220-PE-Cy7 conjugate, CD5-allophycocyanin (APC) and FasL-PE (BD Biosciences) for killer B cells, followed by fixation in Fix-Perm buffer (Biolegend, San Diego, CA) overnight at 4°C. Fixation solution was washed thoroughly followed by labeling of cells with FoxP3-PE, interleukin-17 (IL-17) -APC, or IL-10-PE (EBiosciences, San Diego, CA) for T cells. Flow cytometry was performed on a Cytomics FC 500 flow cytometer (Beckman Coulter, Fullerton, CA), and cell marker analysis was performed using flojo software (TreeStar Inc., Ashland, OR).
Splenocyte cultures for analysis of cytokine expression
Splenocytes were plated in 12-well plates at 5 × 106 cells ml−1 in RPMI-1640 medium supplemented with 10% fetal bovine serum, 1% penicillin/ streptomycin (Gibco), 1% HEPES (Gibco), 1% sodium pyruvate (HyClone, South Logan, UT) and 0.1% 2-mercaptoethanol (Gibco). Cells were treated with PMA/ionomycin (Sigma-Aldrich) for B-cell and T-cell activation. The PMA/ionomycin mimics antigen receptor activation and elicits cytokines from previously activated lymphocytes, with usually higher degrees of expression compared with antigen reactivation. Cell-free supernatants were collected after 48 h of treatment from cultures and evaluated by ELISA for interferon-γ (IFN-γ) for Th1-driven response, IL-17 (BD Biosciences) and IL-6 (R&D Systems, Minneapolis, MN) for Th17-driven response, IL-10 and transforming growth factor-β (TGF-β) (R&D Systems) for Treg cell response, IL-4 (R&D Systems) for Th2 cell response according to the manufacturer’s protocol.
Dendritic cell activation
To evaluate the cytokine production from antigen-presenting cells, bone-marrow-derived dendritic cells were isolated as previously described and activated with different strains of P. gingivalis (Lutz et al., 1999). Briefly, murine hind limbs from non-infected DBA/1 J male mice were collected. Bone marrow was flushed with RPMI-1640 supplemented with 10 µg granulocyte–macrophage colony-stimulating factor, 10% fetal bovine serum, 1% penicillin/streptomycin, and 0.1% 2-mercaptoethanol (Sigma-Aldrich). To obtain dendritic cells with high percentage purity, the flushed bone marrow was cultivated for 10 days and plated at 105 cells per well. Cells were treated with the different strains of P. gingivalis at multiplicity of infection of 0.1, 1 and 10 for 1 h, followed by treatment with gentamycin at 50 µg ml−1. Supernatants were collected after 18 h for cytokine expression analysis. To evaluate the immune response driven by P. gingivalis infection, cytokine expression was evaluated by ELISA for IL-12p40, IL-6 and TGF-β (BD Biosciences) for Th1 and Th17 activation, and Treg cell response, respectively.
Statistical analysis
The preclinical studies consisted of eight animals per group at the 42-day analysis. Dendritic cell experiments were performed in triplicates. The differences among groups were statistically assessed by analysis of variance (significance defined as P < 0.05), with Tukey multiple comparison post hoc test. Groups of infected mice were compared with those given vehicle alone by t-test. Data were analysed using the graphpad prism 5.0 program (GraphPad Software, La Jolla, CA).
RESULTS
Periodontal disease establishment and development
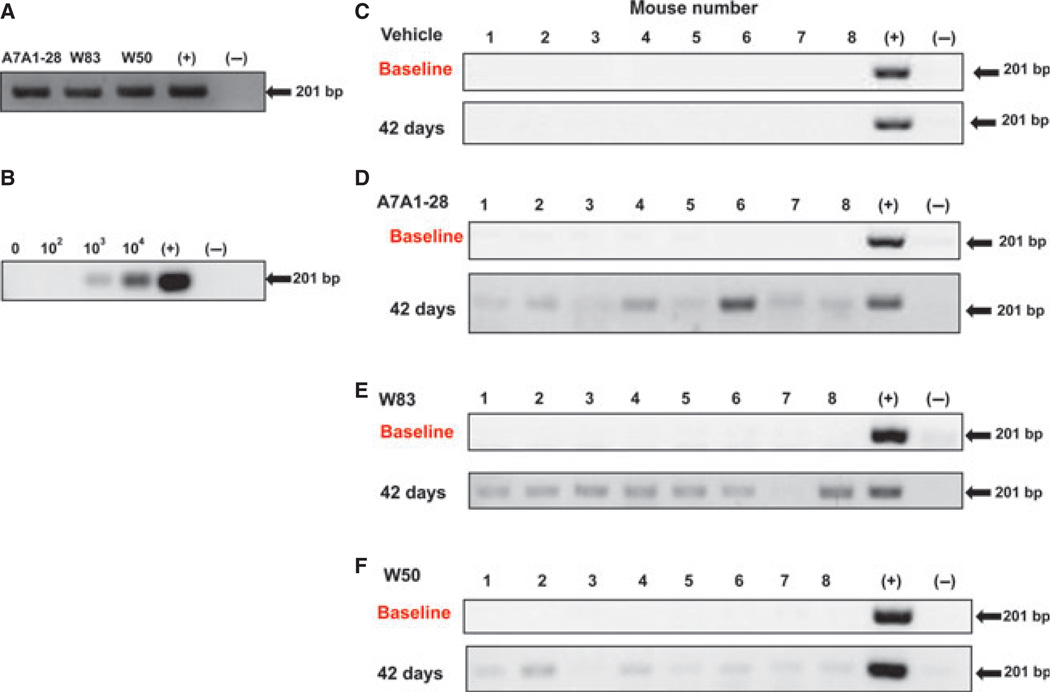
Colonies of P. gingivalis were confirmed by PCR of Arg-gingipain (201 bp) (Fig. 2A) and DNA sequencing. To evaluate the number of P. gingivalis CFU necessary in the oral microflora of mice for their detection by PCR of Arg-gingipain, known amounts of P. gingivalis were added to mouse oral samples at 0, 102, 103, 104 CFU. The presence of 102 CFU P. gingivalis was not detectable in the oral microflora by PCR, but the presence of 103 and 104 CFU was sufficient for P. gingivalis detection (Fig. 2B). At baseline before oral gavage, we evaluated the oral microflora by PCR of Arg-gingipain, and all mice were negative for the presence of P. gingivalis (Fig. 2C). The PCR evaluations of the oral samples demonstrated P. gingivalis infection at all time-points evaluated. At 42 days after the last gavage was performed, 22 of 24 mice were infected with P. gingivalis (Fig. 2D – F). Only one mouse from groups W83 (Fig. 2E) and W50 (Fig. 2F) failed to demonstrate P. gingivalis confirmation at the end of the experiment. When evaluated by band intensity, W83 showed the highest amount of recovery from the oral cavity, followed by A7A1-28 and W50 strains. None of the mice in the vehicle group was positive for P. gingivalis. No differences were found in weight gain by mice among groups, with a mean 44.46% weight gain.
Figure 2.
Polymerase chain reaction (PCR) analysis of Porphyromonas gingivalis oral infection determined by Arg-gingipain (201 bp). (A) Confirmation of P. gingivalis colonies of strains W83, W50 and A7A1-28 before gavage. (B) Porphyromonas gingivalis colony-forming units (CFU) at 0, 102, 103 and 104 were added to a sample collected from the oral microflora of mice to determine the detection limit via PCR. Oral microflora analysis: samples of mice infected with P. gingivalis at baseline (upper panel) and at 42 days after the first gavage (lower panel). Numbers 1 to 8 represent individual mice gavaged with (C) vehicle alone, (D) A7A1-28, (E) W83 and (F) W50.
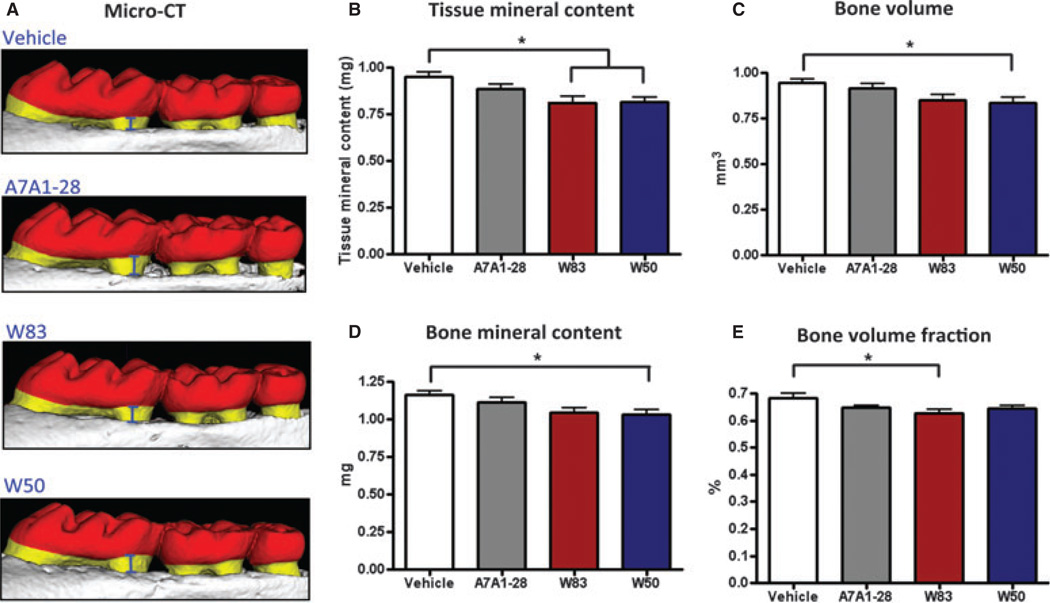
For alveolar bone analysis by micro-CT (Fig. 3A), all four bone-related parameters evaluated showed a significant difference among groups (analysis of variance P < 0.05). For tissue mineral content, bone volume and bone mineral content, a statistical difference was observed between groups W50 and vehicle (Tukey P < 0.05). A higher loss was observed in W50-infected animals by tissue mineral content measurements with 14.1% loss (Fig. 3B), followed by 11.6% loss by bone volume analysis (Fig. 3C), and 11.4% loss of bone mineral content (Fig. 3D) compared with vehicle group. Mice gavaged with W83 strain presented statistically significant differences in bone loss compared with vehicle group (Tukey P < 0.05) when evaluated for tissue mineral content with 14.3% loss (Fig. 3B), and for bone volume fraction with 7.9% loss (Fig. 3E). A7A1-28-infected animals showed a trend of bone loss compared with vehicle among all parameters with a tissue mineral content loss of 6.6%. However, these differences did not reach statistical significance. The vehicle group consistently showed the highest volumetric amount of alveolar bone among all groups when evaluated by all four parameters.
Figure 3.
Alveolar bone loss measured by micro-computed tomography. (A) Representative images of the alveolar bone loss found in mice at 42 days after gavage was performed with the three different strains A7A1-28, W83 and W50. (B) Analysis performed for tissue mineral content, bone mineral content, bone volume and bone volume fraction. Error bars indicate standard error. Asterisks denote differences found among groups by Tukey test (P < 0.05).
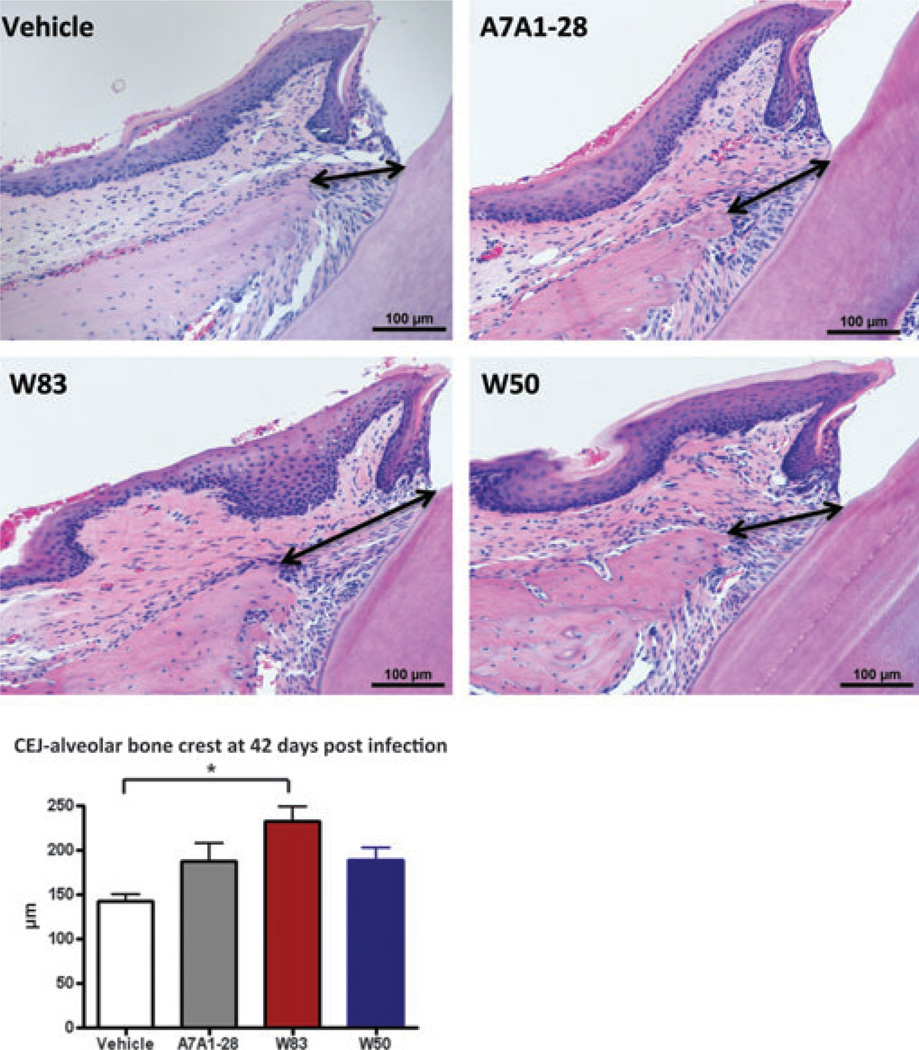
Histomorphometric analysis performed in the mesial region of M1 showed that the distance between the alveolar bone crest and the cementum–enamel junction was significantly higher in W83-infected animals when compared with vehicle (Tukey, P = 0.008) followed by A7A1-28-infected animals and W50-infected animals (Fig. 4). Measurements performed in the interproximal region of M1 and M2 showed W50 with the greatest degree of bone loss (103.47 ± 33.2 µm), followed by A7A1-28 (94.23 ± 25.7 µm), W83 (90.8 ± 24.9 µm) and vehicle (82.6 ± 20.4 µm).
Figure 4.
Histomorphometric image analysis performed in maxillae sections of mice at 42 days after gavage; the distance between the alveolar bone crest to the cementum–enamel junction in M1 was measured in micrometers (depicted with black arrows) (hematoxylin & eosin, 20×)
Systemic immune response
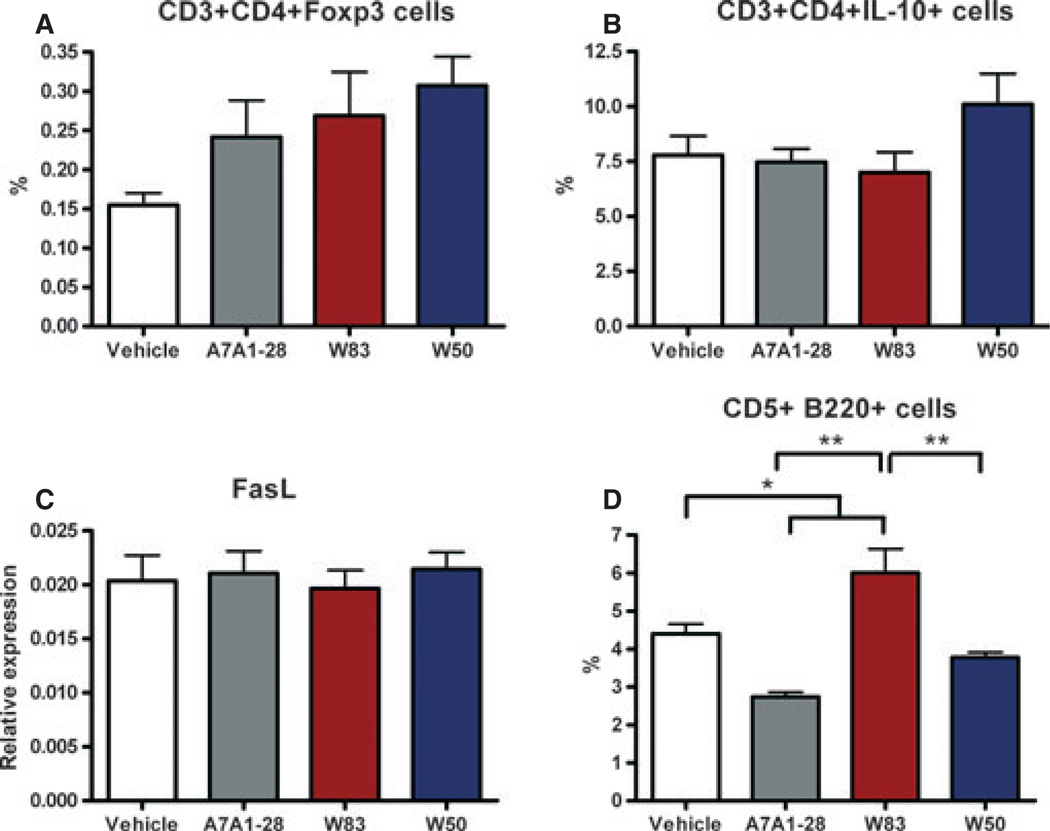
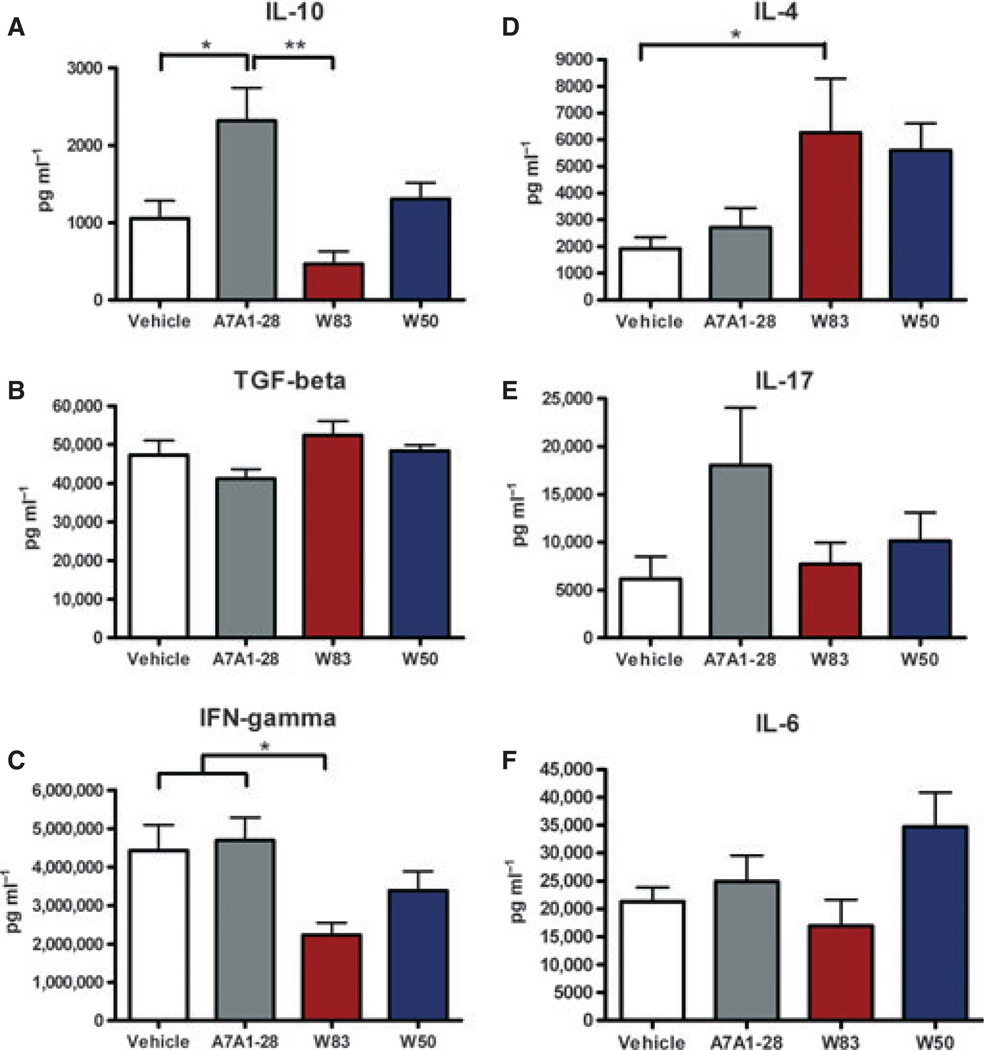
At 42 days post-gavage, splenocyte numbers were similar among groups with an average of 8.9 × 107 cells per mouse. Non-activated splenocytes from control and P. gingivalis-infected mice did not show differences in the mRNA expression of the transcription factors Tbx-21, Gata-3, ROR-γ, Foxp3 or FasL (data not shown). In addition, CD3+ CD4+ non-activated T cells were similar among groups when intracellular staining for Foxp3 and IL-10 for a Treg cell response or FasL expression for a killer cell response was evaluated. A trend towards an increase of CD3+ CD4+ Foxp3+ cell percentage was observed in P. gingivalis-infected mice (Fig. 5). However, a higher percentage of CD3+ CD4+ Foxp3+ was observed in the groups of infected mice compared with vehicle-treated mice (P = 0.019). When splenocytes were evaluated for the expression of CD5+ B220+ cells, a higher percentage was found in mice gavaged with W83 compared with all the other groups (P < 0.0001) (Fig. 5D). Interestingly, splenocytes stimulated with 1% PMA/ionomycin showed a distinct pattern of cytokine expression (Fig. 6). Mice gavaged with the A7A1-28 strain had a significantly higher expression of IL-10 compared with vehicle and mice gavaged with W83 strain (P= 0.0006). A higher expression of IL-4, was found in both W83 and W50 groups with a significantly lower expression in the A7A1-28 group and vehicle (P = 0.04). The expression of IFN-γ was significantly lower in the W83-infected animals when compared with either vehicle or the A7A1-28-infected animals (P = 0.01). No differences were found among groups for the expression of the cytokines IL-17, IL-6 or TGF-β (Fig. 6).
Figure 5.
Flow cytometric analysis of splenocytes derived from mice at 42 days after gavage with Porphyromonas gingivalis for regulatory markers. (A) CD3+ CD4+ Foxp3+ cell percentage, (B) CD3+ CD4+ IL-10+ cell percentage, (C) mRNA FasL relative expression of non-activated splenocytes (towards GAPDH), and (D) CD5+ B220+ cell percentage. Error bars indicate standard error. Asterisks denote differences found among groups by Tukey test (*P < 0.05, **P < 0.001).
Figure 6.
Cytokine expression of 42 days post-gavage murine splenocytes treated with phorbol 12-myristate 13-acetate (PMA)/ionomycin for 48 h in vitro. (A) Differences were found among groups for the expression of interleukin-10 (IL-10; P = 0.0006) and no differences for (B) transforming growth factor- β (TGF-β); (C) interferon-γ (IFN-γ) expression was higher in A7A1-28 strain, and (D) IL-4 expression was higher in W83 strain. No differences were found in expression of (E) IL-17 or (F) IL-6. Experiments were performed in duplicates. Error bars indicate standard error. Asterisks denote differences found among groups by Tukey test (*P < 0.05, **P < 0.001).
Innate immune response
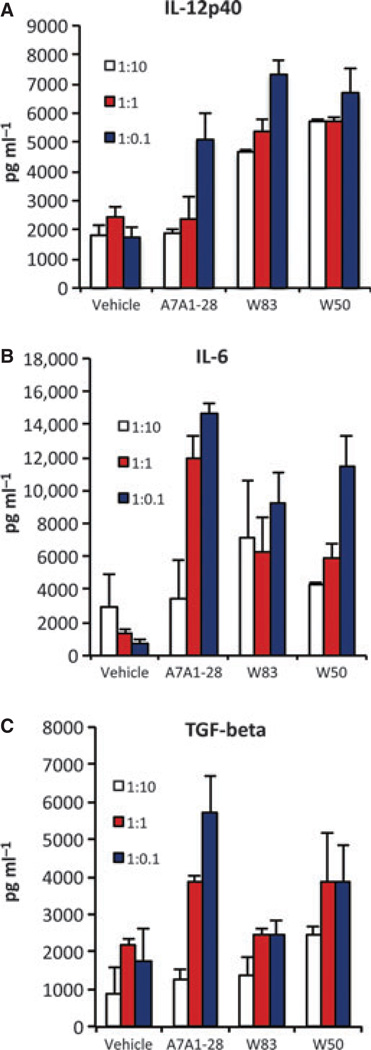
Dendritic cells stimulated in vitro with the three strains of P. gingivalis showed expression of IL-12p40, IL-6 and TGF-β, in a dose-dependent manner with no differences observed among strains (Fig. 7).
Figure 7.
Dendritic cells stimulated with Porphyromonas gingivalis strains A7A1-28, W83 and W50 at multiplicities of infection 1:10, 1:1, 1 : 0.1 showed an expression of (A) interleukin-12 (IL-12) p40, (B) IL-6 and (C) transforming growth factor-β (TGF-β) in a dose-dependent manner. Experiments were performed in duplicates. Error bars indicate standard error.
DISCUSSION
An estimated 48.2% of American adults have periodontal disease, which is the major cause of tooth loss among adults (Albandar, 2011). Porphyromonas gingivalis is an important bacterium involved in disease pathogenesis by inducing a local chronic host inflammatory response that leads to alveolar bone destruction. Our study demonstrated multiple pathogenic differences among different P. gingivalis strains. This could be important for understanding the relationship between oral and systemic diseases affected by P. gingivalis infection. A difference in both alveolar bone resorption and the systemic acquired immune response was found among different strains of P. gingivalis. Mice gavaged with strains W50 and W83 had a predominantly systemic IL-4 response (Fig. 6D) and higher amount of local alveolar bone loss compared with mice given vehicle only (Fig. 3). Mice gavaged with strain A7A1-28 had predominantly systemic production of IL-10 and IFN-γ (Fig. 6) with less induction of local alveolar bone loss (Fig. 3). Interestingly, the genetically closely related strains W83 and W50 displayed more similar results, while strain A7A1-28, which shows a divergence in the genome of 3.5%, had a correspondingly more distinct response (Igboin et al., 2009).
It is interesting to note that the absence of a significant local oral response to A7A1-28 strain did not translate into a lack of an in vitro or systemic response. Although strain A7A1-28 failed to induce bone loss, it resulted in potent dendritic cell responses in vitro, as well as in IL-17 induction. Interestingly, strains W83 and W50 induced significant oral bone loss but less splenic cytokine expression. Based on the results, we speculate that it would be interesting to separate patient samples based on P. gingivalis strains when evaluating the systemic effects of periodontitis.
Interleukin-10 is an important immune regulatory cytokine that has been shown to regulate both IFN-γ-driven and IL-4-driven immune responses (Hoffmann et al., 2000). IL-10−/− mice show severe alveolar bone loss when infected with P. gingivalis (Sasaki et al., 2004). Kobayashi et al. (2011) showed that gingival mononuclear cells stimulated with PMA/ionomycin in vitro had upregulated IL-10 production 30 days after infection with P. gingivalis strain 33277, which was not observed at the early time points of 1, 7 and 15 days. Importantly, IL-10 polymorphisms that may be associated with differing levels of IL-10 expression are related to periodontal disease progression clinically (Cullinan et al., 2008). In alignment with our findings, these studies suggest that IL-10 is an important molecule in the balance between inflammatory, humoral and infection responses with P. gingivalis.
Mice gavaged with strain A7A1-28 displayed less bone loss and higher expression of IL-10 from re-stimulated spleen cells, so we investigated whether regulatory T and B cells were distinctively upregulated in the spleen. All strains of P. gingivalis showed a trend toward induction of splenic Foxp3+ T cells (Fig. 5A). Unexpectedly, the A7A1-28 group had the lowest percentage of CD5+ B220+ cells (Fig. 5D), a population of B cells reported to produce IL-10 (O’Garra & Howard, 1992; Haas et al., 2005; Yanaba et al., 2008). CD5+ B cells have also been demonstrated to express FasL in the presence of IL-4 and IL-10, and to mediate T-cell death in an arthritic model (Lundy & Boros, 2002; Lundy & Fox, 2009). Despite differences in IL-10, IL-4 and CD5+ B cells, we found no difference among groups in the mRNA expression of FasL (Fig. 5C). Therefore, the higher IL-10 expression from splenocytes derived from mice gavaged with A7A1-28 strain could not be directly attributed to either Breg or Treg cells. Further, no change in splenic FasL expression could be attributed to P. gingivalis infection. One possibility is that the expression of IL-10 from splenocytes could be attributed to Foxp3− cells, such as the Treg 1 subset. However, this strain does not express a definite prototypic surface marker for exact identification (Pot et al., 2011).
The importance of the acquired immune response locally in periodontal bone loss has been demonstrated by reduced bone loss in mice lacking both B and T cells (Yamaguchi et al., 2008; Baker et al., 2009). Porphyromonas gingivalis is an important bacterium commonly found in patients with periodontal disease, and is associated with tissue breakdown and disease recurrence (Socransky et al., 1998, 2002). The P. gingivalis strains A7A1-28, W83 and W50 evaluated in this study are among the most prevalent strains found worldwide (Igboin et al., 2009), and are therefore clinically relevant. The mouse DBA1/J is a strain genetically suitable for induction of inflammatory diseases, including arthritis, and parallels the role of genetic loci as an important risk factor for human rheumatoid arthritis. The susceptibility of DBA/1J mice to arthritis is based on the MHC Class II loci and the unique ability to present arthritis-associated antigens to T helper cells. To our knowledge, there have been no studies published that suggest a different response of these mice to toll-like receptor antigens, such as P. gingivalis. Because rheumatoid arthritis has been associated with periodontal disease in patients (De Pablo et al., 2009), the systemic effect of P. gingivalis in this strain of mice is important. We used animals at 4 weeks of age to allow for the development of a co-disease model that considers a younger set of animals.
When P. gingivalis was evaluated for its presence in the oral microflora by PCR analysis, W83 showed higher band intensities, followed by strain A7A1-28 and W50 (Fig. 2). Similar to our findings, recovery of W50 strain from the oral cavity of rodents using direct bacteria cultivation was lower when compared with strain A7A1-28, with only 17% being recovered by 42 days in one of the studies (Baker et al., 2000; Katz et al., 1996). Still, in accordance with Baker et al. (2000), the differences found in the infection assessment did not seem to influence the amount of alveolar bone loss induced by the W50 strain. Our results differ, however, in the amount of alveolar bone loss observed at the end of the experiment by different strains. Baker et al. (2000) found that strain A7A1-28 and W50 induced similar amounts of bone loss, whereas in our study the A7A1-28 strain induced minimal alveolar bone loss (Fig. 3). This finding could perhaps be explained by the different methods used in the studies to assess the alveolar bone level or differences between strains of mice. Although not directly comparable, our micro-CT and histomorphometry data were relatively congruent. The histomorphometry is a classical method for evaluating bone loss and provides histological visualization of the inflammatory process in two dimensions while the micro-CT method provides a three-dimensional analysis of the bone loss.
It is important to compare data generated in vitro with in vivo results to determine the degree to which cells generated in vitro truly reflect the biology of responses generated under physiological, more complex conditions. Our results show that while non-activated splenocytes from control and P. gingivalis-infected mice did not show differences in the expression of transcription factors, splenocytes that were reactivated showed a strong distinct response. This demonstrates that even though there was not an active immune response happening in the spleen, the exposure to P. gingivalis had a systemic effect demonstrated by the ability to change the profile of splenic cytokine expression upon cell reactivation. While the dendritic cells stimulated in vitro had similar responses among strains, with expression of IL-12p40, IL-6 and TGF-β (Fig. 7), the in vivo scenario was distinct (Fig. 6). A potential explanation for the differences found between the in vitro and systemic in vivo results is the changes in oral microflora that occur when P. gingivalis is present, an alteration called dysbiosis (Hajishengallis et al., 2011). Dysbiosis has the potential to impact the systemic immune response and shown to be implicated in a variety of diseases, including obesity, metabolic disorders, and inflammatory bowel disease (Hill and Artis, 2010; Garret et al. 2010). Therefore, while the in vitro data are a direct result of the effect of P. gingivalis in immune cells, the in vivo data could be the result of changes that occur in the amount and composition of the oral microbiota due to P. gingivalis colonization. In accordance with our results, Vernal et al. (2009) also did not find differences in the pattern of cytokines expressed by dendritic cells stimulated with different serotypes of P. gingivalis in vitro, with all strains showing expression of IFN-γ, IL-1β, IL-12, tumor necrosis factor, IL-6 and IL-10. However, we did not observe a higher response to W83 when compared with A7A1-28 (Fig. 7). This could be related to the differences found in mRNA versus protein expression. When the in vivo T-cell activation was evaluated, animals gavaged with P. gingivalis strain 33277 displayed stimulated splenocytes expressing high levels of IFN-γ, IL-2, IL-10 and IL-4, which was consistent with a mixed Th1 and Th2 response (Katz et al., 1999). Co-culture experiments using dendritic cells and T cells also suggest a mixed Th1 and Th2 response (Jotwani & Cutler, 2004; Zeituni et al., 2009). In agreement with our data, these studies suggest that the Th cell type driving the in vivo response of P. gingivalis is more complex than what can be observed with single cell isolates in the in vitro environment. The spleen is a secondary lymphoid organ that contains approximately 55% B cells, 35% T cells and 10% of other leukocytes, including macrophages and natural killer cells. The spleen is a site of interaction between elements of both the innate and adaptive immune responses and is, therefore, an important organ to be evaluated in the context of systemic exposures to bacteria.
In summary, P. gingivalis had the ability to alter the systemic immune response after bacterial exposure. Strains W50 and W83 were shown to induce alveolar bone loss, whereas strain A7A1-28 did not significantly promote bone resorption in mice. Splenocytes derived from mice infected with strains W50 and W83 induced expression of high levels of IL-4, whereas A7A1-28-stimulated splenocytes had increased IL-10. Stimulation of dendritic cells in vitro showed a similar pattern of cytokine expression of IL-12p40, IL-6 and TGF-β among strains. A distinct systemic response in vivo was observed among different strains of P. gingivalis, with IL-10 associated with the least amount of alveolar bone loss. Evaluation of pathogen-driven systemic immune responses associated with periodontal disease pathogenesis may assist in defining how periodontitis may impact other diseases.
ACKNOWLEDGEMENTS
This study was supported by grants DE13397 and DE021934 from the National Institute of Dental and Craniofacial Research.
REFERENCES
- Albandar JM. Underestimation of periodontitis in NHANES surveys. J Periodontal. 2011;82:337–341. doi: 10.1902/jop.2011.100638. [DOI] [PubMed] [Google Scholar]
- Baker PJ, Dixon M, Evans RT, Dufour L, Johnson E, Roopenian DC. CD4+ T cells and the proinflammatory cytokines gamma interferon and interleukin-6 contribute to alveolar bone loss in mice. Infect Immun. 1999;67:2804–2809. doi: 10.1128/iai.67.6.2804-2809.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker PJ, Dixon M, Evans RT, Roopenian DC. Heterogeneity of Porphyromonas gingivalis strains in the induction of alveolar bone loss in mice. Oral Microbiol Immunol. 2000;15:27–32. doi: 10.1034/j.1399-302x.2000.150105.x. [DOI] [PubMed] [Google Scholar]
- Baker PJ, Boutaugh NR, Tiffany M, Roopenian DC. B Cell IgD Deletion Prevents Alveolar Bone Loss Following Murine Oral Infection. Interdiscip Perspect Infect Dis. 2009;2009:864359. doi: 10.1155/2009/864359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne SJ, Dashper SG, Darby IB, Adams GG, Hoffmann B, Reynolds EC. Progression of chronic periodontitis can be predicted by the levels of Porphyromonas gingivalis and Treponema denticola in subgingival plaque. Oral Microbiol Immunol. 2009;24:469–477. doi: 10.1111/j.1399-302X.2009.00544.x. [DOI] [PubMed] [Google Scholar]
- Cullinan MP, Westerman B, Hamlet SM, et al. Progression of periodontal disease and interleukin-10 gene polymorphism. J Periodontal Res. 2008;43:328–333. doi: 10.1111/j.1600-0765.2007.01034.x. [DOI] [PubMed] [Google Scholar]
- De Pablo P, Chapple IL, Buckley CD, Dietrich T. Periodontitis in systemic rheumatic diseases. Nat Rev Rheumatol. 2009;5:218–224. doi: 10.1038/nrrheum.2009.28. [DOI] [PubMed] [Google Scholar]
- Ebersole JL, Kesavalu L, Schneider SL, Machen RL, Holt SC. Comparative virulence of periodon-topathogens in a mouse abscess model. Oral Dis. 1995;1:115–128. doi: 10.1111/j.1601-0825.1995.tb00174.x. [DOI] [PubMed] [Google Scholar]
- Friedewald VE, Kornman KS, Beck JD, et al. The American Journal of Cardiology and Journal of Peri-odontology editors’ consensus: periodontitis and atherosclerotic cardiovascular disease. J Periodontol. 2009;80:1021–1032. doi: 10.1902/jop.2009.097001. [DOI] [PubMed] [Google Scholar]
- Garlet GP. Destructive and protective roles of cytokines in periodontitis: a re-appraisal from host defense and tissue destruction viewpoints. J Dent Res. 2010;89:1349–1363. doi: 10.1177/0022034510376402. [DOI] [PubMed] [Google Scholar]
- Garlet GP, Martins W, Jr, Ferreira BR, Milanezi CM, Silva JS. Patterns of chemokines and chemokine receptors expression in different forms of human periodontal disease. J Periodontal Res. 2003;38:210–217. doi: 10.1034/j.1600-0765.2003.02012.x. [DOI] [PubMed] [Google Scholar]
- Garrett WS, Gallini CA, Yatsunenko T, et al. Enterobacteriaceae act in concert with the gut micro-biota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe. 2010;16:292–300. doi: 10.1016/j.chom.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemmell E, Carter CL, Grieco DA, Sugerman PB, Seymour GJ. P. gingivalis-specific T-cell lines produce Th1 and Th2 cytokines. J Dent Res. 2002;81:303–307. doi: 10.1177/154405910208100503. [DOI] [PubMed] [Google Scholar]
- Haas KM, Poe JC, Steeber DA, Tedder TF. B-1a and B-1b cells exhibit distinct developmental requirements and have unique functional roles in innate and adaptive immunity to S. pneumoniae . Immunity. 2005;23:7–18. doi: 10.1016/j.immuni.2005.04.011. [DOI] [PubMed] [Google Scholar]
- Hajishengallis G, Liang S, Payne MA, et al. Low-abundance biofilm species orchestrates inflammatory periodontal disease through the commensal microbiota and complement. Cell Host Microbe. 2011;10:497–506. doi: 10.1016/j.chom.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill DA, Artis D. Intestinal bacteria and the regulation of immune cell homeostasis. Annu Rev Immunol. 2010;28:623–667. doi: 10.1146/annurev-immunol-030409-101330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann KF, Cheever AW, Wynn TA. IL-10 and the dangers of immune polarization: excessive type 1 and type 2 cytokine responses induce distinct forms of lethal immunopathology in murine schistosomiasis. J Immunol. 2000;164:6406–6416. doi: 10.4049/jimmunol.164.12.6406. [DOI] [PubMed] [Google Scholar]
- Igboin CO, Griffen AL, Leys EJ. Porphyromonas gingivalis strain diversity. J Clin Microbiol. 2009;47:3073–3081. doi: 10.1128/JCM.00569-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igboin CO, Moeschberger ML, Griffen AL, Leys EJ. Porphyromonas gingivalis virulence in a Dro-sophila melanogaster model. Infect Immun. 2011;79:439–448. doi: 10.1128/IAI.00784-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jotwani R, Cutler CW. Fimbriated Porphyromonas gingivalis is more efficient than fimbria-deficient P. gingivalis in entering human dendritic cells in vitro and induces an inflammatory Th1 effector response. Infect Immun. 2004;72:1725–1732. doi: 10.1128/IAI.72.3.1725-1732.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz J, Ward DC, Michalek SM. Effect of host responses on the pathogenicity of strains of Porphyromonas gingivalis . Oral Microbiol Immunol. 1996;11:309–318. doi: 10.1111/j.1399-302x.1996.tb00187.x. [DOI] [PubMed] [Google Scholar]
- Katz J, Black KP, Michalek SM. Host responses to recombinant hemagglutinin B of Porphyromonas gingivalis in an experimental rat model. Infect Immun. 1999;67:4352–4359. doi: 10.1128/iai.67.9.4352-4359.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi R, Kono T, Bolerjack BA, et al. Induction of IL-10-producing CD4+ T-cells in chronic periodontitis. J Dent Res. 2011;90:653–658. doi: 10.1177/0022034510397838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamster IB, Lalla E, Borgnakke WS, Taylor GW. The relationship between oral health and diabetes mellitus. J Am Dent Assoc. 2008;139(Suppl.):19S–24S. doi: 10.14219/jada.archive.2008.0363. [DOI] [PubMed] [Google Scholar]
- Loesche WJ, Lopatin DE, Giordano J, Alcoforado G, Hujoel P. Comparison of the benzoyl-DL-argi-nine-naphthylamide (BANA) test, DNA probes, and immunological reagents for ability to detect anaerobic periodontal infections due to Porphyromonas gingivalis, Treponema denticola, and Bacteroides forsythus . J Clin Microbiol. 1992;30:427–433. doi: 10.1128/jcm.30.2.427-433.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundy SK, Boros DL. Fas ligand-expressing B-1a lymphocytes mediate CD4(+)-T-cell apoptosis during schistosomal infection: induction by interleukin 4 (IL-4) and IL-10. Infect Immun. 2002;70:812–819. doi: 10.1128/iai.70.2.812-819.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundy SK, Fox DA. Reduced Fas ligand-expressing splenic CD5+ B lymphocytes in severe collagen-induced arthritis. Arthritis Res Ther. 2009;11:R128. doi: 10.1186/ar2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz MB, Kukutsch N, Ogilvie AL, et al. An advanced culture method for generating large quantities of highly pure dendritic cells from mouse bone marrow. J Immunol Methods. 1999;223:77–92. doi: 10.1016/s0022-1759(98)00204-x. [DOI] [PubMed] [Google Scholar]
- Neiders ME, Chen PB, Suido H, et al. Heterogeneity of virulence among strains of Bacteroides gingivalis . J Periodontal Res. 1989;24:192–198. doi: 10.1111/j.1600-0765.1989.tb02005.x. [DOI] [PubMed] [Google Scholar]
- O’Garra A, Howard M. IL-10 production by CD5 B cells. Annals N Y Acad Sci. 1992;651:182–199. doi: 10.1111/j.1749-6632.1992.tb24615.x. [DOI] [PubMed] [Google Scholar]
- Park CH, Abramson ZR, Taba M, Jr, et al. Three-dimensional micro-computed tomographic imaging of alveolar bone in experimental bone loss or repair. J Periodontol. 2007;78:273–281. doi: 10.1902/jop.2007.060252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JC, Su C, Jung IH, et al. Mechanism of alveolar bone loss in a collagen-induced arthritis model in mice. J Clin Periodontol. 2011;38:122–130. doi: 10.1111/j.1600-051X.2010.01645.x. [DOI] [PubMed] [Google Scholar]
- Pot C, Apetoh L, Kuchroo VK. Type 1 regulatory T cells (Tr1) in autoimmunity. Semin Immunol. 2011;23:202–208. doi: 10.1016/j.smim.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki H, Okamatsu Y, Kawai T, Kent R, Taubman M, Stashenko P. The interleukin-10 knockout mouse is highly susceptible to Porphyromonas gingiva-lis-induced alveolar bone loss. J Periodontal Res. 2004;39:432–441. doi: 10.1111/j.1600-0765.2004.00760.x. [DOI] [PubMed] [Google Scholar]
- Sasaki H, Suzuki N, Kent R, Jr, Kawashima N, Takeda J, Stashenko P. T cell response mediated by myeloid cell-derived IL-12 is responsible for Porphyromonas gingivalis-induced periodontitis in IL-10-deficient mice. J Immunol. 2008;180:6193–6198. doi: 10.4049/jimmunol.180.9.6193. [DOI] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL., Jr Microbial complexes in subgingival plaque. J Clin Periodontol. 1998;25:134–144. doi: 10.1111/j.1600-051x.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- Socransky SS, Smith C, Haffajee AD. Subgingival microbial profiles in refractory periodontal disease. J Clin Periodontol. 2002;29:260–268. doi: 10.1034/j.1600-051x.2002.290313.x. [DOI] [PubMed] [Google Scholar]
- Vernal R, Leon R, Silva A, van Winkelhoff AJ, Garcia-Sanz JA, Sanz M. Differential cytokine expression by human dendritic cells in response to different Porphyromonas gingivalis capsular serotypes. J Clin Periodontol. 2009;36:823–829. doi: 10.1111/j.1600-051X.2009.01462.x. [DOI] [PubMed] [Google Scholar]
- Yamaguchi M, Ukai T, Kaneko T, et al. T cells are able to promote lipopolysaccharide-induced bone resorption in mice in the absence of B cells. J Periodontal Res. 2008;43:549–555. doi: 10.1111/j.1600-0765.2008.01083.x. [DOI] [PubMed] [Google Scholar]
- Yanaba K, Bouaziz JD, Haas KM, Poe JC, Fujimoto M, Tedder TF. A regulatory B cell subset with a unique CD1dhiCD5+ phenotype controls T cell-dependent inflammatory responses. Immunity. 2008;28:639–650. doi: 10.1016/j.immuni.2008.03.017. [DOI] [PubMed] [Google Scholar]
- Zeituni AE, Jotwani R, Carrion J, Cutler CW. Targeting of DC-SIGN on human dendritic cells by minor fimbriated Porphyromonas gingivalis strains elicits a distinct effector T cell response. J Immunol. 2009;183:5694–5704. doi: 10.4049/jimmunol.0901030. [DOI] [PMC free article] [PubMed] [Google Scholar]