Abstract
We investigated the bacterial community composition of tree holes in relation to the presence and absence of larvae of the mosquito Ochlerotatus triseriatus. Larvae were eliminated from a subset of natural tree holes with Bacillus thuringiensis serovar israelensis, and total bacterial numbers, slow- and fast-growing colony-forming units on minimal media, and 16S rRNA gene sequence data from water column and leaf material were obtained. Total bacterial counts did not change significantly with treatment; however, the number of slow-growing cultivable bacteria significantly increased in the absence of larvae. Sequence classifications and comparisons of sequence libraries using LIBSHUFF indicated that the elimination of larvae significantly altered bacterial community composition. Major groups apparently affected by larvae were Flavobacteriaceae, Rhodobacteraceae, Comamonadaceae, and Sphingomonadaceae. A clear dominance of Flavobacteriaceae in the water column after larval removal suggests members of this group are a major bacterial food source.
Keywords: Ochlerotatus triseriatus larvae, bacterial community, Flavobacteriaceae, Rhodobacteraceae, Comamonadaceae, Sphingomonadaceae
Introduction
Processes operating in larval mosquito environments profoundly affect population regulation of adult mosquitoes and influence their vector capacity (Briegel 1990, Timmerman and Briegel 1999, Briegel et al. 2001). Nearly 40% of all mosquito species utilize small, discrete container habitats for larval development, including many important human disease vectors (Laird 1988). Water-filled tree holes are an example of these breeding sites, and are common and productive habitats for mosquitoes in temperate and tropical areas (Frank and Lounibos 1983, Kitching 2000). In the eastern United States, the larvae of Ochlerotatus triseriatus Say are a prominent, often dominant, member of the macroinvertebrate community in tree holes, and adults are the primary vector of La Crosse encephalitis.
Tree holes are heterotrophic microbial ecosystems because secondary production of insects living in them is dependent upon microbial assimilation and processing of detritus of external origin (Kitching 2001). Mosquito production in tree hole ecosystems is considered primarily a function of microbial-mediated decomposition of senescent plant detritus (Fish and Carpenter 1982, Walker et al. 1988, Walker et al. 1997, Kaufman et al. 2000, 2001), although terrestrial invertebrate detritus inputs are also recognized as important basal resources (Daugherty et al. 2000, Yee and Juliano 2006). Microorganisms provide nutrients directly to mosquito larvae in the form of their own biomass and presumably are also instrumental in larval development via release of nutrients from the leaf material.
Tree hole mosquito production is, therefore, thought to be tightly dependent on the microbial community dynamics, and conversely, microbial groups have been shown to respond to larval grazing pressures (Eisenberg et al. 2000, Kaufman et al. 1999, 2001, 2002). It has been estimated that bacterial biomass can contribute substantially to the growth of Oc. triseriatus larvae in microcosms (Kaufman et al. 2001), and bacterial abundance and productivity, particularly on leaf surfaces, have been shown to be reduced in the presence of larvae (Walker et al. 1988, Kaufman et al. 1999, 2001, 2002, Kaufman and Walker 2006). Other microbial groups, such as free-swimming protozoans, are highly susceptible to grazing (Eisenberg et al. 2000, Kaufman et al. 2002). However, larval grazing effects on fungi remain ambiguous (Kaufman et al. 2001, 2002, Kaufman and Walker 2006).
These previous studies have largely examined microbial groups in tree hole habitats from the broad perspectives of total bacterial numbers or overall bacterial biomass production rates. A few studies have addressed physiological or taxonomic groups of bacteria in simulated larval mosquito habitats (Cochran-Stafira and von Ende 1998, Kaufman et al. 1999). Recently, bacterial communities in tree holes have served as models to address macroecological questions of size–diversity relationships and structure–function questions (Bell et al. 2005a, Bell et al. 2005b). However, there is little information yet available on major taxonomic or physiological groups in natural tree hole systems. In one study (Bell et al. 2005a) addressing bacterial species in natural tree holes, species were enumerated as unique bands in denaturing gradient gel electrophoresis analyses with no accompanying taxonomic information, and the number of bands was linked to tree hole volume with no mention of invertebrate fauna. More recently, our research group has examined leaf-associated bacteria and fungi in natural tree holes containing larval Oc. triseriatus using 16S sequencing methods (Kaufman et al. 2008).
Our interests in investigating microorganisms, specifically bacteria that inhabit tree holes, arise from our goals of understanding larval mosquito development in these systems, and for selection of bacterial species suited for genetic transformation into larvicide (e.g., Bacillus thuringiensis serovar israelensis [Bti]) delivery vehicles. Both goals require an elucidation of bacterial communities residing in the feeding zones of larvae and studies of how the communities change in response to larval feeding pressure. Toward those ends, this study's objectives were to determine changes in bacterial community structure associated with larval mosquito activity in natural tree hole habitats.
Materials and Methods
Experimental design
The study site was in Toumey Woods, Michigan State University campus, East Lansing, MI. Tree holes in this beech–maple woodlot typically contain dense populations of Oc. triseriatus larvae and have been study subjects previously or have served as source material for studies (Walker et al. 1988, 1991, Walker and Merritt 1988, Kaufman et al. 2002). Twenty-four tree holes were selected for manipulation; tree holes with low larval densities (<1 larva/2-ml sample) were excluded from further study. Twelve were randomly selected and treated with 3 granules of a commercial formulation of Bti (Vectobac-G, an aqueous suspension of live cells, spores, and proteinaceous toxin; Valent Biosciences, Libertyville, IL) to kill larvae present in the tree holes, and thus selectively to remove this predator of microorganisms. The remaining 12 tree holes were untreated, as larvae-present controls. Dead 3rd and 4th instars (100–200 total), obtained by heat-killing larvae from our lab colony of Oc. triseriatus, were added to these experimental controls to mimic potential effects of dead larval biomass in the Bti-treated tree holes. After 3 days, the tree holes were sampled again to verify treatment efficacy and to ascertain that larvae were still present and active in untreated holes. At this time, a 10-ml subsample of tree hole water and samples of leaf material were removed aseptically from each tree hole. The samples were transported to the laboratory on ice.
Several small leaf fragments from each tree hole (∼1 cm2) were weighed and placed in 10 ml sterile phosphate buffer. Bacteria were then dislodged from leaf surfaces by sonication following the method in Velgi and Albright (1993), as modified in Kaufman et al. (2001). The sonicate provided material for both direct counts and DNA extraction (below). The remaining leaf fragments were weighed, dried for 2 days at 50°C, and reweighed for wet to dry weight conversion factors.
Bacterial abundance
A 2-ml portion of each water sample and leaf sonicate was preserved with 200 μl formalin for direct microscopic counts (DMCs). The DMCs of bacteria (bacterial but not archaeal domain) were determined with epifluorescence microscopy after hybridization of a carbocyanine-labeled oligonucleotide probe (5′-GCTGCCTCCCGTAGGAGT-3′) to bacterial ribosomes, following methods described in Glöckner et al. (1996). The DMC data were analyzed by 1-way analysis of variance on log10-transformed data separately for water column and leaf surface samples, with the mosquito-present and mosquito-absent variable as the main treatment effect.
Slow- vs. fast-growing phenotypes
A 1-ml aliquot of the water sample from each tree hole was used in a dilution series for plating on minimal essential agar medium, R2A (Reasoner and Geldreich 1985). The R2A medium is less enriched than most media used for general heterotrophic growth of aquatic bacteria and gives higher numbers and diversity of isolates (Reasoner and Geldreich 1985). Because R2A allows slow-growing colonies to develop without overgrowth from faster-growing colonies, the medium has been used to examine changes in broad physiological groups of heterotrophs (e.g., Klappenbach et al. 2000). Plates were incubated at 28°C, and the number of colony-forming units (CFUs) was determined for 9 consecutive days to estimate the distribution of fast- to slow-growing bacteria in the samples (Klappenbach et al. 2000). The CFU data were analyzed as the proportion of the total number of CFUs for the individual replicate. Proportions were transformed by the arcsine of the square root and subjected to multivariate repeated-measures analysis of variance (PROC GLM, SAS System for Windows 8.0; SAS Institute, Cary, NC) in which the main treatment effect was the mosquito-present and mosquito-absent variable. Because repeated measurements taken on the same plates within each treatment are likely to be correlated, thereby affecting standard error calculations, we used a generalized estimating equations approach to Poisson regression (for count data) (PROC GENMOD, SAS System for Windows 8.0; SAS).
16S rDNA sequence analysis
Water and leaf sample sonicate samples remaining after subsampling for direct and cultivable counts were centrifuged at 7,000 rpm for 20 min. The pellet was collected, and microbial DNA was extracted using Ultraclean Soil DNA kits (MO BIO, Solana Beach, CA). The eluted DNA was stored in 100 ml tris-EDTA buffer. The DNA samples from each replicate within a type (water or leaf) and treatment were combined in equal amounts prior to polymerase chain reaction (PCR). An approximately 1,300-bp region of a consensus 16S rRNA gene of bacteria PCR primers were generated by PCR amplification using forward primer 63f (5′-CAG GCC TAA CAC ATG CAA GTC-3′) and reverse primer 1387r (5′-GGG CGG WGT GTA CAA GGC-3′) (Marchesi et al. 1998). Platinum Pfx DNA polymerase and PCR Enhancer (Invitrogen, Carlsbad, CA) were used in the following reactions. The PCR mixture (250 ml) contained 0.25 μM each of primers, 0.3 mM each of deoxyribonucleotide-triphosphate (dNTPs), 2 units of Pfx DNA polymerase, 20% PCR Enhancer, and ∼200 ng of DNA template. The PCR cycle parameters were as follows: denature at 94°C for 2 min, 5 cycles of denaturing at 94°C for 15 sec, annealing at 58°C with 1°C decrease after each cycle for 45 sec, extension at 72°C for 100 sec, then 26 cycles of denaturing at 94°C for 15 sec, annealing at 52°C for 45 sec, and extension at 72°C for 100 sec. Resultant PCR products were subsequently purified by low-melting agarose gel electrophoresis. Purified 16S rDNA fragments were cloned into the pPCR-Script Amp SK vector using PCR-Script Amp Cloning Kit (Stratagene, La Jolla, CA), resulting in a 16S rDNA library. Four such libraries were prepared, respectively, for the microbial communities from the tree hole water column and tree hole leaf surface, with or without mosquito larvae present.
Clones were randomly picked from each 16S rDNA clone library for plasmid preparation. Plasmid DNA was prepared using the Wizard Plus SV Miniprep kit (Promega, Madison, WI). After confirmation that the plasmid contained an insert of the expected size, each insert was subjected to high-throughput sequencing using dideoxy dye terminator chemistry and overlapping sequence primers at the Genomic Technology Support Facility, Michigan State University. For each clone, about 600 bp were sequenced at each end. Sequences were aligned and compared to available databases by using the Ribosomal Database Project (RDP; http://rdp.cme.msu.edu).
Because we pooled DNA from treatment replicate samples and therefore did not have true sample replication within a treatment, we compared sequence libraries using LIBSHUFF, a program designed specifically to compare sequence data from 2 libraries in the form of a similarity data matrix (Singleton et al. 2001, Schloss et al. 2004). The program then calculates the Cramer–von Mises statistic, used in analyses of curve-fitting quality, after repeated randomization of operational taxonomic unit (OTU) accumulation curves from the 1% similarity level to the 50% level. The analysis emphasizes numbers of sequences (OTUs) unique to each library and calculates the probability that libraries are identical using a Monte Carlo approach (Singleton et al. 2001, Schloss et al. 2004).
Results
Bacterial abundance
Results of direct microscopic counts of bacteria on leaf surfaces and in the water column are shown in Fig. 1. There was no significant difference in bacterial densities from leaf sonicates in tree holes with or without mosquito larvae (F = 0.07, df = 1, 13; P = 0.80). Larval presence also did not significantly affect water column bacterial densities (F = 4.30, df = 1, 19; P = 0.052), although the probability value barely exceeded 0.05.
Fig. 1.
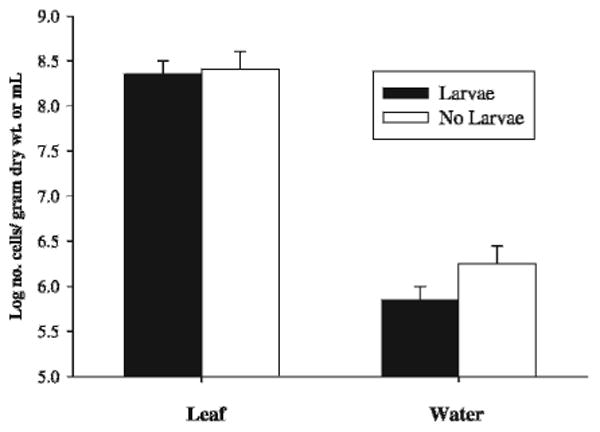
Direct microscopic counts of total bacterial cells in water column and leaf surface samples from tree holes with and without active mosquito larvae. Values are mean ± 6 SE. n = 9–12.
Slow- vs. fast-growing phenotypes
The numbers and proportions of colonies developing on R2A after 7 days of incubation increased in water samples from tree holes without larvae (Fig. 2). Repeated-measures analysis showed that this increase was significant (time × treatment interaction term P < 0.001; F = 7.59, df = 8, 144).
Fig. 2.
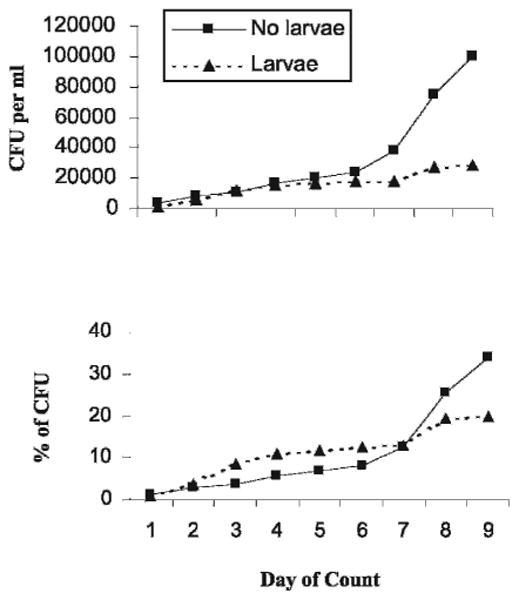
Mean CFU total counts and proportions as a function of R2A plate incubation time of bacteria from tree hole water column samples with and without mosquito larvae. n = 9–12.
Phylogenetic distribution of rDNA sequences
A total of 373 usable sequences, obtained after removal of putative chimeras, were classified with the RDP database. They represented 5 phyla, including Proteobacteria, Bacteroidetes, Firmicutes, Actinobacteria, and Verrucomicrobia, distributed among 29 families (Table 1). In the water column, Flavobacteriaceae and Rhodobacteraceae were dominant groups in the absence of larvae, while Bacteroidetes members other than Flavobacteria, and the Betaproteobacterial group Comamonadaceae, increased in relative proportion when larvae were present. Flavobacteriaceae was also a prominent group in leaf sonicates material from tree holes treated with Bti, but in contrast to the water column, Comamonadaceae and other Betaproteobacteria were also abundant when larvae were absent. Members of the Sphingomonadaceae and Caulobacteriaceae were the dominant groups in leaf samples when larvae were present.
Table 1.
Family-level taxa frequency distributions (percent of sequences per treatment) from 16S rDNA sequence analysis and comparison to Ribosomal Database Project library of bacteria in tree hole larval mosquito habitats.
Percentages of groups above family level reflect numbers of unclassified sequences at that level.
| Taxonomic Category | Bacterial Group | Leaf | Water | ||
|---|---|---|---|---|---|
|
|
|
||||
| Larvae | No larvae | Larvae | No larvae | ||
| Domain | Bacteria | 3 | 4 | 11 | 6 |
| Phylum | Bacteroidetes | 2 | 0 | 10 | 0 |
| Class | Bacteroidetes | ||||
| Order | Bacteroidales | 1 | 0 | 4 | 0 |
| Family | Porphyromonadaceae | 2 | 7 | 7 | 0 |
| Class | Flavobacteria | ||||
| Order | Flavobacteriales | ||||
| Family | Flavobacteriaceae | 13 | 17 | 3 | 62 |
| Class | Sphingobacteria | ||||
| Order | Sphingobacteriales | ||||
| Family | Crenotrichaceae | 0 | 2 | 0 | 0 |
| Phylum | Proteobacteria | 1 | 0 | 1 | 3 |
| Class | Alphaproteobacteria | 1 | 0 | 1 | 1 |
| Order | Rhodobacterales | ||||
| Family | Rhodobacteraceae | 10 | 0 | 4 | 23 |
| Order | Rhodospirillales | ||||
| Family | Acetobacteraceae | 0 | 0 | 1 | 0 |
| Order | Sphingomonadales | ||||
| Family | Sphingomonadaceae | 30 | 6 | 1 | 0 |
| Order | Caulobacterales | ||||
| Family | Caulobacteraceae | 14 | 1 | 1 | 1 |
| Order | Rhizobiales | 2 | 2 | 1 | 0 |
| Family | Rhizobiaceae | 0 | 0 | 1 | 0 |
| Family | Beijerinckiaceae | 2 | 0 | 0 | 0 |
| Family | Bradyrhizobiaceae | 2 | 0 | 1 | 0 |
| Class | Betaproteobacteria | 0 | 12 | 1 | 0 |
| Order | Burkholderiales | 0 | 2 | 1 | 0 |
| Family | Burkholderiaceae | 1 | 0 | 0 | 0 |
| Family | Comamonadaceae | 8 | 18 | 22 | 2 |
| Family | Oxalobacteraceae | 0 | 2 | 4 | 0 |
| Family | Incertae sedis | 6 | 7 | 3 | 0 |
| Order | Rhodocyclales | 0 | 0 | 0 | 0 |
| Family | Rhodocyclaceae | 0 | 10 | 4 | 0 |
| Order | Neisseriales | ||||
| Family | Neisseriaceae | 0 | 1 | 0 | 0 |
| Order | Methylophilales | ||||
| Family | Methylophilaceae | 0 | 2 | 0 | 0 |
| Class | Deltaproteobacteria | ||||
| Order | Myxococcales | ||||
| Family | Polyangiaceae | 0 | 0 | 1 | 0 |
| Order | Desulfobacterales | ||||
| Family | Desulfobulbaceae | 0 | 1 | 1 | 0 |
| Class | Gammaproteobacteria | 0 | 1 | 0 | 0 |
| Order | Pseudomonadales | ||||
| Family | Pseudomonadaceae | 0 | 1 | 1 | 0 |
| Order | Xanthomonadales | ||||
| Family | Xanthomonadaceae | 1 | 0 | 2 | 0 |
| Order | Legionellales | ||||
| Family | Coxiellaceae | 0 | 0 | 1 | 0 |
| Order | Aeromonadales | ||||
| Family | Aeromonadaceae | 0 | 0 | 1 | 0 |
| Order | Methylococcales | ||||
| Family | Methylococcaceae | 0 | 2 | 0 | 0 |
| Order | Enterobacteriales | ||||
| Family | Enterobacteriaceae | 0 | 2 | 0 | 0 |
| Order | Thiotrichales | ||||
| Family | Piscirickettsiaceae | 0 | 0 | 0 | 1 |
| Phylum | Actinobacteria | ||||
| Class | Actinobacteria | ||||
| Order | Actinomycetales | 0 | 0 | 1 | 0 |
| Suborder | Propionibacterineae | 0 | 0 | 1 | 0 |
| Phylum | Verrucomicrobia | ||||
| Class | Verrucomicrobia | ||||
| Order | Verrucomicrobiales | ||||
| Family | Verrucomicrobiaceae | 0 | 0 | 1 | 0 |
| Phylum | Firmicutes | 0 | 0 | 1 | 0 |
| Class | Clostridia | ||||
| Order | Clostridiales | 0 | 1 | 2 | 1 |
| Family | Eubacteriaceae | 0 | 0 | 1 | 0 |
| Family | Clostridiaceae | 0 | 1 | 3 | 1 |
| Phylum | Genera incertae sedis | 0 | 0 | 1 | 0 |
A multiple comparison of sequence libraries using LIBSHUFF techniques indicated that all 4 libraries were different from one another, but that 3 were likely nested subsets of the remaining library. Leaf surface/larvae (LL), water column/larvae (WL), and water column/no larvae (WNL) libraries were likely subsets of the leaf surface/no larvae (NL) library (Table 2). The WL library was also a likely subset of LL and WNL, and LL was a likely subset of WNL.
Table 2.
Results of LIBSHUFF comparisons of all sequence libraries. Values are the probabilities that the library listed in the top row covered the library listed in the left-hand column (i.e., that the left-hand column entry was a subset of the top row entry). The equivalent significance level of 0.05 with Bonferroni corrections for multiple comparisons is 0.003.1
| Library | LN | LL | WL | WNL |
|---|---|---|---|---|
| LN | - | 0.001 | 0.001 | 0.001 |
| LL | 0.867 | - | 0.001 | 0.772 |
| WL | 0.860 | 1.000 | - | 1.000 |
| WNL | 1.000 | 0.000 | 0.001 | - |
LN, leaf/no larvae; LL, leaf/larvae; WL, water column/larvae; WNL, water column/no larvae.
Discussion
Results of this study indicate that Oc. triseriatus larvae can alter bacterial community composition in natural tree holes and that this effect appears to be independent of overall bacterial abundance. Studies of larval mosquito feeding on bacteria have not shown a consistent directional effect on bacterial abundance except for reductions in overall bacterial numbers and productivity associated with leaf material (Kaufman et al. 1999, 2001, 2002). It was therefore not surprising to observe no pronounced mosquito effect on water column total bacterial numbers. The lack of a larval feeding effect on leaf bacterial abundance here was unexpected but may simply reflect the paucity of studies in natural tree hole habitats. In this study, leaf material was collected from the upper layer of detritus in the tree holes, but nothing was known about how long the material had been in the system. Additionally, this study examined bacteria only 3 days after treatment. Previous studies (e.g., Kaufman et al. 2001, 2002) that have documented larval grazing on leaf-associated bacterial abundance have used recently immersed senescent leaf material in lab or field microcosms, and have sampled after a week or longer of larval grazing. It is important to note that our treatment selectively removed mosquito larvae and did not remove other macroinvertebrates in the tree holes, such as scirtid beetle larvae. Scirtid beetle larvae feed principally on leaf and container wall surfaces (Paradise and Dunson 1997) and therefore would have been an important influence on leaf-associated bacterial abundance in both treated and untreated tree holes.
Although we did not detect a change in overall bacterial abundance in this study, CFUs of cultivable slow-growing bacteria and bacterial 16S rRNA sequences indicated a community response to the removal of active mosquito larvae. The significant reduction of slow-growing bacterial forms in the water column would be consistent with metazoan–protist–bacteria trophic cascades seen in planktonic systems. Bacterial communities adapted to Daphnia feeding have shown relatively higher proportions of dividing cells (Degans et al. 2002). Predation by protists (ciliates and flagellates) on bacteria in aquatic systems often results in relative increases of larger, filamentous bacteria that grow relatively slowly (Jurgens and Matz 2002). Because mosquito larvae greatly reduce protozoan abundance in tree hole water columns (Kaufman et al. 2002), removal of larvae would allow protozoan grazing pressures to increase and thereby induce slower-growing forms. Large filaments were not obvious in our direct count preps for this study; however, we have observed them previously (Kaufman et al. 2000) and 3 days beyond larval removal in this experiment may not have been sufficient time to allow development of filaments in the system.
Examination of sequence classifications from water column bacteria (Table 1) did not reveal an obvious shift to slow-growing taxa in the absence of larvae. The trend of increasing relative abundance of Flavobacteriaceae (primarily Flavobacterium spp.) would be consistent with an increase in slow or intermediate growth forms because members of this group of bacteria tend to have low to intermediate rRNA operon copy numbers compared to proteobacterial groups (Klappenbach et al. 2000). However, free-living aquatic bacteria in the Cytophaga–Flavobacteria–Bacteroides (CFB) cluster exhibit a range of growth rates (Kirchman 2002), and the databases for operon copy number are far from comprehensive. Interestingly, the CFB group has been shown to increase in proportion when bacteriovores are eliminated/reduced in freshwater planktonic systems (Hahn and Höfle 2001, Kirchman 2002) and that appears to also be true of tree holes. The CFB group members are known for their abilities to degrade chitin and other polymers (Kirchman 2002) and the availability of this substrate might have increased after Bti treatment with the resultant carcasses of larvae. The percentage of CFB in the water column without larvae treatment (62%; Table 1) reported here was unusually high for an aquatic system (Kirchman 2002), suggesting further investigation of this group in tree hole habitats.
Other groups that appeared to respond positively or negatively to removal of larvae, the Rhodobacteraceae and Caulobacteriacae, and the Comamonadaceae, Rhodocyclaceae, and other Betaproteobacteria, represent diverse ecophysiological categories that make assumptions about their interactions with mosquitoes and other components of the tree hole system tenuous. Although there is much evidence from our previous studies (Kaufman et al. 2001, 2002, Kaufman et al. 2008) and other aquatic systems (King et al. 1991, Jurgens 1994, Hahn and Hofle 2001) indicating direct predation and digestibility of microbial taxa are the mechanisms for microbial community changes, we cannot rule out or separate indirect effects of mosquito larvae on bacterial groups. We have previously noted that microbial nitrogen metabolism is different in microcosms with and without larvae (Kaufman et al. 1999) and that this may be related to larval nitrogen metabolism. We did not address potential chemical changes in the tree holes after larvae were killed, but it is possible that pH, dissolved oxygen, and other solutes were altered by the treatment. For example, Rhodobacteraceae appeared to greatly increase in the water column when larvae were eliminated. Because many Rhodobacteraceae utilize nitrate to oxidize reduced sulfur compounds, the absence of larvae may have stimulated this group indirectly through a change in concentrations or moieties of nitrogen or sulfur compounds.
Additionally, we cannot necessarily rule out indirect effects of the Bti formulation itself. We recovered no Bacillus sequences (Table 1), indicating very low viable cell concentrations in the tree holes, and Bti is not thought to grow after application except possibly in insect cadavers (Siegel et al. 2001). However, it is possible that cellular constituents affected bacterial groups via added nutrients, but general nutrient addition (biomass) from the Bti formulation would have been substantially less than what was added with larval carcasses. Although the dead larvae added to the non-Bti treatment were killed with a combination of boiling water and microwaves, it is possible that some bacteria were added to tree holes in this venue. These would likely be undetectable against the background abundance of natural bacterial populations, but we cannot rule out subsequent growth and influence of any introduced organisms. One would expect these to be spore-formers or Enterobacteriaceae associated with the larval gut, but sequence data indicated these groups were rare (Table 1).
Two previous studies have suggested that Enterobacteriaceae might be important in Oc. triseriatus larval habitats. Kaufman et al. (1999) found that bacterial group to increase in the presence of larvae in a microcosm study, and DeMaio et al. (1996) isolated Klebsiella strains from midguts of 4th instars and other Enterobacteriaceae from guts of adult Oc. triseriatus. Both studies employed fairly general media and would have isolated primarily fast-growing organisms. Our results indicate that Enterobacteriaceae are not prominent members of the tree hole bacterial community and the few sequences found were from the Bti treatment. Interestingly, DeMaio et al. (1996) found Flavobacterium spp. in adult midguts, suggesting the importance of this group continues beyond the larval stage.
Although the LIBSHUFF (Table 2) results indicate that bacterial sequences from the water column are subsets of the leaf surface sequences within a given treatment, bacterial groups on leaves appeared to respond differently. For example, Betaproteobacteria groups were high in relative abundance in the water column with larvae compared to the water column without larvae, but the opposite trend was observed for leaf material. This may indicate some bacterial groups on leaves are being dislodged into the water column. However, many specific sequences of Betaproteobacteria in the water column with larvae were different at the generic level from those seen on leaf material with larvae present, suggesting simple displacement is not the full explanation. Our recent investigations of microbial groups associated with leaves of a known age that had been tethered in natural tree holes (Kaufman et al. 2008) indicated that Betaproteobacteria were more abundant in the presence of larvae and certain Bacteroidetes groups were reduced. In this study, Bacteroidetes on leaves seemed unaffected by larval presence. We believe this illustrates the need for more comprehensive studies that address microbial dynamics in the system across a wider range of conditions. A shortcoming of this study was the sampling of a single time point, and examinations of bacterial communities through the entire larval development cycle are warranted. We also stress the need to examine multiple substrate types in the habitat, as larvae browse on many surfaces and leaf material can vary greatly (Merritt et al. 1992, Walker et al. 1997). We did not, for example, examine microbial communities associated with the larval cadavers in this study. Insect carcasses, however, can be an important detrital resource for tree hole mosquitoes (Daugherty et al. 2000, Yee and Juliano 2006).
We also recognize that our sequence data results as they relate to treatment or substrate effects should be interpreted cautiously for 2 reasons. First, it can be argued that the technique of PCR-based cloning and sequence analysis skews results toward taxa with higher numbers of rRNA gene copies (Amann and Ludwig 2000), and our sequence data would not necessarily match estimates of bacterial group abundance obtained with other methods. The extrapolation of sequence abundance to population density of a bacterial taxon is generally not warranted but is appropriate for examining changes in similar bacterial groups (Amann and Ludwig 2000). Second, because we did not examine sequence variation among individual tree holes, a rigorous statistical test of the larval effect was unavailable. We consider the LIBSHUFF analysis to be strong evidence that sequence libraries generated from pooled samples of the substrate–treatment combinations were sufficiently different to support a hypothesis that larvae altered bacterial communities, but that further study is warranted. In a similar study making use of replicated sequence data, Kaufman et al. (2008) showed that bacterial and fungal groups on leaf material placed in tree holes were affected by larval presence. That study suggests the bacterial taxa trends illustrated here are also likely to be a result of larval effects. Nonetheless, the primary value of the identifications in Table 1 is to provide an initial survey of bacteria in tree holes and to identify potential targets for further investigation.
Because this study represented one of the first to examine tree hole bacteria in relation to larval feeding, our goals were largely exploratory. The evidence that Flavobacteriaceae are likely an important food item has led us to further investigate members of this group and to elucidate promoter and expression system genetics in Flavobacterium species isolated from tree holes (Chen et al. 2007) in order to develop transgenic bacteria capable of expressing larvicidal toxins in natural mosquito habitats. We are continuing our investigations of other microbial groups in the system, particularly slow-growing taxa and those associated with leaf surfaces, in an effort to better understand the relationships between mosquito larvae and their food resources.
Acknowledgments
We thank Tracy Smith and Blair Bullard for technical assistance. This project was funded by National Institutes of Health award AI21884.
References Cited
- Amann R, Ludwig W. Ribosomal RNA-targeted nucleic acid probes for studies in microbial ecology. FEMS Microbiol Rev. 2000;24:555–565. doi: 10.1111/j.1574-6976.2000.tb00557.x. [DOI] [PubMed] [Google Scholar]
- Bell T, Ager D, Song JI, Newman JA, Thompson IP, Lilley AK, van der Gast CJ. Larger islands house more bacterial taxa. Science. 2005a;308:1884. doi: 10.1126/science.1111318. [DOI] [PubMed] [Google Scholar]
- Bell T, Newman JA, Silverman BW, Turner SL, Lilley AK. The contribution of species richness and composition to bacterial services. Nature Lett. 2005b;436:1157–1160. doi: 10.1038/nature03891. [DOI] [PubMed] [Google Scholar]
- Briegel H. Metabolic relationship between female body size, reserves, and fecundity of Aedes aegypti. J Insect Physiol. 1990;36:165–172. [Google Scholar]
- Briegel H, Knüsel I, Timmermann SE. Aedes aegypti: size, reserves, survival, and flight potential. J Vector Ecol. 2001;26:21–31. [PubMed] [Google Scholar]
- Chen S, Kaufman M, Bagdasarian M, Walker E. Characterization of strong promoters from an environmental Flavobacterium hibernum strain. Appl Environ Microbiol. 2007;73:1089–1100. doi: 10.1128/AEM.01577-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochran-Stafira DL, von Ende CN. Integrating bacteria into food webs: studies with Sarracenia purpurea inquilines. Ecology. 1998;79:880–898. [Google Scholar]
- Daugherty MP, Alto BW, Juliano SA. Invertebrate carcasses as a resource for competing Aedes albopictus and Aedes aegypti (Diptera: Culicidae) J Med Entomol. 2000;37:364–372. doi: 10.1093/jmedent/37.3.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degans H, Zöllner E, Van der Gucht K, De Meester L, Jürgens K. Rapid Daphnia mediated changes in microbial community structure: an experimental study. FEMS Microbiol Ecol. 2002;42:137–149. doi: 10.1111/j.1574-6941.2002.tb01003.x. [DOI] [PubMed] [Google Scholar]
- DeMaio J, Pumpui CB, Ken M, Beier JC. The midgut bacterial flora of wild Aedes triseriatus, Culex pipiens, and Psorophora columbiae mosquitoes. Amer J Trop Med Hyg. 1996;54:219–223. doi: 10.4269/ajtmh.1996.54.219. [DOI] [PubMed] [Google Scholar]
- Eisenberg JNS, Washburn JO, Schreiber SJ. Generalist feeding behaviors of Aedes sierrensis larvae and their effects on protozoan populations. Ecology. 2000;81:921–935. [Google Scholar]
- Fish D, Carpenter SR. Leaf litter and larval mosquito dynamics in tree-hole ecosystems. Ecology. 1982;63:283–288. [Google Scholar]
- Frank JH, Lounibos LP, editors. Phytotelmata: terrestrial plants as hosts for aquatic insect communities. Medford, NJ: Plexus; 1983. [Google Scholar]
- Glöckner FO, Amann R, Alfreider A, Pernthaler J, Psenner R, Trebesius K, Schleifer KH. An in situ hybridization protocol for detection and identification of planktonic bacteria. Syst Appl Microbiol. 1996;19:403–406. [Google Scholar]
- Hahn MW, Höfle MG. Grazing of protozoa and its effect on populations of aquatic bacteria. FEMS Microbiol Ecol. 2001;35:113–121. doi: 10.1111/j.1574-6941.2001.tb00794.x. [DOI] [PubMed] [Google Scholar]
- Jürgens K. Impact of Daphnia on planktonic microbial food webs—a review. Mar Microb Food Webs. 1994;8:295–324. [Google Scholar]
- Jürgens K, Matz C. Predation as a shaping force for the phenotypic and genotypic composition of planktonic bacteria. Antonie Van Leeuwenhoek. 2002;81:413–434. doi: 10.1023/a:1020505204959. [DOI] [PubMed] [Google Scholar]
- Kaufman MG, Bland SN, Worthen ME, Walker ED, Klug MJ. Bacterial and fungal biomass responses to feeding by larval Aedes triseriatus (Diptera: Culicidae) J Med Entomol. 2001;38:711–719. doi: 10.1603/0022-2585-38.5.711. [DOI] [PubMed] [Google Scholar]
- Kaufman MG, Chen S, Walker ED. Leaf-associated bacterial and fungal community shifts in response to larvae of the mosquito. Ochlerotatus triseriatus Microb Ecol. 2008 doi: 10.1007/s00248-007-9310-6. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman MG, Goodfriend W, Kohler-Garrigan A, Walker ED, Klug MJ. Soluble nutrient effects on microbial communities and mosquito production in Ochlerotatus triseriatus habitats. Aquat Microb Ecol. 2002;29:73–88. [Google Scholar]
- Kaufman MG, Walker ED. Indirect effects of soluble nitrogen on growth of Ochlerotatus triseriatus larvae in container habitats. J Med Entomol. 2006;43:677–688. doi: 10.1603/0022-2585(2006)43[677:ieosno]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Kaufman MG, Walker ED, Odelson DA, Klug MJ. Microbial community ecology and insect nutrition. Am Entomol. 2000;46:173–184. [Google Scholar]
- Kaufman MG, Walker ED, Smith TW, Merritt RW, Klug MJ. The effects of larval mosquitoes Aedes triseriatus and stemflow on microbial community dynamics in container habitats. Appl Environ Microbiol. 1999;65:2661–2673. doi: 10.1128/aem.65.6.2661-2673.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King CH, Sanders RW, Shotts KG, Jr, Porter KG. Differential survival of bacteria ingested by zooplankton from a stratified eutrophic lake. Limnol Oceanogr. 1991;36:829–845. [Google Scholar]
- Kirchman DL. The ecology of Cytophaga– Flavobacteria in aquatic environments. FEMS Microbiol Ecol. 2002;39:91–100. doi: 10.1111/j.1574-6941.2002.tb00910.x. [DOI] [PubMed] [Google Scholar]
- Kitching RL. Food webs and container habitats. Cambridge, United Kingdom: Cambridge Univ. Press; 2000. [Google Scholar]
- Kitching RL. Food webs in phytotelmata: “bottom-up” and “top-down” explanations for community structure. Annu Rev Entomol. 2001;46:729–760. doi: 10.1146/annurev.ento.46.1.729. [DOI] [PubMed] [Google Scholar]
- Klappenbach JA, Dunbar JM, Schmidt TM. rRNA operon copy number reflects ecological strategies of bacteria. Appl Environ Microbiol. 2000;66:1328–1333. doi: 10.1128/aem.66.4.1328-1333.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird M. The natural history of larval mosquito habitats. San Diego, CA: Academic Press; 1988. [Google Scholar]
- Marchesi JR, Sato T, Weightman AJ, Martin TA, Fry JC, Hiom SJ, Wade WG. Design and evaluation of useful bacterium-specific PCR primers that amplify genes coding for bacterial 16S rDNA. Appl Environ Microbiol. 1998;64:795–799. doi: 10.1128/aem.64.2.795-799.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt RW, Dadd RH, Walker ED. Feeding behavior natural food and nutritional relationships of larval mosquitoes. Annu Rev Entomol. 1992;37:349–376. doi: 10.1146/annurev.en.37.010192.002025. [DOI] [PubMed] [Google Scholar]
- Paradise CJ, Dunson WA. Insect species interactions and resource effects in treeholes: are helodid beetles bottom-up facilitators of midge populations? Oecologia. 1997;109:303–312. doi: 10.1007/s004420050088. [DOI] [PubMed] [Google Scholar]
- Reasoner DJ, Geldreich EE. A new medium for the enumeration and subculture of bacteria from potable water. Appl Environ Microbiol. 1985;49:1–7. doi: 10.1128/aem.49.1.1-7.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Larget BR, Handelsman J. Integration of microbial ecology and statistics: a test to compare gene libraries. Appl Environ Microbiol. 2004;70:5485–5492. doi: 10.1128/AEM.70.9.5485-5492.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel JP, Smith AR, Novak RJ. Recovery of commercially produced Bacillus thuringiensis var. israelensis and Bacillus sphaericus from tires and prevalence of Bacilli in artificial and natural containers. J Am Mosq Cont Assoc. 2001;17:33–41. [PubMed] [Google Scholar]
- Singleton DR, Furlong MA, Rathbun SL, Whitman WB. Quantitative comparisons of 16S rRNA gene sequence libraries from environmental samples. Appl Environ Microbiol. 2001;67:4374–4376. doi: 10.1128/AEM.67.9.4374-4376.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmerman SE, Briegel H. Larval growth and biosynthesis of reserves in mosquitoes. J Insect Physiol. 1999;45:461–470. doi: 10.1016/s0022-1910(98)00147-4. [DOI] [PubMed] [Google Scholar]
- Velgi MI, Albright LJ. Improved sample preparation for enumeration of aggregated aquatic substrate bacteria. In: Kemp PF, Sherr BF, Sherr EB, Cole JJ, editors. Handbook of methods in aquatic microbiology. Boca Raton, FL: Lewis; 1993. pp. 139–142. [Google Scholar]
- Walker ED, Kaufman MG, Ayres MP, Riedel MH, Merritt RW. Effect of variation in quality of leaf detritus on growth of the eastern tree hole mosquito Aedes triseriatus Diptera: Culicidae. Can J Zool. 1997;75:706–718. [Google Scholar]
- Walker ED, Lawson DL, Morgan WT, Klug MJ. Nutrient dynamics bacterial populations and mosquito productivity in tree hole ecosystems. Ecology. 1991;72:1529–1546. [Google Scholar]
- Walker ED, Merritt RW. The significance of leaf detritus to mosquito (Diptera, Culicidae) productivity from treeholes. Env Entomol. 1988;17:199–206. [Google Scholar]
- Walker ED, Merritt RW, Wotton RW. Analysis of the distribution and abundance of Anopheles quadrimaculatus (Diptera, Culicidae) larvae in a marsh. Env Entomol. 1988;17:992–999. [Google Scholar]
- Yee DA, Juliano SA. Consequences of detritus type in an aquatic microsystem: effects on water quality, micro-organisms and performance of the dominant consumer. J Freshwat Biol. 2006;51:448–459. doi: 10.1111/j.1365-2427.2005.01504.x. [DOI] [PMC free article] [PubMed] [Google Scholar]


