Abstract
Recently, synthetic prions with a high level of specific infectivity have been produced from chemically defined components in vitro. A major insight arising from these studies is that various classes of host-encoded cofactor molecules such as phosphatidylethanolamine and RNA molecules are required to form and maintain the specific conformation of infectious prions. Synthetic mouse prions formed with phosphatidylethanolamine exhibit levels of specific infectivity ∼1 million-fold greater than “protein-only” prions (Deleault, N. R., Walsh, D. J., Piro, J. R., Wang, F., Wang, X., Ma, J., Rees, J. R., and Supattapone, S. (2012) Proc. Natl. Acad. Sci. U.S.A. 109, E1938–E1946). Moreover, cofactor molecules also appear to regulate prion strain properties by limiting the potential conformations of the prion protein (see Deleault et al. above). The production of fully infectious synthetic prions provides new opportunities to study the mechanism of prion infectivity directly by structural and biochemical methods.
Keywords: Phosphatidylethanolamine, Phospholipid, Prion, Protein Misfolding, RNA, Cofactor, Infectivity, Synthetic
Introduction
Prions are the unconventional infectious agents of spongiform encephalopathies, a group of deadly neurodegenerative diseases that can be transmitted among humans and other mammals. Although these infectious diseases can be propagated indefinitely by serial passage in wild type hosts, prions do not contain a replicable genome capable of directing self-synthesis, in contrast to all other known infectious agents (2). Instead, infectious prions appear to be formed by the conversion of a host-encoded, membrane-bound glycoprotein termed PrPC2 into several misfolded conformation(s) collectively termed PrPSc. Nonetheless, like other infectious agents, prions can replicate to high titer and exist as self-propagating strains within the same host species, which can be characterized by distinct clinical phenotypes and selective patterns of neurotropism (3, 4).
Several lines of evidence indicate that prion infectivity and strain properties are encoded by specific PrPSc conformation(s), but at present neither the tertiary structure of infectious PrPSc nor the molecular mechanism responsible for generating infectious PrPSc conformation(s) has been fully elucidated, despite decades of intensive investigation. Historically, several factors have conspired to impede attempts to study these questions directly, namely: 1) PrPSc molecules exist as insoluble aggregates, precluding structural determination by conventional methods such as x-ray crystallography; 2) the most accurate method to measure prion infectivity, end-point titration in wild type hosts, is expensive and time-consuming; and 3) no model of prion infection exists that would be amenable to genetic screening.
In recent years, we have learned much about the mechanism of prion infectivity by taking a reductionist biochemical approach, particularly the critical role played by cofactor molecules. Moreover, the insights provided by these studies have enabled the efficient synthesis of high titer infectious prions from chemically defined components in vitro, an advance that should facilitate experimental determination of the tertiary structure of PrPSc by biophysical techniques such as solid-state nuclear magnetic resonance.
In Vitro Formation of PrPSc and Prion Infectivity
Pioneering studies of seeded PrPC-to-PrPSc conversion in vitro using purified substrates indicated that this process recapitulated many of the specific features of in vivo prion transmission, but was relatively inefficient (5–7). Indeed, it was observed that a stoichiometric excess of input PrPSc was required to drive the conformational change of radiolabeled PrPC substrate in this purified system. Interestingly, the efficiency of PrPSc formation in this system could be increased by glycosaminoglycans, providing an early clue that prion conversion might utilize factors other than PrP (8).
Soto and co-workers (9) showed that efficient amplification of PrPSc was achievable through their development of the protein misfolding cyclic amplification (PMCA) technique. In this technique, crude brain homogenates containing PrPC and PrPSc are mixed together and incubated for 1–3 days with intermittent sonication, amplifying both PrPSc and prion infectivity (10). The mixture of crude brain homogenate substrates without sonication also resulted in PrPSc amplification, suggesting (by comparison with the relative inefficiency of converting purified radiolabeled PrPC substrate) that factors present in the brain other than PrP molecules enable efficient PrPSc amplification (11).
Identification of Endogenous PrPSc Propagation Cofactors
In an effort to identify endogenous PrPSc amplification cofactors in crude hamster brain homogenates, my colleagues and I subjected the homogenate substrate to various specific treatments known to degrade specific classes of molecules and assessed the effect of such treatments on PrPSc amplification (12). Remarkably, these experiments indicated that selective degradation of single-stranded RNA within homogenates inhibited the amplification of PrPSc molecules derived from several strains of hamster prions. Moreover, reconstitution of RNA-depleted homogenates with single-stranded nucleic acids restored PrPSc amplification, suggesting that RNA might serve as prion propagation cofactors for various hamster prion strains. Further characterization studies showed that no specific nucleotide sequence was required for cofactor activity, confirming that prions do not contain specific genomes.
Interestingly, similar enzymatic treatment and reconstitution studies using several strains of mouse prions indicated that in vitro PrPSc propagation in this species is generally much less reliant (than hamster) upon RNA as a cofactor (13). This discrepancy suggested that it might be fruitful to search for the presumably novel endogenous factor that facilitates the propagation of mouse prions in vitro. Using a reconstitution PMCA assay in which purified PrPC is mixed with fractions containing cofactor activity to produce a substrate mixture, my colleagues and I were able to purify by classical biochemical methods the novel cofactor activity and identify it as phosphatidylethanolamine (PE) (14). Additional experiments showed that PE could facilitate the propagation of PrPSc molecules derived from multiple mouse strains and even some hamster prion strains, albeit less potently.
Because RNA and PE molecules are so different chemically, it seems likely that there are other major classes of molecules that serve as PrPSc propagation cofactors. Novel classes of cofactors might be required for the efficient propagation of prions derived from different strains or animal species.
In Vitro Generation of Chemically Defined, Highly Infectious Prions
The identification of RNA and PE as prion propagation cofactors provided a unique opportunity to produce chemically defined infectious prions in vitro. Using native PrPC molecules immunopurified from normal hamster brain (note that this preparation was devoid of contaminating proteins, nucleic acids, and metals, but did contain stoichiometric quantities of a co-purified lipid) and synthetic polyadenylic acid as a substrate mixture, my colleagues and I were able to produce and propagate bona fide infectious prions from a minimal set of components (15). End-point titration bioassay in wild type hamsters confirmed that these prions possessed a significant level of specific infectivity (∼2 × 105 LD50 units/μg of PrP). Parallel dropout controls demonstrated that the polyadenylic acid (poly(A) RNA) cofactor was essential for prion propagation. Moreover, using this simple system, it was possible to demonstrate for the first time that PrPSc and prion infectivity could be reproducibly generated de novo (i.e. in the absence of infectious PrPSc seed) in a completely contamination-free (prion-naive) environment (15) (an essential condition for demonstrating de novo prion formation because PMCA can be subject to cross-sample contamination (16)). Thus, the interaction between PrPC and cofactor molecules may provide a useful model for studying the spontaneous formation of infectious PrPSc molecules in the brains of patients with sporadic Creutzfeldt-Jakob disease. Similar results were later obtained using Escherichia coli-expressed recombinant mouse (rec)PrP (rather than hamster PrPC) substrate together with polyadenylic acid and the bacterial lipid phosphatidylglycerol (PG) (17); again, PrPSc and prion infectivity could be produced de novo, albeit with a much lower conversion efficiency, presumably because the ability of PG to substitute for endogenous lipid cofactor(s) such as PE is relatively weak (14) and/or RNA interacts less well with mouse PrPC than with hamster PrPC (13).
To test the ability of endogenous PE to produce high titer prions, my colleagues and I generated synthetic prions using only mouse recPrP and chemically synthesized PE. In these reactions, the PrP conversion efficiency was nearly 100% (14) (as compared with ∼2–10% previously obtained for PG (17) or crude mouse brain homogenates (18)). More significantly, end-point bioassay showed that PE-PrPSc molecules (PrPSc molecules produced by serial PMCA propagation in recPrP and PE mixture) displayed a level of specific infectivity (∼2 × 106 LD50 units/μg of PrP) (1) that is similar to that of brain-derived hamster Sc237 PrP27–30 molecules (∼6.5 × 106 LD50 units/μg of PrP) (15).
A comparison of the levels of specific infectivity associated with various preparations of synthetic prions shows the unique ability of PE to facilitate the production truly high titer prions (Table 1). Notably, all preparations made to date without the addition of cofactor molecules have specific infectivity levels <0.1 LD50 units/μg of PrP (19–22). Because the infectious titers of such preparations are so low (i.e. ∼107-fold lower than brain-derived prions or PE-PrPSc molecules), it is unclear whether these minimal amounts of infectivity observed are due to either the presence of a small amount of bacterially derived cofactor(s) in the purified recPrP preparation or the creation of a minimally infectious conformation that can gradually adapt after inoculation into animals, possibly by interacting with endogenous cofactor molecules (23–25). In the case of “protein-only” samples produced by PMCA (as opposed to amyloid fibrils produced by chemical denaturation), it is also possible that either a correspondingly small fraction of PrPSc molecules truly formed without cofactors or an accidental contamination of the sample with very small quantities of infectious prions occurred.
TABLE 1.
A comparison of the specific infectivity of various preparations of synthetic (chemically defined) prions
| Preparation | Cofactor(s) | Seed | Species | Specific infectivity (LD50/μg of PrP)a | Ability to seed brain homogenate PMCA | References |
|---|---|---|---|---|---|---|
| recPrP amyloid fibrils | No | No | Mouse | 0 | No | 18, 19, 20 |
| recPrP amyloid fibrils | No | No | Hamster | 0 | No | 22, 24, 25 |
| recPrP PMCA | No | Yes | Mouse | 0 | No | 14 |
| recPrP PMCA | No | Yes | Hamster | <0.07 | Not tested | 22 |
| Purified PrPC + synthetic RNA + copurified lipid PMCA | Yes | No | Hamster | ∼2 × 104 | Yes | 15 |
| Purified PrPC + synthetic RNA + copurified lipid PMCA | Yes | Yes | Hamster | ∼2 × 105 | Yes | 15 |
| recPrP + PE PMCA | Yes | Yes | Mouse | ∼2 × 106 | Yes | 14 |
| recPrP + liver RNA + PG PMCA | Yes | No | Mouse | >20b | Yes | 17 |
| recPrP + liver RNA + PG PMCA | Yes | No | Mouse | 0 | No | 27 |
| recPrP + liver RNA + PG PMCA | Yes | No | Mouse | >2b | No | 28 |
| recPrP + liver RNA + PG PMCA | Yes | No | Mouse | 0 | No | 28 |
a Specific infectivity = 50% lethal dose per microgram of PrP inoculated intracerebrally into wild-type recipients.
b End-point titration was not performed.
Essential Role of Cofactor for Maintaining Infectious PrPSc Conformation
Collectively, the results from many different laboratories attempting to make synthetic prions (Table 1) suggest that cofactor molecules are required to produce prions with significant levels of infectivity (1, 15, 17, 19–22). A unique opportunity to test this hypothesis directly in a controlled experiment arose once PE-PrPSc prions were produced (1). Fully infectious PE-PrPSc prions were used as the template seed for two lines of serial PMCA propagation processed in parallel, one using a substrate mixture containing recPrP and PE and the other using a substrate containing recPrP alone (Fig. 1). Interestingly, upon serial PMCA propagation in recPrP alone, autocatalytic protein-only PrPSc molecules were formed exhibiting a conformation distinct from PE-PrPSc, as assessed by the size of its protease-resistant core.
FIGURE 1.
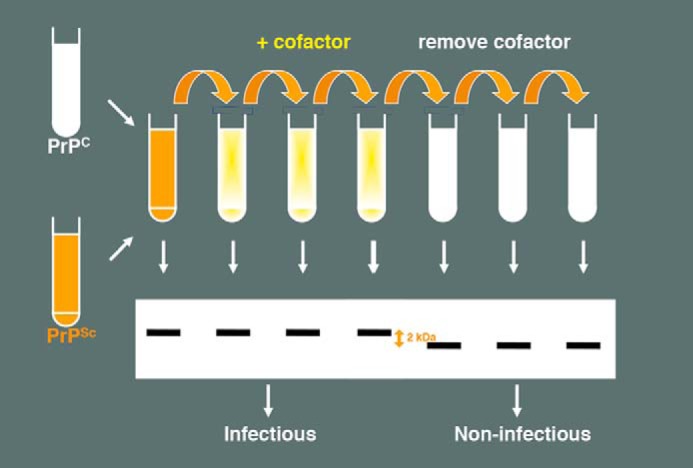
Schematic diagram showing the effect of cofactor withdrawal during serial propagation of purified recombinant PrPSc molecules in vitro. Removal of cofactor from substrate mixture results in PrPSc conformation, as judged by an ∼2-kDa shift in the mobility of the protease-resistant core and ∼106-fold loss of specific infectivity, as judged by end-point dilution bioassay.
End-point titration bioassay showed that protein-only PrPSc molecules were completely noninfectious to wild type mice, and therefore were ∼106-fold less infectious than PE-PrPSc molecules produced in parallel (1). Thus, this internally controlled experiment confirmed that protein molecules alone are unable to maintain the infectious conformation of mammalian prions. Another critical corollary of this experiment is that autocatalytic protein misfolding does not by itself encode infectivity. It is worth noting that the large differences in specific infectivity between various preparations made either with or without cofactor (Table 1) correlate with their ability to seed homogenate PMCA reactions. Therefore, it is unnecessary to invoke other potential mechanisms, such as differential clearance or strain effects, beyond the lack of an infectious conformation to explain the minimal in vivo infectivity of protein-only misfolded PrP preparations.
Interestingly, the infectious conformation of PrPSc appears to be difficult to create and maintain in vitro. Attempts to regenerate the PE-PrPSc conformation by subsequent serial propagation of protein-only PrPSc molecules in a substrate mixture containing both recPrP and PE were not successful (1). These negative results suggest that cofactor molecules do not simply provide a “selective pressure” for the evolution of PrPSc conformers during serial PMCA propagation (26). Rather, it appears more likely that they play a more fundamental role in maintaining the infectious conformation of PrPSc, which is lost irreversibly when cofactor is withdrawn.
Two groups have recently reported that noninfectious PrPSc conformers resembling protein-only PrPSc biochemically could be produced de novo in the presence of PG and RNA (27, 28). However, cofactor withdrawal studies were not performed to distinguish whether these conformers are actually cofactor-dependent or simply represent the formation of protein-only PrPSc in a marginal cofactor mixture.
Strain Properties of Synthetic Prions
A key study by Castilla et al. (29) showed that the biological strain properties of mouse prions could be maintained by serial PMCA propagation in crude brain homogenates. Piro et al. (30) later showed that a reconstituted system containing PrPC and brain homogenates from knock-out mice lacking PrP could also propagate the biological properties of several mouse prion strains in serial PMCA reactions.
Because cofactor molecules are required to maintain the infectious conformation of PrPSc molecules and prion strain properties are potentially encoded by distinctive PrPSc conformations (1), it was logical to employ a chemically defined serial PMCA system to test whether cofactor molecules might play a role in determining strain properties (by directing PrPSc conformation). My colleagues and I serially propagated three different mouse prion strains in a substrate mixture containing only recPrP and PE, inoculated the resulting set of PE-PrPSc molecules into wild type mice, and performed standard strain typing assays to determine whether biological strain properties were maintained or altered by in vitro propagation in the presence of only a single cofactor (Fig. 2) (1). The results showed that propagation in the presence of PE as the sole cofactor caused all three input strains to adapt into a single, apparently novel PE-PrPSc output strain, as judged by incubation time, neurotropism, and biochemical assays. Notably, the dramatic strain shifts observed could not simply be attributed to the use of recPrP substrate lacking post-translational modifications (although glycosylation may influence the biological properties of some mouse prion strains (31)) because one of the input strains used in the experiment consisted of recPrPSc made with different cofactors.
FIGURE 2.
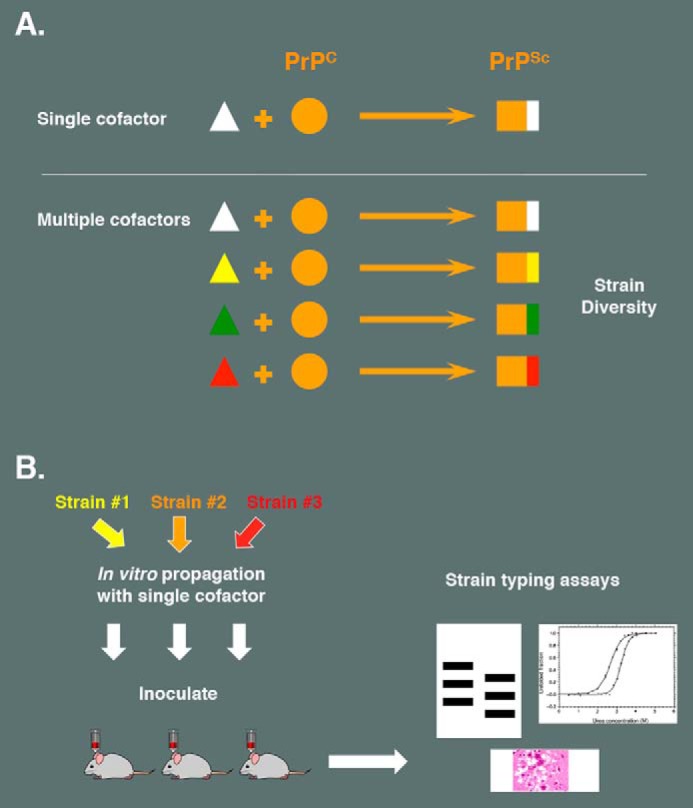
Can a single cofactor maintain prion strain diversity? A, hypothetical model illustrating how the existence of multiple cofactor molecules could encipher prion strain diversity. B, schematic diagram showing the experimental paradigm used to test whether a single cofactor (PE) is able to maintain strain diversity. Strain typing assays showed that three different strains converged to form a single, novel strain after in vitro propagation in the presence of PE alone.
Thus, when only one cofactor is available during prion replication, nascent PrPSc appears to be conformationally constrained, restricting strain diversity (1). These results contrast with those obtained when crude brain homogenate (containing all of the potential endogenous cofactors in brain) is used as substrate, in which case strain diversity was maintained. It is tempting to speculate that prion strain diversity might be encoded by the selective use of different sets of cofactors during the formation of different PrPSc molecules associated with distinct strains. Consistent with this idea, it has been reported that, under specific experimental conditions, PMCA assays of different hamster and mouse prion strains exhibit varying levels of RNA dependence (32, 33). Ultimately, to test this hypothesis rigorously, it will be necessary to isolate and identify additional prion conversion cofactors, especially the components of crude brain homogenate that permit faithful propagation of naturally occurring strains in vitro.
Conclusions and Future Prospects
In summary, studies of infectious prion propagation in vitro have highlighted the important role played by cofactor molecules in this process. Cofactor molecules appear to help maintain the infectious conformation of PrPSc and may also influence strain properties by facilitating specific PrPSc conformation(s).
By using chemically defined substrates to produce purified wild type prions with levels of specific infectivity similar to brain-derived prions, these biochemical studies have formally fulfilled all of Koch's postulates for prions as infectious agents without a nucleic acid genome (1). On the other hand, these studies also raise the possibility that infectious prions may not be composed solely of PrP, i.e. host cofactor molecules may be integral informational components that determine the infectious conformation of the protein scaffold. This possibility could be tested directly by determining the detailed structure of in vitro-generated PrPSc molecules, a scenario that has become more feasible due to our ability to produce high titer recombinant prions. It may be particularly instructive to compare the structures of infectious PrPSc molecules (such as PE-PrPSc) with noninfectious samples prepared without cofactor (such as protein-only PrPSc) to identify the crucial structural determinants of infectivity. It is important to note in this regard that infectious prions formed with cofactors differ in their ultrastructure and seeding specificity from amyloid fibers (18) and that PrP amyloid fibers lack significant prion infectivity (19–21).
Other future prospects include the identification of additional cofactors required for the faithful propagation of various prion strains (29, 30). The selective susceptibility of neuronal populations in vivo (3, 4) and in cultured cells (34) to specific prion strains suggests that strain-specific cofactors may be differentially expressed in various cell types, although other explanations for these phenomena are also possible. Such cofactors could potentially be used as novel therapeutic targets or diagnostic reagents. A prerequisite for the identification of strain-specific cofactors would be the development of a simple and rapid method to confirm the maintenance of specific strain properties during in vitro propagation. Without a doubt, there will be many significant opportunities and challenges ahead in our attempts to understand the unique and remarkable mechanism of prion infectivity.
This work was supported, in whole or in part, by National Institutes of Health Grant 2R01 NS046478 11 (to S. S.).
- PrP
- prion protein
- PrPC
- cellular PrP
- PrPSc
- misfolded PrP
- recPrP
- bacterially expressed recombinant PrP
- PE
- phosphatidylethanolamine
- PG
- phosphatidylglycerol
- PMCA
- protein misfolding cyclic amplification.
REFERENCES
- 1. Deleault N. R., Walsh D. J., Piro J. R., Wang F., Wang X., Ma J., Rees J. R., Supattapone S. (2012) Cofactor molecules maintain infectious conformation and restrict strain properties in purified prions. Proc. Natl. Acad. Sci. U.S.A. 109, E1938–E1946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Prusiner S. B. (1998) Prions. Proc. Natl. Acad. Sci. U.S.A. 95, 13363–13383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bruce M. E. (1993) Scrapie strain variation and mutation. Br. Med. Bull 49, 822–838 [DOI] [PubMed] [Google Scholar]
- 4. Carlson G. A. (1996) Prion strains. Curr. Top. Microbiol. Immunol. 207, 35–47 [DOI] [PubMed] [Google Scholar]
- 5. Bessen R. A., Kocisko D. A., Raymond G. J., Nandan S., Lansbury P. T., Caughey B. (1995) Non-genetic propagation of strain-specific properties of scrapie prion protein. Nature 375, 698–700 [DOI] [PubMed] [Google Scholar]
- 6. Kocisko D. A., Come J. H., Priola S. A., Chesebro B., Raymond G. J., Lansbury P. T., Caughey B. (1994) Cell-free formation of protease-resistant prion protein. Nature 370, 471–474 [DOI] [PubMed] [Google Scholar]
- 7. Kocisko D. A., Priola S. A., Raymond G. J., Chesebro B., Lansbury P. T., Jr., Caughey B. (1995) Species specificity in the cell-free conversion of prion protein to protease-resistant forms: a model for the scrapie species barrier. Proc. Natl. Acad. Sci. U.S.A. 92, 3923–3927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wong C., Xiong L. W., Horiuchi M., Raymond L., Wehrly K., Chesebro B., Caughey B. (2001) Sulfated glycans and elevated temperature stimulate PrPSc-dependent cell-free formation of protease-resistant prion protein. EMBO J. 20, 377–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Saborio G. P., Permanne B., Soto C. (2001) Sensitive detection of pathological prion protein by cyclic amplification of protein misfolding. Nature 411, 810–813 [DOI] [PubMed] [Google Scholar]
- 10. Castilla J., Saá P., Hetz C., Soto C. (2005) In vitro generation of infectious scrapie prions. Cell 121, 195–206 [DOI] [PubMed] [Google Scholar]
- 11. Lucassen R., Nishina K., Supattapone S. (2003) In vitro amplification of protease-resistant prion protein requires free sulfhydryl groups. Biochemistry 42, 4127–4135 [DOI] [PubMed] [Google Scholar]
- 12. Deleault N. R., Lucassen R. W., Supattapone S. (2003) RNA molecules stimulate prion protein conversion. Nature 425, 717–720 [DOI] [PubMed] [Google Scholar]
- 13. Deleault N. R., Kascsak R., Geoghegan J. C., Supattapone S. (2010) Species-dependent differences in cofactor utilization for formation of the protease-resistant prion protein in vitro. Biochemistry 49, 3928–3934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Deleault N. R., Piro J. R., Walsh D. J., Wang F., Ma J., Geoghegan J. C., Supattapone S. (2012) Isolation of phosphatidylethanolamine as a solitary cofactor for prion formation in the absence of nucleic acids. Proc. Natl. Acad. Sci. U.S.A. 109, 8546–8551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Deleault N. R., Harris B. T., Rees J. R., Supattapone S. (2007) Formation of native prions from minimal components in vitro. Proc. Natl. Acad. Sci. U.S.A. 104, 9741–9746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cosseddu G. M., Nonno R., Vaccari G., Bucalossi C., Fernandez-Borges N., Di Bari M. A., Castilla J., Agrimi U. (2011) Ultra-efficient PrPSc amplification highlights potentialities and pitfalls of PMCA technology. PLoS Pathog. 7, e1002370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang F., Wang X., Yuan C. G., Ma J. (2010) Generating a prion with bacterially expressed recombinant prion protein. Science 327, 1132–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Piro J. R., Wang F., Walsh D. J., Rees J. R., Ma J., Supattapone S. (2011) Seeding specificity and ultrastructural characteristics of infectious recombinant prions. Biochemistry 50, 7111–7116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Legname G., Baskakov I. V., Nguyen H. O., Riesner D., Cohen F. E., DeArmond S. J., Prusiner S. B. (2004) Synthetic mammalian prions. Science 305, 673–676 [DOI] [PubMed] [Google Scholar]
- 20. Colby D. W., Wain R., Baskakov I. V., Legname G., Palmer C. G., Nguyen H. O., Lemus A., Cohen F. E., DeArmond S. J., Prusiner S. B. (2010) Protease-sensitive synthetic prions. PLoS Pathog. 6, e1000736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Makarava N., Kovacs G. G., Bocharova O., Savtchenko R., Alexeeva I., Budka H., Rohwer R. G., Baskakov I. V. (2010) Recombinant prion protein induces a new transmissible prion disease in wild-type animals. Acta Neuropathol. 119, 177–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kim J. I., Cali I., Surewicz K., Kong Q., Raymond G. J., Atarashi R., Race B., Qing L., Gambetti P., Caughey B., Surewicz W. K. (2010) Mammalian prions generated from bacterially expressed prion protein in the absence of any mammalian cofactors. J. Biol. Chem. 285, 14083–14087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Makarava N., Kovacs G. G., Savtchenko R., Alexeeva I., Budka H., Rohwer R. G., Baskakov I. V. (2011) Genesis of mammalian prions: from non-infectious amyloid fibrils to a transmissible prion disease. PLoS Pathog. 7, e1002419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Makarava N., Kovacs G. G., Savtchenko R., Alexeeva I., Budka H., Rohwer R. G., Baskakov I. V. (2012) Stabilization of a prion strain of synthetic origin requires multiple serial passages. J. Biol. Chem. 287, 30205–30214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Makarava N., Kovacs G. G., Savtchenko R., Alexeeva I., Ostapchenko V. G., Budka H., Rohwer R. G., Baskakov I. V. (2012) A new mechanism for transmissible prion diseases. J. Neurosci. 32, 7345–7355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Collinge J., Clarke A. R. (2007) A general model of prion strains and their pathogenicity. Science 318, 930–936 [DOI] [PubMed] [Google Scholar]
- 27. Timmes A. G., Moore R. A., Fischer E. R., Priola S. A. (2013) Recombinant prion protein refolded with lipid and RNA has the biochemical hallmarks of a prion but lacks in vivo infectivity. PLoS One 8, e71081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhang Z., Zhang Y., Wang F., Wang X., Xu Y., Yang H., Yu G., Yuan C., Ma J. (2013) De novo generation of infectious prions with bacterially expressed recombinant prion protein. FASEB J. 27, 4768–4775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Castilla J., Morales R., Saá P., Barria M., Gambetti P., Soto C. (2008) Cell-free propagation of prion strains. EMBO J. 27, 2557–2566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Piro J. R., Harris B. T., Nishina K., Soto C., Morales R., Rees J. R., Supattapone S. (2009) Prion protein glycosylation is not required for strain-specific neurotropism. J. Virol. 83, 5321–5328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cancellotti E., Mahal S. P., Somerville R., Diack A., Brown D., Piccardo P., Weissmann C., Manson J. C. (2013) Post-translational changes to PrP alter transmissible spongiform encephalopathy strain properties. EMBO J. 32, 756–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gonzalez-Montalban N., Lee Y. J., Makarava N., Savtchenko R., Baskakov I. V. (2013) Changes in prion replication environment cause prion strain mutation. FASEB J. 27, 3702–3710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Saá P., Sferrazza G. F., Ottenberg G., Oelschlegel A. M., Dorsey K., Lasmézas C. I. (2012) Strain-specific role of RNAs in prion replication. J. Virol. 86, 10494–10504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mahal S. P., Baker C. A., Demczyk C. A., Smith E. W., Julius C., Weissmann C. (2007) Prion strain discrimination in cell culture: the cell panel assay. Proc. Natl. Acad. Sci. U.S.A. 104, 20908–20913 [DOI] [PMC free article] [PubMed] [Google Scholar]


