Background: DNA unwinding by helicases is blocked by proteins bound to duplex DNA.
Results: The single-stranded DNA-binding protein RPA stimulates FANCJ or RECQ1 helicase to disrupt protein-DNA complexes.
Conclusion: Helicases partner with RPA to dislodge proteins bound to duplex DNA.
Significance: Regulation of helicase-catalyzed protein displacement is highly relevant in cellular nucleic acid metabolic processes that require remodeling of chromatinized genomic DNA.
Keywords: DNA-binding Protein, DNA Helicase, DNA Repair, Genetic Disease, Genomic Instability, Telomere, FANCJ, Fanconi Anemia, RECQ1, Replication Protein A
Abstract
Understanding how cellular machinery deals with chromosomal genome complexity is an important question because protein bound to DNA may affect various cellular processes of nucleic acid metabolism. DNA helicases are at the forefront of such processes, yet there is only limited knowledge how they remodel protein-DNA complexes and how these mechanisms are regulated. We have determined that representative human RecQ and Fe-S cluster DNA helicases are potently blocked by a protein-DNA interaction. The Fanconi anemia group J (FANCJ) helicase partners with the single-stranded DNA-binding protein replication protein A (RPA) to displace BamHI-E111A bound to duplex DNA in a specific manner. Protein displacement was dependent on the ATPase-driven function of the helicase and unique properties of RPA. Further biochemical studies demonstrated that the shelterin proteins TRF1 and TRF2, which preferentially bind the telomeric repeat found at chromosome ends, effectively block FANCJ from unwinding the forked duplex telomeric substrate. RPA, but not the Escherichia coli single-stranded DNA-binding protein or shelterin factor Pot1, stimulated FANCJ ejection of TRF1 from the telomeric DNA substrate. FANCJ was also able to displace TRF2 from the telomeric substrate in an RPA-dependent manner. The stimulation of helicase-catalyzed protein displacement is also observed with the DNA helicase RECQ1, suggesting a conserved functional interaction of RPA-interacting helicases. These findings suggest that partnerships between RPA and interacting human DNA helicases may greatly enhance their ability to dislodge proteins bound to duplex DNA, an activity that is likely to be highly relevant to their biological roles in DNA metabolism.
Introduction
Protein bound to single-stranded or double-stranded DNA may impede the progress of replication or transcription or may affect processing steps during DNA repair or recombination; therefore, there has been considerable interest in the cellular mechanism to overcome protein blockades. One of the first pieces of evidence that a helicase may facilitate displacement of protein bound to DNA was provided by Alberts and co-workers (1) where they showed that bacteriophage T4 Dda helicase can dislodge a stationary RNA polymerase from the DNA template. However, biochemical evidence from Yancey-Wrona and Matson (2) demonstrated that the Lac repressor protein bound to DNA inhibited unwinding by various bacterial and bacteriophage helicases to different extents, suggesting that cellular mechanisms may exist to promote the displacement of proteins bound to genomic DNA. Mechanistic studies from Byrd and Raney (3) showed that Dda helicase displaces Escherichia coli Trp repressor from its operator sequence by a cooperative inchworm mechanism. Overall, only a limited number of studies have examined the ability of prokaryotic and eukaryotic DNA helicases to displace proteins bound to DNA (Table 1) (4). For the limited number of helicases that have been shown to catalytically displace protein from DNA, the mechanism(s) are not well understood, particularly in eukaryotes. Understanding how helicase-enacted protein displacement is regulated should be informative in the context of cellular chromosomal DNA metabolism in mammalian cells, which is likely to be much more complex compared with biochemical reactions with naked DNA substrates frequently used in in vitro studies.
TABLE 1.
Roles of DNA helicases that displace proteins bound to DNA in an ATP-dependent manner
| Helicase | Functional activity | Ref. |
|---|---|---|
| E. coli | ||
| UvrD | Dismantle RecQ nucleoprotein filament to regulate HR | 73 |
| Rep | Promote replication of protein-bound DNA at the fork | 47, 48, 74 |
| RecBCD | Dislodge RNA polymerase, lac repressor, and nucleosomes from DNA | 75 |
| Saccharomyces cerevisiae | ||
| Rrm3a | Overcome fork pausing due to non-histone protein-DNA complexes | 76, 77 |
| Srs2 | Disrupt Rad51 presynaptic filament to regulate HR | 54, 57 |
| Pif1 | Negatively regulate telomerase by displacing it from telomeric DNA | 78 |
| Homo sapiens | ||
| RECQ5 | Regulate HR by disrupting RAD51 presynaptic filament | 79 |
| BLM | Regulate HR by disrupting RAD51 presynaptic filament | 80 |
| Helicase-like transcription factorb | Displace RPA, proliferating cell nuclear antigen, and RFC to remodel stalled replication forks | 60 |
| FBH1c | Modulate HR by displacing RAD51 from single-stranded DNA | 68 |
a This is not yet shown to catalytically displace protein bound to DNA in a purified in vitro system.
b This is a helicase-like protein that possesses double-stranded DNA translocase activity but not bona fide helicase activity.
c This is a human recombinant protein used for in vitro studies, but mouse ES cells are used for in vivo studies.
A number of superfamily 2 (SF2) DNA repair helicases from the RecQ and iron-sulfur (Fe-S)2 cluster families are implicated in hereditary disorders characterized by chromosomal instability (5), and it is unclear if and how efficiently these helicases remodel proteins bound to DNA in vivo. Among these helicase proteins, mutations in FANCJ helicase are associated with breast cancer and genetically linked to Fanconi anemia (FA), a progressive bone marrow failure disorder characterized by elevated cancer and a defect in interstrand cross-link repair (6). FANCJ is believed to play a role in homologous recombinational (HR) repair by virtue of its catalytic DNA-unwinding function and interactions with protein partners, including the tumor suppressor BRCA1 (7), the mismatch repair protein MLH1 (8), and the single-stranded DNA-binding protein replication protein A (RPA) (9). Although FANCJ is implicated in downstream events of double strand break repair of interstrand cross-link-induced DNA damage, growing evidence suggests that FANCJ and other FA-linked gene products have roles outside the classical FA pathway. For example, FANCJ resolves G-quadruplex (G4) DNA structures to preserve chromosomal stability (10, 11) and may have a unique role to suppress G4 accumulation or G4-induced DNA damage in vivo (12, 13). G4 DNA (14) and three-stranded T-loops (15) are believed to be prominent structures at telomeres, and recent evidence suggests that FANCJ is found at telomeres in telomerase-negative human cells that preserve their chromosome ends by the alternative lengthening of telomeres (ALT) pathway (16). Telomeres are bound by shelterin proteins (e.g. TRF1, TRF2, and Pot1), and it is generally thought that certain helicases (e.g. WRN, BLM, and RTEL) help to preserve telomere stability by unwinding T-loops and G4 DNA; alternatively, certain helicases (e.g. PIF1) catalytically displace telomerase from DNA to facilitate replication and DNA repair (17–19). Understanding how the catalytic functions of DNA helicases are regulated remains an important question in telomere biology.
Previously, we reported that FANCJ associates with RPA in a DNA damage-inducible manner and that RPA physically binds to FANCJ and stimulates its helicase activity (9). RPA also interacts with several other SF2 RecQ DNA helicases (WRN and BLM (20) and RECQ1 (RECQL and RECQL1) (21)) and stimulates their DNA unwinding activities, suggesting a conservation of the helicase-RPA protein interaction that is important for chromosomal stability. In this study, we have investigated a novel aspect of the functional interaction between RPA and select SF2 DNA repair helicases that enable the helicase to displace protein tightly bound to duplex DNA. Our biochemical results demonstrate that FANCJ and other helicases are strongly blocked from unwinding a forked duplex DNA substrate bound by a catalytically inactive restriction endonuclease or telomere-specific DNA-binding proteins. RPA enables FANCJ or RECQ1 helicase to disrupt the protein-DNA interaction and unwind the double-stranded region to which the protein was bound. These findings suggest that partnerships between RPA and certain helicases may greatly enhance their ability to dislodge proteins bound to duplex DNA, an activity that is likely to be highly relevant to their biological roles in DNA metabolism.
EXPERIMENTAL PROCEDURES
Recombinant Proteins
Baculoviruses encoding FANCJ with a C-terminal FLAG tag (12), His-tagged WRN (22), or His-tagged RECQ1 (23) were used to infect High Five insect cells, and the recombinant proteins were purified as described previously. BamHI-E111A was provided by New England Biolabs. Pot1 (Protection of Telomeres 1) (24), TRF1 (25), and TRF2 (25) were purified as described previously. BLM was kindly provided by Dr. I. Hickson (University of Copenhagen) that was purified as described previously (26). Aro1-RPA and WT-RPA were purified as described previously (27). E. coli SSB (ESSB) was from Promega.
DNA Substrates and Oligonucleotides
PAGE-purified oligonucleotides used for the preparation of forked DNA substrates were purchased from Loftstrand Labs (Gaithersburg, MD). All DNA substrates were prepared by annealing 10 pmol of 32P-end-labeled oligonucleotide to 25 pmol of unlabeled oligonucleotide as described previously (22). The forked DNA substrate with the BamHI consensus sequence was prepared using oligonucleotides BAMHIFORKA (TTTTTTTTTTTTTTTTTTTTTTTTTTTTTTGAATTGCACCGGATCCCTAGGTCGAT) and BAMHIFORKB (ATCGACCTAGGGATCCGGTGCAATTCTTTTTTTTTTTTTTTTTTTTTTTTTTTTTT). This resulted in a substrate with a 26-bp region and 30-nucleotide dT 5′ and 3′ overhangs. The sequences of the oligonucleotides used for the preparation of the forked duplex DNA substrates with two or four telomeric repeats are described elsewhere (28). These substrates contained either 22 or 34 bp flanked by 15-nucleotide 5′ and 3′ overhangs.
Protein Displacement/Helicase Assays
For reactions with RECQ1 (23) and WRN (22), there was an initial 15-min incubation of the forked DNA substrate (0.5 nm) with BamHI-E111A (indicated concentrations) at 24 °C in reaction buffer as described previously, followed by the addition of RECQ1 or WRN in the absence or presence of the indicated concentration of RPA or ESSB. The reactions were then incubated at 37 °C for 15 min. For reactions with FANCJ (29), there was an initial incubation of the indicated forked DNA substrate with BamHI-E111A, TRF1 or TRF2 at 24 °C for 15 min, followed by the addition of FANCJ in the absence or presence of the indicated concentrations of RPA, Pot1, ESSB homotetramer, or BLM proteins. The reactions were then incubated at 30 °C for 15 min. It was noted that the RPA heterotrimer concentration used for helicase experiments with the 22-bp telomeric forked duplex substrates was 5 nm compared with 10 nm RPA for forked 26- or 34-bp duplex substrates because the shorter 22-bp substrate was destabilized by greater RPA concentrations. All reactions were terminated by addition of 20 μl of Stop buffer containing final concentrations of 0.3% SDS, 9 mm EDTA, 0.02% bromphenol blue, 0.02% xylene cyanol, 12.5% glycerol, 5 nm unlabeled oligonucleotide, and 500 μg/ml proteinase K. The reactions were further incubated at 37 °C for 15 min and resolved on nondenaturing 12% (19:1 acrylamide/bisacrylamide) polyacrylamide gels in 1× TBE and quantitated as described previously (11).
Electrophoretic Mobility Shift Assays (EMSA)
Protein/DNA binding mixtures (20 μl) contained the indicated concentrations of BamHI-E111A and 0.5 nm of the 32P-end-labeled forked DNA in binding buffer (10 mm Tris, pH 7.4, 1 mm EDTA, 100 mm potassium acetate, 50 μm DTT, 100 μg/ml BSA, and 10% glycerol). For those experiments in which BamHI-digested calf thymus DNA was included in the binding reaction mixture, the BamHI-digested calf thymus DNA was prepared by incubating 5 μg of calf thymus DNA (Sigma) with 50 units of BamHI in NEBuffer 3 (New England Biolabs). The binding mixtures were incubated at 37 °C for 30 min. After the incubation, 4 μl of Loading dye (74% glycerol, 0.01% xylene cyanol, 0.01% bromphenol blue) was added to each mixture, and samples were loaded onto native 8% (19:1 acrylamide/bisacrylamide) polyacrylamide gels and electrophoresed at 200 V for 2.5 h at 4 °C using 0.5× TBE as the running buffer. The resolved radiolabeled species were visualized using a PhosphorImager and analyzed with ImageQuant software (GE Healthcare).
The reaction mixtures for EMSA experiments with the telomeric 22-bp forked duplex DNA substrate and indicated proteins were the same as those in the protein displacement/helicase assays except that proteinase K digestion of the binding mixture products was not performed. Samples were treated with Loading dye as described above and resolved on native 5% (37.5:1 acrylamide/bisacrylamide) polyacrylamide gels by electrophoresis at 200 V for 2 h at 4 °C using 0.5× TBE as the running buffer.
Restriction Enzyme Protection Experiments
The 26-bp forked duplex DNA substrate containing the BamHI restriction endonuclease recognition sequence was preincubated with BamHI-E111A (38 nm) for 15 min at 30 °C followed by addition of RPA (10 nm) and further incubation at 30 °C for 15 min. BamHI-WT (0.25 units; New England Biolabs) was added to reaction mixtures and incubated for 15 min at 30 °C, followed by addition of proteinase K (500 μg/ml) with Stop buffer and further incubation at 37 °C for 10 min. DNA products were resolved by electrophoresis on nondenaturing 12% polyacrylamide gels. Radiolabeled species were detected and quantitated as described above.
Strand Annealing Experiments
Partially complementary single-stranded oligonucleotides used to prepare 26-bp forked duplex DNA substrates containing the BamHI restriction endonuclease recognition sequence were incubated with the indicated concentrations of RPA or ESSB for 15 min at 30 °C in the presence or absence of BamHI-E111A (38 nm) and subsequent digestion with proteinase K (500 μg/ml) in Stop buffer. Products were resolved on nondenaturing 12% polyacrylamide gels. The annealed 26-bp forked duplex DNA molecule was used as a marker for the annealed product.
RESULTS
Catalytically Inactive BamHI Inhibits DNA Unwinding Activity by Human DNA Repair Helicases
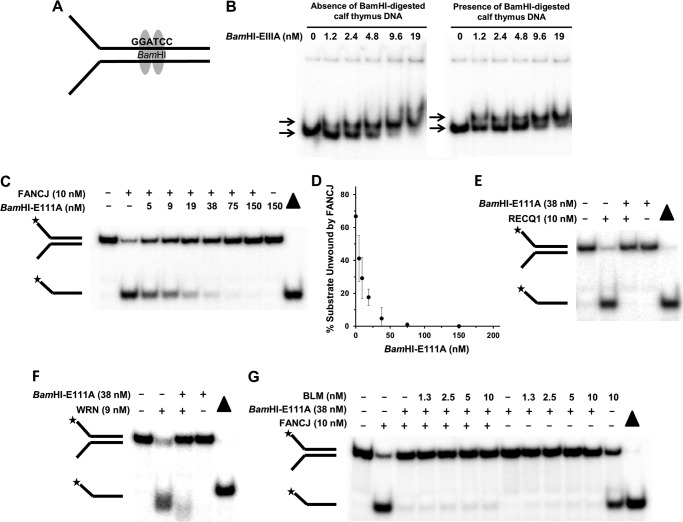
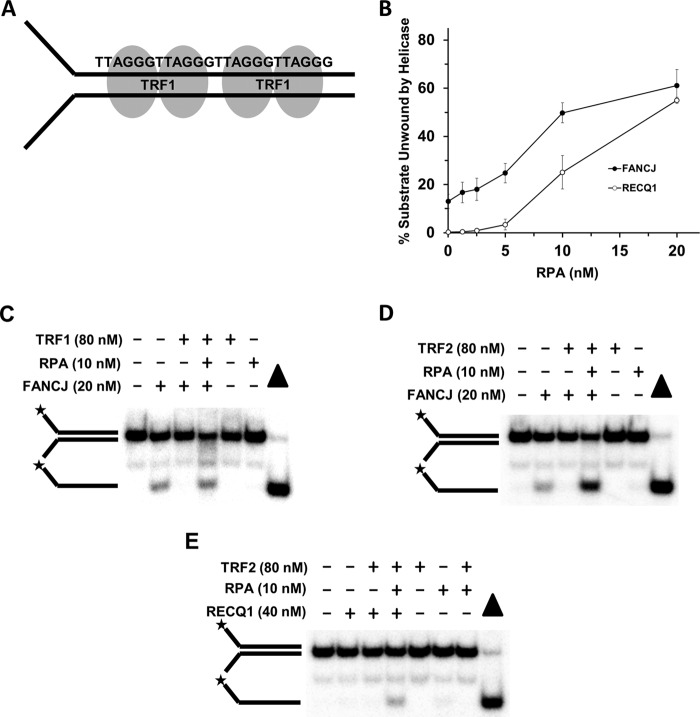
We began our studies by testing whether a catalytically inactive BamHI-E111A restriction endonuclease bound to a forked duplex DNA substrate harboring a cognate palindromic BamHI recognition sequence (Fig. 1A) was able to inhibit helicase-catalyzed unwinding of the DNA substrate. EMSA was used to demonstrate that purified recombinant BamHI-E111A protein bound the radiolabeled forked duplex substrate in a protein concentration-dependent manner in the presence or absence of excess BamHI-digested calf thymus DNA (Fig. 1B). These results are consistent with previously published nitrocellulose filter binding studies (30) and unpublished EMSA data from Jurate Bitinaite (New England Biolabs (Ipswich, MA). BamHI-E111A protein titration experiments revealed that inhibition of FANCJ helicase activity was observed to be dependent on the concentration of BamHI-E111A (Fig. 1C). A concentration of 38 nm BamHI-E111A inhibited FANCJ helicase activity to near completion (Fig. 1D). The greater concentration of BamHI-E111A required for nearly complete FANCJ helicase inhibition compared with the 9 nm BamHI-E111A concentration required for the majority of substrate bound as measured by EMSA may reflect differences in reaction mixtures and conditions for the two assays and/or the effect of native PAGE on the stability of the protein-DNA complex.
FIGURE 1.
BamHI-E111A bound to forked duplex DNA substrate blocks DNA unwinding by human DNA repair helicases. A, schematic representation of BamHI dimer bound to the palindromic recognition sequence harbored within the duplex region of forked DNA substrate. B, representative gel images from EMSA of reaction mixtures containing the indicated concentrations of BamHI-E111A protein and 0.5 nm radiolabeled forked duplex DNA in the absence or presence of 5 μg/ml BamHI-digested calf thymus DNA. The migration of the protein-free radiolabeled forked duplex and BamHI-E111A-bound forked duplex are indicated by the top and bottom arrows, respectively. C, BamHI-E111A inhibits FANCJ helicase activity on forked duplex DNA substrate in a protein concentration-dependent manner. Representative gel of resolved proteinase K-digested products is shown. D, quantitative analysis of BamHI-E111A inhibition of FANCJ helicase activity. E and F, BamHI-E111A (38 nm) inhibits RECQ1 (E) or WRN (F) helicase activity on forked duplex substrate. G, combination of BLM and FANCJ helicases fail to displace BamHI-E111A from the DNA substrate. Proteinase K-digested products from helicase reaction mixtures were resolved by electrophoresis on nondenaturing 12% polyacrylamide gels. Filled triangle, heat-denatured DNA substrate control. Representative gel images from at least three independent experiments are shown. Star denotes 5′-32P end label.
Using this concentration of BamHI-E111A, we observed strong inhibition of RECQ1 (Fig. 1E) or WRN helicase activity as well (Fig. 1F). In the WRN reactions, the unwound product migrated slightly faster than the heat-denatured DNA substrate control (Fig. 1F), consistent with earlier observations that WRN helicase product was slightly degraded by intrinsic WRN nuclease (31). The residual unwound product observed in WRN reactions containing BamHI-E111A migrated even slightly farther than the products from reactions lacking BamHI-E111A, suggesting further degradation by WRN nuclease when the helicase activity was blocked.
Recent studies provided evidence for the coordination of E. coli DNA helicases translocating on opposite strands to displace protein bound to duplex DNA (see under “Discussion”) (32). Based on this paradigm, we tested whether the purified recombinant human FANCJ and BLM helicases, which are known to unwind DNA with opposite polarities yet are physically and functionally interact (29), might collaborate to displace BamHI-E111A from the forked duplex DNA substrate. The results from this experiment proved to be negative. BamHI-E111A failed to be displaced from the DNA substrate in the presence of FANCJ, BLM, or BLM + FANCJ under conditions that each DNA helicase could effectively unwind the naked protein-free forked duplex (Fig. 1G), attesting to the strong blockade imposed by the catalytically inactive restriction endonuclease bound to the double-stranded region of the partial duplex DNA molecule.
RPA Stimulates FANCJ Displacement of BamHI-E111A Bound to Double-stranded DNA Enabling the Helicase to Unwind the Substrate
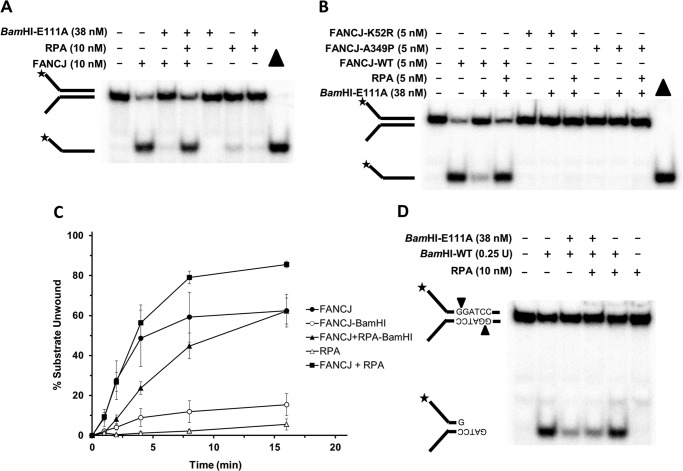
Previously, it was reported that the single-stranded DNA-binding protein RPA physically interacts with FANCJ and stimulates its DNA unwinding reaction (9). This led us to ask if RPA was able to stimulate FANCJ to dislodge BamHI-E111A from the forked duplex DNA substrate and unwind the underlying duplex. As shown in Fig. 2A, the presence of RPA (10 nm) enabled FANCJ to unwind the BamHI-E111A-bound forked duplex to nearly the level of FANCJ helicase activity on the naked forked duplex. In control reactions, RPA alone had only a marginal effect on the stability of the naked forked duplex DNA substrate or the BamHI-E111A-bound forked duplex (Fig. 2A). To ascertain whether the observed RPA-dependent unwinding of the forked duplex DNA substrate bound by BamHI-E111A was dependent on intrinsic FANCJ ATPase/helicase activity, we tested a catalytically inactive FANCJ-K52R mutant protein characterized by an amino acid replacement of the highly conserved lysine 52 in the Walker A (motif I) box with an arginine. The FANCJ-K52R protein was previously shown to be defective as an ATPase or helicase (33). As shown in Fig. 2B, RPA failed to stimulate FANCJ-K52R to unwind the BamHI-E111A-bound forked duplex substrate. We also tested a FANCJ-A349P protein, derived from a clinically relevant FANCJ mutation in the conserved iron-sulfur domain that is genetically linked to FA (34). The FANCJ-A349P mutant protein was previously demonstrated to bind DNA, hydrolyze ATP, and translocate on single-stranded DNA but fail to unwind duplex or G-quadruplex DNA substrates (35). In this study, RPA failed to stimulate FANCJ-A349P to unwind the BamHI-E111A-bound fork duplex substrate, indicating that FANCJ-catalyzed DNA unwinding activity is responsible for RPA-dependent displacement of BamHI-E111A from the forked duplex DNA substrate. Collectively, the studies using catalytic mutants of FANCJ suggest that FANCJ and RPA collaboratively displace BamHI-E111A from the forked duplex as the helicase unwinds the DNA substrate.
FIGURE 2.
RPA stimulates FANCJ to efficiently displace BamHI-E111A from the DNA substrate and unwind it. A, reaction mixtures containing the radiolabeled DNA substrate (0.5 nm) and the indicated proteins were incubated for 15 min at 30 °C, followed by electrophoresis of proteinase K-digested products on 12% polyacrylamide native gels. See “Experimental Procedures” for details. B, reactions mixtures were as described above except the additional presence of a Walker A box ATPase mutant FANCJ-K52R or a patient-derived Fe-S cluster domain mutant FANCJ-A349P as indicated. C, kinetics of FANCJ helicase activity in the presence or absence of RPA on forked duplex substrate pre-bound by BamHI-E111A or not. Proteinase K-digested reaction products were resolved by electrophoresis on nondenaturing 12% polyacrylamide gels. Representative gel images from at least three independent experiments are shown. D, restriction protection analysis suggests RPA does not displace BamHI-E111A bound to DNA substrate. BamHI-E111A was preincubated with DNA substrate for 15 min followed by incubation with RPA for 15 min, subsequent digestion with BamHI-WT, and analysis on nondenaturing 12% polyacrylamide gel as described under “Experimental Procedures.” The BamHI-WT cleavage product is indicated. Star denotes 5′-32P end label.
Kinetic assays were performed to measure DNA unwinding of the BamHI-E111A-bound forked duplex DNA substrate by FANCJ in the presence or absence of RPA. This analysis showed that RPA could stimulate FANCJ to displace BamHI-E111A and unwind the forked duplex in a kinetic manner throughout a 16-min time course (Fig. 2C). By the completion of the time course, FANCJ was able to unwind nearly 60% of the BamHI-E111A-bound forked duplex when RPA was present; however, in the absence of RPA, FANCJ unwound only ∼15% of the forked duplex bound by BamHI-E111A. From a kinetic analysis of the initial rates of DNA unwinding, the percent of BamHI-E111A-bound forked duplex unwound by FANCJ in the presence of RPA was 2-fold greater than that observed for the BamHI-E111A-bound substrate unwound by FANCJ acting alone. For the naked DNA substrate, RPA increased the extent of DNA substrate unwound by FANCJ in the latter time points (8 and 16 min). These results demonstrate that RPA significantly enhanced the rate of FANCJ-catalyzed BamHI-E111A displacement from the forked duplex DNA substrate.
To address whether RPA acting alone was able to displace BamHI-E111A from the forked duplex DNA substrate harboring its cognate recognition sequence element, restriction enzyme protection experiments were performed. BamHI-E111A was preincubated with the DNA substrate followed by addition of RPA for 10 min. The reaction mixture was then incubated with catalytically active BamHI-WT enzyme, and the DNA products were resolved by electrophoresis on native polyacrylamide gels. As shown by a representative gel in Fig. 2D, a similar level of DNA substrate was protected from BamHI-WT digestion by the inactive BamHI-E111A in the presence or absence of RPA (10 nm). These results suggest that RPA alone does not displace BamHI-E111A from the DNA substrate, enabling FANCJ to unwind the duplex DNA.
Specificity and DNA Binding Requirement of RPA to Stimulate FANCJ Protein Displacement
To assess whether the ability of RPA to avidly bind single-stranded DNA was required for it to stimulate FANCJ displacement of BamHI-E111A from the forked duplex substrate, we tested a previously studied RPA mutant heterotrimer (Aro1-RPA), characterized by two amino acid substitutions within the DNA binding domain of the RPA70 subunit (RPA1) (Fig. 3A), that is significantly compromised in single-stranded DNA binding (27). As shown in Fig. 3B, the Aro1-RPA mutant heterotrimer poorly stimulated FANCJ to displace BamHI-E111A from the forked duplex under reaction conditions that the wild-type RPA heterotrimer readily stimulated FANCJ displacement of BamHI-E111A. Based on these results, we conclude that the single-stranded DNA binding activity of RPA is necessary for it to stimulate FANCJ-catalyzed displacement of BamHI-E111A from the DNA substrate during the DNA unwinding reaction.
FIGURE 3.
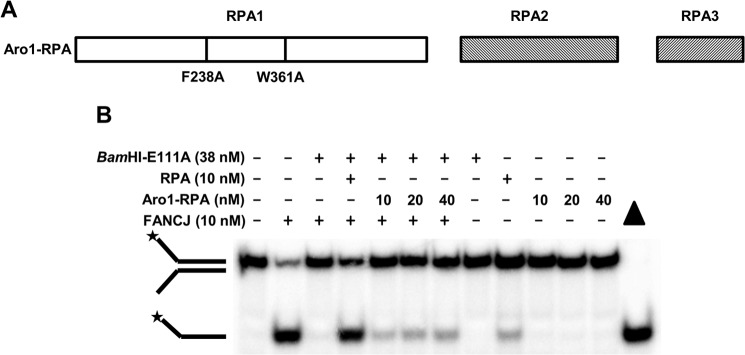
Amino acid substitutions that inactivate DNA binding by RPA negatively affect its ability to stimulate FANCJ disruption of BamHI-E111A protein-DNA substrate interaction. A, schematic representation of Aro1-RPA characterized by two amino acid substitutions in the RPA1 (RPA 70 kDa) subunit of the RPA heterotrimer consisting of RPA1, RPA2, and RPA3. The Aro1-RPA mutant harboring the two amino acid substitutions F238A and W361A was previously shown to inactivate DNA binding by RPA (27). B, reaction mixtures containing BamHI-E111A-bound forked duplex DNA substrate, FANCJ, and either wild-type RPA (RPA) or mutant Aro1-RPA were incubated and analyzed as described under “Experimental Procedures.” Representative gel image showing proteinase K-digested products from at least three independent experiments is shown. Star denotes 5′-32P end label.
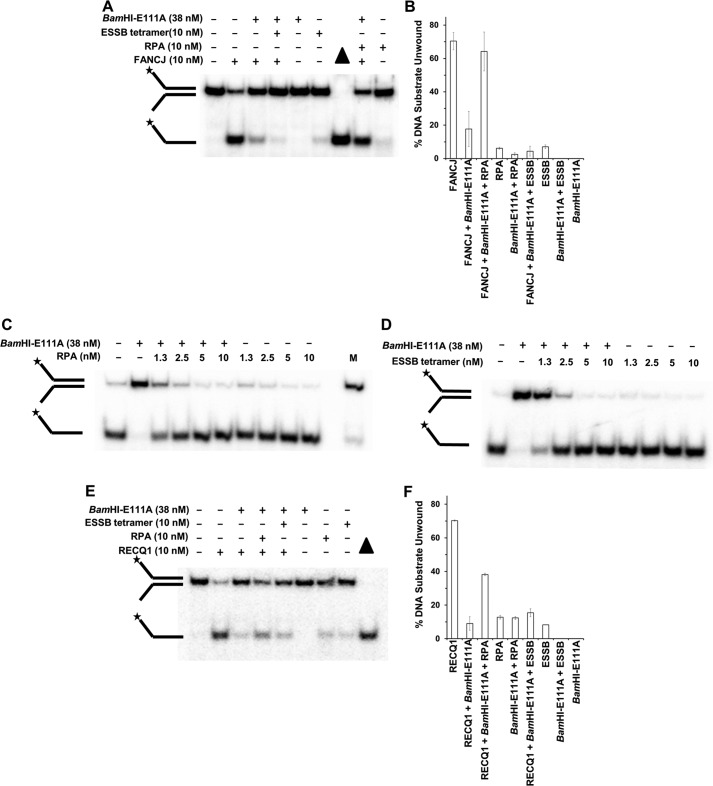
To address whether stimulation of FANCJ to displace BamHI-E111A and unwind the forked duplex was specific, we substituted the ESSB for RPA in the FANCJ reaction mixtures with the BamHI-E111A-bound forked duplex DNA substrate. For these experiments, we used a concentration of ESSB homotetramer the same as RPA heterotrimer (10 nm) because one ESSB homotetramer binds 35 nucleotides (36), and one RPA heterotrimer binds 30 nucleotides (37). ESSB completely failed to stimulate FANCJ to displace BamHI-E111A from the forked duplex and to unwind the DNA substrate under conditions that FANCJ was able to efficiently unwind the naked forked duplex or RPA effectively stimulated FANCJ to displace BamHI-E111A from the DNA substrate (Fig. 4A). By quantitative analysis, we determined that RPA stimulated FANCJ displacement of BamHI-E111A from the DNA substrate 3.5-fold, whereas ESSB actually showed inhibition of FANCJ-catalyzed BamHI-E111A displacement (Fig. 4B), presumably due to ESSB competing with FANCJ for loading onto the DNA substrate. These results suggest that RPA stimulates FANCJ to displace BamHI-E111A from the forked duplex and unwind the DNA substrate in a specific manner because a heterologous SSB failed to stimulate FANCJ protein displacement.
FIGURE 4.
RPA stimulates FANCJ or RECQ1 to disrupt the BamHI-E111A protein-DNA complex in a specific manner. A, reaction mixtures containing the BamHI-E111A-bound or unbound forked duplex DNA substrate were incubated with FANCJ in the presence of RPA or ESSB homotetramer for 15 min, and the proteinase K-digested products were analyzed by nondenaturing 12% PAGE. B, quantitative analyses from A. C and D, strand annealing promoted by BamHI-E111A is inhibited by RPA (C) or ESSB (D). Partially complementary single-stranded oligonucleotides were incubated with the indicated concentration of RPA or ESSB in the presence or absence of BamHI-E111A (38 nm) as described under “Experimental Procedures.” Proteinase K-digested products were resolved on native 12% polyacrylamide gels. M, annealed forked duplex marker. E, reaction mixtures containing the BamHI-E111A-bound or unbound forked duplex DNA substrate were incubated with RECQ1 in the presence of RPA or ESSB for 15 min, and the proteinase K-digested products were analyzed by nondenaturing 12% PAGE. F, quantitative analyses from E. Representative gel images from at least three independent experiments are shown. Star denotes 5′-32P end label.
To further address the mechanism of stimulation, we evaluated the effect of BamHI-E111A on rewinding the complementary strands of the DNA substrate and the effect of single-stranded DNA-binding proteins RPA or ESSB on rewinding. As shown in Fig. 4, C and D, the presence of BamHI-E111A with the unwound complementary strands effectively promoted their annealing. However, the presence of RPA (Fig. 4C) or ESSB (Fig. 4D) inhibited BamHI-E111A-promoted strand annealing at concentrations as low as 1.3 nm single-stranded DNA-binding protein. Based on these results, we conclude that although both RPA and ESSB can prevent rewinding of the complementary strands, only RPA could stimulate FANCJ-catalyzed protein displacement, indicating a specific RPA-FANCJ interaction to promote protein displacement during the DNA unwinding reaction by the helicase.
RPA Stimulates RECQ1 to Disrupt BamHI-E111A-DNA Substrate Interaction
Previously, it was demonstrated that RPA physically and functionally interacts with several SF2 RecQ DNA repair helicases to stimulate their DNA unwinding reactions (38), suggesting that the ability of RPA to stimulate the Fe-S cluster DNA helicase FANCJ to disrupt protein-DNA complexes might also be observed for a human RecQ helicase. To address this, the human RECQ1 helicase, which is known to interact with RPA (21), was tested for its ability to dislodge BamHI-E111A from the forked duplex DNA substrate in the presence of RPA. As shown in Fig. 4E, RPA stimulated RECQ1-catalyzed BamHI-E111A displacement. Quantitative analysis of the products from RECQ1-RPA reaction mixtures with the BamHI-E111A-bound forked duplex revealed a 2-fold stimulation compared with the additive effect of RECQ1 or RPA acting separately on the BamHI-E111A-bound substrate (Fig. 4F). The stimulation of RECQ1-catalyzed protein displacement was specific as evidenced by the inability of ESSB to substitute for RPA in the reaction.
RPA Stimulates FANCJ- or RECQ1-catalyzed Displacement of TRF1 Bound to a Telomeric Duplex Substrate
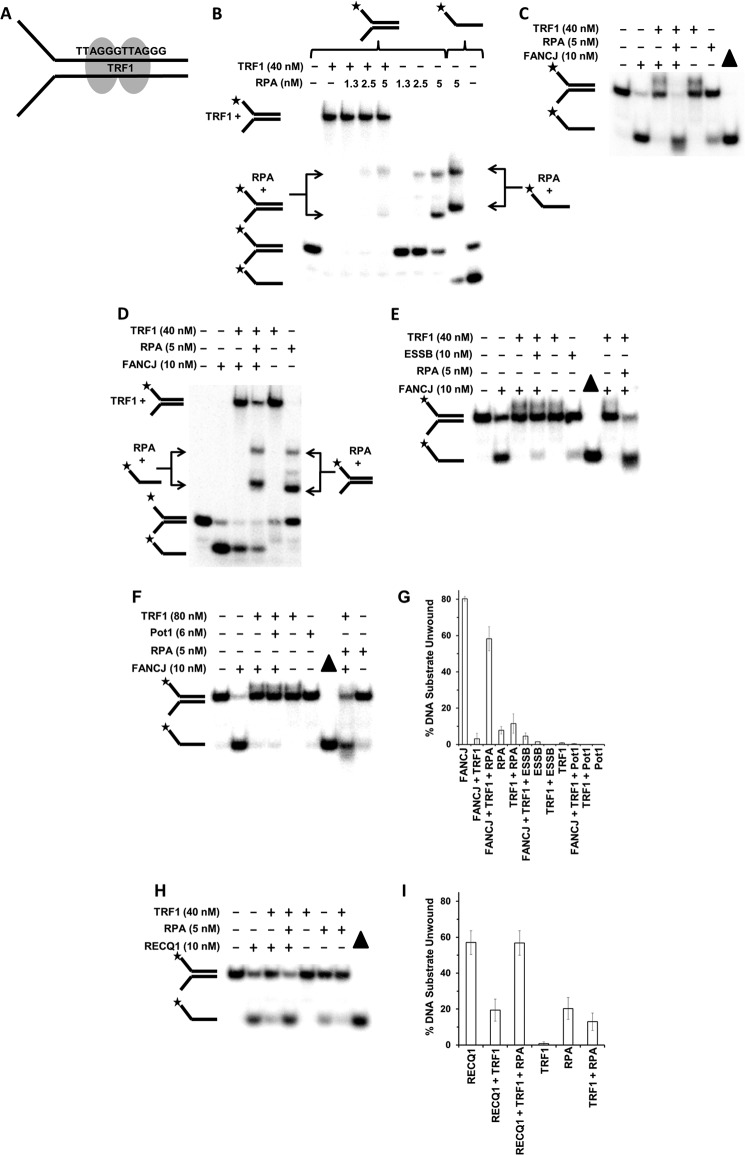
The shelterin protein TRF1 is known to preferentially bind duplex DNA harboring the telomeric sequence repeat TTAGGG/AATCCC and contribute to the protection of the chromosome end (39, 40). However, telomere structural dynamics may require helicase-mediated unwinding of protein-bound t-loops and other DNA structures in order for telomeres to be properly replicated or repaired (17). Previously, it was reported that FANCJ can be found at telomeres of living cells that preserve their chromosome ends by the ALT pathway (16), suggesting a potential role of FANCJ in telomeric DNA metabolism. The ability of RPA to stimulate FANCJ to displace BamHI-E111A and effectively unwind the protein-bound forked duplex raised the experimental question whether RPA would exert a similar effect and enable FANCJ to displace TRF1 from the telomeric forked duplex substrate and unwind the underlying duplex. To perform this experiment, we used a 22-bp forked duplex harboring two direct telomeric repeats (Fig. 5A). TRF1 binds the telomere repeat as a homodimer in which each monomer binds a minimal consensus sequence of 5′-YTAGGGTTR-3′ (41); therefore, the DNA substrate with two direct telomeric repeats would be expected to bind one TRF1 dimer. TRF1 binding to the forked duplex caused the DNA substrate to be strongly retarded upon electrophoresis on a native 5% polyacrylamide gel (Fig. 5B), a result that is consistent with a previous report (41). RPA (5 nm) alone had only a very minor effect on TRF1 displacement (Fig. 5B). Analysis of helicase reaction products that had been proteinase K-treated revealed that the presence of TRF1 in the reaction mixture effectively inhibited FANCJ unwinding of the telomeric forked duplex substrate (Fig. 5C). However, in the presence of RPA (5 nm), FANCJ unwound the telomeric forked duplex to near completion in the 15-min reaction. In control reactions, RPA alone only modestly destabilized the 22-bp telomeric forked duplex, presumably by its ability to bind the single-stranded tails and capture fraying of the short duplex. Taken together, these results strongly suggest that RPA alone poorly displaces TRF1 from the DNA substrate, and the substrate bound by TRF1 is effectively unwound by the concerted action of FANCJ and RPA.
FIGURE 5.
FANCJ or RECQ1 dislodges TRF1 bound to a telomeric forked duplex DNA substrate in an RPA-dependent and -specific manner. A, schematic representation of TRF1 dimer bound to telomeric repeat sequence harbored within 22-bp duplex region of forked DNA substrate. B, RPA does not displace TRF1 bound to telomeric 22-bp forked duplex DNA substrate. TRF1 was preincubated with the DNA substrate (0.5 nm) at 24 °C for 15 min. The indicated concentrations of RPA were added to the binding mixture and further incubated at 30 °C for 15 min. Protein-DNA species were resolved by EMSA as described under “Experimental Procedures.” TRF1 bound to forked duplex or RPA bound to radiolabeled single-stranded oligonucleotide or forked duplex is shown. C, RPA stimulates FANCJ displacement of TRF1 and unwinding of the telomeric DNA substrate. Reaction mixtures containing the radiolabeled 22-bp telomeric forked duplex DNA substrate (0.5 nm) and the indicated proteins were incubated for 15 min at 30 °C, followed by electrophoresis of proteinase K-digested products on 12% polyacrylamide native gels. D, TRF1 is removed from telomeric DNA substrate by FANCJ and RPA. TRF1 was preincubated with the DNA substrate (0.5 nm) at 24 °C for 15 min. Where indicated, RPA (5 nm) and FANCJ (10 nm) were added to the binding mixture with ATP present and further incubated at 30 °C for 15 min. Protein-DNA species were resolved by EMSA as described under “Experimental Procedures.” TRF1 bound to forked duplex or RPA bound to radiolabeled single-stranded oligonucleotide or forked duplex is shown. E and F, RPA stimulates FANCJ displacement of TRF1 from the telomeric DNA substrate in a specific manner. Reaction mixtures containing the radiolabeled 22-bp telomeric forked duplex DNA substrate (0.5 nm) were preincubated with TRF1, followed by addition of the indicated proteins that were incubated for 15 min at 30 °C. Proteinase K-digested products were electrophoresed on native 12% polyacrylamide gels. G, quantitative analyses from E and F. H, RPA stimulates RECQ1 displacement of TRF1 from the telomeric DNA substrate. Reaction mixtures containing the TRF1-bound radiolabeled 22-bp telomeric forked duplex DNA substrate were incubated with the indicated concentrations of RECQ1 and RPA for 15 min at 30 °C, followed by electrophoresis of proteinase K-digested products on 12% polyacrylamide native gels. Representative gel images from at least three independent experiments are shown. I, quantitative analyses from H. Star denotes 5′-32P end label.
We next wanted to ascertain whether TRF1 was actually removed from the DNA substrate by FANCJ and RPA acting together. To address this, we examined the DNA-bound protein species from reaction mixtures containing FANCJ + RPA with the TRF1-bound forked telomeric duplex substrate. In this case, proteinase K was not included during the quench with STOP buffer. Here, we observed that TRF1 inhibited FANCJ helicase activity as before and that the additional presence of RPA with FANCJ and the TRF1-bound DNA substrate during the incubation period resulted in the appearance of RPA-bound single-stranded DNA (Fig. 5D). These experimental data provide evidence that TRF1 was removed from the DNA substrate by the combined presence of FANCJ and RPA in the reaction mixture.
To evaluate whether the effect of RPA on TRF1 displacement from the telomeric forked duplex by FANCJ was specific, we assessed the ability of ESSB to substitute for RPA in this capacity. ESSB failed to stimulate FANCJ displacement of TRF1 under conditions that RPA effectively did so (Fig. 5E), attesting to the specificity of the FANCJ-RPA interaction. To further address the specificity of RPA-FANCJ functional interaction, we tested whether the shelterin protein POT1 was able to stimulate FANCJ to unwind the forked duplex with TRF1 bound to the 22-bp telomeric duplex. Pot1 shares a conserved oligonucleotide/oligosaccharide binding fold also found in RPA and interacts with strong specificity to the telomeric single-stranded DNA sequence 5′-(T)TAGGGTTAG-3′ (42). The experimental results demonstrated that POT1 did not stimulate FANCJ under conditions that RPA did (Fig. 5F), further attesting to the specificity of the FANCJ-RPA interaction. Taking into account the effect of RPA alone on destabilization of the forked duplex, a quantitative analysis demonstrated that RPA stimulated FANCJ displacement of TRF1 ∼10-fold (Fig. 5G).
Recent evidence suggests a potential role of RECQ1 in telomere metabolism (43). The ability of RPA to stimulate RECQ1 removal of the catalytically inactive BamHI mutant protein bound to a DNA substrate suggested RPA may also stimulate RECQ1 displacement of the shelterin proteins TRF1 and TRF2 bound to the telomeric duplex sequence. Using the 22-bp forked duplex with the two direct telomeric repeats described above, we found that TRF1 inhibited RECQ1 unwinding of the telomeric substrate (Fig. 5, H and I). In the presence of RPA (5 nm), RECQ1 unwinding of the TRF1-bound substrate was stimulated ∼2-fold, taking into account the fraction of TRF1-bound substrate destabilized by RPA.
RPA-dependent FANCJ or RECQ1 Displacement of TRF1 or TRF2 Bound to a DNA Substrate Harboring Four Telomeric Repeats
To further examine the effect of RPA on FANCJ displacement of TRF1 bound to telomeric duplex, reactions were performed with a longer 34-bp forked duplex substrate characterized by the presence of four TTAGGG/AATCCC repeats in the duplex region (Fig. 6A). Thus, the 34-bp duplex with four telomeric repeats would be expected to maximally bind two homodimers of TRF1. Because of their low processivity, RECQ1, like FANCJ, alone very poorly unwound the naked DNA substrate; however, in the presence of RPA RECQ1 and FANCJ unwound over 50% of the telomeric forked duplex in an RPA concentration-dependent manner (Fig. 6B). Preincubation of the helicase substrate with RPA followed by the initiation of the helicase reaction with the addition of FANCJ showed a comparable level of DNA unwinding as observed for reactions in which FANCJ and RPA were simultaneously added to reaction mixtures (data not shown), suggesting that RPA does not recruit FANCJ to the helicase substrate. Further studies are warranted to study the mechanism of RPA stimulation of FANCJ and RecQ DNA helicases.
FIGURE 6.
FANCJ or RECQ1 dislodges TRF1 or TRF2 bound to a telomeric forked duplex harboring four telomeric repeats in an RPA-dependent manner. A, schematic representation of two TRF1 dimers bound to telomeric repeat sequences harbored within the duplex region of forked DNA substrate. B, RPA stimulates FANCJ or RECQ1 helicase activity on telomeric forked duplex DNA substrate. The 34-bp telomeric forked duplex substrate was incubated with FANCJ (20 nm) or RECQ1 (20 nm) and the indicated concentrations of RPA for 15 min at 30 °C followed by analysis of proteinase K-digested products on native 12% polyacrylamide gels. Experimental quantitative data represent the mean of at least three independent experiments with standard deviations shown by error bars. C and D, reaction mixtures containing the radiolabeled DNA substrate (0.5 nm) and TRF1 (B) or TRF2 (C) were preincubated for 15 min at 30 °C, followed by addition of FANCJ and RPA for 15 min at 30 °C and subsequent electrophoresis of proteinase K-digested products on 12% polyacrylamide native gels. E, reaction mixtures containing the radiolabeled 34-bp telomeric forked duplex DNA substrate (0.5 nm) and TRF2 were preincubated, followed by incubation with RECQ1 and RPA, and subsequent analysis of proteinase K-digested products as described above. Representative gel images from at least three independent experiments are shown. Star denotes 5′-32P end label.
TRF1 prebound to the 34-bp telomeric forked duplex prevented FANCJ from unwinding the substrate (Fig. 6C). However, in the presence of RPA, FANCJ displaced TRF1 bound to the four consecutive telomeric repeats and unwound the telomeric duplex.
We next tested whether FANCJ could displace TRF2, another shelterin protein that binds telomeric repeats (44), from the forked duplex substrate harboring the 34-bp region with four direct TTAGGG/AATCCC repeats. Like TRF1, TRF2 binds as a homodimer in which each monomer binds a minimal consensus sequence of 5′-YTAGGGTTR-3′ (42, 45). Therefore, the 34-bp duplex with four telomeric repeats would be expected to maximally bind two homodimers of TRF2. TRF2 prebound to the 34-bp telomeric forked duplex prevented FANCJ from unwinding the substrate (Fig. 6D). However, in the presence of RPA, FANCJ was able to unwind the TRF2-bound substrate, displacing TRF2 bound to the four consecutive telomeric repeats.
We next evaluated the ability of RECQ1 with RPA to displace TRF2 bound to the 34-bp telomeric forked duplex substrate. When RPA (10 nm) was present in the reaction mixture containing RECQ1 and the TRF2-bound telomeric substrate, 13% of the DNA substrate was unwound, whereas only ∼0.5% of the TRF2-bound DNA substrate was unwound by RECQ1 alone (Fig. 6E). Based on these results, we conclude that RPA stimulates RECQ1 displacement of TRF1 or TRF2 bound to telomeric substrates.
DISCUSSION
Recent biological data have suggested that protein-DNA complexes are the primary source of replication fork pausing in E. coli (46). Emerging evidence supports a model in which replication collisions are overcome by accessory helicases that assist the replisome to progress through protein-DNA blockades (32). At present, the strongest biochemical and genetic evidence for the role of accessory replicative motors to displace proteins bound to double-stranded DNA ahead of the fork has been garnered from studies in prokaryotic systems. Guy et al. (47) provided evidence that Rep and UvrD are able to promote replisome progression along protein-bound DNA both in vitro and in vivo. Studies from McGlynn and co-workers (47–50) and Michel and co-workers (51–53) have provided both molecular and cellular evidence delineating the roles of accessory helicases in bacteria to disrupt protein-DNA complexes that would impede the replication fork and pose a source of chromosomal instability. Although considerable evidence suggests that eukaryotic DNA helicases (e.g. Srs2) serve to dislodge proteins (Rad51 recombinase) bound to single-stranded DNA to regulate HR (54–57), it has also been proposed that helicases or helicase-like proteins (e.g. Rad54) dislodge proteins bound to double-stranded DNA leading to the disassembly of toxic recombination intermediates or static dead-end complexes (58, 59). The human helicase-like transcription factor coordinates remodeling of stalled replication forks by displacing the clamp proliferating cell nuclear antigen or clamp loader RFC (60). Although these studies suggest a potential role of helicase-like proteins to dislodge proteins bound to DNA, very little is known about the mechanism, regulation, and biological significance of helicase-catalyzed protein displacement, especially in eukaryotes.
In this study, we have carefully examined the effect of protein bound to duplex DNA on the catalytic DNA unwinding function of representative human SF2 DNA repair helicases that are important for helping cells deal with replication stress. BamHI-E111A served as a useful tool for this analysis because the restriction endonuclease with the active site mutation displays a high affinity for its cognate recognition sequence (Kd = 2.95E-11 m),3 ∼300-fold greater than normal BamHI (30). We determined that DNA helicase activity catalyzed by FANCJ, RECQ1, and WRN is potently blocked by a catalytically inactive BamHI-E111A restriction endonuclease bound to a forked duplex DNA substrate with a centrally positioned cognate palindromic hexanucleotide double-stranded DNA sequence recognized by the endonuclease. We next asked whether FANCJ might function with one of its known helicase protein partners to efficiently displace protein bound to a DNA substrate. However, FANCJ failed to collaborate with the 3′ to 5′ DNA repair helicase BLM, with which it is known to interact (29), to dislodge BamHI-E111A bound to the forked duplex. Further studies will elucidate whether certain opposite polarity DNA helicases in eukaryotic cells act together to synergistically disrupt proteins bound to duplex DNA. Up to this point, it is unknown whether eukaryotic cells follow the precedent set by the Rep 3′ to 5′ auxiliary helicase that functionally cooperates with the E. coli DnaB replicative 5′ to 3′ hexameric helicase to facilitate conflicts between replication and transcription (48). It is conceivable that a specific interaction exists in mammalian cells between the replicative mini-chromosome maintenance helicase and an auxiliary helicase yet to be determined.
FANCJ, which interacts with RPA (9), was able to efficiently displace BamHI-E111A from the forked duplex and unwind the underlying duplex. Protein displacement from the DNA substrate was dependent on the intrinsic ATPase activity of FANCJ, and it required a helicase-proficient version of FANCJ because the translocase-proficient but helicase-inactive FANCJ-A349P mutant protein failed to do so. Furthermore, the ability of RPA to promote FANCJ disruption of the BamHI-E119A-DNA interaction required the high affinity DNA binding activity of RPA. The dysfunctional DNA replication and repair phenotypes of human cells expressing the Aro1-RPA mutant (27) may be attributed to, at least in part, the disruption of functional interactions between RPA and DNA helicases responsible for displacing proteins bound to genomic DNA and unwinding the underlying DNA duplexes.
In addition to the high affinity binding of single-stranded DNA by RPA, the protein interaction between RPA and FANCJ (as well as the interaction between RPA and RECQ1) is likely to be important for protein ejection from DNA based on our observation that a heterologous SSB was unable to stimulate FANCJ- or RECQ1-catalyzed protein displacement. This result parallels our previous findings that the physical interaction between RPA and the WRN helicase is required for stimulation of helicase-catalyzed DNA unwinding of protein-free partial duplex DNA substrates (20). These results robustly demonstrate that a protein bound to duplex DNA potently blocks unwinding by representative SF2 Fe-S and RecQ helicases, and this inhibition can be overcome in a specific manner by the collaborative action of RPA and its interacting helicase.
We next evaluated the effect of the shelterin proteins TRF1 or TRF2 bound to a forked duplex substrate harboring direct telomeric repeats on DNA unwinding by FANCJ, a protein that was previously shown to be found at telomeres in ALT cells (16) and was implicated in helping cells deal with replicative stress (29). TRF1 and TRF2 are known to bind exclusively to double-stranded telomeric repeat DNA as homodimers, resulting in highly stable protein-DNA interactions (39, 44, 61). Here, we also observed potent inhibition of FANCJ helicase activity by the biologically relevant interaction of TRF1 or TRF2 with the human telomeric sequence. These findings may be in accord with biochemical data showing that TRF2 protects Holliday junction DNA from unwinding by the human RecQ helicase WRN in part by its ability to bind the telomeric sequence duplex arms (62). In this study, we demonstrated that the presence of RPA in the reaction mixture stimulated FANCJ to dislodge TRF1 or TRF2 from the telomeric duplex and efficiently unwind the DNA substrate. The ability of RPA to stimulate FANCJ in this capacity was specific based on our observations that ESSB or the telomeric SSB Pot1 failed to enhance FANCJ activity on the TRF1-bound DNA substrate. RPA also stimulated FANCJ to displace TRF1 or TRF2 from a forked duplex harboring four direct telomeric repeats, a substrate that would be expected to bind two homodimers under TRF1- or TRF2-saturating conditions. The ability of RPA to stimulate FANCJ displacement of BamHI-E111A, TRF1, or TRF2 from the forked duplex substrates is compelling in light of an earlier observation that RPA does not promote BLM helicase to unwind a forked duplex mononucleosome substrate (63). In the future, it will be of interest to test whether RPA can stimulate FANCJ, or another DNA helicase with which it interacts, to unwind nucleosomal DNA in an efficient manner to regulate the chromatin state.
Both FANCJ and BLM were found to be specifically bound to telomeric DNA in telomerase-negative cells engaged in the ALT maintenance pathway (16), raising the possibility that FANCJ and BLM, two helicases that are known to physically and functionally interact (29), may cooperate to unwind telomeric DNA structures or displace proteins bound to telomeres. Mammalian telomeres display a fragile phenotype that is suppressed by TRF1, which is required for efficient replication of TTAGGG repeats to which it binds (64, 65). However, it was also observed that DNA helicases (BLM (65) and RTEL1 (66)) are required to repress the fragile telomere phenotype, suggesting that specialized DNA unwinding functions are necessary for timely and efficient replication of the telomere repeats. It is conceivable that certain DNA helicases, acting with RPA, dislodge proteins of the shelterin complex once they have fulfilled their functions to repress telomere fragility, providing the opportunity for the telomere sequence to be copied faithfully to preserve chromosome ends. The helicase-RPA interaction would provide a higher level of coordination between events associated with telomere capping by shelterin proteins and telomeric DNA synthesis by the replication machinery.
FANCJ or another DNA repair helicase (e.g. member of the RecQ family) implicated in HR might utilize its motor ATPase function to displace Rad51 from single- or double-stranded DNA. Indeed, the DNA helicases Srs2 (54–57), RECQ5 (67), FBH1 (68), and FANCJ (69) have all been previously shown to strip Rad51 from single-stranded DNA. However, it is also possible that a DNA helicase may disrupt Rad51 double-stranded DNA complexes that would be inhibitory to DNA strand exchange (59). For example, the yeast Srs2 helicase was shown to unwind double-stranded DNA covered with Rad51 (58). Based on our finding that RPA modulates helicase-catalyzed protein displacement in vitro, future studies should address whether RPA promotes DNA helicases to disrupt toxic dead-end complexes such as Rad51 bound to double-stranded DNA. This mechanism of regulation would be distinct from one in which Caenorhabditis elegans DNA helicase HELQ-1 was reported to promote the disassembly of RAD-51 from double-stranded DNA in an ATP-independent manner (70).
Cells from FA-J patients display chromosomal instability and sensitivity to agents that impose replication stress (29). FANCJ was observed to show an enhanced association with chromatin as cells progress through S phase (71) and co-localize with RPA after DNA damage (9). This work suggests that FANCJ performs its catalytic functions in HR repair or replication restart in vivo on chromatinized DNA substrates with its protein partner RPA, which promote FANCJ's ability to displace proteins bound to duplex DNA. Although the emphasis of this work has been on the RPA-dependent ejection of protein from DNA by FANCJ, it is reasonable that FANCJ may help to remodel proteins on DNA to preserve chromatin structure, which perhaps is important for its recently demonstrated role in promoting replication past DNA sequence barriers to suppress heterochromatin spreading (72).
Acknowledgments
We thank Becky Kucera and Dr. Jurate Bitinaite (New England Biolabs) for the BamHI-E111A-purified protein and their helpful suggestions for the EMSA protocol to detect DNA binding by BamHI-E111A. We also thank Dr. Ian Hickson (University of Copenhagen) for the recombinant BLM protein.
This work was supported, in whole or in part, by National Institutes of Health Intramural Research Program of the NIA. This work was also supported by the Fanconi Anemia Research Fund (to R. M. B.).
J. Bitinaite, unpublished data.
- Fe-S
- iron-sulfur
- ESSB
- E. coli single-stranded DNA-binding protein
- FA
- Fanconi anemia
- FANCJ
- FA complementation Group J
- G4
- G-quadruplex
- HR
- homologous recombination
- RPA
- replication protein A
- ALT
- alternative lengthening of telomere
- RFC
- replication factor C.
REFERENCES
- 1. Bedinger P., Hochstrasser M., Jongeneel C. V., Alberts B. M. (1983) Properties of the T4 bacteriophage DNA replication apparatus: the T4 dda DNA helicase is required to pass a bound RNA polymerase molecule. Cell 34, 115–123 [DOI] [PubMed] [Google Scholar]
- 2. Yancey-Wrona J. E., Matson S. W. (1992) Bound Lac repressor protein differentially inhibits the unwinding reactions catalyzed by DNA helicases. Nucleic Acids Res. 20, 6713–6721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Byrd A. K., Raney K. D. (2006) Displacement of a DNA-binding protein by Dda helicase. Nucleic Acids Res. 34, 3020–3029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mackintosh S. G., Raney K. D. (2006) DNA unwinding and protein displacement by superfamily 1 and superfamily 2 helicases. Nucleic Acids Res. 34, 4154–4159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brosh R. M., Jr. (2013) DNA helicases involved in DNA repair and their roles in cancer. Nat. Rev. Cancer 13, 542–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kottemann M. C., Smogorzewska A. (2013) Fanconi anaemia and the repair of Watson and Crick DNA crosslinks. Nature 493, 356–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cantor S. B., Bell D. W., Ganesan S., Kass E. M., Drapkin R., Grossman S., Wahrer D. C., Sgroi D. C., Lane W. S., Haber D. A., Livingston D. M. (2001) BACH1, a novel helicase-like protein, interacts directly with BRCA1 and contributes to its DNA repair function. Cell 105, 149–160 [DOI] [PubMed] [Google Scholar]
- 8. Peng M., Litman R., Xie J., Sharma S., Brosh R. M., Jr., Cantor S. B. (2007) The FANCJ/MutLα interaction is required for correction of the cross-link response in FA-J cells. EMBO J. 26, 3238–3249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gupta R., Sharma S., Sommers J. A., Kenny M. K., Cantor S. B., Brosh R. M., Jr. (2007) FANCJ (BACH1) helicase forms DNA damage inducible foci with replication protein A and interacts physically and functionally with the single-stranded DNA-binding protein. Blood 110, 2390–2398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. London T. B., Barber L. J., Mosedale G., Kelly G. P., Balasubramanian S., Hickson I. D., Boulton S. J., Hiom K. (2008) FANCJ is a structure-specific DNA helicase associated with the maintenance of genomic G/C tracts. J. Biol. Chem. 283, 36132–36139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wu Y., Shin-ya K., Brosh R. M., Jr. (2008) FANCJ helicase defective in Fanconi anemia and breast cancer unwinds G-quadruplex DNA to defend genomic stability. Mol. Cell. Biol. 28, 4116–4128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bharti S. K., Sommers J. A., George F., Kuper J., Hamon F., Shin-ya K., Teulade-Fichou M. P., Kisker C., Brosh R. M., Jr. (2013) Specialization among iron-sulfur cluster helicases to resolve G-quadruplex DNA structures that threaten genomic stability. J. Biol. Chem. 288, 28217–28229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Henderson A., Wu Y., Huang Y. C., Chavez E. A., Platt J., Johnson F. B., Brosh R. M., Jr., Sen D., Lansdorp P. M. (2014) Detection of G-quadruplex DNA in mammalian cells. Nucleic Acids Res. 42, 860–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wu Y., Brosh R. M., Jr. (2010) G-quadruplex nucleic acids and human disease. FEBS J. 277, 3470–3488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. de Lange T. (2002) Protection of mammalian telomeres. Oncogene 21, 532–540 [DOI] [PubMed] [Google Scholar]
- 16. Déjardin J., Kingston R. E. (2009) Purification of proteins associated with specific genomic loci. Cell 136, 175–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chavez A., Tsou A. M., Johnson F. B. (2009) Telomeres do the (un)twist: helicase actions at chromosome termini. Biochim. Biophys. Acta 1792, 329–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Paeschke K., McDonald K. R., Zakian V. A. (2010) Telomeres: structures in need of unwinding. FEBS Lett. 584, 3760–3772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Uringa E. J., Youds J. L., Lisaingo K., Lansdorp P. M., Boulton S. J. (2011) RTEL1: an essential helicase for telomere maintenance and the regulation of homologous recombination. Nucleic Acids Res. 39, 1647–1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Doherty K. M., Sommers J. A., Gray M. D., Lee J. W., von Kobbe C., Thoma N. H., Kureekattil R. P., Kenny M. K., Brosh R. M., Jr. (2005) Physical and functional mapping of the RPA interaction domain of the Werner and Bloom syndrome helicases. J. Biol. Chem. 280, 29494–29505 [DOI] [PubMed] [Google Scholar]
- 21. Cui S., Arosio D., Doherty K. M., Brosh R. M., Jr., Falaschi A., Vindigni A. (2004) Analysis of the unwinding activity of the dimeric RECQ1 helicase in the presence of human replication protein A. Nucleic Acids Res. 32, 2158–2170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Brosh R. M., Jr., Waheed J., Sommers J. A. (2002) Biochemical characterization of the DNA substrate specificity of Werner syndrome helicase. J. Biol. Chem. 277, 23236–23245 [DOI] [PubMed] [Google Scholar]
- 23. Sharma S., Sommers J. A., Choudhary S., Faulkner J. K., Cui S., Andreoli L., Muzzolini L., Vindigni A., Brosh R. M., Jr. (2005) Biochemical analysis of the DNA unwinding and strand annealing activities catalyzed by human RECQ1. J. Biol. Chem. 280, 28072–28084 [DOI] [PubMed] [Google Scholar]
- 24. Wang F., Lei M. (2011) Human telomere POT1-TPP1 complex and its role in telomerase activity regulation. Methods Mol. Biol. 735, 173–187 [DOI] [PubMed] [Google Scholar]
- 25. Chen Y., Yang Y., van Overbeek M., Donigian J. R., Baciu P., de Lange T., Lei M. (2008) A shared docking motif in TRF1 and TRF2 used for differential recruitment of telomeric proteins. Science 319, 1092–1096 [DOI] [PubMed] [Google Scholar]
- 26. Karow J. K., Newman R. H., Freemont P. S., Hickson I. D. (1999) Oligomeric ring structure of the Bloom's syndrome helicase. Curr. Biol. 9, 597–600 [DOI] [PubMed] [Google Scholar]
- 27. Hass C. S., Lam K., Wold M. S. (2012) Repair-specific functions of replication protein A. J. Biol. Chem. 287, 3908–3918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Opresko P. L., Laine J. P., Brosh R. M., Jr., Seidman M. M., Bohr V. A. (2001) Coordinate action of the helicase and 3′ to 5′ exonuclease of Werner syndrome protein. J. Biol. Chem. 276, 44677–44687 [DOI] [PubMed] [Google Scholar]
- 29. Suhasini A. N., Rawtani N. A., Wu Y., Sommers J. A., Sharma S., Mosedale G., North P. S., Cantor S. B., Hickson I. D., Brosh R. M., Jr. (2011) Interaction between the helicases genetically linked to Fanconi anemia group J and Bloom's syndrome. EMBO J. 30, 692–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Engler L. E., Sapienza P., Dorner L. F., Kucera R., Schildkraut I., Jen-Jacobson L. (2001) The energetics of the interaction of BamHI endonuclease with its recognition site GGATCC. J. Mol. Biol. 307, 619–636 [DOI] [PubMed] [Google Scholar]
- 31. Choudhary S., Sommers J. A., Brosh R. M., Jr. (2004) Biochemical and kinetic characterization of the DNA helicase and exonuclease activities of Werner syndrome protein. J. Biol. Chem. 279, 34603–34613 [DOI] [PubMed] [Google Scholar]
- 32. McGlynn P. (2011) Helicases that underpin replication of protein-bound DNA in Escherichia coli. Biochem. Soc. Trans. 39, 606–610 [DOI] [PubMed] [Google Scholar]
- 33. Gupta R., Sharma S., Sommers J. A., Jin Z., Cantor S. B., Brosh R. M., Jr. (2005) Analysis of the DNA substrate specificity of the human BACH1 helicase associated with breast cancer. J. Biol. Chem. 280, 25450–25460 [DOI] [PubMed] [Google Scholar]
- 34. Levran O., Attwooll C., Henry R. T., Milton K. L., Neveling K., Rio P., Batish S. D., Kalb R., Velleuer E., Barral S., Ott J., Petrini J., Schindler D., Hanenberg H., Auerbach A. D. (2005) The BRCA1-interacting helicase BRIP1 is deficient in Fanconi anemia. Nat. Genet. 37, 931–933 [DOI] [PubMed] [Google Scholar]
- 35. Wu Y., Sommers J. A., Suhasini A. N., Leonard T., Deakyne J. S., Mazin A. V., Shin-Ya K., Kitao H., Brosh R. M., Jr. (2010) Fanconi anemia group J mutation abolishes its DNA repair function by uncoupling DNA translocation from helicase activity or disruption of protein-DNA complexes. Blood 116, 3780–3791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lohman T. M., Ferrari M. E. (1994) Escherichia coli single-stranded DNA-binding protein: multiple DNA-binding modes and cooperativities. Annu. Rev. Biochem. 63, 527–570 [DOI] [PubMed] [Google Scholar]
- 37. Wold M. S. (1997) Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu. Rev. Biochem. 66, 61–92 [DOI] [PubMed] [Google Scholar]
- 38. Sharma S., Doherty K. M., Brosh R. M., Jr. (2006) Mechanisms of RecQ helicases in pathways of DNA metabolism and maintenance of genomic stability. Biochem. J. 398, 319–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chong L., van Steensel B., Broccoli D., Erdjument-Bromage H., Hanish J., Tempst P., de Lange T. (1995) A human telomeric protein. Science 270, 1663–1667 [DOI] [PubMed] [Google Scholar]
- 40. Zhong Z., Shiue L., Kaplan S., de Lange T. (1992) A mammalian factor that binds telomeric TTAGGG repeats in vitro. Mol. Cell. Biol. 12, 4834–4843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bianchi A., Stansel R. M., Fairall L., Griffith J. D., Rhodes D., de Lange T. (1999) TRF1 binds a bipartite telomeric site with extreme spatial flexibility. EMBO J. 18, 5735–5744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Palm W., de Lange T. (2008) How shelterin protects mammalian telomeres. Annu. Rev. Genet. 42, 301–334 [DOI] [PubMed] [Google Scholar]
- 43. Popuri V., Hsu J., Khadka P., Horvath K., Liu Y., Croteau D. L., Bohr V. A. (2014) Human RECQL1 participates in telomere maintenance. Nucleic Acids Res. 42, 5671–5688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Broccoli D., Smogorzewska A., Chong L., de Lange T. (1997) Human telomeres contain two distinct Myb-related proteins, TRF1 and TRF2. Nat. Genet. 17, 231–235 [DOI] [PubMed] [Google Scholar]
- 45. Hanaoka S., Nagadoi A., Nishimura Y. (2005) Comparison between TRF2 and TRF1 of their telomeric DNA-bound structures and DNA-binding activities. Protein Sci. 14, 119–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gupta M. K., Guy C. P., Yeeles J. T., Atkinson J., Bell H., Lloyd R. G., Marians K. J., McGlynn P. (2013) Protein-DNA complexes are the primary sources of replication fork pausing in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 110, 7252–7257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Guy C. P., Atkinson J., Gupta M. K., Mahdi A. A., Gwynn E. J., Rudolph C. J., Moon P. B., van Knippenberg I. C., Cadman C. J., Dillingham M. S., Lloyd R. G., McGlynn P. (2009) Rep provides a second motor at the replisome to promote duplication of protein-bound DNA. Mol. Cell 36, 654–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Atkinson J., Gupta M. K., Rudolph C. J., Bell H., Lloyd R. G., McGlynn P. (2011) Localization of an accessory helicase at the replisome is critical in sustaining efficient genome duplication. Nucleic Acids Res. 39, 949–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Atkinson J., Gupta M. K., McGlynn P. (2011) Interaction of Rep and DnaB on DNA. Nucleic Acids Res. 39, 1351–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. McGlynn P., Guy C. P. (2008) Replication forks blocked by protein-DNA complexes have limited stability in vitro. J. Mol. Biol. 381, 249–255 [DOI] [PubMed] [Google Scholar]
- 51. Baharoglu Z., Lestini R., Duigou S., Michel B. (2010) RNA polymerase mutations that facilitate replication progression in the rep uvrD recF mutant lacking two accessory replicative helicases. Mol. Microbiol. 77, 324–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bidnenko V., Lestini R., Michel B. (2006) The Escherichia coli UvrD helicase is essential for Tus removal during recombination-dependent replication restart from Ter sites. Mol. Microbiol. 62, 382–396 [DOI] [PubMed] [Google Scholar]
- 53. De Septenville A. L., Duigou S., Boubakri H., Michel B. (2012) Replication fork reversal after replication-transcription collision. PLoS. Genet. 8, e1002622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Krejci L., Van Komen S., Li Y., Villemain J., Reddy M. S., Klein H., Ellenberger T., Sung P. (2003) DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature 423, 305–309 [DOI] [PubMed] [Google Scholar]
- 55. Le Breton C., Dupaigne P., Robert T., Le Cam E., Gangloff S., Fabre F., Veaute X. (2008) Srs2 removes deadly recombination intermediates independently of its interaction with SUMO-modified PCNA. Nucleic Acids Res. 36, 4964–4974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Qiu Y., Antony E., Doganay S., Koh H. R., Lohman T. M., Myong S. (2013) Srs2 prevents Rad51 filament formation by repetitive motion on DNA. Nat. Commun. 4, 2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Veaute X., Jeusset J., Soustelle C., Kowalczykowski S. C., Le Cam E., Fabre F. (2003) The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature 423, 309–312 [DOI] [PubMed] [Google Scholar]
- 58. Dupaigne P., Le Breton C., Fabre F., Gangloff S., Le Cam E., Veaute X. (2008) The Srs2 helicase activity is stimulated by Rad51 filaments on dsDNA: implications for crossover incidence during mitotic recombination. Mol. Cell 29, 243–254 [DOI] [PubMed] [Google Scholar]
- 59. Symington L. S., Heyer W. D. (2006) Some disassembly required: role of DNA translocases in the disruption of recombination intermediates and dead-end complexes. Genes Dev. 20, 2479–2486 [DOI] [PubMed] [Google Scholar]
- 60. Achar Y. J., Balogh D., Haracska L. (2011) Coordinated protein and DNA remodeling by human HLTF on stalled replication fork. Proc. Natl. Acad. Sci. U.S.A. 108, 14073–14078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Bilaud T., Brun C., Ancelin K., Koering C. E., Laroche T., Gilson E. (1997) Telomeric localization of TRF2, a novel human telobox protein. Nat. Genet. 17, 236–239 [DOI] [PubMed] [Google Scholar]
- 62. Nora G. J., Buncher N. A., Opresko P. L. (2010) Telomeric protein TRF2 protects Holliday junctions with telomeric arms from displacement by the Werner syndrome helicase. Nucleic Acids Res. 38, 3984–3998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Fujimoto S., Tomschik M., Zlatanova J. (2009) Does BLM helicase unwind nucleosomal DNA? Biochem. Cell Biol. 87, 875–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Bosco N., de Lange T. (2012) A TRF1-controlled common fragile site containing interstitial telomeric sequences. Chromosoma 121, 465–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Sfeir A., Kosiyatrakul S. T., Hockemeyer D., MacRae S. L., Karlseder J., Schildkraut C. L., de Lange T. (2009) Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell 138, 90–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Vannier J. B., Pavicic-Kaltenbrunner V., Petalcorin M. I., Ding H., Boulton S. J. (2012) RTEL1 dismantles T loops and counteracts telomeric G4-DNA to maintain telomere integrity. Cell 149, 795–806 [DOI] [PubMed] [Google Scholar]
- 67. Schwendener S., Raynard S., Paliwal S., Cheng A., Kanagaraj R., Shevelev I., Stark J. M., Sung P., Janscak P. (2010) Physical interaction of RECQ5 helicase with RAD51 facilitates its anti-recombinase activity. J. Biol. Chem. 285, 15739–15745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Simandlova J., Zagelbaum J., Payne M. J., Chu W. K., Shevelev I., Hanada K., Chatterjee S., Reid D. A., Liu Y., Janscak P., Rothenberg E., Hickson I. D. (2013) FBH1 disrupts RAD51 filaments in vitro and modulates homologous recombination in mammalian cells. J. Biol. Chem. 288, 34168–34180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Sommers J. A., Rawtani N., Gupta R., Bugreev D. V., Mazin A. V., Cantor S. B., Brosh R. M., Jr. (2009) FANCJ uses its motor ATPase to disrupt protein-DNA complexes, unwind triplexes, and inhibit rad51 strand exchange. J. Biol. Chem. 284, 7505–7517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ward J. D., Muzzini D. M., Petalcorin M. I., Martinez-Perez E., Martin J. S., Plevani P., Cassata G., Marini F., Boulton S. J. (2010) Overlapping mechanisms promote postsynaptic RAD-51 filament disassembly during meiotic double-strand break repair. Mol. Cell 37, 259–272 [DOI] [PubMed] [Google Scholar]
- 71. Kumaraswamy E., Shiekhattar R. (2007) Activation of BRCA1/BRCA2-associated helicase BACH1 is required for timely progression through S phase. Mol. Cell. Biol. 27, 6733–6741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Schwab R. A., Nieminuszczy J., Shin-ya K., Niedzwiedz W. (2013) FANCJ couples replication past natural fork barriers with maintenance of chromatin structure. J. Cell Biol. 201, 33–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Veaute X., Delmas S., Selva M., Jeusset J., Le Cam E., Matic I., Fabre F., Petit M. A. (2005) UvrD helicase, unlike Rep helicase, dismantles RecA nucleoprotein filaments in Escherichia coli. EMBO J. 24, 180–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Boubakri H., de Septenville A. L., Viguera E., Michel B. (2010) The helicases DinG, Rep and UvrD cooperate to promote replication across transcription units in vivo. EMBO J. 29, 145–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Finkelstein I. J., Visnapuu M. L., Greene E. C. (2010) Single-molecule imaging reveals mechanisms of protein disruption by a DNA translocase. Nature 468, 983–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Azvolinsky A., Dunaway S., Torres J. Z., Bessler J. B., Zakian V. A. (2006) The S. cerevisiae Rrm3p DNA helicase moves with the replication fork and affects replication of all yeast chromosomes. Genes Dev. 20, 3104–3116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Ivessa A. S., Zhou J. Q., Schulz V. P., Monson E. K., Zakian V. A. (2002) Saccharomyces Rrm3p, a 5′ to 3′ DNA helicase that promotes replication fork progression through telomeric and subtelomeric DNA. Genes Dev. 16, 1383–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Boulé J. B., Vega L. R., Zakian V. A. (2005) The yeast Pif1p helicase removes telomerase from telomeric DNA. Nature 438, 57–61 [DOI] [PubMed] [Google Scholar]
- 79. Hu Y., Raynard S., Sehorn M. G., Lu X., Bussen W., Zheng L., Stark J. M., Barnes E. L., Chi P., Janscak P., Jasin M., Vogel H., Sung P., Luo G. (2007) RECQL5/Recql5 helicase regulates homologous recombination and suppresses tumor formation via disruption of Rad51 presynaptic filaments. Genes Dev. 21, 3073–3084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Bugreev D. V., Yu X., Egelman E. H., Mazin A. V. (2007) Novel pro- and anti-recombination activities of the Bloom's syndrome helicase. Genes Dev. 21, 3085–3094 [DOI] [PMC free article] [PubMed] [Google Scholar]