Background: Amigos are transmembrane proteins suggested to mediate adhesive interactions of cells.
Results: Down-regulation of Amigo protein expression or inhibition of its function results in defects of fiber pathway development and disturbed locomotor functions in zebrafish.
Conclusion: The neural form of Amigo is required for construction of functional neural circuitries.
Significance: Amigo is identified as a novel factor that regulates formation of neuronal connections.
Keywords: Axon, Brain, Neurite Outgrowth, Neurobiology, Neurodevelopment, Zebrafish, Amigo Protein Family, Axonal Scaffold, Kv2.1 Potassium Channel, Leucine-rich Repeat
Abstract
The Amigo protein family consists of three transmembrane proteins characterized by six leucine-rich repeat domains and one immunoglobulin-like domain in their extracellular moieties. Previous in vitro studies have suggested a role as homophilic adhesion molecules in brain neurons, but the in vivo functions remain unknown. Here we have cloned all three zebrafish amigos and show that amigo1 is the predominant family member expressed during nervous system development in zebrafish. Knockdown of amigo1 expression using morpholino oligonucleotides impairs the formation of fasciculated tracts in early fiber scaffolds of brain. A similar defect in fiber tract development is caused by mRNA-mediated expression of the Amigo1 ectodomain that inhibits adhesion mediated by the full-length protein. Analysis of differentiated neural circuits reveals defects in the catecholaminergic system. At the behavioral level, the disturbed formation of neural circuitry is reflected in enhanced locomotor activity and in the inability of the larvae to perform normal escape responses. We suggest that Amigo1 is essential for the development of neural circuits of zebrafish, where its mechanism involves homophilic interactions within the developing fiber tracts and regulation of the Kv2.1 potassium channel to form functional neural circuitry that controls locomotion.
Introduction
Development of functional neural circuits depends on spatially and temporally precise cellular interactions mediated by a wide variety of secreted and membrane-bound factors. Proteins containing extracellular leucine-rich repeat (LRR)3 domains have recently gained increasing interest as key factors guiding development of neuronal connectivity (for a recent review, see Ref. 1). LRR domains are protein-protein interaction motifs that have been implicated in development and plasticity of fiber tracts, recognition of axon targets, synaptogenesis, and disorders of the nervous system. Slit proteins (Slit 1–3), Trk receptors (TrkA, TrkB, and TrkC), and Nogo receptor 1 (NgR1) are among the most studied LRR proteins in the nervous system development (2, 3).
AMIGO (amphoterin-induced gene and open reading frame) was found by ordered differential display as a transcript up-regulated in rat hippocampal neurons that extend neurites upon ligation of the transmembrane receptor RAGE (receptor for advanced glycation end products) by amphoterin (HMGB1; high mobility group box-1) or by anti-RAGE antibodies (4). Together with two homologous proteins, the three AMIGOs (AMIGO1–3) form a novel family of transmembrane LRR proteins. The three AMIGOs display about 50% sequence similarity compared with each other, and they have the same domain organization, with six extracellular LRR motifs and one immunoglobulin-like (Ig) domain, another protein motif that is widely expressed in neural recognition molecules (5). In BLAST searches using the AMIGO1 ectodomain to identify homologous proteins outside of the AMIGO family, the best fits are found for the axon-guiding Slit proteins and the Nogo-66 receptor (NgR1).
AMIGO1 and AMIGO2 are highly expressed in the mammalian nervous system, whereas AMIGO3 displays a broad expression pattern in different tissues (4, 6, 7). In vitro studies have suggested that AMIGO1 acts as a homophilic adhesion molecule that induces outgrowth and fasciculation of neurites in central neurons (4). The crystal structure of the AMIGO1 dimer has recently revealed that homophilic binding of AMIGO1 occurs through its LRR domains (8). In addition, AMIGO1 and AMIGO2 (also designated as Alivin1) have been reported to enhance survival of neurons in culture (9, 10). Furthermore, AMIGO1 has recently been found to bind to the Kv2.1 potassium channel and to affect excitability of central neurons via the Kv2.1 interactions (11).
Despite several in vitro findings providing clues to developmental roles of the AMIGOs, their in vivo functions remain unknown. In the current study, we have used the zebrafish model to study Amigo functions in nervous system development. We have cloned all three amigo genes of zebrafish and focused our functional studies on amigo1, which was found to be the major member of the gene family expressed in zebrafish during the nervous system development. Using the morpholino knockdown approach and expression of the Amigo1 ectodomain as a dominant negative receptor, we show that Amigo1 is essential for the development of fiber tracts in the zebrafish brain.
EXPERIMENTAL PROCEDURES
Animals
An outbred zebrafish (Danio rerio) strain from a local resource, the Turku line, was used in this study for its steady yield of embryos (12). Fish feeding, breeding, and maintenance were done according to an established protocol (13). The experiment permits were obtained from the University of Helsinki Committee for animal experiments and the National Animal Experiment Board in agreement with the ethical guidelines of the European Convention. Embryos of either sex were staged according to the number of hours postfertilization (hpf) or days postfertilization (dpf). To prevent pigment formation, 0.2 mm 1-phenyl-2-thiourea (Sigma) was added to the medium of embryos after spawning.
Cloning of amigo Genes and Quantitative RT-PCR
Zebrafish orthologs were found by BLASTing the peptide sequences of the mammalian AMIGO proteins using the UCSC genome browser and Ensembl zebrafish ZV9 database. Total RNA from 1–8-dpf larvae was extracted and reverse-transcribed. The subsequent PCR was performed as described previously (12). RNA quality and amount were analyzed spectrophotometrically by a NanoVue Plus spectrophotometer (GE Healthcare). The primers 5′-ATG CCC CCT TCC ATT AAT TG-3′ and 5′-CCG GTC AAA AGA TAC ACA TCC TC-3′ were used for cloning the full-length coding sequence of amigo1 (ENSDARG00000079620). The primers 5′-ATG ACC TCG ACA TCT TGC ATG GTT-3′ and 5′-GAT TCA AAC AAG CAG GAT TTT AAG G-3′ were used for cloning the full-length coding sequence of the second gene that displays clear homology compared with the amigo genes (ENSDARG00000079569; designated here as amigo3a). The primers 5′-ATG CTG TGT GCT CAG GGT GTG GC-3′ and 5′-TCA TCT CTC TGA TTC TAT GTG CTT TCC T-3′ were used for cloning the full-length coding sequence of the third homologous gene (ENSDARG00000074469; designated here as amigo3b). The PCR products were purified with the MinEluteGel Extraction kit (Qiagen, Hilden, Germany), ligated to the pGEM-Teasy plasmid (Promega), and sequenced.
The quantitative RT-PCR (qRT-PCR) primers of amigo1 were 5′-TCG CCG TGA GTG AAT ACC TC-3′ and 5′-TGC CAA GCA ACC CAC CAA A-3′. The qPCR primers of amigo3a were 5′-GGC TGT GTT GTG ACC CTT GT-3′ and 5′-GAT GAG ATG GCT GGA GAT GGA-3′. The qPCR primers of amigo3b were 5′-ACA CTG GCT TCA CCA CAC T-3′ and 5′-AGC GAC AGG GAG TCA AGT AG-3′. The zebrafish β-actin (AF057040.1, GI:3044209), elongation factor 1 α1 (zgc:109885, GI:90652818), and ribosomal protein L13a (BC047855.1, GI:28838761) were set as template quantity controls, and the same primers were used as described previously (12). The PCRs were processed with the Bio-Rad CFX96 real-time PCR machine using the CFX96TM real-time PCR detection system. The three constructed amigo plasmids prepared in different dilutions (from 1 mg/ml to 0.1 μg/ml) in milli-Q water were used as standards for qPCR. The amigo1 expression level normalized to β-actin in 1 dpf larvae was set as 1.
Cloning of amigo1 and kv2.1 mRNAs
The full-length open reading frame mRNA construct encoding Amigo1 was prepared by RT-PCR with primers 5′-ATA AGA TCT ATG CCC CCT TCC ATT AAT TG-3′ and 5′-AAG AAT TCC CGG TCA AAA GAT ACA CAT CCT C-3′. The construct includes BglII and EcoRI restriction sites on both sides of the amigo1 first strand cDNA in the full-length transcript. These restriction sites were used for insertion into the pMC expression vector, which caps the inserted fragment with the sequence elements of the β-globin 5′-UTR and 3′-UTR on both of its sides and in addition the SV40 poly(A) signal at the 3′-tail. This elevates the transcribed mRNA stability and activity about 100-fold in mRNA injection experiments (14). The mRNA encoding the Amigo1 ectodomain was prepared with primers 5′-ATA AGA TCT ATG CCC CCT TCC ATT AAT TG-3′ and 5′-ATG AAT TCT CAG CCA TTG CCG TTG AGG-3′. GFP mRNA was used as another control in mRNA rescue experiments, and it was prepared using the pEGFP-C1 vector. All mRNAs were prepared using the mMessagemMachine kit (Ambion, Austin, TX) according to the manufacturer's instructions.
The full-length kv2.1 cDNA (ZDB-GENE-090831-3) was prepared by RT-PCR with primers 5′-ATA AGC TTC CCT CGG CAG GAA TGA GTA A-3′ and 5′-ATG GAT CCT CAA AGG CCC TTA TCA AAA G-3′ and cloned into pMC expression vector. A cDNA fragment encoding the N terminus and the transmembrane loops of the Kv2.1 protein (432 amino acids) was cloned into pMal-c2E vector (New England Biolabs Inc.) for Kv2.1 ectodomain-maltose-binding protein (MBP) recombinant expression with primers 5′-ATG GAT CCC CCT CGG CAG GAA TGA GTA A-3′ and 5′-ATA AGC TTC TTG ATG GCC TTC TCT TGT-3′.
Antibodies against Amigo1 and Kv2.1
Chicken polyclonal antibodies were produced against the zebrafish Amigo1 (Agrisera, Sweden) using Amigo1 ectodomain-GST (glutathione S-transferase) fusion protein as the antigen. In order to make a high quality antigen, the Amigo1 extracellular coding sequence was cloned into pGEX-2TK GST fusion vector (GE Healthcare) using the primers 5′-ATA GGA TCC TGC GCC AGC AAC ATT GTC AG-3′ and 5′-ATG AAT TCT CAG CCA TTG CCG TTG AGG-3′ to add the BamHI and EcoRI restriction sites on both sides of the coding sequence. The Amigo1 ectodomain-MBP fusion protein was produced by cloning the coding sequence into pMAL-c2E vector. The primers 5′-ATG AAT TCT GCG CCA GCA ACA TTG TCA G-3′ and 5′-ATA GGA TCC TCA GCC ATT GCC GTT GAG G-3′ were used for adding EcoRI and BamHI on both sides of the coding sequence. The plasmids were transformed into Escherichia coli XL-1 blue strain by electroporation (Bio-Rad) for Amigo1 ectodomain-GST/MBP recombinant expression. The recombinant protein was purified using two different affinity columns (GST or MBP tag). The purified Amigo1 ectodomain-GST/MBP fusion protein was analyzed on SDS-PAGE stained with Coomassie Blue (Invitrogen).
The IgY antibodies in the immunized egg yolk were first purified by ammonium sulfate precipitation. In affinity purification, the first step was carried out on an affinity column loaded with Amigo1 ectodomain-GST fusion protein. The second step in affinity purification was carried out on another affinity column loaded with Amigo1 ectodomain-MBP fusion protein to exclude the proteins that bind nonspecifically to the GST tag. The purified zebrafish Amigo1 antibodies were diluted to 1 mg/ml aliquots containing 0.01% NaN3 and stored at −80 °C. Monoclonal anti-Kv2.1 antibodies (K39/25) were obtained from the NeuroMab facility (University of California, Davis/National Institutes of Health).
Western Blotting and Co-immunoprecipitation
Zebrafish Amigo1 antibodies (1:1000 corresponding to 1 μg/ml) and the purified monoclonal Kv2.1 antibodies (2 μg/ml) were used for Western blotting and for co-immunoprecipitation that was carried out according to a previously described protocol (11). Dithiobis(succinimidyl propionate) cross-linking (Pierce) was applied as before (11). Anti-HNK-1 antibodies (C0678, 1:500; mouse monoclonal, Sigma-Aldrich) and anti-acetylated tubulin (6-11B-1 mouse monoclonal, 1:1000; Sigma-Aldrich) were used to detect zebrafish axonogenesis and axonal development in Western blotting and whole mount immunostaining. Mouse monoclonal anti-β-actin antibody (A2228, 1:1000; Sigma-Aldrich) was used as the control of sample loading. The zebrafish tissue preparation and Western blotting were carried out as described previously (12, 13).
Immunocytochemistry
Mouse monoclonal anti-tyrosine hydroxylase (TH) antibody (Diasorin, Stillwater, MN) was used for the whole mount staining of the catecholaminergic system in 3 dpf larval brains as described earlier (12). The mouse anti-3A10 (1:100; Developmental Studies Hybridoma Bank), chicken anti-Amigo1 antibodies (5 μg/ml), and anti-HNK-1 antibodies (1:100) were used for whole mount immunostaining of zebrafish larvae. The Amigo1 ectodomain-MBP fusion protein was purified and used as a competing antigen in antibody binding (100 μg/ml). For immunohistochemistry of the larvae, the samples were fixed in 2% paraformaldehyde for 2 h at room temperature and then washed and preincubated in phosphate-buffered saline containing 0.1% Tween 20 (PBS-T, pH 7.4) with 1% dimethyl sulfoxide (DMSO) and 4% normal goat serum at 4 °C overnight or longer. The specimens were incubated with the primary antibodies in the preincubation solution (PBS-T with 2% normal goat serum) for over 12 h at 4 °C under slow stirring. The samples were then washed thoroughly with PBS-T and incubated with the Alexa®-conjugated goat anti-chicken (Alexa488 and Alexa546) or goat anti-mouse (Alexa488 and Alexa568) secondary antibodies (diluted 1:2000) in the preincubation solution for over 12 h at 4 °C. The samples were washed with PBS-T twice for 30 min, once with PBS for 30 min, and once with 50% glycerol in PBS for 1 h and then infiltrated overnight in 80% glycerol in PBS before mounting.
Whole Mount in Situ Hybridization
Whole mount in situ hybridization was carried out as described previously (15), using the specific probes (16) of pax2a (the ZIRC cb378), pax6a (ZIRC cb280), and krox20 (ZIRC cb427). Larvae at 28 hpf (Prim-5) and 2 dpf (Long-pec) stages were used in the experiments. The probe for the detection of amigo1 expression was obtained from the cDNA clone encoding the Amigo1 ectodomain.
Detection of Apoptosis and Proliferation
A fluorometric TUNEL system (DeadEndTM, Promega) was used for staining of apoptosis in whole mounts, and Edu staining (Click-iT Edu Alexa Fluor 555, Invitrogen) was used for detection of cell proliferation in whole mounts. Apoptosis and proliferation were explored according to the protocol recommended by the manufacturer (12).
Morpholino Oligonucleotide and mRNA Injections
Knockdown experiments of amigo1 were carried out with translation-blocking antisense morpholino oligonucleotides (MOs) (Gene Tools) targeted to the 5′-upstream sequence flanking the translation start site (MO1, 5′-GGC ATT TCT GAC ACG CAG TTA AAA T-3′; MO2, 5′-TGT GGT TGT AGC ACA AGT CAT AAA C-3′) of the amigo1 transcript. To knock down amigo3b expression, the translation-blocking MO was 5′-GGC CAC ACC CTG AGC ACA CAG CAT T-3′. The 5-mispair oligonucleotide 5mis MO, GcC ATT TgT GAg ACc CAc TTA AAA T-3′ (mispairs are in lowercase type), was used as an MO injection control. Cloning of the mRNAs encoding the full-length Amigo1 and the ectodomain of Amigo1 (mRNA and EmRNA, respectively) is explained above.
Microinjections of the MOs and the mRNAs into zebrafish embryos were carried out as described previously (12). An aliquot of 4 nl of the mixture (corresponding to 4 ng of MOs when 100 m solution was used) was injected into the yolk of a 1–4-cell embryo that was allowed to develop at 28.5 °C. For Amigo1 mRNA and GFP mRNA injections, 0.1–0.2 μg/μl mRNA was mixed with injection solution after heating. The injection concentration was tested for eliminating ubiquitous overexpression effects (12). For Kv2.1 mRNA injections, 0.1 μg/μl mRNA was mixed with the injection solution.
Microscopy and Image Analysis
Confocal imaging of larvae stained with anti-TH and anti-Amigo1 antibodies was carried out as described (12). Zeiss LSM710 Pascal confocal microscopy system was used for imaging larval samples stained with anti-3A10 antibodies and anti-HNK1 antibodies. Stacks of images taken at 0.6–1.2-μm intervals were compiled to make maximum intensity projection images. Specimens from in situ hybridization were examined with inverted light microscopy using Olympus IX 70 connected through a CCD camera to the Analysis® software. High resolution images were obtained with a digital MicroFire S99808 camera (Optronics) attached to Olympus BX51 epifluorescence microscope (Olympus, Tokyo, Japan). The acquired images were further processed with CorelDRAW® Graphics Suite X6 (Corel Corp.). Analysis of cell numbers from the whole stack of images scanned through the samples was carried out using ImageJ to count the stained cells (17).
Staining intensity of axonal tracts was measured by Zeiss Efficient Navigation (ZEN 2012) software (Carl Zeiss MicroImaging GmbH) with single maximum intensity z-stacking projections containing the whole image stacks. To quantify the degree of axonal tract development, the axonal tract/cell body ratio was measured on each maximum intensity z-stacking projection (18). For the determination of medial longitudinal fascicle (MLF), average pixel intensity of anti-HNK-1 immunoreactive MLF descending from the ventral caudal cluster (vcc) cells was measured (50 × 2 μm) and compared with that of the vcc cell bodies (5 × 5 μm) in the same z-stacking projection. For the determination of the medial longitudinal catecholaminergic tract (MLCT), average pixel intensity of anti-TH1-immunoreactive MLCT descending from the locus coeruleus was measured (30 × 3 μm) and compared with that of the locus coeruleus cell bodies (3 × 3 μm) in the same z-stacking projection. The average value of 5mis MO larvae was used for normalization in the tract analyses. The background fluorescence was subtracted from each measurement. All image acquisition and analysis was performed blind to the experimental group.
Anatomical structures of larval brain were named and numbered using the neuroanatomical atlas of developing zebrafish brain (19) and the atlas based on location of TH neurons in 5 dpf larval fish (20, 21).
Behavioral Assays
Two behavioral assays were used. First, the locomotor activity of 6-dpf larvae was observed during 10 min in 24-well plates as described previously (20, 22). The analysis included 80 fish (20/group in four independent experiments).
In the second behavioral assay, we assessed the startle response (C-start response) of 9-dpf larvae elicited by electrical stimuli. We chose electrical stimulation because it allowed us to provide a fixed and stable startle stimulus in each group of larvae during constant illumination (23). For the electrical stimulation experiments, recording chambers (60 ml, x = 5 cm, y = 4 cm, z = 3 cm shared from the laboratory of H. A. Burgess) were fitted with stainless steel sidewalls as bipolar stimulating electrodes (5 V, 0.01 s). For each experiment, 20 larvae were acclimated in the recording chamber for 3 min before testing. The MotionPro X3 fast recording camera (Redlake Imaging) was controlled by a script written in Matlab (MathWorks). Recording was started 200 ms before the electrical stimulus was given (via the MotionPro XTM data acquisition hardware module, Redlake Imaging) and was continued for 1 s. This was repeated three times with the same larvae in the chamber, with a 3-min interval set between each electrical stimulus. Sequential images of the C-start response were captured every 1 ms. Latency was defined as the time from the beginning of the stimulation to the C-bend formation. The captured images were analyzed with Flote version 2.1. Short latency C-start (SLC; initiated within 20 ms after stimulus) and long latency C-start (LLC; initiated later than 20 ms after stimulus) share identical movement trajectories and were classified according to the previous report (24). For each group, the experiment was repeated four times with fish from different injections.
Analytical Software
Student's t test and one-way ANOVA analyses were carried out using OriginPro version 8.0. Intensities of the Western blot bands were examined with Quantity One version 4.6.2 (Bio-Rad). Sequence alignments of the AMIGO orthologs of different species were carried out using Geneious Pro R6 (Biomatters Ltd.).
RESULTS
Zebrafish Amigo Proteins and Detection of Zebrafish Amigo1
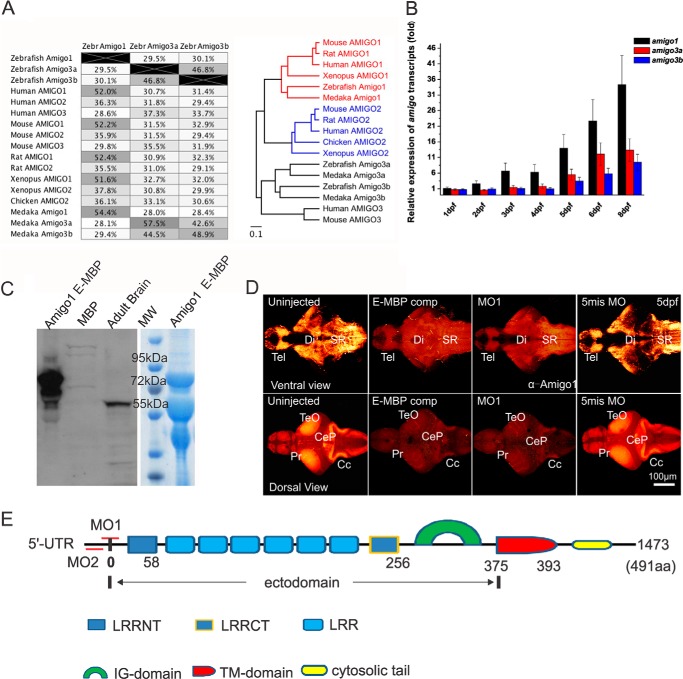
According to the characteristic LRR and Ig domains, three putative amigo transcripts are found in the zebrafish Zv9 database (Ensembl, search zebrafish). The full-length cDNAs were cloned by using mRNA from zebrafish larvae and primers designed according to the putative homologous sequences found in the Zv9 database. Comparison of the deduced amino acid sequences of the zebrafish Amigos with the human, rodent, chicken, medaka, and Xenopus proteins was carried out using the Geneious software. Within the 17 Amigo proteins quoted, the zebrafish Amigo1 shares over 50% identity in its amino acid sequence compared with the Amigo1 protein in all other species (Fig. 1A). The two other zebrafish sequences (designated as Amigo3a and -3b) display a higher degree of similarity to AMIGO3 than to AMIGO2. It appears that no AMIGO2 ortholog has yet been identified in teleost species.
FIGURE 1.
AMIGO proteins in different species, expression of the amigos in zebrafish, and production of antibodies to detect Amigo1 of zebrafish. A, alignment of the amino acid sequences of the zebrafish Amigos with 14 AMIGO proteins found in other species (on the left). Identity of the amino acid sequences is indicated in the table. A phylogenetic tree of 17 AMIGO proteins found in Xenopus, chicken, rat, mouse, human, zebrafish, and medaka was generated by the Clustal multiple-sequence alignment program (on the right). The similarities and identities were computed by Geneious. Scale bar, number of amino acid substitutions/site. B, expression of the three amigos during zebrafish development detected by qRT-PCR. Full-length cDNA plasmid clones of amigo1, amigo3a, and amigo3b were constructed and used for the standardization of each qRT-PCR. Templates were normalized by three reference genes as described under “Experimental Procedures.” amigo1 expression compared with β-actin in 1-dpf larvae was set as 1. amigo1 expression increases over 30-fold during development from 1 to 8 dpf. For each qRT-PCR test, total RNA was extracted from a pool of 20 larvae. The data are based on three independent experiments. Mean values ± S.E. (error bars) are indicated. C, antibodies against zebrafish Amigo1. Specificity of the anti-Amigo1 antibodies is shown by Western blotting on the left. The purified antibodies stain the Amigo1 ectodomain-MBP fusion protein (E-MBP) band (∼75 kDa) clearly with weak background. The antibodies also detect specifically the Amigo1 band (∼60 kDa) in the SDS extract of the adult zebrafish brain. Coomassie Blue staining on the right confirms high expression of the Amigo1 ectodomain-MBP fusion protein used for purification and binding tests of the antibodies. D, whole mount immunostaining of 5-dpf larval brains with the anti-Amigo1 antibodies. Goat anti-chicken Alexa 546 (orange to white) was used as the secondary antibody for fluorescent labeling. The maximum z-stacking projection images (maximum depth, 130 μm) were scanned from both ventral and dorsal sides to show the Amigo1 expression pattern throughout the brain (the thickness of the samples over 200 μm). Compared with the uninjected or the 5mis MO-injected larvae, immunostaining is clearly inhibited in the MO1-injected larvae or in the uninjected larvae in the presence of the competing antigen (E-MBP comp). CC, cerebellar crest; CeP, cerebellar plate; Di, diencephalon; Pr, pretectum; SR, superior raphe; Tel, telencephalon; TeO, tectum opticum. Scale bar, 100 μm. E, domain structure of zebrafish Amigo1 based on the sequence of the transcript. Recognition sites of the designed morpholino oligonucleotides (MO1 and MO2) around the 5′-UTR of the transcript are marked by short red lines. The part of the transcript encoding the Amigo1 ectodomain was used for dominant negative expression (Am1 EmRNA). IG-domain, immunoglobulin-like domain; LRRCT, leucine-rich repeat C-terminal flanking domain; LRRNT, leucine-rich repeat N-terminal flanking domain; TM-domain, transmembrane domain.
A phylogenetic tree view of the alignment result clearly showed that within the Amigo family, Amigo1 has the highest degree of homology when compared with the orthologs expressed in different species (Fig. 1A). Zebrafish Amigo3a and Amigo3b are both similar to mammalian AMIGO3 and grouped into a different branch compared with AMIGO2 of the other species (Fig. 1A). Zebrafish Amigo3a and -3b might be encoded by duplicate copies of the same gene.
In order to detect expression of the Amigos in larval zebrafish, the primers for qRT-PCR of amigo1, amigo3a, and amigo3b were designed and used for expression analysis in larvae at the developmental stages 1–8 dpf. The qRT-PCR results revealed that all three amigo expressions start from the beginning of early developmental stages (Fig. 1B). Expression of amigo1 was found to be the highest of the three amigo transcripts. Expression of amigo1 already started to increase after 24 hpf and was increased in a logarithmic manner during the stages 1–8 dpf. Expression of amigo3a and amigo3b started to increase from 3 dpf and was always lower than expression of amigo1. Expression of amigo3b was found to be the lowest of the three transcripts. Before 4 dpf, amigo1 expression was found to be over 5-fold higher than that of amigo3a or amigo3b. After 4 dpf, amigo1 expression was still over 3-fold higher than expression of the two other transcripts.
Because amigo1 was found to be the major amigo transcript in zebrafish early embryos, we decided to focus on this family member to elucidate putative amigo functions in nervous system development. Because the antibodies that we have previously generated against the mouse AMIGO1 (11) did not specifically detect the zebrafish Amigo1 protein in Western blotting of SDS extracts of larvae, we generated antibodies against the zebrafish Amigo1 sequence. We constructed a fusion plasmid clone encoding Amigo1 ectodomain-GST and purified the expressed protein to be used as an immunogen in chicken (4). Western blotting revealed that the affinity-purified antibodies detect specifically the Amigo1 ectodomain-MBP fusion protein and the endogenously expressed protein in larval SDS extracts that corresponds to the molecular mass of Amigo1 (Fig. 1C).
In addition to the zebrafish Amigo1 specific antibodies, we designed a knockdown approach to study expression and functions of Amigo1 in zebrafish. Two gene-specific antisense MOs (MO1 and MO2; Fig. 1E) were designed for amigo1 translation inhibition (25). Because the amigo1 transcript contains only one integrated exon without any introns, MO1 and MO2 were targeted for binding to the 5′-UTR sites near the start codon (Fig. 1E). They are not expected to inhibit protein translation from capped amigo1 mRNA injected in rescue experiments. Uninjected larvae and larvae injected with 5mis MO (having five non-pairing nucleotides compared with MO1) were used as controls.
To verify the specificity of the Amigo1 immunostaining, we used two different approaches. First, immunostaining using the anti-Amigo1 antibodies was strongly reduced in the knockdown morphants compared with the uninjected or the 5mis MO-injected controls (Figs. 1D and 2A). To restore Amigo1 expression in knockdown experiments, we tested injections of Amigo1 mRNA at the doses ranging from 0.2 to 0.8 ng. Amigo1 was immunostained at an intensity comparable with the endogenous expression in larvae injected with about 0.4 ng of Amigo1 mRNA (Fig. 2A; see also Western blotting results of Fig. 3, A and B). As an additional control, Amigo1 ectodomain-MBP fusion protein was premixed with the antibodies for antigen competition in immunostaining. Amigo1-specific immunostaining was blocked in the prebinding experiments in a similar manner as in the knockdown experiments (Fig. 1D, E-MBP comp).
FIGURE 2.
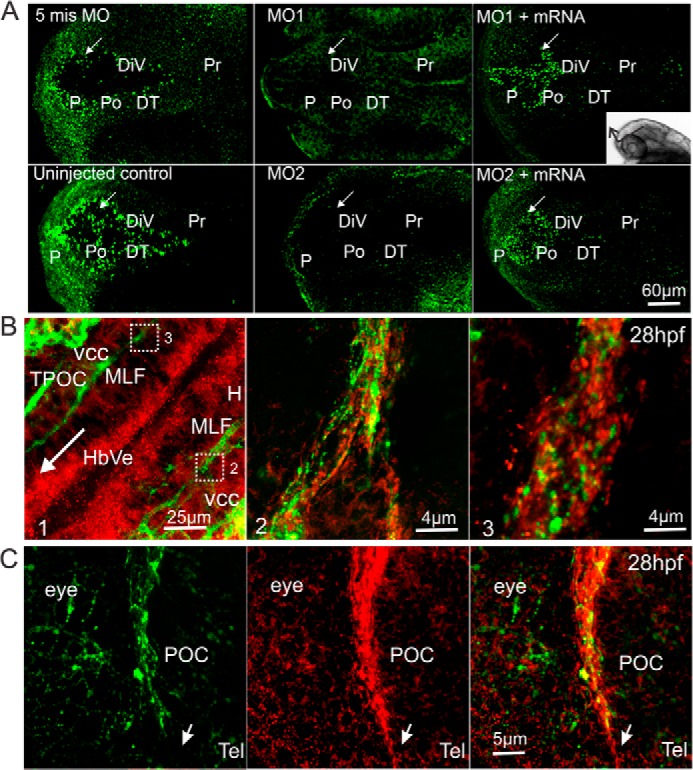
Immunohistochemistry of Amigo1 expression during early development of the zebrafish brain. A, whole mount immunostaining of Amigo1 in 28-hpf larvae. Anti-Amigo1 antibodies were used as the primary antibodies, and goat anti-chicken Alexa 488 antibodies were used as the secondary antibodies for fluorescent labeling. Control groups are the 5mis MO-injected and the uninjected larvae. amigo1 knockdown morphants are marked as MO1 and MO2. Rescued groups are coinjected with the full-length mRNA (MO1 + mRNA and MO2 + mRNA). DT, dorsal thalamus; P, pallium; Pr, pretectum; Po, preoptic area. The confocal images were scanned and stacked as shown by the inset on the right. The arrow in each frame indicates the optic recess of DiV as the landmark. Scale bar, 60 μm. B, double staining of 28-hpf larvae with chicken anti-Amigo1 and mouse anti-HNK1 antibodies; the primary antibodies were detected with anti-chicken Alexa 546 (red; anti-Amigo1 antibodies) and goat anti-mouse Alexa 488 (green; anti-HNK1 antibodies). B1, anti-Amigo1 antibodies showed staining on the superficial layer of the hindbrain ventricle (HbVe). The arrow indicates the direction from posterior to anterior. B2 and B3, Z-stacking confocal images scanned under higher magnification show that anti-Amigo1 and anti-HNK1 antibodies stain the tracts. The rectangles in B1 indicate the areas of B2 and B3. H, hindbrain; TPOC, tract of the postoptic commissure. Scale bars, 25 μm (B1) and 4 μm (B2 and B3). C, the ascending POC in telencephalon (Tel) stained with mouse anti-HNK1 antibodies (left) and chicken anti-Amigo1 antibodies (middle) and overlay of the stainings (right). The arrow shows the leading part of the tract in the direction of its extension. Scale bar, 5 μm.
FIGURE 3.
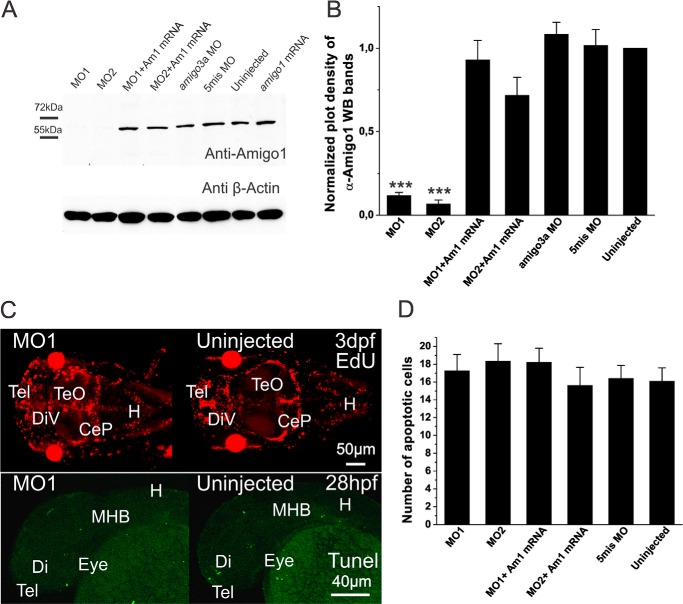
Knockdown of Amigo1 protein expression and phenotypic features of the knockdown larvae. A, Western blotting of 3-dpf larvae lysates with anti-Amigo1 antibodies. No Amigo1 band is detected on the MO1 and MO2 lanes. B, quantification of Amigo1 expression by plot density analyses. In 3-dpf amigo1 knockdown morphants (MO1 and MO2), amigo1 expression is significantly inhibited compared with the 5mis MO-injected controls. The plot density of the bands from uninjected larval SDS extracts was set as 1 for normalization. Western blots were repeated three times in three independent experiments (n = 3; ***, p < 0.001 in one-way ANOVA followed by Tukey's post hoc test). C, Edu staining (red) and fluorescent TUNEL staining (green) do not display differences between the amigo1 knockdown morphants (MO1) and the uninjected control. CeP, cerebellar plate; Di, diencephalon; H, hindbrain; MHB, midbrain hindbrain boundary; Tel, telencephalon; TeO, tectum opticum. Scale bar, 50 μm in the Edu-stained and 40 μm in the TUNEL-stained panel. D, numbers of apoptotic cells based on TUNEL staining in prosencephalon and mesencephalon of 28-hpf larvae. The amigo1 morphants do not display a significant difference compared with the 5mis MO-injected controls. The data are based on three independent experiments; apoptotic cell numbers in 15 larvae of each group were counted (n = 15, one-way ANOVA followed by Tukey's post hoc test). Error bars, S.E.
Amigo1 Expression in Larval Brain
The affinity-purified antibodies detected Amigo1 in whole mount immunostaining already at 28 hpf in the larval brain (Fig. 2A). Amigo1 was intensely stained in the neuronal progenitor cell layers lining the diencephalic ventricle (DiV) and the optic recess of the forebrain (26, 27). Furthermore, the anterior diencephalon was also specifically stained on the alar plate prosomere, from which the anterior thalamic nuclei derive (28), and on the upper layer of posterior tuberculum. Essentially no detection was observed in the amigo1 knockdown morphants compared with the uninjected or the 5mis MO-injected controls (Fig. 2A). Expression of amigo1 was partially rescued by coinjection of the amigo1 full-length mRNA with the MOs, as evidenced by intensive and ubiquitous Amigo1 staining in DiV cells (Fig. 2A, MO1 + mRNA and MO2 + mRNA).
At 28 hpf, Amigo1 was found to be expressed in both cell layers and the early developing fiber tracts. MLF and the postoptic commissure (POC) belong to early developing fiber scaffolds of zebrafish that were immunostained with the anti-Amigo1 and the anti-HNK-1 antibodies. Amigo1 was detected at the leading front and along the fascicle of the developing tracts (Fig. 2, B and C).
The early embryonic brain development in zebrafish is largely accomplished at 5 dpf (29, 30). At this stage, Amigo1 is widely distributed in most nuclei in telencephalon and commissures in diencephalon (for the expression pattern at 5 dpf, see Fig. 1D).
Gross Morphology of amigo1 Knockdown Morphants
In injections with different MO doses, the death rates in larvae injected with MO1, MO2, or 5mis MO were essentially the same (about 5%) at the doses of 2–4 ng but were significantly increased at higher doses (data not shown). We therefore selected 4 ng as the dose for the injections in further experiments because it was found to be the highest dose of the MOs allowing high proportions of live larvae.
We used Western blotting of 3 dpf larval lysates to evaluate the effects of the MOs at a dose of 4 ng on Amigo1 protein expression. Amigo1 expression essentially disappeared in the MO1- and MO2-injected larvae, whereas no major change was observed in amigo3a MO-injected morphants or in larvae injected with 5mis MO. Coinjection of the amigo1 mRNA with MO1 or MO2 displayed a clear rescue effect on the protein expression (Fig. 3A). Quantification of the Western blot results revealed >85% decrease in Amigo1 expression in the MO1-injected morphants and >90% in the MO2-injected morphants (Fig. 3B). The 4-ng dose in the MO injections thus appears optimal to avoid nonspecific effects and to obtain efficient knockdown effects of the Amigo1 protein expression. Immunostaining of Amigo1 in the larvae injected with 4 ng of the MOs (Figs. 1D and 2A) was found to agree with the protein expression data.
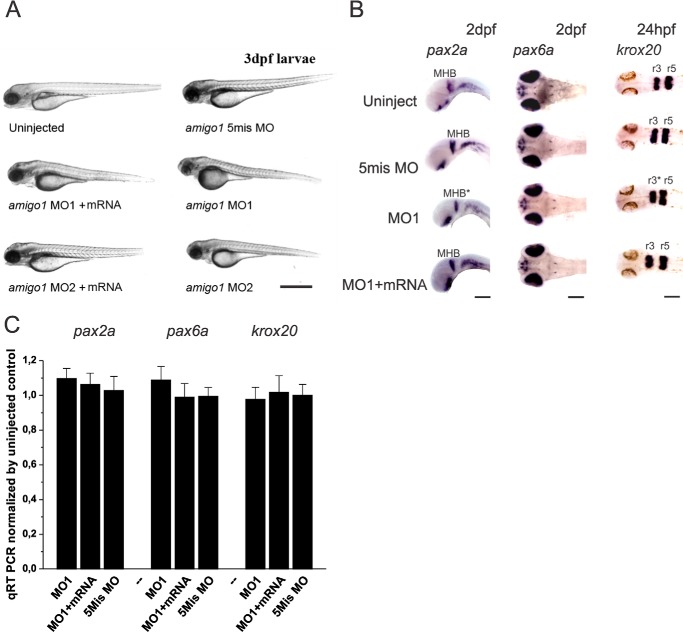
When the endogenous amigo1 expression was inhibited during the development using injections of MO1 and MO2 at a dose of 4 ng, the morphants did not show gross morphological defects during development (Fig. 4A). The knockdown morphants did not either display any difference in expression levels of regional transcription factors (pax2a, pax6a, and krox20) that are known to be important for brain development (Fig. 4, B and C). Closer inspection, however, suggested that the morphants have shorter length and width of head (Fig. 4B, pax6a and krox20 stainings), and smaller tectum opticum (Fig. 3C, TeO) and midbrain-hindbrain boundary (MHB; cell layer between cerebellar plate and midbrain tectum; Figs. 3C and 4B) compared with the controls.
FIGURE 4.
Amigo1 morphants do not display defects in gross morphology or in expression of transcription factors that regulate neuronal development. A, light microscopy images of 3-dpf larvae. Scale bar, 1 mm. B, whole mount in situ hybridization of pax2a and pax6a in 2-dpf larvae (the pharyngula period from High-pec to Long-pec) and krox20 in 24 hpf larvae (the pharyngula period from Prim 5 to Prim 15). The first lane shows a lateral view of pax2a in 2-dpf larvae as a marker of the midbrain-hindbrain boundary (MHB) development. Scale bar, 200 μm. The second lane shows a dorsal view of pax6a in 2-dpf larvae as a marker of forebrain development. Scale bar, 100 μm. The last lane shows a dorsal view of krox20 in 24-hpf larvae as a marker of rhombomere3 (r3) and rhombomere5 (r5) in hindbrain. Scale bar, 80 μm. The asterisks indicate the smaller midbrain-hindbrain boundary and rhombomere3 of MO1 larvae. C, quantification of pax2a, pax6a, and krox20 expression by qRT-PCR using total RNA extracted from 2-dpf larvae. Each qRT-PCR sample was obtained from a pool of 20 larvae. Templates were normalized by three reference genes, as described under “Experimental Procedures.” The data are based on three independent experiments. Mean values ± S.E. (error bars) are indicated.
Amigo1 knockdown might perturb brain development by causing problems in cell proliferation and survival. We therefore carried out EdU and TUNEL stainings in whole mount preparations of larvae but did not find any apparent changes (Fig. 3C). Counting of apoptotic cells in the area covering the whole prosencephalon and mesencephalon (31) did not reveal any differences in different experimental settings used in the study (Fig. 3D).
Amigo1 Regulates Expression of the Kv2.1 Potassium Channel and Binds to Kv2.1
We have recently shown that AMIGO1 binds to Kv2.1 in adult mouse neurons and regulates voltage-gated potassium currents through Kv2.1 (11). We therefore decided to study whether expression of Kv2.1 is changed in the Amigo1 knockdown morphants and whether Amigo1 and Kv2.1 interact in zebrafish neurons.
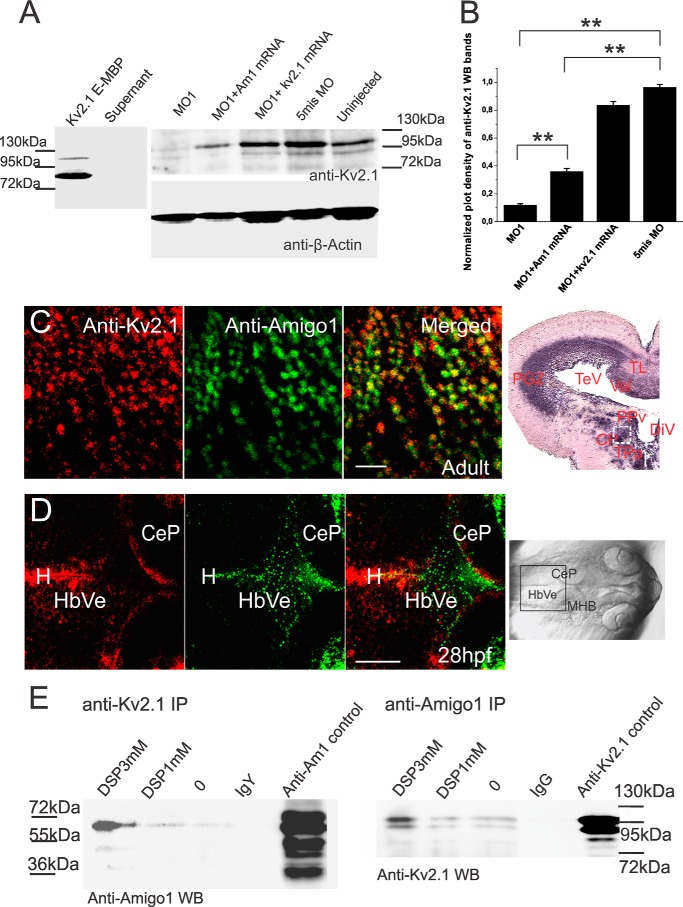
Based on the sequences in the zebrafish Zv9 database (Ensembl, search D. rerio), a putative kv2.1 transcript is expressed in zebrafish displaying 66% identity in its sequence compared with the mouse Kv2.1 transcript. We produced an N-terminal fragment of the zebrafish Kv.2.1 containing the extracellular domain and the transmembrane loops as an MBP fusion protein (∼80 kDa). Monoclonal antibodies against mouse Kv2.1 (K39/25) were found to recognize the recombinant Kv2.1 fragment (Fig. 5A). Western blotting of SDS extracts from 3 dpf larvae showed that the Kv2.1 protein is expressed in the larvae, and its expression is strongly decreased in the amigo1 knockdown larvae (MO1), displaying about 10% of the expression found in control larvae (Fig. 5, A and B). Coinjection of amigo1 mRNA with MO1 (MO1 + Am1 mRNA) restored the expression to 30–40% of that found in the controls. Coinjection of kv2.1 mRNA with MO1 restored Kv2.1 expression to 80% of the level seen in the control larvae (Fig. 5, A and B).
FIGURE 5.
Amigo1 regulates Kv2.1 expression and binds to Kv2.1. A, monoclonal anti-Kv2.1 antibodies detect the ectodomain of zebrafish Kv2.1 (Kv2.1 E-MBP) in Western blotting. Western blotting of 3-dpf larval SDS extracts using the anti-kv2.1 antibodies shows strong down-regulation of Kv2.1 in Amigo1 knockdown morphants (MO1) compared with the 5mis MO-injected or the uninjected larvae. Western blotting suggests that Kv2.1 expression is enhanced by coinjection of amigo1 or kv2.1 mRNA with MO1. B, quantification of Kv2.1 expression in the amigo1 MO1 morphants shows significant down-regulation of the expression compared with the 5mis MO-injected or the uninjected larvae. In the MO1/Amigo1 mRNA-coinjected larvae, Kv2.1 expression is increased to 30% of the uninjected controls, which is still significantly less than the expression in the 5mis MO group. In the MO1/Kv2.1 mRNA-coinjected larvae, Kv2.1 expression is increased to over 60% of the uninjected controls. Western blots were repeated four times in four independent experiments (n = 4; **, p < 0.01 in one-way ANOVA followed by Tukey's post hoc test). C, double staining of adult brain sections with anti-Amigo1 and anti-Kv2.1 antibodies shows their coexpression in the diencephalic neuronal cells in the adult (over 1-year-old) zebrafish. The primary antibodies were detected with goat anti-chicken Alexa 488 (green; anti-Amigo1 antibodies) and goat anti-mouse Alexa 568 (red; anti-Kv2.1 antibodies). The light microscopy image on the right shows Amigo1 whole mount in situ hybridization in a brain sagittal section. Rectangle, the Amigo1-expressing brain region that was used for the double immunostaining of Amigo1 and Kv2.1. Scale bar, 10 μm. CP, central posterior thalamic nucleus; PGZ, periventricular gray zone of optic tectum; PPv, ventral periventricular pretectal nucleus; TeV, tectum ventricle; TL, torus longitudinalis; TPp, periventricular nucleus of posterior tuberculum; Val, vlavula cerebelli. D, double staining of 28-hpf larvae with anti-Amigo1 and anti-Kv2.1 antibodies shows that Kv2.1 and Amigo1 are both expressed in the hindbrain ventricle area from an early development stage but are not expressed in the same cells. The rectangular area in the light microscopy image on the right is the region shown in the fluorescent images. Scale bar, 20 μm. CeP, cerebellar plate; H, hindbrain; HbVe, hindbrain ventricle; MHB, midbrain-hindbrain boundary. E, coimmunoprecipitation (IP) of Kv2.1 and Amigo1 from adult brain SDS extracts using the monoclonal anti-Kv2.1 and the polyclonal chicken anti-Amigo1 antibodies. Dithiobis(succinimidyl propionate) (DSP)-treated SDS extracts show enhanced coimmunoprecipitation. Non-immune IgY and IgG were used as negative controls in Western blotting of Amigo1 and Kv2.1, respectively. In the anti-Amigo1 and anti-Kv2.1 controls, immunoprecipitation and Western blotting were carried out with the same antibodies. Error bars, S.E.
Double immunostaining of brain sections from adult zebrafish showed colocalization of Kv2.1 and Amigo1 throughout the brain, shown for diencephalic neurons in Fig. 5C. Coexpression was also observed in the hindbrain ventricle cell layers (HbVe) and the edge of the cerebellar plate (CeP) in 28-hpf larvae (Fig. 5D), but the staining patterns did not suggest expression in the same cells.
Because the immunostaining experiments suggested colocalization of Amigo1 and Kv2.1 in adult zebrafish neurons, we carried out co-immunoprecipitation experiments using adult brain samples. These experiments suggested binding of Amigo1 to Kv2.1 (Fig. 5E). The binding was more prominent in dithiobis(succinimidyl propionate)-cross-linked samples compared with samples without any cross-linker. The apparent reason for this is that the transmembrane domains of the proteins mediate binding are largely lost in detergent extraction used in the co-immunoprecipitation experiments, which was previously shown in the experiments using brain extracts from mice (11).
Amigo1 Is Required for the Development of Early Axonal Scaffolds in Zebrafish Brain
At 28 hpf, the initial axon scaffold of the zebrafish brain is simple, consisting of a few longitudinal tracts connected by commissures (32, 33). Antibodies against the cell surface marker HNK-1 (34) specifically label neurons and axons in the early embryonic nervous system (35). We used immunocytochemistry with anti-HNK-1 antibodies to visualize the possible role of Amigo1 in the development of early nervous system structures (Fig. 6, A and B).
FIGURE 6.
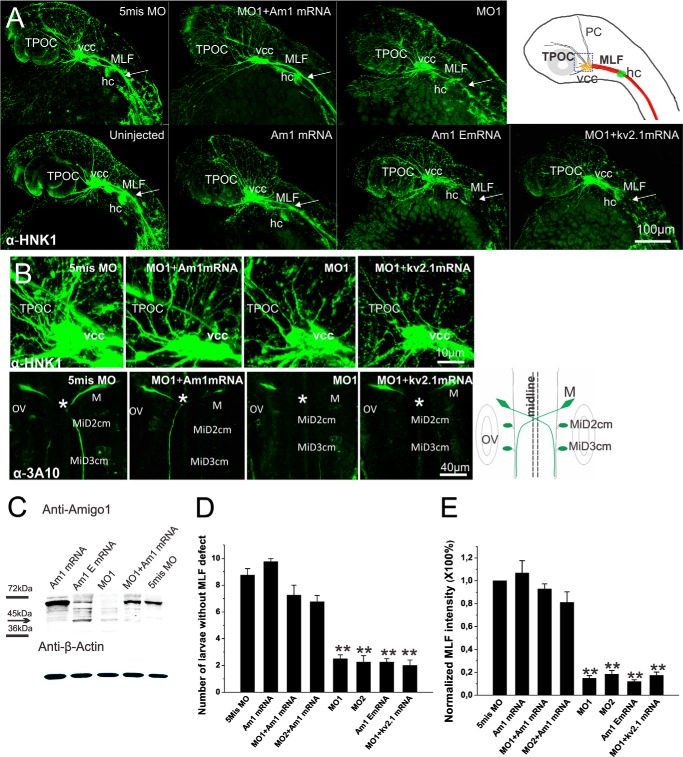
Immunohistochemistry showing decreased fasciculation and fiber outgrowth in amigo1 knockdown morphants (MO1 and MO2) and in larvae expressing the Amigo1 ectodomain (Am1 EmRNA) as an inhibitor of the endogenous full-length protein. A, whole mount anti-HNK-1 immunostaining of 28-hpf larvae. The amigo1 knockdown morphant (MO1) shows decreased staining in MLF, indicated by the arrows. The ectopically expressed amigo1 ectodomain mRNA (Am1 EmRNA) results in a similar MLF defect. Coinjection of the amigo1 mRNA but not the kv2.1 mRNA rescues the tract defect caused by MO1. The schematic map on the right shows the main axonal tracts stained in uninjected 28-hpf larvae. The rectangle in the schematic map indicates the region shown in B. hc, hindbrain cell cluster; PC, posterior commissure; TPOC, tract of the postoptic commissure. Scale bar, 100 μm. B, early axonal development at the area of the vcc shown by whole mount immunostaining using mouse anti-HNK-1 antibodies (top) and mouse anti-3A10 antibodies (bottom) in 28-hpf larvae. The amigo1 knockdown morphant (MO1), the 5mis MO control, and the mRNA rescue effects (MO1 + Am1 mRNA and MO1 + Kv2.1 mRNA) are shown. Fibers in the tract of the postoptic commissure, originating from the vcc, are decreased in the MO1 and the MO1 + kv2.1 mRNA larvae. Anti-3A10 antibodies used in whole mount immunostaining show axon development of Mauthner neurons in the hindbrain of 28-hpf larvae. The schematic map on the right shows the normal staining pattern. The asterisk indicates the defective development of axon projections from Mauthner neurons in the MO1 and the MO1 + kv2.1 mRNA larvae compared with the 5mis MO and the MO1 + Am1 mRNA larvae. M, Mauthner neuron; MiD2cm, middle dorsal 2 contralateral MLF interneuron; MiD3cm, middle dorsal 3 contralateral MLF interneuron; OV, otic vesicle. Scale bars, 10 μm in the top row and 40 μm in the bottom row. C, Western blotting of 2-dpf larval lysates (10 larvae/sample) with anti-Amigo1 antibodies. Expression of the Amigo1 ectodomain in the Am1 EmRNA-injected group is detected as the strong band at around 45 kDa, indicated by the arrow. D, statistics of larvae with normal MLF stained by anti-HNK-1 antibodies. The amigo1 knockdown morphants (MO1 and MO2), the Am1 EmRNA larvae, and the MO1 + Kv2.1 mRNA larvae display significant MLF defects compared with the rescued (MO1 + Am1 mRNA and MO2 + Am1 mRNA) and the 5 misMO-injected groups. For each group, 10 larvae were selected randomly from the pool of 50 larvae, and four independent experiments were included. Mean values ± S.E. (error bars) are indicated (n = 4; **, p ≤ 0.001, one-way ANOVA followed by Tukey's post hoc test). E, average pixel intensity analyses of anti-HNK1 stained MLF from Z-stacking confocal images. The average pixel intensity of MLF in 5mis MO larvae was used for normalization. In larvae injected with MO1, MO2, Am1 EmRNA, or MO1 + Kv2.1 mRNA, MLF staining intensities are significantly lower than in larvae injected with 5mis MO or amigo1 mRNA or coinjected with MO1 or MO2 and the amigo1 mRNA. The results are based on 12 larvae randomly selected from three independent experiments (n = 12; **, p ≤ 0.001, one-way ANOVA followed by Tukey's post hoc test).
Confocal stacking projections of the lateral side of larvae clearly showed disturbed fiber tract development in amigo1 knockdown morphants at 28 hpf (Fig. 6A). The MLF and tract of the postoptic commissure belong to the very early major tracts and only appear as thin fibers in the amigo1 MO1 morphants compared with the wild-type or 5mis MO-injected embryos. Coinjection of the full-length amigo1 mRNA (Am1 mRNA) with MO1 displayed a rescuing effect (Fig. 6, A and B).
We have previously shown that the AMIGO proteins display homophilic binding that appears important for fasciculation of neurites in cultures of rat brain neurons. Furthermore, the ectodomain of AMIGO1 in solution was shown to act as a dominant negative receptor and to inhibit AMIGO1-mediated adhesion and fasciculation in brain neurons in vitro (4). In order to provide an assay that does not depend on MOs, we prepared an mRNA encoding the extracellular part of Amigo1 (Am1 EmRNA, coding for the six LLR domains and one Ig domain of Amigo1; Fig. 1E) and injected it into fertilized embryos. Expression of the dominant negative protein in the injected larvae could be distinguished by Western blotting with anti-Amigo1 antibodies (Fig. 6C). As in the amigo1 MO1 morphants, development of the early tracts was severely disturbed in larvae injected with the Am1 EmRNA. For example, in the confocal stacking projections, MLF could hardly be discerned in the Am1 EmRNA-injected larvae (Fig. 6A). In contrast to the Am1 EmRNA, larvae injected with the full-length mRNA did not display such inhibition (Fig. 6A, Am1 mRNA) but even displayed rescue of axonal growth (Fig. 6A, MO1 + Am1 mRNA). The larvae coinjected with MO1 and kv2.1 mRNA (MO1 + kv2.1 mRNA) showed MLF defects, as did the amigo1 MO1 morphants and the Am1 EmRNA-injected larvae (Fig. 6A).
Statistics of the 28-hpf larvae immunostained with anti-HNK-1 antibodies showed that significantly lower numbers of the amigo1 knockdown morphants and amigo1 EmRNA-injected larvae had intact MLF than 5mis MO-injected or amigo1 mRNA-injected larvae (Fig. 6D). Intensity of the MLF staining confirmed the defective tract development in the knockdown morphants and in the EmRNA-injected larvae (Fig. 6E). A clear rescuing effect in the development of MLF was found in coinjection of the amigo1 mRNA with the MOs (Fig. 6, D and E), which could not be achieved by kv2.1 mRNA coinjection.
The vcc and the hindbrain cell cluster are important for the development of early ascending and descending long tracts (35, 36) and were found to be correctly located in the knockdown morphants (Fig. 6A, vcc and hc). Under higher magnification, the MO1 morphants displayed a clearly reduced HNK-1 staining of axons from the ventral caudal cell clusters (Fig. 6B, top panels). HNK-1 is also present in Mauthner neurons and the long growing trigeminal axons (35). As the largest reticulospinal interneurons in zebrafish, the Mauthner neurons and their long axons contribute to early sensorimotor circuit formation (37). Anti-3A10 antibodies were used for specific labeling of Mauthner neurons and their axonal projections (38). Disturbed axonal growth was also clearly observed in the Mauthner neurons in the hindbrain of the Amigo1 knockdown morphants (Fig. 6B, bottom panels). HNK-1 and 3A10 staining of the axons was partially rescued by coinjection with the amigo1 mRNA but not with the kv2.1 mRNA (Fig. 6B).
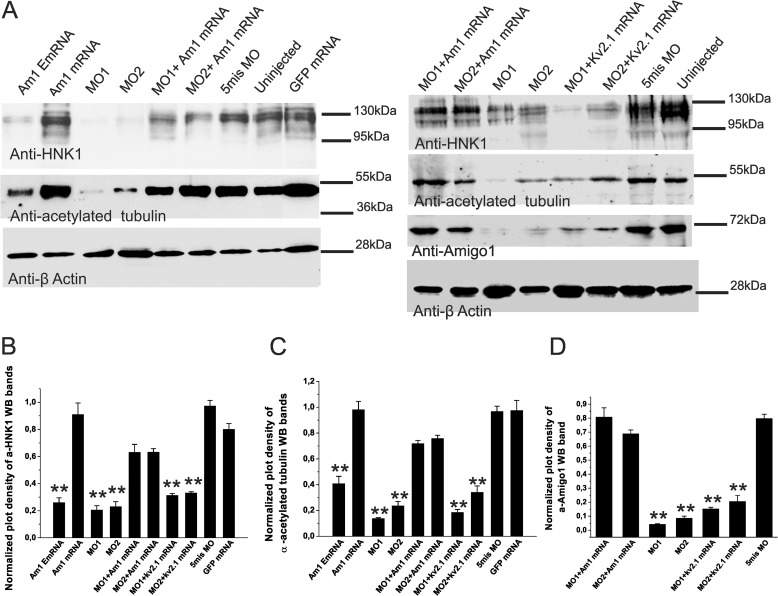
Acetylated tubulin is a component of microtubules, which are the first cytoskeletal elements to appear in axons (39, 40). As another frequently used marker for labeling early developing axon tracts in zebrafish, the anti-acetylated tubulin antibodies show a similar immunostaining pattern as the anti-HNK-1 antibodies (32, 41). Here we used Western blotting to quantify expression of the HNK-1 epitope and acetylated tubulin as markers of the neuronal fiber tract development (Fig. 7). Western blotting of 3-dpf larval samples showed that expression of both HNK-1 and acetylated tubulin is strongly reduced in the amigo1 knockdown (MO1 and MO2) morphants (Fig. 7A). The HNK-1 expression level of the amigo1 MO1 and MO2 morphants was decreased to about 20% of the uninjected controls (Fig. 7B). Coinjection of the full-length amigo1 mRNA (Am1 mRNA) with MO1 or MO2 caused a significant rescue in the HNK-1 expression (Fig. 7, A and B). HNK-1 expression in the Am1 mRNA-MO1-coinjected larvae was elevated to >50% of the uninjected control, and in the Am1 mRNA-MO2-coinjected larvae, the expression was enhanced to >60% of the uninjected control (Fig. 7B). In the same larval samples, Western blotting of acetylated tubulin in the amigo1 MO1 and MO2 morphants showed a similar decrease as the HNK-1 epitope (Fig. 7A). In the mRNA-coinjected rescued groups, acetylated tubulin expression was recovered to >70% of the uninjected control.
FIGURE 7.
Western blotting of larval lysates showing decreased development of neuronal connections in amigo1 knockdown morphants (MO1 and MO2) and in larvae expressing the Amigo1 ectodomain as an inhibitor of the endogenous protein (Am1 EmRNA). A, the left panel shows Western blotting of 3 dpf larval lysates using anti-HNK-1 and anti-acetylated tubulin antibodies as pathway markers. Anti-β-actin antibodies were used for normalizing the loading amount of each sample. The amigo1 knockdown morphants (MO1 and MO2) and the amigo1 EmRNA-injected larvae (Am1 EmRNA) show much weaker bands than the rescued groups (MO1 + Am1 mRNA and MO2 + Am1 mRNA) and the control groups (the 5mis MO-injected, the GFP mRNA-injected and the uninjected controls). The right panel shows the corresponding Western blotting analysis, where the rescue effect of the kv2.1 mRNA is compared with that of the amigo1 mRNA. In addition, expression of Amigo1 in morpholino-injected and in morpholino/mRNA-coinjected larvae is shown. B–D, quantification of the results shown in A. In B–D, the plot density of the bands from the uninjected larvae was set as 1. The experiments were repeated four times with larvae from four independent experiments. Each SDS extract contained 8–10 larvae selected randomly from the pool of 50 larvae. The statistical significance was confirmed by comparison with the 5mis MO-injected larvae. All graphs show mean values ± S.E. (error bars) (n = 4; **, p ≤ 0.001, one-way ANOVA followed by Tukey's post hoc test).
As in the MO-injected larvae, HNK-1 and acetylated tubulin were also decreased in the dominant negative amigo1 EmRNA-injected larvae (Fig. 7A). HNK-1 was reduced to about 25% of the normal control (Fig. 7B), and acetylated tubulin was reduced to about 40% of the normal control (Fig. 7C).
In the amigo1 mRNA-injected larvae, the expression of HNK-1 and acetylated tubulin is >90% of the uninjected larvae. From the statistical results, there is no significant change in the expression of the pathway markers in the mRNA-injected, the 5mis MO-injected, or the GFP mRNA-injected controls (Fig. 7, B and C). In the kv2.1 mRNA and MO1- or MO2-coinjected larvae, Amigo1 expression was also significantly decreased as in the amigo1 knockdown larvae, to 10–20% of the uninjected larvae (Fig. 7D). In contrast to the amigo1 mRNA, the kv2.1 mRNA did not restore HNK-1 and acetylated tubulin expression. HNK-1 expression in the kv2.1 mRNA and MO1- or MO2-coinjected groups was about 30% of the uninjected controls (Fig. 7B). Acetylated tubulin expression in the kv2.1 mRNA and MO1-coinjected group was <20% of the uninjected larvae, and in the kv2.1 mRNA and MO2-coinjected group, the expression was about 30% (Fig. 7C).
Development of Aminergic Circuits in amigo1 Knockdown Morphants
In addition to the early fiber scaffolds, we have analyzed the development of differentiated circuits at later developmental stages in the amigo1 knockdown morphants. To this end, we chose to elucidate the development of the catecholaminergic (CA) system using immunostaining with TH antibodies.
The CA system is the major neuromodulatory system with far ranging projections in the brain, which is important in the modulation of circuit activities in a broad range of behaviors (37, 42). CA development in zebrafish starts quite early, and most of the CA groups and axon tracts found in the adult brain can already be detected in 3-dpf larvae (29, 38, 43). The longitudinal axon tracts from dopaminergic neurons extend from the diencephalon toward the spinal cord in the vicinity of the MLF (see above for the role of Amigo1 in MLF formation) and lateral longitudinal fasciculus (LLF), but their formation may not depend on these early axonal scaffolds (38, 42).
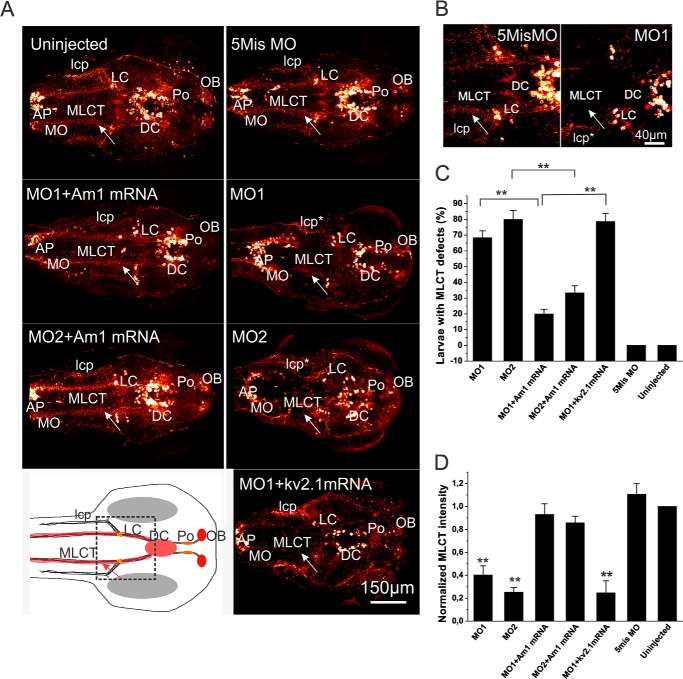
From the confocal z-stacking sections of the 3-dpf whole larvae, the amigo1 knockdown morphants (MO1 and MO2) showed predominant defects in the formation of the MLCT (Fig. 8, A and B). The MLCT projections ascending and descending from the locus coeruleus displayed a disordered pattern. Coinjection of the amigo1 mRNA with MO1 or MO2 (MO1 + Am1 mRNA and MO2 + Am1 mRNA) had a prominent rescue effect on MLCT development, which was not observed in the kv2.1 mRNA and MO1 coinjected larvae (MO1 + kv2.1 mRNA). Statistics of the CA development based on the MLCT formation revealed a developmental defect in a high proportion of the amigo1 knockdown larvae (MO1 and MO2) at 3 dpf compared with the 5mis MO-injected larvae or the uninjected larvae (Fig. 8C). In the MO1 and MO2 morphants, 70–80% of larvae had MLCT defects. In MO1 + Am1 mRNA and MO2 + Am1 mRNA groups, the larvae with MLCT defects decreased to 20–30%. In the MO1 + kv2.1 mRNA groups, over 75% of larvae displayed MLCT defects (Fig. 8C). The intensity analysis of the TH1 antibody staining confirmed the dramatically decreased MLCT development in the MO1 and MO2 morphants, which was rescued by the amigo1 mRNA but not by the kv2.1 mRNA (Fig. 8D).
FIGURE 8.
Whole mount immunostaining with anti-TH antibodies showing disturbed development of catecholaminergic circuits in amigo1 morphants. A, maximum projection of confocal image stacks of 3-dpf whole larval heads. The MLCT staining and its absence in the amigo1 knockdown morphants (MO1 and MO2) is indicated by an arrow. The lateral catecholaminergic projections (lcp), originally lateral to the MLCT in the region between the locus coeruleus (LC) and medulla oblongata (MO), also show a perturbed pattern in the MO1 and MO2 morphants compared with the controls (labeled by an asterisk). The amigo1 but not the kv2.1 mRNA displays a rescue effect on the pathway development. The schematic map shows the normal pattern of anti-TH staining in 3-dpf larvae. The rectangular area was scanned in high resolution and is shown in B. AP, area postrema; DC, diencephalic dopaminergic clusters; lcp, lateral catecholaminergic projections; MO, medulla oblongata; OB, olfactory bulb; Po, preoptic region; Tel, telencephalon. Scale bar, 150 μm. B, Z-stacking confocal images scanned under higher magnification focusing on MLCT ascending and descending from the locus coeruleus in 3-dpf larvae. The amigo1 MO1-injected morphant shows much less staining of MLCT than the 5mis MO-injected control (indicated with an arrow). Scale bar, 40 μm. C, quantification of the MLCT defects in 3-dpf larvae. Over 65% of the MO1 morphants and 80% of the MO2 morphants display defects in the MLCT development. In the amigo1 full-length mRNA-rescued larvae (MO1 + Am1 mRNA and MO2 + Am1 mRNA), the proportion of larvae with MLCT defects is decreased to only 20–30%. In the MO1/Kv2.1-coinjected group (MO1 + Kv2.1 mRNA), around 80% of larvae show MLCT defects. For each group, 10 larvae were picked randomly from the pool of 50 larvae for whole mount immunostaining in three independent experiments. Mean values ± S.E. (error bars) are indicated (n = 3; **, p < 0.01, one-way ANOVA followed by Tukey's post hoc test). D, average pixel intensity analyses of anti-TH1-stained MLCT from Z-stacking confocal images. The average pixel intensity of MLCT staining in uninjected larvae was used for normalization. The MO1, MO2, and MO1 + Kv2.1 morphants show significantly decreased staining intensity of MLCT compared with the 5mis MO-injected control groups. The normalized average pixel intensity of MLCT in the MO1 morphants is only about 40%, and in the MO2 and the MO1 + Kv2.1 mRNA morphants, the staining intensity is even lower than 20% of that found in the control groups. The data are based on five larvae picked randomly for intensity analyses in each group. The experiment was repeated three times (n = 15; **, p < 0.001, one-way ANOVA followed by Tukey's post hoc test).
Behavioral Consequences of amigo1 Knockdown
The remarkable changes observed in neuronal circuitry in the amigo1 knockdown morphants are likely to be reflected in behavior, such as locomotion. We therefore tested motor functions of the amigo1 knockdown morphants using two different behavioral tests.
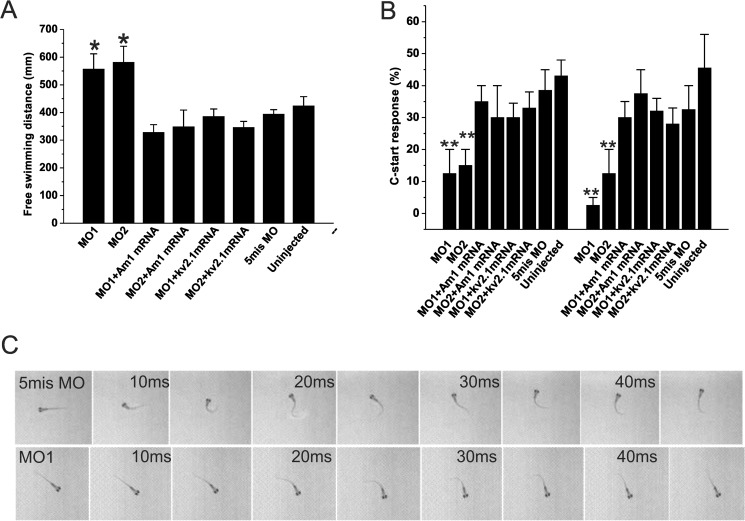
In the first test, free swimming was assayed in 6-dpf larvae to determine the locomotor activity. Surprisingly, the amigo1 MO1 and MO2 morphants showed increased locomotor activity compared with 5mis MO-injected control larvae (Fig. 9A). The amigo1 mRNA-MO1- or mRNA-MO2-coinjected larvae did not show a significant difference in locomotor activity compared with the 5mis MO-injected control larvae (Fig. 9A). These results revealed that coinjection of the amigo1 mRNA with the MOs rescued the hyperactivity observed in both types of amigo1 morphants and that the increase in locomotor activity is a trait specific to the amigo1 knockdown. Interestingly, coinjection of the kv2.1 mRNA with MO1 or MO2 also rescued the hyperactivity phenotype of the morphants (Fig. 9A).
FIGURE 9.
Altered motor functions in amigo1 morphants. A, locomotor activity in 6-dpf larvae during 10 min. The swimming distance was calculated for each larva. The MO1 and MO2 morphants display significantly higher activity compared with the 5mis MO-injected or the uninjected control groups; their swimming distance is enhanced by more than 40% (160 mm). The amigo1 mRNA-coinjected groups (MO1 + Am1 mRNA and MO2 + Am1 mRNA) and the kv2.1 mRNA-coinjected groups (MO1 + kv2.1 mRNA and MO2 + kv2.1 mRNA) do not show a significant difference from the 5mis MO-injected or the uninjected control groups. 10 larvae of each group were selected randomly for the tests, which were repeated four times in independent experiments. The significance was confirmed by one-way ANOVA followed by Fisher's least significant difference post hoc test and Bonferroni correction; n = 40; *, p < 0.05. B, electrically elicited C-start response test in 9-dpf larvae. Under 5 V electrical stimulus for 10 ms, over 90% of the larvae in the uninjected group show C-start response, 40% with SLC and 50% with LLC. The MO1 and MO2 morphants show significantly decreased SLC and LLC compared with the 5mis MO-injected or the uninjected control groups. The amigo1 mRNA-coinjected groups (MO1 + Am1 mRNA and MO2 + Am1 mRNA) and the kv2.1 mRNA-coinjected groups (MO1 + kv2.1 mRNA and MO2 + kv2.1 mRNA) perform similarly to the control groups. There were 20 larvae in each group selected randomly for three tests, which were repeated four times independently. The significance was confirmed by one-way ANOVA followed by Fisher's LSD post hoc test and Bonferroni correction; n = 12; **, p < 0.01. C, real-time recordings of the C-start response. The 5mis MO-injected and the MO1-injected 9-dpf larvae are shown. Pictures were taken from 5 ms after the voltage stimuli. The recording frames are shown every 5 ms and marked every 10 ms. The C-start response captured in the 5-ms MO larvae is defined as the SLC. Error bars, S.E.
Our immunohistochemical analysis showed that Amigo1 is expressed in Mauthner neuron axons (early axonal tracts that are crucial for the functions of the sensorimotor circuits), and the MO experiments indicated that Amigo1 regulates development of connections from the Mauthner neurons in vivo (Fig. 6B, bottom panels). Based on previous reports, we estimated that amigo1 knockdown could therefore perturb startle motor activity (23, 24). Therefore, we decided to study the behavioral repertoire further and investigated the startle response of the amigo1 knockdown morphants elicited by electrical stimuli (10 ms, 5 V) (23, 44). The amigo1 MO1 and MO2 morphants had significant defects in performing both SLC and LLC responses (Fig. 9, B and C). In contrast, the 5mis MO-injected larvae performed C-bend within 20 ms after the stimulation, eliciting an escape response (Fig. 9C, compare the top and bottom rows). In larvae coinjected with amigo1 mRNA-MO1 or amigo1 mRNA-MO2, the SLC and LLC were rescued to the level found in the 5mis MO-injected larvae (Fig. 9B). Similar rescue effects were observed in the larvae coinjected with kv2.1 mRNA-MO1 or kv2.1 mRNA-MO2 (Fig. 9B). Taken together, our results demonstrate that transient knockdown of amigo1 in vivo significantly affects the formation of functionally mature neuronal circuits that are necessary for driving essential behaviors in an intact vertebrate model organism.
DISCUSSION
Amigo Protein Family in Zebrafish
To study the role of the Amigo protein family in the development of functionally active neural circuitry in vivo, we decided to use the zebrafish model in the current study because development of the neuronal connections is more readily analyzed in the zebrafish compared with the mouse. Furthermore, the current results show that the parent form of the protein family (Amigo1) is the main family member expressed until 5 dpf, when several neural circuits, such as those controlling locomotion, have already been formed in zebrafish. In addition, zebrafish does not have a protein that would be clearly similar to AMIGO2 that displays a high expression level in rodent brain and interacts with neurons in a similar manner as AMIGO1. The risk of redundancy should therefore be lower in zebrafish compared with mice, and we expected that knocking down just one family member might produce phenotypic changes.
To follow the expression of the Amigo1 protein, we have produced in the current study affinity-purified antibodies that specifically detect the zebrafish Amigo1 in immunohistochemistry and Western blotting. In 28-hpf larvae, Amigo1 is mainly expressed in the anterior dorsal part of the forebrain area, resembling the expression in embryonic day 10 (E10) mouse embryos (6). In 5-dpf zebrafish larvae, Amigo1 expression is found in most brain areas that already have functionally active neural circuits. This finding also resembles the situation in rodents, in which AMIGO1 expression is increased in postnatal brain during construction of the adult-type circuitry and is found in practically all areas of the CNS in most neurons and their fibers in young adults (4, 7, 9). Such a conserved spatiotemporal expression pattern in vertebrates suggests a conserved function in the CNS development.
Morphological Phenotypes Caused by Down-regulation of Amigo1 Expression or Inhibition of Its Function
Analysis of knockdowns produced by two different morpholino oligonucleotides (MO1 and MO2) shows that the Amigo1 protein essentially disappears, which causes striking defects in the formation of early fiber scaffolds. For example, MLF, which is one of the major early fiber scaffolds, is only composed of thin fibers compared with the robust fascicles seen in uninjected or 5mis MO- injected embryos. Because MOs can sometimes cause unexpected off-target effects, we have used as an alternative approach, injection of the mRNA encoding the protein ectodomain. This approach is similar to the one used in our previous in vitro experiments, where the protein ectodomain was shown to act as a dominant negative receptor that inhibits AMIGO-mediated adhesion and fasciculation of neurites in brain neurons (4). As in studies in vitro, the dominant negative approach causes inhibition of fasciculation in zebrafish in vivo. Furthermore, coinjection of the full-length amigo1 mRNA with the MOs or with the dominant negative construct causes statistically significant rescue in fiber tract development.
It is noteworthy that in addition to the morphological analysis, defective development of neuronal connections in the amigo1 knockdown morphants is also suggested by Western blotting of larval SDS extracts using anti-HNK1 and anti-acetylated tubulin antibodies as fiber pathway markers. Acetylated tubulin has been shown to be a marker of most if not all early axons (32), and the anti-HNK-1 antibodies have been shown to label fiber pathways in a similar manner as the anti-acetylated tubulin antibodies (41). These experiments provide further evidence of the role for Amigo1 in construction of fiber pathways during early development.
Analysis of the amigo1 knockdown morphants at later developmental stages does not show very prominent defects in specific brain structures. This finding is consistent with the finding that the knockdown morphants do not display changes in expression of regional transcription factors, such as Pax2a, Pax6a, and Krox20, that are known to be important for brain development. Analysis at the stages 3–5 dpf, however, displays a modest reduction in head size and disordered development of the aminergic system. This is specifically demonstrated for the catecholaminergic system, but reduced immunostaining of the serotonergic system also appears obvious.4 It seems possible that the defects in the early fiber scaffold development generally disturb development of the aminergic connections, but we cannot exclude specific effects of Amigo1 on the development of neurotransmitter-specific connections.
Behavioral Phenotypes in amigo1 Knockdown Morphants
Disturbed development of neuronal connections observed in the amigo1 knockdown morphants might cause behavioral alterations. Unexpectedly, the amigo1 knockdowns were found to display increased locomotor activity. However, this phenotype is specific because coinjection of the amigo1 mRNA with the morpholinos reduces the enhanced locomotion to the level seen in the misMO-injected and the uninjected larvae.
In addition to spontaneous locomotor activity, we have assessed the escape behavior in the amigo1 morphants. In this analysis, we have used electrical stimulation that has been used to elicit startle responses in adult and larval fish in a range of studies (23, 44). The amigo1 knockdowns display clearly defective responses in both SLC response and LLC response. As in enhanced locomotor activity, the amigo1 mRNA displays a clear rescue effect in both SLC and LLC. It has been reported that the SLC response is mainly mediated by Mauthner neurons (45, 46), whereas the LLC response is not mediated by these neurons (24, 46). Defective development of the Mauthner neuron decussating axons that enter the contralateral MLF (47) may therefore contribute to the defective SLC response. However, a widespread escape network has been identified in the brain stem of zebrafish, containing several descending fiber pathways (46). Maturation of this escape network to form functional connections from the brain to the spinal cord probably regulates the escape responses in zebrafish and is compromised in the amigo1 morphants.
Homophilic and Heterophilic Interactions of Amigo1 in the Development of Neural Circuitry
Analysis of fiber pathway development based on their staining intensity reveals clearly defective tract development in the knockdown morphants and in larvae injected with the mRNA encoding the Amigo1 ectodomain. This suggests that tract development is specifically targeted in the experiments. The finding that the numbers of TUNEL-positive cells are not increased in the experiments agrees with this interpretation and suggests that compromised neuron survival is not the reason why defective tract development is observed.
The mechanism through which Amigo1 specifically affects tract development therefore needs to be considered. Until now, the Robo/Slit signaling pathway has been reported to be crucial for the regulation of longitudinal axonal pathfinding (48). The defects in the development of the early long tracts observed in the amigo1 knockdown morphants appear even more pronounced compared with those reported for manipulation of Robo/Slit signaling. It appears that the role of Amigo1 cannot be explained by Slit/Robo signaling because the phenotypic changes in the amigo1 morphants are different from those found in Slit/Robo studies, and we have not found such changes in Slit or Robo expression that could explain our findings.4
We propose that homophilic binding of Amigo1 within fiber tracts underlies at least partially its remarkable role in the development of long tracts observed in the current study. This inference is supported by our previous studies in which homophilic binding of the Amigo proteins has been demonstrated using pull-down experiments of labeled proteins from cells, bead assays demonstrating aggregation through AMIGO-AMIGO interactions (4), and identification of the LRR region mediating dimerization in the crystal structure (8). Furthermore, inhibition of fasciculation by the Amigo1 ectodomain observed in the current study is strikingly similar to the fasciculation inhibition found in isolated neurons where homophilic interactions and heterophilic interactions within the AMIGO protein family mediate binding to brain neurons (4).
Homophilic interactions within fiber tracts are well known for their essential roles in growth and guidance of axons in simple systems, such as Drosophila, but their roles in complex vertebrate tracts are less clear. However, it has been shown that the zebrafish retinotectal system uses such “isotypic” interactions as an essential strategy for growth and guidance of axon tracts (48, 49). It appears that this fundamental cellular mechanism, acting through pioneer-follower interactions and peer-peer interactions (community effect), is as important as guidance signals outside the tracts and may be used throughout the development of complex nervous systems in vertebrates. Further work on the Amigo family members in different neural systems is required to characterize Amigo-mediated homophilic interactions and cell signaling following such interactions. For example, Amigo-mediated homophilic interactions might affect development of neurotransmitter-specific phenotypes or expression of ion channels required in neural signaling.
We have previously shown that AMIGO1 displays heterophilic interactions with the Kv2.1 potassium channel in brain neurons (11). This inference is based on the finding that the two proteins show a striking colocalization in specific clusters at the plasma membrane. Furthermore, when the Kv2.1 clusters are dispersed at the plasma membrane upon the addition of various stimuli, such as glutamate, AMIGO1 is dispersed with the Kv2.1 channel. These findings, temporal coexpression of the proteins in developing mouse brain and co-immunoprecipitation experiments, have led us to propose that AMIGO1 should be regarded as an auxiliary subunit of the Kv channel (11). From the functional viewpoint, AMIGO1 regulates the channel activity at voltage values close to those required to start action potentials and is therefore expected to play a role in neuronal signaling.
We have therefore considered whether interactions of Amigo1 with Kv2.1 might play a role in the amigo1 morphant phenotypes found in the current study. Surprisingly, the amigo1 morphants were found to lack the Kv2.1 channel almost completely. Expression of the channel in the amigo1 morphants could be restored by coinjection of the kv2.1 mRNA and to a lesser extent by coinjection of the amigo1 mRNA in the morpholino experiments. It thus appears clear that Amigo1 does not only bind to the channel and regulate its activity at the plasma membrane but, in addition, strongly influences Kv2.1 expression in vivo. It is possible that the Kv2.1 protein is not stable in the absence of its auxiliary subunit Amigo1, but further studies will be required to elucidate the biochemical basis of the regulation.
Because the expression of Kv2.1 is strongly inhibited in our amigo1 morphants, we attempted to rescue the phenotypes by restoring the Kv2.1 protein expression through coinjecting the kv2.1 mRNA with the morpholino oligonucleotides. The kv2.1 mRNA did not cause any rescue in the defects of early (1–5 dpf) tract development. In contrast, clear rescue effects were found in the behavioral phenotypes that obviously depend on functionally active neural circuitries. Sufficient anatomical connectivity therefore seems to remain in the morphants to perform normal sensorimotor functions in the case where Kv2.1 is expressed. Interestingly, Amigo1 and Kv2.1 were found to colocalize in mature brain but not in early developing brain, although they were found to be expressed in overlapping anatomical areas in both cases.
During the preparation of this manuscript, a study on Kv2.1 knock-out mice appeared showing that deficiency of the Kv2.1 channel leads to neuronal and behavioral hyperexcitability (50). General locomotor activity in the Kv2.1 knock-out mice was found to be strongly enhanced, clearly resembling the enhanced activity in the amigo1 morphants that we had found in the current study. Because hyperactivity and defective startle responses in the amigo1 knockdown morphants can be rescued by the kv2.1 mRNA, it appears clear that defective Kv2.1 channel function strongly contributes to the behavioral phenotypes caused by knockdown of Amigo 1 expression.
In conclusion, Amigo1 functions independently of Kv2.1 during early fiber tract development. For the construction of neural circuitry with adult-type normal functions, AMIGO1 regulates the expression and function of the Kv2.1 potassium channel of central neurons.
Acknowledgments
The technical assistance of Henri Koivula, Reeta Huhtala, and Erja Huttu is gratefully acknowledged.
This work was supported by the Academy of Finland and the Sigrid Jusélius Foundation.
X. Zhao, P. Panula, and H. Rauvala, unpublished observations.
- LRR
- leucine-rich repeat
- CA
- catecholaminergic
- DiV
- diencephalon ventricle
- dpf
- day(s) postfertilization
- hpf
- hour(s) postfertilization
- Ig domain
- immunoglobulin-like domain
- MBP
- maltose-binding protein
- MLCT
- medial longitudinal catecholaminergic tract
- MLF
- medial longitudinal fasciculus
- MO
- medulla oblongata
- POC
- postoptic commissure
- TH
- tyrosine hydroxylase
- vcc
- ventral caudal cell cluster
- qRT-PCR
- quantitative RT-PCR
- SLC
- short latency C-start
- LLC
- long latency C-start
- ANOVA
- analysis of variance.
REFERENCES
- 1. de Wit J., Hong W., Luo L., Ghosh A. (2011) Role of leucine-rich repeat proteins in the development and function of neural circuits. Annu. Rev. Cell Dev. Biol. 27, 697–729 [DOI] [PubMed] [Google Scholar]
- 2. Söllner C., Wright G. J. (2009) A cell surface interaction network of neural leucine-rich repeat receptors. Genome Biol. 10, R99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Simpson J. H., Bland K. S., Fetter R. D., Goodman C. S. (2000) Short-range and long-range guidance by Slit and its Robo receptors: a combinatorial code of Robo receptors controls lateral position. Cell 103, 1019–1032 [DOI] [PubMed] [Google Scholar]
- 4. Kuja-Panula J., Kiiltomäki M., Yamashiro T., Rouhiainen A., Rauvala H. (2003) AMIGO, a transmembrane protein implicated in axon tract development, defines a novel protein family with leucine-rich repeats. J. Cell Biol. 160, 963–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Maness P. F., Schachner M. (2007) Neural recognition molecules of the immunoglobulin superfamily: signaling transducers of axon guidance and neuronal migration. Nat. Neurosci. 10, 19–26 [DOI] [PubMed] [Google Scholar]
- 6. Homma S., Shimada T., Hikake T., Yaginuma H. (2009) Expression pattern of LRR and Ig domain-containing protein (LRRIG protein) in the early mouse embryo. Gene Expr. Patterns 9, 1–26 [DOI] [PubMed] [Google Scholar]
- 7. Laeremans A., Nys J., Luyten W., D'Hooge R., Paulussen M., Arckens L. (2013) AMIGO2 mRNA expression in hippocampal CA2 and CA3a. Brain Struct. Funct. 218, 123–130 [DOI] [PubMed] [Google Scholar]
- 8. Kajander T., Kuja-Panula J., Rauvala H., Goldman A. (2011) Crystal structure and role of glycans and dimerization in folding of neuronal leucine-rich repeat protein AMIGO-1. J. Mol. Biol. 413, 1001–1015 [DOI] [PubMed] [Google Scholar]
- 9. Chen Y., Hor H. H., Tang B. L. (2012) AMIGO is expressed in multiple brain cell types and may regulate dendritic growth and neuronal survival. J. Cell Physiol. 227, 2217–2229 [DOI] [PubMed] [Google Scholar]
- 10. Ono T., Sekino-Suzuki N., Kikkawa Y., Yonekawa H., Kawashima S. (2003) Alivin 1, a novel neuronal activity-dependent gene, inhibits apoptosis and promotes survival of cerebellar granule neurons. J. Neurosci. 23, 5887–5896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Peltola M. A., Kuja-Panula J., Lauri S. E., Taira T., Rauvala H. (2011) AMIGO is an auxiliary subunit of the Kv2.1 potassium channel. EMBO Rep. 12, 1293–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhao X., Kuja-Panula J., Rouhiainen A., Chen Y. C., Panula P., Rauvala H. (2011) High mobility group box-1 (HMGB1; amphoterin) is required for zebrafish brain development. J. Biol. Chem. 286, 23200–23213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Westerfield M. (2000) The Zebrafish Book: A Guide for the Laboratory Use of Zebrafish (Danio rerio), 4th Ed., University of Oregon Press, Eugene, OR [Google Scholar]
- 14. Fink M., Flekna G., Ludwig A., Heimbucher T., Czerny T. (2006) Improved translation efficiency of injected mRNA during early embryonic development. Dev. Dyn. 235, 3370–3378 [DOI] [PubMed] [Google Scholar]
- 15. Thisse C., Thisse B. (2008) High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat. Protoc. 3, 59–69 [DOI] [PubMed] [Google Scholar]
- 16. Thisse C., Thisse B. (2005) High throughput expression analysis of ZF-Models Consortium clones, ZFIN direct data submission (http://zfin.org)
- 17. Sallinen V., Torkko V., Sundvik M., Reenilä I., Khrustalyov D., Kaslin J., Panula P. (2009) MPTP and MPP+ target specific aminergic cell populations in larval zebrafish. J. Neurochem. 108, 719–731 [DOI] [PubMed] [Google Scholar]
- 18. Rivera J., Chu P. J., Lewis T. L., Jr., Arnold D. B. (2007) The role of Kif5B in axonal localization of Kv1 K+ channels. Eur. J. Neurosci. 25, 136–146 [DOI] [PubMed] [Google Scholar]
- 19. Mueller T., Wullimann M. F. (2005) Atlas of Early Zebrafish Brain Development: A Tool for Molecular Neurogenetics, pp. 29–120, Elsevier, Amsterdam [Google Scholar]
- 20. Sallinen V., Sundvik M., Reenilä I., Peitsaro N., Khrustalyov D., Anichtchik O., Toleikyte G., Kaslin J., Panula P. (2009) Hyperserotonergic phenotype after monoamine oxidase inhibition in larval zebrafish. J. Neurochem. 109, 403–415 [DOI] [PubMed] [Google Scholar]
- 21. Schweitzer J., Lohr H., Filippi A., Driever W. (2012) Dopaminergic and noradrenergic circuit development in zebrafish. Dev. Neurobiol. 72, 256–268 [DOI] [PubMed] [Google Scholar]
- 22. Panula P., Sallinen V., Sundvik M., Kolehmainen J., Torkko V., Tiittula A., Moshnyakov M., Podlasz P. (2006) Modulatory neurotransmitter systems and behavior: towards zebrafish models of neurodegenerative diseases. Zebrafish 3, 235–247 [DOI] [PubMed] [Google Scholar]
- 23. Nissanov J., Eaton R. C., DiDomenico R. (1990) The motor output of the Mauthner cell, a reticulospinal command neuron. Brain Res. 517, 88–98 [DOI] [PubMed] [Google Scholar]
- 24. Burgess H. A., Granato M. (2007) Sensorimotor gating in larval zebrafish. J. Neurosci. 27, 4984–4994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Eisen J. S., Smith J. C. (2008) Controlling morpholino experiments: don't stop making antisense. Development 135, 1735–1743 [DOI] [PubMed] [Google Scholar]
- 26. Wullimann M. F., Puelles L. (1999) Postembryonic neural proliferation in the zebrafish forebrain and its relationship to prosomeric domains. Anat. Embryol. 199, 329–348 [DOI] [PubMed] [Google Scholar]
- 27. Wullimann M. F., Mueller T. (2002) Expression of Zash-1a in the postembryonic zebrafish brain allows comparison to mouse Mash1 domains. Brain Res. Gene. Expr. Patterns 1, 187–192 [DOI] [PubMed] [Google Scholar]
- 28. Mueller T. (2012) What is the thalamus in zebrafish? Front. Neurosci. 6, 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kimmel C. B., Ballard W. W., Kimmel S. R., Ullmann B., Schilling T. F. (1995) Stages of embryonic development of the zebrafish. Dev. Dyn. 203, 253–310 [DOI] [PubMed] [Google Scholar]
- 30. McLean D. L., Fetcho J. R. (2004) Ontogeny and innervation patterns of dopaminergic, noradrenergic, and serotonergic neurons in larval zebrafish. J. Comp. Neurol. 480, 38–56 [DOI] [PubMed] [Google Scholar]
- 31. Cole L. K., Ross L. S. (2001) Apoptosis in the developing zebrafish embryo. Dev. Biol. 240, 123–142 [DOI] [PubMed] [Google Scholar]
- 32. Chitnis A. B., Kuwada J. Y. (1990) Axonogenesis in the brain of zebrafish embryos. J. Neurosci. 10, 1892–1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wilson S. W., Ross L. S., Parrett T., Easter S. S., Jr. (1990) The development of a simple scaffold of axon tracts in the brain of the embryonic zebrafish, Brachydanio rerio. Development 108, 121–145 [DOI] [PubMed] [Google Scholar]
- 34. Kruse J., Mailhammer R., Wernecke H., Faissner A., Sommer I., Goridis C., Schachner M. (1984) Neural cell adhesion molecules and myelin-associated glycoprotein share a common carbohydrate moiety recognized by monoclonal antibodies L2 and HNK-1. Nature 311, 153–155 [DOI] [PubMed] [Google Scholar]
- 35. Metcalfe W. K., Myers P. Z., Trevarrow B., Bass M. B., Kimmel C. B. (1990) Primary neurons that express the L2/HNK-1 carbohydrate during early development in the zebrafish. Development 110, 491–504 [DOI] [PubMed] [Google Scholar]
- 36. Tay T. L., Ronneberger O., Ryu S., Nitschke R., Driever W. (2011) Comprehensive catecholaminergic projectome analysis reveals single-neuron integration of zebrafish ascending and descending dopaminergic systems. Nat. Commun. 2, 171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. McLean D. L., Fetcho J. R. (2004) Relationship of tyrosine hydroxylase and serotonin immunoreactivity to sensorimotor circuitry in larval zebrafish. J. Comp. Neurol. 480, 57–71 [DOI] [PubMed] [Google Scholar]
- 38. Kastenhuber E., Kern U., Bonkowsky J. L., Chien C. B., Driever W., Schweitzer J. (2009) Netrin-DCC, Robo-Slit, and heparan sulfate proteoglycans coordinate lateral positioning of longitudinal dopaminergic diencephalospinal axons. J. Neurosci. 29, 8914–8926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Peters A., Vaughn J. E. (1967) Microtubules and filaments in the axons and astrocytes of early postnatal rat optic nerves. J. Cell Biol. 32, 113–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Piperno G., Fuller M. T. (1985) Monoclonal antibodies specific for an acetylated form of α-tubulin recognize the antigen in cilia and flagella from a variety of organisms. J. Cell Biol. 101, 2085–2094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ross L. S., Parrett T., Easter S. S., Jr. (1992) Axonogenesis and morphogenesis in the embryonic zebrafish brain. J. Neurosci. 12, 467–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kastenhuber E., Kratochwil C. F., Ryu S., Schweitzer J., Driever W. (2010) Genetic dissection of dopaminergic and noradrenergic contributions to catecholaminergic tracts in early larval zebrafish. J. Comp. Neurol. 518, 439–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rink E., Wullimann M. F. (2002) Development of the catecholaminergic system in the early zebrafish brain: an immunohistochemical study. Brain Res. Dev. Brain Res. 137, 89–100 [DOI] [PubMed] [Google Scholar]
- 44. Liu Y. C., Bailey I., Hale M. E. (2012) Alternative startle motor patterns and behaviors in the larval zebrafish (Danio rerio). J. Comp. Physiol. A Neuroethol. Sens. Neural Behav. Physiol. 198, 11–24 [DOI] [PubMed] [Google Scholar]
- 45. Kimmel C., Eaton R., Powell S. (1980) Decreased fast-start performance of zebrafish larvae lacking mauthner neurons. J. Comp. Physiol. 140, 343–350 [Google Scholar]
- 46. Gahtan E., Sankrithi N., Campos J. B., O'Malley D. M. (2002) Evidence for a widespread brain stem escape network in larval zebrafish. J. Neurophysiol. 87, 608–614 [DOI] [PubMed] [Google Scholar]
- 47. Kimmel C. B., Powell S. L., Metcalfe W. K. (1982) Brain neurons which project to the spinal cord in young larvae of the zebrafish. J. Comp. Neurol. 205, 112–127 [DOI] [PubMed] [Google Scholar]
- 48. Dickson B. J., Gilestro G. F. (2006) Regulation of commissural axon pathfinding by slit and its Robo receptors. Annu. Rev. Cell Dev. Biol. 22, 651–675 [DOI] [PubMed] [Google Scholar]
- 49. Pittman A. J., Law M. Y., Chien C. B. (2008) Pathfinding in a large vertebrate axon tract: isotypic interactions guide retinotectal axons at multiple choice points. Development 135, 2865–2871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Speca D. J., Ogata G., Mandikian D., Bishop H. I., Wiler S. W., Eum K., Wenzel H. J., Doisy E. T., Matt L., Campi K. L., Golub M. S., Nerbonne J. M., Hell J. W., Trainor B. C., Sack J. T., Schwartzkroin P. A., Trimmer J. S. (2014) Deletion of the Kv2.1 delayed rectifier potassium channel leads to neuronal and behavioral hyperexcitability. Genes Brain Behav. 13, 394–408 [DOI] [PMC free article] [PubMed] [Google Scholar]